Abstract
Docking methodology aims to predict the experimental binding modes and affinities of small molecules within the binding site of particular receptor targets and is currently used as a standard computational tool in drug design for lead compound optimisation and in virtual screening studies to find novel biologically active molecules. The basic tools of a docking methodology include a search algorithm and an energy scoring function for generating and evaluating ligand poses. In this review, we present the search algorithms and scoring functions most commonly used in current molecular docking methods that focus on protein–ligand applications. We summarise the main topics and recent computational and methodological advances in protein–ligand docking. Protein flexibility, multiple ligand binding modes and the free-energy landscape profile for binding affinity prediction are important and interconnected challenges to be overcome by further methodological developments in the docking field.
Keywords: Protein-ligand docking, Structure-based drug design, Scoring functions, Search algorithms
Introduction
Molecular docking methodologies are of great importance in the planning and design of new drugs. These methods aim to predict the experimental binding mode and affinity of a small molecule within the binding site of the receptor target of interest.
Molecular docking consists of three main connected goals: pose prediction, virtual screening and binding affinity estimation (Jain and Nicholls 2008). A successful docking methodology must be able to correctly predict the native ligand pose within the receptor binding site (i.e. to find the experimental ligand geometry within a certain tolerance limit) and the associated physical–chemical molecular interactions. Furthermore, when investigating large compound libraries, the method must be able to successfully distinguish binding from non-binding molecules and to correctly rank these ligands among the best compounds in the database (Kolb and Irwin 2009).
A search algorithm and an energy scoring function are the basic tools of a docking methodology for generating and evaluating the ligand conformations. The ability to successfully handle the intrinsic molecular flexibility of a system and to correctly describe the energetics of receptor–ligand interactions is critical to the development of predictive docking methodologies that are useful in prospective drug design studies.
In this review, we present the search algorithms and scoring functions most commonly used in current molecular docking methods that focus on protein–ligand applications. We attempt to summarise the main topics and recent computational and methodological advances in protein–ligand docking. We also consider approaches used to include protein flexibility and strategies that aim to improve binding affinity prediction in the context of a docking-based investigation.
Search algorithms
In molecular docking, search algorithms are used to explore the free energy landscape to find the best ligand poses. If the thermodynamics of the system, i.e. the enthalpic and entropic effects, are correctly modelled by the energy function, the global minimum of the energy landscape will correspond to the experimental receptor–ligand conformation, i.e. the native binding mode, and local minima will correspond to alternative binding modes. Unfortunately, considering the entropic effects is not straightforward, and current docking methods employ rough approximations. Therefore, is not guaranteed that the global minimum associated with the energy landscape investigated by a docking methodology will correspond to the native binding mode.
Three main strategies regarding protein and ligand flexibility are employed by docking methods: (1) the protein is considered to be rigid, and only translational and rotational degrees of freedom of the ligand are explored (i.e. the ligand is considered as a rigid body with no internal degrees of freedom), (2) the protein is considered rigid, and all degrees of freedom of the ligand are explored (i.e. translational, rotational and conformational), and (3) the protein is considered totally or partially flexible, and all degrees of freedom of the ligand are also investigated. Most algorithms consider the protein as a rigid entity while the ligand is considered flexible, i.e. sp3 bonds are able to rotate, but bond lengths and bond angles are kept fixed. In this work, we review the main strategies associated with approaches (2) and (3).
Ligand sampling
Search algorithms are classified into the following three main groups according to the methodology employed to explore ligand flexibility: systematic, stochastic and deterministic searches (Brooijmans and Kuntz 2003). Furthermore, some algorithms implement a hybrid approach combining two or all three of these strategies.
Systematic algorithms explore all ligand degrees of freedom during the search. The methods that employ this approach can be further classified as exhaustive, incremental construction and conformational ensemble. Exhaustive searches systematically explore the values of each degree of freedom in a combinatorial manner, rotating all dihedral angles of the ligand according to a predetermined range of values and a set of initial restraints, e.g. geometrical and chemical constraints (Huang and Zou 2010). Evidently, more flexible ligands will have a greater number of rotatable bonds, thereby increasing the complexity of the optimisation problem significantly. The programs Glide (Friesner et al. 2004) and eHiTS (Zsoldos et al. 2007) employ an exhaustive systematic search algorithm. Incremental construction (Kuhl et al. 1984; Smellie et al. 1991) is a fragmentation approach based on the separation of the ligand into smaller fragments, followed by the selection and docking of a base fragment into the receptor binding site. The ligand is then reconstructed incrementally by covalently linking the other fragments to the base group. This strategy is widely employed in de novo ligand design, which aims to discover novel compounds by linking the best fragments docked within the receptor binding site (Nishibata and Itai 1991, 1993; Böhm 1992; Schlosser and Rarey 2009; Lang et al. 2009; Pearce et al. 2009). Docking programs that implement the fragmentation approach include FlexX (Rarey et al. 1995, 1996, 1997) and Hammerhead (Welch et al. 1996). To address the combinatorial explosion problem faced by these approaches, some programs employ the conformational ensemble strategy, which rigidly docks a set of previously generated ligand conformations into the binding site. FLOG (Miller et al. 1994) and DOCK 4.0 (Ewing et al. 2001) are examples of docking programs that use this strategy.
Stochastic methods randomly change all ligand degrees of freedom (translational, rotational and conformational) at each step, generating a great diversity of solutions. These ligand poses are evaluated according to a probabilistic criterion to decide whether or not each will be rejected. Monte Carlo (MC), Evolutionary Algorithms (EAs), Tabu Search (TS) and Swarm Optimisation (SO) are classes of stochastic algorithms (Huang and Zou 2010). The main disadvantage of these heuristic methods is that there is no guarantee of convergence to the optimal solution, and multiple and independent runs of the algorithm are required to maximise the probability of finding the global energy minimum. MC-based methods make random changes in all the ligand degrees of freedom, and energy minimisation is usually carried out for each generated conformation. The poses are then accepted or rejected according to the Boltzmann factor, which considers the energies of the solutions before and after the random changes and the absolute temperature. An important variation of this method, known as Simulated Annealing (Goodsell and Olson 1990), applies temperature variations to increase the probability of finding the global minimum because increasing the temperature may allow some energy barriers to be overcome. EAs are optimisation methods based on the theory of the evolution of biological populations by means of natural selection to obtain optimal solutions for a particular problem (Clark and Westhead 1996; Clark 2000). These methods are divided into three main classes: genetic algorithms, evolutionary programming and evolution strategies. The main advantages of these methods are the ease with which they allow escape from local minima and the possibility of identifying a wide diversity of low energy solutions. The TS strategy explores the search space, making random changes in the ligand degrees of freedom. In addition to maintaining the lowest energy solution, this strategy analyses the similarity of a recently generated, non-lowest energy pose using a “Tabu list”, which contains the lowest energy solutions previously found (Baxter et al. 1998). A pose is only maintained if it is distinct from all solutions previously listed. This approach prevents the algorithm from visiting regions already explored. The SO approach, also known as swarm intelligence, is inspired by the collective behaviour of agents such as birds and ants. According to this algorithm, the changes performed in a current solution are directed to follow the best pose of the population. Particle Swarm Optimisation and Ant Colony Optimisation are two variations of the SO approach. Examples of docking software that implement stochastic algorithms include Prodock (Trosset and Scheraga 1999) and ICM (Abagyan et al. 1994) (MC methods), GOLD (Jones et al. 1995, 1997) and DockThor (de Magalhães et al. 2004a, b) (EA), PRO_LEADS (Baxter et al. 1998) and PSI-DOCK (Pei et al. 2006) (TS methods) and PLANTS (Korb et al. 2006) (SO method).
In deterministic methods, the actual state of the system determines the modifications to be made, leading to its next state. The final result is highly dependent on the initial input structure because, given exactly the same initial system configuration and a particular set of parameters, the final state will always be the same. Energy minimisation (EM) and molecular dynamics (MD) are examples of deterministic methods. EM aims to explore the energy landscape using the direction associated with the potential energy gradient, driving the system to the nearest local minimum. The main problem associated with this strategy is its inability to overcome energy barriers to achieve other local minima, thereby exploring the energy landscape more efficiently. EM is frequently associated, as a local search method, with other docking search strategies, e.g. the energy minimisation of docking solutions obtained using a fragment-based method. MD methods simulate the movements of a system over time by considering some thermodynamic state variables, e.g. temperature and pressure. The main advantages of this strategy are as follows: (1) an explicit solvent can be included in a more natural way and (2) all degrees of freedom of both ligand and protein can be considered during the simulation. The major disadvantages of this approach are its high computational cost and the propensity of the system to become trapped in a local minimum. Some adaptations have been made to address the latter problem, e.g. the use of simulated annealing, soft potentials and an initial set of ligand conformations. MD strategies, similar to EM methods, are employed in conjunction with other docking strategies to improve the predictions. CDOCKER is an example of a docking algorithm that implements MD in association with simulated annealing (Wu et al. 2003).
