Abstract
Antimicrobial resistance represents a significant challenge to future healthcare provision. An acronym ESKAPEE has been derived from the names of the organisms recognised as the major threats although there are a number of other organisms, notably Neisseria gonorrhoeae, that have become equally challenging to treat in the clinic. These pathogens are characterised by the ability to rapidly develop and/or acquire resistance mechanisms in response to exposure to different antimicrobial agents. A key part of the armoury of these pathogens is a series of efflux pumps, which effectively exclude or reduce the intracellular concentration of a large number of antibiotics, making the pathogens significantly more resistant. These efflux pumps are the topic of considerable interest, both from the perspective of basic understanding of efflux pump function, and its role in drug resistance but also as targets for the development of novel adjunct therapies. The necessity to overcome antimicrobial resistance has encouraged investigations into the characterisation of resistance-modifying efflux pump inhibitors to block the mechanisms of drug extrusion, thereby restoring antibacterial susceptibility and returning existing antibiotics into the clinic. A greater understanding of drug recognition and transport by multidrug efflux pumps is needed to develop clinically useful inhibitors, given the breadth of molecules that can be effluxed by these systems. This review discusses different bacterial EPIs originating from both natural source and chemical synthesis and examines the challenges to designing successful EPIs that can be useful against multidrug resistant bacteria.
Keywords: Antimicrobial resistance, Efflux Pump Inhibitors, Rational Drug Design, Medicinal Chemistry Optimisation, Multiple efflux systems, Computational approaches
1. INTRODUCTION
It is now well established that treatment options for infections acquired by community and nosocomial multi-drug resistant (MDR) bacteria are progressively declining at an alarming rate, causing mounting concern for human healthcare around the world. This increasing resistance has rendered ineffective even drugs thought of as the last resort, such as vancomycin, which is administered for the treatment of infections caused by methicillin-resistant Staphylococcus aureus (MRSA), with many instances encountered in the United States [1]. The indiscriminate use of antibiotics also contributes to the fact that these “ESKAPE” pathogens [2], E. faecium, S. aureus, K. pneumoniae,
A. baumannii, P. aeruginosa, and Enterobacter species, extended here to “ESKAPEE” to include E.coli, are showing increased prevalence of multidrug-resistant and in some cases pan-drug resistant isolates, making colonisation among patients more rapid and routine operations life-threatening (Table 1) [3, 4]. Recent analysis of the risks associated with the spread of antibiotic resistant bacteria in North America, highlights the increasing numbers of Gram-negative bacteria which are resistant to more than three classes of front-line therapy, used widely as a definition of multidrug resistance [5]. The report identifies carbapenem-resistant Enterobacteriaceae, used here only in relation to K. pneumoniae and E. coli, as being responsible for around 9000 cases of healthcare-associated infections, leading to in excess of 600 deaths per annum. The report stresses that these strains are often resistant to nearly all available antibiotics, not just carbapenems and as such, these organisms are allocated the highest risk level, urgent, within the classification of potential public health threats. Neisseria gonorrhoeae is also allocated an urgent rating, due to the 30% of isolates which are resistant to all frontline therapies and its common occurrence in the community. In healthcare settings, multidrug resistant A. baumannii, P. aeruginosa and Enterobacteriaceae (excluding carbapenem-resistant strains) isolates are reported in 63%, 13%, and 23% of hospital-associated infections caused by these organisms. These infections are estimated to result in approximately 500, 1700 and 440 deaths, respectively, per annum. The numbers are almost certainly an underestimate, in the absence of mandatory reporting for most of these Gram-negative organisms, and mortality figures reflect only deaths which are directly caused by the infection, not those where it is a significant contributing factor. Whilst this data is specific for the US, there is no reason to believe that this differs fundamentally in other parts of the world, and indeed carbapenem-resistant Enterobacteriaceae are endemic in many parts of the world [6].
Table 1.
Multiple drug-resistant (MDR) bacterial pathogens that are causing major problems in clinic.
| Pathogen | Clinical settings and resistance headlines |
|---|---|
| Enterococcus faecium | Bloodstream, surgical site and urinary tract infections (UTI) |
| Staphylococcus Aureus | Skin and wound infections, pneumonia, bloodstream infections |
| Klebsiella pneumoniae | Bloodstream, UTI, catheter-related infections, ventilator associated pneumonia (VAP). Major issues with resistance to carbapenems; notably through emergence of “Klebsiella pneumoniae carbapenemase” (KPC) producing isolates |
| Acinetobacter baumannii | Bloodstream infections, wounds, VAP At least 63% of Acinetobacter isolates are resistant to >3 classes of front-line antibiotic. |
| Pseudomonas Aeruginosa | Bloodstream, urinary tract and surgical site infections, pneumonia (cystic fibrosis), VAP ~13% of Pseudomonas isolates are resistant to >3 classes of front-line antibiotic. |
| Enterobacter sp | Bloodstream, UTI, catheter-related infections, |
| Escherichia coli | Bloodstream, urinary tract and surgical site infections, pneumonia (cystic fibrosis) |
| Burkholderia cepacia / cenocepacia | Cystic fibrosis, VAP Intrinsically drug resistant and becoming a major problem in conjunction with other pathogens in the lung |
| Stenotrophomonas maltophilia | Various Intrinsically drug resistant and associated with infections in a number of hospital settings. |
A series of high level documents have highlighted the potential dangers if the emerging problem of resistance is not addressed as an urgent priority. For example, the Chief Medical Officer in England, identified the very real potential that “any one of us could go into hospital in 20 years for minor surgery and die because of an ordinary infection that can't be treated by antibiotics” [7]. On a more global scale the UK Government Review on Antimicrobial Resistance, chaired by Lord O’Neill, has suggested that antimicrobial resistance (AMR) may be responsible for 50 million deaths per year by 2050, if the current situation is not resolved [8]. These themes are echoed by high level policy documents throughout the world, a report on emerging antimicrobial resistance threats from the US Centres for Disease Control, and documents from the World Health Organisation and European Union. All of these documents highlight the importance of improving the usage of currently available antibiotics, by exploring new methods for diagnosing infections and developing novel adjunct therapies, alongside the longer term objective of reinvigorating the antibiotic pipeline. With Big Pharma having largely withdrawn from antibacterial research despite the desperate medical need, we are faced with an even greater predicament than was formerly perceived [9]. It is clear that governments and the pharmaceutical sector must work in collaboration to re-incentivise the production of new classes of active antimicrobial medicines and also agents that can reinstate activity of antibiotics, to which bacteria have already become resistant.
Bacterial resistance to antibiotics and other forms of biotic and abiotic stress takes a variety of different forms that enable them to survive in hospital environments. For example, resistance to aminoglycosides, β-lactams, fluoroquinolones and the macrolides, has manifested via target site modification and by inactivation of the antibiotic in a wide variety of different bacterial species [10-13]. In many cases, these resistance mechanisms may function in concert with mechanisms that reduce the ability of drugs to enter the cell (e.g. porin loss) and reduce the intracellular concentration of the drugs by pumping them out of the cell. Targeting these so called efflux pumps is an attractive solution to address the lack of effective antibiotics.
There are a number of clear examples where efflux pumps contribute to the resistance of bacterial cells to antimicrobial agents and likely many more examples where these effects are less evident due to lack of tools to directly analyse efflux pump activity. Recently reviewed by Dreier and Ruggerone (2015), resistance nodulation division (RND) type efflux pumps from P. aeruginosa have an astonishing array of substrates. For example, the MexAB-OprM pump, one of an estimated 12 such systems in P. aeruginosa, transports several clinically-important antibiotics from diverse classes, including beta-lactams (and most carbapenems), aminoglycosides, fluoroquinolones, macrolides and tetracyclines [14, 15]. Such efflux systems may be regulated by cellular responses, including oxidative or nitrosative stress responses, or upregulated directly as a response to antibiotic exposure. For that reason, upregulation of efflux pumps is a common feature of multidrug resistant clinical isolates in a variety of species, including P. aeruginosa (e.g. MexAB-OprM) [16], A. baumannii (e.g. AdeABC) [17, 18] and S. aureus (e.g. NorA) [19]. In the same way that efflux pumps can confer resistance to a wide range of antibiotics, efflux pump inhibitors (EPIs) could potentially provide useful adjunct therapies to be used in conjunction with a number of antibiotic classes.
The development of efflux pump inhibitors that act as competitive and non-competitive adjuvants to reduce resistance has attracted much attention although, as yet, no compounds have successfully reached the clinic [20]. Such inhibitors are capable of successfully interfering with membrane-bound efflux pumps that systematically work together to remove toxic metabolites to promote the survival of the bacterial cell [21]. This approach to targeting efflux pumps may exhibit more evident benefits over others such as dual antibiotic combination therapy. Dual therapy may accentuate resistance further and pharmacokinetic optimisation of the second antibiotic is required in order to achieve a similar desired effect, entailing additional complexity [22, 23].
Investigation towards efflux inhibitors was originally initiated around the same time as the discovery of the first efflux transporter, P-glycoprotein (P-gp) in 1976 [24]. Since then, the few that have reached clinical trials have failed, mostly due to the fact that they are far too toxic at concentrations essential for activity, greatly hindering their medical application [25, 26]. This review attempts to summarise the literature identifying known EPIs as resistance-modulating agents both from natural extracts and those synthesised chemically, and how they can be employed as a new method to prevent and eventually reverse the mechanisms of resistance.
2. THE UNMET NEED IN ANTIBACTERIAL THERAPY: THE IMPORTANCE OF EPIS
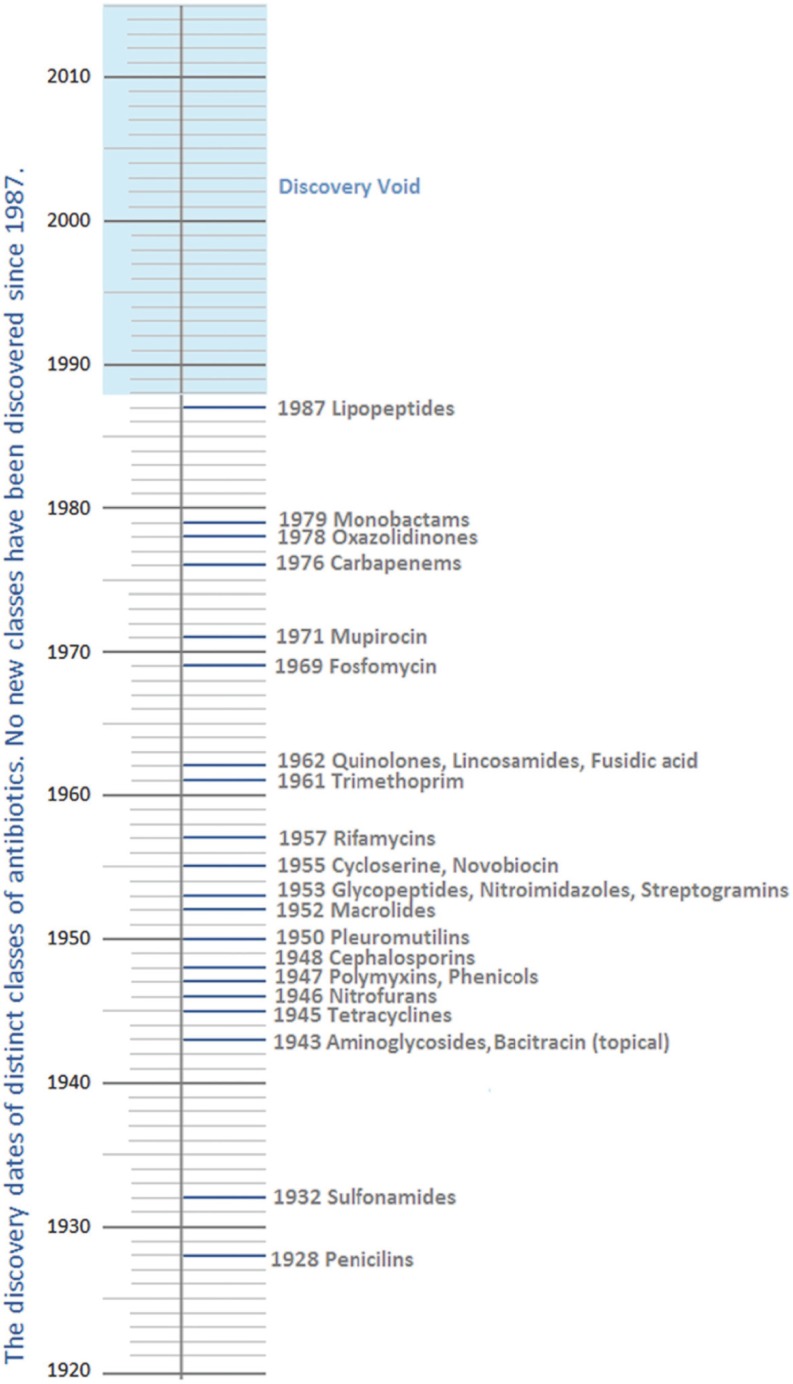
The past two decades have seen an almost empty pipeline of antibacterial drugs in development as the progression of anti-infective research continues to wane. As yet, no new class has emerged since lipopeptides in 1987 to treat Gram-positive infections (Fig. 1) [27].
