Summary
The striking rise of obesity-related metabolic disorders has focused attention on adipocytes as critical mediators of disease phenotypes. To better understand the role played by excess adipose in metabolic dysfunction it is crucial to decipher the transcriptional underpinnings of the low-grade adipose inflammation characteristic of diseases such as type 2 diabetes. Through employing a comparative transcriptomics approach, we identified IRF1 as differentially regulated between primary and in vitro-derived genetically matched adipocytes. This suggests a role as a mediator of adipocyte inflammatory phenotypes, similar to its function in other tissues. Utilizing adipose-derived mesenchymal progenitors we subsequently demonstrated that expression of IRF1 in adipocytes indeed contributes to upregulation of inflammatory processes, both in vitro and in vivo. This highlights IRF1's relevance to obesity-related inflammation and the resultant metabolic dysregulation.
Keywords: human adipocytes, adipose inflammation, metabolic disease, IRF1
Graphical Abstract
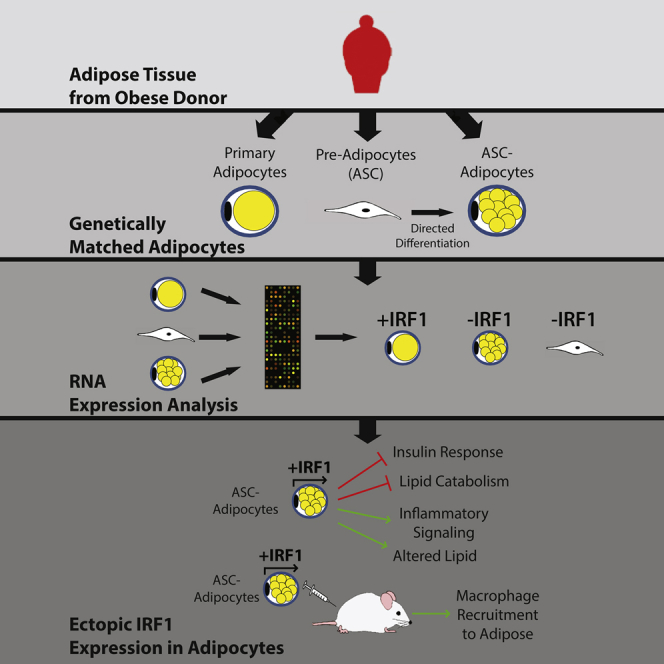
Highlights
-
•
Primary human adipocytes strongly express IRF1 compared with ASC adipocytes
-
•
ASC adipocytes expressing IRF1 exhibit phenotypes associated with metabolic disease
-
•
Adipocyte IRF1 level in vivo leads to recruitment of pro-inflammatory macrophages
-
•
Comparison of in vivo and in vitro adipose inflammation and metabolic phenotypes
In this report, Cowan and colleagues decipher the transcriptional underpinnings of low-grade adipose inflammation. Employing comparative transcriptomics analysis of primary human adipocytes and genetically matched in vitro differentiated counterparts, IRF1 was identified as a mediator of adipocyte inflammatory phenotypes. Both in vitro and in vivo IRF1 expression in adipocytes contributed to upregulation of inflammation.
Introduction
Obesity is a disease in which the energy reserve stored in the adipose tissue has increased excessively to cause adverse health effects. Adipocytes play a critical role in the regulation of triglyceride storage, glucose homeostasis, and energy expenditure. Excess adipose can disrupt this regulation. Obesity-related metabolic diseases, including type 2 diabetes mellitus (T2DM), have reached epidemic proportions and have been recognized as a leading contributor to morbidity and premature mortality (Ogden et al., 2012). More than 85% of people diagnosed with T2DM are overweight or obese (ADA, 2012).
Adipocyte biology has garnered increased attention because of this. Chronic low-grade inflammation of adipose tissue has emerged as an important contributor to obesity-induced insulin resistance (Gregor and Hotamisligil, 2011). Inflammation of adipose tissue is thought to occur due to nutrient overload in adipocytes, activating inflammatory pathways and leading to the recruitment of immune cells. In the context of obesity, adipocytes are a critical source of inflammatory chemokines that contribute to metabolic disease.
Transcriptional profiling of adipocytes has identified several regulators of adipogenesis (transcription factors, long non-coding RNAs, and microRNAs) (Urs et al., 2004). Unfortunately, few transcriptional regulators of adipose inflammation have been identified due to the in vitro nature of these studies. Therefore, the mechanistic underpinnings of how adipocytes contribute to metabolic diseases such as T2DM are underappreciated. To identify determinants of adipose inflammation, we transcriptionally profiled primary human adipocytes isolated from obese donors. We then compared expression data with in vitro-derived adipocytes. By focusing on differentially regulated transcription factors with a known function in immune response, we prioritized interferon regulatory factor 1 (IRF1). Subsequently we examined its role in adipose inflammation.