Protein flexibility
The inclusion of protein flexibility during the search remains a challenge in the field of molecular docking. Proteins in solution experience small to large conformational variations upon ligand binding due to induced fit effects. Moreover, it is now widely accepted that proteins exist as an ensemble of pre-existing conformational states rather than as a unique native state (Changeux and Edelstein 2011). According to this theory, the ligand binds to one of these conformations from the ensemble, shifting the population equilibrium towards a particular ligand-bound conformation (Tsai et al. 1999; Ma et al. 1999; Kar et al. 2010; Changeux and Edelstein 2011; Petukh et al. 2013). The development of search algorithms and sampling strategies that deal effectively with the several degrees of freedom associated with protein flexibility is therefore very challenging. Diverse methodologies have been developed to account for protein flexibility, and these can be divided into five main classes (Teodoro and Kavraki 2003): soft docking, side-chain flexibility, molecular relaxation, ensemble docking and collective degrees of freedom.
Subtle protein movements can have a significant influence on molecular docking results. Rearrangement of the side chain of a single amino acid residue is capable of changing the binding site profile, thereby decreasing the docking accuracy during both pose prediction and binding affinity estimation. The soft docking approach (Jiang and Kim 1991) has been employed to address these small conformational changes (i.e. atomic variations up to 1 Å). In a general way, this strategy softens the repulsive term of the Lennard-Jones potential, allowing small overlaps between the protein and the ligand atoms. Further variations have been developed, including energy minimisation, Monte Carlo simulations and specific treatment of van der Waals interactions (Apostolakis et al. 1998; Mizutani et al. 2006). The main advantage of this method is its speed, but it should only be employed to consider local receptor motions. Ferrari et al. (2004) applied soft-docking in virtual screening studies of potential ligands of the T4 lysozyme and an aldol reductase and obtained better results than using regular docking strategies.
When the induced fit effect is considerable and exceeds the soft-docking limits (but remains a local effect), the side-chain flexibility approach can be employed. In this strategy, the protein backbone is kept fixed while the side-chain conformations are explored by varying their essential torsional degrees of freedom. Since the development of early methods to include side-chain flexibility in docking algorithms, e.g. though the use of a rotamer library to predict the discrete and low-energy conformations of side-chain residues (Leach 1994), several improved variations of this strategy have been developed, including the continuous approach (Abagyan et al. 1994; Desmet et al. 1997; Schaffer and Verkhivker 1998; Schnecke and Kuhn 2000; Frimurer et al. 2003; Meiler and Baker 2006; Nabuurs et al. 2007).
The third strategy consists of molecular relaxation to optimise the protein–ligand complex conformations; this is obtained using rigid-protein docking methods and considers both backbone and side-chain movements of the protein as well as all ligand degrees of freedom. These methods are usually adapted to treat receptor flexibility during docking, and the methods usually employed are energy minimisation, Monte Carlo methods and molecular dynamics (Huang and Zou 2010). Some adjustments to the MD method have been implemented to decrease the search space, thereby making the simulations computationally viable and, at the same time, more efficient. The use of implicit solvent models and the application of geometric constraints to the amino acid residues outside the binding site are some examples (Luty et al. 1995; Mangoni et al. 1999; Huang et al. 2008). Molecular relaxation methods are usually applied during post-processing steps, e.g. to evaluate the stability of docked complexes and as a refinement tool for docking poses (Armen et al. 2009; Sokkar et al. 2011; Maghsoudi et al. 2011; Hao et al. 2011; Nowosielski et al. 2013).
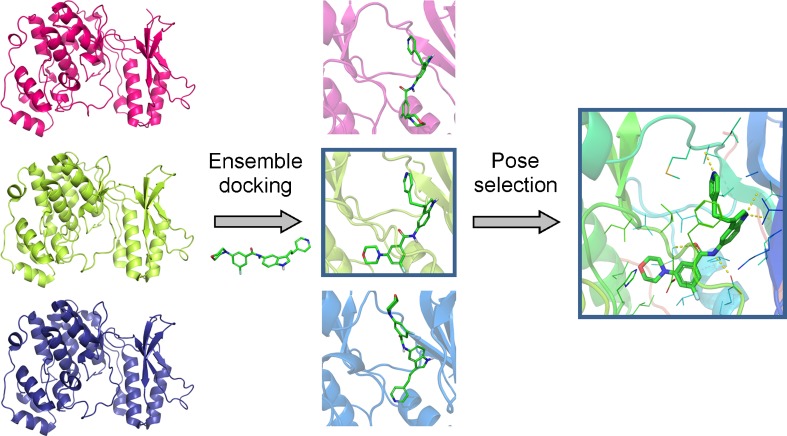
Ensemble docking implicitly considers the receptor flexibility by docking the ligand on a set of protein conformations instead of a single conformation (Fig. 1). Distinct receptor states can be obtained from experimental data (e.g. NMR and X-ray crystallographic structures) or derived from computational techniques, such as comparative modelling (Novoa et al. 2010), normal mode analysis (Sperandio et al. 2010) and molecular dynamics-derived frames (Nichols et al. 2011). Ensemble docking strategies also differ in the way in which the docking results obtained from multiple structures are analysed, e.g. the rescoring process and binding affinity prediction (Carlson 2002; Rueda et al. 2009; Beier and Zacharias 2010; Dietzen et al. 2012; Venkatraman and Ritchie 2012).
Fig. 1.
Scheme of an ensemble docking experiment for the target MAPKp38α with three distinct protein conformations
Whether the ensemble docking strategy leads to real improvements in docking studies remains under debate; successful cases remain restricted to specific cases/targets. Some studies indicate that the improved docking accuracy obtained using the ensemble strategy is not general, and worse performance might be obtained in some cases (Rueda et al. 2010; Craig et al. 2010; Li et al. 2010; Haider et al. 2011). Nevertheless, specific target studies have obtained great improvement in docking accuracy using the ensemble docking strategy (Brooijmans and Humblet 2010; Park et al. 2010; Takaya et al. 2011; Li et al. 2011b). Korb et al. (2011) constructed a metric to evaluate the quality of the ensemble structures (the ensemble performance index, EPI), which may be useful for receptor conformation selection. Regardless of the difficulty in deciding the ideal number of representative protein conformers to be used in an ensemble docking experiment, several studies have found that the use of three structures is generally sufficient to provide more accurate results; increasing the ensemble size to more than four members usually yields lower accuracy (Rueda et al. 2010; Brooijmans and Humblet 2010). Furthermore, the computational cost increases according to the size of the ensemble, and the docking is always biased towards the classes of ligands originally complexed with the representative protein conformations that are considered for the ensemble. Nevertheless, ensemble docking is considered the most promising strategy for incorporating protein flexibility into molecular docking studies (Korb et al. 2012).
The methods based on collective degrees of freedom consider the full protein flexibility. Generally speaking, these methods attempt to reduce the original high-dimensional representation of the protein motion to a lesser representation that captures only the dominant motion modes (Teodoro et al. 2003). Several implementations employ this approach to account for protein flexibility, e.g. normal mode analysis (Zacharias and Sklenar 1999; Kolossváry and Guida 1999; Kolossváry and Keserû 2001; Keserû and Kolossváry 2001) and principal component analysis – PCA (García 1992; Teodoro et al. 2003). The main problem with these methods is that the degrees of freedom considered are not the native ones but rather the collective movements derived from them, which can lead to inaccuracies in binding mode prediction (Teodoro and Kavraki 2003).
Detailed reviews and studies on the development of methods that include protein flexibility during or after docking and a comparative analysis of these methods can be found in the literature (B-Rao et al. 2009; Kokh et al. 2011; Lill 2011; Asses et al. 2012; Flick et al. 2012).
Scoring functions
During the docking process, search algorithms investigate a great number (usually millions) of ligand conformations. The scoring functions used evaluate the quality of these docking poses, guiding the search methods towards relevant ligand conformations. A scoring function must be able to distinguish the experimentally observed binding modes—associating them with the lowest energy values of the energy landscape—from all the other poses explored by the search algorithm (pose prediction). The second goal of the scoring functions is to properly classify active and inactive ligands (virtual screening). The third and most critical issue is that they predict the affinity constants and correctly rank several compounds according to their potency (binding affinity estimation).
