Summary
Hematopoietic stem cell (HSC) defects can cause repopulating impairment leading to hematologic diseases. To target HSC deficiency in a disease setting, we exploited the repopulating defect of Fanconi anemia (FA) HSCs to conduct an in vivo short hairpin RNA (shRNA) screen. We exposed Fancd2−/− HSCs to a lentiviral shRNA library targeting 947 genes. We found enrichment of shRNAs targeting genes involved in the PPARγ pathway that has not been linked to HSC homeostasis. PPARγ inhibition by shRNA or chemical compounds significantly improves the repopulating ability of Fancd2−/− HSCs. Conversely, activation of PPARγ in wild-type HSCs impaired hematopoietic repopulation. In mouse HSCs and patient-derived lymphoblasts, PPARγ activation is manifested in upregulating the p53 target p21. PPARγ and co-activators are upregulated in total bone marrow and stem/progenitor cells from FA patients. Collectively, this work illustrates the utility of RNAi technology coupled with HSC transplantation for the discovery of novel genes and pathways involved in stress hematopoiesis.
Keywords: Fanconi anemia, hematopoietic stem cells, PPARγ, oxidative stress
Highlights
-
•
In vivo screening identifies of deleterious Pparγ effect on HSCs
-
•
Pharmacological activation of Pparγ impaired normal HSC repopulation
-
•
Inhibition of Pparγ improves Fancd2-deficient HSC repopulation ability
Pang and colleagues identified by shRNA screening a deleterious effect of Pparγ activation under replicative/oxidative stress on HSC repopulation activity. These results indicate that inhibition of Pparγ could be a valuable therapeutic approach to improve HSC homeostasis in Fanconi anemia and to improve HSC transplantation.
Introduction
Hematopoietic stem cells (HSCs) are a distinct population of multipotent cells that can self-renew and differentiate into various types of blood cells and thus are responsible for maintenance and homeostasis of a healthy hematopoietic system (Morrison et al., 1995, Orford and Scadden, 2008, Orkin and Zon, 2008). HSCs are exposed daily to internal and external stresses which in turn lead to DNA damage. Accumulation of DNA damage in hematopoietic stem and progenitor cells (HSPCs) during the cell's life span is a factor of hematopoietic system aging and degeneration, and likely contributes to transformation and cancer development (Rossi et al., 2008). Accelerated bone marrow (BM) degeneration leading to BM failure and high risks of leukemia development is frequently observed in diseases with a deficiency in DNA repair pathways such as Fanconi anemia (FA) (Taniguchi and D'Andrea, 2006).
FA is an inherited disease caused by mutations in any of 17 already identified FA DNA repair pathway genes (FANCA-S) (Kottemann and Smogorzewska, 2013, Sawyer et al., 2015). FA proteins have been mainly studied for their role in genomic DNA repair and genome integrity. Upon DNA damage, eight of the FA proteins (FANCA, -B, -C, -E, -F, -G, -L, and -M) interact to form the FA core complex responsible for FANCD2 and FANCI activation by mono-ubiquitination (Kottemann and Smogorzewska, 2013). FANCD2 activation is essential for genome integrity maintenance upon double DNA strand break or interstrand crosslinking by favoring the homologous recombination (HR) DNA repair pathway.
BM failure and leukemia at a young age are the hallmarks of human FA patients, which made the study of FA of high interest for understanding the biology of HSC defect and malignant transformation. Mouse models have been developed for several FA genes (Parmar et al., 2009). Despite the mild phenotypes and absence of BM failure in a steady-state condition, FA mouse models present hypersensitivity to DNA-damaging agents, oxidative stress and a deficiency of HSC repopulation abilities (Parmar et al., 2009). Studies of these mouse models have allowed advances in understanding factors and pathways involved in HSC normal functions and BM failure. For example, the study of FANCD2 interaction partners has demonstrated the importance of FOXO3a/FANCD2 interaction in HSC maintenance and resistance against oxidative stress (Li et al., 2010, Li et al., 2015). Recently, a study emphasizing the relationship between DNA damage and HSC impairment has shown that repeated stresses led to the accumulation of DNA damage and acceleration of functional impairment in FA HSCs (Walter et al., 2015). Despite this accumulation of knowledge about BM failure, potent treatment for FA patients is still needed. Unbiased genomic approaches by short hairpin RNA (shRNA) screens have been successfully used to discover new mechanisms in different pathologies (Mohr et al., 2014), and can be conducted at a stem cell level (Hope et al., 2010, Wang et al., 2012). Using such a strategy, several tumor suppressors and checkpoint factors have been highlighted, such as p53 and p21. Interestingly, several of these proteins have also been implicated in tissue aging and stem cell functions.
In this study, we took advantage of the well-established defective repopulation of FA HSCs and carried out an in vivo shRNA screening in the Fancd2 mouse model to identify genes whose inhibition would improve HSC functions during replicative stress. We demonstrated that deregulated Pparγ activity is a limiting factor for HSC self-renewal and repopulating capacity. Pparγ could be a potent target for improving and delaying FA BM failure and HSC aging in general.
Results
In Vivo shRNA Screening Identifies Proteins Involved in TGF-β and PPARγ Pathways
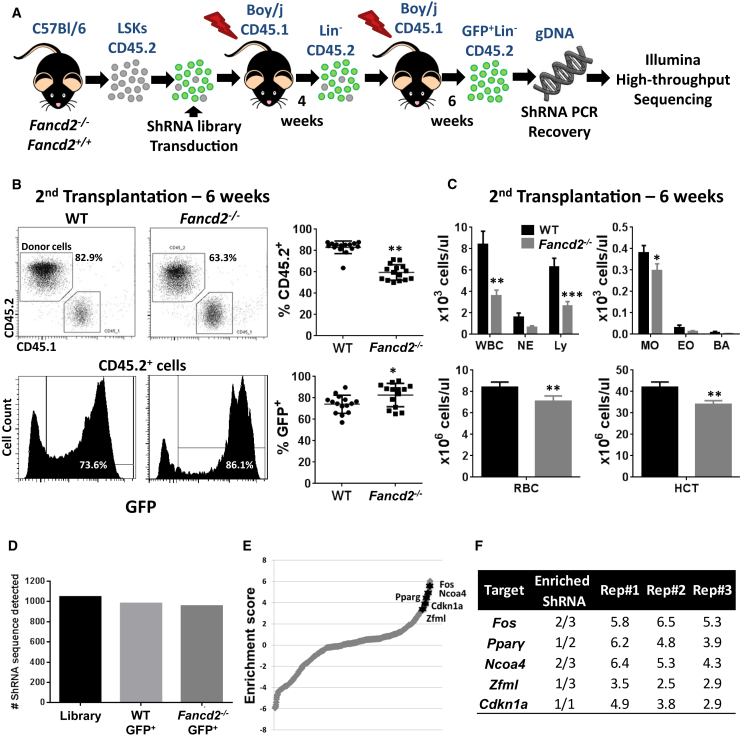
To identify new factors and pathways implicated in impaired functions of HSCs under stress conditions, we conducted an shRNA in vivo screening on transplanted Fancd2+/+ (wild-type [WT]) and Fancd2−/− Lin−Sca1+Kit+ cells (LSKs) (n = 21 donors/group, Figure 1A). To maximize the efficiency of the screening, we used Fancd2-deficient mice as they have the strongest phenotypes (Parmar et al., 2009) and HSPC deficiency compared with other FA mice used in our laboratory (Figures S1A and S1B). The shRNA library used in this screen was derived from previously published studies (Bric et al., 2009, Wang et al., 2012). The shRNA lentiviral vector used for the screening expressed a recombinant GFP protein, allowing a fast evaluation of transduction efficiency of the LSKs and the sorting of shRNA-expressing donor cells after transplantation (Figures S2A and S2B). Pooled LSKs (CD45.2) from WT or Fancd2−/− mice were transduced in vitro with a similar transduction efficiency of 70%–75% in both genotypes (Figure S2) and then transplanted (Figure 1A) into lethally irradiated recipients (CD45.1). A decreased blood donor chimerism (Figure 1B) and different blood parameters (Figure 1C) were observed for the mice transplanted with Fancd2−/− cells at 6 weeks after the second round of transplantation. Thus, the transduction of LSKs with the shRNA library did not rescue by itself the expected phenotype of Fancd2-deficient BM cells, suggesting that the experimental model is suitable for selection of beneficial shRNA. We observed a significant increase of GFP+ cells among donor cells of Fancd2−/− recipient mice at 6 weeks after the second round of transplantation (Figure 1B), indicating a possible selection of cells transduced by beneficial shRNAs.
