Abstract
Anti-terminator proteins are frequently used by bacteria to sense a specific metabolite signal and direct RNA polymerase to either terminate or continue transcription of the genes downstream of an operon. One such protein is HutP, which binds to upstream cis-regulatory sequences to regulate expression of the histidine utilization (hut) operon in Bacillus subtilis. HutP must be activated by L-histidine and divalent metal ions before binding to hut mRNA; binding of activated HutP prevents termination of transcription. Thus, HutP appears to regulate the hut operon in a unique fashion in this class of regulatory proteins. To understand gene (hut operon) regulation by HutP, we performed several biochemical and structural studies. These studies reveal events in the regulatory mechanism, starting with the activation of HutP and ending with the unwinding of hut terminator RNA. In this review, we describe the unique regulatory mechanisms commonly used by many Bacillus species.
Keywords: Anti-termination, hut operon, Bacillus subtilis, Ribonucleoprotein complex, Crystal structure
Introduction
Bacteria utilize a variety of mechanisms for regulating gene expression at the posttranscriptional level in response to changes in the environment. Among these, a common pathway is the stabilization of alternative mRNA secondary structures by RNA-binding proteins. These proteins direct transcription either to pause or to proceed to the synthesis of the full-length transcript. RNA-binding proteins modulate the nascent mRNA in two ways, referred to as termination and anti-termination. Termination proteins bind to the nascent mRNA and prevent the formation of an alternative structure. Anti-termination proteins promote destabilization of the terminator structure. In the latter case, the nascent mRNA forms the terminator structure by default, while in the former case, the mRNA is set for transcription elongation. Several bacterial termination and anti-termination proteins that regulate nascent mRNA have been described in Escherichia coli and Bacillus subtilis, including TRAP, PyrR, LacT, BglG, SacT/SacY, GlpP, and HutP; these mRNAs regulate the trp, pyr, lac, bgl, sac, glpP and hut operons, respectively (Houman et al. 1990; Aymerich and Steinmetz 1992; Oda et al. 1992; Babitzke and Yanofsky 1993; Arnaud et al. 1996; Glatz et al. 1996; Lu et al. 1996; Alpert and Siebers 1997). To facilitate or destabilize terminator structures, regulatory proteins require specific ligands for activation before binding to their cognate RNAs (Table 1). For example, the trp RNA-binding attenuation protein (TRAP) is a well-characterized protein that inhibits the formation of the anti-terminator structure by modulating the leading region of mRNA (Babitzke and Yanofsky 1993). In contrast to the TRAP regulation mechanism, HutP binds directly to the terminator and stabilizes the anti-terminator structure. HutP regulates hut (histidine utilization) operon transcription by an anti-termination mechanism in Bacillus subtilis. HutP is unique in its regulation of mRNA in that it requires not only a specific ligand L-His (L-histidine) but also a divalent metal ion for its activation. In this review, we focus on this unique regulatory mechanism that is unraveled by structural and functional analyses
Table 1.
Proteins that regulate termination and anti-termination of transcription and their cognate ligands required for activation
| Operons | Proteins | Ligands required for activation |
|---|---|---|
| trp | TRAP | Tryptophan |
| pyr | PyrR | Uridine monophosphate |
| lac | LacT | Lactose |
| bgl | BglG | β-Glucosides |
| sac | SacT/SacY | Phosphoenol pyruvate |
| glpP | GlpP | Glycerol-3-phosphate |
| hut | HutP | L-Histidine |
The hut operon and HutP protein
The hut operon is responsible for the degradation of histidine to ammonia, glutamate and a one-carbon compound. Although it is not yet clear just how widely distributed the hut pathway is among bacteria, about 1,164 bacterial species are currently known to possess the histidine utilization (Hut) system (Bender 2012). The regulation of the hut pathway has been extensively studied in enteric bacteria (such as Klebsiella), Pseudomonas species, and Bacillus subtilis. However, the mechanisms of regulation in these bacteria differ significantly, as discussed in a recent review (Bender 2012). In our review, we concentrated primarily on the histidine utilization system that is operating in Bacillus subtilis. The hut genes in B. subtilis are located in a single hut operon which consists of five structural genes, namely, hutH, hutU, hutI, hutG, and hutM, which code for the histidase, urocanase, imidazolone propionate amino hydrolase, formimino l-glutamate formimino hydrolase, and histidine permease enzymes, respectively. In addition, there is a positive regulatory gene, hutP (Fig. 1a) (Chasin and Magasanik 1968; Kimmhi and Magasanik 1970; Oda et al. 1988; Yoshida et al. 1995). These five enzymes are responsible for the degradation of L-histidine and its use as a carbon and nitrogen source. The structural genes are located far downstream of the promoter, whereas the regulatory gene hutP is located much closer to the promoter. The nucleotide sequence located between the structural genes and hutP forms a stem-loop terminator structure (nucleotides +515 to +553) which is known to be modulated by HutP after HutP activation by L-histidine and divalent metal ions (Fig. 1b). Once the terminator structure is attenuated, the RNA polymerase can synthesize a full-length hut mRNA transcript, which is then translated into five structural proteins. HutP is a 16.2-kDa protein consisting of approximately 148 amino acid residues, depending upon the bacterial species. HutP is present in various Bacillus species, including highly pathogenic bacteria such as Bacillus anthracis. The amino acids at the C-terminus of HutP are highly conserved across Bacillus species compared to the N-terminus residues (Kumarevel et al. 2004a), suggesting that the C-terminus residues are important for HutP regulatory functions. HutP has no sequence similarity with other known transcription-regulating proteins, indicating that HutP controls transcription in a unique fashion. The HutP protein is essential for high levels of expression of the hut operon (Oda et al. 1988).
Fig. 1.
