Summary
A point mutation in the BRAF gene, leading to a constitutively active form of the protein, is present in 45%–60% of patients and acts as a key driver in melanoma. Shortly after therapy induction, resistance to MAPK pathway-specific inhibitors develops, indicating that pathway inhibition is circumvented by epigenetic mechanisms. Here, we mimicked epigenetic modifications in melanoma cells by reprogramming them into metastable induced pluripotent cancer cells (iPCCs) with the ability to terminally differentiate into non-tumorigenic lineages. iPCCs and their differentiated progeny were characterized by an increased resistance against targeted therapies, although the cells harbor the same oncogenic mutations and signaling activity as the parental melanoma cells. Furthermore, induction of a pluripotent state allowed the melanoma-derived cells to acquire a non-tumorigenic cell fate, further suggesting that tumorigenicity is influenced by the cell state.
Keywords: melanoma, reprogramming, BRAF, therapy resistance, iPSC, iPCC, cancer, stem cells, induced pluripotency, inhibitor
Highlights
-
•
Human melanoma cells reprogrammed toward an iPSC-like state (iPCCs)
-
•
iPCCs differentiated into neurons and fibroblasts
-
•
iPCC-derived fibroblasts show no tumorigenic potential
-
•
iPCCs and iPCC-derived fibroblasts lose oncogene addiction
Aberrant activation of the MAPK pathway is a major cause of melanoma. Resistance to MAPK pathway-specific inhibitors hampers successful treatment of melanoma. By reprogramming human melanoma cells toward pluripotency and subsequent differentiation, Utikal and colleagues demonstrate that the tumorigenic phenotype and oncogene addiction are linked to a certain differentiation lineage.
Introduction
The discovery that development is not a one-way street but can be reverted by nuclear reprogramming leading to induced pluripotent stem cells (iPSCs) is one of the most promising recent discoveries in translational medicine (Larribere and Utikal, 2014, Tabar and Studer, 2014). Patient-specific iPSCs not only allow for the modeling of distinct diseases and testing of novel drugs, but also provide unique resources for regenerative medicine (Galach and Utikal, 2011). The possibility to differentiate human iPSCs (hiPSCs) into distinct neuronal cells is already beginning to revolutionize research in the neurodegenerative disease field, indicated by the escalating number of publications focusing on hiPSC-derived neurodegenerative disease models (Lojewski et al., 2014, Stanslowsky et al., 2014, Japtok et al., 2015). Nevertheless, the full potential of hiPSCs is not yet utilized. Using hiPSCs to study the influence of the differentiation state on disease-associated mutations is still in its infancy.
Nuclear reprogramming is initiated by ectopic expression of the four transcription factors OCT4, SOX2, KLF4, and MYC. During this process the epigenetic profile of a somatic cell is reverted in stepwise fashion to the profile of pluripotent stem cells (PSCs), which are able to differentiate into any cell of the three germ layers (Takahashi and Yamanaka, 2006, Maherali et al., 2007, Takahashi et al., 2007). Using patient-derived somatic cells, the initiation of even genetically complex diseases such as Alzheimer’s disease (Yagi et al., 2011), multiple sclerosis (reviewed in Di Ruscio et al., 2015), or early events in tumor initiation (Kim et al., 2013) can be modeled. Therefore, iPSC technology enables investigation not only of early events during the onset of a disease but also the influence of the epigenetic status on the disease. Although previous studies successfully demonstrated reprogramming of cancer cells, reprogramming barriers prevent the successful induction of pluripotency in the majority of tumor cells (Utikal et al., 2009, Bernhardt et al., 2012, Kim et al., 2013).
Here, we applied nuclear reprogramming to different human tumor cell lines. Independent of the mutational status or tumor entity, the constitutive overexpression of the three reprogramming factors OCT4, SOX2, and KLF4 was sufficient to generate iPSC-like tumor cells in the presence of human leukemia inhibitory factor (LIF) that show typical characteristics of murine embryonic stem cells (mESCs). Reprogrammed HT-144 melanoma cells acquired a metastable pluripotent state and could be differentiated into cells of all three germ layers in vivo, and generated neuronal and fibroblast-like cell types in vitro. Notably, reprogrammed cells and their differentiated progeny lost typical melanoma markers and failed to initiate novel melanomas. Moreover, reduced tumorigenicity came together with an increased therapy resistance against the mitogen-activated protein kinase (MAPK) kinase inhibitors vemurafenib and trametinib.
Results
Generation and Characterization of iPCCs
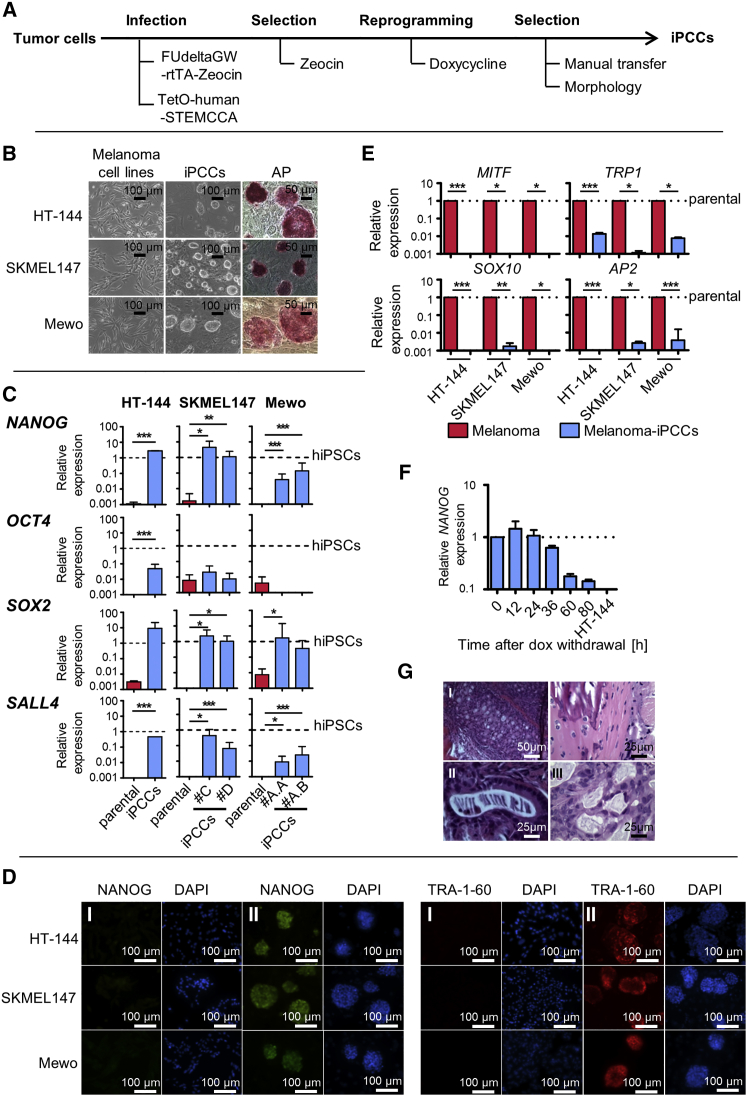
Previously the induction of a pluripotent state in murine melanoma cells by somatic cell nuclear transfer (Hochedlinger et al., 2004) and nuclear reprogramming (Utikal et al., 2009) was reported, demonstrating that a reprogrammed melanoma genome can even give rise to a viable organism. Oncogenes represent barriers impeding the reprogramming process (Liu et al., 2015). Accordingly, several studies reported that cancer cell reprogramming is less effective and more time consuming (Utikal et al., 2009, Lin and Chui, 2012, Lai et al., 2013). To address the question of whether human melanoma cells are amenable to nuclear reprogramming, we used a reverse tetracycline-dependent transactivator (rtTA) and a doxycycline-inducible lentiviral polycistronic vector carrying the reprogramming factors. Since MYC is known to be endogenously expressed in the melanoma cell lines with which we worked, we used OCT4, KLF4, and SOX2 only (Figure S1A) (Kraehn et al., 2001, Sarkar et al., 2006, Bartholomeusz et al., 2007, Zhuang et al., 2008). Tumor cells carrying the rtTA and the reprogramming factors were subjected to doxycycline-induced expression of the transgenes and manually transferred onto feeder cells (Figure 1A). Constitutive expression of the reprogramming factors resulted in the appearance of alkaline phosphatase-positive colonies. These iPSC-like tumor cells showed morphological features of mESCs (Figure 1B) and were similarly resistant to single-cell dissociation without Rho kinase inhibitor (Y-27632), although its addition significantly increased cell survival.