Figure 1.
In Vivo shRNA Screening Reveals Candidate Targets in Pparγ Pathway
(A) Isolated LSKs from 8- to 12-week-old Fancd2−/− or Fancd2+/+ mice (n = 21/genotype) were transduced in vitro with a pooled shRNA lentivirus library. Transduced LSKs along with non-transduced LSKs were transplanted for two rounds into lethally irradiated Boy/J recipient mice (n = 18/groups, three different experiments). Six weeks after the second transplantation, CD45.2+Lin−GFP+ cells were isolated and integrated shRNA sequences were analyzed by deep sequencing.
(B) Plots and graphs represent the percentage of donor cells in blood at 6 weeks after the second transplantation (top) and the percentage of GFP+ cells in these donor cells (bottom).
(C) Bar graphs represent the results of blood analysis by Hemavet (Drew Scientific) of recipient mice 6 weeks after the second transplantation (n = 18 mice/genotype). WBC, white blood cells; NE, neutrophils; Ly, lymphocytes; Mo, monocytes; Eo, eosinophils; Ba, basophils; RBC, red blood cells; HCT, hematocrit.
(D) Numbers of shRNA detected by deep sequencing in the cells used to produce the lentivirus particle (library) and Lin− BM cells after the second round of transplantation.
(E) Graph showing the enrichment score profile (mean of three independent experiments) of all shRNA detected in Lin− BM cells, highlighting the four genes linked to the Pparγ pathway.
(F) This table depicts for each Pparγ linked candidate genes the number of targeting enriched shRNA (enrichment score >2) and the enrichment score of the best shRNA (according to mean value) into the three replicates.
WT, wild-type. Values are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We isolated CD45.2+GFP+Lin− cells from the secondary transplanted recipient mice and performed deep sequencing to determine the integrated shRNA sequences (Sims et al., 2011) (Figure S2). We found that the majority of the shRNAs were still present in the sorted cells of both genotypes (Figure 1D) when compared with the virus-producing cells, indicating no random loss of shRNAs during the experiment. The enrichment analysis conducted using ShRNAseq Pipeline (Sims et al., 2011) indicated the selection or loss of shRNAs in recipient mice of Fancd2−/− cells compared with the recipient of WT cells (Table S1 and Figure 1E). Significantly, we found enrichment of three targeted genes of the transforming growth factor β (TGF-β) pathway, Fos (Zhang et al., 1998), Elf (Chang et al., 2000), and Klf10 (Spittau and Krieglstein, 2012) (Figures S3B and S3C), and four targeted genes of the PPARγ pathway, Fos (Wan et al., 2007), Ncoa4 (Heinlein et al., 1999), Pparγ, and Zfml (Meruvu et al., 2011) (Figures 1E and 1F; Table S1).
Specific shRNA Knockdown of Identified Genes Improves Repopulation Activity of Fancd2−/− HSCs
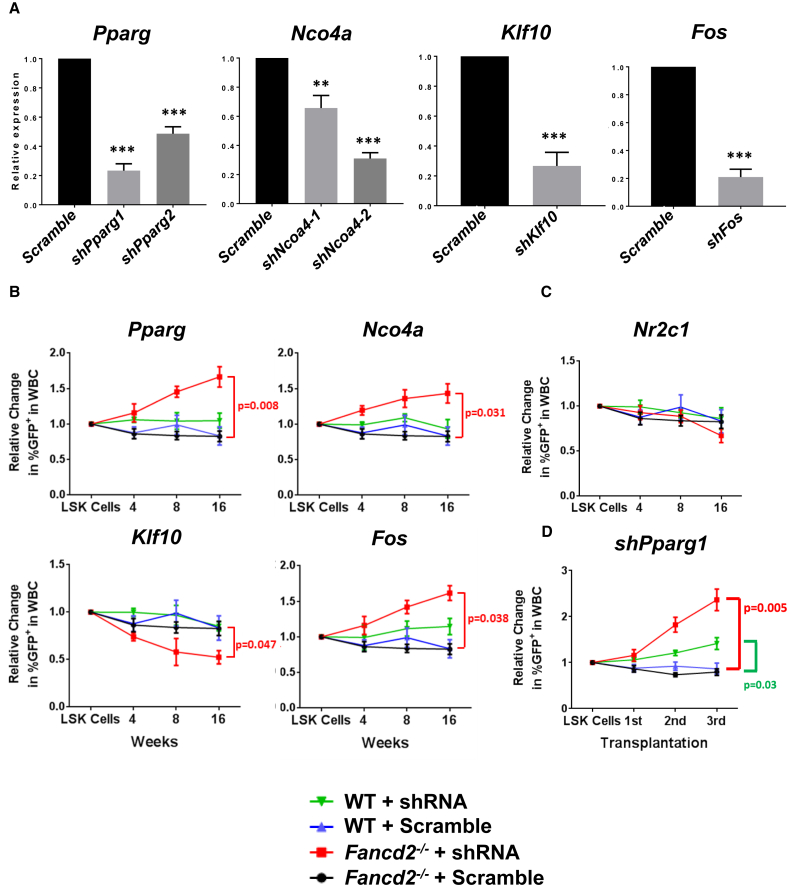
To validate the target genes identified by our screening strategy, we transduced WT and Fancd2−/− LSKs with shRNAs targeting the specific genes identified in our in vivo screen. We decided to focus on the Pparγ-related genes, because the pathway is the most enriched in our screen and because PPARγ is already a therapeutic target that could be interesting for clinical use (Schmidt et al., 2010). LSK cells were infected with Scramble or shRNA for each target gene (Figures 2A and S3A) to reach 20%–30% transduction efficiency and transplanted into lethally irradiated recipient mice. We followed the percentage of GFP+ in donor-derived blood cells as a function of time as an indicator of repopulation efficiency. As shown in Figures 2B and S3D, knocking down Pparγ, Ncoa4 or Fos led individually to an increased GFP+ proportion in Fancd2−/− blood donor-derived cells as a function of time. We confirmed for Pparγ that the shRNA vector was able to sustain a stable knockdown for at least 16 weeks (Figure S3E). We also confirmed the deleterious effect of Klf10 knockdown (Figure 2B and Table S1). As a non-specific control, we used a non-enriched shRNA targeting the nuclear receptor Nr2c1 and found no significant difference of GFP+ proportion 16 weeks after transplantation (Figure 2C).
Figure 2.