Results
Comparative Expression Analysis of Primary versus In Vitro-Derived Adipocytes
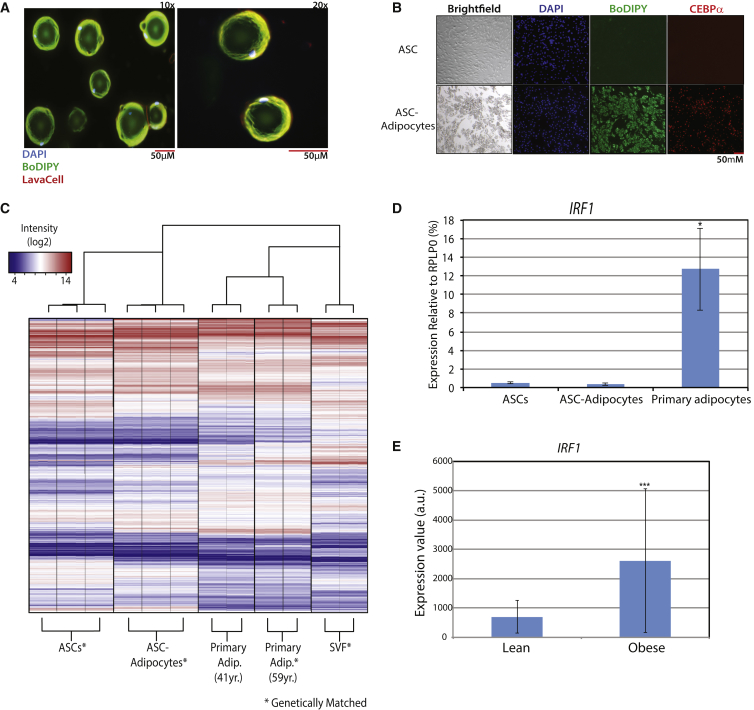
For expression analysis of primary and in vitro-derived adipocytes, we obtained whole subcutaneous adipose tissue from two obese females (body mass index >30; 41 and 59 years old) undergoing elective abdominoplasty. Primary adipocytes were isolated from whole adipose tissue. We verified the integrity of the adipocytes using fluorescence microscopy, showing the large unilocular lipid droplet typical of white adipocytes (Figure 1A). In addition, we isolated adipose stromovascular cells (ASCs) from one of the donors (59 years). The ASCs were subsequently differentiated into adipocytes at >90% efficiency (Figures 1B and S1A), and adipocyte-specific gene expression was measured (Figure S1B).
Figure 1.
Primary and ASC-Adipocyte Integrity and Transcriptional Profiling
(A) Primary adipocytes isolated from whole adipose. Adipocytes were stained with DAPI to visualize the DNA (blue), BoDIPY to stain the lipid droplet (green), and LavaCell to stain the plasma membrane (red), and visualized by wide-field microscopy.
(B) ASCs were differentiated into adipocytes and stained with DAPI to visualize the DNA (blue), BoDIPY to stain lipids (green), and an antibody to visualize C/EBPα (red).
(C) Hierarchical clustering and heatmap representation of transcriptional profiles (mRNA) from ASCs, ASC adipocytes, and primary adipocytes. Probe sets are colored according to expression level.
(D) Gene expression of IRF1 in ASCs (n = 3), ASC adipocytes (n = 3), and primary adipocytes (n = 4).
(E) Gene expression of IRF1 in isolated subcutaneous adipocytes of lean (n = 10) and obese (n = 10) individuals.
For all graphs, error bars represent SD, experiments were performed in biological triplicates and statistically significant p values are denoted by asterisks (∗p ≤ 0.05, ∗∗p ≤ 0.005).
RNA was extracted from the above cell types and subjected to whole-genome microarray analysis. Hierarchical clustering accurately grouped replicates of adipocytes, ASCs, and ASC adipocytes, with significant expression differences between groups (Figure 1C). We identified 1,749 genes with >1.7-fold higher abundance in primary adipocytes versus ASC adipocytes at a 0.1% false discovery rate (FDR) (Table S1). Gene ontology enrichment analysis for the primary adipocyte-enriched genes was consistent with a phenotype of inflammation, with enriched terms including “response to type I interferon,” “immune response,” and “positive regulation of chemotaxis” (Table S2). Genes with increased expression in the aforementioned pathways reflect a spectrum of inflammatory mediators, including interferon-regulated transcription factors (IRF1, IRF8), chemokines (CCL2/MCP1, CCL8), cytokines (IL6, IL8), Toll-like receptors (TLR1, TLR3), and inflammasome components (NLRP3). This inflammatory signature was not observed in undifferentiated ASCs either.
