Abstract
Imaging mass spectrometry (IMS) is two-dimensional mass spectrometry to visualize the spatial distribution of biomolecules, which does not need either separation or purification of target molecules, and enables us to monitor not only the identification of unknown molecules but also the localization of numerous molecules simultaneously. Among the ionization techniques, matrix assisted laser desorption/ionization (MALDI) is one of the most generally used for IMS, which allows the analysis of numerous biomolecules ranging over wide molecular weights. Proper selection and preparation of matrix is essential for successful imaging using IMS. Tandem mass spectrometry, which is referred to MSn, enables the structural analysis of a molecule detected by the first step of IMS. Applications of IMS were initially developed for studying proteins or peptides. At present, however, targets of IMS research have expanded to the imaging of small endogenous metabolites such as lipids, exogenous drug pharmacokinetics, exploring new disease markers, and other new scientific fields. We hope that this new technology will open a new era for biophysics.
Electronic supplementary material
The online version of this article (doi:10.1007/s12551-009-0015-6) contains supplementary material, which is available to authorized users.
Keywords: Imaging mass spectrometry (IMS), Imaging, Matrix assisted laser desorption/ionization (MALDI), Time-of-flight mass spectrometer (TOF-MS), Principal component analysis (PCA)
Introduction
Since the invention of the microscope, scientists have improved the microscope itself and its diverse peripheral technologies, including laser technology and imaging technology, to enhance the contrast and resolution in viewing samples. Morphological abnormalities in tissue samples observed by traditional microscopes do not provide us with any biochemical information. On the other hand, conventional biochemical techniques generally lose the positional information of the tissues. If these two aspects of information can be combined, we can determine the nature of the aberrant structures, thereby elucidating the pathogenesis or identifying the therapeutic targets. Imaging mass spectrometry (IMS) can successfully combine the two characteristics—detecting mass to visualize objects and enabling the determination of both biochemical and positional information of unknown molecules simultaneously. In this review, we briefly present the principle of conventional mass spectrometry (MS) and IMS. In addition, we also mention the recent applications of IMS.
Principle of MS
Mass spectrometry (MS) is an analytical technique that separates ionized particles such as atoms, molecules, and clusters by using differences in the ratios of their charges to their respective masses (mass/charge; m/z), and can be used to determine the molecular weight of the particles. MS instruments consist of the following modules: an ion source, which splits the sample molecules into ions; a mass analyzer, which sorts the ions according to their masses by applying electromagnetic fields; a detector, which measures the value of an indicator quantity and thus provides data for calculating the abundances of each ion present; and a computer, which regulates the mass analyzer and manages the data derived from the detector. In these modules, the prevailing techniques for the ion source and mass analyzer have several variations, and selecting their appropriate combination is particularly important for IMS. Therefore, we discuss here the recommended techniques for these two modules in the following paragraphs.
Among the several ionization methods, MALDI is one of the most generally used techniques for IMS. The basis of MALDI was developed by Koichi Tanaka (Nobel laureate in chemistry in 2002), and sophisticated by Franz Hillenkamp and Michael Karas (Karas and Hillenkamp 1988), allowing the analysis of large numbers of biomolecules ranging from small metabolite molecules (m/z < 1000) (Garrett et al. 2006; Khatib-Shahidi et al. 2006) to much larger proteins with molecular weights of 105 (Chaurand et al. 2006; Stoeckli et al. 2002), by covering them with a matrix and ionizing them using a pulse laser beam. High-molecular weight materials, such as proteins and DNA, are fragile and tend to be fragmented when ionized by conventional ionization methods. A matrix, which consists of crystallized molecules, such as 2,5-dihydroxybenzoic acid (DHB) and sinapinic acid (SA), are used to protect the biomolecules from being destroyed by the direct laser beam and to facilitate vaporization and ionization. An appropriate choice of the matrix is also critical for obtaining good mass spectra.
The most general analyzer for IMS is the time-of-flight (TOF) analyzer. The pulsatile ions generated in the ion source are separated in the vacuum analyzer according to the time of flight, and lighter ions reach the detectors faster than do the heavier ones; therefore, the machine can determine each ion’s atomic or molecular weight. Since extra gas molecules could scatter the sample ions during the flight, the interior of a mass analyzer is generally kept in a high-vacuum state by a turbomolecular pump and a diffusion pump. Tandem mass spectrometry, which are referred to MSn, enables the structural analysis of a molecule. In MS/MS, for example, isolated ionized molecules generated by MS1 are degraded and fragmented by collision with a noble gas or by other methods; the fragments are then analyzed by MS2. Molecular identification of a compound is achieved by comparison of the molecular weights of the compound and the fragmented ions obtained by MSn with those in a preexisting database. A large molecule such as a protein can be analyzed as a composition of the digested fragments, i.e., peptides.
