Abstract
The aggregation of proteins into amyloid fibrils is a topic that has attracted great interest because the process is associated with the pathology of numerous human diseases. Despite considerable progress in the elucidation of the structure of amyloid fibrils and the kinetic mechanism of their formation, knowledge on the thermodynamic aspects underlying the formation and stability of amyloid fibrils is limited. In this review, we summarize recent calorimetric studies of amyloid fibril formation, with the goal of obtaining a better understanding of the causal factors that thermally induce proteins to aggregate into amyloid fibrils. Calorimetric data show that differential scanning calorimetry is a useful technique to study the causative factors that thermally trigger the conversion to the amyloid structure and highlight the physics related to the thermal fluctuation of proteins during this conversion.
Keywords: Amyloid fibril, Aggregation, Calorimetry, Thermal fluctuation, Heat capacity, Amyloid nucleation
Introduction
Amyloid fibrils are highly ordered protein aggregates associated with several neurodegenerative and non-neuropathic diseases (Sipe and Cohen 2000; Caughey and Lansbury 2003; Selkoe 2003; Stefani 2004; Chiti and Dobson 2006; Eisenberg and Jucker 2012). The precursor proteins and peptides implicated in these diseases, regardless of the large variation in the amino acid sequences or the native structures, aggregate into amyloid fibrils that contain extensive β-sheet structures, where the β-strands are oriented perpendicular to the axis of the fibrils (Sunde and Blake 1997; Jahn et al. 2010; Tycko 2011). Some precursors are mostly unstructured proteins and peptides, such as amyloid β-peptide (Aβ), amylin, and α-synuclein, while other precursors are globular proteins, such as β2-microglobulin (β2-m), lysozyme, transthyretin, and immunoglobulin. In vitro experiments have provided evidence supporting the capability of a large variety of proteins and peptides, including those unrelated to any disease, to self-assemble into amyloid fibrils or amyloid-like structures when incubated under appropriate solution conditions. This has led to the suggestion that the ability to form amyloid fibrils is a fundamental property of polypeptide chains (Glenner et al. 1974; Kirschner et al. 1987; Maggio and Mantyh 1996; Fändrich and Dobson 2002; Makin and Serpell 2005; Chiti and Dobson 2006; Goldschmidt et al. 2010).
Research over the past few years has deepened our understanding of the morphological and structural features of amyloid fibrils (Fändrich and Dobson 2002; Saiki et al. 2005; Nelson and Eisenberg 2006; Kodali and Wetzel 2007; Goto et al. 2008; Jahn and Radford 2008). The native conformation of globular proteins is a compact structure that was achieved through an evolutionary process in which the packing and concomitant burial of non-polar side chains and polar backbone groups have been optimized for in vivo functioning (Arai and Kuwajima 2000; Lindorff-Larsen et al. 2005; Dill et al. 2008; Baldwin 2008; Fersht 2008; Cabrita et al. 2010). In contrast, it has been proposed that the amyloid structure of the same polypeptide chains are dominated by the hydrogen-bonded cross β-sheet architecture of the peptide backbones, in which the amino acid side chains are packed differently from those in the native folds (Kardos et al. 2004; Wetzel 2006; Fändrich et al. 2009; Lee et al. 2009; Morel et al. 2010). How the generic cross β-sheet structure is formed and sustained by the differing side chains within the fibrillar structure remains poorly understood.
A nucleation-dependent model has been proposed to explain the general mechanism of amyloid fibril formation in vitro (Harper and Lansbury 1997; Wetzel 2006). The model consists of two processes: nucleation and polymerization. In the nucleation process, several monomeric precursor molecules slowly associate to form a nucleus, which represents the rate-determining step in the formation of amyloid fibrils. Once the nucleus is formed, subsequent polymerization proceeds rapidly through the sequential incorporation of precursor molecules into the nucleated fibrils. Many experiments have demonstrated that fibrillation from various proteins involves a lag period that corresponds to the nucleation process before the formation of mature, well-defined amyloid fibrils (Lomakin et al. 1996; Naiki et al. 1997; Yagi et al. 2005; Hamley 2007; Sasahara et al. 2008; Xue et al. 2008).
In contrast to fibril formation which shows a typical lag phase or nucleation, the assembly of spherical oligomers and other prefibrillar forms has been reported to occur in several proteins without a definite lag phase, leading to the formation of spherical particles or shorter and thinner fibrils than mature amyloid fibrils (Modler et al. 2003; Hurshman et al. 2004; Gosal et al. 2005; Carrotta et al. 2005; Bader et al. 2006; Roychaudhuri et al. 2009; Bhak et al. 2009). The molecular mechanisms underlying the lag phase- and non-lag phase-dependent kinetic routes and morphological features of protein aggregation are not fully understood (Uversky 2010; Eichner and Radford 2011a).
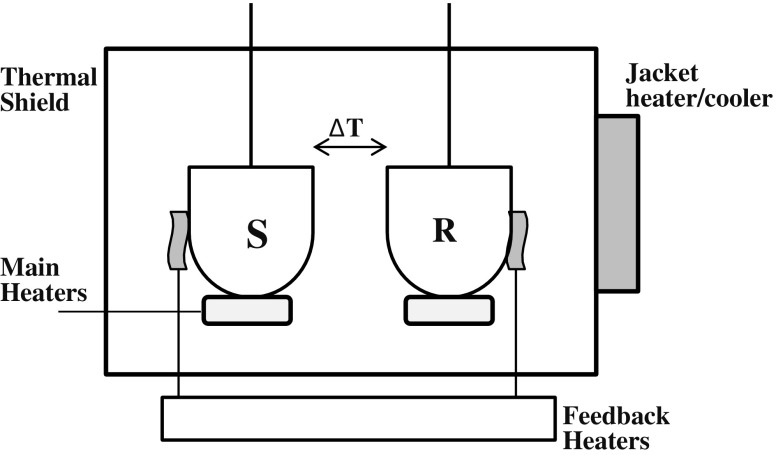
Differential scanning calorimetry (DSC) is an experimental technique to resolve the energetics of conformational transitions of biological macromolecules (Sturtevant 1987; Sanchez-Ruiz 1995; Freire 1995; Spink 2008; Gill et al. 2010; Chiu and Prenner 2011). A general diagram for a differential scanning calorimeter is shown in Fig. 1. In recent decades many of the studies carried out on proteins have used DSC to examine the reversible folding/unfolding transition of globular proteins under equilibrium conditions, focusing on the elucidation of the thermodynamic properties of protein folding and stability (Makhatadze and Privalov 1995; Robertson and Murphy 1997; Privalov and Dragan 2007). In contrast, relatively few attempts have been made to use DSC to characterize the thermodynamics of amyloid fibril formation and stability, possibly due to the irreversibility of protein aggregation, the complex nature of the aggregates, and the lack of a theoretical model to interpret the DSC data. In this review, we present the application and use of DSC for examining protein aggregation into amyloid fibrils. The thermal behaviors of amyloid fibrils formed from several proteins highlight the physics related to the thermal fluctuation of protein during its conversion into amyloid fibrils.
