Abstract
The cytoskeleton framework is essential not only for cell structure and stability but also for dynamic processes such as cell migration, division and differentiation. The F-actin cytoskeleton is mechanically stabilised and regulated by various actin-binding proteins, one family of which are the filamins that cross-link F-actin into networks that greatly alter the elastic properties of the cytoskeleton. Filamins also interact with cell membrane-associated extracellular matrix receptors and intracellular signalling proteins providing a potential mechanism for cells to sense their external environment by linking these signalling systems. The stiffness of the external matrix to which cells are attached is an important environmental variable for cellular behaviour. In order for a cell to probe matrix stiffness, a mechanosensing mechanism functioning via alteration of protein structure and/or binding events in response to external tension is required. Current structural, mechanical, biochemical and human disease-associated evidence suggests filamins are good candidates for a role in mechanosensing.
Keywords: Filamin, Actin, Cytoskeleton, Cellular mechanosensing, Skeletal disease
Introduction
The integration of the cell’s structural framework, the cytoskeleton, with membrane remodelling and cell adhesion is crucial as cells move, change shape, divide and transduce signals in response to their external environment. These cellular properties depend on the dynamic regulation of the filamentous F-actin cytoskeleton by families of actin-binding proteins. In addition to biochemical signals, developmental and cellular processes involving the cytoskeleton are modulated in response to external force and the stiffness of the extracellular (EC) substrate (Alenghat and Ingber 2002). For example, the load-dependent growth of bone has been established for over 100 years (Wolff 1892), with the increased bone mass in professional tennis players’ racquet arms (vs the non-playing arm) (Jones et al. 1977) and the loss of bone density in astronauts exposed to reduced gravity (Carmeliet et al. 2001) being well-documented examples. In addition to the maintenance and growth of mature bone, the appropriate sensing of the EC substrate by embryonic mesenchymal stem cells (MSC) is crucial for cell differentiation during development (Discher et al. 2009). MSCs commit to a bone-forming osteoblast lineage upon interaction with a rigid substrate while intermediate or softer matrices direct MSCs to muscle or neuronal cell fates, respectively (Engler et al. 2006). A mechanosensing system allowing a cell to probe its external substrate involves balancing cell-generated cytoskeletal tensional forces (Fcytoskeletal; Fig. 1), generated from motor protein ATP hydrolysis and active filament polymerisation, against extracellular substrate stiffness (Fmatrix; Fig. 1). Additionally, an elastic internal cytoskeletal network must resist excessive applied force to prevent cell death but remain elastically deformable for signalling responses to force within a physiological range (10–100 pN) and yet still be readily altered to allow motility and cell membrane remodelling (Discher et al. 2005). Actin binding proteins and their functional partners are responsible for the alteration of F-actin properties that allows the cell to adopt varying states of viscoelasticity.
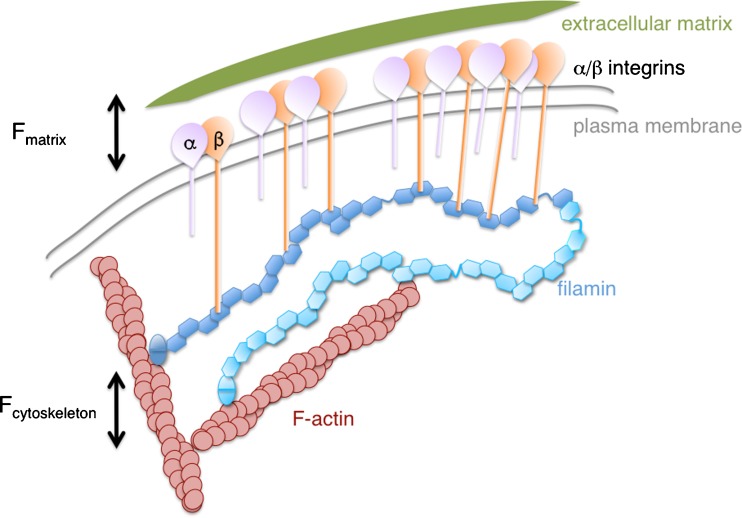
Fig. 1.
Schematic representation of filamin cross-linking F-actin and clustering integrin receptors at the plasma membrane (Ithychanda et al. 2009) contributing to the balancing of external (F cytoskeleton) and extracellular matrix tension (F matrix). Contacts between FLN repeat domains with β-integrins and F-actin are shown on only one monomer for clarity
Filamin domain organisation and structure
The filamins (FLN) are a family of actin binding proteins that can convert, via non-covalent cross-linking, a solution of semi-flexible F-actin filaments into a gel of increased elastic stiffness at a critical concentration of FLN (Wang and Singer 1977). The FLNs are found in animals, amoeba and at least one excavate, suggesting that FLNs arose early in evolutionary terms in ancestral eukaryotes that utilised an actin-based amoeboid motility mechanism (Fritz-Laylin et al. 2010). The human genome contains three FLN genes, filamin A (FLNA), FLNB and FLNC (Stossel et al. 2001). FLNA and FLNB are widely expressed (Xu et al. 1998), while FLNC expression is more restricted in cardiac and skeletal muscle (Maestrini et al. 1993). Human filamins are homodimeric with each ∼280-kDa monomer comprising an N-terminal actin binding domain (ABD) followed by a rod region of 24 immunoglobulin (Ig)-like repeat domains interrupted by two hinges located between repeats 15 and 16 (hinge 1), and between 23 and 24 (hinge 2). Dimerisation occurs in a tail-to-tail manner (Tyler et al. 1980) via the C-terminal repeat 24 (Fig. 1) (Himmel et al. 2003). Filamin structures exhibit a large radius of gyration in which the flexible rod domains can adopt a variety of structures while remaining dimeric.