Principle of IMS
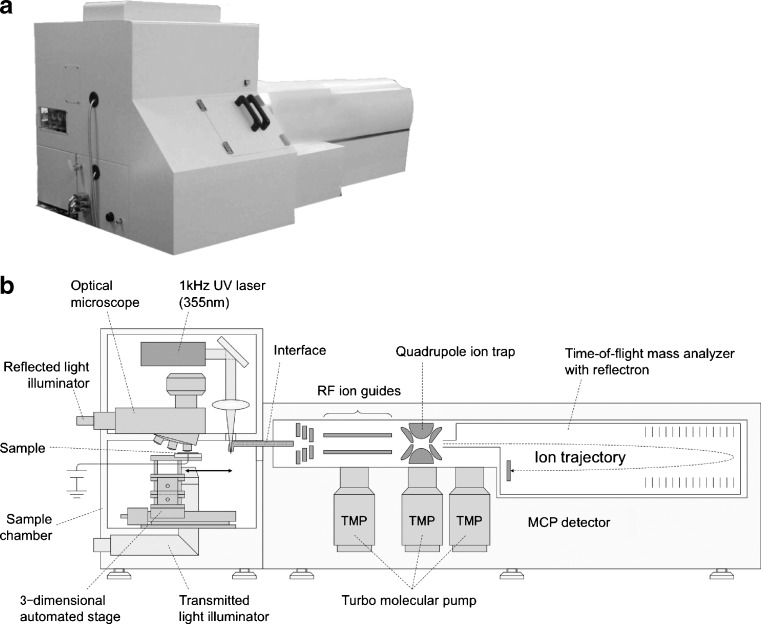
IMS is a technique that visualizes the spatial distribution of molecules and structures by their molecular masses. Tissue samples are sliced into thin sections and sprayed with the MALDI matrix, which forms uniform microcrystals, and are ionized by precise beam exposure to the regions of interest. Two-dimensional laser scanning generates images of numerous signals on the sample surface. In the 1990s, since the primitive IMS techniques had a resolution of only 0.1–1 mm, they were not known as real “microscopes.” In this century, however, IMS microscopes exceeded naked eye resolution, meaning that true IMS was successfully established. At present, our β prototype of the atmospheric pressure IMS (Fig. 1) provides an accuracy of stage movement of less than 1 μm and a spot diameter of 5 μm by independent regulation of the observation optic axis and the laser optic axis (Setou et al. 2008a). Conventional MS requires the separation and purification of the target molecules, resulting in the loss the important positional information, i.e., the distribution and localization of the target molecules. Meanwhile, IMS visualizes the distribution of the target molecule in the tissue section by extracting the mass spectra of the target from thousands of mass spectra obtained from MS analysis. Since the number of mass spectra with positional information is over 10,000 per each sample, it is essential to develop software to analyze the enormous spectra rapidly and efficiently.
Fig. 1.
The β prototype of the atmospheric pressure imaging mass spectrometry (IMS). a Overview of the β prototype. b Schematic diagram of the atmospheric pressure IMS
For accurately determining the distribution of molecules, it is also important to develop more sensitive instruments and appropriate sample preparation and treatment methods for efficient ionization because the amount of a specific molecule in each cell is extremely small. Ionization efficiency is partly dependent on the thickness of tissue sections. Less than 10-μm-thick sections are ideal for high-molecular weight proteins to be ionized efficiently and can be detected with low noise (Sugiura et al. 2006a).
Thin sections are usually prepared on a disposable plastic sheet or glass slide coated with a series of conductive materials. In particular, a plastic sheet with an indium-tin-oxide (ITO) film—a 75- to 125-μm-thick transparent and electric conductive PET basis sheet with vapor deposition on 5–15 nm of ITO (Chaurand et al. 2004; Shimma et al. 2008)—is useful because it has superior optical transparency sufficient for microscopic observation. In addition, an ITO sheet or glass slide provides mass spectra with high quality.
An appropriate sample processing method is necessary to maintain the positional information of biomolecules. For example, proteins have to be denatured and digested on a tissue section to prevent their diffusion. This method is called the on-tissue digestion method (Shimma et al. 2006b). After proteins have been denatured by detergent and heat, small amounts of trypsin solution are spotted by a chemical inkjet printer (Shimadzu Corporation), digesting the proteins into smaller peptides that are detectable by IMS (Setou et al. 2008b). To prepare a sample suitable to this method, a new sample preparation method was devised to transfer tissue sections onto the polyvinylidene difluoride (PVDF) membrane. A PVDF membrane is useful for denaturing and reducing proteins to facilitate trypsin digestion (Shimma et al. 2006a) as well as for removing salt from a tissue (Shimma et al. 2006b).