Fig. 1.
Schematic of a power-compensation differential scanning calorimeter. The solution containing the solute (protein) is loaded into the sample cell (S), and an equal volume of solvent (buffer) is loaded into the reference cell (R). Both cells are identically heated at the same precise controlled rate by the main heaters. As the temperature increases, thermally induced processes occurring in the sample cell that absorb or release heat will result in a thermal imbalance between the sample and reference cells which is compensated for by electrically powered feedback heaters. This electrical power signal provides a direct measure of the heat capacity (C
p) difference between the sample solution and the solvent: C
p
sol−C
p
solv (in units of J K−1). As the heat capacity corresponding to β2-microglobulin (β2-m) amyloid fibril (protein) solution is detected as negative values (Fig. 2a), it is termed the apparent heat capacity (C
p,app). The C
p,app-temperature traces of the fibrils in Fig. 2b are well approximated by: 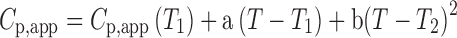
where a and b are constants, and T
1 is a reference temperature. As the heat capacity (C
p,app) at constant pressure is a temperature derivative of the heat quantity from the sample solution, the heat quantity (Q
1−2) needed to induce the exothermic process is represented in the temperature range from T
1 to T
2 by: 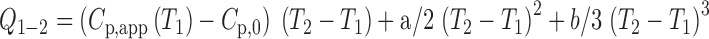
In the calculation of Q 1−2, T 1 and T 2 are set at 20 °C (293.15 K) and 67 °C (340. 15 K), respectively. The C p,app values extrapolated to the zero heating rate at 67 °C were used as C p,0, which corresponds to that of the monomeric state within the heat scale used. With a non-linear least-square fitting program, the calculated DSC curves were fitted to the observed curves so that a and b were determined (Sasahara et al. 2005)
Thermally induced melting of amyloid fibrils
Recent calorimetric studies on protein aggregation are summarized in Table 1. It has been shown that aggregates of several proteins, including amyloid fibrils, can be melted and dissociated by heating at high temperatures, giving rise to characteristic endothermic transitions in DSC experiments (Azuaga et al. 2002; Rezaei et al. 2002; Baxa et al. 2004; Morel et al. 2006, 2010). Conejero-Lara and colleagues have reported the results of detailed DSC analyses of amyloid fibrils formed from an N47A mutant of the α-spectrin SH3 domain (Morel et al. 2010). Their data show that the melting process of the fibrils occurs in a single cooperative transition and can be approximated as a reversible process. These authors provided for the thermodynamic parameters characterizing the stability of amyloid fibrils using a simple equilibrium model of fibrillation. Few calorimetric studies in the literature have reported the thermodynamic parameters of amyloid fibril formation. Using isothermal titration calorimetry (ITC), Kardos et al. (2004) determined that the enthalpy changes that occur during amyloid elongation of β2-m are considerably lower than those of the native protein and suggested a decreased internal packing within the amyloid structure in comparison with the native conformation. The thermodynamic data on the stability of amyloid fibrils obtained by DSC (Morel et al. 2010) and ITC (Kardos et al. 2004) indicate the contribution of entropy increase attained with their assembly, which points toward hydration effects playing a major role in this process.
Table 1.
Recent studies of protein aggregates by differential scanning calorimetry
| Peptide/protein | Morphology | Thermal behavior | Reference |
|---|---|---|---|
| Streptokinase | Aggregatesa | Endothermic transition | Azuaga et al. 2002 |
| Insulin | Aggregatesa | Effect of exothermic/endothermic heat | Dzwolak et al. 2003 |
| Ure2p | Fibrils | Endothermic transition | Baxa et al. 2004 |
| Sheep prion | Fibrils | Endothermic transition or deformed DSC curve | Rezaei et al. 2002 |
| α-Spectrin | Fibrils | Endothermic transition | Morel et al. 2006 |
| β-Lactoglobulin | Aggregatesa | Effect of exothermic heat or deformed DSC curve | Stirpe A et al. 2008 |
| α-Crystallin | Fibrils | Effect of exothermic heat or deformed DSC curve | Attanasio et al. 2009 |
| α-Spectrin | Fibrils | Thermodynamic analysis of the endothermic transition | Morel et al. 2010 |
| PDZ domain | Aggregates/fibrils | Effect of endothermic or exothermic/endothermic heat | Murciano-Calles et al. 2010 |
| β2-M | WL fibrils | Endothermic transition | Sasahara et al. 2006, 2007a |
| β2-M | Fibrils | Effect of exothermic heat before the fibril melting. | Sasahara et al. 2005 |
| Aβ | Fibrils | Effect of exothermic heat before the fibril melting. | Sasahara et al. 2005 |
| Lysozyme | Fibrils | Effect of exothermic heat before the fibril melting. | Sasahara et al. 2007b, 2009 |
Aβ, amyloid β-peptide; β2-M, β2-microglobulin; WL, worm-like
aNo image is given in the reference article
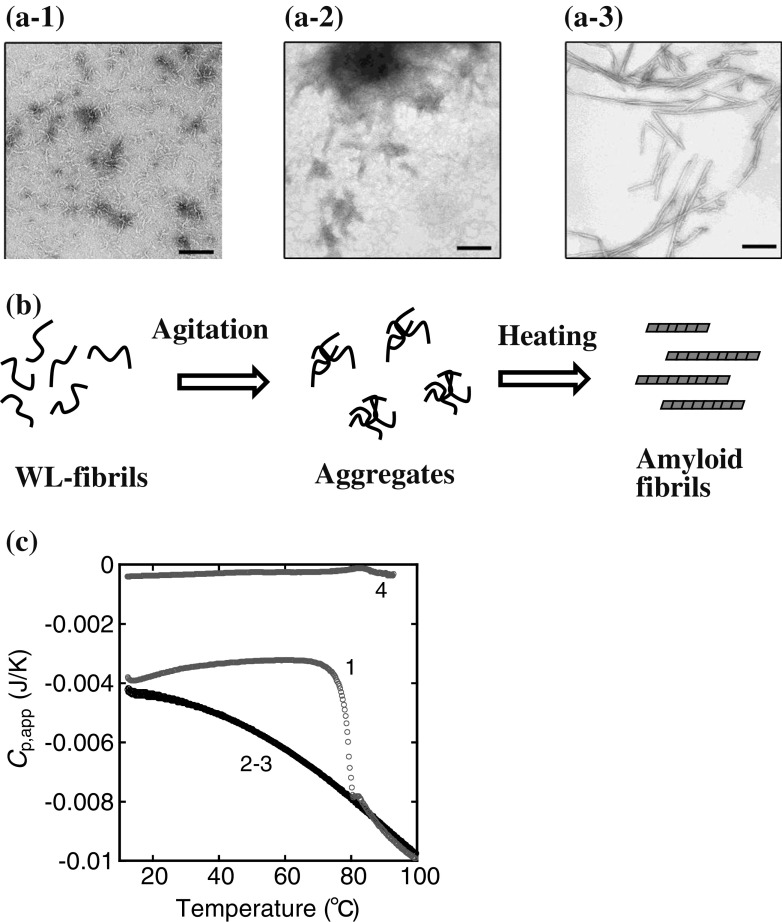
Previous DSC studies have also shown that thermal-induced aggregation of several proteins is accompanied by the exothermic effect of heat (Dzwolak et al. 2003; Stirpe et al. 2008; Attanasio et al. 2009; Murciano-Calles et al. 2010). Such an effect may be explained by a nucleation model, i.e., the formation of a stable nucleus that incorporates additional monomeric proteins into a growing aggregate (Dzwolak et al. 2003), but the exact molecular basis remains elusive. Recently, Goto and colleagues closely studied the thermal behaviors of amyloid fibrils formed from β2-m using DSC (Sasahara et al. 2005, 2006, 2007a, b, 2009). Amyloid fibril formation of β2-m, which is the light chain of the major histocompatibility complex class I molecules (Bjorkman et al. 1987), has been studied extensively because of its clinical importance associated with dialysis-related amyloidosis (Chatani and Goto 2005; Radford et al. 2005; Stoppini et al. 2005; Yamamoto and Gejyo 2005; Eichner and Radford 2011b). It is now known that at pH 2.5, β2-m is acid-unfolded, and two types of fibrils are formed through a lag phase- and a non-lag phase-dependent kinetics, based on a careful choice of salt concentrations (Hong et al. 2002; Gosal et al. 2005; Raman et al. 2005). The lag phase-dependent reaction route produces mature amyloid fibrils with a long (approx. micron scale) and needle-like morphology (diameter 10–20 nm), typical of dialysis-related amyloidosis. In the latter route, β2-m forms short, curved, and thin fibrillar structures (length <600 nm, diameter 2–5 nm) at high salt concentrations (approx. 