Abstract
The classic structure–function paradigm holds that a protein exhibits a single well-defined native state that gives rise to its biological function. Nonetheless, over the past few decades, numerous examples of proteins exhibiting biological function arising from multiple structural states of varying disorder have been identified. Most recently, several examples of ‘metamorphic proteins’, able to interconvert between vastly different native-like topologies under physiological conditions, have been characterised with multiple functions. In this review, we look at the concept of protein metamorphosis in relation to the current understanding of the protein structure–function landscape. Although structural dynamism observed for metamorphic proteins provides a novel source of functional versatility, the dynamic nature of the metamorphic proteins generally makes them difficult to identify and probe using conventional protein structure determination methods. However, as the existence of metamorphic proteins has now been established and techniques enabling the analysis of multiple protein conformers are improving, it is likely that this class will continue to grow in number.
Keywords: Metamorphic proteins, Lymphotactin, Mad2, CLIC1
Introduction
The study of protein structure has classically been based on the assumption that a protein adopts a single, well-defined three-dimensional fold under native conditions. It is assumed that this unique fold is encoded by the protein’s amino acid sequence with, at most, small structural changes available to facilitate function. However, there is now a growing body of proteins shown to possess ambiguous fold characteristics. This includes the identification of several proteins able to adopt more than one native conformation from the same amino acid sequence under physiological conditions. These novel proteins have been termed ‘metamorphic’ (Murzin 2008).
Metamorphic proteins are classified by their ability to undergo reversible conformational transition from one ordered fold topology to a vastly different ordered fold via major structural modification. This ability to interconvert between differing fold topologies has the potential to expand functional utility within the cell. The concept of protein metamorphosis evolutionary intermediates also provides an appealing rationale for the rapid evolution of new divergent folds possessing high sequence homology. The key to understanding the functional role and evolutionary importance of metamorphic proteins lies not only in deciphering the structural transitions involved but also in the kinetics and environmental triggers for conformational change. To date, the majority of identified metamorphic proteins have been structurally classified using the conventional techniques of crystallography and NMR spectroscopy. However, it is becoming increasingly apparent that, in order to identify and isolate individual metamorphic fold topologies or simultaneously probe multiple metamorphic conformational states, the development of novel structural analysis strategies to handle the dynamic nature of this protein class is necessary.
Here, we discuss the rationale for the foundation of the metamorphic protein class in light of current views on both protein folding and structure–function relationships, further exemplified by several illustrative examples. We note the experimental challenges faced by the structural biology community in order to accommodate this unique protein family, both in the development of protein structure prediction strategies and the application of structure determination methods.
Challenging the classic ‘one sequence–one structure’ protein dogma
The first insights into the role of the amino acid sequence in defining the three-dimensional protein structure can be traced back to the early 1900s. The process of protein folding was postulated to be a reversible ‘all-or-nothing’ transition between a denatured state, consisting of biologically latent random coil, and a highly-ordered native fold (Anson and Mirsky 1925; Mirsky and Pauling 1936). The physical process of protein folding could be described by the classic thermodynamic hypothesis of Anfinsen’s dogma (Anfinsen 1972, 1973). In Anfinsen’s dogma, the native state of the protein is said to adopt a single, stable, unique, and well-defined three-dimensional fold. The native state then represents the global minimum of free energy under physiological conditions.
Many of the earliest protein biochemistry studies primarily focused on globular transport proteins and catalytic enzymes. These proteins could be readily purified from tissue materials in large quantities and, as it could be measured simply, their function became relatively well understood. The functional requirement for such proteins to form and maintain an accurate, proficient and specific active site structure likely exerts a strong selective pressure to adopt one stable and conserved fold. This structural rigidity also, quite fortuitously, generally makes globular proteins highly amenable to crystallisation. Hence, the first X-ray crystallography protein structures from the late 1950s and early 1960s (for example, myoglobin Kendrew et al. 1958; haemoglobin, Perutz et al. 1960; and lysozyme, Blake et al. 1965; Johnson and Phillips 1965), reflect this early trend in the study of globular proteins. However, in recent years, both advancements in molecular cloning methods to generate and modify proteins of interest and advancements in protein structure determination methods, such as NMR, have provided us with high-resolution structures of proteins in solution in addition to their crystallised form. These solution structures of proteins have considerably extended our understanding of protein folding to reveal that there are additional structural and functional states that cannot solely be described by Anfinsen’s dogma of ‘one sequence–one structure’.
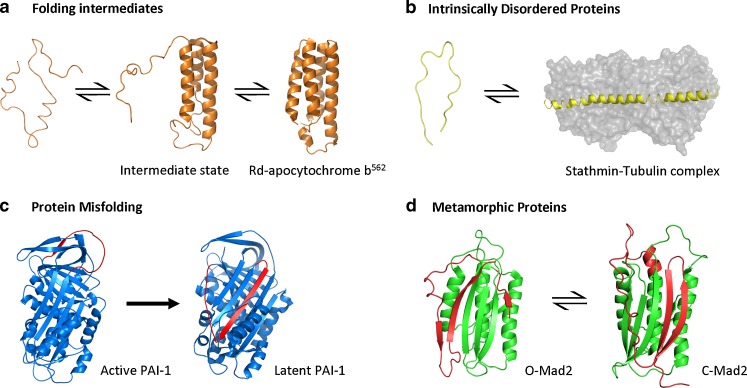
Still today, one of the most puzzling problems in understanding protein structure is the efficiency of protein-folding. Most small proteins fold spontaneously on the millisecond or even microsecond time-scale. Such a rapid rate of protein-folding would not be possible if protein-folding followed a simple stochastic process whereby the protein samples all possible conformations with equal probability. This concept, known as Levinthal’s Paradox (Levinthal 1969; Zwanzig et al. 1992), thus suggests that the protein-folding follows a guided pathway of local interactions or kinetically controlled stable intermediate states. By the late 1960s, there was an increasing accumulation of experimental data detecting the presence of protein folding intermediates and partially folded transition states, primarily through non-simultaneous variation in spectroscopic properties upon protein unfolding (see, for example, Tanford et al. 1966). In light of this experimental evidence challenging the concept of the ‘all-or-nothing’ transition between the unfolded and native state, much debate followed. Initially, it was not clear if these intermediate states were simply incorrectly folded forms obtained under conditions inappropriate for folding or whether they represented true folding intermediates that maintained substructures of the native fold (Baldwin 1975). It was not until the early 1980s that experimental evidence of a third thermodynamically stable class of fold, clearly different from both the native and denatured states, was described (Dolgikh et al. 1981; Ohgushi and Wada 1983). This third equilibrium state, termed the molten globule (MG) (Fig. 1a), describes a compact collapsed state that retains some native-like secondary structure but lacks the tightly packed protein interior characteristic of a globular protein. The concept of the MG intermediate was met with great enthusiasm and its popularity rapidly increased. Experimental evidence for numerous other proteins possessing such MG features as well as identification of other proteins that also exhibited discrete and functionally relevant intermediate folding states have subsequently emerged.