Fig. (1).
The antibiotic discovery timeline. The 2013 World Economic Forum, adapted from L. L. Silver, Clinical Microbiology Reviews, 2011 [28].
Whilst significant advances have been made in the treatment of patients with infections such as MRSA [29], those with critical illness arising from Gram-negative pathogens remain at the highest risk, due to the fact that of the 160 antibiotics approved, few remain successful against them. As an illustration which is particularly relevant to the role of efflux in bacterial resistance, P. aeruginosa is one of the most prevalent types of hospital acquired bacteria. It is the principal cause of higher mortality rates and expenditure on patient management in pulmonary diseases such as cystic fibrosis, and is a major contributor to bacteraemia, meningitis and urinary tract infections [30]. Its notorious versatility has allowed it to swiftly adapt even when under stress in nutrient poor environments, including water systems, with its high intrinsic resistance attributed to a combined effect of poor outer membrane permeability and overexpression of efflux transporters [31]. Evidence of this in the literature has shown that deletion of genes encoding these MDR efflux systems in wild-types of bacteria confers an expectedly high degree of susceptibility to a wide structural variety of antibiotics as well as many other dissimilar substrates such as bile acids and detergents [7]; this will be discussed in more detail below. This poly-substrate specificity has enabled the bacteria to show insensitivity towards novel antibiotics, which were intended to overcome this, even whilst they were still undergoing clinical trials, [32]. Regrettably, many of these compounds were anticipated to be potentially active against a broad spectrum of highly conserved targets in both Gram-negative and Gram-positive pathogens [5] and this has limited their usefulness in the clinic. The broad substrate specificity of the efflux pumps is also a major part of the challenge to develop effective inhibitors that have sufficiently low toxicity to be used as adjunct therapies.
It is apparent from this core information that a global response strategy of a completely novel nature needs to be adopted. While screening for a completely new class of antibiotic that could avoid MDR efflux completely might seem obvious, the physiology of the Gram-negative bacteria in particular, significantly reduces the hit-rate. Such contenders would be unlikely to possess physical properties that would allow passage between two distinctly different membranes [8].
Small molecule efflux pump inhibitors in many ways present themselves as the most promising approach to tackle this looming crisis and remain an expanding topic for research. They could prevent the emergence of new mutant strains and simultaneously weaken intrinsic and acquired resistance [33]. Moreover, inhibitors could be designed to interact with genes that encode for efflux pumps to disrupt pump assembly, obstruct the channel through the pump or even act by collapsing the energy-dependent processes some rely on to pump out such toxic solutes. Most importantly, it has been suggested that efflux inhibitors could also be implemented as weapons to reduce bacterial pathogenicity in vivo, where certain efflux pumps have been found to stimulate the secretion of bacterial toxins as well as defend themselves against other host-derived inhibiting substrates such as bile salts and fatty acids [34].
3. CLASSES OF MDR EFFLUX TRANSPORTERS IN CLINICALLY RELEVANT BACTERIA
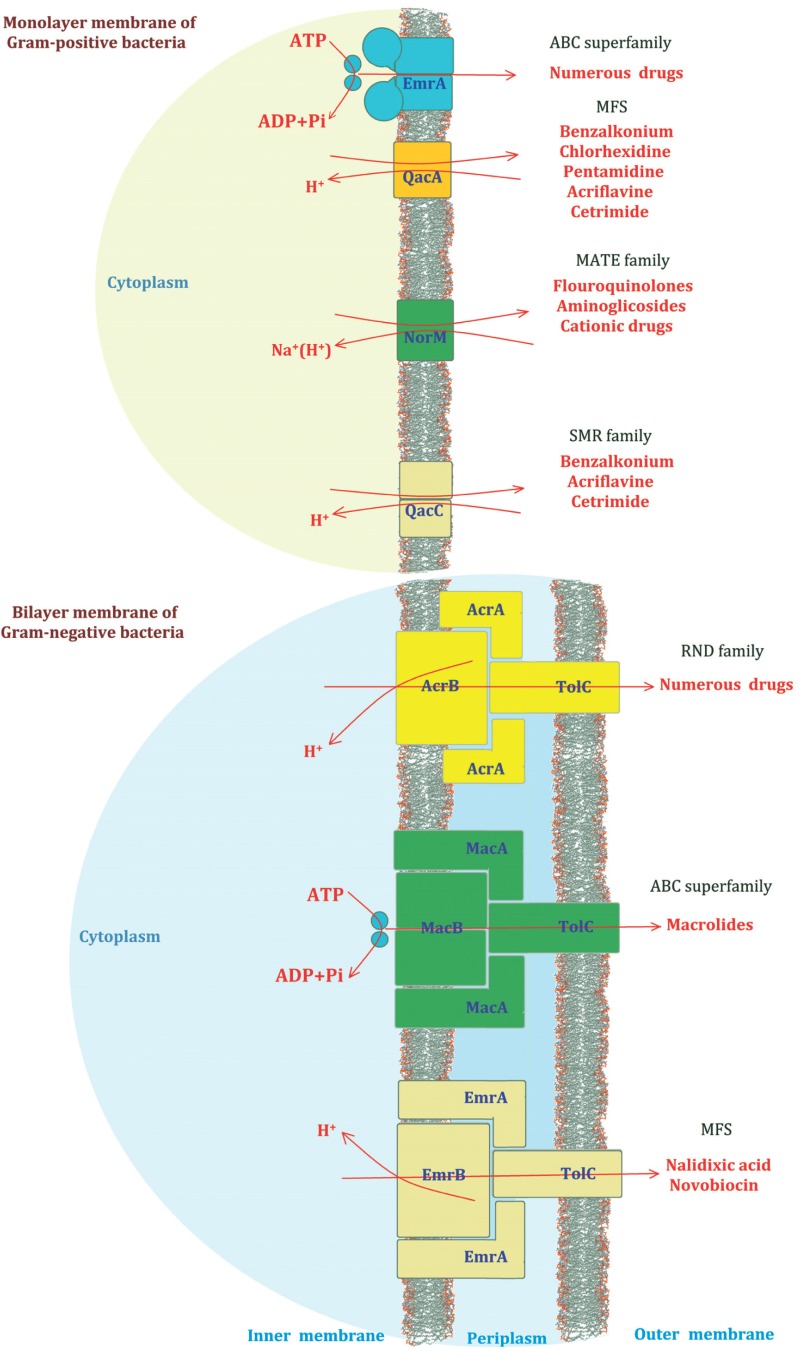
Multidrug resistant efflux systems play a pivotal role in bacterial cell metabolism and are widely distributed in the cytoplasmic and extracellular membranes of all types of bacteria as well as in eukaryotes. These ubiquitous proteins are typically encoded by chromosomal operons and are classified into five distinct groups Fig. (2), where only a single class, the ATP binding cassette (ABC) superfamily, utilises the energy generated by ATP hydrolysis). The others use the energy derived from the electrochemical gradient of the proton motive force across the membrane. These are known as the major facilitator superfamily (MFS), the multidrug and toxic-compound extrusion family (MATE), the small multidrug resistance family (SMR) and finally, the resistance nodulation division (RND) family and are termed secondary transporters [35].
Fig. (2).
A schematic representation of the five common classes of MDR efflux pump. Their general position within the membrane, common substrates and examples of individual and multiple protein components that make up their structures are illustrated.
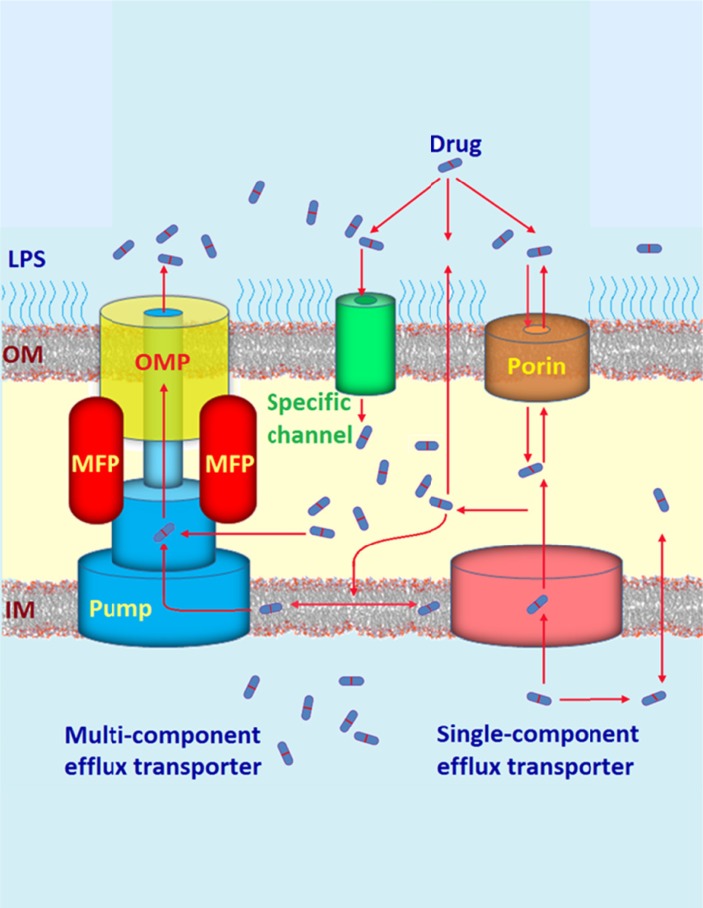
The intricacy with which these proteins are arranged has also provided insight into their structures and mechanisms of transport as well as elucidation of many possible drug binding pockets that would assist in the design of innovative therapies as inhibitors of drug efflux [35, 36]. Efflux transporters are not only characterised by their energy source but also by the substrates they readily export and the number of components that comprises their complete structure. Individual species of bacteria are capable of expressing transporters belonging to more than one family, in addition to different pumps in the same family. An example of this capacity has widely been established in the Gram-negative RND efflux pumps of P. aeruginosa (MexAB-OprM, MexCD-OprJ, and MexEF-OprN) and E. coli (AcrAB-TolC) [37-39]. Due to the second membrane barrier, the majority of efflux pumps found in Gram negative bacteria possess a particular unique tripartite system that is necessary in order to achieve efficient trans-envelope transport, which is the direct removal of substrates into the external medium after capture from the periplasm and inner membrane [40]. However, this may also be accomplished via the slower single-component transporters such as the tetracycline (TET) resistance pumps and other transmembrane proteins such as porins (Fig. 3) [41].
Fig. (3).
An illustration of the competitive pathways of drug uptake and efflux traversing the lipopolysaccharide bilayer and the outer and inner cell membranes of Gram-negative pathogens. The active extrusion of drug molecules occurs via single and multi-component efflux pumps, where the multi-component tripartite complex comprises a membrane fusion protein, an efflux protein pump in the inner membrane and an outer membrane protein.
The tripartite system encompasses an accessory membrane fusion protein (MFP) also known as the periplasmic adaptor protein that bridges between a cytoplasmic membrane efflux pump (that binds the substrate) and an anchored extracellular membrane protein which is usually a TolC-type exit channel [42]. Recent literature has published an enormous amount of structural information on such proteins that make up these tripartite systems [43, 44].
The complexity of the interactions between antibacterial compounds and efflux pumps provides significant challenges to generate effective inhibitors of such systems [14]. The chemical properties of the breadth of substrates that are effluxed through RND pumps, some of which are also competitive or non-competitive inhibitors are staggering [39]. There is no single property, in terms of molecular weight, net charge or logarithm of partition (logP), total polar surface area (TPSA) that defines whether a compound will be an effective substrate and/or inhibitor. This requires these efflux systems to have the capacity to interact with compounds over a large surface area, rather than having a single substrate binding site that would be amenable to the development of high affinity non-competitive inhibitors. For this reason, there are no simple rules to guide the selection of suitable compound libraries for screening of new inhibitors and, as such, known inhibitors have been identified from a wide variety of different sources, both natural and synthetic. Our current knowledge of such compounds is outlined below.
4. CLASSES OF EFFLUX PUMP INHIBITORS
4.1. EPIs Derived from Natural Sources
Plants like other living organisms produce certain beneficial molecules that provide protection against invading species of bacteria, such as cytotoxic phytonutrients that aid vitality and is the main reason behind the few diseases observed in wild plant life [45]. Various articles discuss their inhibitory proficiency when working in synergy with antibiotics, which can potentially offer useful active combination treatments for patients suffering from MDR-associated conditions [46, 47]. Many different chemical structure types have already been identified.