Current docking methods exhibit good performance in identifying experimental binding modes when flexibility does not play a very relevant role for a particular protein. However, the detection of active compounds among a decoy set of ligands and the accurate prediction of binding affinity remain challenging tasks, even when induced fit effects are not important for binding (Yuriev and Ramsland 2013; Damm-Ganamet et al. 2013).
Several methods with various levels of complexity have been developed to predict the free energy of binding (Kim and Skolnick 2008; De Azevedo and Dias 2008; Söderhjelm et al. 2010; Ballester and Mitchell 2010; Stjernschantz and Oostenbrink 2010; Gallicchio et al. 2010; Teramoto and Kashima 2010). The most accurate methods often entail high computational cost, rendering their use unviable in realistic virtual screening experiments in which hundreds or thousands of small molecules are evaluated.
The use of scoring functions remains the main low computational cost alternative for predicting binding affinity in virtual screening experiments. Although no universal scoring function exists that has significant reliability for all molecular systems, some important strategies have improved binding affinity predictions, e.g. the development of target-specific scoring functions. Here, we review the main scoring functions employed during the early stages of the docking process and for binding affinity prediction.
Scoring functions can be divided into three main classes (Wang et al. 2003; Huang et al. 2010): force field-based, empirical and knowledge-based.
Force field-based functions are derived from a classical force field and consist of a sum of energy terms. This type of scoring function usually considers the interaction energies of the protein–ligand complex (non-bonded terms) and the internal ligand energy (bonded and non-bonded terms). The main problems regarding the use of force field-based scoring functions relate to atomic clashes arising from strong repulsion at short distances resulting from the Lennard-Jones potential and the overestimation of interactions between charged atoms. Some variations have been developed to address these undesirable effects, and additional terms have been included to compute other properties, e.g. the solvation effect (Zhang et al. 1997; Shoichet et al. 1999). Examples of docking programs that have implemented force field-based scoring functions include GOLD (GoldScore; Verdonk et al. 2003), AutoDock (Morris et al. 1998) and DockThor (de Magalhães et al. 2004b), which implements dihedral and non-bonded terms using the MMFF94S force field (Halgren 1996).
Empirical scoring functions have been developed to reproduce experimental binding affinity data with high accuracy. These functions are based on the idea that it is possible to correlate the free energy of binding to a weighted sum of non-related variables. The coefficients associated with the functional terms can be obtained through a regression analysis using known binding affinity data of experimentally determined structures. An empirical scoring function may be decomposed into several terms that are related to specific properties of interest, e.g. van der Waals interactions, hydrogen bonds and entropy (De Azevedo and Dias 2008), and can be written as:
| 1 |
where the ci terms are the weighting coefficients obtained from the respective ΔG term. The empirical functions mainly differ with respect to the number and type of terms considered and to the protein–igand complexes of the training set used during the parameterisation process. Examples of empirical scoring functions include ChemScore (Eldridge et al. 1997), ID-Score (Li et al. 2013), PLANTSCHEMPLP (Korb et al. 2009) and GlideScore (Friesner et al. 2004, 2006).
Knowledge-based functions have been developed based on the statistical analysis of interacting atom pairs from protein–ligand complexes with available three-dimensional structures. These pairwise-atom data are converted into a pseudopotential, also known as a mean-force potential, that describes the preferred geometries of the protein–ligand pairwise atoms (Wallqvist and Covell 1996). This strategy tends to capture the physical–chemical complexity of binding effects that are more specific. However, this approach is highly dependent on the number and diversity of the experimental structures used. The use of these functions can be as rapid as the use of empirical functions due to the simplicity of their terms. Examples of knowledge-based scoring functions include DrugScore (Velec et al. 2005) and PMF (Muegge 2006).
It is important to note that no universal scoring function exists that can be successfully applied to all molecular docking studies. To increase the efficiency and reliability of molecular docking experiments, the strategy indicated most often is to identify and use the scoring function that is best adapted to the problem under investigation. It is necessary to know whether all atom types of both ligand and protein are set in the scoring function chosen. The best choice is to use a function that has been parameterised and tested for the particular protein family and ligand class under study; e.g. for the study of carbohydrates, a scoring function should be used that has included this ligand class in its training set during the parameterisation process.
In this context, the consensus scoring approach has been employed to overcome the deficiencies of individual scoring functions (Charifson et al. 1999). The goal of this strategy is to use more than one evaluation function through a consensus scheme to increase the performance of both pose prediction and binding affinity estimation. Various studies and strategies exist concerning consensus scoring that provide notable improvements in docking accuracy (Feher 2006). Consensus scoring is a distinct strategy from the rescoring approach, which aims to evaluate docking poses with different scoring functions in the same docking program. The consensus scoring approach makes use of diverse scoring functions to evaluate the ligands using results obtained from distinct experiments performed using different docking software. Several studies have investigated the consensus efficiency for evaluating ligand poses generated by search methods and have reported significant improvements in pose prediction and affinity estimation when various scoring schemes are used in a consensus manner (Verdonk et al. 2003; Jacobsson et al. 2003; Vigers and Rizzi 2004; Oda et al. 2006; Feher 2006; Cheng et al. 2009; Chang et al. 2010; Kukol 2011; Houston and Walkinshaw 2013). In an interesting study, eight different scoring functions were combined to predict the experimental binding conformation and affinity for 40 matrix metalloproteinase (MMP) inhibitors using PCA (Principal Component Analysis) and PLS (Partial Least-Squares Projections onto Latent Structures), respectively (Terp et al. 2001).
Target-specific scoring functions
Target-specific scoring functions have emerged as an alternative approach to improving the prediction of experimental binding affinity. This type of scoring function has been developed and parameterised by focusing on a specific target family. Hence, it is expected that this scoring function type will be more efficient in accounting for specific interactions and particular binding characteristics associated with a target class of interest.
Several studies have reported improvements in both pose prediction and binding affinity accuracy when employing a target-specific scoring function rather than a universal, non-specific scoring function (Seifert 2009; Li et al. 2011a; Lu and Wang 2012). For instance, Logean et al. (2001) adapted the Fresno empirical scoring function to the class I MHC HLA-B*2705 protein with a significant improvement in affinity prediction over six different traditional scoring functions. Xue et al. (2010) developed a kinase family-specific PMF scoring function that achieved better affinity prediction of diverse ATP-competitive inhibitors compared to eight standard evaluation functions (the rescoring tool is freely available at http://202.127.30.184:8080/scoring/index.jsp). The GOLD program also implements a modified version of the ChemScore function, with an additional term that accounts for weak hydrogen bonds that are important for some kinase-inhibitor binding events (Pierce et al. 2002; Verdonk et al. 2004).
Estimating binding affinity with scoring functions
Some scoring functions have been developed especially to identify native and near-native binding modes. For such functions, affinity data are not considered during the parameterisation process, and it is important to bear this feature in mind prior to a docking study. The GoldScore scoring function (Verdonk et al. 2003), which is implemented in the GOLD software, and the MMFF94S-based function, used by the DockThor program (de Magalhães et al. 2004b), are examples of scoring functions used with the aim of pose prediction because no experimental affinity data were considered during the parameterisation process. When this type of function is used, ideally the top-ranked poses of each ligand are selected, and another scoring function is applied to obtain a more reliable binding affinity prediction.
In addition to predicting the native binding mode of the ligand, a scoring function must also be capable of discriminating active from inactive compounds and of ranking them according to their affinity. The scoring functions developed to date have shown a weak correlation with experimental affinity data (Plewczynski et al. 2011), whereas more sophisticated methods for predicting binding free energies (e.g. free energy perturbation methods, FEP, linear interaction energy, LIE, Aqvist et al. 1994; de Amorim et al. 2008; Gutiérrez-de-Terán and Aqvist 2012; MM-PBSA, ollman et al. 2000; and MM-GBSA, Wang et al. 2001) are computationally too expensive to be applied in virtual screening studies. Recent reviews in the literature have emphasised the importance of developing non-linear scoring functions derived through machine-learning methods and the use of consensus-scoring approaches (Plewczynski et al. 2011; Wang and Lin 2013; Yuriev and Ramsland 2013).
Exploring the energy landscape in docking
Similar to protein folding, the binding process of a ligand to a protein can be associated with an energy landscape in which the different binding modes correspond to energy wells, which are separated by energetic barriers (Rejto and Verkhivker 1996; Miller and Dill 1997; Wang and Verkhivker 2003; Mobley and Dill 2009; Yan and Wang 2012). The native binding mode is represented as the lowest energy pose, and non-native conformations are the local minimum energy poses.
