Summary
Characterization of non-neoplastic and malignant human stem cell populations in their native state can provide new insights into gliomagenesis. Here we developed a purification strategy to directly isolate EGFR+/− populations from human germinal matrix (GM) and adult subventricular zone autopsy tissues, and from de novo glioblastoma (GBM) resections, enriching for cells capable of binding EGF ligand (LBEGFR+), and uniquely compared their functional and molecular properties. LBEGFR+ populations in both GM and GBM encompassed all sphere-forming cells and displayed proliferative stem cell properties in vitro. In xenografts, LBEGFR+ GBM cells showed robust tumor initiation and progression to high-grade, infiltrative gliomas. Whole-transcriptome sequencing analysis confirmed enrichment of proliferative pathways in both developing and neoplastic freshly isolated EGFR+ populations, and identified both unique and shared sets of genes. The ability to prospectively isolate stem cell populations using native ligand-binding capacity opens new doors onto understanding both normal human development and tumor cell biology.
Keywords: neural stem cells, glioma stem cells, germinal matrix, glioblastoma, tumor initiation, neurosphere, FACS, RNA-seq, transcriptome, EGFR
Graphical Abstract
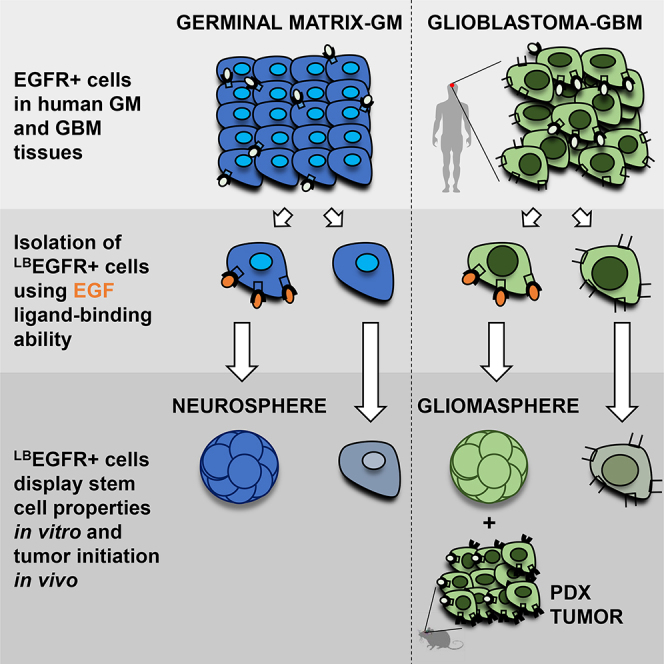
Highlights
-
•
Prospective purification of EGFR+ cells from fresh human neural tissues using EGF
-
•
In vitro stem cell properties are restricted to EGFR+ germinal matrix and GBM cells
-
•
EGFR+ GBM cells are tumor-initiating and re-form high-grade gliomas in mice
-
•
RNA-seq defines developmentally shared pathways in EGFR+ GBM cells
Understanding human brain development and gliomagenesis requires deeper analysis of stem cells in their native state. Here, Tsankova and colleagues employ EGF ligand to prospectively isolate proliferative EGFR+ stem-like populations from fresh germinal matrix and glioblastoma tissues, capturing sphere-forming and tumor-initiating cells in uncultured state. Comparative transcriptome analysis of these populations provides molecular insight into the dysregulated developmental pathways in GBM.
Introduction
Glioblastoma (GBM) is the most common and rapidly fatal adult brain tumor. Several developmental pathways important for the growth and proliferation of normal neural progenitors have been shown to be aberrantly reactivated in GBM and glioma stem cells (Canoll and Goldman, 2008, Chen et al., 2012, Lathia et al., 2015, Sanai et al., 2005), through complex genetic and emerging epigenetic alterations (Flavahan et al., 2016, Mack et al., 2015). Among these is the epidermal growth factor receptor (EGFR) pathway, with activating EGFR genomic alterations defining the most common “classical” GBM molecular signature (Brennan et al., 2013, Verhaak et al., 2010) and chromatin remodeling at its promoter driving EGFR overexpression (Erfani et al., 2015). EGFR is also highly expressed in the human developing germinal matrix (GM), as well as focally in the infant and adult subventricular zone (SVZ) (Erfani et al., 2015, Sanai et al., 2011, Weickert et al., 2000), but the stem cell properties and molecular characteristics of human EGFR-positive (EGFR+) neural cells have not been well characterized nor compared with their EGFR+ GBM counterparts, especially in populations derived from fresh human tissues. Here we prospectively isolated EGFR+ cells from fresh GM, SVZ, and GBM human tissues, based on their ability to bind the cognate EGF ligand, which allowed us to directly compare their acute-state functional properties and whole-transcriptome signatures. We demonstrate that developing EGFR+ GM, but not adult EGFR+ SVZ, populations display proliferative stem cell properties in vitro. EGFR+ GBM cells with ligand-binding capacity (LBEGFR+) recapitulate this developmental phenotype functionally in vitro, show capacity for tumor initiation in vivo, and share transcriptomes related to cell growth and cell-cycle regulation.