Fig. 1.
The classical ‘one sequence–one structure’ protein dogma cannot be used to describe the structural transitions and functional states of all proteins. a Folding intermediates are not only involved in the protein folding pathway, but can also possess unique functional properties distinct from the native state. Folding intermediates of Rd-apocytochrome b562—PDB 1YZC, (Feng et al. 2005)—possess the helical structures of the native four-helix bundle—PDB 1M6T (Chu et al. 2002)—without the tightly packed tertiary interaction of the native core. b Intrinsically disordered proteins possess function despite a lack of define secondary structure along their entire length or in localized regions. Structure may be acquired through interaction with binding partners. For example, stathmin (colored yellow) is an intrinsically disordered protein that forms a ternary complex with tubulin (grey form-filled)—PDB 1SA0, (Ravelli et al. 2004)—which is important in the regulation of microtubule dynamics. c Protein misfolding involves transition from the native state to an alternate structure accompanied by a large increase in stability such that the process is largely irreversible under physiological conditions. The serpin protein PAI-1 can spontaneously undergo an active-to-latent transition - PDB 1DB2, (Nar et al. 2000), and 1DVN (Stout et al. 2000), respectively. Latent PAI-1 can only be partially reactivated by treatment with denaturing reagents followed by renaturation of the unfolded molecule. d Under physiological conditions, metamorphic proteins are able to reversibly transit between multiple stable native states that possess vastly different fold topologies. Mad2 exists in a two-state equilibrium of the open (O-Mad2)—PDB 1DUJ (Luo et al. 2000)—and closed forms (C-Mad2)—PBD 1S2H (Luo et al. 2004). Regions that undergo dramatic structural change are colored red
Over the last decade it has also become evident that a significant fraction of proteins, estimated at over 40% of the human proteome, lack stable structure along either their entire length or within localised region of more than 30–40 residues in length (Uversky and Dunker 2010). Unlike the classical globular proteins, intrinsically disordered proteins (IDPs) exhibit a high degree of structural disorder and, as such, exist in a dynamic ensemble of conformations. Although some IDPs have been shown to remain substantially disordered throughout their entire functional process, others have been shown to adopt ordered structures once in complex with binding partners (Fig. 1b). IDPs also generally appear to possess a distinct functional repertoire compared to highly structured proteins. Many IDPs are involved in functions related to the organisation of complex protein–protein interaction networks such as cell signalling and regulation in which the ability to interact with numerous binding partners may be advantageous (for a further extensive review of the IDP class, see (Tompa 2009)). IDPs demonstrate that a well-defined structure for the IDP segment in the isolated or ‘inactive’ protein is not required for function and perhaps protein structure dynamism is even functionally and evolutionarily advantageous for certain biological processes.
Further, examples challenging the notion of Anfinsen’s dogma also include instances of proteins which exhibit irreversible conformational rearrangements such as protein misfolding. Protein misfolding occurs as a result of large-scale structural rearrangement with an accompanying dramatic increase in protein stability of the misfolded form. As such, the misfolded form represents a lower free energy conformation than the native state. Hence, protein misfolding is essentially an irreversible process under physiological conditions (Fig. 1c). There are several well known and studied protein systems which undergo dramatic and permanent conformational changes, most notably the aggregation of misfolded protein sequences into amyloid fibrils rich in β-structure and eventually into larger aggregated entities. These insoluble fibrous aggregated amyloid structures, also histologically referred to as ‘plaques’, are associated with the pathogenesis of some of the most well-known debilitating neurodegenerative diseases including Alzheimer’s, Parkinson’s and Huntington’s diseases, and prion encephalopathies (Greenwald and Riek 2010). These fibrillar amyloid protein species have long been viewed as pathological structures (or even sometimes simply as experimental artifacts hampering detailed protein structure determination). However, growing evidence now indicates that amyloid formation may also be a highly evolutionarily conserved functional structure necessary for normal physiology including cellular roles such as hormone storage, scaffolding, epigenetic control of polyamines and information transfer (Maury 2009). Additionally, the serpins are an enigmatic group of proteins that also exhibit irreversible structural changes to facilitate protease inhibition in cells. Binding of a serpin protein to its target protease results in cleavage of an exposed amino-acid sequence in the serpin, called the reactive-centre loop (RCL). In its anti-protease form, the RCL is held at the top of the serpin molecule and upon protease binding becomes cleaved. This allows the RCL to become an additional β-stand in the centre of the β-sheet core of itself (Gettins 2002). The metastable nature of the serpins also enables some members to misfold into a latent form in which the RCL is inserted in the core without having been cleaved and also makes them susceptible to polymerisation. However, unlike amyloid, the repeating unit within the serpin polymer essentially adopts a native-like fold. Polymerisation is thought to occur via domain swapping of adjacent serpin molecules (Yamasaki et al. 2008; Huntington and Whisstock 2010). Extensive domain swapping results in a significant increase in protein stability, analogous to protein misfolding, and is consistent with the irreversible nature of the serpinopathy disorders including emphysema, early onset dementia and liver cirrhosis (for further review of this protein class, see Kaiserman et al. 2006). However, it still remains unclear whether domain swapping of the serpins also has a functional role in cells. Further work is required to clarify the relationship between structure and function of such protein ensembles.
While the notion continues to apply that function is exclusively associated with a single, unique and well-defined three-dimensional structure for most protein families, the alternative view that proteins are an ensemble of substructures in equilibrium with the native state is beginning to prevail. Together, the existence of folding intermediates, IDPs and ‘misfolded’ states demonstrates that multiple structures can be accommodated by a single amino acid sequence (Fig. 1a–c). Such a notion poses an entirely new set of questions concerning how protein sequence dictates protein structure and thus functional biology, and invites the possibility that additional protein classes that do not conform to the classic understanding of the relationship between protein structure and function may also exist.