Matrix preparation is also essential for obtaining a good result from IMS. The spray-droplet method, which optimizes the recrystallization of the matrix, is recommended when especially high sensitivity is required (Sugiura et al. 2006b). Since the size of the matrix crystal and the migration of analytes can decrease the resolution in MALDI, a nanoparticle-assisted laser desorption/ionization (nano-PALDI)-based IMS was developed, which visualizes lipids and peptides at a resolution of 15 µm in mammalian tissues (Taira et al. 2008).
Applications
Applications of IMS were initially for studying proteins or peptides (Caprioli et al. 1997; Chaurand et al. 1999; Stoeckli et al. 2001). However, recently, research directed toward the detection and imaging of small organic molecules has been expanding. In the following sections, we introduce IMS applications for studying various metabolites in histological researches from recent studies.
IMS of metabolites
Since phospholipids, glycolipids, and cholesterol are abundant in living organisms and are easily ionized, there are many reports of IMS for these molecules as endogenous metabolites (Garrett et al. 2006; Jackson et al. 2005, 2007; Puolitaival et al. 2008).
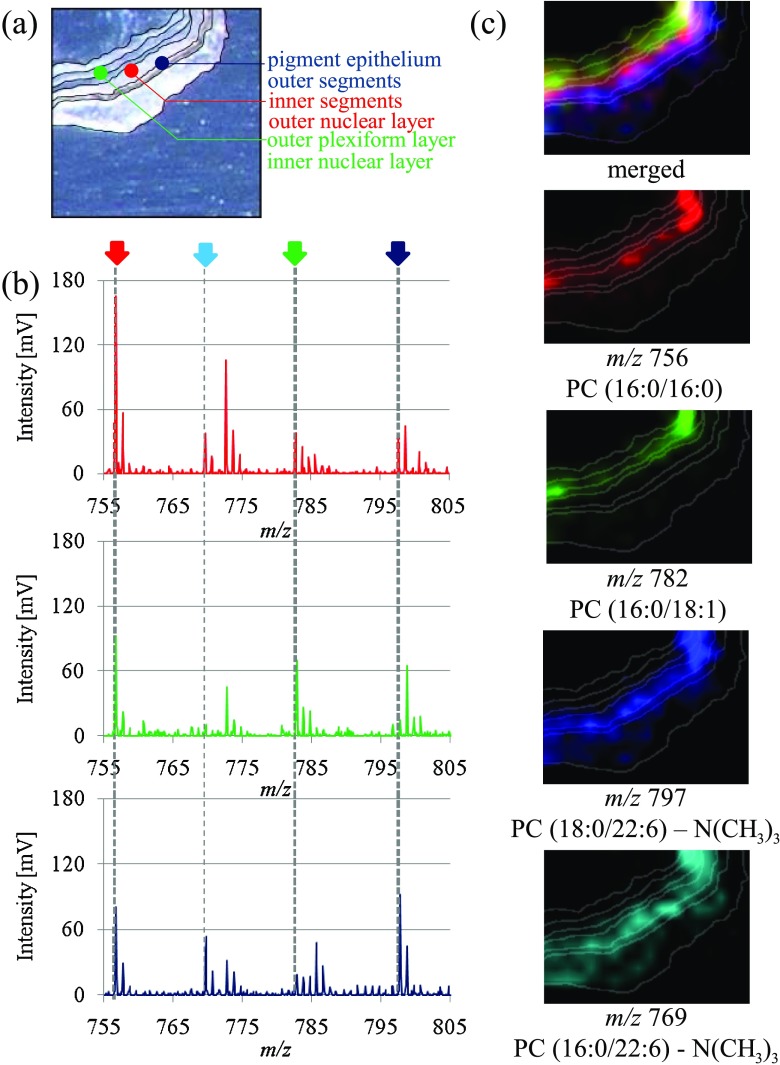
Phospholipids are major components of cell membrane structures and are involved in various biological processes such as cell proliferation, differentiation, cell-cell recognition, and signaling (Hannun and Bell 1989). To understand the distribution and functions of phospholipids is important because many diseases have been shown to be related to phospholipids. Phosphatidylcholines (PCs) are one of the major phospholipids species in the cell membrane and have two fatty acids, whose combination decides their biological activities. The distribution pattern of PCs has been revealed by IMS analysis in mouse brain (Sugiura et al. 2009) and retina (Hayasaka et al. 2008), which has nine layers of distinct membrane structures with a total width of 150 μm. Numerous signals were obtained from a retinal tissue section by laser scanning with a measurement pitch of 50 μm. MS/MS analyses have identified some signals as PCs. We found that each layer has different kinds of fatty acids bound to PCs and succeeded in roughly categorizing them into three groups of layer segments according to their composition of fatty acids, i.e., segment A: the pigment epithelium and outer segment; segment B: the inner segment and outer nuclear layer; and segment C: the outer plexiform layer and inner nuclear layer (Fig. 2). Docosahexaenoate (DHA), dominantly localized in segments A and B, is a major target of lipid peroxidation, which contributes to several diseases (Ford et al. 2008; Pan et al. 2008; Suzuki et al. 2007). In addition, we first reported the colocalization of DHA-PC and rhodopsin in vivo, which has an important role in phototransduction in the retina; their in vitro association has been discussed before (Aveldano 1988). This is a fascinating result that importantly represents the effectiveness of IMS (Hayasaka et al. 2008).