Usually, docking studies are conducted to obtain a single lowest-energy ligand pose (the native binding mode). However, some ligands might assume multiple binding modes in the active site, e.g. due to ligand or protein symmetry, as with some HIV-1 protease inhibitors (Hong et al. 1998; Stoll et al. 2002; Weber et al. 2002) and in cases where the symmetry is less important, e.g. the thiocamphor substrate of cytochrome P450cam (Paulsen and Ornstein 1993). In addition to pose prediction, some studies have shown that the consideration of multiple binding modes significantly improves affinity prediction (Wang et al. 2005; Stjernschantz and Oostenbrink 2010; Yan and Wang 2012).
Stjernschantz and Oostenbrink (2010) developed an iterative scheme for automatic pose selection for use in multiple molecular dynamics simulations that generates weighted ensembles used in the LIE affinity prediction method. This strategy was applied to a set of 12 thioureas, which bind to cytochrome P450 2C9 with known affinity but with unknown experimental binding modes. The authors found significant improvement in the affinity prediction with a RMS error of 2.9 kJ/mol and a Spearman rank of 0.69 when using a model containing only the ensembles derived from molecular docking experiments.
Yan and Wang (2012) developed a knowledge-based scoring function and an optimisation method for simultaneously maximising the specificity and affinity of some COX-2 inhibitors (named SPA; Specificity and Affinity). The study was guided by their specific funnelled energy landscape theory; this theory considers the native binding mode as the lowest energy conformation while non-native poses are represented by a statistical Gaussian distribution. This approach was developed to improve the native binding mode and affinity prediction to distinguish selective from non-selective compounds more accurately. For the purpose of specificity, the authors considered selective inhibitors to be more potent than non-selective inhibitors, making an analogy with the native pose (selective ligand) and non-native poses (non-selective ligands). The traditional virtual screening goals were achieved with a success rate of 84.7 % (RMSD criterion = 2.0 Å) for the native binding mode and a Spearman rank of 0.733 for the affinity prediction. The ability to detect selective compounds was also successfully validated for a COX-2 target based on a set of 37 selective and 20 non-selective inhibitors using the Kolmogorov–Smirnov statistical discrimination test.
Protein flexibility and binding affinity prediction
Despite achieving satisfactory results with respect to pose prediction and virtual screening, affinity prediction remains a challenging task in the docking field (Kolb and Irwin 2009; Kokh et al. 2011; Forrey et al. 2012). As mentioned previously, most docking methods consider the protein as a rigid body; i.e. they do not consider receptor flexibility during docking. Due to the great importance of protein motions for binding, several studies indicate that considering both ligand and protein flexibility in the search process and in the pose evaluation steps is a promising strategy for improving affinity prediction (Yang et al. 2006, 2009; Cavasotto and Singh 2008; Moitessier et al. 2009; Kinnings and Jackson 2009; Tuffery and Derreumaux 2011; Forrey et al. 2012). The treatment of protein flexibility with scoring functions has been addressed by using a wide range of distinct strategies with slight to significant improvements in both pose and affinity predictions (Abagyan et al. 1994; Knegtel et al. 1997; Sotriffer and Dramburg 2005; Sherman et al. 2006; Yang et al. 2006; Meiler and Baker 2006; Morris et al. 2009; Craig et al. 2010; Neves et al. 2012; Shin and Seok 2012).
Despite the encouraging results obtained for binding affinity prediction by considering both ligand and protein as flexible entities, great efforts are necessary to improve the treatment of the enthalpic and entropic contributions during docking to obtain a more realistic and successful estimation of the binding free energies using scoring functions (Beier and Zacharias 2010).
Virtual screening
Pharmaceutical companies and research groups engaged in the discovery of new drug candidates are increasingly looking for faster, more effective and more reliable docking methodologies. The virtual screening (VS) strategy has emerged as an important tool in the search for lead compounds. VS is used to computationally analyse a very large number of compounds to select the most probable active compounds for a particular molecular target according to some pre-defined criteria. This approach can be used to complement the results obtained by experimental high-throughput screening studies (Doman et al. 2002).
The search for ligands in virtual screening studies can be performed using diverse compound libraries that are available online, e.g. the ZINC database (Irwin et al. 2012), BindingDB (Liu et al. 2007), PubChem (Bolton et al. 2008), SuperNatural (Dunkel 2006) and ChEMBL (Gaulton et al. 2011). At these web portals, searches can be conducted using physicochemical properties defined by the user, e.g. the number of rotatable bonds and logP, or by drawing a desired fragment common to all molecules. These filters are frequently used to reduce the number of compounds to be analysed computationally and to specify a particular profile for the compounds. After selecting the list of molecules to be extracted, these web servers usually provide a summary table with the main chemical properties of the ligands. If necessary, as when building an in-house library of compounds, it is possible to use tools that filter and quantify physical–chemical properties, such as FAF-Drugs (Lagorce et al. 2008). Furthermore, the correct assignment of both protein and ligand tautomeric and protonation states, as well as accounting for important water molecules and metal ions, is crucial for virtual screening accuracy and requires careful inspection (Kolb and Irwin 2009; Martin 2009; Kalliokoski et al. 2009; Petukh et al. 2013; Madhavi Sastry et al. 2013).
Several virtual screening approaches are available, which can be broadly classified according to the methodology employed: (1) based on the receptor structure (structure-based virtual screening, SBVS) and (2) based on only the ligand structure (ligand-based virtual screening, LBVS) (Krüger and Evers 2010; Ripphausen et al. 2011; Cheng et al. 2012). Method (1) is most commonly used when a high-quality three-dimensional structure of the receptor is available or when it is possible to obtain such a structure computationally, e.g. by comparative modelling. In this strategy, a molecular docking study is performed for all previously selected compounds. It is also possible, as in traditional molecular docking studies, to consider receptor flexibility through one of the several currently available methods described above. However, the computational cost significantly increases when protein flexibility is considered in virtual screening studies.
Although virtual screening is a widely used technique, the protocol chosen needs to be validated to increase the reliability of the experiments. First, it is important to evaluate whether the search method and the scoring function are able to reproduce the experimental binding mode of the ligand originally complexed with the target. The ability of the protocol to differentiate active from inactive molecules must also be verified. This validation is of great importance in virtual screening because it helps to reduce the number of inactive molecules to be forwarded to experimental tests, which are more expensive and time consuming. The proportion of active and inactive molecules present in a set of compounds with known experimental affinities can be evaluated through enrichment factor (EF) measurements. Molecules that are presumably inactive (decoys) can exhibit similar physical properties to active compounds, e.g. logP, molecular weight and the number of rings present, while being topologically distinct (Huang et al. 2006). Molecular docking is usually carried out with a test set containing both active and decoy ligands; then, the EF can be used to measure the ability of the function to rank a determined fraction of the active ligands at the top positions. The performance of different virtual screening protocols varies significantly according to the validation studies performed and is directly influenced by the methodology used as well as by the test set composition (the class of receptors covered and the ligand profiles included) (Bissantz et al. 2000). Furthermore, when the numbers of active and inactive compounds are similar, the AUC method (area under the ROC curve) is more appropriate for evaluating the performance of virtual screening protocols (Jain 2000).
The selected compounds (known as hits) are then subjected to the experimental step, e.g. chemical synthesis (in the case of theoretical compounds or compounds not available for purchase) and in vitro and in vivo studies of enzymatic and pharmacological activity. Based on these results, the most promising compounds are usually returned to the theoretical phase for optimisation to adjust their potency and/or selectivity as well as their physicochemical properties to improve the pharmacokinetics.
Conclusions
Currently, molecular docking is a standard computational tool that has been successfully employed in drug design and discovery studies. Nonetheless, some theoretical and computational challenges remain to be overcome; doing so would increase the predictive power and widen the applications of this important computational tool.
Satisfactory docking results can be obtained when relatively small ligands with few rotatable bonds are docked against protein binding pockets in which flexibility does not play an important role. However, it is worth noting that even when dealing with relatively simple docking problems, some factors related to ligand and protein preparation can lead to a non-successful docking prediction. Furthermore, current docking predictions for highly flexible ligands are less reliable, and more sophisticated search strategies may be necessary.
Protein flexibility and the fact that protein behaviour is not always related to a single conformation but rather to an ensemble of conformations, are important challenges to be overcome by further docking methodological developments. Moreover, the free energy landscape profile, i.e. multiple ligand binding modes associated with multiple minima with distinct shapes and energy barriers, must be addressed in further scoring function developments to improve binding affinity estimation (Mobley and Dill 2009; Marsh et al. 2012). Thus, the inclusion of the protein flexibility, the identification of multiple ligand binding modes and binding affinity prediction remain as three major, interconnecting docking challenges that require continuous effort to develop more reliable docking methods.