To control for contribution of stromovascular fraction cells (SVFs) to the inflammatory expression signature we isolated the SVF pellet, as contaminating immune cell transcripts should be identified in this fraction as well. Of the 1,749 transcripts identified to be specifically upregulated in primary adipocytes compared with ASC adipocytes, 1,009 were more abundant in the primary adipocytes compared with the SVFs, providing a conservative estimate for the fraction of genes with an adipocyte origin (Table S3). This list of 1,009 genes includes all of the inflammatory mediators listed above. The highest expressed immunoregulatory transcription factor in primary compared with ASC adipocytes was IRF1 (14.3-fold, p = 7.50 × 10−23). We sought to validate this result in two additional datasets. Comparing four additional primary adipocyte samples with three unmatched ASC adipocytes, we found a 33-fold upregulation of IRF1 in the primary adipocytes (Figure 1D). To ascertain that increased IRF1 expression is specific to obesity, we analyzed a publicly available dataset (GEO dataset GDS3602) comparing primary adipocytes from ten lean versus ten obese individuals, and found a 3.72-fold increase in IRF1 expression in adipocytes from obese individuals (Figure 1E; Lee et al., 2005). A large portion of genes overexpressed in obese individuals from this dataset (1.7-fold, 0.1% FDR) (Table S4) overlaps with genes we found upregulated in primary adipocytes (Figure S1C). The list of overlapping genes includes, but is not limited to, inflammatory genes such as the aforementioned IRF1, CCL2, CCL8, IL6, and IL8 (Table S5).
Expression of IRF1 in Adipocytes Activates the Immune Response
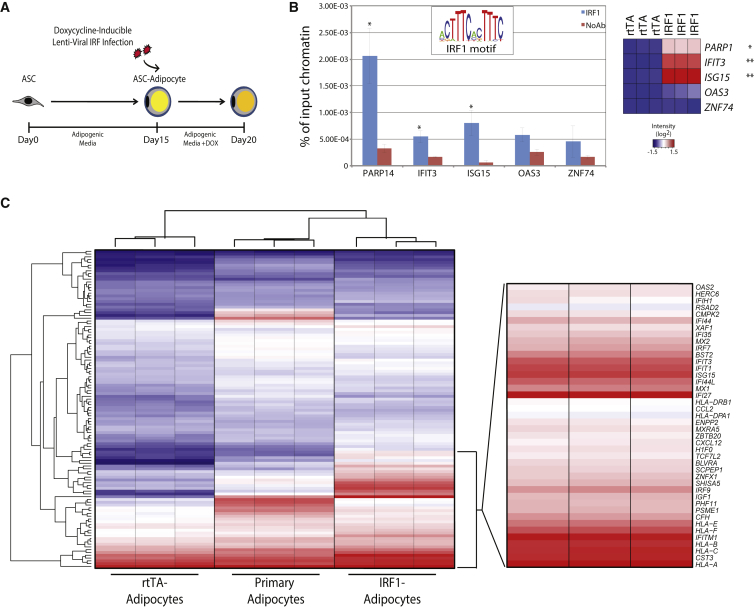
Utilizing lentivirus to express a doxycycline-inducible transgene, we devised a scheme to express IRF1 in ASC adipocytes (IRF1 adipocytes) (Figure 2A). After several days of doxycycline treatment we confirmed the expression of ectopic IRF1 (Figure S2A). Chromatin immunoprecipitation qPCR (ChIP-qPCR) revealed that ectopic IRF1 binds near genes that contain an IRF1-binding motif proximal to their promoter (PARP14, IFIT3, ISG15), and IRF1 binding correlates with increased expression of these genes (Figure 2B). There is no expression difference or IRF1 binding in genes without this motif (OAS3, ZNF74). When IRF1 is not overexpressed, we do not see this effect (Figure S2B). In differentiated adipocytes, IRF1 overexpression affected the mRNA levels of relatively few genes (138 upregulated, 80 downregulated, 1.7-fold, 0.1% FDR) (Figure 2C and Table S5). The majority of upregulated genes in IRF1 adipocytes are associated with innate immunity, including major histocompatibility complex (MHC) class I receptors, interferon-responsive genes, and chemotactic cytokines. Immune response-associated genes found to be upregulated in IRF1 adipocytes are also increased in primary adipocytes compared with ASC adipocytes. Furthermore, 103 of the 138 differentially regulated genes were previously reported as known interferon-responsive genes, confirming that we are observing a bona fide interferon-like response in IRF1 adipocytes (Samarajiwa et al., 2009). This leads us to conclude that ASC adipocytes expressing IRF1 more closely resemble primary human obese adipocytes.
Figure 2.
IRF1 Expression Leads to an Inflammatory Gene Signature
(A) Experimental scheme to ectopically express IRF1 in mature ASC adipocytes.
(B) ChIP-qPCR (n = 3) was performed for IRF1 at putative binding sites (H3K4me1 bound IRF1 motif) (PARP14, IFIT3, ISG15). OAS3 and ZNF74 represent a negative control site where IRF1 is not predicted to bind. Signals are expressed as percentage of total chromatin input. The heatmap represents expression levels of these genes in IRF1 and rtTA adipocytes.