0.5 M NaCl); these are termed worm-like (WL) fibrils (Gosal et al. 2005) (see the images in Fig. 4 for these fibrils). The DSC thermogram of the WL fibrils revealed an endothermic peak at about 85 °C, which represents the fibril melting (Sasahara et al. 2006, 2007a), whereas that of the mature amyloid fibrils revealed that their thermal response is characterized by the effects of exothermic heat; this phenomenon described in more detail below.
Fig. 4.
Thermal response of the agitation-treated worm-like (WL) fibrils of β2-m at pH 2.5. The WL fibrils (0.35 mg/mL) are agitated by the attached cylinder in the cell of an isothermal titration calorimeter and subsequently heated in the DSC cell (Sasahara et al. 2007a). a Electron microscopic observation: a-1 WL fibrils, a-2 agitation-treated WL fibrils, a-3 amyloid fibrils converted from the aggregates in a-2 during heating in the DSC cell. Scale bars 200 nm. b Schematic representation of agitation and heating of the WL fibrils. c DSC thermograms of the WL fibrils before (line 4) and after (lines 1–3) the agitation treatment. Heating rate is 60 °C/h. The numbers 1–3 correspond to the number of DSC scans. Adapted from Sasahara et al. (2007a)
DSC thermograms of amyloid fibrils formed from β2-m and Aβ
As described above, Sasahara et al. reported striking DSC results obtained with amyloid fibrils from proteins such as β2-m, Aβ peptide, and hen egg-white lysozyme (HEWL) (Sasahara et al. 2005, 2006, 2007a, b, 2009). These DSC thermograms show the effects of exothermic heat prior to fibril melting, which are highly dependent upon the heating rate in the DSC scans. The authors investigated in detail the effects of fibril concentration, heating rate, and additives, such as NaCl and polyethylene glycol (PEG), on the DSC thermograms. The resultant data highlight the physics related to the thermal fluctuation of protein during the conversion into amyloid fibrils, as shown below.
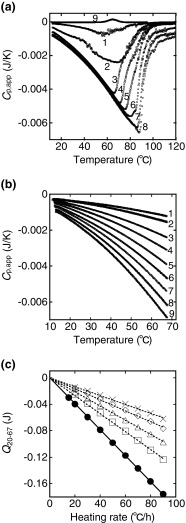
Figure 2a shows the DSC thermograms of β2-m amyloid fibrils at various concentrations (Sasahara et al. 2005). The background scan with a buffer solution is subtracted from the scan of the fibril sample and buffer solutions, and the apparent heat capacity (C p,app) corresponding to the whole sample solution is recorded versus temperature (T) as a DSC thermogram expressed as the C p,app−T trace. Here, it is important to note that the C p,app−T traces are not normalized by the protein concentration in order to show the unique fibril concentration dependence on the C p,app value (Sasahara et al. 2005). The figure includes a DSC thermogram of native β2-m, which shows an endothermic peak representing the thermally induced unfolding of its native fold (line 9). A comparison of this unfolding behavior with an endothermic peak reveals that the amyloid fibrils have remarkable C p,app−T traces, in which the C p,app value gradually becomes more negative as the temperature increases (approx. 80 °C) (lines 1–8). The C p,app value then increases abruptly at about 60–100 °C, with an increase to 120 °C. As a result, a peak minimum is observed at about 60–90 °C in the C p,app−T traces.
Fig. 2.
Thermal response of β2-m amyloid fibrils measured by differential scanning calorimetry (DSC) at pH 2.5. a Representative DSC thermograms of β2-m amyloid fibrils. The fibril concentration varies from 0.015 to 0.28 mg/mL. Lines: 1 0.015, 2 0.025, 3 0.04, 4 0.075, 5 0.125, 6 0.17, 7 0.2, 8 0.28 mg/mL. The heating rate is 60 °C/h. For comparison, a DSC thermogram of native β2-m (0.125 mg/mL) at pH 7.0 is recorded (line 9). b DSC thermograms of the fibrils (0.1 mg/mL) recorded at various heating rates from 10 to 68 °C. The heating rate is varied in repeated consecutive scans from 90 to 15 °C/h: 90 (line 9) → 80 (8) → 70 (7) → 60 (6) → 50 (5) → 40 (4) → 30 (3) → 20 (2) → 15 °C/h (1). Furthermore, heating is carried out twice at each heating rate. c Dependence of the Q 20−67 value on the heating rate. The heat quantity (Q 20−67 ) needed to induce the exothermic process in the temperature 20–67 °C was calculated from DSC thermograms of the fibrils (0.1 mg/mL) in the absence and presence of polyethylene glycol (PEG) 6000 at different concentrations: circles 0 g/L, squares 20 g/L, triangles 40 g/L, diamonds 60 g/L, crosses 80 g/L. Adapted from Sasahara et al. (2005, 2009)
The reversibility of the DSC thermogram was investigated with the fibrils (0.1 mg/mL) by conducting reheating runs (Sasahara et al. 2005). After one cycle of heating to 120 °C and cooling down to 10 °C, the second scan shows a C p,app−T trace that corresponds to that of monomeric β2-m. This result indicates that the amyloid fibrils are thermally and irreversibly depolymerized at the time of the first scan. Figure 2b shows DSC thermograms of β2-m amyloid fibrils recorded at various heating rates (Sasahara et al. 2005). Repeated runs of heating–cooling–reheating are carried out in the temperature region before the peak minimum with the fibrils (0.1 mg/mL) at various heating rates of 90–15 °C/h in a row, in which heating is also repeated twice at each heating rate. After cooling from the first scan, the second thermogram is superimposed on the first one for all heating rates studied, confirming the reversibility of the C p,app traces. The results reveal a distinct heating rate dependence of the C p,app−T trace: as the heating rate increases, the change in C p,app becomes increasingly significant.