Results
EGFR+ Cells Isolated from Human GM Display Stem Cell Properties In Vitro
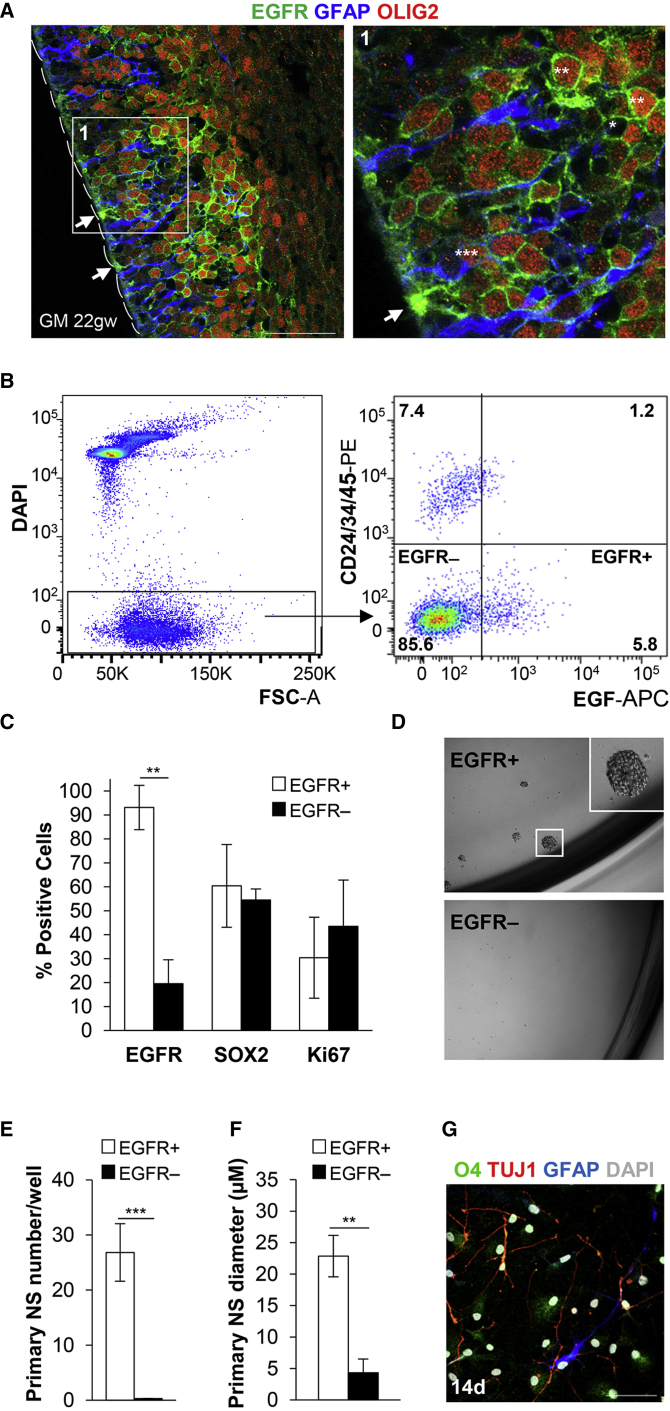
To better define the functional properties of EGFR-expressing cells during human brain development, we first characterized their immunophenotype in vivo in GM and SVZ human postmortem tissues. At 16–22 gestational weeks (gw), many but not all cells within the GM expressed EGFR (Figures 1A, S1, and S2). EGFR+ cells near the ventricular surface displayed radial morphology, and sometimes co-stained with glial fibrillary acidic protein (GFAP), while those in the deeper GM layers frequently co-expressed OLIG2 (Figures 1A and S2A). Both EGFR+ and EGFR− cells expressed Ki67, as well as the stem cell markers SOX2 and Nestin (Figures S1A–S1F). To isolate human EGFR+ and EGFR− populations from unfixed GM and SVZ dissections, we adapted a mouse fluorescence-activated cell sorting (FACS) strategy, which selects for EGFR+ cells based on their native binding to EGF ligand, while simultaneously excluding ependymal cells, endothelium, and inflammatory cells (Figures 1B, S2D, S2H, and S2I; Ciccolini et al., 2005, Codega et al., 2014, Pastrana et al., 2009). Acute immunostaining of the sorted populations from GM tissues demonstrated EGFR expression in more than 93% of cells within the EGFR+ fraction and a similar co-expression pattern of SOX2 and Ki67 as was observed in vivo (Figures 1C and S2G).
Figure 1.
Human EGFR+ GM Cells Isolated by FACS Display Stem Cell Properties In Vitro
(A) Immunofluorescence in human GM tissue shows many EGFR+OLIG2+ (∗∗), scattered EGFR+GFAP+OLIG2+ (∗∗∗), and exclusively EGFR+ (∗) cells, some of which show radial morphology (arrows) next to the developing ependyma (dashes) (see also Figures S1A–S1F and S2A).
(B) Representative FACS isolation of EGFR+/EGFR− cells using EGF-APC for positive selection, and CD24/CD34/CD45-PE and DAPI for exclusion (GM, 21gw).
(C) Acute immunofluorescence of sorted GM EGFR+/− cells (2 hr after FACS) shows predominant distribution of EGFR in the positive fraction (93%) (∗∗p = 0.002), and comparable expression of SOX2 and Ki67 in both fractions (n = 3 independent experiments) (see also Figure S2G).