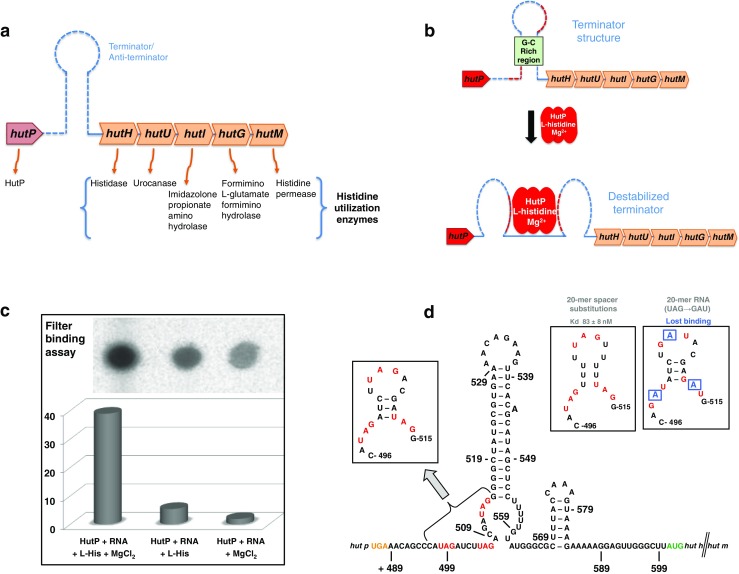
A histidine utilization (hut) operon of Bacillus subtilis and its regulation are mediated by the interaction of HutP (bacterial anti-termination protein which regulates hut) with hut terminator RNA in the presence of L-histidine and metal ions. a A schematic representation of the hut operon of B. subtilis. b The proposed hut mRNA terminator structure is known to be destabilized by activated HutP. Dashed lines Terminator region, red dashed lines within the terminator HutP binding regions. c Binding analyses to evaluate the role of L-histidine and divalent metal ions in the activation of HutP and binding to the hut terminator RNA. The binding assay was carried out with 200 nM of HutP and 1 nM unlabeled RNA (5′-CAUAGAUCUUAGACGAUAGGG-3′, 21-mer) containing the same RNA with 10,000 cpm of 5′ end-labeled product. To this reaction mixture 10 mM of L-histidine and 10 mM of MgCl2 were added in the case of test samples; for controls, either MgCl2 or L-histidine was omitted. Upon completion of a 1-h incubation at 25 °C, the reaction mixture was filtered, washed, and quantitated similar to that reported previously (Kumarevel et al. 2004a). d Identification of activated HutP binding sites within the hut terminator RNA region. Letters in red Important residues (UAG) within the terminator region. Shorter (20 mer) RNA fragments were analyzed for HutP binding to define the binding region and to identify the important nucleotides for HutP recognition
Essential ligands for activating HutP and binding to hut mRNA
Earlier studies showed that HutP required L-histidine for binding to the hut terminator (Oda et al. 2000; Kumarevel et al. 2004a), suggesting that L-histidine controls the binding of HutP to the terminator by modifying the conformational state of HutP. Kumarevel et al. (2004a) observed that 10 mM L-histidine was required for the complete activation of HutP and for efficient binding to hut mRNA. To address which chemical groups of L-histidine were important for the activation of HutP, these authors screened analogs of L-histidine for their ability to activate HutP and induce binding to RNA. Among the analogs tested, L-histidine β-naphthylamide (HBN) and L-histidine benzylester showed tenfold higher affinity than unmodified L-histidine. Other analogs, such as L-histidine methyl ester and L-β-imidazole lactic acid, showed similar activation as L-histidine. In contrast, urocanic acid, histamine, and L-histidinamide showed only weak activation of HutP. The presence of imidazole alone or of several analogs that were modified at the imidazole ring failed to activate HutP for binding to the terminator RNA. These analyses suggested that the imidazole ring and the backbone of L-histidine are essential for activating HutP (Kumarevel et al. 2004a).
To evaluate the divalent metal ion requirement for the HutP–RNA interaction, an important step was to remove the endogenous divalent metal ions by denaturing the protein in 7 M urea followed by refolding HutP in binding buffer without any divalent metal ions. Kumarevel et al. (2005a) used this treated HutP protein to analyze HutP binding to the terminator RNA in the presence and absence of divalent metal ions (Fig. 1c). The results of these binding analyses suggested that HutP requires not only L-histidine but also divalent ions such as MgCl2. To address which other divalent metal ions support the activation of HutP, these authors then screened 15 divalent metal ions, differing in ionic radii and preferred coordination, at a concentration of 10 mM and observed that Mn2+, Zn2+, and Cd2+ were the most efficient ions, followed by Mg2+, Co2+, and Ni2+. The other divalent metal ions, which only weakly supported HutP activation, were Ba2+, Sr2+, Pt2+, Ca2+, Pb2+, and Ag2+. The divalent metal ions Cu2+, Yb2+ and Hg2+ and monovalent metal ions such as NaCl (100 mM) and KCl (100 mM) failed to activate HutP for RNA binding (Kumarevel et al. 2005a).
HutP protein interaction with hut mRNA
The hut mRNA between nucleotides +459 to +572 was found to fold into alternative conformations (Oda et al. 2000). These authors also observed that the hut mRNA appeared to form a stable terminator structure (nucleotides +498 to +572) in the absence of activated HutP, and that the region between nucleotides +459 to +537 seemed to form a destabilized RNA structure (anti-terminator structure) upon binding of activated HutP. To map the HutP binding site between nucleotides +459 to +537, these authors analyzed shorter RNAs of varying lengths. They initially found that nucleotides +459 to +572 were sufficient for recognizing activated HutP, but further deletion analyses revealed the a 20-mer RNA (between nucleotides +496 to +515, 5′-CAUAGAUCUUAGACGAUAGGG-3′) was the shortest that was sufficient for efficient recognition of activated HutP (Oda et al. 2000) (Fig. 1d). Interestingly, in this region, three UAG repeats and four non-conserved spacer residues were found (Kumarevel et al. 2004a). Upon further trimming to UAG, AUAG, UAGA, or UUUAGUU or when UAG was reversed in the 20-mer RNA (5′-CAGAUAUCUGAUACGAGAUGG-3′), the HutP failed to bind. These studies show that the UAG sequence is important and that efficient recognition requires multiple UAG repeats to facilitate cooperative binding (Kumarevel et al. 2004a). To understand the role of the four-spacer nucleotides, Kumarevel et al. (2004b) carried out base substitutions to replace the spacers with four U nucleotides. These authors found that the spacers had only a limited role in the recognition of activated HutP. Upon substitution of other bases for the A and G within the UAG motifs, HutP failed to recognize the mutated RNAs. Taken together, these studies reveal that the activated HutP core recognition motif is XAG, where X indicates any base. To clarify which functional groups of the A and G residues were important, site-specific base modifications were introduced into the 20-mer RNA and analyzed for binding, revealing that the 6-amino moiety of the A base and the 2-amino moiety of the G base at the second and third positions, respectively, were the key functional groups for HutP recognition. Further analyses of the chemical groups within the UAG motif unveiled only one 2′ OH of the A base that was important for HutP recognition (Kumarevel et al. 2004b).
Crystal structures of HutP
To understand regulation of the hut operon by HutP, a number of studies were carried out to resolve a series of crystal structures of HutP and its complexes (Table 2). These complexes included apo-HutP, HutP-L-His, HutP-L-His-Mg2+ (HutP-L-His-Mn2+, HutP-L-His-Ba2+, HutP-L-His-Zn2+), HutP-L-His-Mg2+-RNA (21 mer), and HutP-L-His-Mg2+-RNA (55 mer, a model RNA), as summarized in Table 2. The HutP-Mg2+ complex was also resolved, but no specific binding site for Mg2+ was found, suggesting that the structure of the HutP-Mg2+ complex is the same as that of apo-HutP. Among these crystal structures, the highest resolution structure was 1.48 Å, obtained for the quaternary complex (HutP-L-His-Mg2+-RNA). For clarity, crystal structure details of free HutP and the HutP complexes are described separately in the following sections.