Figure 1.
Generation and Characterization of Metastable Reprogrammed Melanoma Cells
(A) Scheme for tumor cell reprogramming.
(B) Reprogrammed tumor cells form ESC-like alkaline phosphatase-positive colonies on feeder cells.
(C) qPCR measurement shows reactivation of the endogenous loci of the pluripotency markers NANOG, SOX2, and SALL4 but only mild increase in endogenous OCT4 expression. GAPDH was used as endogenous control and hiPSCs as reference sample. Indicated is the mean ± SD. p Values were calculated by two-tailed, unpaired sample t test of technical triplicates in two clones of SKMEL147-and Mewo-iPCCs and in three independent experiments of HT-144-iPCCs. Asterisk indicates t test p value of ≤0.05 in comparison with the respective reference (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.005).
(D) Immunofluorescence staining of NANOG and TRA-1-60 in the parental melanoma cells (I) and melanoma iPCCs (II). DAPI was used for nuclear counterstaining.
(E) qPCR analysis reveals loss of melanocytic markers in iPCCs compared with their parental melanoma cell lines. Gene expression levels were normalized to GAPDH. Error bars indicate 95% confidence intervals. p Values were calculated from three independent experiments by two-tailed, unpaired sample t test. Asterisk indicates t test p value of ≤0.05 in comparison with the respective reference (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.005).
(F) NANOG expression analyzed by qPCR in HT-144-iPCCs at indicated time points after doxycycline withdrawal compared with the parental HT-144. Nanog expression was normalized to internal GAPDH. Error bars indicate 95% confidence intervals. Dotted line, normalization to day 0.
(G) Metastable melanoma iPCCs form teratomas in vivo showing tissue structures of mesodermal (I), ectodermal (II), and endodermal (III) origin. Paraffin-embedded tumor slices were stained with H&E.
See also Figure S1.
As reactivation of the pluripotency network is a hallmark of successfully reprogrammed cells, we quantified the expression levels of pluripotency markers in reprogrammed melanoma cells compared with iPSCs derived from somatic cells (Figures 1C and 1D). Independent of the mutational status, all melanoma cell lines subjected to nuclear reprogramming reactivated the endogenous loci of pluripotency factors such as NANOG, SOX2, SALL4, and TRA-1-60 (Figures 1C, 1D, S1C, and S1D). Furthermore, we included HeLa cells in the study and demonstrated that human cervical carcinoma cells are also amenable to reprogramming. Since HeLa cells are known to have an amplification of chromosomal region 8q24 which carries the MYC locus (Macville et al., 1999) and since there is evidence that the protein is expressed in these cells (Cappellen et al., 2007), we also reprogrammed them without MYC (Figure S2). We draw the conclusion that tumor cells have the ability to reactivate the pluripotency network independent of their origin and mutational load.
We named these iPSC-like tumor cells induced pluripotent cancer cells (iPCCs). Surprisingly, only a slight increase in OCT4 expression was observed (Figure 1C), suggesting that tumor cells harbor barriers impeding the reactivation of OCT4.
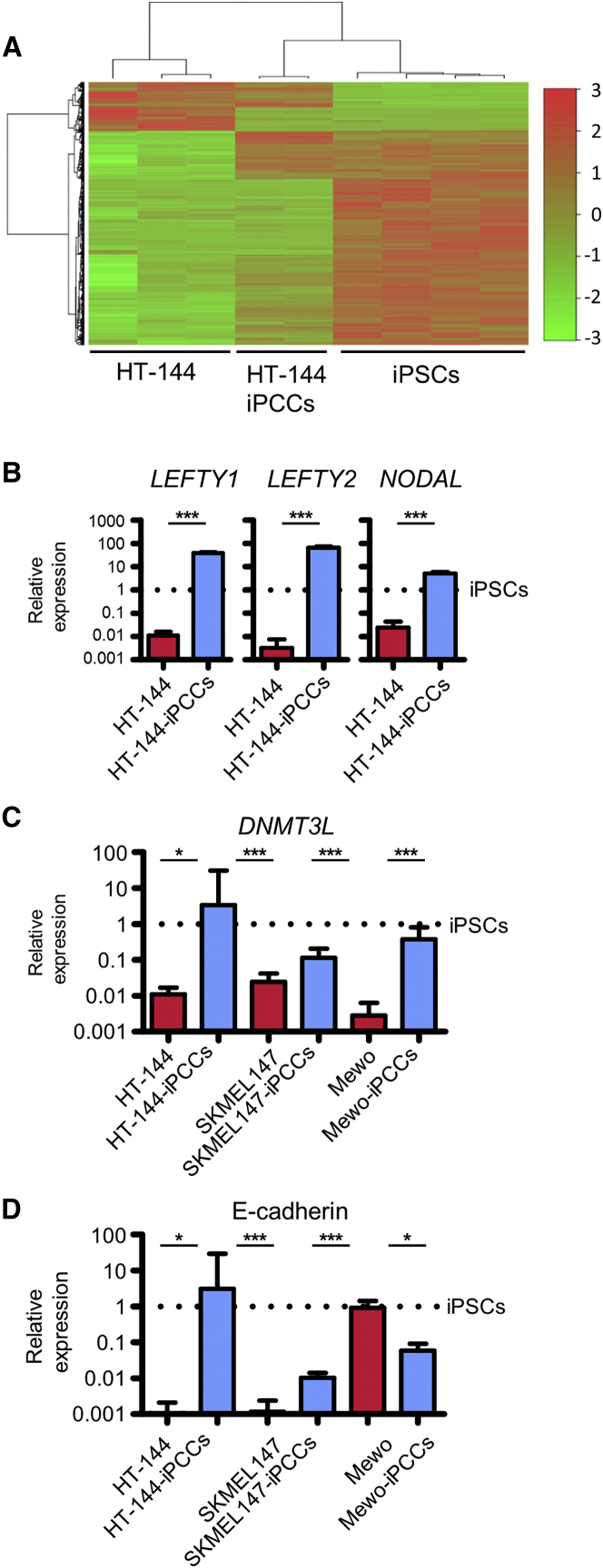
Similar to iPSCs derived from somatic cells (Kim et al., 2010, Bar-Nur et al., 2011, Nukaya et al., 2015), iPCCs lost markers of their lineage of origin. Accordingly, we observed a downregulation of melanocytic markers such as MITF and TRP1 as well as SOX10 and AP2 compared with the parental melanoma cells (Figure 1E). Also, iPCCs cluster closer to iPSCs compared with their parental cells of origin and express development-associated TGF-B superfamily members NODAL, LEFTY1, and LEFTY2 as well as the epigenetic modifier DNMTL3 (Figures 2A–2C).
Figure 2.
Characterization of Melanoma iPCCs
(A) Heatmap and dendrogram generated by unsupervised hierarchical clustering of differentially regulated genes (empirical Bayes moderated t test, p < 0.05) in HT-144, HT-144-iPCCs, and hiPSCs. Red indicates increased expression and green decreased expression relative to the control.
(B–D) qPCR analysis of melanoma iPCCs compared with their reprogrammed progeny. iPSCs were used as reference sample. Data were obtained from three independent experiments. (B) Expression analysis of development-associated TGF-B superfamily members NODAL, LEFTY1, and LEFTY2. (C) Expression analysis of the epigenetic modifier DNMTL3. (D) Expression analysis of the MET marker E-cadherin. GAPDH was used as endogenous control and hiPSCs as reference sample. Error bars indicate 95% confidence intervals. Indicated is the mean + SD. p Values were calculated by two-tailed, unpaired sample t test. Asterisk indicates t test p value of ≤0.05 in comparison with the respective reference (∗∗∗p ≤ 0.005).