Acknowledgments
The authors would like to thank FAPERJ project grant number N. E-26/102.443/2009 and CNPq project grant number N. 307062/2010-4.
Conflict of interest
Isabella Alvim Guedes, Camila Silva de Magalhães, and Laurent Emmanuel Dardenne declare that they have no conflict of interest.
Human & animal studies
This article does not contain any studies with human or animal subjects performed by the any of the authors.
Footnotes
Special Issue: Advances in Biophysics in Latin America
Contributor Information
Isabella A. Guedes, Email: isabella.alvimg@gmail.com
Camila S. de Magalhães, Email: camila.mag@gmail.com
Laurent E. Dardenne, Email: dardenne@lncc.br
References
- Abagyan R, Totrov M, Kuznetsov D. ICM: A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]
- Apostolakis J, Plückthun A, Caflisch A. Docking small ligands in flexible binding sites. J Comput Chem. 1998;19:21–37. [Google Scholar]
- Aqvist J, Medina C, Samuelsson JE. A new method for predicting binding affinity in computer-aided drug design. Protein Eng. 1994;7:385–391. doi: 10.1093/protein/7.3.385. [DOI] [PubMed] [Google Scholar]
- Armen RS, Chen J, Brooks CL. An Evaluation of Explicit Receptor Flexibility in Molecular Docking Using Molecular Dynamics and Torsion Angle Molecular Dynamics. J Chem Theory Comput. 2009;5:2909–2923. doi: 10.1021/ct900262t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asses Y, Venkatraman V, Leroux V, et al. Exploring c-Met kinase flexibility by sampling and clustering its conformational space. Proteins Struct Funct Bioinform. 2012;80:1227–1238. doi: 10.1002/prot.24021. [DOI] [PubMed] [Google Scholar]
- Ballester PJ, Mitchell JBO. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics. 2010;26:1169–1175. doi: 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter CA, Murray CW, Clark DE, et al. Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins. 1998;33:367–382. [PubMed] [Google Scholar]
- Beier C, Zacharias M. Tackling the challenges posed by target flexibility in drug design. Expert Opin Drug Discov. 2010;5:347–359. doi: 10.1517/17460441003713462. [DOI] [PubMed] [Google Scholar]
- Bissantz C, Folkers G, Rognan D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J Med Chem. 2000;43:4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- Böhm HJ. The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J Comput Aided Mol Des. 1992;6:61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- Bolton EE, Wang Y, Thiessen PA, Bryant SH (2008) Chapter 12 in. Annual Reports in Computational Chemistry, vol 4. American Chemical Society, Washington, DC, pp 217–241
- B-Rao C, Subramanian J, Sharma SD. Managing protein flexibility in docking and its applications. Drug Discov Today. 2009;14:394–400. doi: 10.1016/j.drudis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Brooijmans N, Humblet C. Chemical space sampling by different scoring functions and crystal structures. J Comput Aided Mol Des. 2010;24:433–447. doi: 10.1007/s10822-010-9356-2. [DOI] [PubMed] [Google Scholar]
- Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu Rev Biophys Biomol Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- Carlson HA. Protein flexibility is an important component of structure-based drug discovery. Curr Pharm Des. 2002;8:1571–1578. doi: 10.2174/1381612023394232. [DOI] [PubMed] [Google Scholar]
- Cavasotto C, Singh N. Docking and High Throughput Docking: Successes and the Challenge of Protein Flexibility. Curr Comput Aided-Drug Des. 2008;4:221–234. [Google Scholar]
- Chang MW, Ayeni C, Breuer S, Torbett BE. Virtual screening for HIV protease inhibitors: a comparison of AutoDock 4 and Vina. PLoS ONE. 2010;5:e11955. doi: 10.1371/journal.pone.0011955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux J-P, Edelstein S. Conformational selection or induced-fit? 50 years of debate resolved. Biol Rep. 2011 doi: 10.3410/B3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charifson PS, Corkery JJ, Murcko MA, Walters WP. Consensus Scoring: A Method for Obtaining Improved Hit Rates from Docking Databases of Three-Dimensional Structures into Proteins. J Med Chem. 1999;42:5100–5109. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- Cheng T, Li X, Li Y, et al. Comparative Assessment of Scoring Functions on a Diverse Test Set. J Chem Inf Model. 2009;49:1079–1093. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- Cheng T, Li Q, Zhou Z, et al. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 2012;14:133–141. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DE, Westhead DR. Evolutionary algorithms in computer-aided molecular design. J Comput Aided Mol Des. 1996;10:337–358. doi: 10.1007/BF00124503. [DOI] [PubMed] [Google Scholar]
- Clark DE, Wiley InterScience (Online service) Evolutionary algorithms in molecular design. Weinheim: Wiley-VCH; 2000. [Google Scholar]
- Craig IR, Essex JW, Spiegel K. Ensemble Docking into Multiple Crystallographically Derived Protein Structures: An Evaluation Based on the Statistical Analysis of Enrichments. J Chem Inf Model. 2010;50:511–524. doi: 10.1021/ci900407c. [DOI] [PubMed] [Google Scholar]
- Damm-Ganamet KL, Smith RD, Dunbar JB, et al. CSAR Benchmark Exercise 2011–2012: Evaluation of Results from Docking and Relative Ranking of Blinded Congeneric Series. J. Chem. Inf. Model. 2013;53:1853–1870. doi: 10.1021/ci400025f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Amorim HLN, Caceres RA, Netz PA. Linear interaction energy (LIE) method in lead discovery and optimization. Curr Drug Targets. 2008;9:1100–1105. doi: 10.2174/138945008786949360. [DOI] [PubMed] [Google Scholar]
- De Azevedo WF, Jr, Dias R. Computational methods for calculation of ligand-binding affinity. Curr Drug Targets. 2008;9:1031–1039. doi: 10.2174/138945008786949405. [DOI] [PubMed] [Google Scholar]
- de Magalhães CS, Barbosa HJC, Dardenne LE (2004a) Selection-Insertion Schemes in Genetic Algorithms for the Flexible Ligand Docking Problem. In: Deb K (ed) Genetic Evolutionary Computation, GECCO 2004. Springer, Berlin, pp 368–379
- de Magalhães CS, Barbosa HJC, Dardenne LE. A genetic algorithm for the ligand-protein docking problem. Genet Mol Biol. 2004b;27:605–610. [Google Scholar]
- Desmet J, Wilson IA, Joniau M, et al. Computation of the binding of fully flexible peptides to proteins with flexible side chains. FASEB J. 1997;11:164–172. doi: 10.1096/fasebj.11.2.9039959. [DOI] [PubMed] [Google Scholar]
- Dietzen M, Zotenko E, Hildebrandt A, Lengauer T. On the applicability of elastic network normal modes in small-molecule docking. J Chem Inf Model. 2012;52:844–856. doi: 10.1021/ci2004847. [DOI] [PubMed] [Google Scholar]
- Doman TN, McGovern SL, Witherbee BJ, et al. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- Dunkel M. SuperNatural: a searchable database of available natural compounds. Nucleic Acids Res. 2006;34:D678–D683. doi: 10.1093/nar/gkj132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge MD, Murray CW, Auton TR, et al. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- Feher M. Consensus scoring for protein–ligand interactions. Drug Discov Today. 2006;11:421–428. doi: 10.1016/j.drudis.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Ferrari AM, Wei BQ, Costantino L, Shoichet BK. Soft docking and multiple receptor conformations in virtual screening. J Med Chem. 2004;47:5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J, Tristram F, Wenzel W. Modeling loop backbone flexibility in receptor-ligand docking simulations. J Comput Chem. 2012;33:2504–2515. doi: 10.1002/jcc.23087. [DOI] [PubMed] [Google Scholar]
- Forrey C, Douglas JF, Gilson MK. The fundamental role of flexibility on the strength of molecular binding. Soft Matter. 2012;8:6385. doi: 10.1039/C2SM25160D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Frimurer TM, Peters GH, Iversen LF, et al. Ligand-induced conformational changes: improved predictions of ligand binding conformations and affinities. Biophys J. 2003;84:2273–2281. doi: 10.1016/S0006-3495(03)75033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicchio E, Lapelosa M, Levy RM. Binding Energy Distribution Analysis Method (BEDAM) for Estimation of Protein − Ligand Binding Affinities. J Chem Theory Comput. 2010;6:2961–2977. doi: 10.1021/ct1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Olson AJ. Automated docking of substrates to proteins by simulated annealing. Proteins. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-de-Terán H, Aqvist J. Linear interaction energy: method and applications in drug design. Methods Mol Biol (Clifton NJ) 2012;819:305–323. doi: 10.1007/978-1-61779-465-0_20. [DOI] [PubMed] [Google Scholar]