Specific shRNA Knockdown of PPARγ-Related Candidate Genes Ameliorates Repopulation Capacity of Fancd2−/− LSKs
(A) Real-time qPCR validation of shRNA efficiency in transduced LSK cells for the different constructs designed to target the indicated genes. Values are presented as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
(B–D) Isolated LSKs from Fancd2+/+ or Fancd2−/− mice (n = 6 donors/group, two independent experiments) were transduced with a specific shRNA construct or scrambled shRNA control. The transduction efficiency was 25%–33% before transplantation and was normalized to 1. Transduced cells along with untransduced cells were transplanted into lethally irradiated Boy/J (n = 8 mice/group, two independent experiments). The graphs indicate relative changes in GFP+ proportion among donor-derived blood cells (CD45.2) at different time points after transplantation (B and C), or 4 weeks of serial rounds of transplantations (D). Values are presented as mean ± SD.
Furthermore, we performed serial BM transplantation assays and confirmed the increased repopulation activity of shPparγ-transduced Fancd2−/− HSCs in secondary and tertiary recipient mice (Figure 2D). Interestingly, after the third round of transplantation we also found an increase of GFP+ proportion in shPparγ-transduced WT blood donor-derived cells compared with scramble shRNA-transduced cells (Figure 2D). This result indicates a possible benefit of Pparγ knockdown during repeated replicative stress on normal HSCs. Together, these results confirmed that specific knockdown of the targeted genes identified in the in vivo shRNA screen has an impact on the repopulation activity of Fancd2−/− HSCs, and that PPARγ could be a potential target for improvement of repopulation capacity and function of Fancd2−/− HSCs.
PPARγ Activation Impaired Function of Both WT and Fancd2-Deficient HSPCs
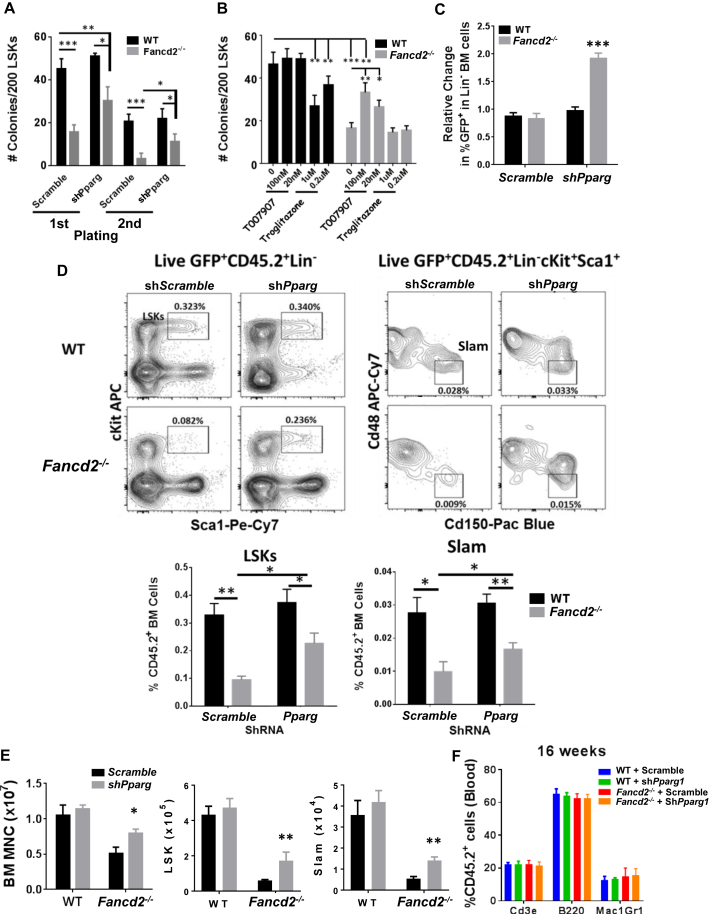
Next, we evaluated the effect of PPARγ inhibition on the function of HSPCs in vitro and in vivo. As expected, we observed with the shScramble a decrease in colony numbers for Fancd2−/− LSKs compared with WT LSKs after the first or second passage (Figure 3A). Targeting WT LSKs by shPparγ did not change the colony number during the first or second passage (Figure 3A). In contrast, Pparγ knockdown in Fancd2−/− LSKs led to a significant increase in colony number compared with shScramble in both the first and second round of culture. The Fancd2−/− LSK-derived colonies remained significantly lower than those of WT LSKs (Figure 3A). To confirm this result and to evaluate a possible pharmacological targeting of PPARγ activity in HSPCs, we used the well-characterized PPARγ antagonist T0070907 (Lee et al., 2002) and agonist troglitazone (Lee and Olefsky, 1995). Inhibition of PPARγ activity did not affect the colony number of WT LSK cells at 20 or 100 nM (Figure 3B). However, treatment by T0070907 significantly increased the colony-forming ability of Fancd2−/− LSKs, indicating a partial rescue when compared with WT LSKs. Significantly, we found a dose-dependent decrease in colonies formed by WT LSKs treated with the PPARγ agonist, indicating that the activation of PPARγ is sufficient to inhibit the colony-forming capacity of WT LSKs in vitro. Interestingly, troglitazone treatment had no effect on Fancd2−/− colony number, indicating that Pparγ activation is probably already at maximum in these cells.
Figure 3.
Inhibition of PPARγ Activity Improves HSC Self-Renewal and BM Repopulation Capacity
(A and B) CFU assay with LSK cells after shRNA knockdown (A) or treatment with the PPARγ agonist (troglitazone) or antagonist (T0070907) (B).
(C) LSKs isolated from Fancd2+/+ or Fancd2−/− mice (n = 5 donors/group, two independent experiments) were transduced with shRNA targeting Pparγ or scrambled shRNA control. The transduction efficiency was 25%–33% before transplantation and was normalized to 1. Transduced cells, along with untransduced cells, were transplanted into lethally irradiated Boy/J (n = 8 mice/group, two independent experiments). The graph shows relative changes in GFP+ proportion among donor-derived Lin− BM cells (CD45.2) at 16 weeks after transplantation.
(D–F) Transduced GFP+ LSKs were sorted and transplanted into lethally irradiated Boy/J recipient mice. (D) Representative plot and histograms depict the LSK (Lin−Sca1+cKit+) and SLAM (CD48−CD150+LSKs) percentage among GFP+ mononuclear donor-derived cells (CD45.2) at 16 weeks after transplantation. (E) Absolute number of BM MNC, LSKs, and SLAM cells at 16 weeks after sorted LSK transplantation. (F) Lymphocytes (Cd3e), B cells (B220), and granulocytes (Gr1Mac1) population percentage into CD45+ cells from the blood of 16-week transplanted mice.
WT, wild-type. Values are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next evaluated the consequence of Pparγ inhibition on HSC function in vivo. We transduced the WT and Fancd2−/− LSKs with shScramble or shPparγ lentivirus and transplanted the cells into lethally irradiated recipient mice. We evaluated the repopulating capacity of the donor HSCs by analyzing the percentage of GFP+ in recipient mice at 4 months post transplantation. We observed a markedly increased proportion of GFP+Lin− cells in the BM of the recipient mice transplanted with the shPparγ-transduced Fancd2−/− LSKs compared with the recipient of shScramble-transduced cells (Figure 3C). shRNA knockdown of Pparγ in WT LSKs did not change the GFP+ proportion of BM cells in recipient mice compared with shScramble-transduced cells. Consistent with previous reports (Ceccaldi et al., 2012, Li et al., 2015, Parmar et al., 2010), recipient mice transplanted with the shScramble-transduced Fancd2−/− LSKs had a decreased total number of donor-derived mononuclear cells as well as a decreased proportion and absolute number of LSKs and HSCs (SLAM, CD150+CD48−LSK) in the BM at 16 weeks after transplantation compared with mice receiving the shScramble-transduced WT LSKs (Figures 3D and 3E). Importantly, shRNA inhibition of Pparγ in Fancd2−/− LSKs led to not only a significant increase in total BM cells but also an augmentation of proportion and absolute number of LSKs and HSCs in the transplanted mice compared with shScramble-transduced Fancd2−/− LSKs (Figures 3D and 3E). The increased repopulation upon Pparγ knockdown did not lead to biased lineage commitment, as the different lineage proportions in the CD45+ blood cells is the same in all the conditions tested (Figure 3F). Altogether, these data indicate a deleterious effect of PPARγ activation on HSC function and suggest the potential benefit of PPARγ inhibition on Fancd2−/− HSCs.