(C) Hierarchical clustering and heatmap representation of transcriptional profiles (mRNA) from rtTA adipocytes, IRF1 adipocytes, and primary adipocytes. Probe sets are colored according to expression level. The highest expressed 30% of genes in IRF-1 adipocytes have been expanded to highlight their inflammatory nature.
Error bars represent SD, experiments were performed in biological triplicates, and statistically significant p values are denoted by asterisks (∗p ≤ 0.05, ∗∗p ≤ 0.005).
IRF1 Alters Metabolism and Lipid Droplet Composition of Adipocytes In Vitro
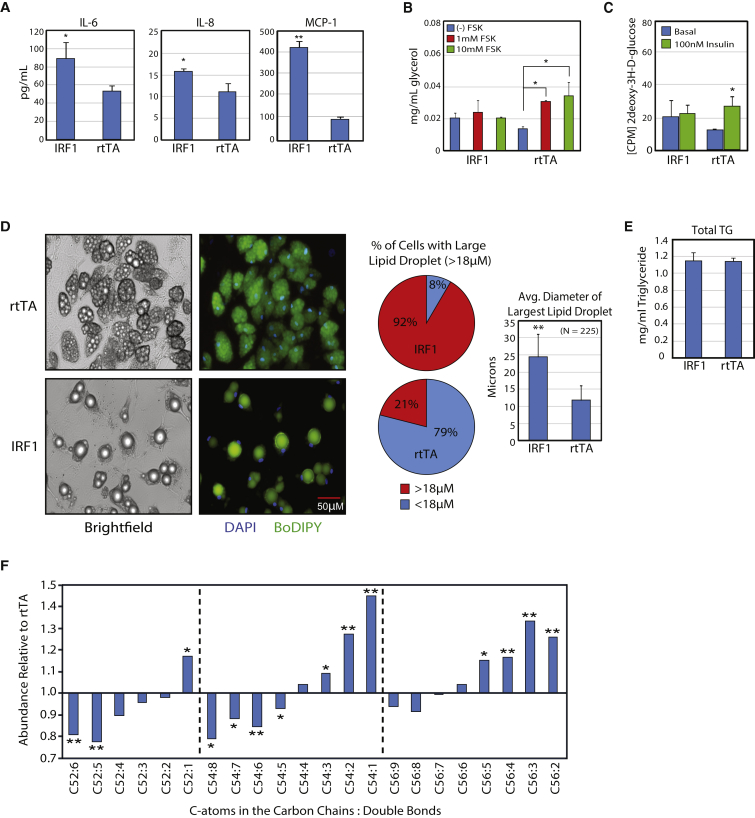
Experimental evidence shows that the interferon response pathway associates with altered adipocyte metabolism, as treatment of in vitro-derived adipocytes with interferon-γ induces insulin resistance (McGillicuddy et al., 2009). The known role of IRF1 in the interferon response and the upregulation of inflammatory genes in IRF adipocytes led us to hypothesize that IRF1 contributes to adipose inflammation. To further characterize IRF1 activation, we performed a multiplex ELISA to determine levels of cytokines and adipokines released by IRF1 adipocytes (Figures 3A and S2C). We found secretion of pro-inflammatory cytokines interleukin-6 (IL-6) and IL-8, as well as monocyte chemotactic protein 1 (MCP-1), were significantly elevated in IRF1 adipocytes concomitant with the increase in RNA. We also found these genes to be upregulated in obese individuals (Figure S2D).
Figure 3.
IRF1 Expression Impairs Adipocyte Metabolism and Affects Lipid Composition
(A) Multiplex ELISA assay was utilized to measure cytokines secreted into medium by IRF1 adipocytes compared with control adipocytes.
(B) Glycerol release into the medium in response to forskolin (FSK)-activated lipolysis was measured in IRF1 and rtTA adipocytes.
(C) Radiolabeled glucose uptake was measured in response to insulin.
(D) After 5 days of IRF1 expression, IRF1 adipocytes become unilocular. Lipid droplet size was measured after staining with the lipid dye BoDIPY (green) and the nuclear dye DAPI (blue). CPM, counts per minute.
(E) Total intracellular triglycerides (TG) were measured in IRF1 and rtTA adipocytes.
(F) Tandem mass spectrometric profiles of triglyceride composition for both IRF1 and rtTA adipocytes.
Error bars represent SD, experiments were performed in biological triplicates, and statistically significant p values are denoted by asterisks (∗p ≤ 0.05, ∗∗p ≤ 0.005). n = 3 for each experiment.
Since obese adipocytes display reduced lipolysis in vivo, we performed a lipolysis assay to measure the ability of IRF1 adipocytes to catabolize intracellular lipids. In response to β-adrenergic stimulation, IRF1 adipocytes were unable to increase triglyceride hydrolysis, unlike control cells (Figures 3B and S2E). To assess insulin sensitivity, we performed a glucose uptake assay. Expression of IRF1 abrogates insulin-mediated glucose uptake in ASC adipocytes, consistent with the increase in pro-inflammatory signaling and altered lipid metabolism (Figure 3C).