- Haider MK, Bertrand H-O, Hubbard RE. Predicting Fragment Binding Poses Using a Combined MCSS MM-GBSA Approach. J Chem Inf Model. 2011;51:1092–1105. doi: 10.1021/ci100469n. [DOI] [PubMed] [Google Scholar]
- Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- Hao M, Li Y, Zhang S-W, Yang W. Investigation on the binding mode of benzothiophene analogues as potent factor IXa (FIXa) inhibitors in thrombosis by CoMFA, docking and molecular dynamic studies. J Enzyme Inhib Med Chem. 2011;26:792–804. doi: 10.3109/14756366.2011.554414. [DOI] [PubMed] [Google Scholar]
- Hong L, Hartsuck JA, Foundling S, et al. Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant. Protein Sci. 1998;7:300–305. doi: 10.1002/pro.5560070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DR, Walkinshaw MD. Consensus Docking: Improving the Reliability of Docking in a Virtual Screening Context. J Chem Inf Model. 2013;53:384–390. doi: 10.1021/ci300399w. [DOI] [PubMed] [Google Scholar]
- Huang S-Y, Zou X. Advances and Challenges in Protein-Ligand Docking. Int J Mol Sci. 2010;11:3016–3034. doi: 10.3390/ijms11083016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Shoichet BK, Irwin JJ. Benchmarking Sets for Molecular Docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Wong CF, Wheeler RA. Flexible protein-flexible ligand docking with disrupted velocity simulated annealing. Proteins. 2008;71:440–454. doi: 10.1002/prot.21781. [DOI] [PubMed] [Google Scholar]
- Huang S-Y, Grinter SZ, Zou X. Scoring functions and their evaluation methods for protein-ligand docking: recent advances and future directions. Phys Chem Chem Phys. 2010;12:12899–12908. doi: 10.1039/c0cp00151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, et al. ZINC: A Free Tool to Discover Chemistry for Biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson M, Lidén P, Stjernschantz E, et al. Improving Structure-Based Virtual Screening by Multivariate Analysis of Scoring Data. J Med Chem. 2003;46:5781–5789. doi: 10.1021/jm030896t. [DOI] [PubMed] [Google Scholar]
- Jain AN. Morphological similarity: a 3D molecular similarity method correlated with protein-ligand recognition. J Comput Aided Mol Des. 2000;14:199–213. doi: 10.1023/a:1008100132405. [DOI] [PubMed] [Google Scholar]
- Jain AN, Nicholls A. Recommendations for evaluation of computational methods. J Comput Aided Mol Des. 2008;22:133–139. doi: 10.1007/s10822-008-9196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Kim SH. “Soft docking”: matching of molecular surface cubes. J Mol Biol. 1991;219:79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, et al. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kalliokoski T, Salo HS, Lahtela-Kakkonen M, Poso A. The effect of ligand-based tautomer and protomer prediction on structure-based virtual screening. J Chem Inf Model. 2009;49:2742–2748. doi: 10.1021/ci900364w. [DOI] [PubMed] [Google Scholar]
- Kar G, Keskin O, Gursoy A, Nussinov R. Allostery and population shift in drug discovery. Curr Opin Pharmacol. 2010;10:715–722. doi: 10.1016/j.coph.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keserû GM, Kolossváry I. Fully flexible low-mode docking: application to induced fit in HIV integrase. J Am Chem Soc. 2001;123:12708–12709. doi: 10.1021/ja0160086. [DOI] [PubMed] [Google Scholar]
- Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. J Comput Chem. 2008;29:1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnings SL, Jackson RM. LigMatch: a multiple structure-based ligand matching method for 3D virtual screening. J Chem Inf Model. 2009;49:2056–2066. doi: 10.1021/ci900204y. [DOI] [PubMed] [Google Scholar]
- Knegtel RM, Kuntz ID, Oshiro CM. Molecular docking to ensembles of protein structures. J Mol Biol. 1997;266:424–440. doi: 10.1006/jmbi.1996.0776. [DOI] [PubMed] [Google Scholar]
- Kokh DB, Wade RC, Wenzel W. Receptor flexibility in small-molecule docking calculations. Wiley Interdiscip Rev Comput Mol Sci. 2011;1:298–314. [Google Scholar]
- Kolb P, Irwin JJ. Docking screens: right for the right reasons? Curr Top Med Chem. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman PA, Massova I, Reyes C, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Kolossváry I, Guida WC. Low-mode conformational search elucidated: Application to C39H80 and flexible docking of 9-deazaguanine inhibitors into PNP. J Comput Chem. 1999;20:1671–1684. [Google Scholar]
- Kolossváry I, Keserû GM. Hessian-free low-mode conformational search for large-scale protein loop optimization: application to c-jun N-terminal kinase JNK3. J Comput Chem. 2001;22:21–30. [Google Scholar]
- Korb O, Stützle T, Exner TE, et al. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. In: Dorigo M, Gambardella LM, Birattari M, et al., editors. Ant Colony Optimisation Swarm Intelligence. Berlin: Springer; 2006. pp. 247–258. [Google Scholar]
- Korb O, Stützle T, Exner TE. Empirical Scoring Functions for Advanced Protein − Ligand Docking with PLANTS. J Chem Inf Model. 2009;49:84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- Korb O, McCabe P, Cole J. The Ensemble Performance Index: An Improved Measure for Assessing Ensemble Pose Prediction Performance. J Chem Inf Model. 2011;51:2915–2919. doi: 10.1021/ci2002796. [DOI] [PubMed] [Google Scholar]
- Korb O, Olsson TSG, Bowden SJ, et al. Potential and Limitations of Ensemble Docking. J Chem Inf Model. 2012;52:1262–1274. doi: 10.1021/ci2005934. [DOI] [PubMed] [Google Scholar]
- Krüger DM, Evers A. Comparison of structure- and ligand-based virtual screening protocols considering hit list complementarity and enrichment factors. Chem Med Chem. 2010;5:148–158. doi: 10.1002/cmdc.200900314. [DOI] [PubMed] [Google Scholar]
- Kuhl FS, Crippen GM, Friesen DK. A combinatorial algorithm for calculating ligand binding. J Comput Chem. 1984;5:24–34. [Google Scholar]
- Kukol A. Consensus virtual screening approaches to predict protein ligands. Eur J Med Chem. 2011;46:4661–4664. doi: 10.1016/j.ejmech.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Lagorce D, Sperandio O, Galons H, et al. FAF-Drugs2: Free ADME/tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinform. 2008;9:396. doi: 10.1186/1471-2105-9-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang PT, Brozell SR, Mukherjee S, et al. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15:1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach AR. Ligand docking to proteins with discrete side-chain flexibility. J Mol Biol. 1994;235:345–356. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu Z, Wang R. Test MM-PB/SA on True Conformational Ensembles of Protein − Ligand Complexes. J Chem Inf Model. 2010;50:1682–1692. doi: 10.1021/ci100036a. [DOI] [PubMed] [Google Scholar]
- Li L, Khanna M, Jo I, et al. Target-Specific Support Vector Machine Scoring in Structure-Based Virtual Screening: Computational Validation, In Vitro Testing in Kinases, and Effects on Lung Cancer Cell Proliferation. J Chem Inf Model. 2011;51:755–759. doi: 10.1021/ci100490w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kim DJ, Ma W, et al. Discovery of Novel Checkpoint Kinase 1 Inhibitors by Virtual Screening Based on Multiple Crystal Structures. J Chem Inf Model. 2011;51:2904–2914. doi: 10.1021/ci200257b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-B, Yang L-L, Wang W-J, et al. ID-Score: A New Empirical Scoring Function Based on a Comprehensive Set of Descriptors Related to Protein–Ligand Interactions. J Chem Inf Model. 2013;53:592–600. doi: 10.1021/ci300493w. [DOI] [PubMed] [Google Scholar]
- Lill MA. Efficient Incorporation of Protein Flexibility and Dynamics into Molecular Docking Simulations. Biochemistry (Mosc) 2011;50:6157–6169. doi: 10.1021/bi2004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Lin Y, Wen X, et al. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logean A, Sette A, Rognan D. Customized versus universal scoring functions: application to class I MHC-peptide binding free energy predictions. Bioorg Med Chem Lett. 2001;11:675–679. doi: 10.1016/s0960-894x(01)00021-x. [DOI] [PubMed] [Google Scholar]