Elevated Expression of PPARγ Co-activators and Target Genes in Fancd2−/− HSPCs
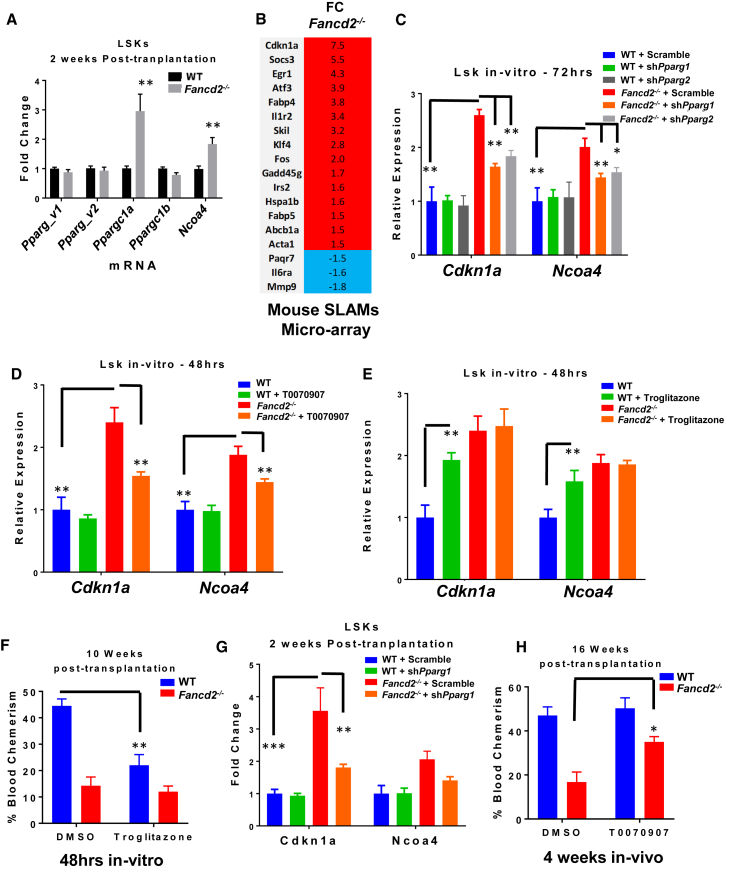
To determine the mechanism underlying deregulated PPARγ activity in Fancd2-deficient HSPCs, we evaluated the expression of PPARγ co-activators and target genes in donor-derived LSK cells 2 weeks after transplantation. At that time, the HSPCs were under a high proliferative and inflammatory stress because of the need to reconstitute the damaged hematopoietic system (Roy et al., 2012). Surprisingly, we observed no difference in the expression for the two Pparγ variant mRNAs in sorted CD45.2+ WT and Fancd2−/− LSKs from transplanted mice (Figure 4A). Since it has been described that Pparγ expression is not always a good indicator of activity, we evaluated the expression of the PPARγ co-activators Ncoa4 (Heinlein et al., 1999), Pparγc1a (Puigserver et al., 1998), and Pparγc1b (Lin et al., 2002). We observed significantly increased expression of Pparγc1a and Ncoa4 in donor-derived Fancd2−/− LSKs at 2 weeks post transplantation (Figure 4A). Interestingly, Ncoa4 was one of the hits in our shRNA screening (Figure 2B). We also analyzed our microarray data obtained with freshly isolated phenotypic HSCs (CD150+CD48−LSK; SLAM) from WT and Fancd2−/− mice (Li et al., 2015) (GEO: GSE64215). Several Pparγ target genes are deregulated in the Fancd2−/− SLAM population (Figure 4B). We did not observe in this microarray the deregulation of Ncoa4 or Pparγc1a, suggesting the possible requirement for higher stress conditions to upregulate their expression. As previously reported in FA-deficient cells (Barroca et al., 2012, Ceccaldi et al., 2012), we found upregulation of Cdkn1a (p21) in Fancd2−/− SLAM cells in the Slam microarray (Figure 4B). Moreover, shRNA targeting Cdkn1a was enriched in our shRNA screen (Figure 1 and Table S1).
Figure 4.
Deregulated Expression of Pparγ and PPARγ Co-activators in Fancd2−/− HSPCs
(A) Real-time qPCR measurement of indicated genes in sorted LSKs 2 weeks after transplantation of LSKs from 8- to 12-week-old Fancd2−/− or Fancd2+/+ mice (n = 4/genotype) into lethally irradiated Boy/J recipient mice (n = 6 mice/group, two independent experiments).
(B) Deregulated Pparγ target genes from previously published Fancd2−/− and Fancd2+/+ SLAM microarray.
(C–E) Real-time qPCR mRNA measurement of indicated genes in Fancd2−/− or Fancd2+/+ LSKs (n = 4/genotype) transduced by shScramble or shPparγ (C), or in presence or absence of 100 nM T0070907 (D) or 1 μM troglitazone (E).
(F) LSKs from 8- to 12-week-old Fancd2−/− or Fancd2+/+ mice (n = 4 mice/genotype) were treated for 48 hr with 1 μM troglitazone, and 2,000 live cells were transplanted into lethally irradiated Boy/J mice along with CD45.1 competitor BM recipient cells. The bar graph depicts the blood chimerism (CD45.2+ cells) of recipient mice 10 weeks after transplantation.
(G) Real-time qPCR measurement in sorted LSKs 2 weeks after transplantation of LSKs from 8- to 12-week-old Fancd2−/− or Fancd2+/+ mice transduced with shScramble or shPparγ into lethally irradiated Boy/J recipient mice (n = 4 mice/group).
(H) LSKs were sorted from 8 to 12 weeks old Fancd2−/− or Fancd2+/+ mice (n = 4 mice/genotype), and 2,000 sorted LSKs (CD45.2) were transplanted into lethally irradiated Boy/J mice along with 2,000 competitor LSKs (CD45.1). Mice were injected intraperitoneally every other day with T0070907 (5 mg/kg) or control vehicle (DMSO) (n = 4 mice/group) during the first 4 weeks after transplantation. The bar graph depicts the blood chimerism (CD45.2+ cells) of recipient mice 16 weeks after transplantation.
WT, wild-type. Values are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To validate the regulation of Cdkn1a and Ncoa4 expression by PPARγ, we inhibited or activated PPARγ activity by genetic or pharmacological approaches. Under steady-state conditions, Fancd2−/− LSKs showed elevated expression of Cdkn1a and Nco4 compared with WT LSKs (Figures 4C–4E). Inactivation of PPARγ by either shRNA or by treatment with the antagonist T0070907 led to significant reduction of Cdkn1a and Ncoa4 in Fancd2-deficient LSKs compared with WT LSKs (Figures 4C and 4D). We treated WT LSK cells with troglitazone and found that activation of Pparγ increased the expression of Cdkn1a and Ncoa4 (Figure 4E). In accordance with the colony assay, we were unable to find a difference in Cdkn1a and Ncoa4 expression after treatment in Fancd2−/− LSKs. These data support the notion that PPARγ regulates Cdkn1a and Ncoa4 expression in HSPCs.