4.1.1. Plant Alkaloids
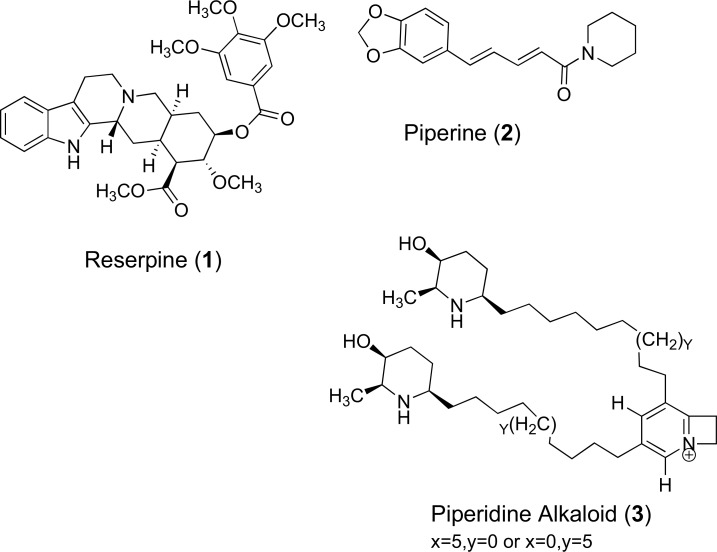
Reserpine (1) is an example of an indole plant alkaloid used for its antipsychotic and antihypertensive activity. It is extracted from the roots of Rauwolfia serpentina and R. vomitoria and is an effective EPI of tetracycline efflux via the BMR pump of Gram-positive Bacillus subtilis [48]. The mechanism of reserpine involves the direct binding to a BMR target site involving two phenylalanine residues and one valine at positions 143, 306 and 286 respectively. It has been reported that reserpine demonstrates a 4-fold decrease in the Minimum Inhibitory Concentration (MIC) for tetracycline in two MRSA isolates containing Tet(K) and can eradicate resistance conferred by the S. aureus NorA efflux transporter, which shares 44% sequence homology to BMR. Expression of the BMR protein pump in B. subtilis thus potentiates susceptibility to fluoroquinolones and other structurally diverse molecules after the administration of reserpine [49]. Although reserpine has widely been prescribed as a treatment for hypertension, clinical application with antibiotics cannot be achieved due to the nephrotoxic concentrations needed for effective inhibition of NorA [50]. Another isolated NorA alkaloid inhibitor (from black pepper (Piper nigrum)) known as piperine (2) has the ability to reverse ciprofloxacin resistance in mutant strains of MRSA that also possessed a high MIC against the reference substrate ethidium bromide [51]. These along with a piperidine alkaloid (3) are noticeably large, basic nitrogenous compounds with both hydrophilic and lipophilic constituents. Their chemical structures are displayed in (Fig. 4). Despite showing early promise there has been limited medicinal chemistry research on these two alkaloidal scaffolds to develop them as clinically useful efflux pump inhibitors.
Fig. (4).
Chemical structures of EPIs 1 – 3.
4.1.2. Phenolic Metabolites
4.1.2.1. Flavolignans
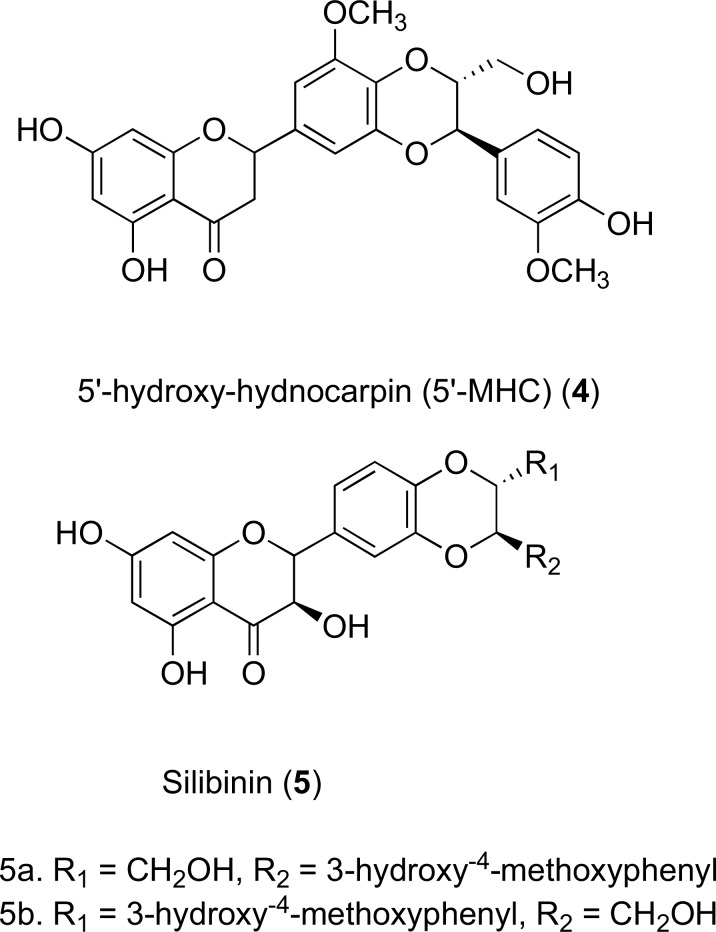
The flavolignan 5’-methoxy-hydnocarpin (5’-MHC) (4) found in the Berberis plant has also been identified as a successful efflux inhibitor of the NorA protein pump. It restores the activity and accumulation of the antibiotic berberine, which is produced by the same species of plant. The weak antibacterial activity of berberine is conveyed by its high MIC (256 mg/L). It has been reported in a synergistic study, that this may be significantly reduced to 16 mg/L when in combination with typical NorA substrates including 5’-MHC and norfloxacin [52]. A similar combination effect has been visualised in two diastereoisomers of the flavolignan silibinin (5), which are inhibitors isolated from the mediterranean milk thistle species, Silybum marianum acting against NorA in wild-type S. aureus. Currently, silibinin has only been clinically evaluated for its inhibition of cell cycle progression in various cancers, already passing phase I trials [53]. Although much potential has been confirmed, no pre-clinical or clinical studies were carried out to determine the efficacy of silibinin as an inhibitor of other efflux proteins in drug-resistant bacteria. However, silibinin has shown promising pre-clinical efficacy as a chemopreventive agent against skin cancer [54]. Structures 4 and 5 can be found in (Fig. 5).
Fig. (5).
Chemical structures of EPIs 4 and 5.
4.1.2.2. Methoxylated Flavones and Isoflavones
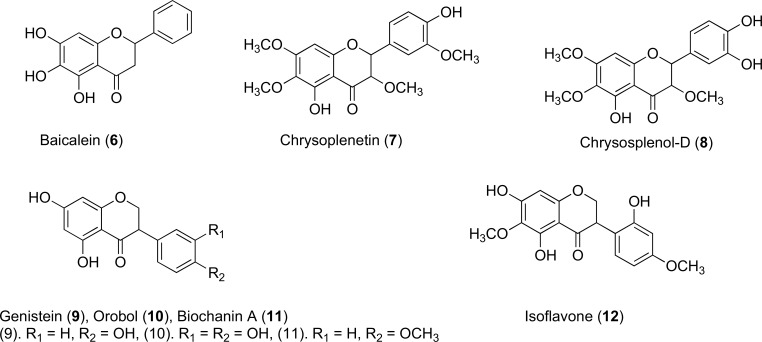
Trihydroxy compounds such as Baicalein (6) is an example of a weak antibacterial flavone (MIC 100mg/L) that is extracted from thyme leaves, a frequently used culinary herb. Baicalein improves susceptibility of certain MRSA isolates when administered in combination with tetracycline or several of the β-lactam antibiotics. It can be noticed that many inhibitors and other potentiators of efflux proteins in MDR–bacteria also have a tendency to exert a relative antibacterial character, which may cause complications when interpreting assay results of antibiotic activity [55]. Chrysoplenetin (7) and chrysosplenol-D (8) are two examples of flavones Fig. (6) that display this weak effect. Structurally related active Isoflavones 9-11, derived from Lupinus argenteus and 12 from Dalea spinosa all boost berberine activity via NorA, reducing the MIC up to a remarkable 16-fold [56]. It is interesting to note that these flavones and isoflavones have shown inhibitory activity against efflux pumps that operate in Gram-positive bacteria, but their ability to interact and inhibit efflux pumps that operate in the MDR Gram-negative bacteria has not been studied in detail. Both flavone and isoflavone chemical scaffolds are amenable to med-chem modifications and molecular docking studies with RND type efflux pumps may help the researchers working in this field to identify analogues that interacts with these pumps and can potentially inhibit them. Recently, a similar approach was adopted by docking phyotchemicals to the E. coli AcrB protein to predict the bioactivity of these phytochemicals, and three phytochemicals were identified that were able to sensitize drug-resistant E. coli to five currently used antibiotics [57].
Fig. (6).
Chemical structures of EPIs 6 – 12.
4.1.2.3. The Catechin Gallates
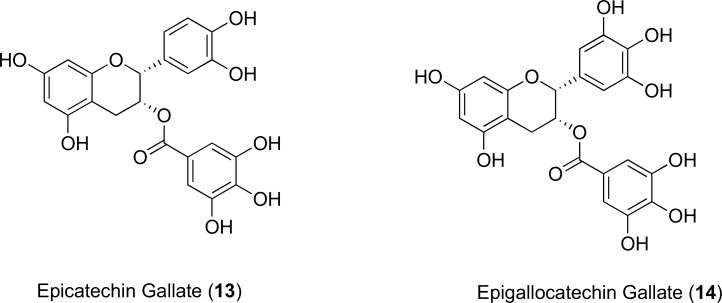
Polyphenol inhibitors isolated from green tea extracts, known as the catechin gallates Fig. (7) have generated much interest as they have been proven to reverse MRSA resistance [58]. Kaatz and Gibbons showed that when incorporated each at 20 mg/L there was a 4-fold decrease in MIC of norfloxacin against the 1199B isolate of S. aureus as well as in S. epidermis. It was concluded in an ethidium bromide inhibition assay that both epicatechin gallate (13) and epigallocatechin gallate (14) possess a weak inhibitory action towards the NorA transporter, with 13 acting with a slightly higher potency than 14 [59]. In their study, they also mentioned that administration of 13 and 14 at concentrations less than or equal to 20 μM, efflux was actually stimulated, which had not previously been encountered for multidrug resistant efflux pump inhibitors [59]. Further investigations would be necessary in order to determine an understanding of how these pumps might be regulated. The authors propose that there is a possibility of two binding sites on the NorA pump with different affinity. At low concentrations, 13 and 14 would only occupy the high affinity sites, leading to increased efflux activity [59]. Resistant staphylococci overexpressing the Tet(K) pump also showed moderately improved tetracycline activity in the presence of 14 [60]. So far, clinical application of 14 has been hindered by toxicity concerns of its carcinogenic potential [60]. The synthetic B-ring modified analogues of epicatechin gallate modulated β-lactamase resistance in MRSA strains and potentiated the activity of oxacillin, but it is not known whether the observed synergistic activity is related to efflux pump inhibition [61-63].
Fig. (7).
Chemical structures of EPIs 13 and 14.
4.1.2.4. Phenolic Diterpenes
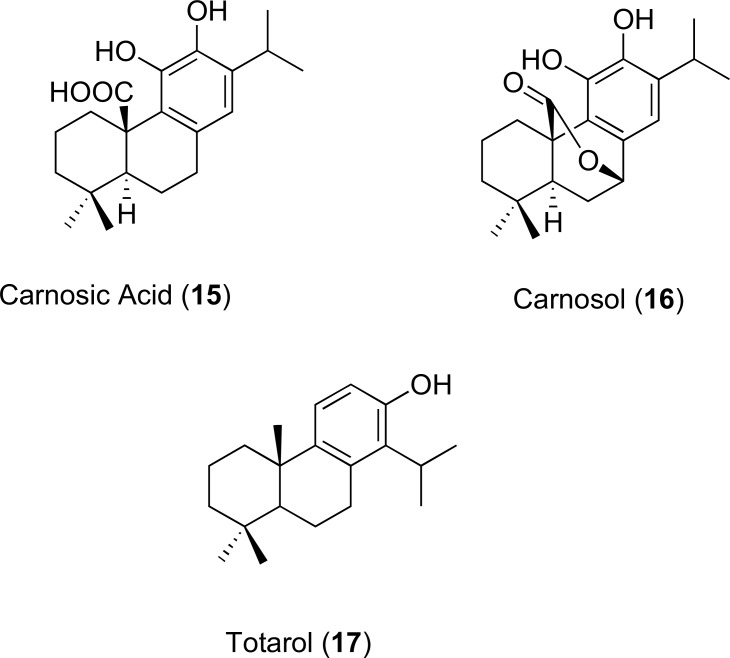
The structures of 15 and 16 represent carnosic acid and carnosol, which are abietane diterpenes of the herb rosemary (Rosmarinus officinalis). They act as potentiators of erythromycin and tetracycline against the respective S. aureus containing Msr(A) and Tet(K) pumps. Both compounds in combination with 10 mg/L of tetracycline enhanced antibiotic activity in an S. aureus strain containing Tet(K). Only carnosic acid decreased the MIC of erythromycin from 256 to 32 mg/L in strains that express the Msr(A) efflux transporter such as RN4220 [64]. Furthermore, it was concluded by an inhibition assay (using ethidium bromide as a reference substrate) that carnosic acid possesses similar EPI-action against the norfloxacin efflux transporter protein, NorA, in the SA-1199B isolate of S. aureus. Another example of an active phenolic diterpene with EPI activity against MDR S. aureus is totarol (17). A study by Smith et al. revealed that 17 produced marked decreases in ethidium bromide efflux at 15 μM inside a totarol-resistant mutant that overexpresses NorA. The MIC of ethidium was also reduced when used in a combination study of these two molecules suggesting that totarol is an effective EPI of NorA as well as a moderately active antibacterial agent [65]. The structures of the diterpenes 15-17 are shown in (Fig. 8).