Fig. 2.
Distribution of PC molecular species in a mouse retinal section. a Each area can be roughly distinguished in this optical image of the mouse retinal section, and three colored dots are shown. b Three spectra between m/z 755 and 805 from the measurement areas at the blue dot (segment A: outer segment and pigment epithelium), green dot (segment B: inner nuclear layer and outer plexiform layer), and red dot (segment C: outer nuclear layer and inner segment) in the mouse retinal section are compared. The signal intensities at m/z 756.5 [PC diacyl-(16:0/16:0)] and m/z 782.7 [PC diacyl-(16:0/18:1)] were the highest in the red dot and green dot, respectively. The signal intensities at m/z 797.4 [PC diacyl-(18:0/22:6)] and m/z 769 [PC diacyl-(16:0/22:6)] were the highest in the blue dot. c The ion image merged from [PC diacyl-(16:0/16:0)] (red), [PC diacyl- (16:0/18:1)] (green), and [PC diacyl-(18:0/22:6)] (blue) revealed the three-zone distribution of the retinal section. [PC diacyl-(16:0/22:6)] (light blue) was distributed in the same region as [PC diacyl- (18:0/22:6)]. DHA, which consists of both [PC diacyl-(16:0/22:6)] and [PC diacyl-(18:0/22:6)] is dominantly localized in segments A and B. Reprinted from Hayasaka et al. (2008) with permission from Wiley InterScience
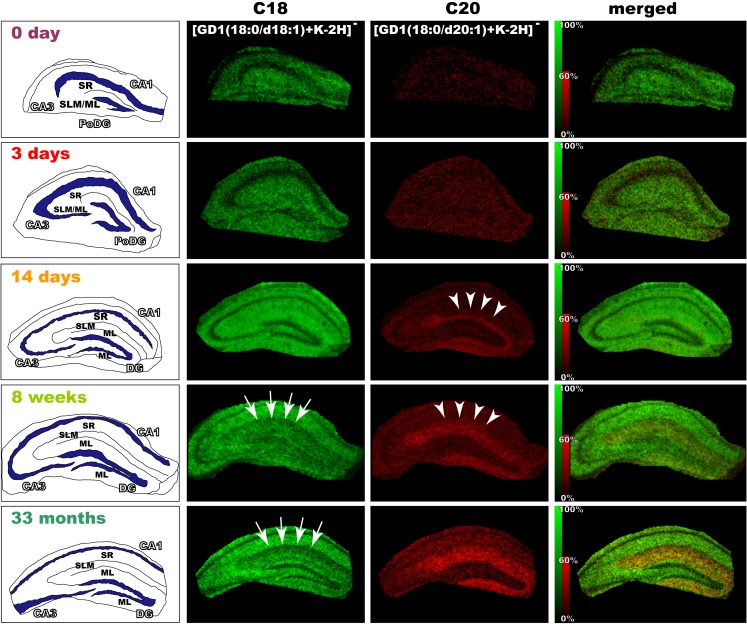
Ganglioside, a glycosphingolipid consisting of sialyated oligosaccharide and ceramide, is difficult to visualize in its entirety by a traditional light microscope, which is able to distinguish the different species of certain oligosaccharides by using antibodies; however, immunological methods cannot detect the differences in the ceramide structure. IMS, however, identifies both the oligosaccharide and ceramide moieties at the same time. IMS observation has revealed that there are two kinds of gangliosides with different fatty acid lengths in the ceramide moiety in the mouse brain. These two molecules show different distributions in the hippocampal formation and in the entorhinal cortex (Sugiura et al. 2008); moreover, a ganglioside with a longer fatty acid chain showed aging-related accumulation in the mouse hippocampal formation (Sugiura et al. 2008) (Fig. 3).
Fig. 3.