The FLN ABD (30 kDa), composed of two calponin homology (CH) domains, is required for F-actin binding (Bresnick et al. 1990) and has a structure conserved with related F-actin binding proteins including dystrophin, α-actinin, β-spectrin, fimbrin and utrophin (Clark et al. 2009; Sawyer et al. 2009).
The filamin repeat domains (11 kDa) adopt an immunoglobulin domain β-sandwich fold comprising stacked three-stranded and four-stranded β-sheets (Fucini et al. 1997). The structures of FLNA repeat 21 and β7-integrin peptide (Kiema et al. 2006), and repeat 17 in complex with the GPIbα peptide (Nakamura et al. 2006), have defined the molecular interactions between FLNA repeat domains and these membrane receptors. The structure of FLNA repeats 19–21 strikingly revealed that the repeats do not necessarily fold linearly as distinct domains (Lad et al. 2007). Domain 21 is positioned between repeats 19 and 20 (Fig. 2), with repeat 20 partially unfolded contributing its N-terminal β-strand (strand A) to the β-integrin binding site on repeat 21 (Kiema et al. 2006). A mechanism proposed for mechanosensing, supported by molecular dynamics calculations, involves a force (Kesner et al. 2009; Pentikainen and Ylanne 2009) and/or phosphorylation-induced (Chen et al. 2009) conformational change displacing this domain-swapped β-strand uncovering this cryptic β-integrin binding site recruiting FLN to adhesion sites (Lad et al. 2007). Sequence and structural analysis has revealed that FLN repeats exist as domain pairs e.g. 16/17, 18/19 and 20/21 (Lad et al. 2007; Heikkinen et al. 2009) with multiple β-integrin interaction sites functionally identified (Ithychanda et al. 2009).
Fig. 2.
Structural cartoon representation of filamin A repeat domains 19–21 (Lad et al. 2007) showing the non-sequential nature of the repeat domains and the N-terminal strand A of repeat 20 contributing to the domain structure of repeat 21 covering the β-integrin binding site. The figure was prepared with PYMOL (DeLano 2002)
In addition to F-actin binding, FLN dimerisation via its C-terminal repeat domains is central to actin cross-linking ability (Davies et al. 1980; Himmel et al. 2003). The dimerisation domain is required to rescue a defective chemotaxis phenotype in dictyostelium FLN (ddFLN) null mutants (Khaire et al. 2007), and a truncating mutation within human FLNC repeat 24 is associated with myofibrillar myopathy (Vorgerd et al. 2005) owing to protein aggregation from reduced dimerisation and stability (Lowe et al. 2007). The issue of whether FLNs can heterodimerise is not fully resolved; FLNB/C repeat 24 heterodimers were observed by in vitro cross-linking assays, but the combinations of FLNA/B or FLNA/C were not (Himmel et al. 2003). In contrast, longer FLNA and FLNB constructs can heterodimerise by yeast-two-hybrid assay, immunoprecipitation and colocalisation experiments in neuronal precursor cells (Sheen et al. 2002).
Filamin cellular functions
Over 70 binding partners for FLN have been identified (reviewed in Sarkisian et al. 2008), with the most well characterised being the interaction with F-actin with the FLN ABD (Hartwig et al. 1980) and a second site within repeats 9–15 of the rod domain (Nakamura et al. 2007). F-actin polymerisation is a local phenomenon in cells prevalent at the cell membrane and around extracellular matrix contact focal adhesion sites (Pollard and Cooper 2009) where FLN is also localised (Langanger et al. 1984; Nikki et al. 2002). FLN binds F-actin with moderate affinity Kd 0.13–3.2 μM (Nomura et al. 1987; Ohta and Hartwig 1995), an interaction that can be regulated by inositol phospholipids (Furuhashi et al. 1992), Ca2+-calmodulin (Nakamura et al. 2005) and phosphorylation (Ohta and Hartwig 1995). Most actin binding proteins have affinities for F-actin in the μM range suggesting that having many interaction partners with moderate binding affinity conveys a high degree of dynamic elasticity upon the F-actin cytoskeleton (Goldmann et al. 1997).
The interaction of FLN C-terminal domains with membrane bound receptors provides a link between the F-actin cytoskeleton and focal adhesions important for a role in mechanotransducing substrate stiffness and/or externally applied force to biochemical signalling pathways. The cytoskeleton bears loads applied externally to cells, and tension generated intracellularly by motor proteins, via EC matrix attached integrins that bind FLN and other cytoskeletal proteins (Sharma et al. 1995; Loo et al. 1998) mediating extracellular signalling and adhesion with cytoskeletal organisation. FLNA is required for cell membrane structure and stability, protecting cells from force-induced cell death via an integrin-dependent mechanism (Glogauer et al. 1998) for which the FLN ABD is essential (Kainulainen et al. 2002). Additionally, FLNA targets FLNA-binding RhoGTPase-activating protein (FilGAP) to sites of force application resisting force-induced cell extension that can lead to cell death (Shifrin et al. 2009). Intracellular connectivity through integrins and adaptor proteins like FLN are implicated in mechanisms by which cells sense the rigidity of their anchoring matrix and respond by rearranging their actin cytoskeleton central to cellular mechanotaxis (Choquet et al. 1997). When external force is applied to integrins, forces are transmitted via cytoskeletal filaments leading to physical rearrangements at the nucleus (Maniotis et al. 1997) with upregulation of FLNA expression (D'Addario et al. 2001). FLN shows differential affinities for different β-integrin isoforms (Pfaff et al. 1998), and the location of multiple integrin binding sites along the rod domain suggests that FLN may be responsible for integrin clustering (Ithychanda et al. 2009) leading to the inhibition of cell migration and polarisation via reduced membrane remodelling (Calderwood et al. 2001).
