Abstract
Wnt signaling pathways are required for a wide variety of biological processes ranging from embryonic development to tissue repair and regeneration. Dickkopf-2 (DKK2) is classically defined as a canonical Wnt inhibitor, though it may play a role in activating non-canonical Wnt pathways in the context of endothelial network formation after acute injury. Here we report the discovery of a fusion partner for a DKK2 polypeptide that significantly improves the expression, biochemical properties and pharmacokinetics (PK) of the DKK2 polypeptide. Specifically, human serum albumin (HSA) was identified as a highly effective fusion partner. Substitution of selected amino acid residues in DKK2 designed to decrease heparan sulfate binding by HSA-DKK2 variants, further improved the PK properties of the molecule in rodents. The HSA-DKK2 variants were monomeric, as thermally stable as wild type, and active as measured by their ability to bind to and prevent phosphorylation of the Wnt coreceptor LRP6. Our engineering efforts resulted in potent long-lived variants of the canonical Wnt inhibitor DKK2, applicable for Wnt pathway manipulation either by systematic delivery or focused administration at sites of tissue injury.
Keywords: Dickkopf-2, heparan sulfate, human serum albumin, tissue injury, Wnt signaling
Introduction
The Wnt signaling pathway is critical for multiple aspects of development, including organogenesis, stem cell renewal and cell fate decisions, as well as for adult tissue homeostasis and metabolism. Dysregulation of Wnt signaling can result in disease pathologies such as cancer and a loss of cell and organ integrity (Clevers and Nusse, 2012). Wnt signals are transduced by the Frizzled (Fzd) family of transmembrane proteins. Frizzled cell-surface receptors play essential roles in both canonical and non-canonical Wnt signaling. In the canonical pathway, Wnt proteins form a ternary complex with Fzd and the low-density-lipoprotein receptor-related proteins 5 and 6 (LRP5 and LRP6). The activation of Fzd and LRP5/6 by Wnt ligands generates a signal that prevents the phosphorylation and subsequent degradation of β-catenin. This allows β-catenin to translocate and accumulate in the nucleus where it activates T-cell factor/lymphoid enhancer factor (TCF/LEF) target genes (MacDonald et al., 2009). The non-canonical Wnt signaling pathway is less well defined. There are at least two non-canonical Wnt signaling pathways that have been proposed, including the planar cell polarity pathway and the Wnt/Ca++ pathway (van Amerongen, 2012).
Dickkopf-2 (DKK2) is a secreted polypeptide originally discovered as an antagonist of the canonical Wnt signaling pathway (Glinka et al., 1998; Bafico et al., 2001; Mao et al., 2001; Semenov et al., 2001). DKK2 contains two cysteine-rich domains, C1 and C2, each containing 10 conserved cysteines (Li et al., 2002; Chen et al., 2008). The C1 domain of human DKK2 protein is between amino acid positions 78 and 127 and the C2 domain of human DKK2 protein is between amino acid positions 183 and 256. Wnt3a-specific antagonism by DKK2 requires the binding of the C-terminal cysteine-rich domain to the Wnt coreceptor LRP5/6 (Li et al., 2002; Bourhis et al., 2010). The DKK2-LRP5/6 complex antagonizes canonical Wnt signaling by inhibiting LRP5/6 interaction with Wnt, and by forming a ternary complex with the transmembrane protein Kremen that promotes clathrin-mediated internalization of LRP5/6 (Mao et al., 2002). In addition to its role in regulating canonical Wnt signaling, DKK2 has been shown to promote repair after acute ischemic injury (Lin et al., 2010) by acting through the non-canonical Wnt pathway to induce endothelial cell migration and blood vessel formation (Min et al., 2011).
Like many low molecular weight proteins, DKK2 has a very short serum half-life, which is due in part to heparan sulfate (HS) binding (see below). HS is a sulfated polysaccharide covalently attached to proteoglycans present on the surface of most cells that mediates interactions between different proteins (Sarrazin et al., 2011). HS binding by proteins is conferred by patches of positively charged amino acids that bind to negatively charged sulfate groups on HS (Silvian et al., 2006). Interactions through HS decrease serum exposure of proteins, resulting in reduced serum half-life (Hartmann et al., 1998; Pepinsky et al., 2002; Datta-Mannan et al., 2013). Consistent with this, twice daily injections of DKK2 cysteine-rich domain 2 (DKK2C2) were required in a mouse kidney ischemic reperfusion model to gain therapeutic effects in order to compensate for its poor exposure (Lin et al., 2010). In the studies by Min et al. (2011), this issue was circumvented by local administration of DKK2 at the injury site. Many strategies have been developed to extend serum half-life of proteins, including conjugation with high molecular weight chemical polymers such as polyethylene glycol (PEG), insertion of glycosylation sites to add carbohydrate or genetic fusions with the Fc domain of antibodies, human serum albumin (HSA) or polyamino acids such as XTEN (Strohl, 2015; Kontermann, 2016). There is no single strategy that can be applied successfully to all proteins, but rather it often requires extensive optimization to develop a molecule with the desired pharmacological properties. For HS-binding proteins, added size of the engineered proteins is often not sufficient and further modifications are needed to partially eliminate or sterically block the HS sites (Pepinsky et al., 2002; Shapiro et al., 2003; Datta-Mannan et al., 2013). Site-directed mutagenesis has been used successfully to disrupt HS-binding sites (Hartmann et al., 1998; Silvian et al., 2006; Runeberg-Roos et al., 2016). Alternatively, glycosylation or PEGylation sites can be incorporated at positions where they can interfere sterically with unwanted properties such as HS binding (Pepinsky et al., 2002; Sinclair and Elliott, 2005).
To address potential therapeutic applications, we generated a potent long-acting variant of DKK2 that could be administered either for modulation of non-canonical Wnt signaling to stimulate repair and revascularization following tissue injury or for the inhibition of canonical Wnt signaling to modulate cell proliferative responses. For instance, overriding the down-regulation of DKK2 by microRNAs overexpressed in gliomas and squamous cell carcinomas, by way of exogenous DKK2 delivery, could prove effective for the treatment of activated canonical Wnt signaling in these cancers (Li et al., 2013; Kawakita et al., 2014). Since the C-terminal C2 domain of DKK2 is both necessary and sufficient for its function (Li et al., 2002; Mao and Niehrs, 2003), we engineered a series of recombinant versions of this molecule containing mutations that reduce or eliminate HS binding as HSA fusion proteins and analyzed the effects of mutations on protein stability, efficacy in canonical Wnt signaling assays and serum half-life in rodents. We generated a potent and selective version of DKK2C2 with comparable stability and LRP6 binding as wild-type DKK2C2. The HSA fusion protein exhibited an extended pharmacokinetic (PK) profile in rodents, being detectable in serum after 2 days, whereas the DKK2C2 protein alone could only be detected for 30 min. Administration of this molecule either systemically or at the site of tissue injury is predicted to substantially augment the ability of endogenous DKK2 to impact Wnt signaling.