The area representing the negative changes in C p,app observed in Fig. 2b, i.e., the heat (Q 20−67) needed to induce the exothermic process, was calculated in the temperature range 20–67 °C at various heating rates according to Eq. (2) in Fig. 1. The resultant Q 20−67 values are plotted versus the heating rate, resulting in a linear dependence of Q 20−67 on the heating rate, the extrapolation of which to the heating rate of zero reaches zero within the experimental error (solid line in Fig. 2c).
The authors repeated the DSC experiments using the β2-m fibril solution containing PEG 6000 (molecular weight 6,000) at several concentrations (0, 20, 40, 60 and 80 g/L) (Sasahara et al. 2009). PEG, which is a neutral and linear polymer, was expected to inhibit the inter-fibrillar association during heating due to its steric constraint. The increase in PEG concentration was found to lessen the extent of the exothermic effect. The Q 20−67 values were calculated in the temperature range 20–67 °C at various heating rates from the thermograms obtained at all the PEG concentrations studied (dotted lines in Fig. 2c). The data in Fig. 2c show that all of the Q 20−67 values are extrapolated to zero at the heating rate of zero and that C p,app values of β2-m amyloid fibrils and its monomeric form are indistinguishable at equilibrium (at zero heating rate) within the heat scale presented. These results imply that the effect of exothermic heat results from the heat-dependent property of β2-m amyloid fibrils.
Similarly, DSC thermograms of β2-m amyloid fibrils in the presence of PEG 400 (mean molecular weight 400), in which ≤30 % (vol/vol) of solvent water was replaced by the organic solvent PEG 400, were recorded at various heating rates (Sasahara et al. 2009). The extent of the exothermic effect decreases according to the solution exchange. The heat (Q 20−60) required to induce the exothermic process was estimated in the range 20–60 °C at various heating rates. Plots of Q 20−60 values against the heating rate reveal a behavior very similar to that seen in Fig. 2c. The results demonstrate that the presence of PEG lessens the negative C p,app change and suggest a causal link between the exothermic effect and the inter-fibrillar association during heating.
The authors also conducted DSC experiments with amyloid fibrils formed from two types of peptides, Aβ(1–40) and Aβ(25–35) (Sasahara et al. 2005). The thermal responses of Aβ amyloid fibrils were characterized by the effects of exothermic heat with a definite heating rate dependence. Plots of the calculated Q 20−67 values against the heating rate revealed the linear dependence of Q 20−67 on the heating rate, as was the case of β2-m amyloid fibrils.
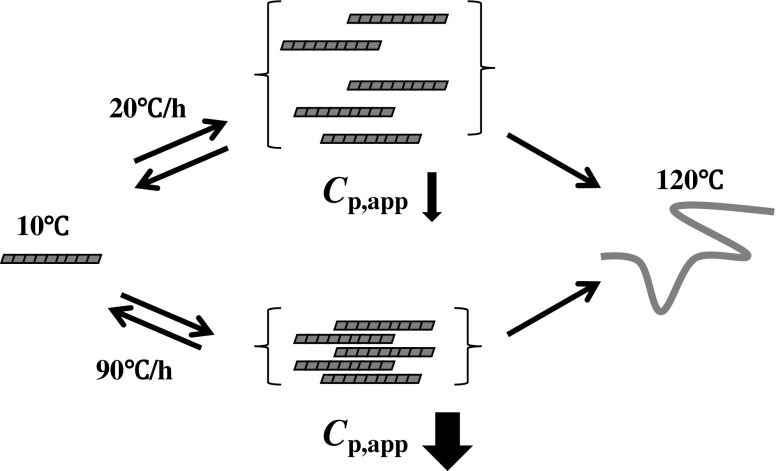
A model of inter-fibrillar association that is transiently enhanced with the increase in heating rate has been proposed to explain the C p,app−T traces observed for β2-m and Aβ amyloid fibrils (Fig. 3) (Sasahara et al. 2005, 2009). In the model, the transient association is thermally induced by hydrophobic interaction due to hydrophobic residues packed regularly along the fibril structure. Hydrophobic interactions usually play an important role in thermally induced aggregation of protein molecules, the basic features of which are assumed to be the spatial approach and dehydration of the exposed hydrophobic regions to solvent water (Kauzmann 1959; Tanford 1978; Baldwin 1986; Israelachvili and Wennerström 1996; Chandler 2005). In the amyloid fibrils formed from β2-m and Aβ, the hydrophobic and hydrophilic residues may be patterned on the fibril surface (Broome and Hecht 2000; Mandel-Gutfreund and Gregoret 2002; Hecht et al. 2004; Saiki et al. 2005; Fändrich et al. 2009; Bowerman and Nilsson 2012), resulting in transient association upon heating. The dynamic feature of hydration in biomolecular systems causes the remarkably varied entropic and energetic responses of water molecules in the vicinity of hydrophobic, polar, and charged solutes that are frequently reflected in the entropy–enthalpy compensation (Lumry and Rajender 1970; Liu et al. 2000). In light of the experimental data in Fig. 2c, which show that Q 20−67 values are extrapolated to zero at the heating rate of zero, the authors suggest that there is a high likelihood that the structure of amyloid fibrils fluctuates through gain and loss in the entropy of water molecules adjacent to the hydrophobic surface that is caused by thermal energy (Sasahara et al. 2009).
Fig. 3.
A model representing the transient inter-fibrillar association during heating. The association, which is reversibly heating rate-dependent, results in a decrease in the surface area of the fibrils accessible to the solvent water and, concomitantly, the decrease in C p,app. Amyloid fibrils thermally depolymerize upon heating up to 120 °C
Effects of exothermic heat are not necessarily common in fibrillar structures
DSC studies on amyloid fibrils of β2-m and Aβ have revealed that their thermal response is characterized by the effects of exothermic heat (Fig. 2). In remarkable contrast, the WL fibrils of β2-m show no effect of exothermic heat prior to the endothermic peak that represents the melting of the fibrils. Similar effects of heat have been observed in DSC measurements in other studies (Azuaga et al. 2002; Razaei et al. 2002; Baxa et al. 2004; Morel et al. 2006, 2010). These contrasting results may be ascribed to the structural diversity of amyloid fibrils formed from a protein (Kreplak and Aebi 2006; Pedersen et al. 2010) and the solution conditions used. Regarding β2-m, which forms the two types of fibrils under acidic solution conditions (i.e., amyloid fibrils and WL fibrils), it is presumed that fibrillar organization by a different reaction route (i.e., lag phase-dependent and non-lag phase-dependent kinetics, respectively) is a key factor in the molecular mechanism leading to the exothermic effects observed in the DSC measurements. More comprehensive data by DSC and other experimental techniques will be essential to substantiate the concept of thermal fluctuation of the amyloid fibrils formed from β2-m and Aβ.