FLNA integrates cytoskeletal organisation and signalling events through further protein–protein interactions mainly with repeat domains 15–24. FLN is associated with the GTPases RhoA, Rac, Ral and Cdc42 (Marti et al. 1997), and upstream/downstream effectors guanine nucleotide-exchange factors and activators, e.g. FilGAP (Ohta et al. 2006; Bellanger et al. 2000; Pi et al. 2002) involved in the dynamic regulation of the cytoskeleton. FLNA is a substrate for multiple kinases (reviewed in Sarkisian et al. 2008) depending on whether or not its actin bound (Kawamoto and Hidaka 1984), with its cross-linking activity itself regulated by phosphorylation (Cukier et al. 2007). FLN is a substrate for the Ca2+-activated protease calpain (Gorlin et al. 1990) and for the ASB2 ubiquitin ligase (Heuze et al. 2008) which provide irreversible regulatory mechanisms for severing FLN cross-links in processes requiring membrane and cytoskeleton remodelling such as cell migration (Marzia et al. 2006; Baldassarre et al. 2009) and receptor cycling (Seck et al. 2003). FLN’s role in cell motility and spreading appears to be cell-type dependent. Melanoma cell lines (M2) that lack FLNA show impaired motility with cytoskeletal blebbing defects and reduced cellular elasticity that can be rescued by the transfection of FLNA (Cunningham et al. 1992) increasing β-integrin localisation at the membrane (Meyer et al. 1998). FLNA null mouse embryonic fibroblasts (Hart et al. 2006), neural crest cells (Feng et al. 2006) and FLNA missense mutant human fibroblasts (Clark et al. 2009) show no motility phenotype, whereas downregulation of FLNA expression leads to increased breast tumour cell line motility via a mechanism in which FLNA regulates focal adhesion disassembly in a calpain-dependent manner (Xu et al. 2010). Simultaneous knockdown of both FLNA and FLNB, or increased proteasomal degradation of all three FLNs, results in a defect in initiation of HT1080 cell motility and spreading (Baldassarre et al. 2009).
Filamin biomechanics
The hypothesis that FLN acts as a molecular spring was initially formulated upon sequence analysis of FLN domain structure (Gorlin et al. 1990) with subsequent analysis of its biomechanical and structural properties showing it suited to a cellular role regulating both the organization and mechanics of F-actin networks. FLN is found in the F-actin rich region near cell membranes where fast remodelling of the cytoskeleton is required, e.g. the leading edge during cell motility, but these regions also require reasonable stiffness for cell adhesion traction and actin polymerisation against the cell membrane (Pollard and Borisy 2003).
The defining activity of FLN is the ability to convert a population of non-linked fluid-like F-actin filaments into an orthogonal network structure or gelated state of rigidity proportional to the molar ratio of FLN to F-actin (Wang and Singer 1977). FLN is the most powerful F-actin cross-linking protein characterised, requiring a minimal concentration to cause gelation with the threshold determined as a relative FLN:actin monomer molar concentration of ∼1:200 (Tseng et al. 2004) compared to a physiological ratio of ∼1:50–100 (with actin concentration 25–72 uM (Gardel et al. 2006)). Tighter F-actin bundles and decreased actin filament turnover are observed when the FLNA concentration is increased (Nakamura et al. 2002; Schmoller et al. 2009). F-actin/FLN networks can show differing rheological properties depending on the concentration of FLN. At higher relative molar FLN concentrations (FLN:actin > 1:200), FLN-F-actin networks undergo stress stiffening under applied force with increased elastic modulus enabling cells to respond to substrate of increasing stiffness, e.g. up to 40 kPa for bone (Engler et al. 2006). In contrast, at lower relative FLN cross-link concentrations, F-actin networks are more dynamic and can be considered as weakly elastic solids that can soften in response to stress (Tseng et al. 2004). M2 FLNA null cells are softer and lose the ability to actively tension their cytoskeleton for mechanosensing external substrate stiffness compared to equivalent cells expressing FLNA (Kasza et al. 2009). The mechanical properties of the cross-linked F-actin cytoskeleton rely on the length and conformational flexibility of FLN molecular structure in contrast to more rigid cross-links (Kasza et al. 2010). By cross-linking F-actin into bundles, FLN acts to increase the apparent length of F-actin filaments with increased networking and shear resistance (Cortese and Frieden 1990). Other actin binding proteins, e.g. gelsolin, can antagonise the effect of FLN by shortening F-actin filaments and reducing network elasticity (Kasza et al. 2010). The ability to tune the elasticity of the cytoskeleton appears to be crucial for the maintenance of cell morphology under different conditions and also for key cellular processes such as differentiation, polarisation, phagocytosis, cell division and motility. Differential splicing provides an alternate mechanism for producing FLN isoforms of altered biomechanical properties; FLNB can be differentially spliced so that only the hinge I region is excluded from the translated protein (Xie et al. 1998). FLNA and FLNB respond similarly during in vitro rheological measurements showing resistance to 30- and 10-Pa shear stress, respectively, but in contrast, FLNA/B isoforms missing the first hinge region showed decreased elasticity in response to applied stress breaking at lower applied stress, 0.1 Pa, even though the cross-linking gelation concentration was equivalent to full-length FLN (Gardel et al. 2006). The absence of hinge I also affects regulation of cross-linking by proteolysis since this hinge contains the calpain cleavage site (Gorlin et al. 1990).