Table 2.
Summary of the crystallographic data collected for HutP in the presence and absence of different ligands
| HutP complex | Space group | Unit cell (Å, °) | Number of molecules In ASU | Resolution (Å) | PDB code | Reference |
|---|---|---|---|---|---|---|
| Apo-HutP | P213 | a = b = c = 96.82 | 2 | 2.78 | 1WPS | Kumarevel et al. 2005b |
| HutP- Mg2+ | P213 | a = b = c = 95.41 | 2 | 2.6 | 1WPT | Kumarevel et al. 2005a |
| HutP-HBN | P213 | a = b = c = 95.41 | 2 | 2.8 | 1VEA | Kumarevel et al. 2004a |
| HutP-L-His-Mg2+ | P21212 | a = 77.79, b = 80.88, c = 75.17 | 3 | 1.70 | 1WPV | Kumarevel et al. 2005b |
| HutP-L-His-Mn2+ | P21212 | a = 77.76, b = 81.40, c = 76.04 | 3 | 2.2 | 1WRN | Kumarevel et al. 2005a |
| HutP-L-His-Ba2+ | P21212 | a = 78.22, b = 81.14, c = 75.97 | 3 | 2.35 | 1WRO | Kumarevel et al. 2005a |
| HutP-L-His-Zn2+ | P3 | a = b = 129.33, c = 76.67, γ = 120.0 | 12 | 2.5 | 4H4L | Balasundaresan et al. 2013 |
| HutP-L-His-Mg2+ -RNA (21 mer) | H3 | a = b = 75.80, c = 133.35, γ = 120.0 | 2 | 2.20 | 1WRQ | Unpublished |
| HutP-L-His-Mg2+ -RNA (21 mer) | R3 | a = b = 76.21, c = 133.56, γ = 120.0 | 2 | 1.60 | 1WMQ | Kumarevel et al. 2005b |
| HutP-L-His-Mg2+ -RNA (21 mer) | H3 | a = b = 76.06, c = 133.99, γ = 120.0 | 2 | 1.48 | 1WPU | Unpublished |
| HutP-L-His-Mg2+ -RNA (55 mer) | C2 | a = 99.64, b = 76.47, c = 62.79, β = 109.06 | 3 | 1.70 | 3BOY | Gopinath et al. 2008 |
ASU Asymmetric Unit; PDB Protein Data Bank; HBN L-histidine β-naphthylamide
Apo-HutP
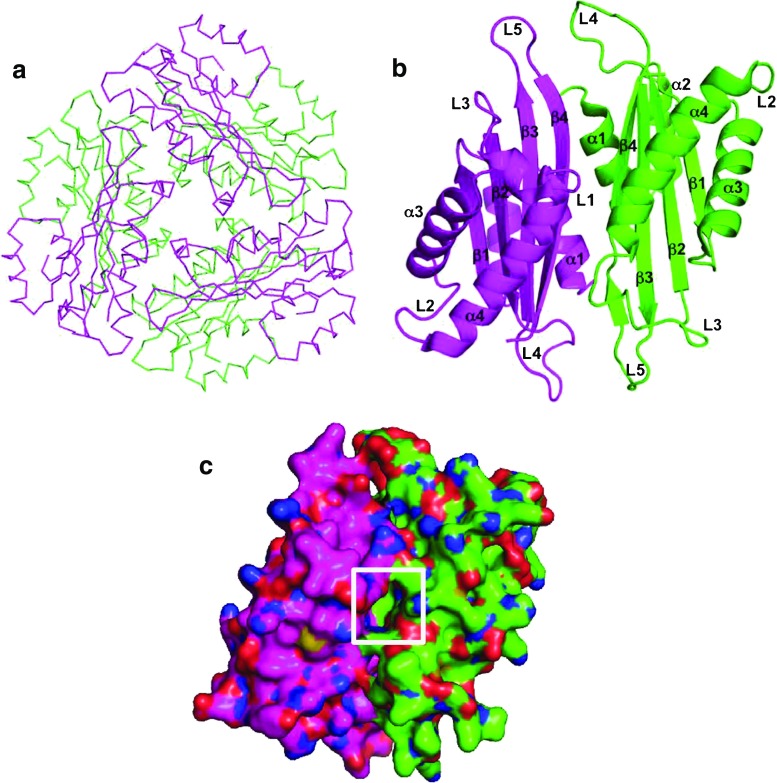
Analysis of the crystal structure of apo-HutP (in the absence of any ligand) revealed that two HutP monomers form a dimer with a non-crystallographic twofold axis, and three dimmers are symmetrically arranged along a crystallographic threefold axis to form a hexamer (Fig. 2a). The hexamer structure resembles a flattened cylinder approximately 83 Å wide and 48 Å thick along the threefold axis. A close-up view in Fig. 2b shows the HutP dimer. The monomer belongs to the α/β family of proteins whose structure consists of four α helices and four β strands arranged in the order ααβααβββ in the primary structure. Four antiparallel strands form a β sheet in the order β1β2β3β4 with two α helices each at the front and back of the β sheet. A hydrophobic pocket is visible in the vicinity of the twofold axis (Fig. 2c), and this pocket is employed for binding of L-His as shown in the following section.
Fig. 2.