The observation that activation of PPARγ by troglitazone increased the expression of Cdkn1a in WT LSKs prompted us to ask whether activation of PPARγ inhibited HSPC repopulation capacity. To this end, we treated WT and Fancd2−/− LSKs ex vivo with troglitazone and evaluated the hematopoietic repopulating capacity of the treated HSPCs using a BM transplantation assay. At 10 weeks after transplantation, troglitazone-treated WT HSPCs were less efficient in repopulating the irradiated recipients than untreated WT LSKs (Figure 4F). Interestingly, activation of PPARγ in Fancd2−/− HSCs did not further impair the repopulation activity. No difference in apoptosis was observed in LSKs treated with troglitazone during 48 hr (data not shown). Thus, the impaired repopulation does not seem to be linked to increased cell death before transplantation. To validate in vivo our in vitro data, we transplanted lethally irradiated mice with WT or Fancd2−/− LSKs transduced by shScramble or shPparγ, along with recipient competitor LSKs. As observed in vitro, inactivation of PPARγ by shRNA led to a decreased expression of Cdkn1a and Ncoa4 at 2 weeks post transplantation (Figure 4G). We further confirmed the beneficial role of PPARγ inhibition on Fancd2−/− HSPCs by treating transplanted mice with the antagonist T0070907 during the first 4 weeks after irradiation, when the donor cells are subjected to a high level of replicative stress due to rigorous BM repopulation. As observed with Pparγ shRNA knockdown, in vivo inhibition of PPARγ activity by T0070907 led to a partial rescue of Fancd2−/− HSC repopulation capacity (Figure 4H). Long-term evaluation of transplanted mice showed an extended life span of mice receiving shPparγ-transduced Fancd2−/− LSKs compared with mice receiving shScramble-transduced Fancd2−/− LSKs (Figure S3F). Notably, after 1 year no sign of leukemia was observed as depicted by the white blood cell count at 50 weeks (Figure S3G). Altogether, these data indicate that HSPCs are able to respond to aberrant PPARγ activity and that PPARγ activation under replicative stress impairs FA HSPC function, likely through upregulating Cdkn1a expression.
Upregulated PPARγ and Its Co-activators in FA Patient-Derived Cell Lines and Primary FA Patient Samples
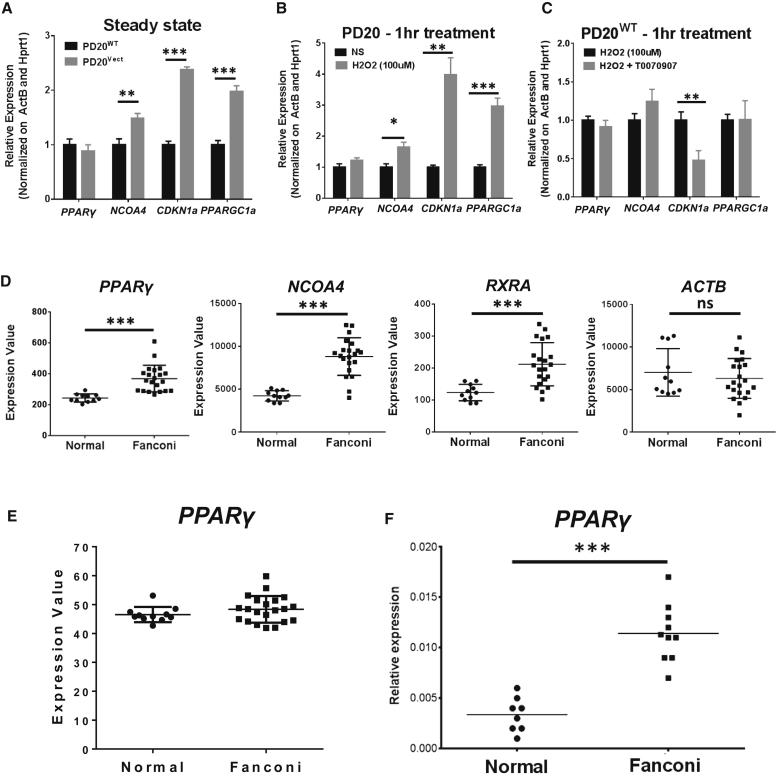
We next evaluated whether our findings in the mouse model were extendable to humans. We first compared the expression of PPARγ, NCOA4, CDKN1a, and PPRγC1a in human FANCD2-deficient lymphoblastic cell lines genetically complemented (PD20WT) or not (PD20Vect) by WT human FANCD2 cDNA. We observed a significant increase in the expression of NCOA4, CDKN1a, and PPARγC1a in FANCD2-deficient cells compared with the complemented cells (Figure 5A). Since antioxidant treatment is able to ameliorate PD20 FA phenotypes (Li et al., 2012, Li et al., 2014, Ponte et al., 2012), we decided to evaluate the expression of the PPARγ-related genes in response to treatment with hydrogen peroxide (H2O2). Oxidative stress increased expression of CDKN1a and PPARγC1a (Figure 5B). Interestingly, treatment with H2O2 elevated the expression of NCOA4, indicating a link between oxidative stress and this PPARγ co-activator. However, H2O2 treatment did not increase the expression of PPARγ in PD20WT cells (Figure 5B). It is possible that oxidative stress affects the activity of PPARγ, not its gene expression. To test this notion, we inhibited PPARγ activity using T0070907 in PD20WT cells exposed to H2O2. Inhibition of PPARγ significantly reduced the expression of CDKN1a upon H2O2 treatment (Figure 5C), indicating that PPARγ participates in the regulation of this gene in response to oxidative stress. PPARγ inhibition did not change the expression of NCOA4 and PPARγC1a (Figure 5C).
Figure 5.
Deregulated PPARγ in Human FA-Deficient Cells
(A) Real-time qPCR mRNA measurement of indicated genes in FANCD2-deficient (PD20vect) or FANCD2-complemented (PD20WT) human lymphoblastic cell lines (three independent experiments).
(B) Real-time qPCR mRNA measurement of indicated genes in PD20WT treated with or without 100 μM H2O2 (three independent experiments).
(C) Real-time qPCR mRNA measurement of indicated genes in PD20WT treated with or without 100 μM H2O2 in the presence or absence of 100 nM T0070907 (three independent experiments).
(D and E) Analysis of PPARγ-related genes and ACTB control in previously published microarray on BM cells from normal donors or FA patients.
(F) Real-time qPCR measurement of PPARγ mRNA in BM CD34+ cells from normal (n = 8) or FA patients (n = 10).
Values are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ns, not significant.
To substantiate the human relevance further, we analyzed the published transcriptome data derived from BM cells of a large cohort of FA patients (GEO: GSE16334) (Vanderwerf et al., 2009) for PPARγ-related genes. Interestingly, we found significantly increased expression of CDKN1a and NCOA4 in FA patients (Figure 5D). While we did not find upregulation of PPARγ (Figure 5E) and PPARγC1a (data not shown), we found upregulation of RXRa (Figure 5D), a gene encoding another co-activator of PPARγ (Mangelsdorf and Evans, 1995). Furthermore, we observed by real-time qPCR a significant elevation in the expression of PPARγ in BM HSPCs (CD34+) isolated from a small cohort of FA patients compared with the samples from healthy donors (Figure 5F). Together, these data suggest that consistent with its role in Fancd2−/− HSPCs, deregulated PPARγ activity may contribute to BM pathology in FA patients.