Fig. (8).
Chemical structures of EPIs 15-17.
4.1.3. Polyacylated Neohesperidosides
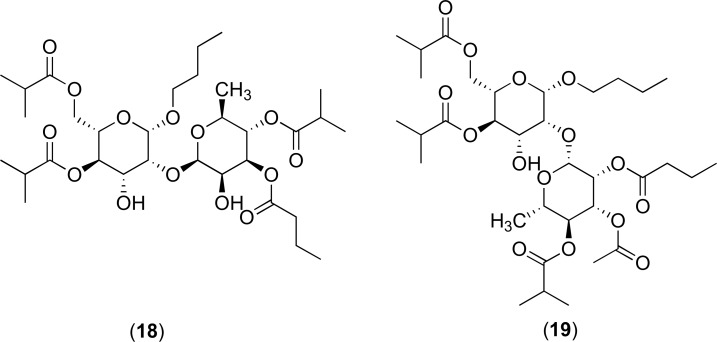
Further putative inhibitors of the NorA protein isolated from Geranium caespitosum have been documented and these include the polyacylated neohesperidosides. The complex 19 containing one additional ester group can reinforce the activities of many fluoroquinolone antibacterial agents such as norfloxacin and ciprofloxacin as well as an antibacterial constituent of rhubarb, named cassic acid. However, these inhibitors did not possess direct inhibitory action against the 8325-4 wild-type strain of S. aureus when in combination with berberine (at sub-inhibitory doses). The structures of 18 and 19 are shown in (Fig. 9) [66].
Fig. (9).
Chemical structures of EPIs 18 and 19.
4.1.4. Fermentation Products
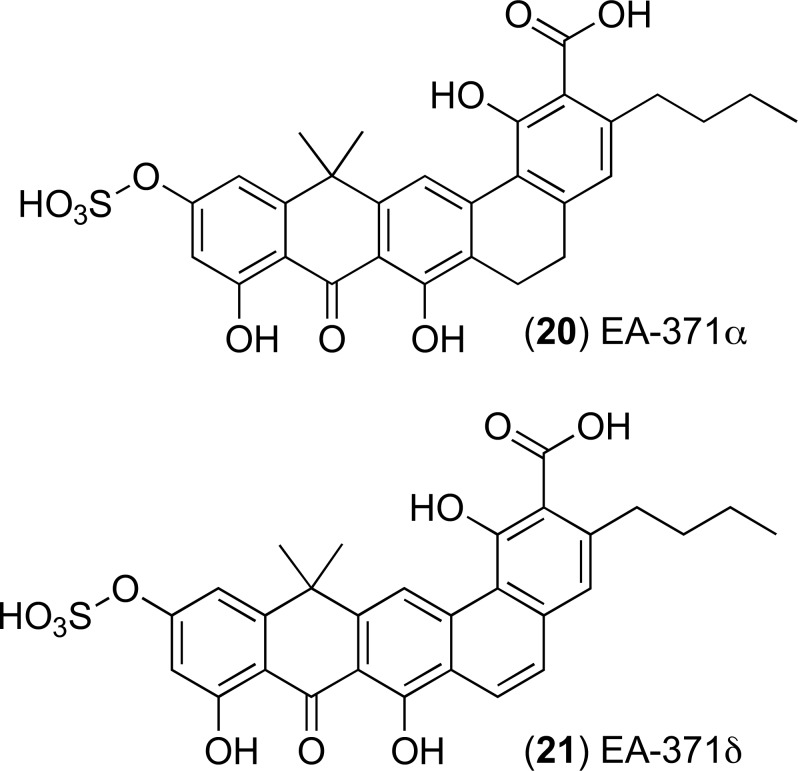
Screening for novel efflux pump inhibitors originating from microbial fermentation has yielded compounds top one: (20) EA-371α and bottom one: EA-371δ (21), Fig. (10) respectively. They were first extracted from a strain of Streptomyces bacteria and have been recognised as effective inhibitors of the P. aeruginosa MexAB-OprM system. EA-371α and EA-371δ did not display activity against other strains of P. aeruginosa with selected triple deletions in this tripartite efflux pump. The MIC of levofloxacin can be reduced 4-fold at a concentration of 0.625 mg/L for both 20 and 21 [67]. These two compounds are of particular importance due to their ability to inhibit tripartite efflux pumps in an MDR Gram-negative pathogen. There has been no report of total synthesis of these two compounds and they were not developed further towards pre-clinical evaluations. The structures of these compounds offer opportunities to carry out medicinal chemistry optimisation, but this is only possible after determining the binding site of these compounds within the MexAB-OprM efflux system. The crystal structure of MexAB-OprM efflux pump has been published [68, 69] and this data can be used to develop molecular models of these pumps to obtain valuable insight about the interactions of these molecules with the efflux pump system.
Fig. (10).
Chemical structures of EPIs 20 and 21.
4.1.5. Heterocyclic Macrocycles
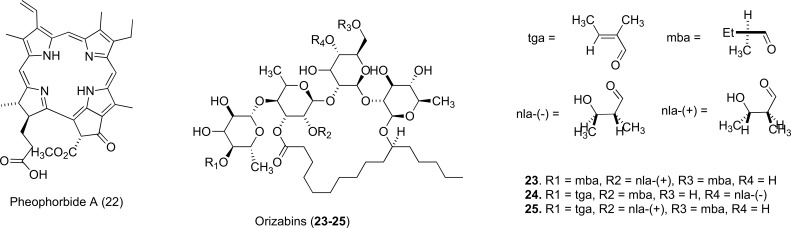
The natural porphyrin pheophorbide A (22), represents a very different structural class of inhibitors against NorA. Pheophorbide A is also found in the Berberis plant. It comprises 4 modified pyrrole subunits and is a product of cholesterol metabolism. These compounds can sensitize drug-resistant S. aureus to berberine, but the difficulties associated with synthesis and purification affected their further development. Unpredictable toxicity concerns also may have caused reduced research interest [70].
Heterocyclic oligosaccharides such as the orizabins (23-25) suppress resistance by inducing a 4-16 fold increase in activity of norfloxacin against the SA119B strain of S. aureus that repeatedly expresses NorA, showing greater potency when administered at low concentration. It was interestingly reported that cellular uptake by the MDR target transporter is mediated by acylation of the free hydroxyl and lipophilic side chains [71]. Orizabins have been shown to exhibit weak cytotoxic and antitumor activity and thus appear to be promising leads for development as antibacterial adjuvants. Illustrations of these macrocycles are shown in (Fig. 11).
Fig. (11).
Chemical structures of EPIs 22-25.
4.2. EPIs Derived from Synthetic Sources
4.2.1. Peptidomimetic Compounds
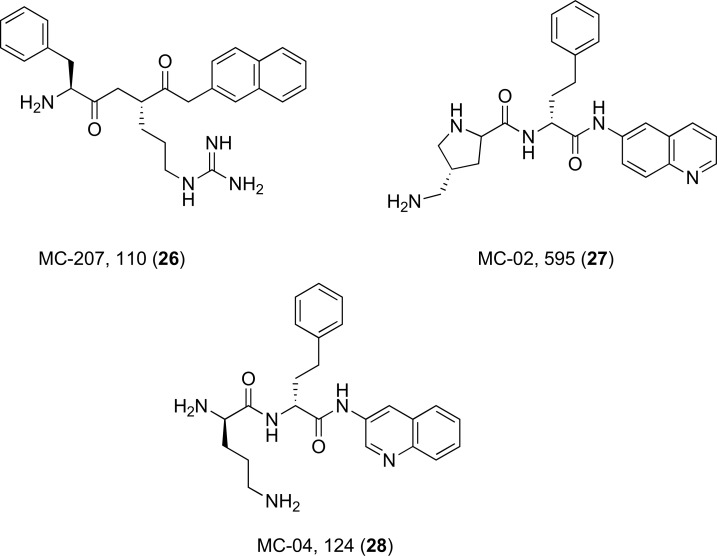
The EPI lead compound named Phe-Arg-β-naphthylamide (MC-207, 110) (26) is a dipeptide amide and is depicted with some of its derivatives (27 and 28) in (Fig. 12). It is more commonly known as PAβN and has been found to potentiate or restore activity of various classes of antibiotics including 4-fluoroquinolones, macrolides and chloramphenicol by inhibiting efflux pumps in a wide range of clinical pathogens [32, 72-74]. The addition of this EPI also decreases the selection of fluoroquinolone-resistant mutants. They remain to be the most widely used inhibitors for strains of P. aeruginosa that overexpress MexAB-OprM proteins and exhibit a competitive mechanism of action, where the target transporter is activated upon recognition of the inhibitor as a substrate rather than the quinolone antibiotic [75]. PAβN induces an 8-fold increase in activity for levofloxacin and has also been experimented upon in combination with a wide variety of antibiotics against the MexCD-OprJ and MexEF-OprN efflux transporters of P. aeruginosa. The attractiveness of these molecules comes from their versatile inhibitory activity to potentiate such a broad spectrum of antibacterial agents, which greatly lessens the emergence of new resistant strains of Gram-negative bacteria. In spite of this, these compounds could not be further developed due to their cytotoxic properties [73]. PAβN is routinely used as a research tool compound to study antimicrobial resistance and evaluation of newly identified EPIs from both natural and synthetic sources.
Fig. (12).
Chemical structures of EPIs 26-28.
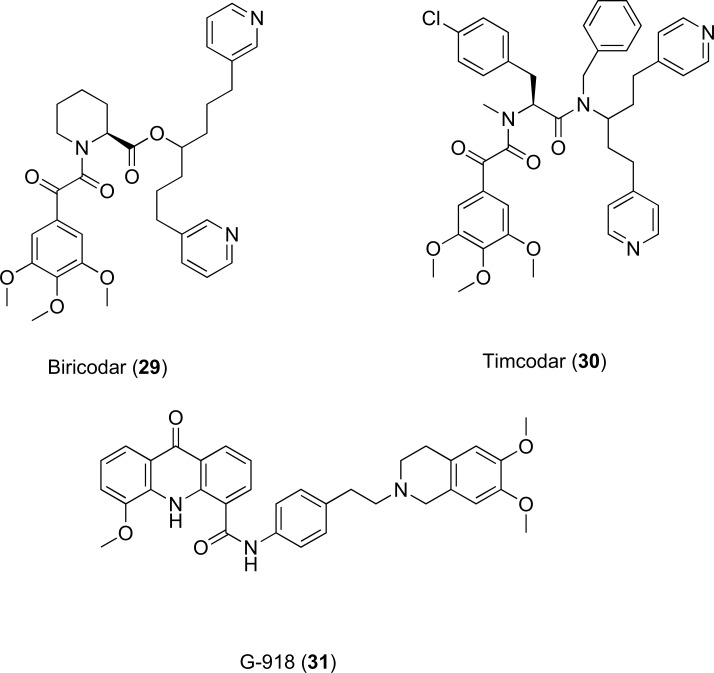
4.2.2. G-918, Biricodar (VX-710) and Timcodar (VX-853)
These three multi-cyclic compounds (29-31, Fig. 13) are capable of restoring drug sensitivity to mammalian cells that express the MRP-1 and P-gp efflux proteins while generating minimal in vivo toxicity [76, 77]. Biricodar has been developed by Vertex Pharmaceutical and approved as an adjunct therapy in ovarian cancer [78]. These compounds have been shown to work synergistically with fluoroquinolones against S. aureus, S. pneumoniae and E. faecalis [76]. Inhibition assays with the model substrate ethidium bromide demonstrated a 2-31 fold decrease in MIC for all three species of bacteria with compounds 29 and 30 displaying a direct inhibition effect in S. aureus. Recently Grossman et al. has reported that timcodar increases the potency of antituberculosis (anti-TB) drugs isoniazid and rifampicin against Mycobacterium tuberculosis in both in vitro and in vivo combination studies [79]. This is an interesting observation and warrants further study to understand the mechanism of drug-potentiating activity, as timcodar has an acceptable toxicity profile and could be developed as an adjunct therapy in MDR TB.
Fig. (13).