To assess whether iPCCs acquired a stable pluripotent state, we withdrew doxycycline, thereby stopping reprogramming factor expression. Within 80 hr after withdrawal, NANOG expression levels were reduced by 90% (Figure 1F), followed by morphological changes and loss of alkaline phosphatase activity (Figure S3). This indicated that the tumor cells could not acquire a stable pluripotent state. To exclude the possibility that the reprogramming process is particularly impeded in tumor cells, we transferred fully reprogrammed melanocyte-derived iPSCs to feeder cells after transgene expression was induced. After two to three passages in the presence of doxycycline on dense feeder cells, the iPSCs formed colonies indistinguishable from the iPCCs (Figure S1B). Together, these data indicate that the metastable pluripotent state is an effect based on the constitutive expression of reprogramming factors and dense feeder cells serving as substrate. Thereby, the partial pluripotent state is not restricted to cancer cells but can also be induced in already fully reprogrammed iPSCs.
To further characterize the cells, we injected HT-144-iPCCs subcutaneously in the flanks of NOD/SCID mice. In all cases tumors developed after 10–12 weeks. Excised tumors stained with H&E demonstrated that the iPCCs differentiated into tissues derived from all three germ layers (Figure 1G).
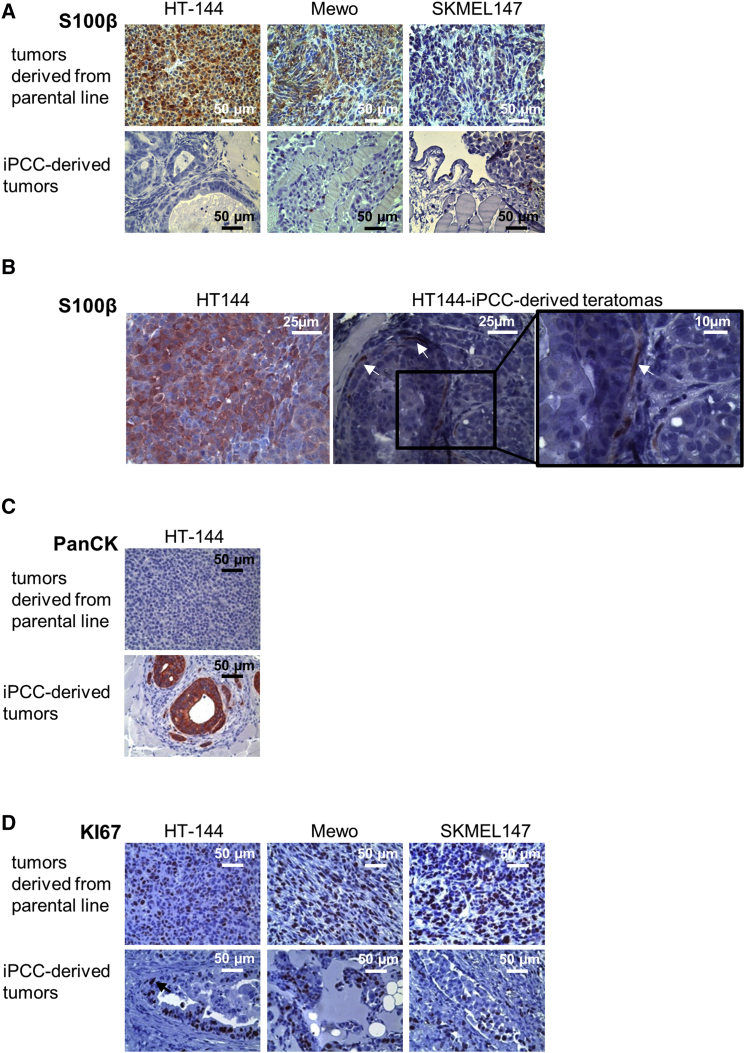
A previous publication demonstrated that tumor-iPSCs resemble early stages of tumor development in vivo (Kim et al., 2013). Hence, we analyzed the expression of typical melanoma and standard tumor markers in tumors derived from iPCCs and the parental melanoma cells (Figures 3A–3D). Parental tumor lines generated homogeneous S100B-positive melanomas with high expression of the proliferation marker Ki67, and were negative for epithelial cytokeratins. In contrast, iPCC-derived teratomas showed multiple areas of differentiated foci that were architecturally organized and contained irregularly shaped cells with enlarged cytoplasm. In addition, formation of gland-like structures was observed in most of the tumors that developed from iPCCs. Independent of the mutational status, iPCC-derived tumors rarely generated any S100B-positive melanoma-like structures. Furthermore, we observed epithelial structures in iPCC-derived tumors. According to the heterogeneous pattern of cellular differentiations, the proliferation marker Ki67 was present in distinct tissue structures but was rarely expressed in the tumor mass.
Figure 3.
Reduced Tumor Marker Expression in Reprogrammed Melanoma Cells
(A) Histological staining for the melanoma marker S100B in melanomas derived from HT-144, Mewo, and SKMEL147 cells and their respective reprogrammed progeny.
(B) In contrast to melanomas, S100B expression is restricted to neuronal-like cells in HT-144-iPCC-derived teratomas (indicated by arrows).
(C) Histological staining for epithelial cytokeratins (PanCK) in HT-144-iPCC-derived tumors and melanomas derived from the parental HT-144 cells.
(D) Histological analysis of the expression of the proliferation marker KI67 in melanomas derived from HT-144, Mewo, and SKMEL147 cells and their respective reprogrammed progeny.
iPCCs Can Be Terminally Differentiated into Different Cell Lineages
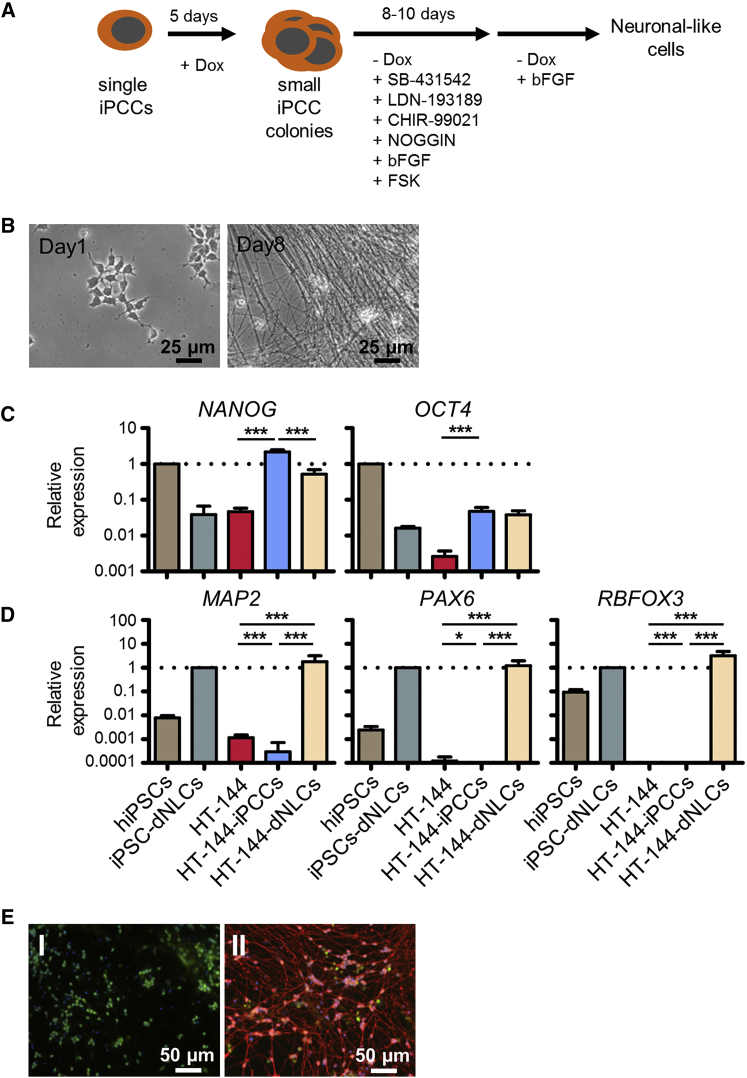
Next, we assessed whether iPCCs can be terminally differentiated in vitro into cell lineages different from that of the parental cells. We applied a previously published neuronal differentiation strategy based on dual SMAD inhibition using small molecules inhibiting SMAD and GSK3β signaling (Figure 4A) in combination with a low dose of Noggin and the bone morphogenetic protein (BMP) inhibitor LDN-193189 (Chambers et al., 2009, Ladewig et al., 2012). Within 3 days after induction of neuronal differentiation, the first axon-like structures appeared and elongated with continued differentiation (Figure 4B). In accordance with the differentiation, pluripotency markers NANOG and OCT4 were downregulated. Nevertheless, expression levels of both transcription factors were clearly higher than in the parental cell line, suggesting that the differentiation was not yet complete, although early neuronal markers such as microtubule-associated protein 2 (MAP2) and PAX6 were increased. After 20–50 days, the late post-mitotic neuronal marker RBFOX3 was significantly upregulated in HT-144-derived neuronal-like cells while parental HT-144 cells were negative for all neuronal markers (Figures 4C and 4D). Immunofluorescence staining of TUJ1 in neuronal differentiated cells derived from HT-144-iPCCs constitutively expressing GFP confirmed the successful neuronal differentiation (Figure 4E).