Materials and methods
Expression of DKK2C2 in Escherichia coli
The C2 domain of murine DKK2C2 was produced in E. coli using the methods described in US Patent 8,470,554 with slight modifications to the purification protocol.
The amino acid sequence encoded by the construct is provided below (thioredoxin (TRX) bolded, hexa-his tag underlined, S-tag italicized, thrombin cleavage sites bolded and underlined, enterokinase cleavage site italicized and underlined, and DKK2C2 in lower case):
MSDKIIHLTDDSFDTDVLKADGAILVDFWAEWCGPCKMIAPILDEIADEYQGKLTVAKLNIDQNPGTAPKYGIRGIPTLLLFKNGEVAATKVGALSKGQLKEFLDANLAGSGSGHMHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTDDDDKALVPRGSmphikghegdpclrssdcidgfccarhfwtkickpvlhqgevctkqrkkgshgleifqrcdcakglsckvwkdatysskarlhvcqki
The TRX-DKK2C2 fusion expression vector was transformed into an ORIGAMI™ B strain of E. coli (Invitrogen). Cells were grown in Luria-Bertani media. Protein expression was induced with 0.2 mM isopropyl-1-thio-β-D-galactoside. SDS-PAGE was used to verify protein expression. Cells were harvested by centrifugation at 5000×g for 20 min at 4°C. The cell pellet was resuspended in lysis buffer (25 mM Bis-Tris, pH 6.8, 500 mM NaCl, 5 mM MgCl2 and 2% Glycerol) containing protease inhibitor cocktail (Roche). The cell suspension was passed through a Microfluidizer Processor (Model M110L, Microfluidics, Newton, MA) twice at 10 000 psi. Clarified lysate, generated from centrifugation at 13 000 rpm for 20 min at 4°C, was loaded by gravity onto a Ni-NTA agarose (Qiagen) column using 1 ml resin/10 ml lysate. The column was washed sequentially with 5-column volumes of lysis buffer followed by lysis buffer containing 50 mM imidazole. The fusion protein was eluted with 5-column volumes of lysis buffer containing 250 mM imidazole.
DKK2C2 was cleaved from the TRX–DKK2C2 fusion with thrombin from human plasma (Sigma) at a ratio of 100 units/l of original culture for 4 h at room temperature (RT) and then overnight at 4°C. Thrombin was removed with benzamidine-sepharose 4 FF (GE Healthcare) at 2 ml/1000 units.
Acetic acid was added to a final concentration of 5% and the sample centrifuged at 5000×g for 20 min. The supernatant was filtered and loaded onto a C8 SepPak column (Waters) equilibrated in 0.1% trifluoroacetic acid (TFA). A 5 g column was used for a 10-l preparation. The column was washed with 5-column volumes of 0.1% TFA, and DKK2C2 was eluted with 5-column volumes of 0.1% TFA with 20%, acetonitrile. The sample was lyophilized and dissolved in phosphate buffered saline (PBS): 20 mM Na2HPO4, 150 mM sodium chloride pH 6.0. Mass spectrometry of reduced and non-reduced samples indicated the protein contained 5 disulfide bonds; however, further disulfide analysis identified significant scrambling (Supplementary Table SI).
Expression and purification of HSA fusions of DKK2C2
HSA-DKK2C2 from five constructs (ACE461: HSA-huDKK2 (M172-I259); ACE463: HSA-huDKK2 (M172-I259 S173P); ACE464: HSA-huDKK2 (H174-I259); ACE465: HSA-huDKK2 (K176-I259) and ACE466: HSA-huDKK2 (H178-I259)) was purified from 300 ml of transient culture medium. For preparation of the conditioned medium, transfected Chinese hamster ovary (CHO) cells were expanded in serum-free media, grown to high density, fed with supplements and then shifted to a reduced temperature for up to 14 days or until cell viability started to drop. Conditioned media were harvested by centrifugation and clarified by 0.45-μm filtration. The HSA-DKK2C2 fusion proteins were purified on a CaptureSelect™ HSA (ThermoFisher Scientific) affinity column using 0.5 M arginine/1 M NaCl as the elution buffer, followed by gel filtration on Superdex 200 (GE Healthcare) in 10 mM sodium succinate pH 5.5, 75 mM NaCl, 100 mM arginine. Samples were analyzed for absorbance at 280 nm, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing and non-reducing conditions, analytical size exclusion chromatography (SEC), mass spectrometry and potency in the Super TopFlash (STF) assay. The amino acid sequences of the above-noted HSA-DKK2C2 fusions are indicated in Supplementary Table SII.
ACE464 was purified from 5 l of culture medium from a stable CHO cell line by cation exchange chromatography on SP-Sepharose Fast Flow (GE Healthcare) and SEC on Sephacryl S200 (GE Healthcare). The clarified culture medium was directly loaded onto a SP-Sepharose column. The column was washed with 20 mM Na2HPO4 pH 7.0, 150 mM NaCl and ACE464 was eluted with 20 mM Na2HPO4 pH 7.0, 1 M NaCl. The SP elute was loaded onto a Sephacryl S200 gel filtration column equilibrated in 10 mM sodium succinate pH 5.5, 75 mM NaCl, 100 mM arginine. Approximately 400 mg of ACE464 was recovered. The product was >95% pure by SDS-PAGE and contained <0.25% aggregate by analytical SEC. This large-scale preparation of HSA-DKK2C2 was used in rat and mouse PK studies.
A reengineered version of ACE464, ACE486: (HSA (D25-L609)-DKK2C2 (H174-I259)), was produced to eliminate the heterogeneity of the product caused by the partial retention of the pro-domain in HSA. ACE486 from 600 ml clarified culture medium from a stable CHO cell line was purified on SP-Sepharose and SEC on Sephacryl S200. The culture medium was loaded by gravity onto a SP-Sepharose Fast Flow column (GE Healthcare). The column was washed sequentially with 20 mM Na2HPO4 pH 7.0, 50 mM NaCl; 20 mM Na2HPO4 pH 7.0, 100 mM NaCl and 20 mM Na2HPO4 pH 7.0, 150 mM NaCl. HSA-DKK2C2 was eluted from the column with 20 mM Na2HPO4 pH 7.0, 300 mM NaCl. Fractions were analyzed for absorbance at 280 nm and by SDS-PAGE. Peak fractions containing 120 mg of ACE486 were pooled, filtered through a 0.2-μm membrane, and concentrated. The protein was loaded onto a HiPrep 26/60 Sephacryl S200 high-resolution column (GE Healthcare) in 10 mM sodium succinate pH 5.5, 75 mM NaCl, 100 mM arginine. Samples in the effluent were analyzed for absorbance at 280 nm and by SDS-PAGE.