The metamorphic protein class
Over the past decade, several metamorphic proteins have been shown to interconvert between multiple native-like functional states consisting of the same amino acid sequence under physiological conditions. Whilst the cases of anomalous protein folding previously discussed share the ability to mediate multiple functions via different structural conformations, protein metamorphosis appears to be a distinct phenomenon. Unlike protein folding intermediates, metamorphic proteins possess the ability to undergo global structural transitions in order to switch between forms that are not precursors to a single native topology. The well-defined secondary and tertiary structure interactions exhibited in the alternate forms of metamorphic proteins also distinguishes them from the IDP protein class. Additionally, whilst the large-scale structural rearrangement required to switch between alternate metamorphic fold topologies is akin to protein misfolding, protein metamorphosis is not accompanied by an insurmountable increase in protein stability. Thus, despite the dramatic repacking of the hydrophobic protein core and exposure of new binding surfaces, the most notable feature of this protein class is that the metamorphosis event is reversible under physiological conditions (Fig. 1d). Hence, for a metamorphic protein, the free energy landscape can be characterised by the presence of two or more local minima representing each of the stable three-dimensional structures that define the different native topology states. Each minimum may be separated by relatively low energy barriers. Hence, the alternate metamorphic protein folds are often found to co-exist in a dynamic equilibrium under physiological conditions.
The existence of multiple folded conformations is not prohibited by the principles of physics or chemistry. However, the application of traditional protein structure determination techniques, such as X-ray crystallography and NMR, have traditionally been biased towards the detection of the single most populated conformation and thus inadvertently select against the presence of multiple topologies. As such, the existence of additional metamorphic proteins may have been unintentionally overlooked and alternate conformations selected against. Thus, the ability for a protein to undergo structural metamorphosis may not be as rare as currently thought. It is also not fully understood whether protein metamorphosis is a unique characteristic specific to particular proteins or a more universal phenomenon. Anfinsen himself stated that the native protein is “the one in which the Gibbs free energy of the whole system is lowest; that is, the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment” (Anfinsen 1973). Hence, local changes within a protein’s environment may be sufficient to shift the equilibrium between alternate metamorphic states. On the surface, this notion may appear energetically unfavourable as stable alternative folds are likely to act as kinetic traps, slowing the rate of protein folding and thus not likely to have been favoured by evolution (Onuchic and Wolynes 2004). However, the ability to stabilise areas of inherent flexibility or instability, may in fact facilitate repacking of the hydrophobic core and exposure of new binding surfaces required to transit between the different metamorphic fold topologies, whilst avoiding kinetic traps which may lead to misfolding. Hence, the unusual folding behaviour of metamorphic proteins may be viewed as having evolved for a biological purpose. The biological purpose may be either for duality of function by mutually exclusive activities which could not be accommodated by a single fold or structural dynamism to enable conformational selection (Boehr and Wright 2008). Hence, the metamorphic transition between the varying structural forms appears to allow the cell to use a unique mechanism of post-translational regulation, as a result of changes in local physiological conditions, to enable access to the alternate functional structures.
Recently identified examples of metamorphic proteins arise from a diverse range of functional biology, while the recognised triggers for the metamorphic transitions of these proteins also involve a broad range of environmental factors. For example, T7 RNA polymerase converts between the initiation and elongation states upon formation of a growing RNA polymer (Tahirov et al. 2002; Yin and Steitz 2002) and the spindle assembly checkpoint protein Mad2 adopts two distinct conformations in a ligand free state, both of which are required for correct attachment of microtubules to kinetochores necessary for accurate DNA replication in mitosis (Luo et al. 2004). This protein is explored in further detail later. Two other metamorphic proteins, also described in further detail, appear to possess a conserved canonical native state and an alternate form. The first, lymphotactin, converts between a monomeric form that binds the G-protein coupled receptor, and a unique dimeric form that binds to heparin (Tuinstra et al. 2008; Camilloni and Sutto 2009). The second example is the CLIC1 chloride intracellular channel which undergoes a redox induced conversion between a glutathione-S-transferase like canonical monomer form and a soluble dimer form but can also insert into the membrane to form an ion channel (Littler et al. 2004, 2010b).
The above-mentioned metamorphic proteins have mostly been found serendipitously. Hence, the majority of cases of proteins now classified as metamorphic were previously treated as unique exceptions to the notion that function arises from a single sequence dictating a single native structure. From such a limited number of representative examples, there does not appear to be any obvious common structural or functional elements or an approach to predict whether a protein will exhibit the ability to undergo metamorphosis. However, a closer inspection of the best characterised conformational switches for this novel class reveals that the metamorphic proteins do have some common properties. These properties include regions of inherent flexibility as well as the ability to avoid kinetic trapping in a misfolded state during conversion between the alternate metamorphic forms, often through the protein-protein interactions of an oligomeric species (Bryan and Orban 2010). The following section further elaborates and illustrates the major secondary structure modifications required to convert between the alternate metamorphic forms for three examples of metamorphic proteins. We describe how each fold achieves a novel function for a range of biological processes. From these selected examples, it is evident how relatively ‘simple’ changes in environmental conditions can effectively shift the equilibrium between alternate metamorphic states.