Chemical structures of EPIs 29-31
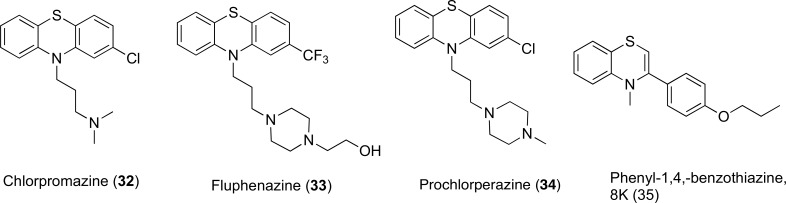
4.2.3. Phenothiazines and Thioxanthene Derivatives
The effect of inhibition by phenothiazines Fig. (14) against S. aureus strains possessing unique MDR-efflux proteins showed that phenothiazines had a particular intrinsic antibacterial effect [80]. In the SA-1199B isolate of S. aureus, that overexpresses NorA, the IC50 for efflux of ethidium bromide was found to be between 4 and 15% of the respective MICs of the 4 compounds. It was also concluded that 34 could disrupt the proton motive force after the active reduction of potential across the membrane. Chan et al. reported that phenothiazines potentiated the activity of range of antibiotics including erythromycin, levofloxacin and azithromycin in Burkholderia pseudomallei by interfering with the proton gradient at the inner membrane of the bacteria [81]. Chlorpromazine (32) also inhibited AcrB in Salmonella enterica, but it was suggested that phenothiazines do not directly inhibit efflux pumps and they exhibit synergistic activity by modulating the expression of the acrB gene [82]. Sabatini et al. synthesized phenyl-1,4-benzothiazine derivatives (35) as minimized structural templates of phenothiazine scaffold and the synthesized derivatives restored the activity of ciprofloxacin in norA-overexpressing S. aureus strain SA-K2378 [83]. These studies suggest that phenothiazines and derivatives of phenothiazine scaffold can inhibit a range of multidrug efflux transporters by a combination of mechanisms, and are a useful chemical scaffold to carry out further modifications.
Fig. (14).
Chemical structures of EPIs 32-35.
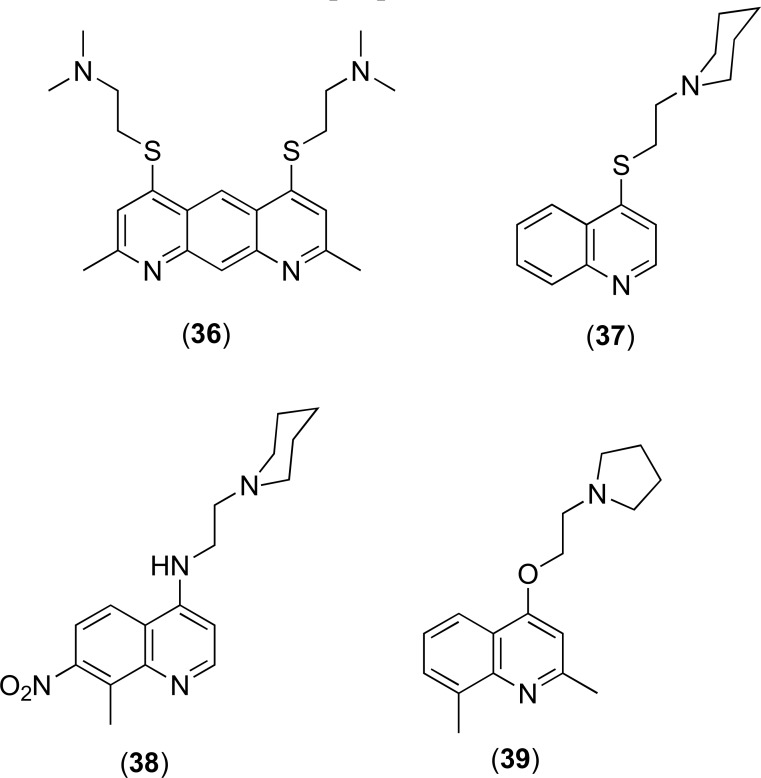
4.2.4. Quinoline Derivatives
Quinolines have shown inhibition of antibiotic efflux in isolates of the multidrug resistant bacteria such as E. aerogenes [84]. Pyridoquinolines such as 36 can restore norfloxacin activity in E. aerogenes by acting as substrate competitors on the pumps extruding fluoroquinolones [85]. A number of quinolone derivatives including those shown in Fig. (15) are capable of potentiating activity of antibiotics via inhibition of the AcrAB-ToIC efflux transporter [84, 86-88]. Experimentation on analogues 37-39 showed that they were able to recover susceptibility of tetracycline, norfloxacin and chloramphenicol in isolates of the Gram-negatives, K. pneumoniae and E. aerogenes [89]. More recently, Sabatini and co-workers developed 2-(4-Propoxy-phenyl)quinolone derivatives as inhibitors of the NorA efflux pump in S. aureus by modifying the natural flavone scaffold [90].
Fig. (15).
Chemical structures of EPIs 36-39.
4.2.5. Arylpiperazine Derivatives
N-heterocyclic compounds including phenyl piperazine derivatives have shown activity against both AcrAB and AcrEF efflux pumps present in E. coli [91, 92]. One of the leading aryl piperazine compounds 1-(1- Naphthylmethyl)-piperazine (NMP, Fig. (16) has shown notable EPI activity and inhibited both AcrAB and AcrEF efflux pumps [92, 93]. It was able to reverse drug resistance in E. coli clinical isolates and make them susceptible to fluoroquinolones [94]. It also demonstrated a partial reversal in species of Enterobacteriaceae, e.g. E. aerogenes and K. pneumonia [95] and Acinetobacter baumannii [92]. Interestingly, NMP and PAβN showed a differential preference for different antibacterial agents in potentiating their activities, suggesting non-overlapping mechanisms of action [92, 95, 96]. It has been suggested that NMP inhibits the AcrB efflux pump by interfering with its functional assembly and movement of the G-loop that plays an important role in the extrusion of certain substrates [97]. Arylpiperazines have enjoyed limited success as EPIs due to their relatively low potency, and the structural similarity with serotonin agonists has further hindered their development. [70].
Fig. (16).
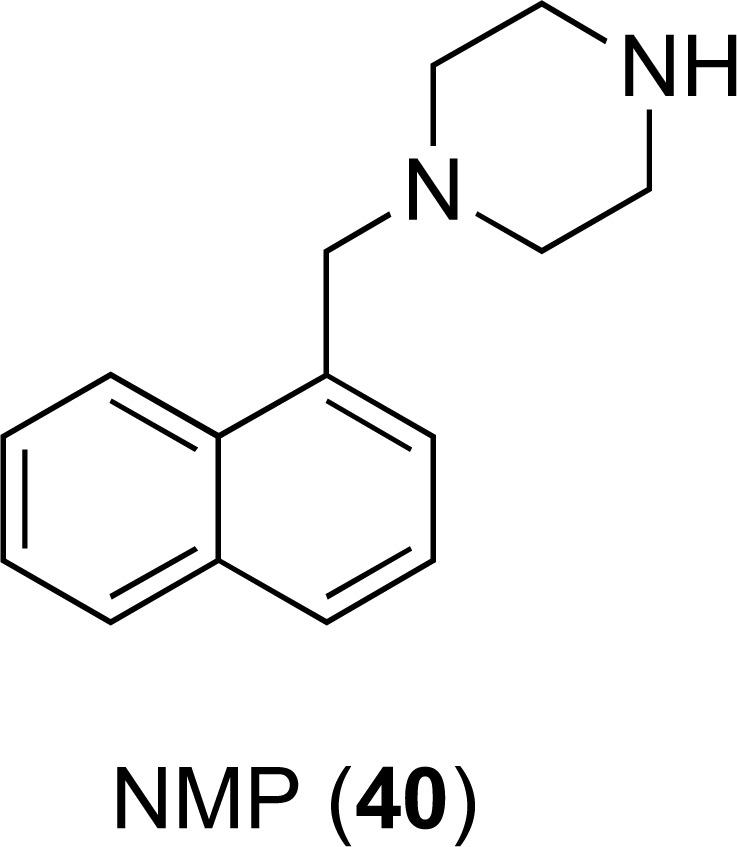
Chemical structures of EPI 40.
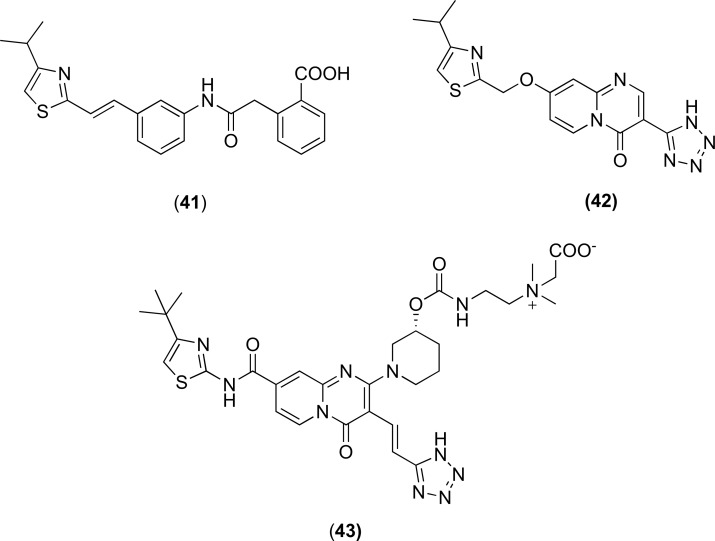
4.2.6. Pyridopyrimidine Derivatives
Pyridopyrimidine analogues were first reported in 2003 and showed notable activity against the MexAB-OprM pump in P. aeruginosa. The compounds progressed to the pre-clinical development and two analogues D13-9001 (43) and D2 (42) Fig. (17) showed both in vitro and in vivo activity [98, 99]. D13-9001 is the current lead compound and crystal structures of D13-9001 bound to AcrB (E. coli) and MexB (P. aeruginosa) have been recently published [35]. It has been proposed that D13-9001 binds tightly to the hydrophobic trap and prevents the conformational changes that are needed for the proper activity of the pump [35]. Additionally, the compound has a hydrophilic component that interacts with the substrate binding channel of both AcrB and MexB, and thereby prevents substrate binding to the pumps. Despite the early promise in the lead optimisation and preclinical studies [101-104], the compounds are yet to progress to the clinic for clinical evaluation.
Fig. (17).
Chemical structures of EPIs 41-43.
4.2.7. Pyranopyridine Derivatives
Recently a pyranopyridine compound MBX2319 Fig. (18) has been reported as a potent inhibitor of the AcrAB efflux pump of E. coli, but did not show any notable activity in P. aeruginosa [99, 105-107]. It significantly potentiated the activity of antibiotics that are known as substrates of the AcrAB efflux pump [105]. The addition of polymyxin B nonapeptide (PMBN), a derivative of the membrane active compound polymixin B, increased the EPI potency of this molecule in P. aeruginosa suggesting the barrier function of the outer membrane as the key obstacle to its EPI activity. A detailed computational study suggested that MBX2319 interacted with the “hydrophobic trap” [35] of the AcrB pump and the pyridine ring formed a ring stacking interaction with the amino acid residues [99, 107]. There has been no report of in vivo activity of this chemical series to date. The knowledge obtained from the computational study may help the researchers to develop a more potent pyranopyrdine analogue and take this chemical scaffold towards pre-clinical development.
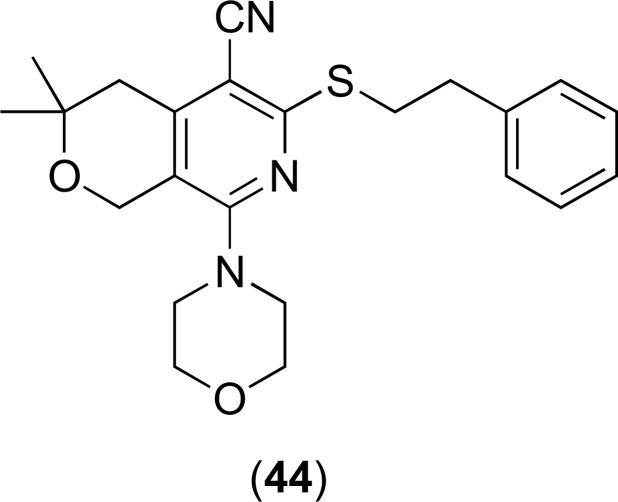
Fig. (18).
Chemical structures of EPIs 44.
5. STRUCTURAL AND COMPUTATIONAL APPROACHES TO DEVELOP EFFLUX PUMP INHIBITORS
While a number of structurally diverse compounds exhibiting significant efflux inhibition activity have been cited in the literature, significant concerns to do with potency, pharmacokinetics and toxicity have prevented them from being tested in a clinical setting [108]. Another major challenge that today’s researchers face is the apparent dearth of biochemical, computational and structural methods that has restricted the discovery of new, more efficient classes of efflux inhibitors. Implementing tools such as these would provide a greater understanding of not only the MDR efflux pump-inhibitor binding site but also a highly detailed mechanism of drug transport that could explain the mode of inhibition [109]. The invaluable data would generate a wealth of information that could be used to direct the rational design of the next generation of EPI analogues to address and eventually reverse resistance in bacterial pathogens.