The amino acid sequence for ACE464 is (signal peptide underlined, 7 residues removed in ACE486 crossed out, and human DKK2C2 in lower case):
METDTLLLWVLLLWVPGAHASRGVFRRDAHKSEVAHRFKDLGEENFKALVLIAFAQYLQQCPFEDHVKLVNEVTEFAKTCVADESAENCDKSLHTLFGDKLCTVATLRETYGEMADCCAKQEPERNECFLQHKDDNPNLPRLVRPEVDVMCTAFHDNEETFLKKYLYEIARRHPYFYAPELLFFAKRYKAAFTECCQAADKAACLLPKLDELRDEGKASSAKQRLKCASLQKFGERAFKAWAVARLSQRFPKAEFAEVSKLVTDLTKVHTECCHGDLLECADDRADLAKYICENQDSISSKLKECCEKPLLEKSHCIAEVENDEMPADLPSLAADFVESKDVCKNYAEAKDVFLGMFLYEYARRHPDYSVVLLLRLAKTYETTLEKCCAAADPHECYAKVFDEFKPLVEEPQNLIKQNCELFEQLGEYKFQNALLVRYTKKVPQVSTPTLVEVSRNLGKVGSKC CKHPEAKRMPCAEDYLSVVLNQLCVLHEKTPVSDRVTKCCTESLVNRRPCFSALEVDETYVPKEFNAETFTFHADICTLSEKERQIKKQTALVELVKHKPKATKEQLKAVMDDFAAFVEKCCKADDKETCFAEEGKKLVAASQAALGLGShikghegdpclrssdciegfccarhfwtkickpvlhqgevctkqrkkgshgleifqrcdcakglsckvwkdatysskarlhvcqki
Of note, human DKK2C2 (H174-I259) differs from murine DKK2C2 sequence, used to generate material in E. coli, by a single amino acid: E191D.
Other human DKK2C2 product modalities that were expressed and characterized included DKK2C2 (M172-I259) with a human CD169 signal sequence produced by CHO cells; DKK2C2 (H174-I259) as an XTEN fusion protein produced by CHO cells with DKK2C2 attached to the C-terminus of XTEN-144, and DKK2C2 (M172-I259) as Fc fusion proteins attached to the N- and C-terminus of the human IgG1 hinge and Fc by CHO and human embryonic kidney HEK293 cells. Observed titers were <0.1 mg/l for DKK2C2 alone, ~100 mg/l for XTEN-DKK2C2 and 30–100 mg/l for the four preparations of Fc fusion proteins. XTEN-DKK2C2 was purified from clarified conditioned medium on Q-Sepharose, and DKK2C2 Fc-fusions on Protein A Sepharose followed by Heparin Sepharose.
Expression and purification of HSA-DKK2C2 mutants
A total of 15 different variants of DKK2C2 were designed, engineered into the ACE486 construct, and expressed by CHO cells. The calculated isoelectric point (pI) for the HSA-DKK2C2 fusion proteins is a composite of the different pI values for HSA, which is ~5, and for DKK2C2, which is >9. The pI of the DKK2C2 portion was impacted by the charge mutations thus influencing the chromatographic properties of the fusion proteins, which had to be experimentally determined for each. Three of the constructs (ACE502, ACE506 and ACE507) retained tight binding for SP-Sepharose at pH 6.5 and were purified following the same protocol used for the wild-type proteins. Five constructs (BKM225, BKM226, BKM227, ACE506 and ACE507) failed to bind at pH 6.5, but bound at pH 5.5 following a 1:1 dilution of the culture medium with water. Five constructs (ACE504, BKM230, BKM231, BKM232 and BKM233) failed to bind at pH 6.5 or 5.5, but bound at pH 5.0 following a 1:2 dilution of the culture medium with water. Four constructs (ACE503, ACE505, BKM228 and BKM229) failed to bind to SP-Sepharose under any of the test conditions and were purified on Fractogel EMD TMAE (M) resin (Merck Millipore) at pH 7.0. Because additional medium components bind to SP-Sepharose at the lower pH values and to Fractogel TMAE resin, additional salt steps were included in the wash and elution steps and SDS-PAGE was used to select peak elution fractions based on their purity. Elution fractions were scanned for absorbance at 280 nm using a Nanodrop 2000c (ThermoFisher Scientific) and relevant fractions were pooled, filtered with a 0.2-μm filter device and dialyzed overnight into PBS pH 7.04. For SP gravity purifications, 4 ml of resin was used per 300 ml of cell supernatant. Each supernatant was loaded onto a freshly poured column that was equilibrated in wash buffer. Purification quality of samples was examined by SDS-PAGE and analytical SEC on a Superdex 200 5/150 column at a flow rate of 0.2 ml/min with PBS. For SP-based purifications at pH 6.5, columns were washed with 10 column volumes of equilibration buffer and protein eluted with 10 mM sodium citrate, 1 M NaCl pH 6.5 in 8 fractions of 2 ml. For SP-based purifications at pH 5.5, columns were washed with two column volumes of 15 mM sodium citrate 50 mM NaCl pH 5.5, then two column volumes of PBS. The protein was eluted with 20 mM Na2HPO4 300 mM NaCl in five fractions of 3 ml. For SP-based purifications at pH 5.0, columns were washed with eight column volumes of 10 mM sodium citrate, 15 mM NaCl pH 5.0. The protein was eluted with steps containing increasing concentrations of NaCl up to 300 mM in 25 mM sodium citrate pH 6.0, in eight fractions of 3 ml (two fractions per concentration). For TMAE gravity purifications, 10 ml of resin was used per 100 ml of supernatant. The column was washed with two column volumes of 20 mM Na2HPO4 and 50 mM NaCl pH 7.0. The protein was eluted with increasing concentrations of NaCl up to 450 mM in 20 mM Na2HPO4, in 10 fractions of 5 ml (2 fractions per concentration). For two of the constructs (BKM231 and ACE503), there was extensive aggregation of the expressed HSA-DKK2C2 that was evident in the SDS-PAGE analysis of conditioned medium, indicated by staining in the higher molecular weight region of the gel. The aggregated forms of BKM231 and ACE503 were removed from monomer during the ion exchange chromatography purification step. Only the monomeric fraction was characterized in subsequent studies. The amino acid sequences of the HSA-DKK2C2 mutants are presented in Supplementary Table SII.