(D) Representative primary NS growth at 6 days.
(E and F) Quantification of primary NS growth (n = 12 independent experiments; ∗∗∗p = 2.9 × 10−5) and (F) NS size (n = 5 independent experiments; ∗∗p = 0.01) at 6 days (EGF + FGF).
(G) Under differentiating conditions, EGFR+-derived cells show tri-lineage differentiation toward astrocytic (GFAP+), oligodendroglial (O4+), and neuronal (TUJ1+) fates (representative example of three independent samples).
Scale bars, 50 μm. Magnification of NS images, 10×. Bar graphs show mean ± SEM.
We then functionally characterized the in vitro stem cell properties of the freshly isolated EGFR+ and EGFR− populations, by examining their ability to form proliferative and self-renewing neurospheres (NS) and their potency for tri-lineage differentiation. Under standard NS medium conditions with EGF + fibroblast growth factor (FGF) ligand supplementation, EGFR+ cells (EGFR+DAPI−CD24−CD34−CD45−) isolated from prenatal GM showed NS formation by 6 days (Figures 1D–1F) and could be passaged serially (Figures S2D and S2F). To assess whether our ligand-binding isolation strategy selects for proliferating cells dependent on EGF for growth, we also cultured EGFR+ cells in the absence of exogenous EGF, supplementing the medium with FGF only or without any ligand. FACS-isolated EGFR+ cells showed similar number of primary NS with EGF + FGF, FGF only, and in the absence of any ligand, and formed self-renewing NS in the absence of EGF (Figure S2F). EGFR expression was maintained during NS passaging, including in clonal NS derived from single-cell seeding (1 cell/well) (Figure S1G). Under differentiation conditions, only EGFR+ GM-derived NS showed potential for tri-lineage differentiation (Figures 1G and S2D). In contrast, EGFR− cells (EGFR−DAPI−CD24−CD34−CD45−) showed little or no NS proliferation, and lacked the capacity for self-renewal in serial passaging (Figures 1D–1F, S2D, and S2E). We also studied the properties of EGFR+ cells in the infant and adult human SVZ. Immunofluorescence revealed focal and weaker EGFR expression in the postnatal SVZ in vivo, with drastic decline in Ki67 cell-cycle labeling (Figures S1H, S2B, and S2C), consistent with previous reports (Erfani et al., 2015, Sanai et al., 2011). Accordingly, EGFR+ cells isolated from infant SVZ gave rise to primary NS only (Figure S2H), and adult SVZ EGFR+ cells did not show any growth (n = 10) (Figure S2I). Therefore, EGFR+ populations derived acutely from human GM encompassed the cells with in vitro stem cell properties.
EGFR+ GBM Populations with Ligand-Binding Capacity Display Stem Cell Properties In Vitro
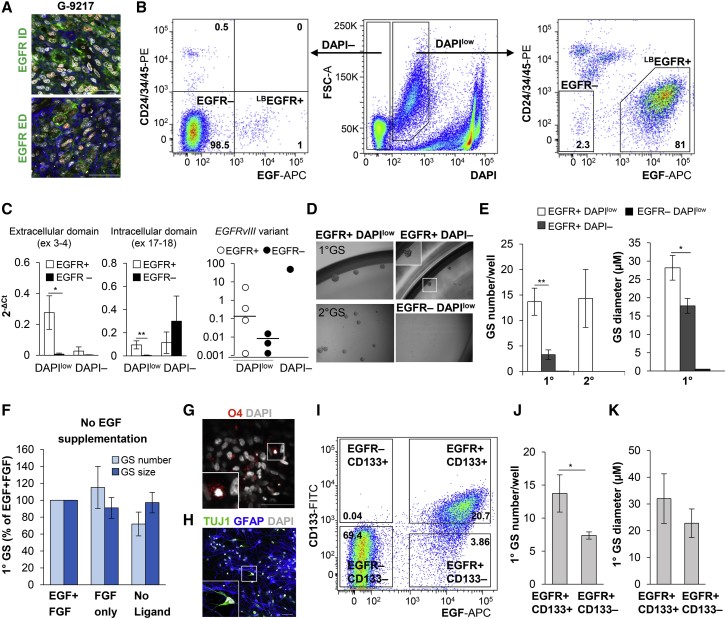
To assess whether EGFR+ cells in freshly dissociated GBM samples also exhibit stem cell properties in vitro, we employed the same isolation methodology and culture conditions used for normal neural tissues. All GBM samples were de novo resections, with different genomic alterations, including EGFR amplification and/or chromosome 7 polysomy, EGFRvIII mutation, and isocitrate dehydrogenase 1 (IDH1) mutation (Table S1). In vivo analysis of EGFR expression in these GBM samples revealed widespread expression of its intracellular domain (EGFR-ID) and more variable expression of its extracellular domain (EGFR-ED), consistent with EGFR-ED's focal loss in EGFRvIII and/or other truncating mutations (Figure 2A; Brennan et al., 2013). Expression of SOX2 and Nestin in GBM resembled the immunophenotype seen during GM development (Figures S1I–S1K).
Figure 2.