However, due to difficulties in producing co-crystals coupled with the biochemical complexity of these transporters, scientists are increasingly leaning towards computational techniques to decipher the similarities and differences between their hypothesised structures and mechanisms of action (MOA) [106]. Efflux pump inhibitors have previously been developed via random screening of libraries of synthetic chemical agents or derived from natural sources, for instance, plant metabolites [110]. Molecular dynamics simulation (MDS), advanced three-dimensional structure resolution and molecular modelling can aid the identification of pharmacophores of a putative inhibitor that are able to detect a specific binding site located on the efflux pump [110]. In particular, MDS is a system that utilises computer algorithms that are designed to simulate the physical, molecular movement of assemblies of atoms or molecules comprising a structure in the presence of the solvent, their microscopic interactions and changes in conformation.
One way in which the data generated by MDS can be exploited is to introduce a series of compounds that compete with the flux of antibiotics in both competitive and non-competitive fashions so as to ensure the maximum possible inhibition. For example, agents that interact non-competitively, those that cause steric hindrance by covalently bonding at the binding site and those that directly plug or block the efflux channel represent the most attractive approaches when designing new inhibitors as an adjunctive therapy [110]. It has been shown that targeted strategies towards the inhibition of specific efflux pumps can be developed by MDS data that allows the researcher to distinguish between molecular interactions caused by the binding of substrates from the binding of inhibitors [35].
Although MDS methodologies currently in use are limited by i) expensive computational cost, ii) an inability to determine the effect of the local environment surrounding the binding site on a charged substrate and iii) are based upon approximations of the force fields generated by the atoms in the model (making the estimation of binding affinity more difficult), it has proved to be beneficial when multiple high quality 3D structures are available [110]. Longer MD simulations of more intricate membrane-bound protein structures are rapidly emerging that will generate quantitative data on the microsecond timescale that remains undetected by more traditional computational experiments [111]. These advances improve the predictive accuracy of the model and permit the scientist to gather vital atomic level information, creating a more realistic picture, including the contributions of specific residues involved in ligand to pump binding [112]. In silico experiments also essentially require validation by wet-laboratory studies and functional testing.
All-atom MD simulations, molecular docking and other well-established computational tools have also been applied to elucidate further information on the molecular interactions between EPIs and bacterial pumps, particularly those belonging to the RND family found in Gram-negative pathogens [35, 36, 97, 113-115]. Molecular level knowledge of interactions between an EPI and the efflux pump can be very helpful to design new and effective EPIs that can be used to break efflux mediated antimicrobial resistance [35].
Among the inhibitors mentioned in the literature displaying notable potency, peptidomimetics such as PAβN and its derivatives, as well as derivatives of pyridopyrimidine have widely been investigated, both having advanced to the preclinical stages of drug development [30, 32, 33, 98, 116]. These structural groups show significant differences relating to the specific modes of binding of an inhibitor to the substrate binding pockets. Unlike PAβN, more promising pyridopyrimidine inhibitors such as ABI-PP do not induce membrane dysfunction and do not show intrinsic antimicrobial activity [35, 117]. This relatively water-soluble inhibitor binds to the distal substrate pocket on AcrB and MexB proteins, competitively inhibiting distal binding drugs. The lack of interaction of ABI-PP with the proximal region correlates with the fact that proximal binding drugs remained unaffected [100, 118]. In this instance, MD simulations were used to depict the tight attachment of ABI-PP to a narrow, hydrophobic cavity consisting of phenylalanine residues that are situated directly adjacent to the substrate translocation channel. The orientation with which the inhibitor binds sterically prevents the rotation of AcrB and MexB and so potentiates the activities of antibiotics that are effluxed by these inner membrane transporters [35]. Inhibitor specificity is restricted to the slight variances in the size of the hydrophobic trap [35]. Molecular docking studies have also suggested that ABI-PP has the ability to efficiently interact and bind to the distal pockets of AcrB and MexB pumps of differing species with a desirable binding affinity and energy (unpublished results). Data derived from these experiments would significantly influence our understanding of other MDR efflux families.
A 2012 study conducted by Collu et al. aimed to examine and discriminate between the binding properties of the two highly potent carbapenem antibiotics imipenem and meropenem to MexB, the major RND transporter found primarily in isolates of P. aeruginosa. Their experiments produced an insight into the mechanism that MDR efflux pumps adopt in order to differentiate between their substrates from their inhibitors or modulating agents [119]. The transporter’s periplasmic domain identified two affinity binding sites that appeared to share multiple resemblances with the proximal and distal binding regions found in that of AcrB of E. coli, a close homologue of MexB. Although both antibiotic drugs indicated a relatively weak affinity for the proximal binding region, imipenem displayed a weaker binding affinity to the distal binding pocket compared to meropenem. This behaviour can be explained by the non-pharmacophore regions of the two drugs, where imipenem has a greater hydrophobic, bulky character. Applying MDS, along with free energy estimation methods and molecular docking produced results in good agreement with the experimental evidence of the carbapenems. It also highlights the important role played by the surrounding solvent in antibiotic transport as both imipenem and meropenem showed a high degree of solvation in both binding pockets [119].
Finally, in another EPI investigation, Vargiu and Nikaido implemented MD simulations in order to inspect the binding modes of thirteen structurally diverse ligands to the distal binding site of the AcrB pump. Nine substrates, two inhibitors and two non-substrate compounds were included [120]. These studies were performed due to the scarcity of compounds that have been co-crystallised with AcrB, emphasising how essential computational approaches are in delivering a more realistic view of binding compared to docking experiments. They noted a high degree of flexibility in the distal binding pocket of the AcrB secondary transporter that underwent major conformational changes and that explicit water molecules undoubtedly contributed to the docking of the compounds [120]. Remarkably, it was also reported that the inhibitors 1-(1-naphtylmethyl)-piperazine (NMP) and PAβN had a tendency to partially move out of the pocket in order to engage contact with a glycine rich loop (also known as the switch loop) that is thought to separate the proximal region from the distal binding site. On the other hand, non-inhibitor substrates remained completely inside the pocket for the entire duration of the MD simulation. This G-loop must, therefore, discriminate on which substrates are permitted to access the distal binding pocket. The MD prediction illuminates features of the inhibition pathways taken by NMP and PAβN that concur with a study performed by Feng et al. also in the same year, that explains how mutations in the G-loop (G616Pro/ G619Pro) of AcrB interferes with the extrusion of drug molecules, likely arising from the reduction in the loops flexibility [36, 97, 121]. It is known that diverse EPIs display subtle differences in their inhibition pathways, however, most are able to deform the distal binding cavity and disturb proper substrate binding.
It is of crucial importance that the binding processes and efflux mechanisms used by these intricate molecular machines are well characterised and understood. In silico simulations offer an ideal method by which molecular hypotheses for the mechanism of action of EPIs can be made. It provides the physiochemical reasoning for the differences observed in the transport efficiency of structurally diverse antibiotics and other drugs. Moreover, computational approaches could also define transporter sensitivity to inhibition that might be determined from the primary and tertiary structures of the proteins comprising the efflux pumps, thus expediting the rational design of new EPIs with enhanced stability and greater targeting affinity for the binding site.
6. CHALLENGES IN DEVELOPING NEW EFFLUX PUMP INHIBITORS: FUTURE PERSPECTIVES
Efflux pump inhibitors can play a major role in tackling antimicrobial resistance by reviving antibiotics to which many clinically relevant pathogenic microorganisms have become resistant. They can be also used as a chemical tool to understand the molecular mechanisms of antimicrobial resistance that are mediated by efflux, particularly in Gram-negative bacteria. Efflux pump activities can be blocked by interfering with the functional assemblies of efflux pumps' components. Efflux pump activity may be bypassed or inhibited by employing a range of diverse approaches [118] including i) modifying the chemical structure of antibiotics to decrease their binding affinity to the transporter cavities; an approach which has been used for tetracycline antibiotics [122], ii) using permeabilizers of the bacterial membrane to artificially increase intracellular antibiotic concentration; this approach has been applied for MexAB-OprM and MexXY-OprM efflux pump of P. aeruginosa [123, 124], iii) decreasing the number of active efflux complexes in the envelope of bacteria, by downregulating efflux pump gene expression or destabilising the protein component; like what has been applied for OmpF and OmpC [125], iv) the destruction of the drug transporter energy source, using potassium cyanide and carbonyl cyanide m-chlorophenylhy- drazone (CCCP) affect the energy level of the bacterial membrane and reduce the efflux of various agents, v) blocking the functional assembly of the components of efflux systems [86], vi) designing inhibitors that bind covalently to the substrate-binding cavities or block the channel of antibiotic transporter pumps; various natural products [126] and nanoparticles like zinc oxide [127] have been shown to inhibit bacterial efflux pumps by blocking them (a “molecular plug”) and vii) applying a decoy substrate as a competitive inhibitor for antibiotic transport inside the pump.
There has been very limited success in developing efflux pump inhibitors that can be used clinically as an adjunct to antimicrobial therapy. Although there have been some encouraging results for EPIs derived from natural sources [55], there has been limited effort to optimize the vast number of chemical compounds that has been identified from natural sources through medicinal-chemistry optimisation due to the lack of reliable model systems. This failure in the development of EPIs and associated model systems can be attributed to the gap of knowledge in the functional assemblies of various efflux pumps and the lack of reliable biochemical, computational and structural models of efflux pump functions [107]. It has been particularly problematic to develop EPIs that successfully inhibit RND type pumps due to their structural complexity and the polyspecific nature of these pumps. It is urgent to study the mechanisms of action of the current EPIs as well as a fuller understanding of the structure of efflux pumps to develop successful EPIs.
Although experimental methods like site directed mutagenesis and X-ray crystallography are very useful approaches to understanding the functional assemblies of efflux pumps and mechanism of action of efflux pump inhibitors (EPIs), they often fail to pick up interactions between various components of efflux pumps and EPIs at the molecular level. X-ray crystallography coupled with computational methods can be a very powerful approach to understanding the mechanisms of efflux pumps and developing a rational drug design programme to designing novel EPIs. Understanding of the mechanisms of inhibition of efflux pumps by running long-term (micro seconds) molecular dynamics (MD) simulations and quantum molecular dynamics (QMD) simulations could have a major impact in the field of EPI discovery. The binding sites that are currently unknown could be identified by performing long-term MD simulations of already developed EPIs like PAβN and different bacterial efflux pumps that can be inhibited by these EPIs. These simulations can also shed important lights in identifying the binding protomers of different transporters. More efforts should be made to co-crystallize EPIs with efflux pumps and use these crystal structures to develop reliable and functional molecular models to take advantage of the recent advances in powerful computing. By using the rational drug design approach, and exploring both natural and synthetic compounds, it may be possible to develop clinically useful EPIs that can be a major weapon in our fight against antimicrobial resistance.
ACKNOWLEDGEMENTS
Declared none.