Unlike their in vivo counterparts, in vitro-derived adipocytes typically display multilocular lipid droplets. After several days of IRF1 expression, ASC adipocytes demonstrate a transition from a multilocular to unilocular state and the average size of the largest lipid droplet increases significantly (Figure 3D). The total intracellular triglyceride (TG) content in IRF1 adipocytes is similar to that of ASC adipocytes, indicating no lipid uptake or synthesis phenotype (Figure 3E). These data suggest that IRF1 has a specific effect on lipid droplet coalescence. Next we used tandem mass spectroscopy to assess lipid composition. We found that intracellular TGs in IRF1 adipocytes exhibit a greater degree of saturation, with fewer double bonds (p = 7 × 10−6) and longer carbon chains (p = 8 × 10−6) (Figures 3F and S2F).
IRF1 Expression in Adipose Leads to Localized Inflammation In Vivo
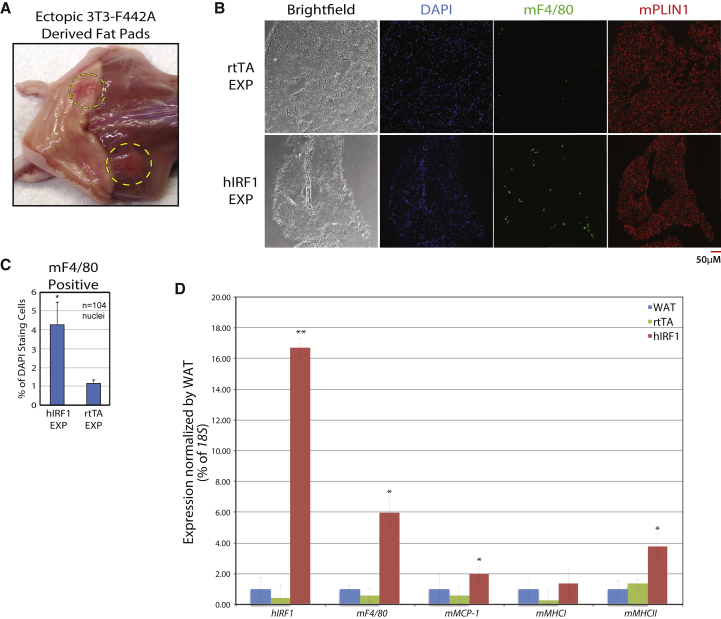
IRF1-adipocyte phenotypes in vitro are consistent with those reported for inflamed primary adipose tissue; hence, we tested whether IRF1 also contributes to adipose inflammation in vivo. We transfected the adipogenic mouse cell line 3T3-F442A with a human IRF1 (hIRF1) overexpression construct or control (Figure S3A). These cells were injected subcutaneously onto the flanks of nude mice. After 6 weeks of engraftment, ectopic fat pads were excised and assayed for inflammatory phenotypes (Figure 4A). We observed a significant increase in the number of macrophages localizing to implants expressing IRF1 compared with control implants or autologous white adipose tissue (WAT) (Figures 4B, 3C, S3B, and S3C). IRF1 implants exhibited increased expression of MHC class II and MCP-1 compared with autologous WAT, as well as an increase in the macrophage-specific gene F4/80, in line with the immunostaining shown above (Figure 4D). Inexplicably, we did find noticeable differences in gene expression between autologous WAT of hIRF1 and rtTA (reverse tetracycline-controlled transactivator) mice despite using biological triplicates. While this is worrying, there is consistency with the staining data. Altogether these data are consistent with both the transcriptional analysis of primary adipocytes and in vitro assays performed in ASC adipocytes, and imply that IRF1 contributes to adipose inflammation.
Figure 4.
IRF1 Leads to Increased Inflammation In Vivo
(A) 3T3-F442A cells were transduced with either rtTA alone, or human IRF1 (hIRF1) containing lentiviral particles, and were injected into 6-week-old nude mice (n = 4). Six weeks after injection, ectopic subcutaneous fat pads (dashed circles) were excised.
(B) Ectopic fat explants (EXP) were stained with DAPI (blue) and probed with α-F4/80 (green) and α-PLIN1 (red).
(C) F4/80-positive cells were counted relative to DAPI staining foci in hIRF1 explants versus rtTA explants and autologous WAT (see Figure S3C). A minimum of 104 DAPI foci were counted for each experiment.
(D) qPCR was used to assess expression of hIRF1 and inflammation-associated mouse genes in both explants and autologous WAT (n = 3).
Error bars represent SD, experiments were performed in biological triplicates, and statistically significant p values are denoted by asterisks (∗p ≤ 0.05, ∗∗p ≤ 0.005).