Figure 4.
HT-144-iPCCs Efficiently Differentiate into Neuronal Cells
(A) Schematic illustration of the neuronal differentiation protocol.
(B) Morphology of HT-144-iPCCs subjected to neuronal induction at indicated time points.
(C and D) qPCR analysis of pluripotency marker (C) as well as early and late neuronal marker expression (D). Data were pooled from three independent experiments. NANOG and OCT4 were normalized to hiPSCs. The neuronal markers MAP2, PAX6, and RBFOX3 were normalized to hiPSC-derived neuronal cells. GAPDH served as internal control. Error bars indicate 95% confidence intervals. p Values were calculated by two-tailed, unpaired sample t test. Asterisk indicates t test p value of ≤0.05 in comparison with the respective reference (∗∗∗p ≤ 0.005).
(E) Immunofluorescence staining of B3-tubulin in HT-144-iPCCs (I) and iPCC-derived neuronal cells (II). iPCCs used for the staining constitutively express GFP.
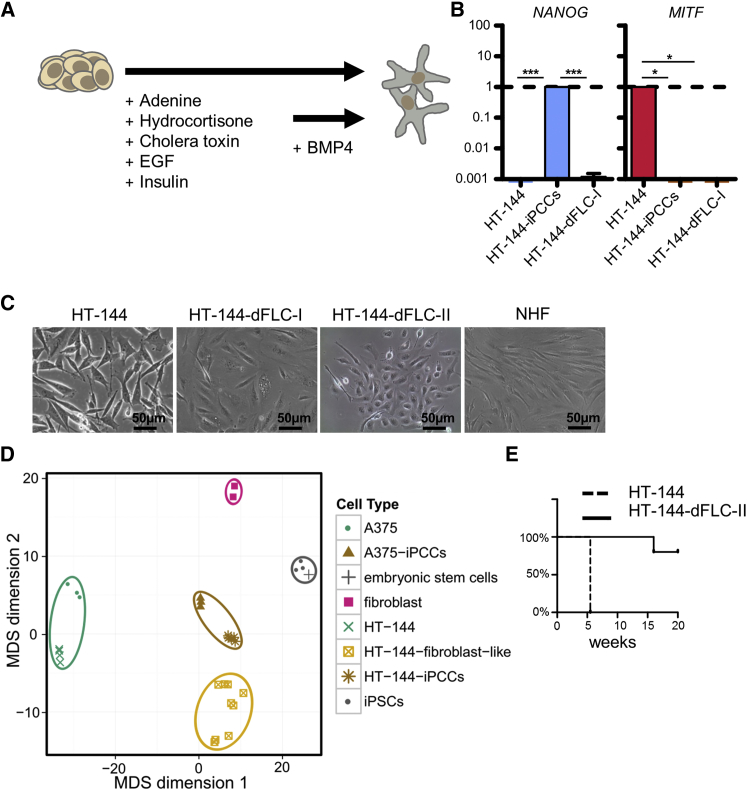
Since iPCC-derived neuronal cells became post-mitotic, we applied a differentiation protocol that guided cells toward mesodermal lineage by stimulation with epidermal growth factor (EGF), insulin, and at later stages with additional BMP-4 (Figure 5A). Loss of melanoma and pluripotency markers indicated a non-melanocytic cell identity (Figure 5B). Due to the formation of spindle-like cells resembling normal human fibroblasts (NHFs) (Figure 5C), we named the cells HT-144-derived fibroblast-like cells (HT-144-dFLCs). These cells were clonally selected from two independent differentiations (HT-144-dFLCs-I and -II) and could be cultured for more than 40 passages.
Figure 5.
Melanoma iPCCs Gain the Potential to Terminally Differentiate into Non-tumorigenic Cells
(A) Schematic protocol for the differentiation of iPCC-derived fibroblast-like cells.
(B) qPCR analysis of NANOG and MITF expression in HT-144, reprogrammed HT-144-iPCCs, and iPCC-derived fibroblast-like cells (HT-144-dFLC-I). GAPDH served as endogenous control. Error bars indicate 95% confidence intervals. p Values were calculated from three independent experiments by two-tailed, unpaired sample t test. Asterisk indicates t test p value of ≤0.05 in comparison with the respective reference (∗p ≤ 0.05, ∗∗∗p ≤ 0.005).
(C) Morphological comparison of the parental cell line HT-144 and fibroblast-like cells from two differentiations (HT-144-dFLC) with NHFs.
(D) Global methylation profiles of A375 and HT-144 compared with the profiles of their respective iPCCs and HT-144-iPCC-derived fibroblast-like cells from three independent differentiations were used for a multidimensional scaling. Human fibroblasts (pink squares represent preparations from two different individuals), hiPSCs, and a publicly available dataset of hESCs were added as controls.
(E) Survival of mice after subcutaneous injection of 1 × 106 parental HT-144 melanoma cells or HT-144-iPCC-derived fibroblasts, respectively. Parental HT-144 melanoma cells gave rise to tumors in all five cases while no tumor growth was observed in five mice injected with HT-144-dFLC-II even after 20 weeks (one mouse was euthanized for other reasons).
The Global Methylation Profile of iPCCs Resembles the Profiles of PSCs
To further assess the epigenetic similarities between the different cell types, we performed a global methylation analysis and grouped the cells according to their methylation status (Figures 5D and S4). Cells with a similar methylation profile clustered together in a multidimensional scale. As expected, the melanoma cell lines formed one cluster, as well as iPCCs and differentiated progeny. Using published methylation data from human embryonic stem cells (hESCs) and hiPSCs, we showed that the iPCCs moved toward stable PSCs but formed a separate cluster, indicating that the majority of methylation sites showed a similar profile to that of the PSCs. In contrast, the methylation profile of HT-144-dFLCs was distinct from the profile of NHFs.
iPCC-Derived Fibroblasts Show No Tumorigenic Potential
Previous publications demonstrated that reprogrammed tumor cells can acquire a non-tumorigenic phenotype (Utikal et al., 2009, Stricker et al., 2013, Fehrenbach et al., 2016). Nevertheless, so far there is no evidence of how epigenetic modifications take shape in cells harboring constitutively active oncogenes. Therefore, we investigated the tumor-initiating potential of the parental cell line and HT-144-dFLC-II by subcutaneous injection in NSG mice. We observed no tumor formation in any of the HT-144-dFLC-II injected animals over a time period of 18 weeks. In contrast, all mice injected with the parental cells were euthanized after 5.5 weeks because of the formation of large tumors (Figure 5E).