FLN cross-linked F-actin networks have mechanical properties that depend on the attachment kinetics and elasticity of FLN. Deformation of the FLN cross-linked F-actin network in response to applied stress can be achieved by detachment of the FLN cross-links from F-actin thus being dependent on the F-actin binding affinity (Kd) or the FLN dimerisation dissociation constant. An alternative mechanism is that the F-actin network remains cross-linked with FLN domains unfolding and hinge regions undergoing conformational rearrangement in response to applied tension (Fig. 3). The relative force required to break the attachment of FLN to F-actin and/or FLN dimerisation compared to the force required to unfold individual FLN domains is critical for evaluating these alternative but not mutually exclusive mechanisms. Optical tweezer force microscopy of FLN cross-linked F-actin at different force loading rates gave rupture forces of ∼40–60 ± 20 pN leading to the conclusion that unbinding events were more common than unfolding events at low loading rates though potential unfolding events were also observed (Ferrer et al. 2008). This observation infers that F-actin unbinding and FLN domain unfolding events are close in energy providing alternate or complementary mechanisms for force responses. Atomic force microscopy has lead to a minimal estimate for the force required to rupture ddFLN dimerisation of 200 pN suggesting that domain unfolding is likely to occur before dimer dissociation (Schwaiger et al. 2004). The FLN F-actin Kd is in the low μM range while the Kd for dimerisation is undetermined for full-length FLN but is likely to be low as FLN is normally isolated as dimers with no monomeric species seen by analytical ultracentrifugation at 0.7–2 μM (Hartwig and Stossel 1981). The FLNC dimerisation Kd for isolated repeat 24 is 2.5 μM, but it is likely that dimerisation is augmented through interactions with hinge II (Himmel et al. 2003). Atomic force microscopy force extension of FLNA dimers show reversible unfolding of FLN repeat domains with a force range of 50–220 pN and a stretching distance distribution around 30 nm meaning that FLN can be extended to several times its native length (Yamazaki et al. 2002). Molecular dynamics calculations yield similar forces broadly within the same extension range (Kolahi and Mofrad 2008). It has been proposed that a common folding pathway is shared between domains of the immunoglobulin fold superfamily (Clarke et al. 1999), but it remains to be determined if forced unfolding properties are conserved with structure. Single molecule AFM revealed that most ddFLN repeat domains unfold via a two-state mechanism but repeat 4 exhibited a stable intermediate state consisting of the five C-terminal β-strands (Schwaiger et al. 2004) with unfolding of the N-terminal two strands the first event. The first event during forced unfolding of titin immunoglobulin domain 27 also occurs via the dissociation of the N-terminal two β-strands (Marszalek et al. 1999). Modelling FLN-actin networks has shown that under strain a large fraction of FLN involved in cross-links is on the brink of unfolding (DiDonna and Levine 2007) consistent with a hypothesis of force-induced FLN unfolding to uncover cryptic binding sites to facilitate mechanosensing. It has been shown that forces as low as 2–12 pN applied to the cytoskeletal protein talin activates vinculin binding (del Rio et al. 2009). The biomechanical properties of FLN are consistent with a protein that must exhibit a degree of plasticity; deforming to resist/sense external force transducing force/stiffness signals into biochemical cues for downstream cellular events.
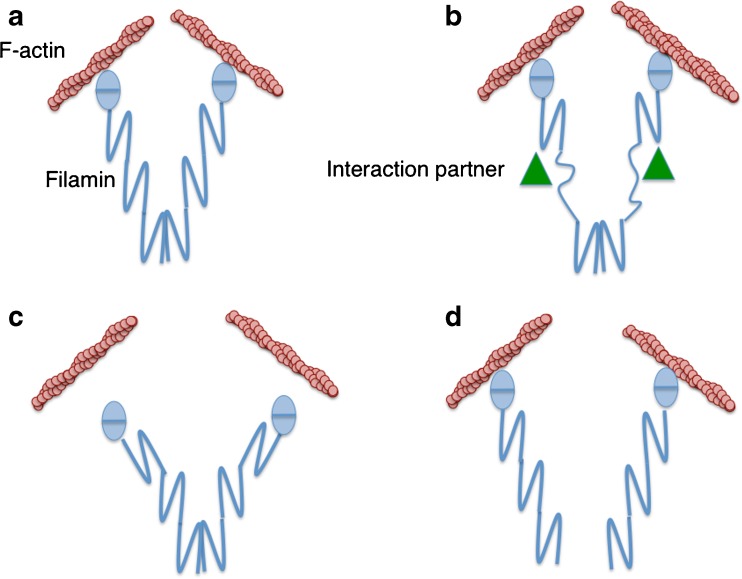
Fig. 3.
Models for filamin mediated mechanosensing responses. a A schematic representation of dimeric filamin cross-linked to F-actin. b Filamin force response mechanism involving repeat domain unfolding leading to cryptic binding sites being made accessible for interactions with protein binding partners (triangles). c Filamin force response mechanism via release from F-actin. d Filamin force response via dimer dissociation
Filamin associated diseases
Given the near-ubiquitous distribution of FLNA and FLNB and the large number of interaction partners, it is perhaps not unexpected that absence or alteration of FLNA/B function results in abnormal cellular events leading to human diseases, as previously mentioned for FLNC. Loss-of-function mutations in X-linked FLNA are associated with the brain malformation disorder, periventricular nodular heterotopia (PH) (Fox et al. 1998), typically male embryonic lethal and underlying abnormal neuronal migration during brain development causing seizures in females (Moro et al. 2002). FLNA missense mutations are associated with the otopalatodigital spectrum malformation disorders (OPD) primarily affecting skeletal development, clinically distinct from PH (Robertson et al. 2003). In parallel, loss-of-function mutations in FLNB are associated with spondylocarpotarsal synostosis syndrome, and dominant gain-of-function missense mutations are associated with the atelosteogenesis group of skeletal malformation disorders (Krakow et al. 2004). The clinical distinction, mode of inheritance of the FLN gain-of-function phenotypes, FLN expression and increased actin binding activity of disease-associated mutant FLNs are consistent with a gain/alteration of FLN function mechanism for these disorders (Robertson et al. 2003; Sawyer et al. 2009; Clark et al. 2009). FLNB is highly localised along the human growth plate and in the cartilage of developing vertebrae (Krakow et al. 2004), though the role that FLN plays in bone development remains unclear. The challenge now is to identify what properties of FLN are altered and the downstream consequences that lead to these diseases. The final differentiation of bone-forming osteoblast cells to bone remodelling osteocytes is accompanied by alterations to the distribution of FLN, so how FLN’s molecular properties facilitates this redistribution would appear crucial for correct bone patterning (Kamioka et al. 2004). The cytoskeleton is central to the mechanism by which osteocytes sense fluid shear stress and transduce mechanical signals via an integrin-mediated mechanism to downstream signalling events that impact on bone growth (Burra et al. 2010; Wang et al. 2007). Mechanisms may involve force-sensitive control of transcription factor activity by retaining regulatory partners in the cytosol; FLN associates with Smads (Sasaki et al. 2001; Zheng et al. 2007) and PEBP2β (Yoshida et al. 2005), proteins that regulate RUNX2 an important transcription factor regulating bone formation (Huang et al. 2001).