Animal studies
All animal studies were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, 1996) and approved by the Biogen institutional animal care and use committee. PK assessments in 12–16 week (20–25 g) C57BL/6 J mice and 3-month-old (250–320 g) Sprague–Dawley rats were performed following intravenous dosing through the tail vein (three animals per group). Blood was drawn from mice and serum prepared after 0.08, 0.25, 0.5, 1, 3, 6, 10 and 24 h. Rat serum was prepared after 0.25, 1, 3, 7 and 24 h. DKK2C2 and HSA-DKK2C2 levels in serum samples were analyzed both by enzyme-linked immunosorbent assay (ELISA) and western blotting. PK parameters were calculated by non-compartmental analysis using WinNonlin Software (WinNonlin professional version 5.0.1, Pharsight Inc, Mountain View, CA). While the ELISA was very sensitive and reproducible for detection of the unmodified DKK2C2, results for the HSA-DKK2C2 were highly variable and the method could not be established as a reliable readout. Conversely, the western blot analysis was not sensitive enough for detection of the unmodified DKK2C2. Consequently, different assays were used to quantify the unmodified and HSA-DKK2C2 products. Lung and kidney tissue lysates were generated and characterized using the western method.
Analysis of PK samples by ELISA
Levels of DKK2C2 in serum were measured using an ELISA protocol against a standard curve of DKK2C2. Specifically, serum samples were diluted 1:10 in PBS and coated onto a Nunc clear flat-bottom immuno non-sterile 96-well plate (ThermoFisher Scientific) blocked with fish gelatin blocking buffer (PBS, 0.5% fish gelatin, 0.1% Triton X-100 pH 7.4). A standard curve of E. coli DKK2C2 spiked into 10% mouse serum/PBS in a concentration series starting at 2 µg/ml, preceded by seven 3-fold dilutions, was included on the same plate. Following three washes with PBST (PBS, 0.1% Tween-20), wells were incubated at RT for 1 h with a biotinylated version of an affinity-purified anti-DKK2C2 antibody at 2 µg/ml in blocking buffer. Following three washes with PBST, wells were incubated at RT for 15 min with streptavidin-HRP (ThermoFisher Scientific) in a 1:8000 dilution in blocking buffer. Following three washes with PBST, wells were incubated at RT for 4 min with TMB substrate (0.1 M sodium acetate—citric acid pH 4.9, 0.42 mM TMB, 0.004% H2O2). Developed ELISAs were stopped by the addition of 2 N H2SO4 and plates were scanned at 450 nm using a Molecular Devices SpectraMax M5 microplate reader and data analyzed using Softmax Pro v5.4.4 software. The limit of quantitation (LOQ) in the ELISA assay for E. coli derived DKK2C2 was ~10 ng/ml.
Analysis of PK samples by western blotting
Levels of DKK2 in serum were measured using a quantitative western blot protocol against a standard curve of HSA-DKK2C2 in serum. Specifically, samples were diluted 1:10 in PBS and 7.5 μl was loaded onto a 4–12% Bis-Tris NuPAGE gel in 2-(N-morpholino)-ethanesulfonic acid (MES) buffer under non-reducing conditions. Gels were run at 200 V for 35 min and then transferred to nitrocellulose for 7 min at 20 V using a Life Technologies iBlot apparatus. Following 1 h blocking in Pierce protein-free T20 (PBS) blocking buffer (ThermoScientific) with 0.05% Tween-20, blots were incubated 1 h in 1:1000 Abcam rabbit anti-DKK2 (ab95274) and 1:1000 Abcam mouse anti-HSA (ab10241) in blocking buffer with 0.2% Tween-20. After four washes for 5 min in blocking buffer with 0.2% Tween-20, blots were incubated for 45 min with 1:5000 dilution of IRDye800CW donkey anti-rabbit IgG (LI-COR Biosciences) and 1:5000 dilution of IRDye680CW donkey anti-mouse IgG (LI-COR Biosciences) in blocking buffer with 0.2% Tween-20. After four washes for 5 min with PBST, blots were washed quickly four times with PBS and scanned on the Odyssey CLx reader (LI-COR Biosciences). Levels were quantified using ImageStudio software (LI-COR Biosciences) and concentrations determined by interpolation against the standard curve. Antibodies against HSA and DKK2 gave similar values. The LOQ by western blotting for HSA-DKK2C2 was ~1 µg/ml.
Antibody generation for detection of ACE504
The C-terminal DKK2 antibody Abcam ab95274 fails to detect ACE504 since the introduced amino acids substitutions overlap with the epitope recognizing DKK2C2. A high titer polyclonal antibody targeting DKK2C2 was produced in rabbits immunized with DKK2C2 material from E. coli. The DKK2 specific antibody was affinity purified on a DKK2C2 affinity column.
Native polyacrylamide gel electrophoresis
Samples (5 µg/lane) were analyzed by native PAGE under non-reducing conditions on a 4–20% PAGE gradient gel (Invitrogen) and stained with SimplyBlue SafeStain (ThermoFisher Scientific). SDS was omitted from both the running buffer (50 mM acetic acid pH 5.0, adjusted with Tris base) and sample buffer (50 mM acetic acid pH 5.0, adjusted with Tris base, 25% glycerol). The gel was run at 150 V for 4 h.
Differential scanning fluorimetry
Fifty microliters of protein at 2 mg/ml in 20 mM sodium citrate, 20 mM Na2HPO4, 0.1 M NaCl pH 7.5 was added to 5 µl of 100x SYPRO Orange Protein Gel Stain (ThermoFisher Scientific #S6650) diluted in buffer. Fifty microliters were delivered to each well of an ABI Prism 96-Well Optical Reaction Plate (Applied Biosystems #4 306 737) and analyzed with MXPro qPCR software on a Stratagene Mx3005P Real-time System (Agilent Technologies). The thermal denaturation method involved ramping temperature from 25°C to 95°C in 0.5°C increments for 142 cycles.
Dynamic light scattering
Filtered proteins, at 13 mg/ml in PBS pH 7.5, were diluted stepwise to 11, 9, 7, 5, 3, 1 and 0.5 mg/ml into seven adjacent wells of a Corning 384-well flat clear bottom black polystyrene microplate (Cat# 3540). The plate was loaded into a DynaPro PlateLoader II (Wyatt) at 25°C and analyzed with Dynamics software at a laser wavelength of 829 nm. Ten acquisitions were collected at 25°C for each sample where each acquisition was averaged over 5 s. Sample replicate data were averaged. The mutual diffusion coefficient, Dm, was determined for each molecule at protein concentrations between 0.5 and 13 mg/ml in PBS at pH 7.5. Values of the diffusion interaction parameter (KD) were calculated from the plot of Dm versus protein concentration, where KD is a ratio of the slope and the y-intercept (Hanlon et al., 2010; Lehermayr et al., 2011).