- Lu I-L, Wang H. Protein-specific Scoring Method for Ligand Discovery. J Comput Biol. 2012;19:1215–1226. doi: 10.1089/cmb.2012.0188. [DOI] [PubMed] [Google Scholar]
- Luty BA, Wasserman ZR, Stouten PFW, et al. A molecular mechanics/grid method for evaluation of ligand-receptor interactions. J Comput Chem. 1995;16:454–464. [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Madhavi Sastry G, Adzhigirey M, Day T, et al. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Maghsoudi AH, Khodagholi F, Hadi-Alijanvand H, et al. Homology modeling, docking, molecular dynamics simulation, and structural analyses of coxsakievirus B3 2A protease: an enzyme involved in the pathogenesis of inflammatory myocarditis. Int J Biol Macromol. 2011;49:487–492. doi: 10.1016/j.ijbiomac.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Mangoni M, Roccatano D, Di Nola A. Docking of flexible ligands to flexible receptors in solution by molecular dynamics simulation. Proteins. 1999;35:153–162. doi: 10.1002/(sici)1097-0134(19990501)35:2<153::aid-prot2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Teichmann SA, Forman-Kay JD. Probing the diverse landscape of protein flexibility and binding. Curr Opin Struct Biol. 2012;22:643–650. doi: 10.1016/j.sbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Martin YC. Let’s not forget tautomers. J Comput Aided Mol Des. 2009;23:693–704. doi: 10.1007/s10822-009-9303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- Miller DW, Dill KA. Ligand binding to proteins: The binding landscape model. Protein Sci. 1997;6:2166–2179. doi: 10.1002/pro.5560061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Kearsley SK, Underwood DJ, Sheridan RP. FLOG: a system to select “quasi-flexible” ligands complementary to a receptor of known three-dimensional structure. J Comput Aided Mol Des. 1994;8:153–174. doi: 10.1007/BF00119865. [DOI] [PubMed] [Google Scholar]
- Mizutani MY, Takamatsu Y, Ichinose T, et al. Effective handling of induced-fit motion in flexible docking. Proteins. 2006;63:878–891. doi: 10.1002/prot.20931. [DOI] [PubMed] [Google Scholar]
- Mobley DL, Dill KA. Binding of Small-Molecule Ligands to Proteins: “What You See” Is Not Always “What You Get. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitessier N, Englebienne P, Lee D, et al. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go: Docking/scoring methods-a review. Br J Pharmacol. 2009;153:S7–S26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge I. PMF scoring revisited. J Med Chem. 2006;49:5895–5902. doi: 10.1021/jm050038s. [DOI] [PubMed] [Google Scholar]
- Nabuurs SB, Wagener M, de Vlieg J. A Flexible Approach to Induced Fit Docking. J Med Chem. 2007;50:6507–6518. doi: 10.1021/jm070593p. [DOI] [PubMed] [Google Scholar]
- Neves MAC, Totrov M, Abagyan R. Docking and scoring with ICM: the benchmarking results and strategies for improvement. J Comput Aided Mol Des. 2012;26:675–686. doi: 10.1007/s10822-012-9547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SE, Baron R, Ivetac A, McCammon JA. Predictive Power of Molecular Dynamics Receptor Structures in Virtual Screening. J Chem Inf Model. 2011;51:1439–1446. doi: 10.1021/ci200117n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibata Y, Itai A. Automatic creation of drug candidate structures based on receptor structure. Starting point for artificial lead generation. Tetrahedron. 1991;47:8985–8990. [Google Scholar]
- Nishibata Y, Itai A. Confirmation of usefulness of a structure construction program based on three-dimensional receptor structure for rational lead generation. J Med Chem. 1993;36:2921–2928. doi: 10.1021/jm00072a011. [DOI] [PubMed] [Google Scholar]
- Novoa EM, de Pouplana LR, Barril X, Orozco M. Ensemble Docking from Homology Models. J Chem Theory Comput. 2010;6:2547–2557. doi: 10.1021/ct100246y. [DOI] [PubMed] [Google Scholar]
- Nowosielski M, Hoffmann M, Kuron A, et al. The MM2QM tool for combining docking, molecular dynamics, molecular mechanics, and quantum mechanics†. J Comput Chem. 2013;34:750–756. doi: 10.1002/jcc.23192. [DOI] [PubMed] [Google Scholar]
- Oda A, Tsuchida K, Takakura T, et al. Comparison of Consensus Scoring Strategies for Evaluating Computational Models of Protein − Ligand Complexes. J Chem Inf Model. 2006;46:380–391. doi: 10.1021/ci050283k. [DOI] [PubMed] [Google Scholar]
- Park S-J, Kufareva I, Abagyan R. Improved docking, screening and selectivity prediction for small molecule nuclear receptor modulators using conformational ensembles. J Comput Aided Mol Des. 2010;24:459–471. doi: 10.1007/s10822-010-9362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen MD, Ornstein RL. Substrate mobility in thiocamphor-bound cytochrome P450cam: an explanation of the conflict between the observed product profile and the X-ray structure. Protein Eng. 1993;6:359–365. doi: 10.1093/protein/6.4.359. [DOI] [PubMed] [Google Scholar]
- Pearce BC, Langley DR, Kang J, et al. E-novo: an automated workflow for efficient structure-based lead optimization. J Chem Inf Model. 2009;49:1797–1809. doi: 10.1021/ci900073k. [DOI] [PubMed] [Google Scholar]
- Pei J, Wang Q, Liu Z, et al. PSI-DOCK: towards highly efficient and accurate flexible ligand docking. Proteins. 2006;62:934–946. doi: 10.1002/prot.20790. [DOI] [PubMed] [Google Scholar]
- Petukh M, Stefl S, Alexov E. The role of protonation states in ligand-receptor recognition and binding. Curr Pharm Des. 2013;19:4182–4190. doi: 10.2174/1381612811319230004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AC, Sandretto KL, Bemis GW. Kinase inhibitors and the case for CH…O hydrogen bonds in protein-ligand binding. Proteins. 2002;49:567–576. doi: 10.1002/prot.10259. [DOI] [PubMed] [Google Scholar]
- Plewczynski D, Łaźniewski M, Augustyniak R, Ginalski K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J Comput Chem. 2011;32:742–755. doi: 10.1002/jcc.21643. [DOI] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T. Time-efficient docking of flexible ligands into active sites of proteins. Proc Int Conf Intell Syst Mol Biol. 1995;3:300–308. [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T. Multiple automatic base selection: protein-ligand docking based on incremental construction without manual intervention. J Comput Aided Mol Des. 1997;11:369–384. doi: 10.1023/a:1007913026166. [DOI] [PubMed] [Google Scholar]
- Rejto PA, Verkhivker GM. Unraveling principles of lead discovery: from unfrustrated energy landscapes to novel molecular anchors. Proc Natl Acad Sci USA. 1996;93:8945–8950. doi: 10.1073/pnas.93.17.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripphausen P, Nisius B, Bajorath J. State-of-the-art in ligand-based virtual screening. Drug Discov Today. 2011;16:372–376. doi: 10.1016/j.drudis.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Rueda M, Bottegoni G, Abagyan R. Consistent Improvement of Cross-Docking Results Using Binding Site Ensembles Generated with Elastic Network Normal Modes. J Chem Inf Model. 2009;49:716–725. doi: 10.1021/ci8003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda M, Bottegoni G, Abagyan R. Recipes for the Selection of Experimental Protein Conformations for Virtual Screening. J Chem Inf Model. 2010;50:186–193. doi: 10.1021/ci9003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer L, Verkhivker GM. Predicting structural effects in HIV-1 protease mutant complexes with flexible ligand docking and protein side-chain optimization. Proteins. 1998;33:295–310. doi: 10.1002/(sici)1097-0134(19981101)33:2<295::aid-prot12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Schlosser J, Rarey M. Beyond the virtual screening paradigm: structure-based searching for new lead compounds. J Chem Inf Model. 2009;49:800–809. doi: 10.1021/ci9000212. [DOI] [PubMed] [Google Scholar]
- Schnecke V, Kuhn L. Virtual screening with solvation and ligand-induced complementarity. Perspect Drug Discov Des. 2000;20:171–190. [Google Scholar]