Structure of apo-HutP (PDB code: 1WPS). Different HutP monomers are shown in different colors. a Cα backbone chain presentation of the HutP hexamer, viewed along the threefold symmetry axis, b ribbon presentation of the HutP dimer, viewed along the pseudo twofold axis, c alternative view of the HutP dimer surface reveals the hydrophobic pocket (boxed) in the center of dimer interface
HutP-L-His and -L-histidine analogs
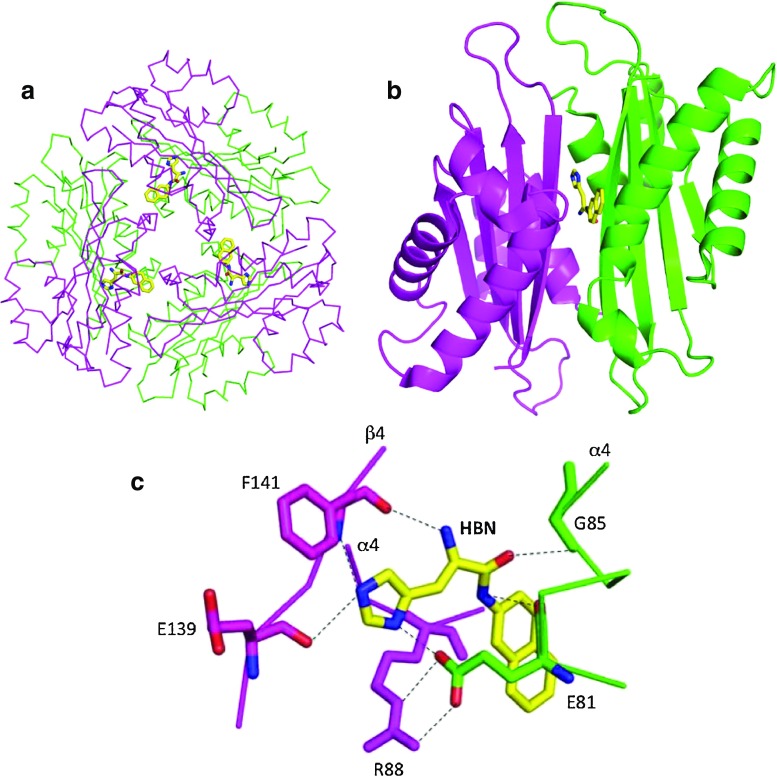
Of all the HutP series structures, HutP-L-His was the first structure to be solved by crystallography. Phasing information was obtained from multi-wavelength anomalous dispersion (MAD) experiments (Kumarevel et al. 2004a). All other crystal structures of the HutP series were solved by molecular replacement methods. HBN was employed as an L-histidine analog for the X-ray study of this binary complex and showed the highest activation of HutP and subsequent RNA binding among all of the L-histidine analogs tested (Kumarevel et al. 2004a). In the crystal structure of HutP-L-histidine analog (HutP–HBN), HBN is bound in the hydrophobic pocket (Fig. 2c), resulting in three L-histidine analogs in the HutP hexamer (Fig. 3a). As described in Table 2, crystals of the HutP-L-histidine analog are isomorphic with apo-HutP. Indeed, HutP in the binary complex is very similar to the apo form of HutP, indicating that HBN fits in the hydrophobic pocket without inducing significant structural changes (Fig. 3b). It is clear that the existence of the hydrophobic pocket is a prerequisite for the recognition of the L-His imidazole group and its discrimination from other amino acid residues. This is the necessary step to verify the incoming residue in the pocket because L-His in the following step does not employ the imidazole group but rather the backbone amino and carboxyl groups of the L-His to coordinate with Mg2+, as described later in this review. The details of the L-His binding site are shown in Fig. 3c. Arg88 and Glu81 of another monomer form a salt bridge at the gateway to the hydrophobic pocket, which may provide stabilizing interactions. Mutational analyses of Arg88 and Glu81 suggest that these residues are important for L-His binding. The imidazole side-chain of HBN is buried in the hydrophobic pocket and hydrogen bonds with Glu139, Phe141, and Glu81. The backbone nitrogen of HBN bonds with Phe141 and the backbone oxygen of HBN bonds with Gly85 of another monomer. The naphthyl ring of HBN is at the gateway to the hydrophobic pocket, placing it in the vicinity of the methylene chains of Arg88 and Glu81 that are involved in the salt bridge mentioned above (Kumarevel et al. 2004a). When the structure of HutP, co-crystallized with L-histidine benzylester and L-histidine, was also solved, the mode of binding was found to be the same as in the case of HBN, and the HutP structure was also similar to that in the HBN complex (Kumarevel et al. 2004a).
Fig. 3.
Structure of HutP-L-His (PDB code: 1VEA). The L-histidine analog (L-histidine β-naphthylamide; HBN) is shown as a stick model. Different HutP monomers are shown in different colors. A yellow-colored stick model represents HBN. a Cα backbone chain presentation of the HutP hexamer and HBN, viewed along the threefold symmetry axis, b ribbon presentation of the HutP dimer and HBN, c L-His binding site in the HutP-L-His complex. Dotted lines Hydrogen bonds
In biochemical experiments, a filter-binding assay showed that HutP failed to bind to RNA in the absence of Mg2+ ions, despite the presence of L-His (Fig. 1c; Kumarevel et al. 2005a, b). When Mg2+ ions were added to the binding buffer, formation of the RNA-bound quaternary complex was observed (Fig. 1c). These studies suggest that the L-His-bound HutP is not activated to bind RNA and that L-His and Mg2+ ions are required to activate HutP. The next step to understanding the role of divalent metal ions in the activation of HutP was to be resolved a ternary complex structure of HutP (HutP-L-His-Mg2+).
HutP-L-His-Mg2+
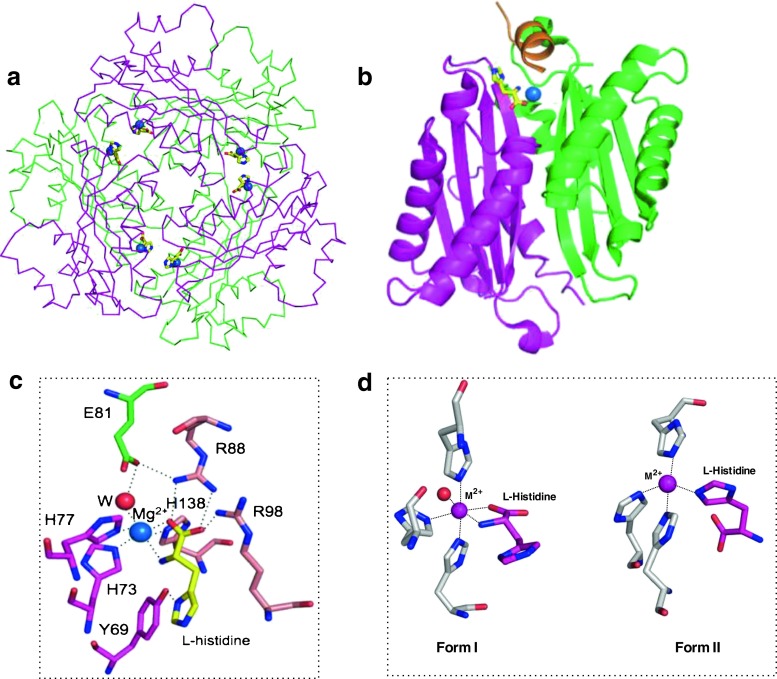
Crystals of the HutP-L-His-Mg2+ complex provide better atomic resolution than those of apo-HutP and HutP-L-His and improve the accuracy of the structure. Indeed, in HutP-L-His-Mg2+, the entire HutP peptide chain (Leu2-Ile148) is visible (the N-terminal Met1 must be fMet and may be removed during translation) except for some side-chain atoms that are likely exposed to solvent regions. In contrast, N-terminus residues 1–5 of one chain and 1–4 of another chain were not visible in the HutP-L-His structure. Analysis of the crystal structure of HutP-L-His-Mg2+ showed that the Mg2+ ion and L-His interact. This interaction site is at the interface of three HutP monomers (Fig. 4a, b; Kumarevel et al. 2005b); thus, the HutP hexamer has six L-His-Mg2+ binding sites, as shown in Fig. 4a. A close-up view of the L-His-Mg2+ binding site is shown in Fig. 4c. The bound Mg2+ ion is located at the interface between HutP monomers, forming a six-way coordination with the L-His ligand and the histidine cluster of the HutP protein. Of the six coordination interactions, two are with the amino and carboxyl groups of the L-His ligand, and three others are with the imidazole nitrogens of His73, His77, and His138. The sixth coordination site of the Mg2+ ion is with a water molecule that is anchored by a hydrogen bond to the side-chain of Glu81. When the structures of the HutP-L-His-Mg2+ complex and the HutP-HBN complex are superimposed, the L-His-Mg2+ binding site is approximately 12 Å from the L-His binding pocket in the HutP-HBN complex. This result suggests that when the Mg2+ ion is added to the HutP-L-His complex, the ion binds to the amino and carboxyl groups of the L-His ligand and induces the conformational change of the L-His imidazole which in turn loosens the binding of L-His to HutP, resulting in L-His being pulled out of the hydrophobic pocket. In this way, Mg2+-bound L-His can move along the dimer interface of HutP to a new binding site (Kumarevel et al. 2005b). This movement is linked to the movement of Arg88 in the opposite direction, leading to the disruption of the hydrophobic pocket; the salt bridge between Arg88 and Glu81 in the HutP-HBN complex is disrupted, and a different type of salt bridge is newly formed between Arg88 and Glu81, as shown in Fig. 4c. The carboxyl group of the L-His ligand also forms salt bridges with Arg88 and Arg98. The L-His-Mg2+ binding site is thus created by the drastic rearrangement of these amino acid residues as they become involved in hydrogen bonds, salt bridges, and Mg2+ ion coordination, as mentioned above. The rearrangement of Arg98 (C α position shifted by 5.4 Å) is accompanied by a large change (5.0 Å) in the C α position of the next residue, Thr99. Consequently, the Thr99 side-chain forms two hydrogen bonds with the N3 and 2′ OH of the A4 base of the RNA, as described below (Fig. 5).
Fig. 4.
Structure of HutP-L-His-Mg2+ (PDB code: 1WPV). L-His and Mg2+ are shown as a yellow-colored stick model and a blue sphere model, respectively. Different HutP monomers are shown in different colors. a Cα backbone chain presentation of the HutP hexamer, L-His, and Mg2+, viewed along the threefold symmetry axis, b ribbon presentation of the HutP dimer, c L-His-Mg2+ binding site in the HutP-L-His-Mg2+ complex. Dotted lines Coordination interactions and hydrogen bonds, red sphere water molecule. d Alternative-binding modes of L-His with respect to the metal ions
Fig. 5.
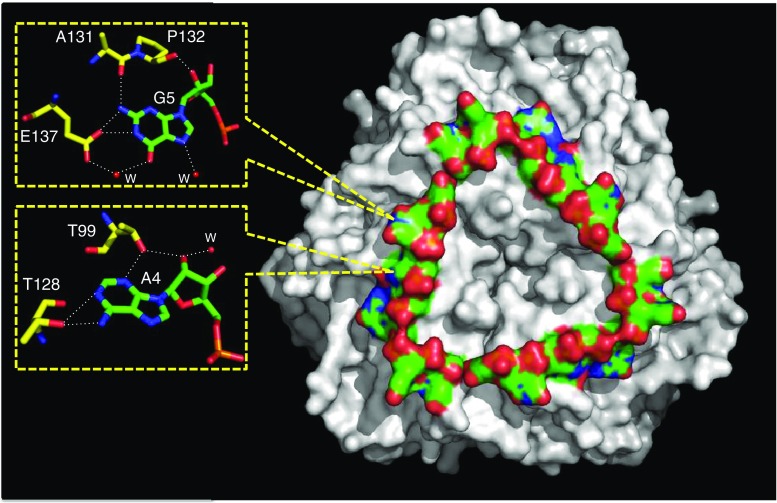
Crystal structure of the HutP-L-His-Mg2+-RNA (21 mer) complex (PDB code: 1WPU). The HutP hexamer complex is presented as a surface model (gray), as is the RNA (color). Close-up views of specific HutP-RNA nucleotide (A and G) interactions are shown in the box
HutP-L-His complexes with different divalent metal ions
In a series of biochemical analyses Kumarevel et al. (2004a) demonstrated that several divalent metal ions were able to influence the activation of HutP. Among these, Mn2+, Zn2+, and Cd2+ were the most efficient, followed by Mg2+, Co2+, and Ni2+ ions. Other divalent metal ions that weakly supported activation were Ba2+, Sr2+, Pt2+, Ca2+, Pb2+, and Ag2+. To clarify the importance of these divalent metal ions in the activation of HutP, three of these divalent cations (Mn2+, Ba2+ and Zn2+) were also used for crystallographic studies (Kumarevel et al. 2004a; Balasundaresan et al. 2013). The crystallization conditions for the HutP-L-His-Mn2+ and HutP-L-His-Ba2+ complexes were essentially the same as those for HutP-L-His-Mg2+. The obtained crystals were isomorphic with HutP-L-His-Mg2+, as shown in Table 2, and their structures were very similar to that of HutP-L-His-Mg2+. These similarities occurred despite slight changes in ion coordination; the coordination distances between the Ba2+ ions and the N atoms of the histidine residues were approximately 0.3 Å larger than the Mg2+ and Mn2+ distances, thereby widening the dimer interface (Kumarevel et al. 2004a).
However, in the case of the HutP-L-His-Zn2+ complex, crystals were obtained under conditions different from those used for the Mg2+, Mn2+, and Ba2+ complexes. The structure was solved at a resolution of 2.5 Å (Balasundaresan et al. 2013), and the structure of HutP was very similar to that of the HutP-L-His-Mg2+ complex. Differences were observed in the active site where Zn2+ formed a distorted four-coordinate complex, and the orientation of the L-His ligand coordinated to Zn2+ was reversed relative to its orientation with Mg2+, Mn2+, and Ba2+. This resulted in the coordination of Zn2+ with the N ε atom of the imidazole ring of L-His. The coordination structure observed in the complexes with the Mg2+, Mn2+, and Ba2+ ions is Form I, and the structure in the complex with the Zn2+ ion is Form II (Fig. 4d). Interestingly, the crystal structure of the HutP-L-His-Zn2+ complex has 12 HutP monomers in an asymmetric unit (Table 2). Therefore, it may be possible for the orientation of L-His in some monomers to be Form I, however, the coexistence of Form I and Form II is possible in the HutP active site; the imidazole side chain and the nitrogen-carboxyl oxygen backbone chain of the L-His are similar in size and metal coordination ability and are thus exchangeable. In this context, it is possible to speculate that Zn2+ is the best metal ion to mediate the activation of RNA binding (Kumarevel et al. 2004a). As described later Thr99 is an important residue that specifically interacts with the adenosine of the UAG motif. The next residue Arg98 binds tightly to L-His through the salt bridge in Form I as seen in Fig. 7c, but weakly to L-His in Form II. The positional flexibility of Arg98 is transmitted to Thr99 and the side chain of Thr99 is adjusted to a preferable position for better binding to RNA (Balasundaresan et al. 2013).