Heparin-sepharose chromatography
Approximately 100 µg of material in PBS (diluted ~20-fold) was loaded onto a 1 ml HiTrap Heparin HP column (GE Healthcare) in binding buffer (5 mM Na2HPO4, pH 6.5) on an AKTA Avant (GE Healthcare). The resin was washed with 5-column volumes of binding buffer followed by elution over 20 column volumes using a linear salt gradient to 1 M NaCl. Protein was monitored by absorbance at 280 nm and conductance in milliSieverts (mS).
Heparin-binding ELISA
Wild-type HSA-DKK2C2 (ACE464) and each of the HS-binding variants were examined for binding to heparin–biotin using ELISA. Nunc clear flat-bottom immuno non-sterile 96-well plates (ThermoFisher Scientific) were coated with 15 µg/ml of each of the HSA-DKK2C2 variants and incubated overnight at 4°C. Following three washes with PBST (20 mM Na2HPO4, 150 mM NaCl, 0.05% Tween-20), wells were incubated with ELISA blocking buffer HBSS (25 mM HEPES pH 7.0, 1% bovine serum albumin (BSA), 0.1% ovalbumin, 0.1% non-fat dry milk (NFDM), 0.001% NaN3) at RT for 1 h. Following three washes with PBST, wells were incubated at RT for 1 h with heparin–biotin sodium salt (Sigma-Aldrich) in a concentration series starting at 50 µg/ml (~4 µM), preceded by eight 5-fold dilutions in PBST, 0.05% BSA. Following three washes with PBST, wells were incubated at RT for 10 min with streptavidin-HRP (ThermoFisher Scientific) in a 1:8000 dilution in PBST, 0.05% BSA. Following two washes with PBST, wells were incubated at RT for 20 min with 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (0.1 M sodium acetate citric acid pH 4.9, 0.42 mM TMB, 0.004% H2O2). Developed ELISAs were stopped by the addition of 2 N H2SO4 and plates were scanned at 450 nm using a Molecular Devices SpectraMax M5 microplate reader. IC50 values were calculated with Softmax Pro v5.4.4 software.
LRP6 binding assay
HSA-DKK2C2 proteins were diluted (final concentration ranging from 2.5 to 15 µM) in cold Fluorescent-Activated Cell Sorting (FACS) buffer (1% fetal calf serum, 20 mM Na2HPO4 pH 7.0, 150 mM NaCl, 0.05% NaN3) in a Nunc 96-well conical bottom polypropylene plate (ThermoFisher Scientific) and eleven 3-fold serial dilutions were generated. Human LRP6-expressing BaF3 cells (50 000/well) suspended in cold FACS buffer were distributed to each well and incubated at 4°C for 1 h. Anti-LRP6 antibody generated in house using published sequence for YW211.31.57 human IgG1 agly (Bourhis et al., 2014) at 0.75 nM in cold FACS buffer was added to each well and incubated at 4°C for 10 min. Cells were washed twice with cold FACS buffer after centrifugation at 1500 rpm for 2 min at 4°C. Cell pellets were resuspended in goat anti-human kappa-phycoerythrin (Southern Biotech) diluted 1:300 in cold FACS buffer and incubated at 4°C for 1 h. Cells were pelleted by centrifugation at 1500 rpm for 2 min and washed once with cold FACS buffer. Cells were fixed with fixation buffer (1% paraformaldehyde, 20 mM Na2HPO4 pH 7.0, 150 mM NaCl) for 10 min at RT and then pelleted by centrifugation. Cells were resuspended in FACS buffer for analysis on a FACSCalibur Cell Analyzer (BD Biosciences).
STF assay
DKK2C2 molecules were assessed for their ability to inhibit canonical Wnt signaling utilizing the STF cell line (Xu et al., 2004). For each assay, STF cells were seeded into 96-well Purecoat amine plates (BD Biosciences) at a density of 4 × 104 cells/well in media. After 24 h, the media were aspirated and replaced with (i) control conditioned medium (negative control); (ii) Wnt3a conditioned medium (positive control) or (iii) Wnt3a conditioned medium and the DKK2 molecule (0–1000 nM). Samples were tested in triplicate. Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) + 10% fetal bovine serum (FBS). Wnt3a conditioned medium was generated using mouse L cells stably transfected with a full-length mouse Wnt3a construct (Xu et al., 2004). Control conditioned medium was derived from wild-type mouse L cells.
Luciferase activity was measured using the Luciferase Dual Glo kit (Promega) per manufacturer's instructions. Luciferase was measured on a Synergy H1 plate reader (BioTek). Data were normalized to the negative control to derive Wnt3a-induced fold changes. For PK studies, animal serum was diluted 1:5 in DMEM and added to Wnt3a conditioned medium. Standard curves were generated in pooled normal serum in an identical manner. Standard curve equations were generated using Prism (GraphPad).
Assessment of phospho-LRP6 levels
STF cells were seeded into 6 cm tissue culture plates at a density of 1 × 106 cells per well. Once cells reached 90% confluency, the medium was replaced with the following: (i) control conditioned medium (negative control); (ii) medium + 200 ng/ml recombinant human Wnt3a (R&D systems) (positive control) or (iii) medium + 200 ng/ml recombinant human Wnt3a + DKK2 molecule (at 250, 500 or 1000 nM). After 24 h, cells were lysed with cold RIPA buffer (Millipore) containing HALT protease + phosphatase inhibitors (ThermoFisher Scientific). An aliquot of the lysate was taken for a BCA assay (ThermoFisher Scientific) and the remaining lysate was denatured and reduced using Bolt 10X Sample Reducing Agent and 4X Bolt LDS Sample Buffer (ThermoFisher Scientific). Samples were heated at 95°C for 5 min and passed through a QIAShredder column (Qiagen). Protein (20 µg/lane) was subjected to SDS-PAGE on Bolt 4–12% Bis-Tris gradient gels (ThermoFisher Scientific) in Bolt MES gel running buffer. Protein was transferred onto nitrocellulose using the iBlot2 system (ThermoFisher Scientific). Duplicate blots were blocked in a 1:1 mix of Tris-buffered saline (TBS):Odyssey TBS blocking buffer (LI-COR Biosciences) for 1 h and then probed 16 h at 4°C with rabbit anti-pLRP6 Ser1490 (Cell Signaling), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling) or rabbit anti-LRP6 (Cell Signaling, clone C47E12) + mouse anti-GAPDH (Cell Signaling) in diluted blocking buffer. Blots were washed 4× for 5 min in TBS + 0.1% Tween-20 (TBST) at RT, then incubated for 2 h in 1:10 000 dilutions of IRDye800CW donkey anti-rabbit IgG (LI-COR Biosciences) and IRDye680CW donkey anti-mouse IgG (LI-COR Biosciences). Blots were then washed 4× in TBST at RT, imaged on an Odyssey CLx Imager (LI-COR Biosciences) and appropriate bands were quantified using ImageStudio v 4.0 (LI-COR Biosciences). pLRP6 and LRP6 values were normalized to GAPDH values. The pLRP6 values were divided by total LRP6 values to determine the proportion of pLRP6/LRP6, then normalized to pLRP6/LRP6 in control medium and further normalized to the 0 nM DKK2 construct, such that 0 nM had a value of 1 for each construct series.