Thermally induced conversion of WL fibrils into amyloid fibrils
Under acidic solution conditions (pH 2.5) including high salt concentrations (approx. 0.5 M NaCl), β2-m forms spontaneously WL fibrils without the definite nucleation step characterized by a lag period (Fig. 4a-1) (Hong et al. 2002; Raman et al. 2005; Gosal et al. 2005). The C p,app trace of the WL fibrils shows an endothermic peak at about 85 °C, which represents fibril depolymerization (line 4 in Fig. 4c). Sasahara et al. (2007a) introduced the combined agitation–heating method to thermally convert the WL fibrils to amyloid fibrils; the aggregates of WL fibrils were produced intentionally by agitation under controlled conditions (Fig. 4a-2), followed by DSC measurements (Fig. 4b). Heating runs of the agitation-treated WL fibrils show an abrupt decrease in C p,app (lines 1∼3 in Fig. 4c), followed by the heating rate dependence of the C p,app trace at various heating rates, as shown in the case of β2-m amyloid fibrils. Electron microscopy studies reveal that the agitation-treated WL fibrils are converted to mature amyloid fibrils during the first heating run, showing a large decrease in C p,app (Fig. 4a-3). These results indicate that two fibrillar structures, the WL and amyloid fibrils of β2-m, can be linked by a certain mechanism and that the change in the heat capacity of the fibril solution is an important clue for elucidating the link. Heating alone is not sufficient to produce amyloid fibrils from the WL fibrils, but in combination with agitation, heating does effectively trigger the conversion to amyloid fibrils (Sasahara et al. 2007a).
Thermally induced aggregation of proteins
Protein aggregation is often triggered by destabilization and partial unfolding of the native structure caused by low pH, high temperature, enzymatic cleavage, the addition of denaturants, or other processes (Uversky and Fink 2004; Ellis and Minton 2006). The application of heat is a well-established method that allows protein aggregation to occur on a biochemically feasible time scale. In fact, the thermally induced unfolding of many proteins has been shown to be occasionally followed by an irreversible process that induces aggregation (Sanchez-Ruiz et al. 1988; Azuaga et al. 2002; Rezaei et al. 2002; Weijers et al. 2003; Michnik et al. 2005). Generally, such aggregation has been modeled as shown in Scheme 1, where N, U, and A are native, unfolded, and irreversible unfolded protein forms, respectively (Lumry and Eyring 1954; Sanchez-Ruiz et al. 1988; Azuaga et al. 2002; Rezaei et al. 2002; Weijers et al. 2003; Michnik et al. 2005). The model consists of the reversible folding/unfolding transition (N ⇌ U) of the protein and the subsequent aggregation of non-native species that has been traditionally considered an essentially irreversible process. Only A (i.e., aggregation-competent species) may form insoluble aggregates Am composed of m protein monomers.
| Scheme 1 |
However, the conformational and energetic aspects of thermally induced aggregation into amyloid fibrils have not been fully characterized due to the intricate kinetics during the heating process. In particular, little is known about the nature of structural fluctuation of proteins by heating, which triggers the conversion to the amyloid structure (Scheme 2).
| Scheme 2 |
The combined agitation–heating method, which enables the conversion of WL fibrils to amyloid fibrils, was applied to produce amyloid fibrils from β2-m and HEWL in the native states (Sasahara et al. 2007b, 2009) (see below).
Thermally induced conversion of β2-m and HEWL to amyloid fibrils under physiological pH conditions
In the DSC study of Sasahara et al. (2007b), after the solutions of β2-m containing NaCl at various concentrations were agitated under neutral pH conditions where the protein was in the native state, DSC measurements of these agitation-treated aggregates were conducted at repeated scans (Fig. 5a–c). At lower concentrations of NaCl (0–0.4 M), the DSC thermograms show an endothermic peak centered at 63 °C in the first run, corresponding to the unfolding of the native structure (line 1 in Fig. 5a, b). After cooling from the first run, the subsequent thermograms show relatively similar C p,app traces compared to the first one, in which the endothermic peaks become less notable, probably due to the irreversible aggregation of the protein molecules (lines 2–6). However, the samples at high concentrations of NaCl (0.6–1.4 M) reveal a marked negative change in the C p,app trace in the first run. The repeated scans clearly provide the superimposable C p,app traces that show the effect of exothermic heat over the temperature range scanned, indicative of the generation of a different type of aggregate after the abrupt decrease in C p,app during the first run. In contrast, the C p,app−T trace of the same sample without agitation at 0.6–1.4 M NaCl shows an endothermic peak typical of the unfolding transition of a small globular protein (line 7 in Fig. 5c). These results indicate that the aggregation of β2-m by agitation needs to increase by a certain amount under high ionic strength conditions in order to reveal a specific effect of exothermic heat during heating. In a more recent study, Sasahara et al. (2008) have examined the potential of agitation-treated aggregates of β2-m to produce a fibril nucleus in detail.
Fig. 5.