Fig. 7.
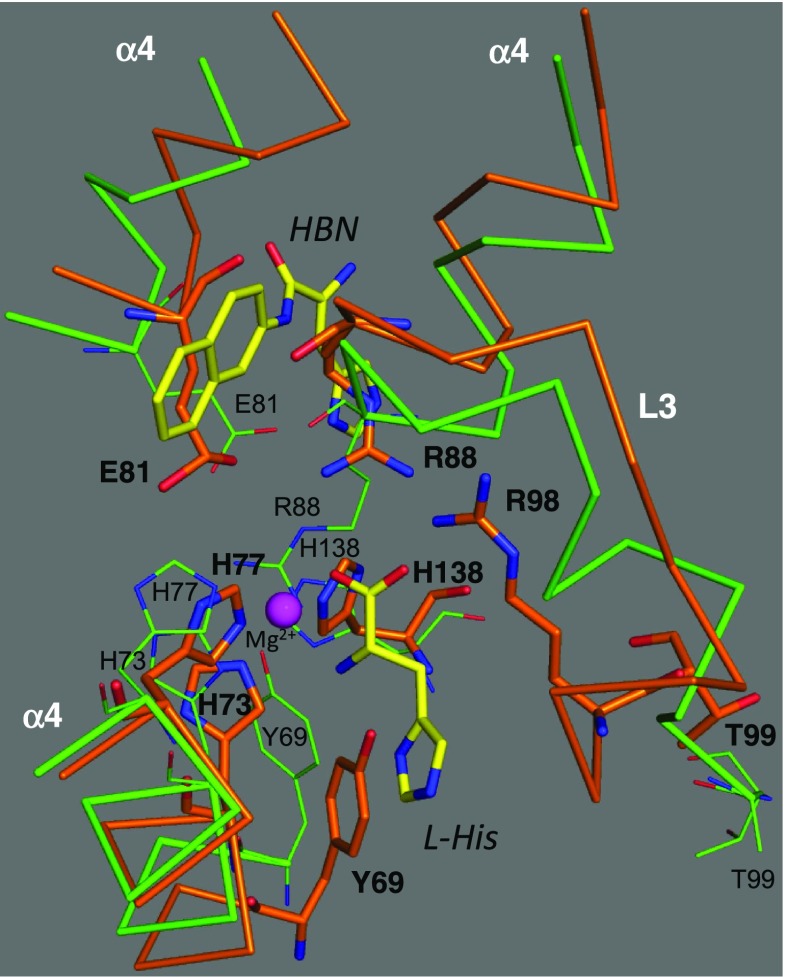
Superposition between HutP-L-His (PDB code: 1VEA) and HutP-L-His-Mg2+-RNA (PDB code: 3BOY) showing structural rearrangement of HutP. C backbone chain models of the HutP of 1VEA and 3BOY are shown in orange and green, respectively. HBN and L-His are represented by stick models with labels. Important amino acid residues involved in structural rearrangement are shown by thick stick models with bold labels in 3BOY, and by thin stick models with small labels in 1VEA
HutP-L-His-Mg2+-RNA
A minimal 20-base region for HutP binding to hut mRNA was identified as the region between nucleotides +496 to +515 (Fig. 1d). In this region, three UAG repeats and four non-conserved spacer residues were found to be important for RNA binding (Kumarevel et al. 2004b). In the quaternary complex structural analysis, a 21-mer RNA (5′-UUUAGUUUUUAGUUUUUAGUU-3′) was employed. The resolved structure (Kumarevel et al. 2005b) shows that the orientation of HutP, L-His, and the Mg2+ ions in the quaternary complex with RNA is very similar to that of the HutP-L-His-Mg2+ complex, indicating that the L-His-Mg2+-bound form of HutP is able to bind RNA. Therefore, it is natural to reason that the L-His-Mg2+-bound form is the active form of HutP. In contrast, apo-HutP and HutP-L-His are inactive forms of HutP that correspond to pre-structural rearrangements. The 21-mer RNA is bound on each side of the HutP hexamer with a triangle shape (Fig. 5). Each of the bound 21-mer RNAs has three repeated seven-nucleotide sequences (UUUAGUU) arranged with a crystallographic threefold symmetry. The central A4-G5 of the UAG motif forms an extensive hydrogen-bonding network with HutP (Fig. 5), and bases A4 and G5 are stacked on top of each other. Figure 5 shows that A4 hydrogen bonds to side-chains of Thr99 and Thr128 and that G5 bonds with a side-chain of Glu137. These amino acid residues are critical at the RNA-binding site, as shown by the failure of the mutant proteins Thr99Ala, Thr128Ala, and Glu137Ala to bind the RNA (Kumarevel et al. 2004b). Thus, the specific interactions between the bases A4 and G5 and HutP demonstrate the importance of these nucleotides within the UAG motif. On the other hand, the bases U1, U2, U3, and U6 do not interact with HutP, and U7 interacts with the neighboring dimer within the hexamer. These U nucleotides most likely serve as spacers; their basic function is to appropriately place the next UAG binding site on the HutP hexamer. The backbone conformation at the U2 nucleotide is unusual, with the RNA bent to form a triangular shape on the surface of the HutP hexamer. In Fig. 5, the negatively charged phosphate backbone of the RNA is directed outwards toward the solvent region, and the RNA bases bury into HutP for hydrogen-bonding interactions. This orientation readily explains the RNA sequence-dependent nature of the recognition by HutP.