Discussion
The current study provides several pieces of evidence that abnormal Pparγ activation in HSCs leads to HSC function impairment under replicative stress. (1) shRNA against Pparγ and Pparγ-related genes were enriched in Fancd2-deficient HSPCs undergoing hematopoietic repopulation. (2) Specific knockdown of Pparγ improved the repopulation capacity of Fancd2-deficient HSCs. (3) Genetic or pharmacological inhibition of Pparγ decreased expression of Cdkna1a in mouse and human FA-deficient cells and improved the repopulation capacity of Fancd2-deficient HSCs.
PPARγ is a central transcription factor regulating adipocyte differentiation and energy metabolism (Siersbaek et al., 2010). Interestingly, increased population of adipocytes in the BM has been observed in FA patients and mouse models (Pulliam-Leath et al., 2010), suggesting a possible dysregulation of PPARγ. Moreover, it has been shown that adipocytes are deleterious for HSC self-renewal (Naveiras et al., 2009). These observations are in accordance with our current study demonstrating that pharmacological or intrinsic activation of PPARγ impaired hematopoietic repopulation of Fancd2-deficient HSCs as well as WT HSCs. In this context, it is noteworthy that our recent study (Amarachintha et al., 2015) with Fanca- and Fancd2-deficient BM mesenchymal stromal cells revealed upregulation of PPARγ activity and Pparγc1a expression and adipocyte differentiation in FA, suggesting that hyperactivation of PPARγ could be a common feature of FA cells. In accordance with a deleterious effect of PPARγ on the hematopoietic system, development of anemia in humans has been shown to be a side effect after the introduction of a PPARγ agonist drug on the market (Werner and Travaglini, 2001). In addition, FA patients are frequently treated with androgens, which delay BM failure (Dokal, 2003). Interestingly, androgen and PPARγ signaling inhibit each other (Dokal, 2003, Du et al., 2009, Singh et al., 2006). Thus, the effectiveness of androgen treatment in FA patients may be in part related to PPARγ inhibition, although this hypothesis has to be investigated.
Since the p53/p21 axis has been described as an impairment mechanism of FA-deficient HSCs and also plays important roles in other models of HSC aging (Choudhury et al., 2007), we evaluated the effect of PPARγ on CDKN1a expression. We found that inhibition of PPARγ downregulated the expression of CDKNA1a in FANCD2-deficient human lymphoblast cell line (PD20) and mouse Fancd2−/− HSPC (LSK) cells. PPARγ interacts and activates p53 to upregulate p21 expression (Han et al., 2003) and can bind directly to the TP53 promoter to increase p53 expression (Bonofiglio et al., 2006). Interestingly, the levels of the p53 protein are known to be upregulated in FANCD2-deficient PD20 cells and Fancd2−/− mouse LSK cells (Ceccaldi et al., 2012). In connection with PPARγ and p53 interaction, PPARγ activation leads to an increase in cell-cycle arrest and apoptosis in different cancer cell types (Burstein et al., 2003, Mueller et al., 2000, Theocharisa et al., 2003). In this context, we speculate a possible linkage between PPARγ activation and upregulation of p21 through p53 upregulation. Since direct targeting of p53 or p21 is not a safe therapeutic strategy, treatments that modulate the p53/p21 activity might improve HSC functions while maintaining vital tumor-suppressive functions. In supporting this notion, we kept lethally irradiated recipient mice reconstituted with Scramble or shPparγ -transduced Fancd2−/− LSK cells for 1 year without detecting leukemia development.
Surprisingly, we did not observe an alteration in PPARγ expression in FANCD2-deficient PD20 cells and HSPCs or the published FA patient BM microarray. However, we detected overexpression of PPARγ in human FA CD34+ BM cells, suggesting that modulation of PPARγ expression might be more important in primitive HSCs. Since PPARγ level in either gene expression or total protein is not always a good indicator of its activity, we evaluated the expression of PPARγ co-activators. We found significantly increased expression of NCOA4, a known co-activator of PPARγ (Heinlein et al., 1999) and a hit in our screen, in mouse Fancd2-deficient LSKs after transplantation and in BM cells of an FA patient. This similarity in the mouse model and human samples suggests a possible deleterious role of PPARγ in humans. Nevertheless, no functional assessment has been conducted on human HSPCs in this study, and further studies are needed to confirm the relevance of PPARγ signaling in primary human cells and patients.
Little is known about the relationship between NCOA4 and PPARγ beyond transcriptional co-activation. In this study, we showed that PPARγ regulated the expression of NCOA4 under oxidative or replicative stress. Interestingly, it has been recently reported that inhibition of NCOA4 confers resistance to oxidative stress (Mancias et al., 2014). We and others have described the hypersensitivity of FA-deficient cells to oxidative stress (Pagano et al., 2012). Moreover, we have shown that treatment of FA mice with the antioxidant quercetin can ameliorate the FA phenotype (Li et al., 2014), and a pilot study on the use of quercetin in FA patients is in progress at the Cincinnati Children's Hospital. Thus, it is possible that the upregulation of NCOA4 in FA-deficient cells has a direct impact on PPARγ activation and sensitivity to oxidative stress. Another link between PPARγ and oxidative stress comes from the direct activation of PPARγ by oxidative stress (Polvani et al., 2012). In fact, oxidative stress is a known inducer of PPARγ activity. PPARγ is believed to play a protective role against oxidative stress under physiological conditions. The increased levels of reactive oxygen species (ROS) in FA cells could be responsible for an increased and sustained activation of PPARγ that could impair HSC functions. In support of this notion, we found that treatment of human cells with H2O2 elevated the expression of the PPARγ-related genes such as NCOA4, CDKN1a, and PPARγC1a, and that activation of PPARγ in WT murine LSKs impaired their repopulation activities. Finally, we observed an upregulation of PPARγC1a in Fancd2-deficient mouse LSKs as well as in FANCD2-deficient PD20 cells. PPARγC1a can act as co-stimulator of PPARγ (Puigserver et al., 1998), which has an important role in mitochondrial biogenesis (Wu et al., 1999). Emerging evidence shows that FA deficiency leads to damaged mitochondria (Pagano et al., 2012, Pagano et al., 2014). It is conceivable that when repopulating the BM upon stress, HSCs would have to enter the cell cycle that requires enhanced mitochondrial activity to meet high energy demand, which would increase ROS production. We speculate that because of damaged and less efficient mitochondria, the FA-deficient HSCs need to increase mitochondrial biogenesis to meet energy demand. Therefore, upregulation of PPARγC1a in FA may be due to mitochondrial damages leading to increased accumulation of ROS, which in turn activates PPARγ.
In summary, using an in vivo shRNA screen in donor HSCs, we identified a deleterious effect of deregulated Pparγ activity on HSC functions. Modulation by pharmaceutical compounds of PPARγ signaling is potentially a therapeutic modality in a large proportion of patients with blood diseases, including FA.
Experimental Procedures
Mice
Mice were maintained in a pathogen-free environment in the Cincinnati Children's Hospital Medical Center (CCHMC) mouse housing facility. The protocols of animal experiments were approved by the Institutional Animal Care and Use Committee at CCHMC. Fancd2+/− mice were provided by Dr. Markus Grompe (Oregon Health & Sciences University) (Houghtaling et al., 2003). When indicated, mice were injected intraperitoneally with the PPARγ inhibitor T0070907 (5 mg/kg) or with control vehicle (DMSO) as indicated in the figure legends.
Bone Marrow Transplantation
Congenic CD45.1 mice were lethally irradiated (12 Gy) and intravenously injected on the same day with whole mononuclear BM cells or isolated Lin− or Lin−Sca1+cKit+ (LSK) BM cells from donor mice. For competitive transplantation, 500 LSKs (CD45.2) were transplanted with 0.3 million congenic BM cells.