Discussion
The mechanistic underpinnings of obesity's contribution to chronic adipose inflammation are poorly understood. Few transcriptional regulators have been identified that activate adipose inflammatory cascades in vivo. Progress is impeded by the lack of direct experimentation on primary adipocytes and reliance on in vitro systems that cannot recapitulate obesity, endocrine regulation, and immune system interactions. However, previous studies have implicated the major inflammatory transcription factors nuclear factor κB and activator protein 1 as possible mediators of adipose inflammation associated with obesity, but the mechanism for activation is unclear (Suganami et al., 2007, Osborn and Olefsky, 2012). This study found several inflammatory mediators differentially expressed in primary versus differentiated adipocytes, most notably IRF1. This result was verified in an independent dataset showing an increase of IRF1 in obese individuals when compared with lean individuals. IRF1 has been identified as a transcriptional activator of type I interferon gene activation in response to viral infection (Harada et al., 1989). Additionally it has been observed that IRF4 represses early stages of adipogenesis (Eguchi et al., 2008) and modulates adipocyte lipid handling in vitro (Eguchi et al., 2011), suggesting a significant role for IRFs in adipose biology. However, there has been no study of IRF1 in human adipocytes. Expression of IRF1 in ASC adipocytes is sufficient to activate genes in the interferon response and inflammation pathways. Despite few transcriptional changes, IRF1 overexpression leads to phenotypes associated with metabolic disease, including insulin resistance and attenuated lipolysis. Further supporting its role in adipose inflammation, expression of IRF1 in 3T3-derived mouse adipose in vivo increases macrophage infiltration. We also demonstrate elevated MCP-1, which leads to inflammation and is associated with obesity, insulin resistance, and altered adipocyte metabolism (Kanda et al., 2006, Kim et al., 2006).
Lipidomic profiling of human subjects has revealed that the level of circulating saturated TGs is strongly correlated with insulin resistance and is a predictor of T2DM development (Rhee et al., 2011, Würtz et al., 2012). One striking phenotype of IRF adipocytes is the change in length and saturation level of TGs. The finding that expression of an immunoregulatory transcription factor leads to changes in composition of lipid droplet TGs is surprising. This finding in conjunction with our lipidomics data suggests that inflammation triggers production of more highly saturated fatty acids, in turn driving ER stress and further inflammation in a feedback loop (Wang et al., 2006). IRF1 adipocytes also acquire a unilocular lipid droplet, characteristic of primary adipocytes but absent in all in vitro models. Longer-chain, saturated TGs in IRF1 adipocytes could increase hydrophobicity of the intracellular lipids, potentiating the observed coalescence of lipids into a single large droplet.
Our data suggest that IRF1 expression in human adipocytes contributes to adipose inflammation and insulin resistance.
Experimental Procedures
Primary Adipocyte Isolation and ASC Cell Culture/Differentiation
Primary human adipocytes were obtained from discarded adipose tissue from two female elective abdominoplasty patients. Whole adipose was chopped up and digested with Liberase Blendzyme (Roche) for 1 hr at 37°C, forced through a 250-μm filter, and centrifuged. The floating adipocyte fraction was washed with PBS and re-collected. The stromovascular cell pellet was washed twice with PBS and plated onto gelatin-coated plates in ASC growth medium (DMEM, 10% fetal bovine serum [FBS], 1% penicillin-streptomycin, and 2.5 ng/mL basic fibroblast growth factor [Aldevron]). ASCs were brought to homogeneity by passaging and when ∼95% confluent, adipocyte medium (DMEM, 10% FBS, 1% penicillin-streptomycin, 0.1 μM dexamethasone, 10 μg/mL insulin, 0.5 μM rosiglitazone, 250 μM 3-isobutyl-1-methylxanthine [IBMX]) was added for 5 days. On day 6, ASCs were switched to adipocyte medium without IBMX for 14 additional days.
RNA Extraction/qRT-PCR/Transcriptional Profiling
Total RNA from all cell preparations was extracted using TRIzol (Invitrogen). RNA quality was determined using an RNA nanochip on a Bioanalyzer (Agilent Technologies). RNA was converted to cDNA using the Superscript First-Strand III kit (Invitrogen) and qRT-PCR was carried out using a Realplex Mastercycler (Eppendorf) with Quantifast-SYBR Green Mastermix (Qiagen). Primer sets are listed in supplemental Experimental Procedures.
RNA samples were processed by Asuragen and microarray analysis was performed on the Affymetrix Human Genome U133 Plus 2.0 Array. Arrays were hybridized overnight at 50°C, washed, stained, and scanned on a GeneChip Scanner 3000. Microarray data was analyzed using R/Bioconductor (http://www.bioconductor.org). Raw data were normalized by robust multi-array averaging (Bolstad et al., 2003) using a custom Chip Description File from the Michigan Microarray Lab (http://brainarray.mbni.med.umich.edu, version 13). Primary adipocyte RNA is from Zenbio, cat. #RNAmi-Q10-1, RNAmi-Q10-2, and RNA-Q10-3. GEO data were taken from Lee et al. (2005), dataset record GEO: GDS3602.