FACS-Isolated GBM LBEGFR+ Populations Display Stem Cell Properties In Vitro and Encompass All Sphere-Forming Cells
(A) Representative GBM specimen showing widespread expression of intracellular domain (ID) and focal expression of extracellular domain (ED) of EGFR, with co-localization for GFAP (blue) and OLIG2 (red).
(B) Cell sorting based on EGF binding allows the purification of EGFR+ cells with ligand-binding capacity (LBEGFR+) from fresh GBM samples, while excluding non-neoplastic endothelium and inflammatory cells (CD24+/CD34+/CD45+-PE) and dead cells (high DAPI+, 104–105). A significant fraction of LBEGFR+ live cells shows a distinct forward scatter shift (FSC-A) and dim DAPI fluorescence (DAPIlow) (see also Figure S3B).
(C) qRT-PCR confirms selectivity for EGFR with intact ligand-binding ED transcript (exons 3–4) in the positive fraction (LBEGFR+) (∗p = 0.049; n = 3 independent experiments). EGFR-ID transcript (exons 17–18) and mutant EGFRvIII transcript (exons 1–8 junction) are variably present in both fractions (n = 3 independent experiments; ∗∗p = 0.009).
(D) GS formation is seen exclusively in LBEGFR+ populations (day 12).
(E) GS derived from LBEGFR+DAPIlow cells are more numerous than LBEGFR+DAPI− cells (∗∗p = 0.003) and larger in size (∗p = 0.05) (n = 16 independent experiments). Only LBEGFR+DAPIlow cells form secondary (2°) GS (day 12) (n = 10 independent experiments).
(F) LBEGFR+DAPIlow cells form similar number and size of 1° GS when cultured with EGF + FGF, FGF only, or without any ligand (n = 3 independent experiments) (see also Figures S2J and S2K).
(G and H) EGFR+ GS show the ability for multipotent lineage differentiation (representative example from three independent tumors).
(I–K) Combined FACS for CD133 and EGFR shows both CD133+ and CD133− GS-forming populations to be EGFR+ (I), with greater number of GS derived from LBEGFR+CD133+ cells (∗p = 0.03) (n = 3 independent experiments) (J), of equal size to LBEGFR+CD133− cells (K) (n = 3 independent experiments; 12 days) (see also Figure S3C).
Scale bars, 50 μm. Magnification of GS images, 10×. Bar graphs show mean ± SEM.
In contrast to GM, GBM FACS revealed two populations of EGFR+ cells. The main population (defined here as EGFR+DAPIlow) showed a unique shift in DAPI fluorescence and greater forward scatter (Figures 2B [right] and S3B). The minor EGFR+ population was DAPI− (Figure 2B, left), similar to that in GM. qRT-PCR analysis revealed that ligand-binding EGFR-ED transcript is significantly overexpressed in EGFR+DAPIlow populations only, while EGFR-ID and EGFRvIII were variable (Figure 2C). This suggested that our technique is selective for cells with preserved EGF ligand-binding ability and EGFR-ED expression (annotated hereafter as LBEGFR+), regardless of their EGFRvIII status.
We next assessed the ability of each different GBM FACS-purified population to grow and form self-renewing gliomaspheres (GS) in vitro. Similar to GM, GBM GS only arose from LBEGFR+ populations (Figure 2D), with LBEGFR+DAPIlow cells forming more and larger primary GS than LBEGFR+DAPI− cells at 12 days (Figure 2E, n = 16), and were the only ones to show self-renewal in serial passaging (Figures 2D and 2E). In contrast, EGFR−DAPI− cells did not divide (Figure S3B) and EGFR−DAPIlow cells divided initially in one sample only, but did not form GS even after 4 weeks of growth (Figures 2D and 2E). Importantly, populations from our exclusion channels, CD24-PE, CD34-PE, and CD45-PE, cultured under identical conditions, attached to the plate but did not proliferate into GS (Figure S3D). As with GM, sphere formation in LBEGFR+DAPIlow glioma cells was not dependent on EGF being present in the culture medium. LBEGFR+DAPIlow GBM populations formed similar numbers of primary and self-renewing GS under EGF + FGF, FGF only, and no ligand conditions (Figures 2F, S2J, and S2K). They also displayed capacity for tri-lineage differentiation (Figures 2G and 2H). Extreme limiting dilution analysis (ELDA) revealed comparable stem cell frequencies in GM and GBM LBEGFR+ populations, regardless of whether EGF was added to the culture medium (Table S2). Expression of EGFR was maintained in clonogenic spheres (1 cell/well) (Figure S1N), and was upregulated in GS formed without exogenous EGF in the culture medium (Figure S1O).
Of note, two GBM samples in our study did not show any expression of EGFR, and thus also lacked a defined LBEGFR+ population. In one of them, G-10416, GS formations were present in the EGFR− tumor population (Figure S2L) while in the other, G-11702, GS were not formed (data not shown). Overall, the data indicate that for GBM tumors that express EGFR, with or without EGFR amplification/vIII mutation, EGF-based isolation enriches for LBEGFR+ populations with in vitro stem cell properties.