- Seifert MHJ. Targeted scoring functions for virtual screening. Drug Discov Today. 2009;14:562–569. doi: 10.1016/j.drudis.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Sherman W, Day T, Jacobson MP, et al. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Shin W-H, Seok C. GalaxyDock: Protein–Ligand Docking with Flexible Protein Side-chains. J Chem Inf Model. 2012;52:3225–3232. doi: 10.1021/ci300342z. [DOI] [PubMed] [Google Scholar]
- Shoichet BK, Leach AR, Kuntz ID. Ligand solvation in molecular docking. Proteins. 1999;34:4–16. doi: 10.1002/(sici)1097-0134(19990101)34:1<4::aid-prot2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Smellie AS, Crippen GM, Richards WG. Fast drug-receptor mapping by site-directed distances: a novel method of predicting new pharmacological leads. J Chem Inf Model. 1991;31:386–392. doi: 10.1021/ci00003a004. [DOI] [PubMed] [Google Scholar]
- Söderhjelm P, Kongsted J, Genheden S, Ryde U. Estimates of ligand-binding affinities supported by quantum mechanical methods. Interdiscip Sci Comput Life Sci. 2010;2:21–37. doi: 10.1007/s12539-010-0083-0. [DOI] [PubMed] [Google Scholar]
- Sokkar P, Sathis V, Ramachandran M. Computational modeling on the recognition of the HRE motif by HIF-1: molecular docking and molecular dynamics studies. J Mol Model. 2011;18:1691–1700. doi: 10.1007/s00894-011-1150-0. [DOI] [PubMed] [Google Scholar]
- Sotriffer CA, Dramburg I. “In situ cross-docking” to simultaneously address multiple targets. J Med Chem. 2005;48:3122–3125. doi: 10.1021/jm050075j. [DOI] [PubMed] [Google Scholar]
- Sperandio O, Mouawad L, Pinto E, et al. How to choose relevant multiple receptor conformations for virtual screening: a test case of Cdk2 and normal mode analysis. Eur Biophys J EBJ. 2010;39:1365–1372. doi: 10.1007/s00249-010-0592-0. [DOI] [PubMed] [Google Scholar]
- Stjernschantz E, Oostenbrink C. Improved ligand-protein binding affinity predictions using multiple binding modes. Biophys J. 2010;98:2682–2691. doi: 10.1016/j.bpj.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll V, Qin W, Stewart KD, et al. X-ray crystallographic structure of ABT-378 (lopinavir) bound to HIV-1 protease. Bioorg Med Chem. 2002;10:2803–2806. doi: 10.1016/s0968-0896(02)00051-2. [DOI] [PubMed] [Google Scholar]
- Takaya D, Yamashita A, Kamijo K, et al. A new method for induced fit docking (GENIUS) and its application to virtual screening of novel HCV NS3-4A protease inhibitors. Bioorg Med Chem. 2011;19:6892–6905. doi: 10.1016/j.bmc.2011.09.023. [DOI] [PubMed] [Google Scholar]
- Teodoro ML, Kavraki LE. Conformational flexibility models for the receptor in structure based drug design. Curr Pharm Des. 2003;9:1635–1648. doi: 10.2174/1381612033454595. [DOI] [PubMed] [Google Scholar]
- Teodoro ML, Phillips GN, Jr, Kavraki LE. Understanding protein flexibility through dimensionality reduction. J Comput Biol. 2003;10:617–634. doi: 10.1089/10665270360688228. [DOI] [PubMed] [Google Scholar]
- Teramoto R, Kashima H. Prediction of protein–ligand binding affinities using multiple instance learning. J Mol Graph Model. 2010;29:492–497. doi: 10.1016/j.jmgm.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Terp GE, Johansen BN, Christensen IT, Jørgensen FS. A new concept for multidimensional selection of ligand conformations (MultiSelect) and multidimensional scoring (MultiScore) of protein-ligand binding affinities. J Med Chem. 2001;44:2333–2343. doi: 10.1021/jm001090l. [DOI] [PubMed] [Google Scholar]
- Trosset J-Y, Scheraga HA. Prodock: Software package for protein modeling and docking. J Comput Chem. 1999;20:412–427. [Google Scholar]
- Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proc Natl Acad Sci USA. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffery P, Derreumaux P. Flexibility and binding affinity in protein-ligand, protein-protein and multi-component protein interactions: limitations of current computational approaches. J R Soc Interface. 2011;9:20–33. doi: 10.1098/rsif.2011.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velec HFG, Gohlke H, Klebe G. DrugScore(CSD)-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J Med Chem. 2005;48:6296–6303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- Venkatraman V, Ritchie DW. Flexible protein docking refinement using pose-dependent normal mode analysis. Proteins. 2012;80:2262–2274. doi: 10.1002/prot.24115. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn MJ, et al. Improved protein-ligand docking using GOLD. Proteins Struct Funct Bioinform. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, Berdini V, Hartshorn MJ, et al. Virtual Screening Using Protein-Ligand Docking: Avoiding Artificial Enrichment. J Chem Inf Model. 2004;44:793–806. doi: 10.1021/ci034289q. [DOI] [PubMed] [Google Scholar]
- Vigers GPA, Rizzi JP. Multiple active site corrections for docking and virtual screening. J Med Chem. 2004;47:80–89. doi: 10.1021/jm030161o. [DOI] [PubMed] [Google Scholar]
- Wallqvist A, Covell DG. Docking enzyme-inhibitor complexes using a preference-based free-energy surface. Proteins. 1996;25:403–419. doi: 10.1002/prot.1. [DOI] [PubMed] [Google Scholar]
- Wang J-C, Lin J-H. Scoring functions for prediction of protein-ligand interactions. Curr Pharm Des. 2013;19:2174–2182. doi: 10.2174/1381612811319120005. [DOI] [PubMed] [Google Scholar]
- Wang J, Verkhivker GM. Energy landscape theory, funnels, specificity, and optimal criterion of biomolecular binding. Phys Rev Lett. 2003;90:188101. doi: 10.1103/PhysRevLett.90.188101. [DOI] [PubMed] [Google Scholar]
- Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- Wang R, Lu Y, Wang S. Comparative Evaluation of 11 Scoring Functions for Molecular Docking. J Med Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- Wang J, Kang X, Kuntz ID, Kollman PA. Hierarchical database screenings for HIV-1 reverse transcriptase using a pharmacophore model, rigid docking, solvation docking, and MM-PB/SA. J Med Chem. 2005;48:2432–2444. doi: 10.1021/jm049606e. [DOI] [PubMed] [Google Scholar]
- Weber J, Mesters JR, Lepsík M, et al. Unusual binding mode of an HIV-1 protease inhibitor explains its potency against multi-drug-resistant virus strains. J Mol Biol. 2002;324:739–754. doi: 10.1016/s0022-2836(02)01139-7. [DOI] [PubMed] [Google Scholar]
- Welch W, Ruppert J, Jain AN. Hammerhead: fast, fully automated docking of flexible ligands to protein binding sites. Chem Biol. 1996;3:449–462. doi: 10.1016/s1074-5521(96)90093-9. [DOI] [PubMed] [Google Scholar]
- Wu G, Robertson DH, Brooks CL, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER?A CHARMm-based MD docking algorithm. J Comput Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- Xue M, Zheng M, Xiong B, et al. Knowledge-Based Scoring Functions in Drug Design. 1. Developing a Target-Specific Method for Kinase − Ligand Interactions. J Chem Inf Model. 2010;50:1378–1386. doi: 10.1021/ci100182c. [DOI] [PubMed] [Google Scholar]
- Yan Z, Wang J. Specificity quantification of biomolecular recognition and its implication for drug discovery. Sci Rep. 2012 doi: 10.1038/srep00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-Y, Wang R, Wang S. M-Score: A Knowledge-Based Potential Scoring Function Accounting for Protein Atom Mobility. J Med Chem. 2006;49:5903–5911. doi: 10.1021/jm050043w. [DOI] [PubMed] [Google Scholar]
- Yang C-Y, Sun H, Chen J, et al. Importance of Ligand Reorganization Free Energy in Protein − Ligand Binding-Affinity Prediction. J Am Chem Soc. 2009;131:13709–13721. doi: 10.1021/ja9039373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuriev E, Ramsland PA. Latest developments in molecular docking: 2010-2011 in review. J Mol Recognit JMR. 2013;26:215–239. doi: 10.1002/jmr.2266. [DOI] [PubMed] [Google Scholar]
- Zacharias M, Sklenar H. Harmonic modes as variables to approximately account for receptor flexibility in ligand-receptor docking simulations: Application to DNA minor groove ligand complex. J Comput Chem. 1999;20:287–300. [Google Scholar]
- Zhang C, Vasmatzis G, Cornette JL, DeLisi C. Determination of atomic desolvation energies from the structures of crystallized proteins. J Mol Biol. 1997;267:707–726. doi: 10.1006/jmbi.1996.0859. [DOI] [PubMed] [Google Scholar]
- Zsoldos Z, Reid D, Simon A, et al. eHiTS: a new fast, exhaustive flexible ligand docking system. J Mol Graph Model. 2007;26:198–212. doi: 10.1016/j.jmgm.2006.06.002. [DOI] [PubMed] [Google Scholar]