iPCCs and iPCC-Derived Fibroblasts Lose Their Oncogene Addiction
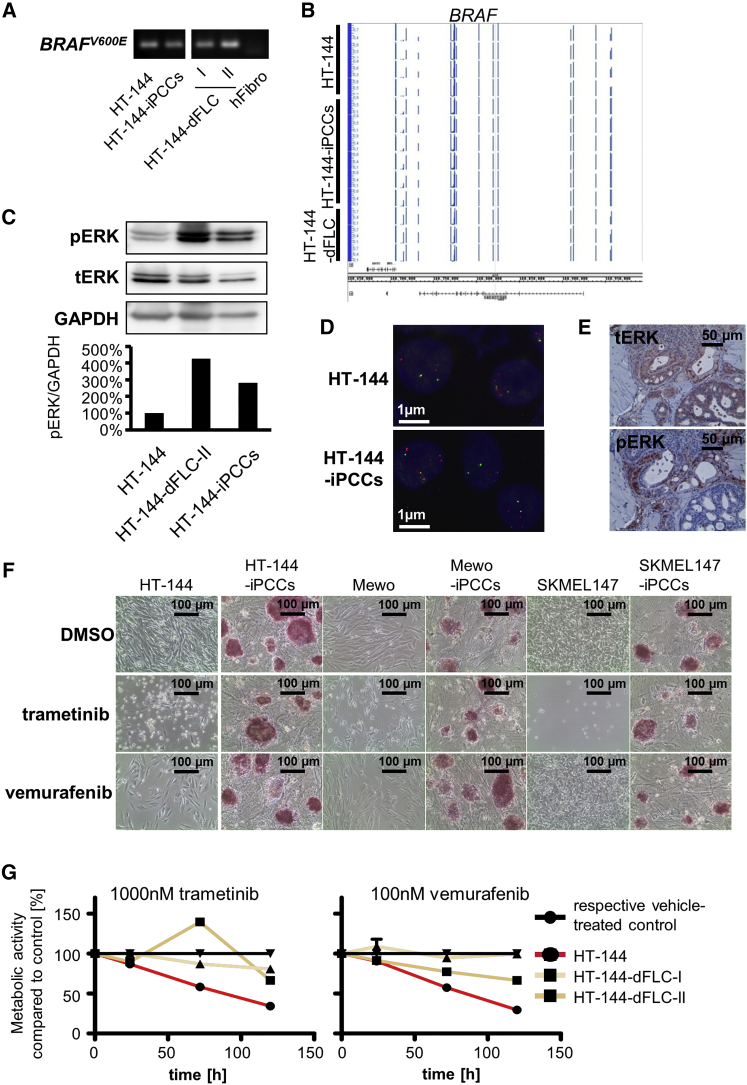
It has recently been demonstrated that malignant leukemia cells lose their oncogene dependence by nuclear reprogramming into iPSCs (Carette et al., 2010). Due to the activating V600E mutation in HT-144 cells as a key driver of melanoma initiation and progression, we focused on MAPK signaling before and after the reprogramming process. Previous data demonstrated that the reprogramming process selects for cells harboring a low mutational load (Lai et al., 2013). Therefore, as a first step we confirmed that all HT-144-derived cells harbored the BRAFV600E mutation (Figure 6A) using BRAFV600E-specific primers. Moreover, we found that the BRAFV600E-mutated HT-144, HT-144-iPCCs, and HT-144-dFLCs showed a methylation profile similar to that of the BRAF locus (Figure 6B). In line with this, we found high levels of phosphorylated ERK in all three cell types (Figure 6C). These results indicate that reprogramming of BRAFV600E-positive melanoma cells and in vitro differentiation of the iPCCs lead neither to the loss of the oncogene mutation nor to an epigenetic remodeling process that silences the gene. Fluorescence in situ hybridization (FISH) analysis also shows no gain or loss of gene copy numbers related to genes of the BRAF-MEK-ERK signaling pathway (Figure 6D). However, in vivo differentiation of HT-144-iPCCs generated tumors containing structures without ERK activity (Figure 6E), suggesting that MAPK activity is restricted to cells of a specific tissue type.
Figure 6.
Epigenetic Modifications Induced by Nuclear Reprogramming Lead to Therapy Resistance against MAPK Inhibitors in Melanoma Cells
(A) Oncogene-specific PCR analysis of BRAFV600E in the parental HT-144 cells, HT-144-iPCCs, and iPCC-derived fibroblast-like cells. Human fibroblasts were used as a negative control.
(B) Methylation analysis of the BRAF locus in HT-144, HT-144-iPCCs, and HT-144-dFLCs.
(C) ERK activity in reprogrammed HT-144-iPCCs and fibroblast-like cells compared with the parental cell line determined by western blot analysis of phosphorylated ERK. GAPDH was used as an internal reference control for semi-quantitative protein analysis.
(D) FISH of HT144 and HT144-iPCCs shows no gain or loss of gene copy numbers related to genes of the BRAF-MEK-ERK signaling pathway such as BRAF (shown are nuclei with signals for the BRAF gene locus [red] and a centromeric reference probe [green]).
(E) HT-144-iPCCs give rise to tumors with reduced ERK activity in distinct differentiated structures. Histological staining of total ERK (tERK) and phosphorylated ERK (pERK) in HT-144-iPCC-derived tumors.
(F) Treatment of parental melanoma cells and their reprogrammed counterparts with the MEK inhibitor trametinib and the BRAFV600E-specific inhibitor vemurafenib.
(G) Therapy response of MAPK inhibitor-treated HT-144 and two fibroblast-like in vitro differentiations. Cells were incubated with 1,000 nM trametinib and 100 nM vemurafenib and analyzed for their metabolic activity at indicated time points.
See also Figure S5.
Next, we investigated the response of reprogrammed melanoma cells and differentiated daughter cells to targeted melanoma therapies. Parental melanoma cells and the reprogrammed iPCCs were treated with 1 μM of the MEK inhibitor trametinib and with the oncogene-specific BRAF inhibitor vemurafenib. Treatment with trametinib reduced cell proliferation in all melanoma cell lines and resulted in the appearance of floating, dead cells. In accordance with their mutational status, HT-144 cells were sensitive to vemurafenib treatment, unlike the BRAF wild-type cell lines Mewo and SKMEL147. Compared with the parental cell lines, iPCCs showed increased therapy resistance against MAPK inhibition without affecting the expression of the pluripotency marker alkaline phosphatase (Figure 6F).
To exclude that the ectopic expression of the pluripotency factors facilitates the therapy resistance, we investigated the therapy response in HT-144-dFLCs. Concentrations of 1,000 nM trametinib and 100 nM vemurafenib, which effectively killed HT-144 melanoma cells, showed no significant effect on HT-144-dFLCs (Figures 6G and S5). These data suggest that despite the presence of the mutated oncogene and its signaling activity, epigenetic modifications can facilitate a loss of oncogene addiction, which in turn results in resistance to targeted therapies.
Discussion
Here, we present a method to induce a pluripotent-like state even in tumor cells with a high mutational load. Melanoma cells harboring BRAFV600E or NRAS mutations were amenable to reprogramming similarly to wild-type cells. In contrast to the “classical” reprogramming protocol, we constitutively overexpressed OCT4, SOX2, and KLF4 and cultivated the cells similar to mESCs in the presence of human LIF on dense feeder cells. Previous studies in fibroblasts described similar murine-like ESCs upon ectopic expression of OCT4, SOX2, KLF4, MYC, and NANOG when supplemented with LIF. Like our iPCCs, these cells formed tightly packed colonies and could not stabilize the maintenance of the pluripotent state (Buecker et al., 2010). In contrast to our study, those cells did not reactivate the expression of endogenous pluripotency markers. Recently it was demonstrated that ectopic expression of reprogramming factors can generate an alternative NANOG-positive cell state. Although these so-called F-class cells share many features with our iPCCs in terms of gene expression and transgene dependence, F-class cells did not undergo mesenchymal-to-epithelial transition (MET) (Tonge et al., 2014), an early event during the reprogramming progress (Li et al., 2010, Samavarchi-Tehrani et al., 2010). On a molecular level, the successfully completed MET manifests itself by an upregulation of E-cadherin (Chen et al., 2010). This indicates that iPSC-like tumor cells generated in this study proceeded further in the reprogramming process than the F-class cells (Figure 2D). Similarly to early reports, we found that endogenous expression of reprogramming genes can compensate for ectopic expression (Utikal et al., 2009, Montserrat et al., 2012). This allowed us to reprogram the melanoma cells with OCT4, SOX2, and KLF4 only, without using the oncoprotein MYC.