Precedents exist for disease mechanisms involving altered biomechanical properties of F-actin binding cytoskeletal proteins. α-actinin 4 missense mutations underlie the autosomal dominant kidney disorder, focal segmental glomerulosclerosis (Kaplan et al. 2000), and are associated with increased F-actin affinity leading to cross-linking at a lower concentration of α-actinin 4 (Weins et al. 2007). The viscoelastic properties of mutant α-actinin 4 cross-linked F-actin networks show relaxation frequencies lowered by an order of magnitude owing to a reduced off rate of the mutant compared to wild-type (Ward et al. 2008). The networks cross-linked by mutant α-actinin 4 also displayed a more prominent elastic plateau resulting in a more solid network, which could be replicated by increasing concentration of wild-type α-actinin 4. In another example, mutations in the polymeric keratin 5 and 14 intermediate protein filaments, associated with epidermolysis bullosa simplex, reduce the length and the elasticity of keratin filament networks in response to large deformations (Ma et al. 2001).
A model with FLN as part of a molecular mechanosensor seems appropriate with reference to FLN gain-of-function missense mutations. Mechanosensor functioning can be affected by changing either the attachment of FLN to F-actin and/or the tension response within the FLN structure (altered repeat domain unfolding). OPD mutant FLNA is expressed at wild-type levels in patient cells and ABD mutations, associated with skeletal disease, result in increased affinity for F-actin (Clark et al. 2009; Sawyer et al. 2009). An elevated FLN affinity for F-actin will change the energetic balance between FLN F-actin dissociation versus FLN domain unfolding, disrupting cytoskeletal network force responses to applied stress leading to abnormal cell behaviour. Increased cross-linking activity could lead to greater force-induced FLN unfolding resulting in increased stress stiffening of F-actin networks (for the same FLN concentration). Gain-of-function mutations in FLN repeat domains, with no identified function, are associated with an identical disease phenotype to those found within the ABD, suggesting that these mutations also affect the biomechanical parameters of FLN/F-actin networks either by abnormal force-induced conformational rearrangement and/or altering F-actin interaction kinetics, thus disrupting downstream signalling events. FLNA and FLNB are widely expressed in all tissues, but it is skeletal development that is primarily affected by gain-of-function disease-associated mutations. Bone is the stiffest tissue, and as such represents an upper limit for cell substrate mechanosensing consistent with FLN playing a key role in cell stiffening via the active generation of cytoskeletal tension and cellular responses to external force. Countering this hypothesis is the unusual dual phenotype FLNA mutation, L2349M, that does not alter measurable mechanical properties (Nakamura et al. 2009) but yet is not able to replace missing FLNA function (Zenker et al. 2004); the further biomechanical characterisation of other FLN disease-associated mutations is required.
Conclusions
As connectors between F-actin, the integrins and other signalling proteins, FLNs are key to coordinating biomechanical responses to force, EC matrix stiffness and/or adhesive properties, with intracellular consequences. It has long been established that FLN mediates essential cellular processes via mechanisms involving the dynamic cross-linking of F-actin filaments into elastic networks anchored to EC matrix adhesion sites and the scaffolding of signaling proteins to these networks. Recent accumulated evidence suggests the FLNs act as essential components of a finely tuned mechanosensor system associated with F-actin, membrane EC receptors and signalling proteins to facilitate force and matrix stiffness sensing. Mechanisms by which FLN responds to external force and EC matrix stiffness likely involve differential actin attachment and/or domain conformational rearrangement, influencing downstream cellular responses. With such a central role coordinating mechanical and signalling responses, it is perhaps not unexpected to find inherited mutations that alter FLN function associated with human disease. FLN has been termed a ‘finely-tuned rheology regulator’ (DiDonna and Levine 2007), and so it seems reasonable to propose that FLN gain-of-function mutations alter the tuning of this regulator. The exact mechanism of detuning by disease-associated mutations, whether it is via altered actin affinity and/or conformational properties, remains to be conclusively determined.
Acknowledgements
I would like to acknowledge the Royal Society of New Zealand Marsden Fund and Massey University Research Fund for financial support. I thank M.A.K. Williams and S.P. Robertson for critical evaluation of the manuscript.