Development and age-related accumulation of di-sialylated ganglioside with C20-sphingosine (C20-GD1) in mouse hippocampal formation. We visualized the ion corresponding to GD1 (m/z 1874 and 1902) in the mouse hippocampus at the indicated time points (P0, P3, P14, 1 month, and 33 months). In the P14 mouse hippocampus, C20-GD1 was concentrated in the narrow area of the dentate gyrus-stratum lacunosum moleculare (DG-SLM) and began to spread over the medial edge of the region (arrow heads). In contrast, the concentration of the C-18 species decreased in the molecular layer (ML)/SLM with aging (arrows). Reprinted from Sugiura et al. (2008)
Although IMS for tissue sections is suitable for the analysis of principal components, it is difficult to detect minor constituents or non-easily ionizable molecules because numerous molecules exist in the crude mixture of tissue samples. We succeeded in highly sensitive detection and detailed structural analysis of glycosphingolipids by using thin-layer chromatography (TLC) followed by IMS (TLC-IMS) (Goto-Inoue et al. 2008). This technique has achieved a 10-times higher sensitivity than the conventional primuline staining technique, and characterized the differences in ceramide structures. We have shown that a combination of IMS and a conventional analytical technology would be a powerful tool for the sensitive identification of the structures of minor molecules.
Principal component analysis (PCA)
PCA is a mathematical procedure that reduces a large set of variables to a small set of variables called principal factors, which are linear combinations of the original variables. PCA provides information regarding which molecules are altered in tissue microdomains and is very helpful in analyzing large datasets of imaging MS (Altelaar et al. 2007; Denkert et al. 2006; Lapolla et al. 2007; McCombie et al. 2005; Zaima et al. 2009b).
We have reported that a novel E3 ligase, Scrapper (SCR), regulates synaptic activity through protein degradation (Yao et al. 2007); further, based on the PCA of mass spectra from some brain regions of interest, we analyzed Scrapper-knockout mouse brains by IMS. We were able to find various alterations in the knockout mouse brain and obtain information on where the alterations occurred (Yao et al. 2008) (Fig. 4).
Fig. 4.
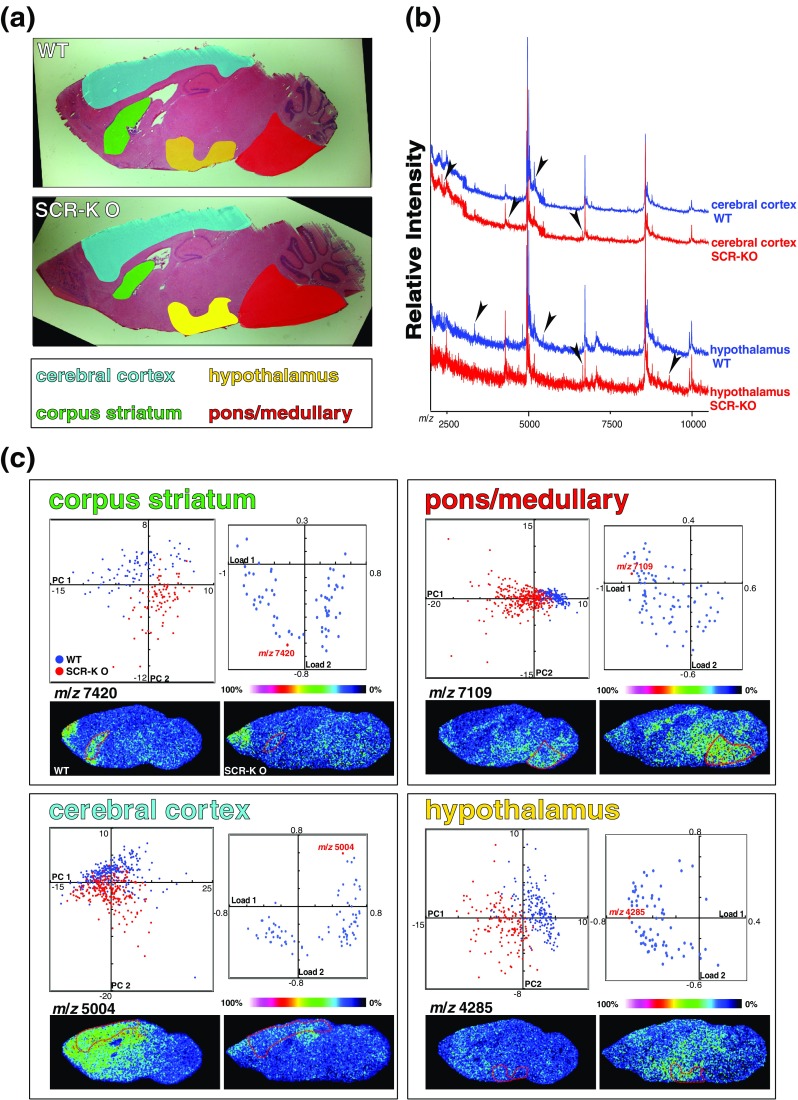
In situ proteomics of the SCR-KO mouse brain using imaging MS and PCA. a HE-stained images of the WT and SCR-KO mouse brain. The regions focused on in the imaging MS analyses are indicated in colors. b Mass spectra obtained from each region of the WT or SCR-KO mouse brain sections. Specific signals of the regions are indicated by arrowheads. c Distributions of principal component scores of mass spectra from various brain regions (left spray graphs; WT, blue; KO, red) and the loading factors plot (right graphs). The signal intensities of mass spectra of the substances with indicated m/z are shown in the reconstructed images of the mouse brain analyzed by imaging MS. Reprinted from Yao et al. 2008 with permission from Wiley-VCH
Our group recently reported another example of PCA using IMS, which was applied to the analysis of metabolites in starvation-induced fatty liver sections—a model of fatty liver caused by non-alcoholic steatohepatitis (NASH) (Zaima et al. 2009a). This approach is very convenient and can be applied to the analysis of human biopsy samples because it does not require complex pretreatments.