Sphere-Forming Cells Are Fully Captured within LBEGFR+ GBM Populations
To further validate our technique, we compared EGF-based FACS against several established glioma stem cell isolation markers, including CD133 (Prominin-1), CD171 (L1CAM), CD44, and CD140a (PDGFRA) (Anido et al., 2010, Bao et al., 2008, Flavahan et al., 2016, Singh et al., 2004). Simultaneous sorting with CD133 and EGF revealed three main populations: LBEGFR+CD133+, LBEGFR+CD133−, and EGFR−CD133− (Figures 2I, S3A, and S3B). Upon culturing, GS formation was seen in LBEGFR+CD133+ and LBEGFR+CD133− populations, the former being more numerous, consistent with previous reports (Figures 2J and 2K; Beier et al., 2007). No GS were present in EGFR−CD133− cells (Figure S3B). Again, GS grew in similar numbers with and without the addition of exogenous EGF to the medium (Figure S3C). Similarly to CD133, LBEGFR+ GBM cells were present within both positive and negative CD171, CD140a, and CD44 populations, encompassing all sphere-forming populations (Figures S3E–S3G). These results underscore the utility of our ligand-based methodology for the selective yet inclusive isolation of glioma cells with in vitro proliferative stem cell properties from fresh EGFR-expressing GBM tumors, capturing all sphere-forming populations.
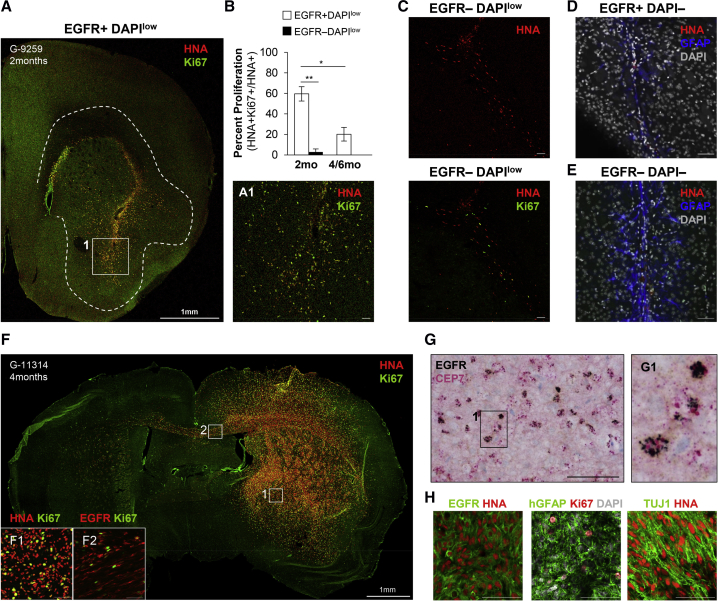
Transplanted LBEGFR+DAPIlow GBM Cells Display Robust Tumorigenic Abilities In Vivo
To test the tumorigenic capacity of purified LBEGFR+ GBM cells in vivo, we injected acutely sorted LBEGFR+DAPIlow, EGFR−DAPIlow, LBEGFR+DAPI−, and EGFR−DAPI− cells, without any prior culture, intracranially into 2-month-old immunocompromised mice, and assessed their ability for tumor initiation at an early, subclinical time point of 60 days. Only LBEGFR+DAPIlow populations showed capacity for tumor initiation, displaying robust proliferation (60% HNA+Ki67+ co-localization) (Figures 3A and 3B) and migration along striatal white matter fibers (Figure 3A). In contrast, mice transplanted with EGFR−DAPIlow, EGFR−DAPI−, or LBEGFR+DAPI− cells showed minimal proliferation in one case only of EGFR−DAPIlow transplantation (Figure 3C) or no engraftment (Figures 3D and 3E). By 4–6 months, mice injected with LBEGFR+DAPIlow cells exhibited large and diffusely infiltrative high-grade gliomas (Figure 3F), retaining the EGFR amplification status of their original resection (Figure 3G). The phenotype in the well-formed tumors at 4–6 months was less proliferative than at 2 months (Figure 3B), with expression of differentiating markers such as GFAP and TUJ1 (TUBB3) (Figure 3H), recapitulating human GBM heterogeneity (Figures S1L and S1M). Overall, these results demonstrate that LBEGFR+DAPIlow GBM cells not only define sphere-forming populations in vitro but are also capable of tumor initiation and diffuse migration in vivo, underscoring the utility of EGFR-binding capacity as a powerful approach to isolate GBM populations with tumorigenic properties.
Figure 3.
Transplanted LBEGFR+ GBM Cells Are Robustly Tumor-Initiating In Vivo
(A) Intracranial xenotransplantation yields robust tumor growth and infiltration at 60 days in LBEGFR+DAPIlow populations only (1 × 105).
(B) Quantification of proliferation (Ki67+) in human nuclear antigen (HNA+) glioma cells from LBEGFR+DAPIlow and EGFR−DAPIlow transplantations (∗∗p = 0.001 at 2 months, ∗p = 0.014 at 4–6 months) (n = 3 independent experiments).
(C) EGFR−DAPIlow (1 × 105) glioma cells show no or minimal proliferation close to the needle track at 60 days (n = 3 tumors; example shown is the only one with minimal proliferation).
(D and E) LBEGFR+DAPI− (0.5 × 105) and EGFR−DAPI− (1 × 105) populations show only debris around the needle track surrounded by a GFAP+ astroglial scar at 60 days (representative examples of three independent tumors).