Mad2
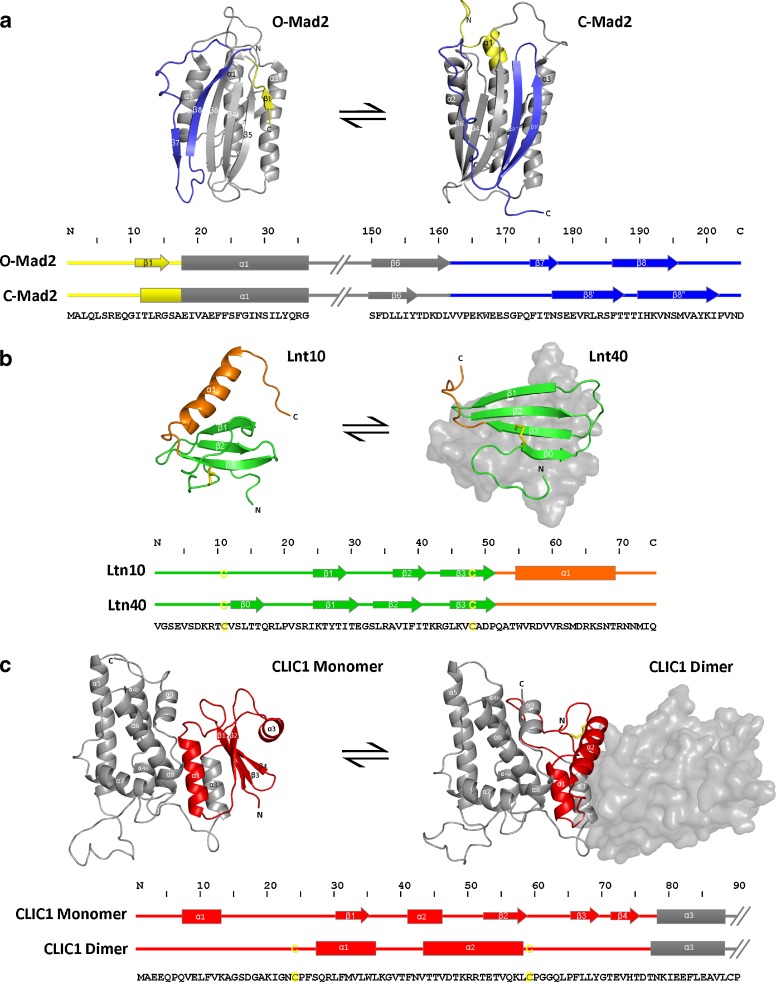
Mitotic arrest deficient 2 (Mad2) is a spindle assembly checkpoint protein and one of the most extensively studied members of the metamorphic protein class. Previously, it was thought that the alternate ‘open’ and ‘closed’ folds structurally characterised for Mad2 represented two different binding states: one the free protein and the other the ligand (Cdc20/Mad1) bound form (Luo et al. 2002). However, both the inactive open (O-Mad2) and active closed (C-Mad2) conformations have now been shown to exist in equilibrium in a ligand-free state (Luo et al. 2004) (Fig. 2a). Structural comparison of the two forms reveals a central core structure, comprising approximately 70% of the Mad2 sequence, which remains unchanged between the two states. The remaining residues, localised within regions at the C- and N- termini, undergo major structural rearrangement involving the refolding and translocation of the C-terminal β-strand from one end of the main β-sheet to the other.
Fig. 2.
Metamorphic protein structural transitions showing regions that undergo dramatic structural change (highlighted in color in both structures and the secondary structure alignments). a Interconversion between the ‘open’ O-Mad2 form—PDB 1DUJ (Luo et al. 2000)—and ‘closed’ Mad2-C—PDB 1S2H (Luo et al. 2004)—involves major tertiary structural rearrangement of the N-terminus (yellow) and C-terminus (blue) b Interconversion between the monomeric Ltn10—PDB 2HDM (Tuinstra et al. 2007) and dimeric Ltn40—2JP1 (Tuinstra et al. 2008)—forms involves a shift in the hydrogen bonding network between strands β1-β2 and β2-β3 by one amino acid; reorientation of many of the amino acids that form the core of the Ltn10 state to form the Ltn40 dimer interface; and, loss of the C22-59 disulfide and helical structure c Interconversion between reduced monomeric CLIC1—PDB 1k0M (Harrop et al. 2001)—and the CLIC1 oxidised dimer form—PDB 1RK4 (Littler et al. 2004)—involves major secondary and tertiary structural rearrangement of the N-terminus (red). Rearrangement of the sheet structures of the monomer to form the large hydrophobic dimer interface is accompanied by formation of the C24-59 disulfide (colored yellow and represented by ‘C’ on the structure alignment). Grey form-filled represents the equivalent dimer partner
Switching between O-Mad2 and C-Mad2 is essential for full function in vivo. By transiting between the open and closed conformations, Mad2 mediates the correct attachment of microtubules to kinetochores, a process essential in ensuring the fidelity of chromosome inheritance during mitosis (Luo and Yu 2008). Briefly, unattached kinetochores contain the checkpoint protein Mad1, which recruits C-Mad2 to provide a surface for the conversion of the available soluble pool of inactive O-Mad2 to active C-Mad2. It has been proposed that a partially unfolded intermediate state of O-Mad2, consisting of the conserved core structure but with more flexible N- and C-terminal regions, is stabilised by dimerising with the C-Mad2 complex where the latent binding site blocked by the C-terminal of O-Mad2 is then revealed. (Skinner et al. 2008). This results in conversion of the soluble pool of inactive O-Mad2 to checkpoint active C-Mad2 which can then bind and inhibit Cdc20 of the mitotic checkpoint complex, halting progression to anaphase. Additionally, the newly formed C-Mad2-Cdc20 complex is able to facilitate conversion of O-Mad2 to C-Mad2 independent of the C-Mad2-Mad1 complex (Lad et al. 2009).
The spontaneous conversation between O-Mad2 and C-Mad2 in vitro is very slow, with a lifetime of 9 h and the reverse reaction is even six-times slower again (Tuinstra et al. 2008; Camilloni and Sutto 2009). This is far too slow to account for the short half-life described for Mad2 in living cells (24–28 s; Howell et al. 2000) and indicates that a relatively large energy barrier exists for this structural interconversion. In vivo, it is likely that interaction between C-Mad2 and Mad1 or Cdc20 makes the structural transition of the subsequently bound O-Mad2 kinetically more favourable than when the O-Mad2 dimerises with C-Mad2 alone (Lad et al. 2009); however, this mechanism is not currently well understood.
Lymphotactin
A second representative of the metamorphic protein family is Lymphotactin (Ltn) for which the two distinct folds identified at equilibrium are a monomeric canonical chemokine fold (Ltn10) and a novel dimeric-β-sandwich fold (Ltn40) (Fig. 2b). Under near physiological conditions (37°C and 150 mM NaCl), the NMR spectra of lymphotactin showed evidence of more than one conformation in solution. Under this condition, two Ltn forms were found in approximately equal amounts. The equilibrium between these two forms could be shifted completely from one to the other by small variations in the salt and temperature conditions alone (Volkman et al. 2009). Inter-conversion between these two alternate Ltn structures involved almost complete rearrangement of the hydrogen bonding network (Tuinstra et al. 2008; Camilloni and Sutto 2009). In the Ltn40 dimer form, a β-sandwich is formed between two subunits in which the positioning of the β-sheets hydrogen-bonding register is shifted by a single residue such that strands β1 and β3 are rotated along the lengthwise axes by 180° in comparison to the Ltn10 monomer structure. A substantial number of hydrophobic side-chains at the dimer interface are also key residues forming the core of the canonical Ltn10 monomeric chemokine form. All the α-helical elements present in the Ltn10 state appear to unfold in the Ltn40 form, and a significant number of positively-charged residues become exposed to solution. Interconversion between the alternate Ltn states is thought to take place via interaction between two subunits in a monomeric, partially unfolded form which exhibits residual structure involving local contacts common to both the Ltn10 and Ltn40 states (Camilloni and Sutto 2009).