A defined pattern of epigenetic signatures determines a cellular fate. Nuclear reprogramming allows us to reset a cell’s specific profile of epigenetic marks to direct its cell fate using differentiation protocols. Resetting the epigenetic profile of melanoma cells into a pluripotent-like state facilitated the differentiation of melanoma iPCCs into terminally differentiated cells. Although all melanoma cell lines investigated in this study were sensitive to MEK inhibition and in the case of HT-144 additionally to BRAF inhibition, their respective melanoma iPCCs as well as iPCC-derived in vitro differentiations lost their oncogene dependence, indicated by the resistance to targeted therapy. The same phenomenon was observed in reprogrammed human myeloid leukemia cells, which lost their dependence on the BCR-ABL oncogene upon reprogramming or after terminal differentiation into non-hematopoietic lineages (Carette et al., 2010, Kumano et al., 2012).
Reprogramming toward pluripotency induces a stepwise increase in the developmental potential. This allows tumor cells to acquire a terminal differentiation other than its origin (Zhang et al., 2013). Fully reprogrammed murine R545 melanoma cells even gained the potential to give rise to a viable mouse (Utikal et al., 2009). Accordingly, we observed that BRAF mutant melanoma iPCCs can be differentiated into neurons and fibroblast-like cells in vitro. In vivo, the majority of iPCC-derived tumors did not contain melanoma cells. In contrast to our results, other studies showed that reprogrammed pluripotent cells tend to differentiate into the cell type of their origin. Reprogrammed pancreatic cancer cells predominately differentiate into pancreatic tissue, recapitulating early and late events of carcinoma development (Kim et al., 2013). This phenomenon is based on epigenetically anchored marks that generate an epigenetic memory (Bar-Nur et al., 2011, Kim et al., 2011). Here, we did not observe that the melanoma-derived iPCCs preferentially differentiate into a melanocytic lineage in vivo. Furthermore, no differences between Mewo-iPCCs, lacking BRAF or NRAS mutations, and other melanoma iPCCs, harboring mutations in members of the MAPK signaling, were observed. These results suggest that mutations of components of the MAPK pathway do not interfere with the epigenetic memory and hence do not influence differentiation toward the melanocytic lineage.
One major drawback of current melanoma therapy is the development of resistance mechanisms against novel therapeutics targeting the MAPK members RAF or MEK. Upon MAPK inhibition, additionally acquired mutations reactivate the MAPK pathway by gain of RAF gene copy numbers (Shi et al., 2012, Villanueva et al., 2013), and mutations in RAS (Poulikakos et al., 2011) or receptor tyrosine kinases (Nazarian et al., 2010), leading to therapy resistance. However, recent data revealed that a considerable amount of resistant melanomas show no genomic but rather transcriptomic alterations as drivers of therapy resistance caused by epigenetic modifications (Hugo et al., 2015). Here, we demonstrated that epigenetic modifications in melanoma cells induced by nuclear reprogramming can lead to therapy resistance against BRAF and MEK inhibitors. This supports the notion that therapy response is linked to the cellular differentiation state. A dedifferentiation of melanoma cells was already linked to the resistance to adoptive T cell therapy (Landsberg et al., 2012), indicating a clinical relevance of melanoma cell state for therapy outcome.
Global epigenetic remodeling processes are well-known hallmarks of tumor development and also play an import role in melanomagenesis (reviewed in Lee et al., 2014). Nevertheless, the vast majority of melanomas shares common mutational events. In most cases, mutations leading to constitutively active RAS or RAF molecules combined with loss of tumor suppressors are key drivers of melanomagenesis (Hodis et al., 2012). Thus, nuclear reprogramming provides a tool to study the influence of a tumor genome on tumorigenesis in the context of specific cellular differentiation states. In this study, melanoma cells harboring BRAF mutations lost their tumor-initiating potential during the reprogramming process. Furthermore, dedifferentiation of melanoma cells is assumed to enhance their malignant potential (Landsberg et al., 2012). Expression of single stem cell factors such as OCT4 or SOX2 increases melanoma stem cell properties, leading to a more aggressive tumor phenotype (Kumar et al., 2012, Santini et al., 2014, Weina et al., 2016). In contrast, the simultaneous expression of three pluripotency markers leading to a semi-stable pluripotent state reduces the tumor-initiating potential.
In the majority of melanomas, oncogenic mutations of genes of the MAPK pathway drive tumor development. The well-known BRAFV600E mutation is responsible for constitutively active BRAF leading to hyperactive MAPK signaling. Targeting the tumor-specific mutation with small molecules provided a breakthrough in melanoma therapy, although resistance mechanisms leading to disease relapse rapidly damped expectations. We demonstrated that melanoma cells can acquire a metastable pluripotent state independent of BRAF or NRAS mutations. The cells reactivate expression of endogenous pluripotency markers and show further characteristics of PSCs, such as the potential to generate teratomas after subcutaneous injection into the flanks of immunocompromised mice. Interestingly, OCT4 expression was not upregulated, which might prevent the acquisition of a stable pluripotent state. In accordance, reprogrammed tumor cells remained dependent on the ectopic overexpression of the reprogramming factors.
Here, we demonstrated that a melanoma genome could be reprogrammed into a metastable mESC-like state of pluripotency. We could also show that subsequent differentiation of these pluripotent cells toward the mesodermal lineage was consistent with significantly impaired tumorigenicity. Therefore, we demonstrated a direct correlation between the tumorigenic potential of a cancer cell and its differentiation status.
Experimental Procedures
Ethics Statement
Experiments with primary human material were conducted with patients’ informed consent and were approved by the Medical Ethics Committee of the Medical Faculty Mannheim, Heidelberg University. Animal experiments were approved by the Animal Experiments Committee.
Cell Culture of Tumor Cells and Murine Embryonic Fibroblasts
The BRAFV600E mutant melanoma cell lines HT-144 and A375, the BRAFV600D mutant cell line WM266.4, the NRAS mutant cell line SK-MEL147, the BRAF and NRAS wild-type cell line Mewo, and murine embryonic fibroblasts were cultivated in DMEM (Gibco Life Technologies) with 4,500 mg/L glucose and 4 mM L-alanyl-L-glutamine supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Biochrom), 1% (v/v) 100× non-essential amino acids (NEAA; Sigma-Aldrich), 100 units/mL penicillin (Sigma-Aldrich), 100 μg/mL streptomycin (Sigma-Aldrich), and 0.1 mM 2-mercaptoethanol (Gibco Life Technologies). When 80% confluence was reached, cells were passaged using a 21-mM trypsin solution (Sigma-Aldrich). All cells were grown in a humidified atmosphere at 37°C and 5% CO2. Trametinib and vemurafenib (both Selleck Chemicals) were dissolved in DMSO (Carl Roth) and administered as indicated.
Generation of iPSCs and iPCCs
For reprogramming of human tumor cells, fibroblasts and melanocytes (105 cells per cm2) were seeded on gelatin-coated plates and transduced with FUdeltaGW-rtTA-zeocin. Cells were selected with 100 μg/mL zeocin (Invivogen) in complete medium and then co-infected with tetO-hSTEMCCA-puro-loxP encoding for OCT4, SOX2, KLF4, and a puromycin resistance gene or alternatively with tetO-hSTEMCCA-loxP encoding for OCT4, SOX2, KLF4, and MYC. The next day, superinfection was performed to reach higher efficiencies. All transductions were conducted for 24 hr at 37°C in DMEM supplemented with 10 μg/mL polybrene (Sigma-Aldrich). Twenty-four hours after the last infection, 105 cells in complete medium were plated onto 6-well plates coated with gelatin. After the cells attached, doxycycline (Sigma-Aldrich) was added to the medium to induce transgene expression. From here on the medium was changed every second day. After 30–40 days the first colony-forming cells originated. To create reprogrammed clones derived from single cells, we manually transferred individual colonies onto fresh feeder cells in DMEM/F12 (Gibco Life Technologies) with 20% (v/v) knockout serum replacement (KOSR) (Gibco Life Technologies), 2 mM L-glutamine (Sigma-Aldrich), 1% (v/v) NEAA, 100 units/mL penicillin, 100 μg/mL streptomycin, 0.1 mM 2-mercaptoethanol, and 1 μg/mL doxycycline, supplemented with 10 ng/mL human LIF (Sigma-Aldrich).