ABBREVIATIONS AND DEFINITIONS
- AMR
= Antimicrobial Resistance
- Antibiotic
= A chemical compound produced by one microorganism that selectively inhibits the growth of another microorganism
- Antimicrobials
= Chemical compounds that kill or inhibit growth of microorganisms
- EPIs
= Efflux Pump Inhibitors
- MD
= Molecular Dynamics
- MDR
= Multidrug Resistance
- MES
= Multiple Efflux Systems
- OMP
= Outer Membrane Protein
- RND
= Resistance Nodulation Division
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Ruef C. Epidemiology and clinical impact of glycopeptide resistance in Staphylococcus aureus. Infection. 2004;32(6):315–327. doi: 10.1007/s15010-004-4124-7. [DOI] [PubMed] [Google Scholar]
- 2.Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008;197(8):1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 3.Piddock L.J. The crisis of no new antibiotics—what is the way forward? Lancet Infect. Dis. 2012;12(3):249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- 4.Collins A.S. 2008. Preventive Health Care-Associated Infections. [Google Scholar]
- 5.Fair R.J., Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta N., Limbago B.M., Patel J.B., Kallen A.J. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 2011;53(1):60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 7.Poole K., Krebes K., McNally C., Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 1993;175(22):7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. R.: MMBR. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., Scheld M., Spellberg B., Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 10.Wright G.D., Berghuis A.M., Mobashery S. Aminoglycoside antibiotics. Structures, functions, and resistance. Adv. Exp. Med. Biol. 1998;456:27–69. [PubMed] [Google Scholar]
- 11.Thomson K.S., Smith Moland E. Version 2000: the new beta-lactamases of Gram-negative bacteria at the dawn of the new millennium. Microbes Infect. 2000;2(10):1225–1235. doi: 10.1016/s1286-4579(00)01276-4. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis.: an official publication of the Infect. Dis. Soc. Am. 2005;41(Suppl. 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 13.Leclercq R., Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 1991;35(7):1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreier J., Ruggerone P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 2015;6:660. doi: 10.3389/fmicb.2015.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister P.D., Wolter D.J., Hanson N.D. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riera E., Cabot G., Mulet X., García-Castillo M., del Campo R., Juan C., Cantón R., Oliver A. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J. Antimicrob. Chemother. 2011;66(9):2022–2027. doi: 10.1093/jac/dkr232. [DOI] [PubMed] [Google Scholar]
- 17.Lin M-F., Lin Y-Y., Tu C-C., Lan C-Y. Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J. Microbiol. Immunol. Infect. 2015 doi: 10.1016/j.jmii.2015.04.004. [Epub ahead of Print]. [DOI] [PubMed] [Google Scholar]
- 18.Nowak J., Seifert H., Higgins P.G. Prevalence of eight resistance-nodulation-division efflux pump genes in epidemiologically characterized Acinetobacter baumannii of worldwide origin. J. Med. Microbiol. 2015;64(6):630–635. doi: 10.1099/jmm.0.000069. [DOI] [PubMed] [Google Scholar]
- 19.Kosmidis C., Schindler B.D., Jacinto P.L., Patel D., Bains K., Seo S.M., Kaatz G.W. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents. 2012;40(3):204–209. doi: 10.1016/j.ijantimicag.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Lamers R.P., Cavallari J.F., Burrows L.L. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAbetaN) permeabilizes the outer membrane of gram-negative bacteria. PLoS One. 2013;8(3):e60666. doi: 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquez B. Bacterial efflux systems and efflux pumps inhibitors. Biochimie. 2005;87(12):1137–1147. doi: 10.1016/j.biochi.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Tangden T. Combination antibiotic therapy for multidrug-resistant Gram-negative bacteria. Ups. J. Med. Sci. 2014;119(2):149–153. doi: 10.3109/03009734.2014.899279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischbach M.A. Combination therapies for combating antimicrobial resistance. Curr. Opin. Microbiol. 2011;14(5):519–523. doi: 10.1016/j.mib.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juliano R.L., Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta. 1976;455(1):152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 25.Lomovskaya O., Zgurskaya H.I., Totrov M., Watkins W.J. Waltzing transporters and 'the dance macabre' between humans and bacteria. Nat. Rev. Drug Discov. 2007;6(1):56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- 26.Kourtesi C., Ball A.R., Huang Y.Y., Jachak S.M., Vera D.M., Khondkar P., Gibbons S., Hamblin M.R., Tegos G.P. Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol. J. 2013;7:34–52. doi: 10.2174/1874285801307010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pirri G., Giuliani A., Nicoletto S., Pizzuto L., Rinaldi A. Lipopeptides as anti-infectives: a practical perspective. Cent. Eur. J. Biol. 2009;4(3):258–273. [Google Scholar]
- 28.Silver L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24(1):71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurosu M., Siricilla S., Mitachi K. Advances in MRSA drug discovery: where are we and where do we need to be? Expert Opin. Drug Discov. 2013;8(9):1095–1116. doi: 10.1517/17460441.2013.807246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Askoura M., Mottawea W., Abujamel T., Taher I. Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J. Med. 2011:6. doi: 10.3402/ljm.v6i0.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X.Z., Livermore D.M., Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 1994;38(8):1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomovskaya O., Bostian K.A. Practical applications and feasibility of efflux pump inhibitors in the clinic--a vision for applied use. Biochem. Pharmacol. 2006;71(7):910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Lomovskaya O., Watkins W. Inhibition of efflux pumps as a novel approach to combat drug resistance in bacteria. J. Mol. Microbiol. Biotechnol. 2001;3(2):225–236. [PubMed] [Google Scholar]
- 34.Sumithra T.G., Chaturvedi V.K., Cherian S., Krishnan B.B., Jacob S.S. Effux pump inhibitors for antibacterial therapy. JIVA. 2012;10(1):69–75. [Google Scholar]
- 35.Nakashima R., Sakurai K., Yamasaki S., Hayashi K., Nagata C., Hoshino K., Onodera Y., Nishino K., Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 36.Yu E.W., Aires J.R., McDermott G., Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187(19):6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikaido H., Pages J.M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012;36(2):340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruggerone P., Murakami S., Pos M.K., Vargiu A.V. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 2013;13(24):3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 39.Venter H., Mowla R., Ohene-Agyei T., Ma S. RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 2015;6:377. doi: 10.3389/fmicb.2015.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amaral L., Martins A., Spengler G., Molnar J. Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front. Pharmacol. 2013;4:168. doi: 10.3389/fphar.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee A., Mao W., Warren M.S., Mistry A., Hoshino K., Okumura R., Ishida H., Lomovskaya O. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 2000;182(11):3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinchliffe P., Symmons M.F., Hughes C., Koronakis V. Structure and operation of bacterial tripartite pumps. Annu. Rev. Microbiol. 2013;67:221–242. doi: 10.1146/annurev-micro-092412-155718. [DOI] [PubMed] [Google Scholar]
- 43.Blair J.M., Piddock L.J. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 2009;12(5):512–519. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T., Venter H., Chiu W., Luisi B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509(7501):512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowan M.M. Plant products as antimicrobial agents. Clin. Microbiol. 1999;12(4):564–582. doi: 10.1128/cmr.12.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rana T., Singh S., Kaur N., Pathania K., Farooq U. A review of Efflux Pump Inhibitors of Medically Important Bacteria from Plant Sources. Int. J. Pharm. Sci. Rev. Res. 2014;26(2):101–111. [Google Scholar]
- 47.Sibanda T., Okoh A.I. The challenges of overcoming antibiotic resistance: Plant extracts as potential sources of antimicrobial and resistance modifying agents. Afr. J. Biotechnol. 2007;6(25):2886–2896. [Google Scholar]
- 48.Neyfakh A.A., Bidnenko V.E., Chen L.B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA. 1991;88(11):4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neyfakh A.A., Borsch C.M., Kaatz G.W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993;37(1):128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeifer H.J., Greenblatt D.K., Koch-Wester J. Clinical toxicity of reserpine in hospitalized patients: a report from the Boston Collaborative Drug Surveillance Program. Am. J. Med. Sci. 1976;271(3):269–276. doi: 10.1097/00000441-197605000-00002. [DOI] [PubMed] [Google Scholar]
- 51.Khan I.A., Mirza Z.M., Kumar A., Verma V., Qazi G.N. Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50(2):810–812. doi: 10.1128/AAC.50.2.810-812.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stermitz F.R., Lorenz P., Tawara J.N., Zenewicz L.A., Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5'-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA. 2000;97(4):1433–1437. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deep G., Agarwal R. New combination therapies with cell-cycle agents. Curr. Opin. Investig. Drugs. 2008;9(6):591–604. [PMC free article] [PubMed] [Google Scholar]
- 54.Singh R.P., Agarwal R. Mechanisms and preclinical efficacy of silibinin in preventing skin cancer. Eur. J. Cancer. 2005;41(13):1969–1979. doi: 10.1016/j.ejca.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 55.Stavri M., Piddock L.J., Gibbons S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007;59(6):1247–1260. doi: 10.1093/jac/dkl460. [DOI] [PubMed] [Google Scholar]
- 56.Morel C., Stermitz F.R., Tegos G., Lewis K. Isoflavones as potentiators of antibacterial activity. J. Agric. Food Chem. 2003;51(19):5677–5679. doi: 10.1021/jf0302714. [DOI] [PubMed] [Google Scholar]
- 57.Ohene‐Agyei T., Mowla R., Rahman T., Venter H. Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. MicrobiologyOpen. 2014;3(6):885–896. doi: 10.1002/mbo3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stapleton P.D., Shah S., Anderson J.C., Hara Y., Hamilton-Miller J.M., Taylor P.W. Modulation of β-lactam resistance in Staphylococcus aureus by catechins and gallates. Int. J. Antimicrob. Agents. 2004;23(5):462–467. doi: 10.1016/j.ijantimicag.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 59.Gibbons S., Moser E., Kaatz G.W. Catechin gallates inhibit multidrug resistance (MDR) in Staphylococcus aureus. Planta Med. 2004;70(12):1240–1242. doi: 10.1055/s-2004-835860. [DOI] [PubMed] [Google Scholar]
- 60.Sudano Roccaro A., Blanco A.R., Giuliano F., Rusciano D., Enea V. Epigallocatechin-gallate enhances the activity of tetracycline in staphylococci by inhibiting its efflux from bacterial cells. Antimicrob. Agents Chemother. 2004;48(6):1968–1973. doi: 10.1128/AAC.48.6.1968-1973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson J.C., Headley C., Stapleton P.D., Taylor P.W. Synthesis and antibacterial activity of hydrolytically stable (−)-epicatechin gallate analogues for the modulation of β-lactam resistance in Staphylococcus aureus. Bioorg. Med. Chem. Lett. 2005;15(10):2633–2635. doi: 10.1016/j.bmcl.2005.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson J.C., Headley C., Stapleton P.D., Taylor P.W. Asymmetric total synthesis of B-ring modified (−)-epicatechin gallate analogues and their modulation of β-lactam resistance in Staphylococcus aureus. Tetrahedron. 2005;61(32):7703–7711. doi: 10.1016/j.tet.2005.05.086. [DOI] [PubMed] [Google Scholar]
- 63.Hamilton-Miller J., Shah S. Activity of the tea component epicatechin gallate and analogues against methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2000;46(5):852–853. doi: 10.1093/jac/46.5.852. [DOI] [PubMed] [Google Scholar]
- 64.Oluwatuyi M., Kaatz G.W., Gibbons S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry. 2004;65(24):3249–3254. doi: 10.1016/j.phytochem.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 65.Smith E.C., Kaatz G.W., Seo S.M., Wareham N., Williamson E.M., Gibbons S. The phenolic diterpene totarol inhibits multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51(12):4480–4483. doi: 10.1128/AAC.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stermitz F.R., Cashman K.K., Halligan K.M., Morel C., Tegos G.P., Lewis K. Polyacylated neohesperidosides from Geranium caespitosum: bacterial multidrug resistance pump inhibitors. Bioorg. Med. Chem. Lett. 2003;13(11):1915–1918. doi: 10.1016/s0960-894x(03)00316-0. [DOI] [PubMed] [Google Scholar]
- 67.Lee M.D., Galazzo J.L., Staley A.L., Lee J.C., Warren M.S., Fuernkranz H., Chamberland S., Lomovskaya O., Miller G.H. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco. 2001;56(1-2):81–85. doi: 10.1016/s0014-827x(01)01002-3. [DOI] [PubMed] [Google Scholar]
- 68.Akama H., Kanemaki M., Yoshimura M., Tsukihara T., Kashiwagi T., Yoneyama H., Narita S-i., Nakagawa A., Nakae T. Crystal Structure of the Drug Discharge Outer Membrane Protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004;279(51):52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 69.Akama H., Matsuura T., Kashiwagi S., Yoneyama H., Narita S-i., Tsukihara T., Nakagawa A., Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279(25):25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 70.Zechini B., Versace I. Inhibitors of multidrug resistant efflux systems in bacteria. Recent Pat. Antiinfect. Drug Discov. 2009;4(1):37–50. doi: 10.2174/157489109787236256. [DOI] [PubMed] [Google Scholar]