It is interesting to note that the monomeric Ltn10 protein is also missing a highly conserved disulfide bond found in other chemokines (linking α1 to β1). The monomeric Ltn10 state can be viewed as effectively being destabilised so as to aid the transition to the dimeric Ltn40 form. If the absent α1 to β1 linking disulfide is experimentally engineered back into the Ltn10 protein, it locks the protein into the monomeric state which cannot then interconvert to the dimeric form (Tuinstra et al. 2007). The complementary experiment where a destabilising point mutation (W55D) was introduced into the core of the Ltn also prevented reversibility between the two states, but this time locked the protein into the non-canonical Ltn40 dimeric conformation (Tuinstra et al. 2008). Thus, in order to determine the Ltn10 canonical lymphotactin structure by NMR, engineering in the second chemokine conserved disulfide bond or acquiring data under very specific conditions of low temperature and high salt (10°C, 200 mM NaCl) were necessary (Tuinstra et al. 2007).
Binding interactions of the two Ltn states are quite distinct: only the Ltn10 monomer can bind and activate the XCR1 G-coupled protein receptor, while Ltn40 binds heparin, a polysaccharide component of the extracellular matrix. Tuinstra et al. (2008) suggest that the ability to bind completely unrelated surfaces are actually functionally complimentary and enable a novel mechanism to regulate the Ltn biological activity. Ltn40 sequestered to the extracellular matrix, and thus rendered inactive, must dissociate and convert to the Ltn10 form in order to stimulate the XCR1 receptor and induce cell migration in vivo. However, a mechanistic understanding of the interconversion between bound Ltn40 and the Ltn10 state in this functional context is still lacking.
CLIC1
Our own interest in metamorphic proteins came about through structural studies of the chloride intracellular channel (CLIC) protein family. The first member of the CLIC family, p64 (later renamed as bovine CLIC5B), was initially identified in 1987 in the search for the then elusive cystic fibrosis ion channel protein (Landry et al. 1987; Redhead et al. 1992). Since then, six vertebrate CLIC members (CLIC1–CLIC6) have been identified. In addition to their function as ion channels, the CLICs have also been described to be redox proteins, enzymes and scaffolding proteins coupling the membrane to the cytoskeleton. However, the exact cellular role of the CLIC proteins is still not clearly defined or understood (for further review, see Littler et al. 2010b).
The CLICs were first viewed to contain significant sequence homology to the glutathione-S-transferase (GST) family, a family of cytosolic enzymes not previously known to possess ion channel activity (Dulhunty et al. 2001). The first X-ray crystal structure of a CLIC family member, CLIC1, was obtained under reducing conditions and confirmed CLIC1 to consist of a GST-like fold possessing a thioredoxin N-domain and a larger all α-helical C-domain (Harrop et al. 2001). Subsequent crystal structures of several other CLIC family members all share the same monomeric GST-like canonical CLIC fold (Littler et al. 2005, 2008, 2010a; Cromer et al. 2007). However, upon exposure to oxidative conditions in solution, CLIC1 was also shown by X-ray crystallography to adopt an alternative non-covalent dimer form (Littler et al. 2004) (Fig. 2c). In this oxidised CLIC1 dimer form, an intramolecular disulfide can be seen to form between cysteine residues 24 and 59, whilst the entire N-domain becomes completely rearranged into a helical bundle. This large-scale rearrangement results in the loss of structural integrity of the conserved N-domain glutathione binding site and exposure of a large hydrophobic surface that forms the interface between the two helical N-domains in the stable dimer structure. Additionally, this property of inherent plasticity exhibited by the CLIC1 N-domain has also been demonstrated by hydrogendeuterium exchange experiments triggered by low pH conditions (Fanucchi et al. 2008; Stoychev et al. 2009).
The ability of CLIC1 to dimerise upon oxidation has only been demonstrated for CLIC1 and cannot solely be attributed to the formation of the intramolecular disulfide. The cysteine 24 residue involved in the disulfide of the CLIC1 dimer is conserved in all CLIC family members. However, the second cysteine, residue 59, is unique to CLIC1. Efforts to engineer a cysteine residue in the equivalent position as CLIC1 C59 in other CLIC homologues has proven unsuccessful in inducing the ability to form a structure equivalent to the CLIC1 oxidised dimer form (Cromer et al. 2007). However, mutation of either C24 or C59 in CLIC1 does eliminate the ability to form the CLIC1 dimer (Littler et al. 2004).
The physiological relevance of the alternative CLIC1 structures is still not clear. Neither of the soluble CLIC1 forms, based purely on their structures, appears to have a direct connection to ion channel function. However, sequence analysis predicts some enzymatic function may be present for the CLIC1 GST-like monomer fold (Littler et al. 2010b). The function of the soluble dimer is also contentious, although it has been proposed that the interfacial hydrophobic surface may act as a membrane docking intermediate prior to membrane insertion (Littler et al. 2004; Goodchild et al. 2009). Further structural transitions of CLIC1 have also been shown to take place in a lipid environment, with the N-terminal transmembrane containing domain capable of transcending the bilayer in order to form the pore required for ion channel conductance (Goodchild et al. 2009, 2010). Again, the key environmental trigger for this transition is oxidation which appears to promote interaction of the CLIC1 with the membrane bilayer (Goodchild et al. 2009).