Exploration for biomedical markers
Analyzing molecular changes related to disease progression has been one of the primary purposes of IMS (Cornett et al. 2006; Schwartz et al. 2005; Skold et al. 2006; Stoeckli et al. 2006). By analyzing pathological tissues, we can explore principal markers that importantly influence pathological conditions. For example, biomarkers specific for various tumor tissues or neurological disorders could be made available along with their distribution patterns. We have reported the limited localization of sphingomyelin in the tumor area of colon cancer liver metastasis (Shimma et al. 2007). In contrast, Hem B, constituting hemoglobin, has showed low ion intensity in a cancerous region, consistent with more abundant blood supply in the normal liver (Shimma and Setou 2007).
Future perspective
We plan to visualize the subcellular distribution of proteins with post-translational modifications. IMS is the only technique that can analyze the status of post-translational modifications of proteins, such as ubiquitination (Hatanaka et al. 2006; Yao et al. 2007), polyglutamylation (Ikegami et al. 2007), polyglycylation (Ikegami et al. 2008; Ikegami and Setou 2009), tyrosination (Konishi and Setou 2009), and acetylation (Kahyo et al. 2008), because by using conventional MS, the positional information is lost. Moreover, we also plan to identify the constituents of inclusion bodies observed in neurodegenerative diseases such as Parkinson’s disease and Alzheimer’s disease.
IMS can be applied in small experimental animals such as the medaka (Oryzias latipes) (Zaima et al. 2009a), indicating that this powerful tool has already been connected to model organisms such as the fruit fly and nematode, allowing us to analyze every life process genetically in a rapid manner.
Since IMS can monitor both quantitative and positional information of numerous unidentified molecules simultaneously, pharmaceutical companies have already begun to use this technique for determining pharmacokinetics and pharmacological actions in drug development. In fact, some advanced researches of pharmacokinetics have already been reported (Atkinson et al. 2007; Stoeckli et al. 2006). Furthermore, since IMS provides us with various quantitative information as well as positional information, it will be applied not only to the biomedical field but also to the biophysical field in the future.
Conclusion
We have developed IMS and reported several applications for the same. IMS is an extremely powerful technique that allows us to directly detect morphological information of unidentified molecules without losing positional information or biochemical information. We hope that IMS will be widely applied in a variety of areas and will help in resolving numerous outstanding problems in the biological field.
Electronic supplemetary materials
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to our collaborators in the Hamamatsu University School of Medicine—Dr. S. Koizumi, Dr. T. Hayasaka, Dr. N. Goto-Inoue, Dr. I. Yao, Dr. N. Zaima, Dr. K. Ikegami, Dr. H. Tanaka, and Dr. Y. Morita. This work was supported by a Grant-in-Aid for SENTAN from the Japanese Science and Technology Agency (JST) and Young Scientists S (2067004) by Japan Society for the Promotion of Science (JSPS) to Mitsutoshi Setou.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12551-009-0015-6) contains supplementary material, which is available to authorized users.