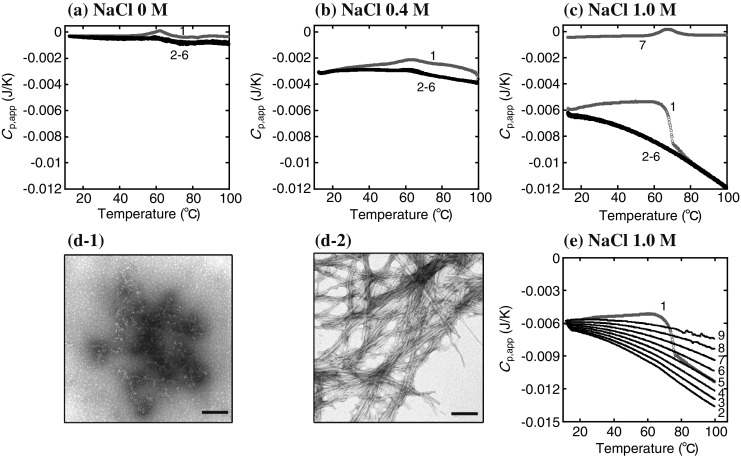
Thermally induced fibrillation of agitation-treated β2-m (0.35 mg/mL) at physiological pH. a–c DSC thermograms of the agitation-treated β2-m at different concentrations of NaCl: a 0 M, b 0.4 M, c 1.0 M. Heating–cooling–reheating cycles are performed consecutively. The numbers 1–6 correspond to the number of DSC scans. For comparison, a DSC thermogram of the same sample before agitation is presented (line 7 in c). The heating rate is 60 °C/h. d Electron microscopic images of agitation-treated β2-m (sample in c) before (d-1) and after (d-2) heating from 10 to 100 °C by DSC. Scale bars 200 nm. e Heating rate-dependent thermograms of agitation-treated β2-m at 1.0 M NaCl. The heating rate is varied in repeated, consecutive runs from 10 to 100 °C: 60 (line 1) → 90 (2) → 80 (3) → 70 (4) → 60 (5) → 50 (6) → 40 (7) → 30 (8) → 20 °C/h (9). Adapted from Sasahara et al. (2007b)
The changes in the morphology of the protein subjected to the agitation and heating treatments were observed by electron microscopy (Fig. 5d). The aggregates of β2-m that formed by agitation at 1.0 M NaCl show an amorphous-like morphology without ordered structures (Fig. 5d-1). However, the presence of amyloid fibrils in the aggregates subjected to a large decrease in C p,app in the first run were revealed (Fig. 5d-2). Consequently, DSC data and electron microscopy images demonstrate the thermally induced conversion of the agitation-treated β2-m to amyloid fibrils, which is accompanied by the negative change in C p,app (i.e., an exothermic effect). The heating rate dependence of the C p,app trace after the conversion to amyloid fibrils was investigated; after the first run at 60 °C/h, repeated heating was carried out nine times consecutively at different rates of 90–20 °C/h. The results show that the heating rate dependence of C p,app traces becomes marked as the concentration of NaCl is increased. The representative data obtained at 1.0 NaCl is presented in Fig. 5e, which shows a similar thermal response to that observed for β2-m amyloid fibrils (Fig. 2b).
The same agitation–heating method as performed with β2-m was applied to HEWL (Sasahara et al. 2007b, 2009). Several agitation-treated aggregates were produced with the solutions of HEWL containing 1.0 M NaCl at a pH range 2–6. Heating runs at 60 °C/h for these aggregates show an abrupt decrease in C p,app in the first scan under all solution conditions studied. After the first run at 60 °C/h, the heating rate dependence of the C p,app trace were observed by conducting repeated heating runs at various heating rates, as shown in the case of β2-m. The conversion from the aggregated states to amyloid fibrils after the abrupt decrease in C p,app in the first run was confirmed by electron microscopy.
Thermally induced fluctuation of amyloid fibrils
As described above, it is presumed that the effects of exothermic heat observed for amyloid fibrils formed from β2-m and Aβ result from the specific arrangement of the hydrophobic and hydrophilic residues along the fibrillar structures, which induce transient inter-fibrillar association during heating (Fig. 3). The increase in temperature triggers amyloid fibril formation for agitation-treated β2-m (including WL fibrils) and HEWL, as seen in Fig. 5, and these thermally induced fibrillations are accompanied by the effects of exothermic heat, which are very similar to the thermal response observed for β2-m amyloid fibrils (Fig. 2). Based on these data, it is postulated that the fibrillar structures produced during the heating are stabilized by gain and loss in the entropy of water molecules around the formed hydrophobic/hydrophilic surfaces, resulting in a transient inter-fibrillar association by thermal energy. In other words, conformational trapping in the intermolecular interactions of proteins as they convert to an amyloid structure during heating may be an aggregation mediated by solvent water. This might produce a unique hydration pattern around the exposed hydrophobic–hydrophilic residues along the fibrillar surface, giving rise to a specific fluctuation of the aggregates by thermal energy. This stabilization mechanism is in contrast to that of globular proteins with hydrophobic side chains located in the interior of the native conformation.
Concluding remarks
Some amyloid fibrils (e.g., fibrils of N47A Spc-SH3 domain and WL-fibrils of β2-m) have shown single cooperative transitions corresponding to the fibril melting in DSC experiments. On the other hand, the DSC data for amyloid fibrils formed from β2-m, HEWL, and Aβ reveal the effects of exothermic heat which are highly heating rate-dependent. These DSC data suggest that such effects of heat are attributed to the transient inter-fibrillar association during heating, which is induced by the specific arrangement of the hydrophobic/hydrophilic residues of amyloid fibrils. The data also highlight the physics related to the effects of exothermic heat in terms of interactions between the specific structure of amyloid fibrils and water molecules, pointing toward the fluctuation of the amyloid structure by thermal energy, which may play a major role in amyloid fibril formation.
Acknowledgments
Conflict of interest
None.
Contributor Information
Kenji Sasahara, Email: sasahara@med.kobe-u.ac.jp.
Yuji Goto, Email: ygoto@protein.osaka-u.ac.jp.
References
- Arai M, Kuwajima K. Role of the molten globule state in protein folding. Adv Protein Chem. 2000;53:209–282. doi: 10.1016/S0065-3233(00)53005-8. [DOI] [PubMed] [Google Scholar]
- Attanasio F, Cataldo S, Fisichella S, Nicoletti S, Nicoletti VG, Pignataro B, Savarino A, Rizzarelli E. Protective effects of L-and D-carnosine on α-crystallin amyloid fibril formation: implication for cataract disease. Biochemistry. 2009;48:6522–6531. doi: 10.1021/bi900343n. [DOI] [PubMed] [Google Scholar]
- Azuaga AI, Dobson CM, Mateo PL, Conejero-Lara F. Unfolding and aggregation during the thermal denaturation of streptokinase. Eur J Biochem. 2002;269:4121–4133. doi: 10.1046/j.1432-1033.2002.03107.x. [DOI] [PubMed] [Google Scholar]
- Bader R, Bamford R, Zurdo J, Luisi BF, Dobson CM. Probing the mechanism of amyloidogenesis through a tandem repeat of the PI3-SH3 domain suggests a generic model for protein aggregation and fibril formation. J Mol Biol. 2006;356:189–208. doi: 10.1016/j.jmb.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin RL. The search for folding intermediates and the mechanism of protein folding. Annu Rev Biophys. 2008;37:1–21. doi: 10.1146/annurev.biophys.37.032807.125948. [DOI] [PubMed] [Google Scholar]
- Baxa U, Ross PD, Wickner RB, Steven AC. The N-terminal prion domain of Ure2p converts from an unfolded to a thermally resistant conformation upon filament formation. J Mol Biol. 2004;339:259–264. doi: 10.1016/j.jmb.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Bhak G, Choe Y-J, Paik SR. Mechanism of amyloidogenesis: nucleation-dependent fibrillation versus double-concerted fibrillation. BMB Rep. 2009;42:541–551. doi: 10.5483/BMBRep.2009.42.9.541. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Bowerman CJ, Nilsson BL. Self-assembly of amphipathic β-sheet peptides: insights and applications. Biopolymers. 2012;98:169–184. doi: 10.1002/bip.22058. [DOI] [PubMed] [Google Scholar]