Another interesting feature of the CLIC family is the presence of a nuclear localisation signal (NLS) sequence which is highly conserved across all vertebrate CLICs (CLIC1-6) with the exception of CLIC3 (Mynott et al. 2011). Under conditions of cellular stress, CLIC4 was found to be transported to the nucleus via the classical nuclear import pathway by binding to the import receptor, importin-α (Suh et al. 2004). An X-ray crystal structure of a CLIC4 peptide containing the NLS motif clearly shows binding to the major site of importin-α in an extended fashion. The extended nature of the NLS within the binding pocket suggests that, for the interaction between CLIC4 and importin-α to occur in vivo, CLIC4 must undergo significant conformational rearrangement to expose the NLS binding site which may be controlled by S-nitrosylation of CLIC4 (Mynott et al. 2011). While no structure of an importin–α CLIC4 complex has been determined to date, the intrinsic flexibility of the N-domain together with S-nitrosylation appear to contribute to the exposure of the NLS binding site in cells. Therefore, a further CLIC functional and structural state may be assigned to the CLIC family in addition to the GST-like and ion channel forms. The CLIC family clearly demonstrates a metamorphic protein system in which inherent flexibility facilitates multiple functional properties. However, clarification of the role of the different CLIC forms in vivo needs to be sought to determine the precise functional role of the CLIC protein in the cell.
Implications of protein structure evolution via metamorphic intermediates
Evidence for the evolutionary adaptability of proteins is compelling not only in recent evolutionary events such as the emergence of drug resistance but also in the vast range of proteins that have presumably diverged from very few common ancestors. The traditional view that proteins possess absolute functional specificity and a single native structure generally correlates with a lack of versatility, and thus markedly conflicts with the ability to rapidly evolve new protein structures and functions. The key question is then, what role does the existence of metamorphic proteins have for the enrichment of pre-existing functional and structural diversity or the evolution of new protein folds?
It is well known that many amino acids can be mutated without significantly changing a protein’s overall structure and thus function. However, there are several examples within the literature that highlight the possibility that only a few point mutations or truncations can take a protein to a novel fold or through a new folding pathway (for example, see Alexander et al. 2009; Yadid et al. 2010). As discussed earlier, the demonstrated loss of a single conserved disulfide in lymphotactin is sufficient to facilitate Ltn-like metamorphic behaviour in a homologous chemokine, whilst the introduction of a single destabilising point mutation within lymphotactin is sufficient to lock Ltn in the non-canonical Ltn40 dimer fold (Tuinstra et al. 2008). An analogous series of mutations in vivo could lead to the formation of a protein lineage with a new functionality and unrelated fold, whilst maintaining significant sequence homology. Several examples of abrupt structural changes within a protein lineage that may be attributed to such metamorphic evolutionary intermediates have previously been identified. These include members of the Cro family of bacteriophage transcription factors (Murzin 2008; Roessler et al. 2008) and proteins of the Sfri0567-like family from different Shewanella species identified serendipitously through sequencing pipelines (Andreeva and Murzin 2010). Whilst we do not include proteins with high sequence similarity but distinctly different folds within our definition of the metamorphic protein class, these examples of abrupt structural differences within these protein lineages may have been mediated by metamorphic ancestor proteins without the loss of necessary ‘foldedness’ and function. This carries important implications for the current structural genomics approach and, in particular, homology modelling. It follows that protein homologues may not necessarily have the same three-dimensional structure despite the high conservation of ‘core’ residues andthus, sequence homology does not necessarily infer structural homology. However, such a role of metamorphic intermediates in mediating evolutionary transitions of structure and function still remains largely unexplored.
Identifying and characterising metamorphic proteins
Identifying and probing the structural transition of metamorphic proteins are key to understanding their functional and evolutionary importance. Structural biology has its roots in the study of globular proteins and until recently has primarily been targeted towards molecules exhibiting high levels of structural homogeneity. Hence, structural analysis typically starts with efforts to minimise conformational heterogeneity of proteins through trimming of flexible regions and purifying monodispersed species to aid in crystal formation. This has proven a very useful approach for studying structured proteins through the many large structural genomic consortiums. While conventional X-ray crystallography has successfully been used to solve the alternate structures of several metamorphic proteins, as already alluded to, this method relies on the ability to isolate and crystallise a single conformation. For example, for the CLIC1 protein, reducing and oxidising conditions used for protein purification were sufficient to trap the monomer and dimer folds, respectively, in near homogeneity. However, due to the very nature of metamorphic protein, i.e. their inherent flexibility and dynamism, metamorphic proteins will not naturally be amenable to crystal formation unless conditions favouring or trapping one conformation are established.
So, how do we identify metamorphic proteins and probe metamorphic structural transitions? At present, we lack any clear method from either a bioinformatics or structural approach to identify metamorphic proteins, and current examples of metamorphic proteins have only been discovered through good fortune. In light of the current concerted effort in structural genomics, new structures for proteins exhibiting high sequence homology but divergent structures associated with metamorphic behaviour may continue to be revealed through high-throughput structure determination pipeline studies. However, many more are likely to be by-passed and thus discarded early in the pipeline due to inherent disorder. For those proteins that are identified as possibly belonging to the metamorphic class, we need means of probing multiple conformations simultaneously.
Recent developments in NMR, including relaxation dispersion spectroscopy and paramagnetic relaxation enhancement, may be used as an alternate approach to characterise multiple metamorphic structures. The presence of more than one structure in solution can be revealed by these NMR approaches, in much the same way as they are currently used to render short-lived, sparsely populated states of the same protein (Clore 2011). However, methods which enable the ‘tracking’ of changes in structure between metamorphic conformations will perhaps prove more useful to probe metamorphic protein structure. Low resolution biophysical techniques, including fluorescence resonance energy transfer (FRET) and pulsed Electron Paramagnetic Resonance (EPR), can measure long-range distances and distance distributions within a single sample thereby enabling multiple fold information to be obtained simultaneously (Fajer et al. 2007). Alternatively, the observation of individual molecules by methods such as single-molecule FRET avoids this averaging and allows, in principle, microscopic distributions of conformations and folding to be identified (Hwang et al. 2009). However, a technique to obtain high resolution snapshots of single molecules in motion has not yet been practically realized. Hence, despite the above approaches for pursuing alternate metamorphic structures, at present it appears that the ability to isolate a significant proportion of a single conformer is still essential for a full and accurate high-resolution assignment of alternate structures.
So where are we in understanding the structure and function of metamorphic proteins?