References
- Altelaar AF, Taban IM, McDonnell LA, Verhaert PDEM, de Lange RP, Adan RAH, Mooi WJ, Heeren RM, Piersma SR. High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass Spectrom. 2007;260:293–211. [Google Scholar]
- Atkinson SJ, Loadman PM, Sutton C, Patterson LH, Clench MR. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1271–1276. doi: 10.1002/rcm.2952. [DOI] [PubMed] [Google Scholar]
- Aveldano MI. Phospholipid species containing long and very long polyenoic fatty acids remain with rhodopsin after hexane extraction of photoreceptor membranes. Biochemistry. 1988;27:1229–1239. doi: 10.1021/bi00404a024. [DOI] [PubMed] [Google Scholar]
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Stoeckli M, Caprioli RM. Direct profiling of proteins in biological tissue sections by MALDI mass spectrometry. Anal Chem. 1999;71:5263–5270. doi: 10.1021/ac990781q. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- Cornett DS, Mobley JA, Dias EC, Andersson M, Arteaga CL, Sanders ME, Caprioli RM. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol Cell Proteomics. 2006;5:1975–1983. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Konsgen D, Dietel M, Fiehn O. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- Ford DA, Monda JK, Brush RS, Anderson RE, Richards MJ, Fliesler SJ. Lipidomic analysis of the retina in a rat model of Smith-Lemli-Opitz syndrome: alterations in docosahexaenoic acid content of phospholipid molecular species. J Neurochem. 2008;105:1032–1047. doi: 10.1111/j.1471-4159.2007.05203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TJ, Prieto-Conaway MC, Kovtoun V, Bui H, Izgarian N, Stafford G, Yost RA. Imaging of small molecules in tissue sections with a new intermediate-pressure MALDI linear ion trap mass spectrometer. Int J Mass Spectrom. 2006;260:166–176. [Google Scholar]
- Goto-Inoue N, Hayasaka T, Sugiura Y, Taki T, Li YT, Matsumoto M, Setou M. High-sensitivity analysis of glycosphingolipids by matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight imaging mass spectrometry on transfer membranes. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;870:74–83. doi: 10.1016/j.jchromb.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Bell RM. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science. 1989;243:500–507. doi: 10.1126/science.2643164. [DOI] [PubMed] [Google Scholar]
- Hatanaka T, Hatanaka Y, Tsuchida J, Ganapathy V, Setou M. Amino acid transporter ATA2 is stored at the trans-Golgi network and released by insulin stimulus in adipocytes. J Biol Chem. 2006;281:39273–39284. doi: 10.1074/jbc.M604534200. [DOI] [PubMed] [Google Scholar]
- Hayasaka T, Goto-Inoue N, Sugiura Y, Zaima N, Nakanishi H, Ohishi K, Nakanishi S, Naito T, Taguchi R, Setou M. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun Mass Spectrom. 2008;22:3415–3426. doi: 10.1002/rcm.3751. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Setou M. TTLL10 can perform tubulin glycylation when co-expressed with TTLL8. FEBS Lett. 2009;583:1957–1963. doi: 10.1016/j.febslet.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, Campbell PK, Yuasa S, Janke C, Macgregor GR, Setou M. Loss of α-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci USA. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Horigome D, Mukai M, Livnat I, MacGregor GR, Setou M. TTLL10 is a protein polyglycylase that can modify nucleosome assembly protein 1. FEBS Lett. 2008;582:1129–1134. doi: 10.1016/j.febslet.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SN, Wang HY, Woods AS. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal Chem. 2005;77:4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Albert Schultz J, Woods AS. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahyo T, Mostoslavsky R, Goto M, Setou M. Sirtuin-mediated deacetylation pathway stabilizes Werner syndrome protein. FEBS Lett. 2008;582:2479–2483. doi: 10.1016/j.febslet.2008.06.031. [DOI] [PubMed] [Google Scholar]
- Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10, 000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Lapolla A, Ragazzi E, Andretta B, Fedele D, Tubaro M, Seraglia R, Molin L, Traldi P. Multivariate analysis of matrix-assisted laser desorption/ionization mass spectrometric data related to glycoxidation products of human globins in nephropathic patients. J Am Soc Mass Spectrom. 2007;18:1018–1023. doi: 10.1016/j.jasms.2007.02.011. [DOI] [PubMed] [Google Scholar]
- McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal Chem. 2005;77:6118–6124. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- Pan HZ, Zhang H, Chang D, Li H, Sui H. The change of oxidative stress products in diabetes mellitus and diabetic retinopathy. Br J Ophthalmol. 2008;92:548–551. doi: 10.1136/bjo.2007.130542. [DOI] [PubMed] [Google Scholar]
- Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SA, Weil RJ, Thompson RC, Shyr Y, Moore JH, Toms SA, Johnson MD, Caprioli RM. Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 2005;65:7674–7681. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- Setou M, Matsumoto M, Hosokawa N, Sugiura Y, Hayasaka T. Development of mass microscopy. Microscope. 2008;43:24–28. [Google Scholar]
- Setou M, Hayasaka T, Shimma S, Sugiura Y, Matsumoto M. Protein denaturation improves enzymatic digestion efficiency for direct tissue analysis using mass spectrometry. Appl Surf Sci. 2008;255:1555–1559. doi: 10.1016/j.apsusc.2008.05.120. [DOI] [Google Scholar]
- Shimma S, Setou M. Mass microscopy to reveal distinct localization of Heme B (m/z 616) in colon cancer liver metastasis. J Mass Spectrom Soc Jpn. 2007;55:230–238. [Google Scholar]
- Shimma S, Furuta M, Ichimura K, Yoshida Y, Setou M. A novel approach to in situ proteome analysis using chemical inkjet printing technology and MALDI-QIT-TOF tandem mass spectrometer. J Mass Spectrom Soc Jpn. 2006;54:133–140. [Google Scholar]
- Shimma S, Furuta M, Ichimura K, Yoshida Y, Setou M. Direct MS/MS analysis in mammalian tissue sections using MALDI-QIT-TOFMS and chemical inkjet technology. Surf Int Anal. 2006;38:1712–1714. doi: 10.1002/sia.2389. [DOI] [Google Scholar]
- Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855:98–103. doi: 10.1016/j.jchromb.2007.02.037. [DOI] [PubMed] [Google Scholar]
- Shimma S, Sugiura Y, Hayasaka T, Zaima N, Matsumoto M, Setou M. Mass imaging and identification of biomolecules with MALDI-QIT-TOF-based system. Anal Chem. 2008;80:878–885. doi: 10.1021/ac071301v. [DOI] [PubMed] [Google Scholar]
- Skold K, Svensson M, Nilsson A, Zhang X, Nydahl K, Caprioli RM, Svenningsson P, Andren PE. Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J Proteome Res. 2006;5:262–269. doi: 10.1021/pr050281f. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Staab D, Staufenbiel M, Wiederhold KH, Signor L. Molecular imaging of amyloid beta peptides in mouse brain sections using mass spectrometry. Anal Biochem. 2002;311:33–39. doi: 10.1016/S0003-2697(02)00386-X. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole body tissue sections. Int J Mass Spectrom. 2006;260:195–202. [Google Scholar]
- Sugiura Y, Shimma S, Setou M. Thin sectioning improves the peak intensity and signal-to-noise ratio in direct tissue mass spectrometry. J Mass Spectrom Soc Jpn. 2006;54:45–48. [Google Scholar]
- Sugiura Y, Shimma S, Setou M. Two-step matrix application technique to improve ionization efficiency for matrix-assisted laser desorption/ionization in imaging mass spectrometry. Anal Chem. 2006;78:8227–8235. doi: 10.1021/ac060974v. [DOI] [PubMed] [Google Scholar]
- Sugiura Y, Shimma S, Konishi Y, Yamada MK, Setou M. Imaging mass spectrometry technology and application on ganglioside study; visualization of age-dependent accumulation of C20-ganglioside molecular species in the mouse hippocampus. PLoS ONE. 2008;3:e3232. doi: 10.1371/journal.pone.0003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y, Konishi Y, Zaima N, Kajihara S, Nakanishi H, Taguchi R, Setou M (2009) Visualization of the cell-selective distribution of PUFA-containing phosphatidylcholines in mouse brain by imaging mass spectrometry. J Lipid Res (in press) [DOI] [PMC free article] [PubMed]
- Suzuki M, Kamei M, Itabe H, Yoneda K, Bando H, Kume N, Tano Y. Oxidized phospholipids in the macula increase with age and in eyes with age-related macular degeneration. Mol Vis. 2007;13:772–778. [PMC free article] [PubMed] [Google Scholar]
- Taira S, Sugiura Y, Moritake S, Shimma S, Ichiyanagi Y, Setou M. Nanoparticle-assisted laser desorption/ionization based mass imaging with cellular resolution. Anal Chem. 2008;80:4761–4766. doi: 10.1021/ac800081z. [DOI] [PubMed] [Google Scholar]
- Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, Fukuda Y, Chiba T, Morone N, Yuasa S, Inokuchi K, Ohtsuka T, Macgregor GR, Tanaka K, Setou M. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao I, Sugiura Y, Matsumoto M, Setou M. In situ proteomics with imaging mass spectrometry and principal component analysis in the Scrapper-knockout mouse brain. Proteomics. 2008;8:3692–3701. doi: 10.1002/pmic.200701121. [DOI] [PubMed] [Google Scholar]
- Zaima N, Hayasaka T, Goto-Inoue N, Setou M. Imaging of metabolites by MALDI mass spectrometry. J Oleo Sci. 2009;58:415–419. doi: 10.5650/jos.58.415. [DOI] [PubMed] [Google Scholar]
- Zaima N, Matsuyama Y, Setou M. Principal component analysis of direct matrix-assisted laser desorption/ionization mass spectrometric data related to metabolites of fatty liver. J Oleo Sci. 2009;58:267–273. doi: 10.5650/jos.58.267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.