Flow Cytometry and Cell Sorting
BM cells isolated from the femurs and tibias of mice were labeled on ice (for antibody details see Supplemental Experimental Procedures). Propidium iodide (PI) was used to detect dead cells. Live Lin− or LSK cells (PI−Lin−Sca1+cKit+) were sorted from mice BM using a FACSAria II (BD Bioscience). Lin−, LSK, and Slam cells (CD48−CD150+ LSK cells) were detected by flow cytometry on a BD Fortessa cytometer (BD Biosciences). CD45.1 and CD45.2 labeling were used to distinguish recipient and donor-derived BM or blood cells.
Cell Culture
Sorted LSKs were cultured in Stem Span SFEM medium (STEMCELL Technologies) supplemented with 100 ng/mL murine stem cell factor and 50 ng/mL murine thrombopoietin (Preprotech). The Pparγ agonist troglitazone or antagonist T0070907 (Cayman) were added at the initiation of culture at the concentration indicated on the figures. For colony-forming assay, 200 LSKs or 5 × 103 MNC BM cells were cultured into cytokines supplemented Methocult3434 medium (STEMCELL). Colonies were counted on day 7 and replated for the second round in the same condition.
shRNA Library and Vector Construction
The shRNA library pool was provided by Dr. Lenhard Rudolph (Max-Planck-Research Institute) (Wang et al., 2012). Construction details about SFFVΔEcoR1-Egfp-shRNA vector use in specific gene silencing are provided in Supplemental Experimental Procedures.
LSK Transduction and In Vivo Screening
A schematic experimental flow of the LSK transduction for shRNA screening is depicted in Figure S1. In brief, sorted Fancd2−/− or Fancd2+/+ LSKs were cultured in 96-well round-bottomed low-attachment plates (BD Biosciences). A total of 20,000 LSKs/well were transduced 12 hr after culture ignition to obtain 70%–75% (MOI = 20) or 20%–30% (MOI = 5) transduction efficiency. For in vivo screening, 20,000–25,000 pooled transduced LSK cells were transplanted into lethally irradiated recipient mice. Four weeks after transplantation, CD45.2+Lin− BM cells were sorted and transplanted for a second round into recipient mice. Six weeks later, CD45.2+GFP+Lin− BM cells were sorted for genomic DNA extraction using the Gentra PureGene Blood Kit (QIAGEN).
Integrated shRNA Detection and Enrichment Analysis
We adapted the protocol and primers previously described (Sims et al., 2011) (Figure S2 and Supplemental Experimental Procedures). FASTQ files generated after sequencing of screening were mapped to the shRNA library sequence using the ShAlign Perl program (Sims et al., 2011). The enrichment score of each shRNA sequence was calculated using the ShRNAseq R package (Sims et al., 2011).
Real-Time qPCR
Total RNA was isolated from cells in culture or freshly sorted LSKs with the RNeasy plus mini kit (QIAGEN). Reverse transcription of total mRNA was obtained using The High Capacity cDNA kit (Applied Biosystems). qPCR was conducted on an ABI 7900 (Applied Biosystems). Primers for qPCR are listed in Figure S4.
Statistical Analysis
Student's t test was used for two-group comparison and one-way ANOVA for more than two-group comparison using the Tukey post hoc test to correct for multiple comparisons with GraphPad Prism software. Values of p < 0.05 were considered statistically significant. Results are presented as mean ± SD. Values in the figures are depicted as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Author Contributions
M.S.: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing. W.D.: Collection and assembly of data, data analysis and interpretation. S.A.: Collection and assembly of data. A.W.: Collection of data. Q.P.: Conception and design, manuscript writing, final approval of manuscript.
Acknowledgments
We thank Drs. Lenhard Rudolph and Jianwei Wang (Germany Research Group on Stem Cell Aging, Germany) for the shRNA library and helpful suggestions about this work, and Dr. Markus Grompe (Oregon Health & Sciences University) for Fancd2+/− mice, the Comprehensive Mouse and Cancer Core of the Cincinnati Children's Research Foundation (CCHMC) for bone marrow transplantation service, and the Viral Vector Core of Cincinnati Children's Research Foundation (CCHMC) for the preparation of lentiviruses. We also thank members of the Q.P. laboratory for helpful discussions and Ms. Samantha Losekamp for editing the manuscript. This work was supported by NIH grants R01 HL076712 and R01 CA157537. Q.P. is supported by a Leukemia and Lymphoma Scholar award.
Published: April 13, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and one table, and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.03.008.
Supplemental Information
References
- Amarachintha S., Sertorio M., Wilson A., Li X., Pang Q. Fanconi anemia mesenchymal stromal cells-derived glycerophospholipids skew hematopoietic stem cell differentiation through toll-like receptor signaling. Stem Cells. 2015;33:3382–3396. doi: 10.1002/stem.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroca V., Mouthon M.A., Lewandowski D., Brunet de la Grange P., Gauthier L.R., Pflumio F., Boussin F.D., Arwert F., Riou L., Allemand I. Impaired functionality and homing of Fancg-deficient hematopoietic stem cells. Hum. Mol. Genet. 2012;21:121–135. doi: 10.1093/hmg/ddr447. [DOI] [PubMed] [Google Scholar]
- Bonofiglio D., Aquila S., Catalano S., Gabriele S., Belmonte M., Middea E., Qi H., Morelli C., Gentile M., Maggiolini M. Peroxisome proliferator-activated receptor-gamma activates p53 gene promoter binding to the nuclear factor-kappaB sequence in human MCF7 breast cancer cells. Mol. Endocrinol. 2006;20:3083–3092. doi: 10.1210/me.2006-0192. [DOI] [PubMed] [Google Scholar]
- Bric A., Miething C., Bialucha C.U., Scuoppo C., Zender L., Krasnitz A., Xuan Z., Zuber J., Wigler M., Hicks J. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein H.J., Demetri G.D., Mueller E., Sarraf P., Spiegelman B.M., Winer E.P. Use of the peroxisome proliferator-activated receptor (PPAR) gamma ligand troglitazone as treatment for refractory breast cancer: a phase II study. Breast Cancer Res. Treat. 2003;79:391–397. doi: 10.1023/a:1024038127156. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R., Parmar K., Mouly E., Delord M., Kim J.M., Regairaz M., Pla M., Vasquez N., Zhang Q.S., Pondarre C. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Lee C., Hahm K.B., Yi Y., Choi S.G., Kim S.J. Over-expression of ERT(ESX/ESE-1/ELF3), an ets-related transcription factor, induces endogenous TGF-beta type II receptor expression and restores the TGF-beta signaling pathway in Hs578t human breast cancer cells. Oncogene. 2000;19:151–154. doi: 10.1038/sj.onc.1203252. [DOI] [PubMed] [Google Scholar]
- Choudhury A.R., Ju Z., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Dokal I. Inherited aplastic anaemia. Hematol. J. 2003;4:3–9. doi: 10.1038/sj.thj.6200215. [DOI] [PubMed] [Google Scholar]
- Du J., Zhang L., Wang Z. Testosterone inhibits the activity of peroxisome proliferator-activated receptor gamma in a transcriptional transaction assay. Pharmazie. 2009;64:692–693. [PubMed] [Google Scholar]
- Han C., Demetris A.J., Michalopoulos G.K., Zhan Q., Shelhamer J.H., Wu T. PPARgamma ligands inhibit cholangiocarcinoma cell growth through p53-dependent GADD45 and p21 pathway. Hepatology. 2003;38:167–177. doi: 10.1053/jhep.2003.50296. [DOI] [PubMed] [Google Scholar]
- Heinlein C.A., Ting H.J., Yeh S., Chang C. Identification of ARA70 as a ligand-enhanced coactivator for the peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 1999;274:16147–16152. doi: 10.1074/jbc.274.23.16147. [DOI] [PubMed] [Google Scholar]