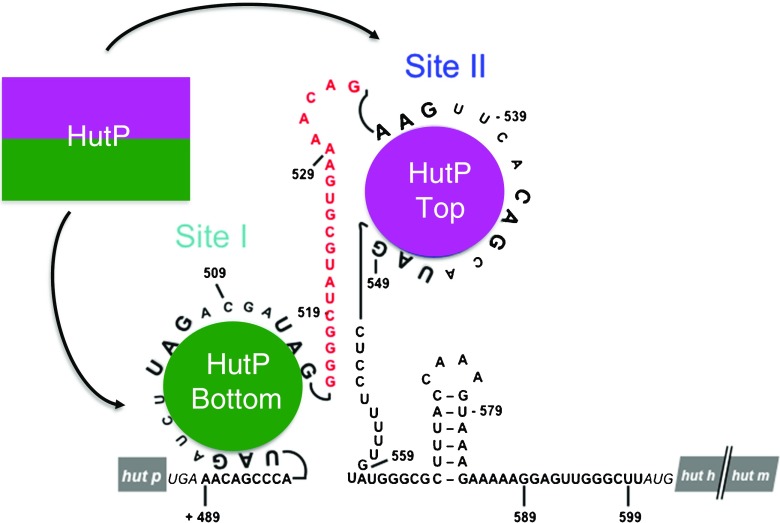
There is one RNA-binding site on each surface of the HutP hexamer. In addition, there are two UAG-rich regions (highlighted in red letters in Fig. 6a) within the hut terminator structure. These are considered to be potential HutP binding sites and are flanked by a 20-nucleotide spacer region (site I, +498 to +514; site II, +535 to +549). Each binding site consists of three XAG motifs (X denotes any base). Of these two potential binding sites, the first recognizes one surface (site I) of the HutP hexamer, and the second recognizes the other surface (site II). Between these two binding sites, there is a 17- to 20-nucleotide spacer region (depending on the first XAG motif count at site II; cyan-highlighted region in Fig. 6a), which is most likely sufficient to reach the RNA on the other surface of the hexamer. To evaluate whether the two XAG motifs separated by the spacer region are important for HutP binding, a model RNA was designed, synthesized, and tested for its binding to the activated HutP (Fig. 6b; Gopinath et al. 2008). These binding studies revealed that the model 55-mer RNA binds in a cooperative manner to HutP. To understand this mode of HutP-RNA recognition, the quaternary complex (HutP-L-His-Mg2+-55 mer RNA) was resolved at 1.70 Å. As predicted, the structure revealed that the XAG motifs in site I (nucleotides 1–20) bind in a triangular fashion to one surface of HutP. The following linker nucleotides carry the remaining XAG motif to site II (nucleotides 37–55) on the other surface of HutP, where binding occurs in the same fashion as in site I (Fig. 6c). The structural analysis was further confirmed by in-line mapping, mutational, and in vivo hut anti-termination studies which demonstrated that the model RNA bound to the both surfaces of HutP, as shown in Fig. 6c.
Fig. 6.
Presence of two potential RNA-binding regions within the hut terminator RNA, and the structure of the model 55-mer bound to HutP (PDB code: 3BOY). a hut terminator regions showing two potential binding sites for the HutP protein separated by the spacer region, highlighted in cyan. Important residues for hut binding are shown in red letters. b A model 55-mer RNA sequence mimicking the hut terminator RNA for HutP binding. The spacer region flanking sites I and II is shown in red letters. c Molecular surface representation of the quaternary complex structure showing the model 55-mer RNA binding the two surfaces (site I and site II) of HutP
Structural rearrangement of HutP
A large structural change in the HutP protein is observed between the structures of apo-HutP and the HutP-L-His-Mg2+ complex [root-mean-square deviation (RMSD) 3.1 Å], as mentioned earlier. However, the structure of the HutP-L-His-Mg2+ complex closely resembles that of the HutP-L-His-Mg2+-RNA complex (RMSD 0.69 Å), suggesting that divalent metal ions and L-His, but not RNA, cause structural rearrangement of the HutP. Structural differences between the HutP-L-His-Mg2+ complex and the apo-HutP complex are observed mainly in the region of the L-His-Mg2+-binding site of HutP. Figure 7 shows a structural comparison between the two complexes. It appears that His73, His77, and His138 are rearranged to coordinate with the Mg2+ ion. The rearrangement of His138 is linked to the movement of the following residue, Glu137, whose side-chain binds specifically with G5 of the UAG motif (Fig. 5). Arg98 rearranges to form a salt bridge with L-His, and this rearrangement is linked to the movement of the next residue, Thr99, whose side chain binds specifically with A4 of the UAG motif (Fig. 5). Arg88 is also rearranged to form a salt bridge with L-His, and this arrangement is linked to the movement of Glu81, whose side chain oxygen forms both a salt bridge with Arg88 and water-mediated coordination with a Mg2+ ion (Fig. 4c). After rearrangement, the above residues are located at the interface between three HutP monomers, and they appear to contribute directly or indirectly to the formation of the L-His-Mg2+-binding site, which is necessary for RNA binding.
Destabilization of hut terminator structure
The above section describes how the HutP protein is activated through structural rearrangement caused by divalent metal ions and the L-His ligand. The activated HutP is able to bind to RNA, and the bound RNA is arranged in a novel triangular fashion (Fig. 5). Before HutP binding, the hut mRNA forms a stem-loop terminator structure in between the HutP and HutH coding regions. After HutP binding, the terminator structure is destabilized, allowing the RNA polymerase to synthesize a full-length hut mRNA transcript. A simple destabilization pathway is shown in Fig. 2. Taking into account the structural, biochemical, and in vivo studies described in preceding section, we propose a detailed mechanism of hut terminator structure destabilization, as shown in Fig. 8. There are two potential HutP binding sites (site I, +498 to +514; site II, +535 to +549). Each binding site consists of three XAG motifs. The first binding site recognizes one surface of the HutP hexamer, and the second site recognizes the other surface. Between these two binding sites, there is a 20-nucleotide spacer region (Fig. 8), which is sufficient to reach the RNA on the other surface. The quaternary complex was tested using model RNA, which consisted of a 17-nucleotide spacer (U17) region flanked by two UAG-rich regions (Fig. 8). In this structure, the first binding site, consisting of 21-mer RNA (U1–U21) forms a triangle shape on one surface of the HutP hexamer. The second binding site (U35–U55) forms another triangle shape on the other surface via the spacer region (U22–U34). In vivo analyses using the native hut terminator (nucleotides +489 to +599 in Fig. 8; Gopinath et al. 2008) suggest that HutP recognizes the first two XAG motifs in site I and then rolls over the RNA towards the third XAG motif. The adjacent G–C-rich base-pair stem will start to melt. Consequently, the XAG motif in site II is exposed and binds to the other surface of HutP, resulting in the disruption of the stable stem region. Taken together, in vitro, in vivo, and structural analyses reveal that HutP destabilizes the hut terminator RNA structure by accessing two XAG-rich sites.
Fig. 8.