References
- Alenghat FJ, Ingber DE. Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002;2002(119):pe6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- Baldassarre M, Razinia Z, Burande CF, Lamsoul I, Lutz PG, Calderwood DA. Filamins regulate cell spreading and initiation of cell migration. PLoS ONE. 2009;4(11):e7830. doi: 10.1371/journal.pone.0007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2(12):888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- Bresnick AR, Warren V, Condeelis J. Identification of a short sequence essential for actin binding by Dictyostelium ABP-120. J Biol Chem. 1990;265(16):9236–9240. [PubMed] [Google Scholar]
- Burra S, Nicolella DP, Francis WL, Freitas CJ, Mueschke NJ, Poole K, et al. Dendritic processes of osteocytes are mechanotransducers that induce the opening of hemichannels. Proc Natl Acad Sci USA. 2010;107(31):13648–13653. doi: 10.1073/pnas.1009382107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, et al. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3(12):1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- Carmeliet G, Vico L, Bouillon R. Space flight: a challenge for normal bone homeostasis. Crit Rev Eukaryot Gene Expr. 2001;11(1–3):131–144. [PubMed] [Google Scholar]
- Chen HS, Kolahi KS, Mofrad MRK. Phosphorylation facilitates the integrin binding of filamin under force. Biophys J. 2009;97(12):3095–3104. doi: 10.1016/j.bpj.2009.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88(1):39–48. doi: 10.1016/S0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- Clark AR, Sawyer GM, Robertson SP, Sutherland-Smith AJ. Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders. Hum Mol Genet. 2009;18(24):4791–4800. doi: 10.1093/hmg/ddp442. [DOI] [PubMed] [Google Scholar]
- Clarke J, Cota E, Fowler SB, Hamill SJ. Folding studies of immunoglobulin-like beta-sandwich proteins suggest that they share a common folding pathway. Structure. 1999;7(9):1145–1153. doi: 10.1016/S0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- Cortese JD, Frieden C. Effect of filamin and controlled linear shear on the microheterogeneity of F-actin/gelsolin gels. Cell Motil Cytoskeleton. 1990;17(3):236–249. doi: 10.1002/cm.970170310. [DOI] [PubMed] [Google Scholar]
- Cukier IH, Li Y, Lee JM. Cyclin B1/Cdk1 binds and phosphorylates Filamin A and regulates its ability to cross-link actin. FEBS Lett. 2007;581(8):1661–1672. doi: 10.1016/j.febslet.2007.03.041. [DOI] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255(5042):325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- D'Addario M, Arora PD, Fan J, Ganss B, Ellen RP, McCulloch CA. Cytoprotection against mechanical forces delivered through beta 1 integrins requires induction of filamin A. J Biol Chem. 2001;276(34):31969–31977. doi: 10.1074/jbc.M102715200. [DOI] [PubMed] [Google Scholar]
- Davies PJ, Wallach D, Willingham M, Pastan I, Lewis MS. Self-association of chicken gizzard filamin and heavy merofilamin. Biochemistry. 1980;19(7):1366–1372. doi: 10.1021/bi00548a015. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- DiDonna BA, Levine AJ. Unfolding cross-linkers as rheology regulators in F-actin networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;75(4 Pt 1):041909. doi: 10.1103/PhysRevE.75.041909. [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006;103(52):19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer JM, Lee H, Chen J, Pelz B, Nakamura F, Kamm RD, et al. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc Natl Acad Sci USA. 2008;105(27):9221–9226. doi: 10.1073/pnas.0706124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21(6):1315–1325. doi: 10.1016/S0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140(5):631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA. The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol. 1997;4(3):223–230. doi: 10.1038/nsb0397-223. [DOI] [PubMed] [Google Scholar]
- Furuhashi K, Inagaki M, Hatano S, Fukami K, Takenawa T. Inositol phospholipid-induced suppression of F-actin-gelating activity of smooth muscle filamin. Biochem Biophys Res Commun. 1992;184(3):1261–1265. doi: 10.1016/S0006-291X(05)80018-X. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc Natl Acad Sci USA. 2006;103(6):1762–1767. doi: 10.1073/pnas.0504777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273(3):1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- Goldmann WH, Tempel M, Sprenger I, Isenberg G, Ezzell RM. Viscoelasticity of actin-gelsolin networks in the presence of filamin. Eur J Biochem. 1997;246(2):373–379. doi: 10.1111/j.1432-1033.1997.00373.x. [DOI] [PubMed] [Google Scholar]
- Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, et al. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111(3):1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15(16):2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Stossel TP. Structure of macrophage actin-binding protein molecules in solution and interacting with actin filaments. J Mol Biol. 1981;145(3):563–581. doi: 10.1016/0022-2836(81)90545-3. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Tyler J, Stossel TP. Actin-binding protein promotes the bipolar and perpendicular branching of actin filaments. J Cell Biol. 1980;87(3 Pt 1):841–848. doi: 10.1083/jcb.87.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen OK, Ruskamo S, Konarev PV, Svergun DI, Iivanainen T, Heikkinen SM, et al. Atomic structures of two novel immunoglobulin-like domain-pairs in the actin cross-linking protein filamin. J Biol Chem. 2009;284(37):25450–2548. doi: 10.1074/jbc.M109.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuze ML, Lamsoul I, Baldassarre M, Lad Y, Leveque S, Razinia Z, et al. ASB2 targets filamins A and B to proteasomal degradation. Blood. 2008;112(13):5130–5140. doi: 10.1182/blood-2007-12-128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel M, Van Der Ven PF, Stocklein W, Furst DO. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry. 2003;42(2):430–439. doi: 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20(4):723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithychanda SS, Hsu D, Li H, Yan L, Liu D, Das M, et al. Identification and characterization of multiple similar ligand-binding repeats in filamin: implication on filamin-mediated receptor clustering and cross-talk. J Biol Chem. 2009;284(50):35113–35121. doi: 10.1074/jbc.M109.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HH, Priest JD, Hayes WC, Tichenor CC, Nagel DA. Humeral hypertrophy in response to exercise. J Bone Joint Surg Am. 1977;59(2):204–208. [PubMed] [Google Scholar]