- 71.Babayan A., Nikaido H. In Pseudomonas aeruginosa ethidium bromide does not induce its own degradation or the assembly of pumps involved in its efflux. Biochem. Biophys. Res. Commun. 2004;324(3):1065–1068. doi: 10.1016/j.bbrc.2004.09.146. [DOI] [PubMed] [Google Scholar]
- 72.Renau T.E., Léger R., Flamme E.M., Sangalang J., She M.W., Yen R., Gannon C.L., Griffith D., Chamberland S., Lomovskaya O. Inhibitors of Efflux Pumps in Pseudomonas aeruginosa Potentiate the Activity of the Fluoroquinolone Antibacterial Levofloxacin. J. Med. Chem. 1999;42(24):4928–4931. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 73.Lomovskaya O., Warren M.S., Lee A., Galazzo J., Fronko R., Lee M., Blais J., Cho D., Chamberland S., Renau T., Leger R., Hecker S., Watkins W., Hoshino K., Ishida H., Lee V.J. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lynch A.S. Efflux systems in bacterial pathogens: an opportunity for therapeutic intervention? An industry view. Biochem. Pharmacol. 2006;71(7):949–956. doi: 10.1016/j.bcp.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 75.Tohidpour A., Peerayeh S.N., Mehrabadi J.F., Yazdi H.R. Determination of the efflux pump-mediated resistance prevalence in Pseudomonas aeruginosa, using an efflux pump inhibitor. Curr. Microbiol. 2009;59(3):352–355. doi: 10.1007/s00284-009-9444-5. [DOI] [PubMed] [Google Scholar]
- 76.Mullin S., Mani N., Grossman T.H. Inhibition of antibiotic efflux in bacteria by the novel multidrug resistance inhibitors biricodar (VX-710) and timcodar (VX-853). Antimicrob. Agents Chemother. 2004;48(11):4171–4176. doi: 10.1128/AAC.48.11.4171-4176.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Minderman H., O’Loughlin K.L., Pendyala L., Baer M.R. VX-710 (biricodar) increases drug retention and enhances chemosensitivity in resistant cells overexpressing P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Clin. Cancer Res. 2004;10(5):1826–1834. doi: 10.1158/1078-0432.ccr-0914-3. [DOI] [PubMed] [Google Scholar]
- 78.Seiden M.V., Swenerton K.D., Matulonis U., Campos S., Rose P., Batist G., Ette E., Garg V., Fuller A., Harding M.W. A phase II study of the MDR inhibitor biricodar (INCEL, VX-710) and paclitaxel in women with advanced ovarian cancer refractory to paclitaxel therapy. Gynecol. Oncol. 2002;86(3):302–310. doi: 10.1006/gyno.2002.6762. [DOI] [PubMed] [Google Scholar]
- 79.Grossman T.H., Shoen C.M., Jones S.M., Jones P.L., Cynamon M.H., Locher C.P. The Efflux Pump Inhibitor Timcodar Improves the Potency of Antimycobacterial Agents. Antimicrob. Agents Chemother. 2015;59(3):1534–1541. doi: 10.1128/AAC.04271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaatz G.W., Moudgal V.V., Seo S.M., Kristiansen J.E. Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003;47(2):719–726. doi: 10.1128/AAC.47.2.719-726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan Y.Y., Ong Y.M., Chua K.L. Synergistic interaction between phenothiazines and antimicrobial agents against Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2007;51(2):623–630. doi: 10.1128/AAC.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bailey A.M., Paulsen I.T., Piddock L.J., Ram A. Confers Multidrug Resistance in Salmonella enterica via Increased Expression of acrB, Which Is Inhibited by Chlorpromazine. Antimicrob. Agents Chemother. 2008;52(10):3604–3611. doi: 10.1128/AAC.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sabatini S., Kaatz G.W., Rossolini G.M., Brandini D., Fravolini A. From Phenothiazine to 3-Phenyl-1,4-benzothiazine Derivatives as Inhibitors of the Staphylococcus aureus NorA Multidrug Efflux Pump. J. Med. Chem. 2008;51(14):4321–4330. doi: 10.1021/jm701623q. [DOI] [PubMed] [Google Scholar]
- 84.Mahamoud A., Chevalier J., Davin-Regli A., Barbe J. Quinoline derivatives as promising inhibitors of antibiotic efflux pump in multidrug resistant Enterobacter aerogenes isolates. Curr. Drug Targets. 2006;7(7):843–847. doi: 10.2174/138945006777709557. [DOI] [PubMed] [Google Scholar]
- 85.Chevalier J., Atifi S., Eyraud A., Mahamoud A., Barbe J., Pagès J-M. New Pyridoquinoline Derivatives as Potential Inhibitors of the Fluoroquinolone Efflux Pump in Resistant Enterobacter aerogenes Strains. J. Med. Chem. 2001;44(23):4023–4026. doi: 10.1021/jm010911z. [DOI] [PubMed] [Google Scholar]
- 86.Pagès J-M., Masi M., Barbe J. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol. Med. 2005;11(8):382–389. doi: 10.1016/j.molmed.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 87.Malléa M., Mahamoud A., Chevalier J., Alibert-Franco S., Brouant P., Barbe J. Alkylaminoquinolines inhibit the bacterial antibiotic efflux pump in multidrug-resistant clinical isolates. Biochem. J. 2003;376:801–805. doi: 10.1042/BJ20030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chevalier J., Bredin J., Mahamoud A., Malléa M., Barbe J., Pagès J-M. Inhibitors of antibiotic efflux in resistant Enterobacter aerogenes and Klebsiella pneumoniae strains. Antimicrob. Agents Chemother. 2004;48(3):1043–1046. doi: 10.1128/AAC.48.3.1043-1046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pradel E., Pages J.M. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 2002;46(8):2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sabatini S., Gosetto F., Manfroni G., Tabarrini O., Kaatz G.W., Patel D., Cecchetti V. Evolution from a Natural Flavones Nucleus to Obtain 2-(4-Propoxyphenyl)quinoline Derivatives As Potent Inhibitors of the S. aureus NorA Efflux Pump. J. Med. Chem. 2011;54(16):5722–5736. doi: 10.1021/jm200370y. [DOI] [PubMed] [Google Scholar]
- 91.Bohnert J.A., Kern W.V. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005;49(2):849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pannek S., Higgins P.G., Steinke P., Jonas D., Akova M., Bohnert J.A., Seifert H., Kern W.V. Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J. Antimicrob. Chemother. 2006;57(5):970–974. doi: 10.1093/jac/dkl081. [DOI] [PubMed] [Google Scholar]
- 93.Kern W.V., Steinke P., Schumacher A., Schuster S., von Baum H., Bohnert J.A. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 2006;57(2):339–343. doi: 10.1093/jac/dki445. [DOI] [PubMed] [Google Scholar]
- 94.Schumacher A., Steinke P., Bohnert J.A., Akova M., Jonas D., Kern W.V. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 2006;57(2):344–348. doi: 10.1093/jac/dki446. [DOI] [PubMed] [Google Scholar]
- 95.Bina X.R., Philippart J.A., Bina J.E. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J. Antimicrob. Chemother. 2009;63(1):103–108. doi: 10.1093/jac/dkn466. [DOI] [PubMed] [Google Scholar]
- 96.Hannula M., Hänninen M-L. Effect of putative efflux pump inhibitors and inducers on the antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 2008;57(7):851–855. doi: 10.1099/jmm.0.47823-0. [DOI] [PubMed] [Google Scholar]
- 97.Eicher T., Cha H.J., Seeger M.A., Brandstatter L., El-Delik J., Bohnert J.A., Kern W.V., Verrey F., Grutter M.G., Diederichs K., Pos K.M. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA. 2012;109(15):5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoshida K., Nakayama K., Ohtsuka M., Kuru N., Yokomizo Y., Sakamoto A., Takemura M., Hoshino K., Kanda H., Nitanai H., Namba K., Yoshida K., Imamura Y., Zhang J.Z., Lee V.J., Watkins W.J. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg. Med. Chem. 2007;15(22):7087–7097. doi: 10.1016/j.bmc.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 99.Vargiu A.V., Ruggerone P., Opperman T.J., Nguyen S.T., Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014;58(10):6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 101.Nakayama K., Ishida Y., Ohtsuka M., Kawato H., Yoshida K-i., Yokomizo Y., Hosono S., Ohta T., Hoshino K., Ishida H. MexAB-OprM-specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 1: discovery and early strategies for lead optimization. Bioorg. Med. Chem. Lett. 2003;13(23):4201–4204. doi: 10.1016/j.bmcl.2003.07.024. [DOI] [PubMed] [Google Scholar]
- 102.Nakayama K., Ishida Y., Ohtsuka M., Kawato H., Yoshida K-i., Yokomizo Y., Ohta T., Hoshino K., Otani T., Kurosaka Y. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 2: achieving activity in vivo through the use of alternative scaffolds. Bioorg. Med. Chem. Lett. 2003;13(23):4205–4208. doi: 10.1016/j.bmcl.2003.07.027. [DOI] [PubMed] [Google Scholar]
- 103.Nakayama K., Kuru N., Ohtsuka M., Yokomizo Y., Sakamoto A., Kawato H., Yoshida K-i., Ohta T., Hoshino K., Akimoto K. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 4: Addressing the problem of poor stability due to photoisomerization of an acrylic acid moiety. Bioorg. Med. Chem. Lett. 2004;14(10):2493–2497. doi: 10.1016/j.bmcl.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 104.Yoshida K-i., Nakayama K., Yokomizo Y., Ohtsuka M., Takemura M., Hoshino K., Kanda H., Namba K., Nitanai H., Zhang J.Z. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 6: exploration of aromatic substituents. Bioorg. Med. Chem. 2006;14(24):8506–8518. doi: 10.1016/j.bmc.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 105.Opperman T.J., Kwasny S.M., Kim H-S., Nguyen S.T., Houseweart C., D'Souza S., Walker G.C., Peet N.P., Nikaido H., Bowlin T.L. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 2014;58(2):722–733. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vargiu A.V., Ruggerone P., Opperman T.J., Nguyen S.T., Nikaido H. Inhibition of E. coli AcrB multidrug efflux pump by MBX2319: molecular mechanism and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014 doi: 10.1128/AAC.03283-14. [AAC. 03283-03214]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Opperman T.J., Nguyen S.T. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015;6:421. doi: 10.3389/fmicb.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mandal S.M., Roy A., Ghosh A.K., Hazra T.K., Basak A., Franco O.L. Challenges and future prospects of antibiotic therapy: from peptides to phages utilization. Front. Pharmacol. 2014;5:105. doi: 10.3389/fphar.2014.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nikaido H., Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta. 2009;1794(5):769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pages J.M., Amaral L., Fanning S. An original deal for new molecule: reversal of efflux pump activity, a rational strategy to combat gram-negative resistant bacteria. Curr. Med. Chem. 2011;18(19):2969–2980. doi: 10.2174/092986711796150469. [DOI] [PubMed] [Google Scholar]
- 111.Tanaka Y., Hipolito C.J., Maturana A.D., Ito K., Kuroda T., Higuchi T., Katoh T., Kato H.E., Hattori M., Kumazaki K., Tsukazaki T., Ishitani R., Suga H., Nureki O. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature. 2013;496(7444):247–251. doi: 10.1038/nature12014. [DOI] [PubMed] [Google Scholar]
- 112.Lindahl E., Sansom M.S. Membrane proteins: molecular dynamics simulations. Curr. Opin. Struct. Biol. 2008;18(4):425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 113.Nakashima R., Sakurai K., Yamasaki S., Nishino K., Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480(7378):565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 114.Husain F., Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol. Microbiol. 2010;78(2):320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van Bambeke F., Pages J.M., Lee V.J. Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent Pat. Antiinfect. Drug Discov. 2006;1(2):157–175. doi: 10.2174/157489106777452692. [DOI] [PubMed] [Google Scholar]
- 116.Nakayama K., Ishida Y., Ohtsuka M., Kawato H., Yoshida K., Yokomizo Y., Hosono S., Ohta T., Hoshino K., Ishida H., Yoshida K., Renau T.E., Leger R., Zhang J.Z., Lee V.J., Watkins W.J. MexAB-OprM-specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 1: discovery and early strategies for lead optimization. Bioorg. Med. Chem. Lett. 2003;13(23):4201–4204. doi: 10.1016/j.bmcl.2003.07.024. [DOI] [PubMed] [Google Scholar]
- 117.Matsumoto Y., Hayama K., Sakakihara S., Nishino K., Noji H., Iino R., Yamaguchi A. Evaluation of multidrug efflux pump inhibitors by a new method using microfluidic channels. PLoS One. 2011;6(4):e18547. doi: 10.1371/journal.pone.0018547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vargiu A.V., Collu F., Schulz R., Pos K.M., Zacharias M., Kleinekathofer U., Ruggerone P. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J. Am. Chem. Soc. 2011;133(28):10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 119.Collu F., Vargiu A.V., Dreier J., Cascella M., Ruggerone P. Recognition of imipenem and meropenem by the RND-transporter MexB studied by computer simulations. J. Am. Chem. Soc. 2012;134(46):19146–19158. doi: 10.1021/ja307803m. [DOI] [PubMed] [Google Scholar]
- 120.Vargiu A.V., Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2012;109(50):20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Feng Z., Hou T., Li Y. Unidirectional peristaltic movement in multisite drug binding pockets of AcrB from molecular dynamics simulations. Mol. BioSys. 2012;8(10):2699–2709. doi: 10.1039/c2mb25184a. [DOI] [PubMed] [Google Scholar]
- 122.Chopra I., Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [second page, table of contents.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li X.Z., Zhang L., Poole K. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;45(4):433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 124.Iino R., Nishino K., Noji H., Yamaguchi A., Matsumoto Y. A microfluidic device for simple and rapid evaluation of multidrug efflux pump inhibitors. Front. Microbiol. 2012;3:40. doi: 10.3389/fmicb.2012.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dupont M., De E., Chollet R., Chevalier J., Pages J.M. Enterobacter aerogenes OmpX, a cation-selective channel mar- and osmo-regulated. FEBS Lett. 2004;569(1-3):27–30. doi: 10.1016/j.febslet.2004.05.047. [DOI] [PubMed] [Google Scholar]
- 126.Molnár J., Engi H., Hohmann J., Molnár P., Deli J., Wesolowska O., Michalak K., Wang Q. Reversal of multidrug resistance by natural substances from plants. Curr. Top. Med. Chem. 2010;10(17):1757–1768. doi: 10.2174/156802610792928103. [DOI] [PubMed] [Google Scholar]
- 127.Banoee M., Seif S., Nazari Z.E., Jafari-Fesharaki P., Shahverdi H.R., Moballegh A., Moghaddam K.M., Shahverdi A.R. ZnO nanoparticles enhanced antibacterial activity of ciprofloxacin against Staphylococcus aureus and Escherichia coli. J. Biomed. Mater. Res. B Appl. Biomater. 2010;93(2):557–561. doi: 10.1002/jbm.b.31615. [DOI] [PubMed] [Google Scholar]