(F–H) Four to six months after injection, LBEGFR+DAPIlow (1 × 105) transplants display large and diffusely infiltrative high-grade gliomas, (G) retain EGFR amplification status by chromogenic in situ hybridization, and (H) express EGFR as well as differentiating markers GFAP and TUJ1 (representative examples, n = 3 independent tumors for F–H).
Scale bars, 50 μm. Bar graphs show mean ± SEM.
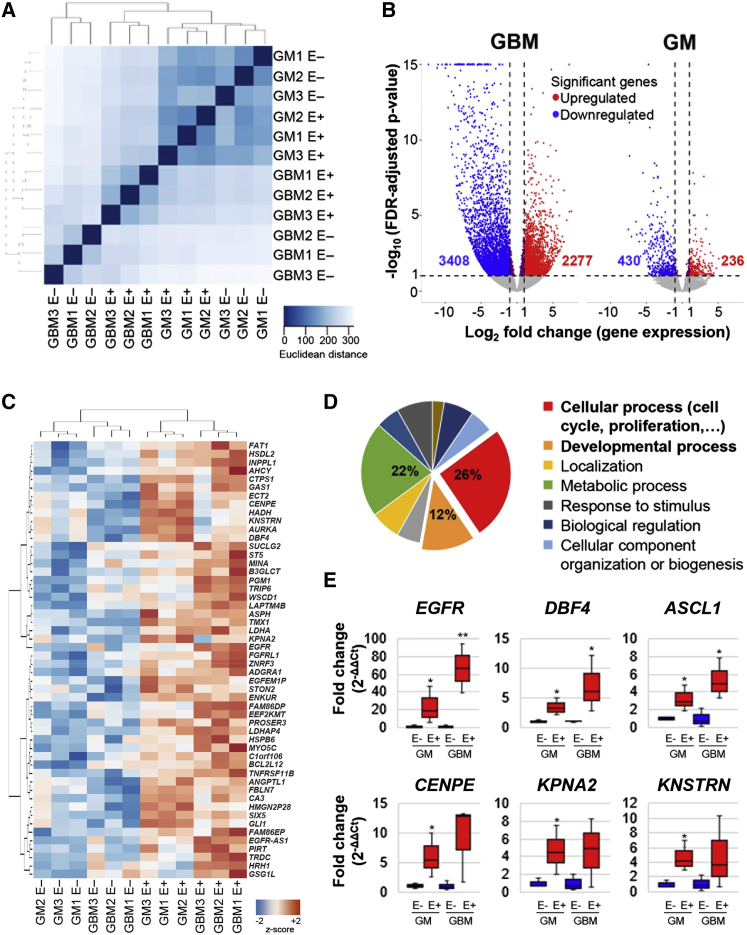
Comparative Transcriptome Profiling of LBEGFR+ GM and GBM Populations
To define the transcriptional signature in our FACS-purified human germinal matrix and glioma-derived EGFR+ populations, we performed deep transcriptome sequencing analysis via RNA sequencing (RNA-seq) on acutely isolated LBEGFR+ and EGFR− GM and GBM (DAPIlow) populations. Developing GM EGFR+ and EGFR− transcriptomes appeared most similar to one another by hierarchical clustering and principal component analysis, and interestingly, GBM LBEGFR+ transcriptomes were more similar to GMs than to their neoplastic EGFR− counterpart (Figures 4A and S4A). Differential expression analysis revealed well-defined large subsets of upregulated and downregulated genes between GBM LBEGFR+ and EGFR− populations, and smaller ones between GM EGFR+ and EGFR− populations (Figure 4B). Differential gene set analysis revealed strong enrichment in pathways related to cell proliferation and mitotic cell cycle in both GM and GBM LBEGFR+ populations (Figures S4B–S4D and Table S3). Further analysis defined subset of 50 developmentally regulated genes to be uniquely shared between human GBM and GM EGFR+ populations, including several implicated in cell-cycle regulation: CENPE, DBF4, KPNA2, and KNSTRN (Figures 4C–4E and Table S3).
Figure 4.
Differential Transcriptome Analysis of EGFR+ GM and GBM Populations Defines a Shared Gene Set
(A) Unsupervised hierarchical clustering of whole-transcriptome RNA-seq data, using Euclidean distance as the metric, of acutely isolated EGFR+/EGFR− populations from three GM (19–22 gw) and three GBM independent samples, shows higher resemblance of LBEGFR+ GBMs with GMs than with EGFR− GBMs (see also Figure S4A).
(B) Differential gene expression analysis in LBEGFR+ versus EGFR− populations, performed independently for all three GBM and GM samples, defines a subset of significantly upregulated and downregulated genes in both datasets.
(C–E) Analysis of the intersection between the two upregulated gene datasets (2,277 and 236, red) in EGFR+ GM and GBM. (C) Heatmap of row-normalized expression values for upregulated shared genes (false discovery rate adjusted p < 0.1, z = [x − mean]/SD) (see also Table S3). (D) Pie chart depicts distribution of top biological processes in the shared gene set, analyzed by PANTHER database. (E) Validation of gene expression by qRT-PCR in upregulated EGFR+ GM/GBM genes related to cell proliferation (n = 3 independent experiments). Plots show the median (black horizontal line), 25th and 75th percentiles (boxes), and range (whiskers) of the data. One-tailed unpaired Students’ t test and Mann-Whitney U test for non-parametric test were used to calculate statistical significance (EGFR GM, ∗p = 0.024, GBM, ∗∗p = 0.007; DBF4 GM, ∗p = 0.023, GBM, ∗p = 0.046; ASCL1 GM, ∗p = 0.03, GBM, ∗p = 0.02; CENPE GM, ∗p = 0.039; KPNA2 GM, ∗p = 0.04; KNSTRN GM, ∗p = 0.02).