- Broome BM, Hecht MH. Nature disfavors sequences of alternating polar and non-polar amino acids: implications for amyloidogenesis. J Mol Biol. 2000;296:961–968. doi: 10.1006/jmbi.2000.3514. [DOI] [PubMed] [Google Scholar]
- Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Carrotta R, Manno M, Bulone D, Martorana V, San Biagio PL. Protofibril formation of amyloid β-protein at low pH via a non-cooperative elongation mechanism. J Biol Chem. 2005;280:30001–30008. doi: 10.1074/jbc.M500052200. [DOI] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- Chatani E, Goto Y. Structural stability of amyloid fibrils of β2-microglobulin in comparison with its native fold. Biochim Biophys Acta. 2005;1753:64–75. doi: 10.1016/j.bbapap.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiu MH, Prenner EJ. Differential scanning calorimetry: an invaluable tool for a detailed thermodynamic characterization of macromolecules and their interactions. J Pharm Bioallied Sci. 2011;3:39–59. doi: 10.4103/0975-7406.76463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzwolak W, Ravindra R, Lendermann J, Winter R. Aggregation of bovine insulin probed by DSC/PPC calorimetry and FTIR spectroscopy. Biochemistry. 2003;42:11347–11355. doi: 10.1021/bi034879h. [DOI] [PubMed] [Google Scholar]
- Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid folds. Mol Cell. 2011;43:8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Eichner T, Radford SE. Understanding the complex mechanisms of β2-microglobulin amyloid assembly. FEBS J. 2011;278:3868–3883. doi: 10.1111/j.1742-4658.2011.08186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol Chem. 2006;387:485–497. doi: 10.1515/BC.2006.064. [DOI] [PubMed] [Google Scholar]
- Fändrich M, Dobson CM. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fändrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Aβ and other amyloid fibrils. Prion. 2009;3:89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht AR. From the first protein structures to our current knowledge of protein folding: delights and scepticisms. Nat Rev Mol Cell Biol. 2008;9:650–654. doi: 10.1038/nrm2446. [DOI] [PubMed] [Google Scholar]
- Freire E. Differential scanning calorimetry. Methods Mol Biol. 1995;40:191–218. doi: 10.1385/0-89603-301-5:191. [DOI] [PubMed] [Google Scholar]
- Gill P, Mogbadam TT, Ranjbar B. Differential scanning calorimetry techniques: applications in biology and nanoscience. J Biom Tech. 2010;21:167–193. [PMC free article] [PubMed] [Google Scholar]
- Glenner GG, Eanes ED, Bladen HA, Linke RP, Termine JD. β-pleated sheet fibrils. A comparison of native amyloid with synthetic protein fibrils. J Histochem Cytochem. 1974;22:1141–1158. doi: 10.1177/22.12.1141. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Reik R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosal WS, Morten IJ, Hewitt EW, Smith DA, Thomson NN, Radford SE. Competing pathways determine fibril morphology in the self-assembly of β2-microglobulin into amyloid. J Mol Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Goto Y, Yagi H, Yamaguchi K, Chatani E, Ban T. Structure, formation and propagation of amyloid fibrils. Curr Pharm Des. 2008;14:3205–3218. doi: 10.2174/138161208786404146. [DOI] [PubMed] [Google Scholar]
- Hamley IW. Peptide fibrillization. Angew Chem Int Ed. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- Harper JD, Lansbury PT. Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- Hecht MH, Das A, Go A, Gradley L, Wei Y. Do novo proteins from designed combinatorial libraries. Protein Sci. 2004;13:1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D-P, Gozu M, Hasegawa K, Naiki H, Goto Y. Conformation of β2-microglobulin amyloid fibrils analyzed by reduction of the disulfide bond. J Biol Chem. 2002;277:21554–21560. doi: 10.1074/jbc.M200188200. [DOI] [PubMed] [Google Scholar]
- Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- Israelachvili J, Wennerström H. Role of hydration and water structure in biological and colloidal interactions. Nature. 1996;379:219–225. doi: 10.1038/379219a0. [DOI] [PubMed] [Google Scholar]
- Jahn TR, Radford SE. Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The common architecture of cross-β amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- Kardos J, Yamamoto K, Hasegawa K, Naiki H, Goto Y. Direct measurement of the thermodynamic parameters of amyloid formation by isothermal titration calorimetry. J Biol Chem. 2004;279:55308–55314. doi: 10.1074/jbc.M409677200. [DOI] [PubMed] [Google Scholar]
- Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/S0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kirschner DA, Inouye H, Duffy LK, Sinclair A, Lind M, Selkoe DJ. Synthetic peptide homologous to β protein from Alzheimer disease forms amyloid-like fibrils in vivo. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Kreplak L, Aebi U (2006) From the polymorphism of amyloid fibrils to their assembly mechanism and cytotoxicity. 73:217−233 [DOI] [PubMed]
- Lee Y-H, Chatani E, Sasahara K, Naiki H, Goto Y. A comprehensive model for packing and hydration for amyloid fibrils of β2-microglobulin. J Biol Chem. 2009;284:2169–2175. doi: 10.1074/jbc.M806939200. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Røgen P, Paci E, Vendruscolo M, Dobson CM. Protein folding and the organization of the protein topology universe. Trends Biochem Sci. 2005;30:13–19. doi: 10.1016/j.tibs.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Liu L, Yang C, Guo Q-X. A study on the enthalpy–entropy compensation in protein unfolding. Biophys Chem. 2000;84:239–251. doi: 10.1016/S0301-4622(00)00130-7. [DOI] [PubMed] [Google Scholar]
- Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid β-protein fibrils: detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumry R, Eyring H. Conformation changes of proteins. J Phys Chem. 1954;58:110–120. doi: 10.1021/j150512a005. [DOI] [Google Scholar]
- Lumry R, Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- Maggio JE, Mantyh PW. Brain amyloid–a physicochemical perspective. Brain Pathol. 1996;6:147–162. doi: 10.1111/j.1750-3639.1996.tb00797.x. [DOI] [PubMed] [Google Scholar]
- Makhatadze GI, Privalov PL. Energetics of protein structure. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/S0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- Makin OS, Serpell LC. Structures for amyloid fibrils. FEBS J. 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y, Gregoret LM. On the significance of alternating patterns of polar and non-polar residues in beta-strands. J Mol Biol. 2002;323:453–461. doi: 10.1016/S0022-2836(02)00973-7. [DOI] [PubMed] [Google Scholar]
- Michnik A, Drzazga Z, Kluczewska A, Michalik K. Differential scanning microcalorimetry study of the thermal denaturation of haemoglobin. Biophys Chem. 2005;118:93–101. doi: 10.1016/j.bpc.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Modler AJ, Gast K, Lutsch G, Damaschun G. Assembly of amyloid protofibrils via critical oligomers–a novel pathway of amyloid formation. J Mol Biol. 2003;325:135–148. doi: 10.1016/S0022-2836(02)01175-0. [DOI] [PubMed] [Google Scholar]