Conformational changes in the hut terminator RNA upon binding to activated HutP
Summary
The histidine utilizing (Hut) pathway is highly conserved among bacteria, but the mechanisms of regulation in these bacteria differ . In Bacillus subtilis, the Hut pathway is known to be regulated by a positive regulatory protein, HutP. Several studies have shown that HutP binds to the hut mRNA terminator and destabilizes the terminator structure to allow polymerase to synthesize the full-length hut mRNA. A series of structural analyses were carried out to unravel the overall events starting from activation of HutP to binding to the terminator RNA. These studies revealed snapshots of the conformational changes at each stage, and these snapshots have enabled clarification of the entire activation process of HutP in the presence of two ligands (L-histidine and a divalent metal ion), as summarized in Fig. 9. These conformational changes appear to be important for binding to the terminator RNA in order to facilitate the unwinding of the hut terminator RNA. This kind of regulation appears to be unique among the known termination and anti-termination proteins, where a divalent metal ion renders help in this process. We suggest that such a metal-ion dependent anti-termination regulatory mechanism is also being utilized by other species of bacteria.
Fig. 9.
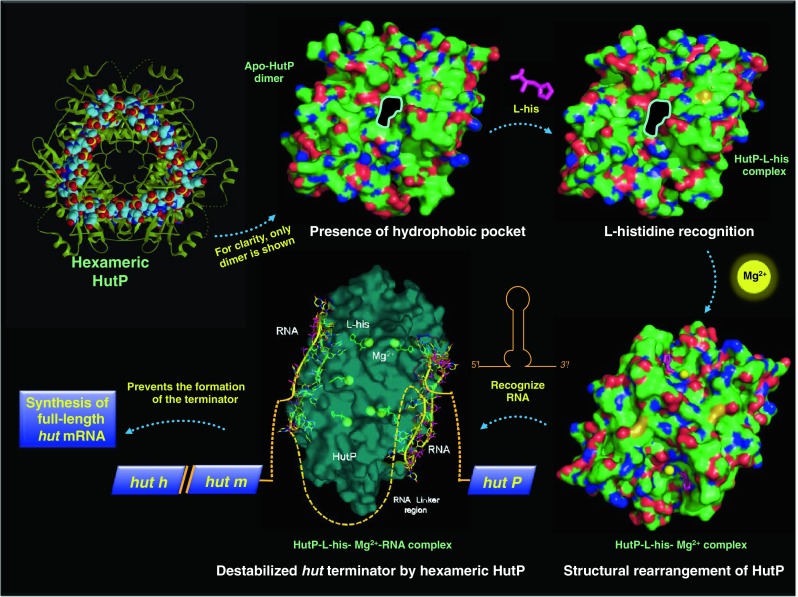
Anti-termination of the hut operon mediated by activated HutP
Acknowledgments
The authors would like to acknowledge the former members who participated in the HutP project (Drs. T. Kumarevel, Z. Fujimoto, M. Oda, S.C.B. Gopinath, T. Misono, and D. Balasundaresan) and the staff at beam-lines 6A and NW-12 at the Photon Factory for their help during data collection. This work was supported by funds from the National Institute of Advanced Industrial Science and Technology (AIST) and the Japanese Society for the promotion of Science (JSPS) to P.K.R.K.
Conflict of Interest
The authors certify that there is no actual or potential conflict of interest in relation to this article.
Footnotes
This article does not present data from any studies with human or animal subjects performed by the authors.
References
- Alpert CA, Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the Bg1G family of transcriptional antiterminators. J Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud MD, Debarbouille M, Rapoport G, Saier MH, Reizer J. In vitro reconstitution of transcriptional attenuation by the SacT and SacY proteins of Bacillus subtilis. J Biol Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaresan D, Mizuno H, Kumar PKR. Alternative binding modes of L-histidine guided by metal ions for the activation of the antiterminator protein HutP of Bacillus subtilis. J Struct Biol. 2013;183:512–518. doi: 10.1016/j.jsb.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Bender RA. Regulation of the histidine utilization (Hut) system in bacteria. Microbiol. Mol Biol. Rev. 2012;76:565–584. doi: 10.1128/MMBR.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin LA, Magasanik B. Induction and repression of histidine-degrading enzymes of Bacillus subtili. J Biol Chem. 1968;243:5165–5178. [PubMed] [Google Scholar]
- Glatz E, Nilsson RP, Rutberg L, Rutberg B. A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol Microbiol. 1996;19:319–328. doi: 10.1046/j.1365-2958.1996.376903.x. [DOI] [PubMed] [Google Scholar]
- Gopinath SCB, Balasundaresan D, Kumarevel T, Misono TS, Mizuno H, Kumar PKR. Insight into anti-termination regulation of the hut operon in Bacillus subtilis: importance of the dual RNA-binding surfaces of HutP. Nucleic Acids Res. 2008;36:3463–3473. doi: 10.1093/nar/gkn199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houman F, Diaz-Torres MR, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-R. [DOI] [PubMed] [Google Scholar]
- Kimmhi Y, Magasanik B. Genetic basis of histidine degradation in Bacillus subtili. J Biol Chem. 1970;245:3545–3548. [PubMed] [Google Scholar]
- Kumarevel T, Fujimoto Z, Karthe P, Oda M, Mizuno H, Kumar PKR. Crystal structure of activated HutP: An RNA binding protein that regulates transcription of the hut operon in Bacillus subtili. Structure. 2004;12:1269–1280. doi: 10.1016/j.str.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Kumarevel T, Gopinath SCB, Nishikawa S, Mizuno H, Kumar PKR. Identification of important chemical groups of the hut mRNA for HutP interactions that regulates the hut operon in Bacillus subtili. Nucleic Acids Res. 2004;32:3904–3912. doi: 10.1093/nar/gkh725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarevel T, Mizuno H, Kumar PKR. Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtili. Nucleic Acids Res. 2005;33:5494–5502. doi: 10.1093/nar/gki868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarevel T, Mizuno H, Kumar PKR. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature. 2005;434:183–191. doi: 10.1038/nature03355. [DOI] [PubMed] [Google Scholar]
- Lu Y, Turner RJ, Switzer RL. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc Natl Acad Sci USA. 1996;93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Sugishita A, Furukawa K. Cloning and nucleotide sequence of histidase and regulatory genes in the Bacillus subtili hut operon and positive regulatory operon. J Bacteriol. 1988;170:3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Katagai T, Tomura D, Shoun H, Hoshino T, Furukawa T. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated with catabolite repression downstream from the site of transcription. Mol Microbiol. 1992;6:2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- Oda M, Kobayashi N, Ito A, Kurusu Y, Taira K. Cis-acting regulatory sequences for antitermination in the transcript of the Bacillus subtili hut operon and histidine-dependent binding of HutP to the transcript containing regulatory sequences. Mol Microbiol. 2000;35:1244–1254. doi: 10.1046/j.1365-2958.2000.01795.x. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sano H, Seki S, Oda M, Fujimura M, Fujita Y. Cloning and sequencing of a 29 kb region of the Bacillus subtili genome containing the hut and wapA loci. Microbiology. 1995;141:337–343. doi: 10.1099/13500872-141-2-337. [DOI] [PubMed] [Google Scholar]