Results
Characterization of recombinant E. coli derived DKK2C2
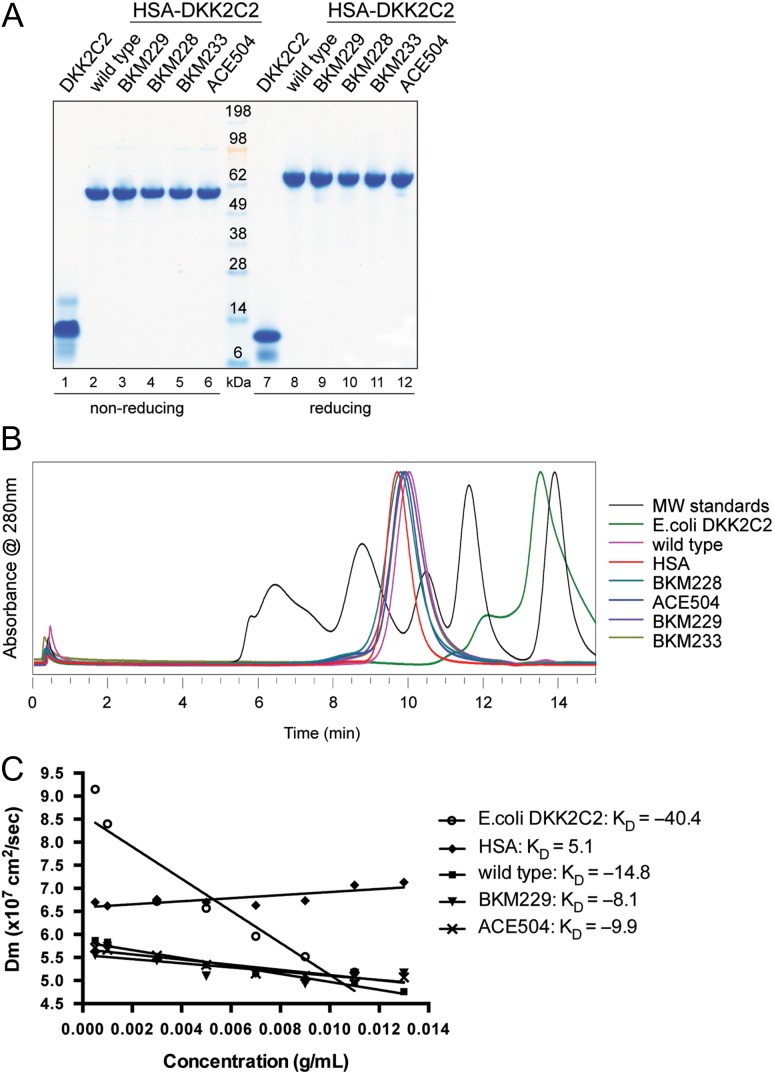
DKK2C2 was produced in E. coli, as previously described (Lin et al., 2010). The TRX-6xHis-S-tag-DKK2C2 fusion protein was subjected to a Ni-NTA agarose-based purification. After thrombin cleavage to remove the tag, the DKK2C2 protein was separated from proteolytic fragments using reverse phase chromatography and SEC. SDS-PAGE analysis of DKK2C2 revealed a prominent band of ~10 kDa with slightly faster mobility under reducing conditions (Fig. 1A; Lanes 1 and 7). The identity of the product was confirmed by mass spectrometry (calculated mass 10 109.8 Da; observed mass 10 109 Da). Observed titers of the TRX-DKK2C2 fusion protein were 5 mg/l and ~1 mg of purified DKK2C2 was recovered per liter of culture. The E. coli derived DKK2C2 was heterogeneous by SEC, with ~15% total contribution from aggregation (Fig. 1B). This is consistent with an observed propensity to self-associate, as measured by an assessment of diffusion interaction coefficients by dynamic light scattering (DLS, Fig. 1C), as well as extensive disulfide scrambling, as determined by mass spectrometry analysis (Supplementary Fig. S1 and Table SI).
Fig. 1.
Characterization of untagged DKK2C2 and HSA-DKK2C2 fusion proteins. (A) SDS-PAGE stained with SimplyBlue SafeStain examining SEC purified DKK2C2 proteins under both non-reducing (left) and reducing conditions (right). Lanes 1 and 7: E. coli derived untagged DKK2C2; Lanes 2 and 8: wild type HSA-DKK2C2 ACE464; Lanes 3 and 9: BKM229; Lanes 4 and 10: BKM228; Lanes 5 and 11: BKM233; Lanes 6 and 12: ACE504. (B) Graphical depiction of the elution profile (absorbance at 280 nm versus time in min) of E. coli derived untagged DKK2C2 (green); wild type HSA-DKK2C2 ACE464 (pink); HSA (red); BKM228 (cyan); ACE504 (blue); BKM229 (purple) and BKM233 (brown) by analytical SEC. The elution profile of molecular weight standards is shown in black. From left to right, the five peaks are 670, 158, 44, 17 and 1.4 kDa. (C) DLS analysis of DKK2C2 variants represented as a linear plot of mutual diffusion coefficient (Dm) versus concentration of DKK2C2. Variants are displayed with different markers: circle is E. coli derived untagged DKK2C2; diamond is HSA; square is wild type HSA-DKK2C2 ACE464; triangle is BKM229; cross is ACE504.
Since DKK2 is a known inhibitor of canonical Wnt signaling, we employed the STF assay (Xu et al., 2004) to assess the inhibitory activity of E. coli derived DKK2C2. The assay involves a HEK293 cell line stably transfected with a luciferase reporter under the control of seven TCF/LEF binding sites. As the binding of TCF/LEF to its target genes is a hallmark of active canonical Wnt signaling, STF is a robust system to measure a transcriptional readout of canonical Wnt signaling. Despite some degree of aggregation, E. coli derived DKK2C2 potently inhibited Wnt signaling, with an IC50 of 20 nM. PK analysis in mice revealed that this version of DKK2C2 was rapidly cleared. Intravenously administered DKK2C2 at 2 mg/kg was detected in serum at 5 ± 1.1 µg/ml after 5 min, and at 0.4 ± 0.3 µg/ml after 15 min, but then fell either at or below the LOQ of the assay at all later time points, both by ELISA (Fig. 2A) and in a cellular assay assessing Wnt signaling inhibition using the STF reporter (Fig. 2B) with no detectable protein present in serum after as little as 30 min. The rapid clearance of DKK2C2 from the bloodstream is likely attributable to its highly basic charge (pI of 9.1) and small size (~10 kDa).