Together, our data provide functional and molecular evidence for stem cell properties in ligand-binding EGFR+ cells during both neural development and in GBM.
Discussion
Comparative analyses of non-neoplastic and neoplastic human neural populations, isolated ex vivo from fresh pathology samples, are technically challenging but can yield important insight into both normal biology and gliomagenesis. Here we provide a useful ligand-based methodology to purify EGFR+ GM progenitors and GBM cells from fresh human tissues, and elaborate on their phenotypic, functional, and molecular comparative properties. Importantly, we discover that EGFR+ GM and GBM populations encompass all sphere-forming cells in vitro, displaying self-renewal and multi-lineage differentiation stem cell properties, and that freshly isolated LBEGFR+ GBM cells form high-grade and diffusely infiltrating gliomas by 4 months after orthotopic transplantation. We confirm that this ligand-based isolation strategy does not simply select for populations dependent on EGF ligand for growth, demonstrating the ability of LBEGFR+ populations for sphere formation in vitro and tumor initiation in vivo in the absence of exogenous EGF ligand supplementation.
Several important markers have been utilized to identify and characterize glioma stem cells (GCS) in fresh human GBM tissues and patient-derived neurosphere cultures, but many of them have not captured all sphere-forming cells in vitro (Lathia et al., 2015). By combining our EGF ligand-based isolation methodology with CD133, CD140a, CD171, or CD44, we show that EGF ligand binding captures all GS-forming colonies. This underscores the ability of our EGF ligand-based isolation methodology to recapitulate the in vitro GSC phenotypes previously characterized by others, as well as to capture additional sphere-forming populations with potential functional and molecular significance.
Although GBM tumors with diffuse EGFR amplification displayed the most pronounced LBEGFR+ population in FACS, we also captured all sphere-forming cells in EGFR-expressing GBM tumors without EGFR amplification (7 out of 19), one of which harbored IDH1 mutation. Thus, we believe this methodology will be useful for isolating stem cell populations in both EGFR-amplified “classical” and IDH1-mutant “proneural” GBM tumors, and perhaps others, as long as they express EGFR, but may not be applicable for occasional tumors that lack a defined LBEGFR+ population. Given the bias of neurosphere assays and xenograft tumor initiation models for proliferation, our methodology is particularly useful for isolating populations with active, but not necessarily quiescent, stem cell properties.
The similarities between the transcriptome of GM and GBM LBEGFR+ stem cell populations for markers of cell-cycle regulation suggest that the proliferative tumor phenotype of LBEGFR+ GBM populations may be at least partially driven by co-opted transcriptional programs important for cell division during germinal matrix development (Flavahan et al., 2016, Liu et al., 2015, Mack et al., 2015, Suva et al., 2014, Tsankova and Canoll, 2014). The interplay between genomic alterations and developmental remodeling of transcriptional networks in tumor cells is an intriguing question to be explored in future studies, and to this end we have here validated a useful and clinically relevant methodology for the prospective isolation of neural and GBM populations with proliferative stem cell properties.
Experimental Procedures
Detailed methods are provided in Supplemental Experimental Procedures.
Sample Collection
All specimen collection was performed de-identified in accordance with the policies and regulation at the Icahn School of Medicine at Mount Sinai (ISMMS) and its institutional review board (IRB#AAAJ9652-Y1M00, HS#14-01007).
Fluorescence-Activated Cell Sorting
Single-cell suspension was obtained from fresh tissue (dissected GM/SVZ autopsy or GBM surgical material) by mechanical and enzymatic papain dissociation, and incubated with EGF-Alexa Fluor 647(APC) complex for positive selection of EGFR cells (1 μg per 106 live cells), and with CD24/CD34/CD45 antibodies and DAPI for exclusions (Codega et al., 2014).
Cell Culture
Cells were cultured on 96-well low-adherence plates in freshly made NS medium (see recipe in Supplemental Experimental Procedures), with addition of EGF (20 ng/mL) and/or basic FGF (bFGF) (20 ng/mL), or with no ligand, at a density of 10 cells/μL or 1 cell/μL (for 1° or 2° NS/GS formation, respectively). For ELDA, acutely sorted cells were seeded at 1, 10, 50, and 100 cells/well. For differentiation, NS/GS were seeded on PDL/laminin-coated coverslips and grown in NS medium without B27, EGF, and bFGF for 14 days.
Orthotopic Transplantation
Mouse studies were performed in accordance with the Institutional Animal Care and Use Committee (IACUC), protocol number IACUC-2014-0183. Acutely FACS-sorted cells (0.5–1 × 105) from fresh GBM samples were injected into the striatum of 2-month-old SCID male mice (2 mm right lateral to bregma and 3 mm deep). Mice were euthanized for histological examination of tumor initiation at 60 days and of endpoint tumor formation when clinically symptomatic.