Human iPSC Culture
Stable clones of human iPSCs were cultivated under xeno-free cell-culture conditions using a synthetic surface matrix. One day in advance, 6-well plates were coated with Matrigel (Stemgent) for 1 hr at room temperature and stored at 4°C. Human iPSCs were washed and undifferentiated parts were manually dissociated into cell clusters of 50–100 cells. These small cell aggregates were transferred to Matrigel-coated plates in mTeSR1 medium (STEMCELL Technologies) containing 20% (v/v) mTeSR1 supplements of BSA, recombinant human basic fibroblast growth factor (bFGF), recombinant human TGF-B, lithium chloride, pipecolic acid, and G-aminobutyric acid (STEMCELL Technologies). Every other day medium was changed and the differentiated parts manually removed. Alternatively, human iPSCs were cultivated on feeder cells in human ES medium, which consists of DMEM/F12 supplemented with 20% (v/v) KOSR, 2 mM L-glutamine, 1% (v/v) NEAA, 100 units/mL penicillin, 100 μg/mL streptomycin, 0.1 mM 2-mercaptoethanol, and 10 ng/mL bFGF (Promokine).
Culture of Human iPCCs
Growth-arrested feeder cells were plated on gelatin-coated 6-well plates in complete medium and incubated for 2 days to ensure proper attachment and spread. iPCCs were then transferred onto feeder cells and medium was changed to DMEM/F12 with 20% (v/v) KOSR, 2 mM L-glutamine, 1% (v/v) NEAA, 100 units/mL penicillin, 100 μg/mL streptomycin, 0.1 mM 2-mercaptoethanol, and 1 μg/mL doxycycline, supplemented with 10 ng/mL human LIF. For passaging, cells were harvested every 4–7 days using trypsin and replated at 1:30 to 1:100 dilutions in medium containing 10 μM ROCK inhibitor (Y-27632, Stemgent). For separation of iPCCs from feeder cells by preplating, cells were harvested using trypsin, dissociated into single cells, washed, and resuspended in medium containing ROCK inhibitor. The cell suspension was then transferred onto gelatin-coated tissue culture plates and incubated for 2 hr at 37°C. Undifferentiated cells floating in the supernatant were collected and prepared for further experiments.
Fibroblast Differentiation
For the differentiation into fibroblast-like cells, HT-144-iPCCs were seeded onto 80% confluent mitotically inactivated feeder cells in naive human ES medium with 10 μM ROCK inhibitor and 1 μg/mL doxycycline, and cultivated for 2–5 days until small colonies were formed. Different protocols were followed to establish the clones I–III. Clone I was generated by changing the medium to DMEM/F12 1:1 with Neurobasal medium (Gibco Life Technologies) containing 1% B27 (Gibco Life Technologies) and 0.5% (v/v) N2 supplement (Gibco Life Technologies) for 3 days. The medium was then also switched to complete medium with 20% (v/v) fetal calf serum (FCS). For clone II, iPCC colonies were cultivated in DMEM/F12 3:1 supplemented with 10% (v/v) FCS, 0.18 mM adenine (Sigma-Aldrich), 0.5 μg/mL hydrocortisone (Sigma-Aldrich), 100 pM cholera toxin (Sigma-Aldrich), 10 ng/mL EGF (Gibco Life Technologies), and 5 μg/mL insulin (Sigma-Aldrich) for 10 days and supplemented on days 4–10 with 0.5 nM BMP-4 (Promokine). Afterward, fibroblast-like cells were split and maintained in T75 cell-culture flasks with complete medium.
Neuronal Differentiation
For neuronal induction, 2 × 104 cells per cm2 were seeded on Matrigel-coated dishes in DMEM/F12 with 20% (v/v) KOSR, 2 mM L-glutamine, 1% (v/v) NEAA, 100 units/mL penicillin, 100 μg/mL streptomycin, 0.1 mM 2-mercaptoethanol, and 1 μg/mL doxycycline, supplemented with 10 ng/mL human LIF and 10 μM ROCK inhibitor. When small colonies of five to ten cells appeared, medium was changed to DMEM/F12 and Neurobasal mixed at a 1:1 ratio with 1% (v/v) B27 and 0.5% (v/v) N2, 100 ng/mL Noggin (R&D Systems), 0.5 μM LDN-193189 (Stemgent), 10 μM SB-431542 (Tocris), 2 μM CHIR-99021 (Selleckchem), 10 μM forskolin (Tocris), and 10 ng/mL bFGF for 3–10 days. Then, cells were cultivated for an additional 5–10 days without small compound inhibitors but in the presence of 10 ng/mL bFGF.
Methylation Array Analysis
Genome-wide methylation analysis using Illumina Infinium HumanMethylation450 BeadChips according to the manufacturer’s instructions was performed at the German Cancer Research Center (DKFZ) Genomics and Proteomics Core Facility. The data discussed in this publication have been deposited in the NCBI GEO (Edgar et al., 2002) and are accessible through series accession number GEO: GSE95816 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95816). Raw-intensity data files of the following publicly available HumanMethylation450 samples from the ENCODE project (Kellis et al., 2014) were downloaded from http://www.encodeproject.org: ENCBS111ENC. The software RnBeads (Assenov et al., 2014) was used for quality assessment, normalization, dimension reduction, and other analyses of the combined dataset.
Cell Viability Assay
Cell viability was analyzed using the alamarBlue Cell viability assay (Thermo Fisher Scientific). Cells were plated in black 96-well plates in phenol-free DMEM (Gibco Life Technologies). After 24 hr at 37°C, 100 μL of medium was added and supplemented with DMSO or inhibitors for final concentrations ranging from 1 nM to 1 μM. At indicated time points 1/10 alamarBlue was added followed by fluorescence measurement 2–4 hr later at 37°C using a SpectraMax M5 microplate reader with an excitation wavelength of 540 nm and an emission wavelength of 590 nm. Cell viability was calculated from the resulting change in fluorescence intensity normalized to cells exposed to vehicle only.
Animal Experiments
For analysis of tumor initiation and in vivo differentiation potential, 1 × 106 cells were resuspended in 50% Matrigel and subcutaneously injected in either NOD/SCID mice or NSG mice according to the German Animal Protection Law. Mice were euthanized and developing tumors were isolated when reaching the size of 1 cm in diameter.
Author Contributions
Conception and Design: M.B., N.K., K.W., and J.U.; Collection and/or Assembly of Data: M.B., D.N., E.O., M.R., and J.U.; Data Analysis and Interpretation: M.B., Y.A., L.L., C.G., C.P., V.U., and J.U.; Provision of Study Material or Patients: J.U.; Manuscript writing: M.B., D.N., N.K., and J.U.; Final approval of manuscript: M.B., D.N., Y.A., E.O., N.K., K.W., M.R., L.L., C.G., C.P., V.U., and J.U.; Financial support: J.U.; Administrative support: J.U.
Acknowledgments
The authors thank Daniel Roth, Jennifer Dworacek, and Sayran Arif-Said for excellent technical assistance. We thank the Genomics & Proteomics Core Facility and the Imaging and Cytometry Core facility of the DKFZ for expression profiling and provision of FACS devices. This work was supported by a grant from the German Cancer Aid (Max-Eder Research Group, no. 784395, to J.U.).
Published: April 6, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.03.007.
Accession Numbers
The accession number for the gene expression data reported in this paper is GEO: GSE95281. The accession number for the methylation data reported in this paper is GEO: GSE95816.