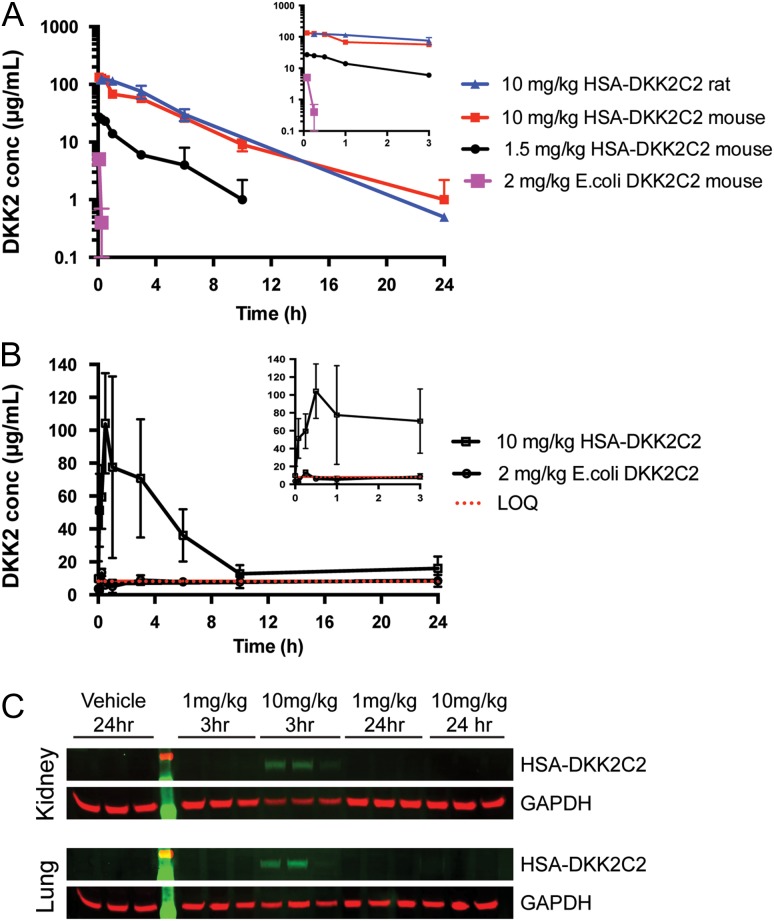
Fig. 2.
PK assessment of untagged DKK2C2 and HSA-DKK2C2. (A) A plasma-drug concentration–time curve from three cohorts of three mice intravenously dosed with 2 mg/kg of E. coli derived DKK2C2, 1.5 or 10 mg/kg of wild-type HSA-DKK2C2 (ACE464). A plasma-drug concentration–time curve for one cohort of three rats intravenously dosed with 10 mg/kg ACE464 is also illustrated. ACE464 was detected by quantitative western blotting, while E. coli derived DKK2C2 was measured by ELISA. (B) A graphical depiction of serum levels of ACE464 and E. coli derived DKK2C2 in mouse samples described in A, as measured by activity in the STF assay. The LOQ of the assay was 8.25 µg/ml (red dashed line). (C) Western blots examining ACE464 in lysates of mouse kidneys (top) and lungs (bottom) 3 or 24 h post intravenous dosing at 1 or 10 mg/kg. GAPDH was considered as a loading control.
We also attempted to produce the DKK2C2 protein in CHO cells as an alternative to E. coli expression, but observed titers in conditioned medium were <0.1 mg/l. SDS-PAGE/western blot analysis of the conditioned medium showed an immunoreactive band at the expected molecular weight under reduced conditions, but disulfide-linked aggregates were present when analyzed under non-reduced conditions and we were unable to purify the DKK2C2 from the conditioned medium (data not shown).
Expression and characterization of HSA fusions of DKK2C2
The poor expression, significant degree of aggregation and disulfide scrambling, and poor PK of E. coli derived DKK2C2 prompted an investigation into an alternative fusion partner for the DKK2 polypeptide. Based on a careful and extensive experimental analysis of different strategies to augment DKK2 as a protein therapeutic (see Discussion), HSA was chosen as a preferred fusion partner. Key findings that prompted the selection of the HSA fusion are summarized in the Discussion. HSA has many desirable pharmaceutical properties. These include a serum half-life of 19–20 days; solubility of ~300 mg/ml; good stability; ease of expression; no effector function; low immunogenicity and circulating serum levels of ~45 mg/ml (Rogers et al., 2015). The crystal structure of HSA with and without ligands, including biologically important molecules such as fatty acids and drugs, or in complex with other proteins is well documented (He and Carter, 1992; Sugio et al., 1999). According to X-ray crystallographic studies of HSA, this polypeptide forms a heart-shaped protein with approximate dimensions of 80 × 80 × 80 Å and a thickness of 30 Å. It has ~67% α-helix but no β-sheet and can be divided into three homologous domains (I–III). Each of these three domains comprised two subdomains (A and B). The A and B subdomains have six and four α-helices, respectively, connected by flexible loops. The principal regions of ligand binding are located in cavities in subdomains IIA and IIIA, which are formed mostly of hydrophobic and positively charged residues and exhibit similar chemistry. All but 1 of the 35 cysteine residues in the molecule are involved in the formation of 17 stabilizing disulfide bonds.
Selection of an appropriate linker length is an important step in the design of fusion proteins. If the linker is too short, the fusion partner can interfere with folding and/or activity of the protein, and if it is too long, it can be a potential target for degradation. To assess if the linker length connecting the HSA and DKK2C2 domain impacted productivity and/or product quality, five versions of HSA-DKK2C2 (ACE461 (M172-I259); ACE463 (M172-I259, S173P); ACE464 (H174-I259); ACE465 (K176-I259) and ACE466 (H178-I259)), each carrying different lengths of the natural spacer sequence that connects the C1 and C2 domains of DKK2, were expressed by CHO cells, affinity purified and subjected to gel filtration (see Materials and methods). All molecules were monomeric, stable and recovered at levels >100 mg/l. Mass spectrometry analysis confirmed intact molecules with no clipping. Furthermore, all molecules exhibited dose-dependent inhibition of canonical Wnt signaling in the STF assay. The HSA fusion did not alter the activity of the DKK2C2 molecule in the STF assay, as all HSA fusions were as equally potent as their untagged counterpart (data not shown). IC50 values ranged from 25 to 50 nM for the five different versions of HSA-DKK2C2. ACE466, the shortest version tested, showed a small 2-fold reduction in activity. Slightly better expression prompted us to select ACE464 as the lead candidate. ACE464 was very stable with no evidence of degradation after storage for >4 months at 4°C, incubation for 3 days at 37°C or after multiple freeze–thaw cycles. By SDS-PAGE, ACE464 ran as a homogeneous band (>95% purity) with molecular weight of 55 and 60 kDa under non-reducing and reducing conditions, respectively (Fig. 1A, Lanes 2 and 8), and it eluted as a single peak with molecular weight of 75 kDa by SEC (Fig. 1B). The disulfide connectivity in ACE464 was determined by mass spectrometry under reducing and non-reducing conditions following proteolytic digestion of the protein (Supplementary Fig. S1 and Table SIII). The disulfides were as predicted with low-level scrambling. Scrambling measurements for each of the 10 cysteines in the DKK2C2 domain ranged from 0% to 2% for the HSA fusion, as compared to 15–65% for E. coli derived DKK2C2 (SupplementaryFig. S1C). Also as expected, in the HSA region of the fusion protein, the single unpaired cysteine (Cys61) was greater than 90% cysteinylated (~8% free).