- Morel B, Casares S, Conejero-Lara F. A single mutation induces amyloid aggregation in the α-spectrin SH3 domain: analysis of the early stages of fibril formation. J Mol Biol. 2006;356:453–468. doi: 10.1016/j.jmb.2005.11.062. [DOI] [PubMed] [Google Scholar]
- Morel B, Varela L, Conejero-Lara F. The thermodynamic stability of amyloid fibrils studied by differential scanning calorimetry. J Phys Chem B. 2010;114:4010–4019. doi: 10.1021/jp9102993. [DOI] [PubMed] [Google Scholar]
- Murciano-Calles J, Cobos ES, Mateo PL, Camara-Artigas A, Martinez JC. An oligomeric equilibrium intermediate as the precursory nucleus of globular and fibrillar supramacromolecular assemblies in a PDZ domain. Biophys J. 2010;99:263–272. doi: 10.1016/j.bpj.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki H, Hashimoto N, Suzuki S, Kimura H, Nakakuki K, Gejyo F. Establishment of a kinetic model of dialysis-related amyloid fibril extension in vitro. Amyloid. 1997;4:223–232. doi: 10.3109/13506129709003833. [DOI] [Google Scholar]
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- Pedersen J, Andersen CB, Otzen DE. Amyloid structure–one but not the same: the many levels of fibrillar polymorphism. FEBS J. 2010;277:4591–4601. doi: 10.1111/j.1742-4658.2010.07888.x. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Dragan AI. Microcalorimetry of biological macromolecules. Biophys Chem. 2007;126:16–24. doi: 10.1016/j.bpc.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Radford SE, Gosal WS, Platt GW. Towards an understanding of the structural molecular mechanism of β2-microglobulin amyloid formation in vitro. Biochim Biophys Acta. 2005;1753:51–63. doi: 10.1016/j.bbapap.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Raman B, Chatani E, Kihara M, Ban T, Sakai M, Hasegawa K, Naiki H, Rao CM, Goto Y. Critical balance of electrostatic and hydrophobic interactions is required for β2-microglobulin amyloid fibril growth and stability. Biochemistry. 2005;44:1288–1299. doi: 10.1021/bi048029t. [DOI] [PubMed] [Google Scholar]
- Rezaei H, Choiset Y, Eghiaian F, Treguer E, Mentre P, Debey P, Grosclaude J, Haertle T. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J Mol Biol. 2002;322:799–814. doi: 10.1016/S0022-2836(02)00856-2. [DOI] [PubMed] [Google Scholar]
- Robertson AD, Murphy KP. Protein structure and the energetics of protein stability. Chem Rev. 1997;97:1251–1267. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid β-protein assembly and Alzheimer disease. J Biol Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki M, Honda S, Kawasaki K, Zhou D, Kaito A, Konakahara T, Morii H. Higher-order molecular packing in amyloid-like fibrils constructed with linear arrangements of hydrophobic and hydrogen-bonding side-chains. J Mol Biol. 2005;348:983–998. doi: 10.1016/j.jmb.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz JM. Differential scanning calorimetry of proteins. Subcell Biochem. 1995;24:133–176. doi: 10.1007/978-1-4899-1727-0_6. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz JM, Lopez-Lacomba JL, Cortijo M, Mateo PL. Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry. 1988;27:1648–1652. doi: 10.1021/bi00405a039. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Naiki H, Goto Y. Kinetically controlled thermal response of β2-microglobulin amyloid fibrils. J Mol Biol. 2005;352:700–711. doi: 10.1016/j.jmb.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Naiki H, Goto Y. Exothermic effects observed upon heating of β2-microglobulin monomers in the presence of amyloid seeds. Biochemistry. 2006;45:8760–8769. doi: 10.1021/bi0606748. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Yagi H, Naik H, Goto Y. Heat-triggered conversion of protofibrils into mature amyloid fibrils of β2-microglobulin. Biochemistry. 2007;46:3286–3293. doi: 10.1021/bi602403v. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Yagi H, Naiki H, Goto Y. Heat-induced conversion of β2-microglobulin and hen egg-white lysozyme into amyloid fibrils. J Mol Biol. 2007;372:981–991. doi: 10.1016/j.jmb.2007.06.088. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Yagi H, Sakai M, Naiki H, Goto Y. Amyloid nucleation triggered by agitation of β2-microglobulin under acidic and neutral pH conditions. Biochemistry. 2008;47:2650–2660. doi: 10.1021/bi701968g. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Yagi H, Naiki H, Goto Y. Thermal response with exothermic effects of β2-microglobulin amyloid fibrils and fibrillation. J Mol Biol. 2009;389:584–594. doi: 10.1016/j.jmb.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- Sipe JD, Cohen AS. Review: history of the amyloid fibril. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- Spink CH. Differential scanning calorimetry. Methods Cell Biol. 2008;84:115–141. doi: 10.1016/S0091-679X(07)84005-2. [DOI] [PubMed] [Google Scholar]
- Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Stirpe A, Rizzuti B, Pantusa M, Bartucci R, Sportelli L, Guzzi R. Thermally induced denaturation and aggregation of BLG-A: effect of the Cu2+ and Zn2+ metal ions. Eur Biophys J. 2008;37:1351–1360. doi: 10.1007/s00249-008-0346-4. [DOI] [PubMed] [Google Scholar]
- Stoppini M, Mangione P, Monti M, Giorgetti S, Marchese L, Arcidiaco P, Verga L, Segagni S, Pucci P, Merlini G, Bellotti V. Proteomics of β2-microglobulin amyloid fibrils. Biochim Biophys Acta. 2005;1753:23–33. doi: 10.1016/j.bbapap.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Sturtevant J. Biochemical applications of differential scanning calorimetry. Annu Rev Phys Chem. 1987;38:463–488. doi: 10.1146/annurev.pc.38.100187.002335. [DOI] [Google Scholar]
- Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv Protein Chem. 1997;50:123−159. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Mysterious oligomarization of the amyloidogenic proteins. FEBS J. 2010;277:2940–2953. doi: 10.1111/j.1742-4658.2010.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Weijers M, Barneveld PA, Cohen Stuart MA, Visschers RW. Heat-induced denaturation and aggregation of ovalbumin at neutral pH described by irreversible first-order kinetics. Protein Sci. 2003;12:2693–2703. doi: 10.1110/ps.03242803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel R. Kinetics and thermodynamics of amyloid fibril assembly. Acc Chem Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- Xue W-F, Homans SW, Radford SE. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc Natl Acad Sci USA. 2008;105:8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Kusaka E, Hongo K, Mizobata T, Kawata Y. Amyloid fibril formation of α-synuclein is accelerated by preformed amyloid seeds of other proteins: implications for the mechanism of transmissible conformational diseases. J Biol Chem. 2005;280:38609–38616. doi: 10.1074/jbc.M508623200. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Gejyo F. Historical background and clinical treatment of dialysis-related amyloidosis. Biochim Biophys Acta. 2005;1753:4–10. doi: 10.1016/j.bbapap.2005.09.006. [DOI] [PubMed] [Google Scholar]