Gene Expression Analysis
Whole-transcriptome analysis was performed by RNA-seq on RNA extracted from freshly FACS-sorted EGFR+/− GBM and GM cells (125 bp pair-end sequencing, 38–50 million paired-end reads/sample, on Illumina HiSeq 2500). cDNA was generated using Clontech SMART-seq v4 (634888) and libraries using Nextera XT (FC-131-1024). The expression of selected genes was validated by qRT-PCR.
Immunofluorescence
Floating vibratome sections (40–60 μm), formalin-fixed paraffin-embedded (FFPE) sections (4 μm), NS/GS, and acutely fixed FACS-sorted cells were blocked in 10% normal donkey serum for 0.5–1 hr, incubated with primary antibody overnight at 4°C, incubated with secondary antibodies for 2–4 hr at room temperature, and visualized on a confocal Zeiss LSM710 microscope. Modified protocols were used for EGFR and O4 staining (see Supplemental Experimental Procedures).
Author Contributions
Conception of project: F.D. and N.M.T. Experimental design and data interpretation: J.T.-G. and N.M.T. Development of EGF FACS technique: V.S.-V., F.D., J.T.-G., and N.M.T. In vitro assays, library preparation, qPCR, and histology: J.T.-G. Orthotopic transplantations: R.T. and R.H.F. Sequencing and bioinformatics: G.N., E.Z., M.W., and R.S. Tissue procurement: H.W., R.L.Y., M.M., M.F., and N.M.T. Manuscript preparation: J.T-G., F.D., and N.M.T. Editing: all authors. R.T. and G.N. contributed equally.
Acknowledgments
We thank members of the Pathology Department and Biorepository Core (director M. Donovan) at ISMMS for facilitating tissue collection and procurement; the ISMMS Flow Cytometry CORE for expert advice and accommodation of fresh human tissue sorts 24hr/day; members of the F.D., J. Goldman, and J. Silva laboratories for assistance with reagents and equipment; P. Codega for advice on NS differentiation; H. Zou for scientific advice; and G. Petrov and M. Olabarria for graphical assistance. The research was supported by Mount Sinai seed grant funds (N.M.T., R.H.F.) and R01NS092735 (R.H.F.).
Published: April 20, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.03.019.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE96682.
Supplemental Information
References
- Anido J., Saez-Borderias A., Gonzalez-Junca A., Rodon L., Folch G., Carmona M.A., Prieto-Sanchez R.M., Barba I., Martinez-Saez E., Prudkin L. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Bao S., Wu Q., Li Z., Sathornsumetee S., Wang H., McLendon R.E., Hjelmeland A.B., Rich J.N. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D., Hau P., Proescholdt M., Lohmeier A., Wischhusen J., Oefner P.J., Aigner L., Brawanski A., Bogdahn U., Beier C.P. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoll P., Goldman J.E. The interface between glial progenitors and gliomas. Acta Neuropathol. 2008;116:465–477. doi: 10.1007/s00401-008-0432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., McKay R.M., Parada L.F. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccolini F., Mandl C., Holzl-Wenig G., Kehlenbach A., Hellwig A. Prospective isolation of late development multipotent precursors whose migration is promoted by EGFR. Dev. Biol. 2005;284:112–125. doi: 10.1016/j.ydbio.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A.R., Deleo A.M., Pastrana E., Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfani P., Tome-Garcia J., Canoll P., Doetsch F., Tsankova N.M. EGFR promoter exhibits dynamic histone modifications and binding of ASH2L and P300 in human germinal matrix and gliomas. Epigenetics. 2015;10:496–507. doi: 10.1080/15592294.2015.1042645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suva M.L., Bernstein B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Hon G.C., Villa G.R., Turner K.M., Ikegami S., Yang H., Ye Z., Li B., Kuan S., Lee A.Y. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol. Cell. 2015;60:307–318. doi: 10.1016/j.molcel.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack S.C., Hubert C.G., Miller T.E., Taylor M.D., Rich J.N. An epigenetic gateway to brain tumor cell identity. Nat. Neurosci. 2015;19:10–19. doi: 10.1038/nn.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E., Cheng L.C., Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc. Natl. Acad. Sci. USA. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N., Alvarez-Buylla A., Berger M.S. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- Sanai N., Nguyen T., Ihrie R.A., Mirzadeh Z., Tsai H.H., Wong M., Gupta N., Berger M.S., Huang E., Garcia-Verdugo J.M. Corridors of migrating neurons in the human brain and their decline during infancy. Nature. 2011;478:382–386. doi: 10.1038/nature10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Suva M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., Riggi N., Chi A.S., Cahill D.P., Nahed B.V. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N.M., Canoll P. Advances in genetic and epigenetic analyses of gliomas: a neuropathological perspective. J. Neurooncol. 2014;119:481–490. doi: 10.1007/s11060-014-1499-x. [DOI] [PubMed] [Google Scholar]
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert C.S., Webster M.J., Colvin S.M., Herman M.M., Hyde T.M., Weinberger D.R., Kleinman J.E. Localization of epidermal growth factor receptors and putative neuroblasts in human subependymal zone. J. Comp. Neurol. 2000;423:359–372. doi: 10.1002/1096-9861(20000731)423:3<359::aid-cne1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.