Microarray Gene and Sample Clustering
To select a minimally biased set of genes for hierarchical clustering, we used the R package limma (Smyth, 2005) to test for differential expression across all ten pairwise comparisons between biologically distinct samples: primary adipocytes (59 and 41 years of age), ASCs (59 years), ASC adipocytes (59 years), and the pelleted SVF from disassociated adipose tissue (59 years). Hierarchical clustering was performed on the 2,489 probe sets with five or more significantly different pairwise comparisons (FDR = 0.001). The hmap function in the seriation R package (Hahsler et al., 2008) was used to generate the heatmap.
Production of Inducible Transgenes/Lentivirus Transduction
Doxycycline-inducible lentiviral Gateway (Invitrogen) vector containing IRF1 was constructed by PCR amplification from verified cDNA clones (GenBank: NM_002198) (GeneCopoeia), and standard TOPO cloning (Invitrogen) techniques were used to generate entry clones. The IRF1 construct was transduced via calcium chloride transfection into HEK293T cells as previously described (al Yacoub et al., 2007). Media containing viral supernatant was collected, filtered, and used to infect ASC adipocytes. Infection efficiency was measured as compared by co-infection with a GFP lentiviral construct. Expression of IRF1 was induced by addition of doxycycline (1 μg/mL final) to the adipogenic medium.
ChIP
ASC adipocytes were crosslinked in 1% formaldehyde and quenched with glycine (200 mM final). Cells were washed several times in PBS. ChIP-qPCR was performed with an antibody against IRF1 (Santa Cruz Biotechnology, sc-497) as previously described (Camahort et al., 2007). All ChIPs were graphed as percentage of total chromatin input. qPCR was performed as described above.
Protein Normalization
Protein concentration used to normalize other assays was measured using the Bradford protein assay (Bio-Rad).
Metabolic Assays
For glucose uptake, after 5 days of IRF1 expression ASC adipocytes were serum starved in low-glucose DMEM containing 0.2% BSA overnight. Cells were incubated in KRH buffer (120 mM NaCl, 5 mM KCl, 1 mM MgSO4, 0.3 mM CaCl2 and 10 mM HEPES [pH 7.4]) in the presence or absence of 100 nM insulin for 30 min at 37°C. Non-specific uptake was measured in the presence of 10 μM cytochalasin B. Glucose uptake was measured by incubating cells with 0.5 μCi/mL−1 2-deoxy-D-[3H]glucose (Perkin-Elmer) for 5 min at 37°C.
For glycerol release, after 5 days of IRF1 expression ASC-adipocyte cells were starved in DMEM with 1% FBS for 1 hr and then incubated in Hank's balanced salt solution with 2% fatty acid-free BSA with forskolin for 1 hr (0 μM, 1 μM, and 10 μM). The incubation medium was collected for glycerol measurement using a colorimetric assay kit (Sigma).
All assays were performed in biological triplicates and normalized to protein.
Multiplex ELISA Assay
Medium was collected 5 days after IRF1 expression and ELISA was performed on a Luminex 200 analyzer using the Human Adipocyte Multiplex Panel (Millipore, HADCYMAG-61K) as per manufacturer's instructions. The assay was performed in biological triplicate and normalized to protein.
Triglyceride Analysis/Lipidomics
Total TG levels were measured using a colorimetric assay kit (Cayman Chemicals). Protein concentration was used to normalize intracellular TG content. For lipidomics, after 5 days of transgene expression ASC adipocytes were washed with fresh medium. Ice-cold isopropanol was added, and cells were scraped and collected into cold tubes. Extracts were incubated for 1 hr at 4°C, than vortexed and centrifuged at 2,300 × g for 10 min. The supernatant was used for mass spectroscopic analysis. All data were acquired using a Sciex 4000 QTRAP mass spectrometer as previously described (Rhee et al., 2011). MultiQuant software (version 1.1; Applied Biosystems/Sciex) was used for automated peak integration, and peaks were manually reviewed for quality of integration. Internal standard peak areas were monitored for quality control and used to normalize analyte peak areas.
For the transduced ASCs we averaged six replicates for metabolite abundance of 44 TG species and computed an abundance ratio of the two (IRF1/rtTA). We built a regression model, predicting the abundance ratio using number of double bonds and total number of carbon atoms as variables as well as an interaction term to assess whether the relationship with number of double bonds varies with carbon number. Statistical significance of regression coefficients was assessed using the F test.
Transplantation
Differentiated 3T3-F442A preadipocytes were transduced with hIRF-1 or rtTA lentivirus at day 7 and injected subcutaneously on the backs of 6-week-old female NCr nude mice (n = 4, Taconic). The cells developed into mature adipocytes over the next 6 weeks. Mice were euthanized to harvest fat pads which were fixed, embedded in paraffin, and stained after sectioning. Harvard University Animal Care and Use Committee approved all animal procedures.