- Kainulainen T, Pender A, D'Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin A in connective tissues after challenge by applied tensile forces. J Biol Chem. 2002;277(24):21998–22009. doi: 10.1074/jbc.M200715200. [DOI] [PubMed] [Google Scholar]
- Kamioka H, Sugawara Y, Honjo T, Yamashiro T, Takano-Yamamoto T. Terminal differentiation of osteoblasts to osteocytes is accompanied by dramatic changes in the distribution of actin-binding proteins. J Bone Miner Res. 2004;19(3):471–478. doi: 10.1359/JBMR.040128. [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24(3):251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- Kasza KE, Nakamura F, Hu S, Kollmannsberger P, Bonakdar N, Fabry B, et al. Filamin A is essential for active cell stiffening but not passive stiffening under external force. Biophys J. 2009;96(10):4326–4335. doi: 10.1016/j.bpj.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza KE, Broedersz CP, Koenderink GH, Lin YC, Messner W, Millman EA, et al. Actin filament length tunes elasticity of flexibly cross-linked actin networks. Biophys J. 2010;99(4):1091–1100. doi: 10.1016/j.bpj.2010.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Hidaka H. Ca2 + -activated, phospholipid-dependent protein kinase catalyzes the phosphorylation of actin-binding proteins. Biochem Biophys Res Commun. 1984;118(3):736–742. doi: 10.1016/0006-291X(84)91456-6. [DOI] [PubMed] [Google Scholar]
- Kesner B, Ding F, Temple B, Dokholyan N. N-terminal strands of filamin Ig domains act as a conformational switch under biological forces. Proteins. 2009;78(1):12–24. doi: 10.1002/prot.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaire N, Muller R, Blau-Wasser R, Eichinger L, Schleicher M, Rief M, et al. Filamin-regulated F-actin assembly is essential for morphogenesis and controls phototaxis in Dictyostelium. J Biol Chem. 2007;282(3):1948–1955. doi: 10.1074/jbc.M610262200. [DOI] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21(3):337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Kolahi KS, Mofrad MR. Molecular mechanics of filamin's rod domain. Biophys J. 2008;94(3):1075–1083. doi: 10.1529/biophysj.107.118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, Bertolotto C, et al. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet. 2004;36(4):405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- Lad Y, Kiema T, Jiang P, Pentikainen OT, Coles CH, Campbell ID, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26(17):3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langanger G, de Mey J, Moeremans M, Daneels G, de Brabander M, Small JV. Ultrastructural localization of alpha-actinin and filamin in cultured cells with the immunogold staining (IGS) method. J Cell Biol. 1984;99(4 Pt 1):1324–1334. doi: 10.1083/jcb.99.4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo DT, Kanner SB, Aruffo A. Filamin binds to the cytoplasmic domain of the beta1-integrin. Identification of amino acids responsible for this interaction. J Biol Chem. 1998;273(36):23304–23312. doi: 10.1074/jbc.273.36.23304. [DOI] [PubMed] [Google Scholar]
- Lowe T, Kley RA, van der Ven PF, Himmel M, Huebner A, Vorgerd M, et al. The pathomechanism of filaminopathy: altered biochemical properties explain the cellular phenotype of a protein aggregation myopathy. Hum Mol Genet. 2007;16(11):1351–1358. doi: 10.1093/hmg/ddm085. [DOI] [PubMed] [Google Scholar]
- Ma L, Yamada S, Wirtz D, Coulombe PA. A 'hot-spot' mutation alters the mechanical properties of keratin filament networks. Nat Cell Biol. 2001;3(5):503–506. doi: 10.1038/35074576. [DOI] [PubMed] [Google Scholar]
- Maestrini E, Patrosso C, Mancini M, Rivella S, Rocchi M, Repetto M, et al. Mapping of two genes encoding isoforms of the actin binding protein ABP-280, a dystrophin like protein, to Xq28 and to chromosome 7. Hum Mol Genet. 1993;2(6):761–766. doi: 10.1093/hmg/2.6.761. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, Schulten K, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402(6757):100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- Marti A, Luo Z, Cunningham C, Ohta Y, Hartwig J, Stossel TP, et al. Actin-binding protein-280 binds the stress-activated protein kinase (SAPK) activator SEK-1 and is required for tumor necrosis factor-alpha activation of SAPK in melanoma cells. J Biol Chem. 1997;272(5):2620–2628. doi: 10.1074/jbc.272.5.2620. [DOI] [PubMed] [Google Scholar]
- Marzia M, Chiusaroli R, Neff L, Kim NY, Chishti AH, Baron R, et al. Calpain is required for normal osteoclast function and is down-regulated by calcitonin. J Biol Chem. 2006;281(14):9745–9754. doi: 10.1074/jbc.M513516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SC, Sanan DA, Fox JE. Role of actin-binding protein in insertion of adhesion receptors into the membrane. J Biol Chem. 1998;273(5):3013–3020. doi: 10.1074/jbc.273.5.3013. [DOI] [PubMed] [Google Scholar]
- Moro F, Carrozzo R, Veggiotti P, Tortorella G, Toniolo D, Volzone A, et al. Familial periventricular heterotopia: missense and distal truncating mutations of the FLN1 gene. Neurology. 2002;58(6):916–921. doi: 10.1212/wnl.58.6.916. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Osborn E, Janmey PA, Stossel TP. Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. J Biol Chem. 2002;277(11):9148–9154. doi: 10.1074/jbc.M111297200. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Hartwig JH, Stossel TP, Szymanski PT. Ca2+ and calmodulin regulate the binding of filamin A to actin filaments. J Biol Chem. 2005;280(37):32426–32433. doi: 10.1074/jbc.M502203200. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpelainen I, Munday AD, et al. The structure of the GPIb-filamin A complex. Blood. 2006;107(5):1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179(5):1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Heikkinen O, Pentikainen OT, Osborn TM, Kasza KE, Weitz DA, et al. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS ONE. 2009;4(3):e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikki M, Merilainen J, Lehto VP. FAP52 regulates actin organization via binding to filamin. J Biol Chem. 2002;277(13):11432–11440. doi: 10.1074/jbc.M111753200. [DOI] [PubMed] [Google Scholar]