Supplemental Information
References
- Assenov Y., Müller F., Lutsik P., Walter J., Lengauer T., Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O., Russ H.A., Efrat S., Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz G., Talpaz M., Bornmann W., Kong L.Y., Donato N.J. Degrasyn activates proteasomal-dependent degradation of c-Myc. Cancer Res. 2007;67:3912–3918. doi: 10.1158/0008-5472.CAN-06-4464. [DOI] [PubMed] [Google Scholar]
- Bernhardt M., Galach M., Novak D., Utikal J. Mediators of induced pluripotency and their role in cancer cells—current scientific knowledge and future perspectives. Biotechnol. J. 2012;7:810–821. doi: 10.1002/biot.201100347. [DOI] [PubMed] [Google Scholar]
- Buecker C., Chen H.H., Polo J.M., Daheron L., Bu L., Barakat T.S., Okwieka P., Porter A., Gribnau J., Hochedlinger K. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 2010;6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellen D., Schlange T., Bauer M., Maurer F., Hynes N.E. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 2007;8:70–76. doi: 10.1038/sj.embor.7400849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Pruszak J., Varadarajan M., Blomen V.A., Gokhale S., Camargo F.D., Wernig M., Jaenisch R., Brummelkamp T.R. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Di Ruscio A., Patti F., Welner R.S., Tenen D.G., Amabile G. Multiple sclerosis: getting personal with induced pluripotent stem cells. Cell Death Dis. 2015;6:e1806. doi: 10.1038/cddis.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbach S., Novak D., Bernhardt M., Larribere L., Boukamp P., Umansky V., Utikal J. Loss of tumorigenic potential upon transdifferentiation from keratinocytic into melanocytic lineage. Sci. Rep. 2016;6:28891. doi: 10.1038/srep28891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galach M., Utikal J. From skin to the treatment of diseases–the possibilities of iPS cell research in dermatology. Exp. Dermatol. 2011;20:523–528. doi: 10.1111/j.1600-0625.2011.01282.x. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., Blelloch R., Brennan C., Yamada Y., Kim M., Chin L., Jaenisch R. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P., Nickerson E., Auclair D., Li L., Place C. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W., Shi H., Sun L., Piva M., Song C., Kong X., Moriceau G., Hong A., Dahlman K.B., Johnson D.B. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japtok J., Lojewski X., Naumann M., Klingenstein M., Reinhardt P., Sterneckert J., Putz S., Demestre M., Boeckers T.M., Ludolph A.C. Stepwise acquirement of hallmark neuropathology in FUS-ALS iPSC models depends on mutation type and neuronal aging. Neurobiol. Dis. 2015;82:420–429. doi: 10.1016/j.nbd.2015.07.017. [DOI] [PubMed] [Google Scholar]
- Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Zhao R., Doi A., Ng K., Unternaehrer J., Cahan P., Huo H., Loh Y.H., Aryee M.J., Lensch M.W. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hoffman J.P., Alpaugh R.K., Rhim A.D., Reichert M., Stanger B.Z., Furth E.E., Sepulveda A.R., Yuan C.X., Won K.J. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraehn G.M., Utikal J., Udart M., Greulich K.M., Bezold G., Kaskel P., Leiter U., Peter R.U. Extra c-myc oncogene copies in high risk cutaneous malignant melanoma and melanoma metastases. Br. J. Cancer. 2001;84:72–79. doi: 10.1054/bjoc.2000.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K., Arai S., Hosoi M., Taoka K., Takayama N., Otsu M., Nagae G., Ueda K., Nakazaki K., Kamikubo Y. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- Kumar S.M., Liu S., Lu H., Zhang H., Zhang P.J., Gimotty P.A., Guerra M., Guo W., Xu X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31:4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J., Mertens J., Kesavan J., Doerr J., Poppe D., Glaue F., Herms S., Wernet P., Kögler G., Müller F.J. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Lai J., Kong C.M., Mahalingam D., Xie X., Wang X. Elite model for the generation of induced pluripotent cancer cells (iPCs) PLoS One. 2013;8:e56702. doi: 10.1371/journal.pone.0056702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg J., Kohlmeyer J., Renn M., Bald T., Rogava M., Cron M., Fatho M., Lennerz V., Wölfel T., Hölzel M. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–416. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- Larribere L., Utikal J. De- and re-differentiation of the melanocytic lineage. Eur. J. Cell Biol. 2014;93:30–35. doi: 10.1016/j.ejcb.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Lee J.J., Murphy G.F., Lian C.G. Melanoma epigenetics: novel mechanisms, markers, and medicines. Lab. Invest. 2014;94:822–838. doi: 10.1038/labinvest.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lin F.K., Chui Y.L. Generation of induced pluripotent stem cells from mouse cancer cells. Cancer Biother. Radiopharm. 2012;27:694–700. doi: 10.1089/cbr.2012.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Han Q., Peng T., Peng M., Wei B., Li D., Wang X., Yu S., Yang J., Cao S. The oncogene c-Jun impedes somatic cell reprogramming. Nat. Cell Biol. 2015;17:856–867. doi: 10.1038/ncb3193. [DOI] [PubMed] [Google Scholar]
- Lojewski X., Staropoli J.F., Biswas-Legrand S., Simas A.M., Haliw L., Selig M.K., Coppel S.H., Goss K.A., Petcherski A., Chandrachud U. Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum. Mol. Genet. 2014;23:2005–2022. doi: 10.1093/hmg/ddt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macville M., Schröck E., Padilla-Nash H., Keck C., Ghadimi B.M., Zimonjic D., Popescu N., Ried T. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Montserrat N., Ramírez-Bajo M.J., Xia Y., Sancho-Martinez I., Moya-Rull D., Miquel-Serra L., Yang S., Nivet E., Cortina C., González F. Generation of induced pluripotent stem cells from human renal proximal tubular cells with only two transcription factors, OCT4 and SOX2. J. Biol. Chem. 2012;287:24131–24138. doi: 10.1074/jbc.M112.350413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.K., Attar N., Sazegar H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukaya D., Minami K., Hoshikawa R., Yokoi N., Seino S. Preferential gene expression and epigenetic memory of induced pluripotent stem cells derived from mouse pancreas. Genes Cells. 2015;20:367–381. doi: 10.1111/gtc.12227. [DOI] [PubMed] [Google Scholar]
- Poulikakos P.I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M.T. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Santini R., Pietrobono S., Pandolfi S., Montagnani V., D'Amico M., Penachioni J.Y., Vinci M.C., Borgognoni L., Stecca B. SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene. 2014;33:4697–4708. doi: 10.1038/onc.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D., Park E.S., Fisher P.B. Defining the mechanism by which IFN-beta dowregulates c-myc expression in human melanoma cells: pivotal role for human polynucleotide phosphorylase (hPNPaseold-35) Cell Death Differ. 2006;13:1541–1553. doi: 10.1038/sj.cdd.4401829. [DOI] [PubMed] [Google Scholar]
- Shi H., Moriceau G., Kong X., Lee M.K., Lee H., Koya R.C., Ng C., Chodon T., Scolyer R.A., Dahlman K.B. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanslowsky N., Haase A., Martin U., Naujock M., Leffler A., Dengler R., Wegner F. Functional differentiation of midbrain neurons from human cord blood-derived induced pluripotent stem cells. Stem Cell Res. Ther. 2014;5:35. doi: 10.1186/scrt423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker S.H., Feber A., Engström P.G., Carén H., Kurian K.M., Takashima Y., Watts C., Way M., Dirks P., Bertone P. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabar V., Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat. Rev. Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tonge P.D., Corso A.J., Monetti C., Hussein S.M., Puri M.C., Michael I.P., Li M., Lee D.S., Mar J.C., Cloonan N. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516:192–197. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- Utikal J., Maherali N., Kulalert W., Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J., Infante J.R., Krepler C., Reyes-Uribe P., Samanta M., Chen H.Y., Li B., Swoboda R.K., Wilson M., Vultur A. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–1099. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weina K., Wu H., Knappe N., Orouji E., Novak D., Bernhardt M., Hüser L., Larribère L., Umansky V., Gebhardt C. TGF-β induces SOX2 expression in a time-dependent manner in human melanoma cells. Pigment Cell Melanoma Res. 2016;29:453–458. doi: 10.1111/pcmr.12483. [DOI] [PubMed] [Google Scholar]
- Yagi T., Ito D., Okada Y., Akamatsu W., Nihei Y., Yoshizaki T., Yamanaka S., Okano H., Suzuki N. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- Zhang X., Cruz F.D., Terry M., Remotti F., Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32:2249–2260. doi: 10.1038/onc.2012.237. 2260.e1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang D., Mannava S., Grachtchouk V., Tang W.H., Patil S., Wawrzyniak J.A., Berman A.E., Giordano T.J., Prochownik E.V., Soengas M.S. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–6634. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.