Immunostaining
The following antibodies and dilutions were used: α-CEBPA 1:100 (Santa Cruz, sc-61), α-PLIN1 1:200 (Sigma, P1873), and α-F4/80 1:200 (eBioscience, 14-4801-81). Lipid droplets were stained using BoDIPY neutral lipid dye (Thermo Fisher, D-3922), cell membranes using LavaCell dye (Active Motif, 15004), and cell nuclei with DAPI. Images were acquired using a Nikon Eclipse Ti-S microscope. Both NIS-Elements and ImageJ software packages were used for image analysis.
Author Contributions
M.F. and R.C. performed the research and wrote the manuscript. Y.-K.L. performed the research with support of F.X. R.E.G. and E.P.R. performed the lipidomics. R.C.D. carried out the bioinformatics analysis. C.A.C. designed the study, supported the finance, and approved the final manuscript.
Acknowledgments
We thank Greg Mower, and former and current members of the C.A.C. laboratory for technical assistance and discussion. We would also like to thank Alexa Nicholls of the Division of Plastic & Reconstructive Surgery at Massachusetts General Hospital for coordinating acquisition of human adipose tissue. R.C.D. acknowledges support from NIH/NHLBI K08 HL098361.
Published: April 13, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.03.014.
Accession Numbers
All microarray data were deposited in the GEO under GEO: GSE96062.
Supplemental Information
References
- ADA American Diabetes Association—standards of medical care in diabetes. Diabetes Care. 2012;35:S11–S63. doi: 10.2337/dc12-s011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al Yacoub N., Romanowska M., Haritonova N., Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. J. Gene Med. 2007;9:579–584. doi: 10.1002/jgm.1052. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Eguchi J., Yan Q.W., Schones D.E., Kamal M., Hsu C.H., Zhang M.Q., Crawford G.E., Rosen E.D. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7:86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J., Estall J.L., Klein U., Maratos-Flier E., Rosen E.D. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- Hahsler M., Hornik K., Buchta C. Getting things in order: an introduction to the R package seriation. J. Stat. Software. 2008;25:1–34. [Google Scholar]
- Harada H., Fujita T., Miyamoto M., Kimura Y., Maruyama M., Furia A., Miyata T., Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- Kanda H., Tateya S., Tamori Y., Kotani K., Hiasa K., Kitazawa R., Kitazawa S., Miyachi H., Maeda S., Egashira K. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Invest. 2006;116:1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.S., Park H.S., Kawada T., Kim J.H., Lim D., Hubbard N.E., Kwon B.S., Erickson K.L., Yu R. Circulating levels of MCP-1 and IL-8 are elevated in human obese subjects and associated with obesity-related parameters. Int. J. Obes. 2006;30:1347–1355. doi: 10.1038/sj.ijo.0803259. [DOI] [PubMed] [Google Scholar]
- Lee Y.H., Nair S., Rousseau E., Allison D.B., Page G.P., Tataranni P.A., Bogardus C., Permana P.A. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillicuddy F.C., Chiquoine E.H., Hinkle C.C., Kim R.J., Shah R., Roche H.M., Smyth E.M., Reilly M.P. Interferon γ attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J. Biol. Chem. 2009;284:31936–31944. doi: 10.1074/jbc.M109.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden C.L., Carroll M.E., Kit B.K., Flegal K.M. Prevalence of obesity in the United States, 2009-2010. NCHS Data Brief. 2012:1–8. [PubMed] [Google Scholar]
- Osborn O., Olefsky J.M. The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- Rhee E.P., Cheng S., Larson M.G., Walford G.A., Lewis G.D., McCabe E., Yang E., Farrell L., Fox C.S., O’Donnell C.J. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarajiwa S.A., Forster S., Auchettl K., Hertzog P.J. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 2009;37:D852–D857. doi: 10.1093/nar/gkn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Limma: linear models for microarray data. In: Gentlemen R., Carey V., Huber W., Irizarry R., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. pp. 397–420. [Google Scholar]
- Suganami T., Tanimoto-Koyama K., Nishida J., Itoh M., Yuan X., Mizuarai S., Kotani H., Yamaoka S., Miyake K., Aoe S. Role of the toll-like receptor 4/NF-κB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler. Thromb. Vasc. Biol. 2007;27:84–91. doi: 10.1161/01.ATV.0000251608.09329.9a. [DOI] [PubMed] [Google Scholar]
- Urs S., Smith C., Campbell B., Saxton A.M., Taylor J., Zhang B., Snoddy J., Jones Voy B., Moustaid-Moussa N. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J. Nutr. 2004;134:762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- Wang D., Wei Y., Pagliassotti M.J. Saturated fatty acids promote endoplasmic reticulum stress and liver injury in rats with hepatic steatosis. Endocrinology. 2006;147:943–951. doi: 10.1210/en.2005-0570. [DOI] [PubMed] [Google Scholar]
- Würtz P., Mäkinen V.-P., Soininen P., Kangas A.J., Tukiainen T., Kettunen J., Savolainen M.J., Tammelin T., Viikari J.S., Rönnemaa T. Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes. 2012;61:1372–1380. doi: 10.2337/db11-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.