- Nomura M, Yoshikawa K, Tanaka T, Sobue K, Maruyama K. The role of tropomyosin in the interactions of F-actin with caldesmon and actin-binding protein (or filamin) Eur J Biochem. 1987;163(3):467–471. doi: 10.1111/j.1432-1033.1987.tb10892.x. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Hartwig JH. Actin filament cross-linking by chicken gizzard filamin is regulated by phosphorylation in vitro. Biochemistry. 1995;34(20):6745–6754. doi: 10.1021/bi00020a020. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8(8):803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- Pentikainen U, Ylanne J. The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. J Mol Biol. 2009;393(3):644–657. doi: 10.1016/j.jmb.2009.08.035. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Liu S, Erle DJ, Ginsberg MH. Integrin beta cytoplasmic domains differentially bind to cytoskeletal proteins. J Biol Chem. 1998;273(11):6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- Pi M, Spurney RF, Tu Q, Hinson T, Quarles LD. Calcium-sensing receptor activation of Rho involves filamin and Rho-guanine nucleotide exchange factor. Endocrinology. 2002;143(10):3830–3838. doi: 10.1210/en.2002-220240. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33(4):487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- Sarkisian MR, Bartley CM, Rakic P. Trouble making the first move: interpreting arrested neuronal migration in the cerebral cortex. Trends Neurosci. 2008;31(2):54–61. doi: 10.1016/j.tins.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates transforming growth factor-beta signaling. J Biol Chem. 2001;276(21):17871–17877. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: Insights from the crystal structures of filamin B actin binding domains. J Mol Biol. 2009;390(5):1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Schmoller KM, Lieleg O, Bausch AR. Structural and viscoelastic properties of actin/filamin networks: cross-linked versus bundled networks. Biophys J. 2009;97(1):83–89. doi: 10.1016/j.bpj.2009.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11(1):81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- Seck T, Baron R, Horne WC. Binding of filamin to the C-terminal tail of the calcitonin receptor controls recycling. J Biol Chem. 2003;278(12):10408–10416. doi: 10.1074/jbc.M209655200. [DOI] [PubMed] [Google Scholar]
- Sharma CP, Ezzell RM, Arnaout MA. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J Immunol. 1995;154(7):3461–3470. [PubMed] [Google Scholar]
- Sheen VL, Feng Y, Graham D, Takafuta T, Shapiro SS, Walsh CA. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum Mol Genet. 2002;11(23):2845–2854. doi: 10.1093/hmg/11.23.2845. [DOI] [PubMed] [Google Scholar]
- Shifrin Y, Arora PD, Ohta Y, Calderwood DA, McCulloch CA. The role of FilGAP-filamin A interactions in mechanoprotection. Mol Biol Cell. 2009;20(5):1269–1279. doi: 10.1091/mbc.E08-08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2(2):138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Tseng Y, An KM, Esue O, Wirtz D. The bimodal role of filamin in controlling the architecture and mechanics of F-actin networks. J Biol Chem. 2004;279(3):1819–1826. doi: 10.1074/jbc.M306090200. [DOI] [PubMed] [Google Scholar]
- Tyler JM, Anderson JM, Branton D. Structural comparison of several actin-binding macromolecules. J Cell Biol. 1980;85(2):489–495. doi: 10.1083/jcb.85.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorgerd M, van der Ven PF, Bruchertseifer V, Lowe T, Kley RA, Schroder R, et al. A mutation in the dimerization domain of filamin C causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77(2):297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Singer SJ. Interaction of filamin with F-actin in solution. Proc Natl Acad Sci USA. 1977;74(5):2021–2025. doi: 10.1073/pnas.74.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, McNamara LM, Schaffler MB, Weinbaum S. A model for the role of integrins in flow induced mechanotransduction in osteocytes. Proc Natl Acad Sci USA. 2007;104(40):15941–15946. doi: 10.1073/pnas.0707246104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Weins A, Pollak MR, Weitz DA. Dynamic viscoelasticity of actin cross-linked with wild-type and disease-causing mutant alpha-actinin-4. Biophys J. 2008;95(10):4915–4923. doi: 10.1529/biophysj.108.131722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weins A, Schlondorff JS, Nakamura F, Denker BM, Hartwig JH, Stossel TP, et al. Disease-associated mutant alpha-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci USA. 2007;104(41):16080–16085. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J (1892) The law of bone remodelling (trans: Maquet P, Furlong R). Springer, Berlin
- Xie Z, Xu W, Davie EW, Chung DW. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem Biophys Res Commun. 1998;251(3):914–919. doi: 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- Xu W, Xie Z, Chung DW, Davie EW. A novel human actin-binding protein homologue that binds to platelet glycoprotein Ibalpha. Blood. 1998;92(4):1268–1276. [PubMed] [Google Scholar]
- Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, et al. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010;207(11):2421–2437. doi: 10.1084/jem.20100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Furuike S, Ito T. Mechanical response of single filamin A (ABP-280) molecules and its role in the actin cytoskeleton. J Muscle Res Cell Motil. 2002;23(5–6):525–534. doi: 10.1023/A:1023418725001. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Ogata T, Tanabe K, Li S, Nakazato M, Kohu K, et al. Filamin A-bound PEBP2beta/CBFbeta is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Mol Cell Biol. 2005;25(3):1003–1012. doi: 10.1128/MCB.25.3.1003-1012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker M, Rauch A, Winterpacht A, Tagariello A, Kraus C, Rupprecht T, et al. A dual phenotype of periventricular nodular heterotopia and frontometaphyseal dysplasia in one patient caused by a single FLNA mutation leading to two functionally different aberrant transcripts. Am J Hum Genet. 2004;74(4):731–737. doi: 10.1086/383094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Baek HJ, Karsenty G, Justice MJ. Filamin B represses chondrocyte hypertrophy in a Runx2/Smad3-dependent manner. J Cell Biol. 2007;178(1):121–128. doi: 10.1083/jcb.200703113. [DOI] [PMC free article] [PubMed] [Google Scholar]