It is clear from the current examples of metamorphic proteins described here that the ability to reversibly transit between different, well-defined topologies under physiological conditions has evolved to fulfil a variety of biological functions. Currently, little is understood about the mechanism that drives the process of metamorphosis and the folding dynamics involved. The presented case studies of metamorphic proteins, although limited in number, are suggestive of this novel class possessing regions of inherent flexibility as well as an ability to avoid kinetic trapping in a misfolded state during conversion between the alternate metamorphic forms. It remains to be firmly established whether these identified features are indeed defining properties of the metamorphic protein family. Only through increasing our understanding of this metamorphic protein behaviour will we learn more about what is possible at the boundaries of protein structure and function.
Abbreviations
- CLIC
Chloride intracellular channel
- EPR
Electron paramagnetic resonance
- FRET
Fluorescence resonance energy transfer
- IDP
Intrinsically disordered proteins
- Ltn
Lymphotactin
- Mad2
Mitotic arrest deficient 2
- MG
Molten globule
- RCL
Serpin reactive centre loop
References
- Alexander PA, He Y, Chen Y, Orban J, Bryan PN. A minimal sequence code for switching protein structure and function. Proc Natl Acad Sci USA. 2009;106(50):21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Murzin AG. Structural classification of proteins and structural genomics: new insights into protein folding and evolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 10):1190–1197. doi: 10.1107/S1744309110007177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. The formation and stabilization of protein structure. Biochem J. 1972;128(4):737–749. doi: 10.1042/bj1280737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(96):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Anson ML, Mirsky AE. On some general properties of proteins. J Gen Physiol. 1925;9(2):169–179. doi: 10.1085/jgp.9.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin RL. Intermediates in protein folding reactions and the mechanism of protein folding. Annu Rev Biochem. 1975;44:453–475. doi: 10.1146/annurev.bi.44.070175.002321. [DOI] [PubMed] [Google Scholar]
- Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206(986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Wright PE. Biochemistry. How do proteins interact? Science. 2008;320(5882):1429–1430. doi: 10.1126/science.1158818. [DOI] [PubMed] [Google Scholar]
- Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20(4):482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni C, Sutto L. Lymphotactin: how a protein can adopt two folds. J Chem Phys. 2009;131(24):245105. doi: 10.1063/1.3276284. [DOI] [PubMed] [Google Scholar]
- Chu R, Takei J, Knowlton JR, Andrykovitch M, Pei W, Kajava AV, Steinbach PJ, Ji X, Bai Y. Redesign of a four-helix bundle protein by phage display coupled with proteolysis and structural characterization by NMR and X-ray crystallography. J Mol Biol. 2002;323(2):253–262. doi: 10.1016/S0022-2836(02)00884-7. [DOI] [PubMed] [Google Scholar]
- Clore GM. Exploring sparsely populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Sci. 2011;20(2):229–246. doi: 10.1002/pro.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Littler DR, Brown LJ, Mazzanti M, Breit SN, Curmi PM, Dulhunty AF, Board PG, Parker MW. Structure of the Janus protein human CLIC2. J Mol Biol. 2007;374(3):719–731. doi: 10.1016/j.jmb.2007.09.041. [DOI] [PubMed] [Google Scholar]
- Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov S, Ptitsyn OB. Alpha-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981;136(2):311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- Dulhunty A, Gage P, Curtis S, Chelvanayagam G, Board P. The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J Biol Chem. 2001;276(5):3319–3323. doi: 10.1074/jbc.M007874200. [DOI] [PubMed] [Google Scholar]
- Fajer PG, Brown LJ, Song L. Chapter 4. Practical pulsed dipolar ESR (DEER) In: Hemminga MA, Berliner LJ, editors. ESR spectrocopy in membrane biophysics. New York: Springer; 2007. [Google Scholar]
- Fanucchi S, Adamson RJ, Dirr HW. Formation of an unfolding intermediate state of soluble chloride intracellular channel protein CLIC1 at acidic pH. Biochemistry. 2008;47(44):11674–11681. doi: 10.1021/bi801147r. [DOI] [PubMed] [Google Scholar]
- Feng H, Zhou Z, Bai Y. A protein folding pathway with multiple folding intermediates at atomic resolution. Proc Natl Acad Sci USA. 2005;102(14):5026–5031. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102(12):4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Goodchild SC, Howell MW, Cordina NM, Littler DR, Breit SN, Curmi PM, Brown LJ. Oxidation promotes insertion of the CLIC1 chloride intracellular channel into the membrane. Eur Biophys J. 2009;39(1):129–138. doi: 10.1007/s00249-009-0450-0. [DOI] [PubMed] [Google Scholar]
- Goodchild SC, Howell MW, Littler DR, Mandyam RA, Sale KL, Mazzanti M, Breit SN, Curmi PM, Brown LJ. Metamorphic response of the CLIC1 chloride intracellular ion channel protein upon membrane interaction. Biochemistry. 2010;49(25):5278–5289. doi: 10.1021/bi100111c. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18(10):1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Harrop SJ, DeMaere MZ, Fairlie WD, Reztsova T, Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Jankova L, Warton K, Bauskin AR, Wu WM, Pankhurst S, Campbell TJ, Breit SN, Curmi PM. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution. J Biol Chem. 2001;276(48):44993–45000. doi: 10.1074/jbc.M107804200. [DOI] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150(6):1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington JA, Whisstock JC. Molecular contortionism - on the physical limits of serpin 'loop-sheet' polymers. Biol Chem. 2010;391(8):973–982. doi: 10.1515/BC.2010.085. [DOI] [PubMed] [Google Scholar]
- Hwang LC, Hohlbein J, Holden SJ, Kapanidis AN. Chapter 5. Single-molecule FRET: methods and biological applications. In: Hinterforfer P, van Oijen A, editors. Handbook of single-molecule biophysics. New York: Springer; 2009. [Google Scholar]
- Johnson LN, Phillips DC. Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Angstrom resolution. Nature. 1965;206(986):761–763. doi: 10.1038/206761a0. [DOI] [PubMed] [Google Scholar]
- Kaiserman D, Whisstock JC, Bird PI. Mechanisms of serpin dysfunction in disease. Expert Rev Mol Med. 2006;8(31):1–19. doi: 10.1017/S1462399406000184. [DOI] [PubMed] [Google Scholar]
- Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181(4610):662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- Lad L, Lichtsteiner S, Hartman JJ, Wood KW, Sakowicz R. Kinetic analysis of Mad2-Cdc20 formation: conformational changes in Mad2 are catalyzed by a C-Mad2-ligand complex. Biochemistry. 2009;48(40):9503–9515. doi: 10.1021/bi900718e. [DOI] [PubMed] [Google Scholar]
- Landry DW, Reitman M, Cragoe EJ, Jr, Al-Awqati Q. Epithelial chloride channel. Development of inhibitory ligands. J Gen Physiol. 1987;90(6):779–798. doi: 10.1085/jgp.90.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal C. How to fold graciously. In: DeBrunner JTP, Munck E, editors. Mossbauer spectroscopy in biological systems: proceedings of a meeting held at Allerton House, Monticello. Illinois: University of Illinois Press; 1969. pp. 22–24. [Google Scholar]
- Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, Mazzanti M, Breit SN, Curmi PM. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004;279(10):9298–9305. doi: 10.1074/jbc.M308444200. [DOI] [PubMed] [Google Scholar]
- Littler DR, Assaad NN, Harrop SJ, Brown LJ, Pankhurst GJ, Luciani P, Aguilar MI, Mazzanti M, Berryman MA, Breit SN, Curmi PM. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J. 2005;272(19):4996–5007. doi: 10.1111/j.1742-4658.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- Littler DR, Harrop SJ, Brown LJ, Pankhurst GJ, Mynott AV, Luciani P, Mandyam RA, Mazzanti M, Tanda S, Berryman MA, Breit SN, Curmi PM. Comparison of vertebrate and invertebrate CLIC proteins: the crystal structures of Caenorhabditis elegans EXC-4 and Drosophila melanogaster DmCLIC. Proteins. 2008;71(1):364–378. doi: 10.1002/prot.21704. [DOI] [PubMed] [Google Scholar]
- Littler DR, Brown LJ, Breit SN, Perrakis A, Curmi PM. Structure of human CLIC3 at 2 A resolution. Proteins. 2010;78(6):1594–1600. doi: 10.1002/prot.22675. [DOI] [PubMed] [Google Scholar]
- Littler DR, Harrop SJ, Goodchild SC, Phang JM, Mynott AV, Jiang L, Valenzuela SM, Mazzanti M, Brown LJ, Breit SN, Curmi PM. The enigma of the CLIC proteins: ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010;584(10):2093–2101. doi: 10.1016/j.febslet.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16(11):1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7(3):224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9(1):59–71. doi: 10.1016/S1097-2765(01)00435-X. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11(4):338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Maury CP. The emerging concept of functional amyloid. J Intern Med. 2009;265(3):329–334. doi: 10.1111/j.1365-2796.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- Mirsky AE, Pauling L. On the structure of native, denatured, and coagulated proteins. Proc Natl Acad Sci USA. 1936;22(7):439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320(5884):1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- Mynott AV, Harrop SJ, Brown LJ, Breit SN, Kobe B, Curmi PM. Crystal structure of importin-alpha bound to a peptide bearing the nuclear localisation signal from chloride intracellular channel protein 4. FEBS J. 2011;278(10):1662–1675. doi: 10.1111/j.1742-4658.2011.08086.x. [DOI] [PubMed] [Google Scholar]
- Nar H, Bauer M, Stassen JM, Lang D, Gils A, Declerck PJ. Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation. J Mol Biol. 2000;297(3):683–695. doi: 10.1006/jmbi.2000.3604. [DOI] [PubMed] [Google Scholar]
- Ohgushi M, Wada A. ‘Molten-globule state’: a compact form of globular proteins with mobile side-chains. FEBS Lett. 1983;164(1):21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14(1):70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Rossman MG, Cullis AF, Muirhead H, Will G, North ACT. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A resolution, obtained by x-ray analysis. Nature. 1960;185(4711):416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Redhead CR, Edelman AE, Brown D, Landry DW, al-Awqati Q. A ubiquitous 64-kDa protein is a component of a chloride channel of plasma and intracellular membranes. Proc Natl Acad Sci USA. 1992;89(9):3716–3720. doi: 10.1073/pnas.89.9.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci USA. 2008;105(7):2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JJ, Wood S, Shorter J, Englander SW, Black BE. The Mad2 partial unfolding model: regulating mitosis through Mad2 conformational switching. J Cell Biol. 2008;183(5):761–768. doi: 10.1083/jcb.200808122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout TJ, Graham H, Buckley DI, Matthews DJ. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry. 2000;39(29):8460–8469. doi: 10.1021/bi000290w. [DOI] [PubMed] [Google Scholar]
- Stoychev SH, Nathaniel C, Fanucchi S, Brock M, Li S, Asmus K, Woods VL, Jr, Dirr HW. Structural dynamics of soluble chloride intracellular channel protein CLIC1 examined by amide hydrogen-deuterium exchange mass spectrometry. Biochemistry. 2009;48(35):8413–8421. doi: 10.1021/bi9010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh KS, Mutoh M, Nagashima K, Fernandez-Salas E, Edwards LE, Hayes DD, Crutchley JM, Marin KG, Dumont RA, Levy JM, Cheng C, Garfield S, Yuspa SH. The organellular chloride channel protein CLIC4/mtCLIC translocates to the nucleus in response to cellular stress and accelerates apoptosis. J Biol Chem. 2004;279(6):4632–4641. doi: 10.1074/jbc.M311632200. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420(6911):43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- Tanford C, Pain RH, Otchin NS. Equilibrium and kinetics of the unfolding of lysozyme (muramidase) by guanidine hydrochloride. J Mol Biol. 1966;15(2):489–504. doi: 10.1016/S0022-2836(66)80123-7. [DOI] [PubMed] [Google Scholar]
- Tompa P. Structure and function of intrinsically disordered proteins. Boca Raton: Chapman & Hall/CRC Press; 2009. [Google Scholar]
- Tuinstra RL, Peterson FC, Elgin ES, Pelzek AJ, Volkman BF. An engineered second disulfide bond restricts lymphotactin/XCL1 to a chemokine-like conformation with XCR1 agonist activity. Biochemistry. 2007;46(10):2564–2573. doi: 10.1021/bi602365d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci USA. 2008;105(13):5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman BF, Liu TY, Peterson FC. Chapter 3. Lymphotactin structural dynamics. Methods Enzymol. 2009;461:51–70. doi: 10.1016/S0076-6879(09)05403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proc Natl Acad Sci USA. 2010;107(16):7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455(7217):1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298(5597):1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- Zwanzig R, Szabo A, Bagchi B. Levinthal’s paradox. Proc Natl Acad Sci USA. 1992;89(1):20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]