- Hope K.J., Cellot S., Ting S.B., MacRae T., Mayotte N., Iscove N.N., Sauvageau G. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 2010;7:101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Houghtaling S., Timmers C., Noll M., Finegold M.J., Jones S.N., Meyn M.S., Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.K., Olefsky J.M. Acute effects of troglitazone on in vivo insulin action in normal rats. Metabolism. 1995;44:1166–1169. doi: 10.1016/0026-0495(95)90010-1. [DOI] [PubMed] [Google Scholar]
- Lee G., Elwood F., McNally J., Weiszmann J., Lindstrom M., Amaral K., Nakamura M., Miao S., Cao P., Learned R.M. T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J. Biol. Chem. 2002;277:19649–19657. doi: 10.1074/jbc.M200743200. [DOI] [PubMed] [Google Scholar]
- Li J., Du W., Maynard S., Andreassen P.R., Pang Q. Oxidative stress-specific interaction between FANCD2 and FOXO3a. Blood. 2010;115:1545–1548. doi: 10.1182/blood-2009-07-234385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sipple J., Maynard S., Mehta P.A., Rose S.R., Davies S.M., Pang Q. Fanconi anemia links reactive oxygen species to insulin resistance and obesity. Antioxid. Redox Signal. 2012;17:1083–1098. doi: 10.1089/ars.2011.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li L., Li J., Sipple J., Schick J., Mehta P.A., Davies S.M., Dasgupta B., Waclaw R.R., Pang Q. Concomitant inactivation of foxo3a and fancc or fancd2 reveals a two-tier protection from oxidative stress-induced hydrocephalus. Antioxid. Redox Signal. 2014;21:1675–1692. doi: 10.1089/ars.2013.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li J., Wilson A., Sipple J., Schick J., Pang Q. Fancd2 is required for nuclear retention of Foxo3a in hematopoietic stem cell maintenance. J. Biol. Chem. 2015;290:2715–2727. doi: 10.1074/jbc.M114.619536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta ), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Meruvu S., Hugendubler L., Mueller E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638. J. Biol. Chem. 2011;286:26516–26523. doi: 10.1074/jbc.M110.212506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S.E., Smith J.A., Shamu C.E., Neumüller R.A., Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Uchida N., Weissman I.L. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Mueller E., Smith M., Sarraf P., Kroll T., Aiyer A., Kaufman D.S., Oh W., Demetri G., Figg W.D., Zhou X.P. Effects of ligand activation of peroxisome proliferator-activated receptor gamma in human prostate cancer. Proc. Natl. Acad. Sci. USA. 2000;97:10990–10995. doi: 10.1073/pnas.180329197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveiras O., Nardi V., Wenzel P.L., Hauschka P.V., Fahey F., Daley G.Q. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K.W., Scadden D.T. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G., Talamanca A.A., Castello G., Pallardó F.V., Zatterale A., Degan P. Oxidative stress in Fanconi anaemia: from cells and molecules towards prospects in clinical management. Biol. Chem. 2012;393:11–21. doi: 10.1515/BC-2011-227. [DOI] [PubMed] [Google Scholar]
- Pagano G., Shyamsunder P., Verma R.S., Lyakhovich A. Damaged mitochondria in Fanconi anemia—an isolated event or a general phenomenon? Oncoscience. 2014;1:287–295. doi: 10.18632/oncoscience.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K., D'Andrea A., Niedernhofer L.J. Mouse models of Fanconi anemia. Mutat. Res. 2009;668:133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K., Kim J., Sykes S.M., Shimamura A., Stuckert P., Zhu K., Hamilton A., Deloach M.K., Kutok J.L., Akashi K. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28:1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polvani S., Tarocchi M., Galli A. PPARγ and oxidative stress: Con(β) catenating NRF2 and FOXO. PPAR Res. 2012;2012:641087. doi: 10.1155/2012/641087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte F., Sousa R., Fernandes A.P., Gonçalves C., Barbot J., Carvalho F., Porto B. Improvement of genetic stability in lymphocytes from Fanconi anemia patients through the combined effect of α-lipoic acid and N-acetylcysteine. Orphanet J. Rare Dis. 2012;7:28. doi: 10.1186/1750-1172-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Pulliam-Leath A.C., Ciccone S.L., Nalepa G., Li X., Si Y., Miravalle L., Smith D., Yuan J., Li J., Anur P. Genetic disruption of both Fancc and Fancg in mice recapitulates the hematopoietic manifestations of Fanconi anemia. Blood. 2010;116:2915–2920. doi: 10.1182/blood-2009-08-240747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Jamieson C.H., Weissman I.L. Stem cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Roy S., Javed S., Jain S.K., Majumdar S.S., Mukhopadhyay A. Donor hematopoietic stem cells confer long-term marrow reconstitution by self-renewal divisions exceeding to that of host cells. PLoS One. 2012;7:e50693. doi: 10.1371/journal.pone.0050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S.L., Tian L., Kähkönen M., Schwartzentruber J., Kircher M., Majewski J., Dyment D.A., Innes A.M., Boycott K.M., Moreau L.A. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.V., Brüne B., von Knethen A. The nuclear hormone receptor PPARγ as a therapeutic target in major diseases. ScientificWorldJournal. 2010;10:2181–2197. doi: 10.1100/tsw.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R., Nielsen R., Mandrup S. PPARgamma in adipocyte differentiation and metabolism—novel insights from genome-wide studies. FEBS Lett. 2010;584:3242–3249. doi: 10.1016/j.febslet.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Sims D., Mendes-Pereira A.M., Frankum J., Burgess D., Cerone M.A., Lombardelli C., Mitsopoulos C., Hakas J., Murugaesu N., Isacke C.M. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Artaza J.N., Taylor W.E., Braga M., Yuan X., Gonzalez-Cadavid N.F., Bhasin S. Testosterone inhibits adipogenic differentiation in 3T3-L1 cells: nuclear translocation of androgen receptor complex with beta-catenin and T-cell factor 4 may bypass canonical Wnt signaling to down-regulate adipogenic transcription factors. Endocrinology. 2006;147:141–154. doi: 10.1210/en.2004-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittau B., Krieglstein K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012;347:65–72. doi: 10.1007/s00441-011-1186-6. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., D'Andrea A.D. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- Theocharisa S., Margeli A., Kouraklis G. Peroxisome proliferator activated receptor-gamma ligands as potent antineoplastic agents. Curr. Med. Chem. Anticancer Agents. 2003;3:239–251. doi: 10.2174/1568011033482431. [DOI] [PubMed] [Google Scholar]
- Vanderwerf S.M., Svahn J., Olson S., Rathbun R.K., Harrington C., Yates J., Keeble W., Anderson D.C., Anur P., Pereira N.F. TLR8-dependent TNF-(alpha) overexpression in Fanconi anemia group C cells. Blood. 2009;114:5290–5298. doi: 10.1182/blood-2009-05-222414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- Wan Y., Chong L.W., Evans R.M. PPAR-gamma regulates osteoclastogenesis in mice. Nat. Med. 2007;13:1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- Wang J., Sun Q., Morita Y., Jiang H., Gross A., Lechel A., Hildner K., Guachalla L.M., Gompf A., Hartmann D. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Werner A.L., Travaglini M.T. A review of rosiglitazone in type 2 diabetes mellitus. Pharmacotherapy. 2001;21:1082–1099. doi: 10.1592/phco.21.13.1082.34615. [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng X.H., Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.