To test if the HSA fusion strategy extended PK compared to untagged E. coli derived DKK2C2, mice were intravenously injected with either 1.5 or 10 mg/kg ACE464 and serum was collected at 5 min, 15 min, 30 min, 1 h, 3 h, 6 h, 10 h and 24 h postinjection. The HSA fusion strategy greatly extended PK relative to E. coli derived DKK2C2, as measured by quantitative western blotting (Fig. 2A and SupplementaryFig. S2) and Wnt pathway inhibition in the STF cellular assay (Fig. 2B). ACE464 was detectable at the LOQ of 1 µg/ml out to 24 h following a single 10 mg/kg intravenous dose, or up to 7 h following a single 1.5 mg/kg intravenous dose. PK parameters calculated for the 10 mg/kg dose group revealed an area under the curve (AUC) value of 471 ± 52 µg × h/ml, a terminal half-life of 3.2 ± 0.8 h, clearance rate (CL) of 21 ± 2 ml/h/kg and a volume of distribution at steady state (Vss) of 94 ± 15 ml/kg. The shorter than anticipated serum half-life of the fusion protein is almost certainly due to binding attributes of DKK2C2, since HSA alone has a half-life in rodents of several days. From analysis of the samples by SDS-PAGE followed by western blotting with anti-HSA and anti-DKK2 C-terminal peptide antibodies, there appeared no evidence for protein breakdown in the serum (Supplementary Fig. S2). HSA-DKK2C2 serum levels measured in the STF bioassay were indistinguishable from those detected by western blot, indicating that the administered protein retained activity. Because of the rapid clearance of the E. coli derived DKK2C2, we were not able to calculate terminal half-life or volume of distribution parameters; however, we estimated an AUC value of 2.5 µg x h/ml assuming a serum volume of 40 ml/kg and a maximum concentration (Cmax) at time zero of 50 µg/ml. The presence of the mass of the HSA in the fusion protein (76.3 kDa) complicates a direct comparison of the data with the E. coli derived DKK2C2 (10 kDa). However, since DKK2C2 and HSA-DKK2C2 are equipotent when activity is expressed in molar equivalents, by first adjusting dose to moles (2 mg/kg DKK2C2 = 200 nmol/kg, 10 mg/kg HSA-DKK2C2 = 130 nmol/kg) and then converting AUC to mole equivalents at a dose of 200 nmol/kg, we estimate that the HSA fusion (AUC = 9.8 nmol x h/ml) would result in about a 40-fold increase in AUC over the unmodified protein (AUC = 0.25 nmol × h/ml). A similar PK profile was observed for ACE464 in rats (Fig. 2A and Supplementary Fig. S3). The lack of breakdown products in the western blot analysis from the rat serum PK samples (SupplementaryFig. S3A) and similarity in the serum levels from titer and activity measurements (SupplementaryFig. S3B) provide further confirmation of the in vivo stability of the HSA-DKK2C2 fusion protein.
A potent long-acting variant of DKK2 could enhance epithelial repair and revascularization following acute lung and kidney injury. With the ultimate goal of delivering functional protein to these organs to support efficacy studies, in a separate study ACE464 was intravenously injected into mice at 1 or 10 mg/kg, and perfused lungs and kidneys were examined for the presence of ACE464 by western blot at both 3 and 24 h post-dosing. ACE464 could be detected in tissues, but only at 3 h after the higher dose (Fig. 2C). This finding reveals that there is no accumulation of HSA-DKK2C2 in these tissues. Since levels in these target organs track with serum levels, they indicate a need for longer acting forms beyond what we achieved with ACE464, to provide continuous exposure for efficacy studies.
Incomplete processing of the pro-domain of HSA was noted with HSA-DKK2C2 (H174-I259), resulting in a seven amino acid addition at the N-terminus of ~30% of the protein (calculated mass of intact HSA-DKK2C2 (H174-I259) protein, 76 360.1 Da; observed mass, 76 360 Da: calculated mass of +7 amino acid version of HSA-DKK2C2 (H174-I259), 77 219.1 Da; observed mass, 77 221 Da). This heterogeneity was abrogated by engineering a new molecule containing the wild-type DKK2C2 sequence (ACE486): HSA (D25-L609)-DKK2C2 (H174-I259) in which the seven amino acid proHSA sequence was deleted. Like the original construct, ACE486 was monomeric, and stable. From 600 ml of culture medium, we recovered 120 mg of fusion protein. Mass spectrometry confirmed the expected mass of the molecule (calculated mass, 76 360.1 Da; observed mass, 76 363 Da) and the protein was active in the STF assay (Table I). ACE464 and ACE486 were comparable in their binding affinities for LRP6 (Table I). The ACE486 HSA (D25-L609)-DKK2C2 (H174-I259) framework was incorporated into the engineering design of all subsequent mutants.
Table I.
Properties of HSA-DKK2C2 fusion proteins
| Construct | Substitutions | Heparin-Sepharose elution mM NaCl (mS/cm)a | IC50 (µg/ml) biotin–heparin bindingb | IC50 (nM) LRP6 bindingc | IC50 (nM) Wnt inhibitiond |
|---|---|---|---|---|---|
| ACE464 | – | 650 (44) | 0.26 | 72 | 51 |
| ACE486 | – | NA | NA | 59 | 92 |
| ACE502 | R185N | 590 (42) | 0.13 | 120 | 96 |
| BKM231 | K202E | 580 (42) | 0.41 | 400 | 300 |
| BKM230 | R197E | 560 (40) | 0.50 | 900 | NA |
| BKM227 | H223E | 560 (40) | 0.60 | 21 | 34 |
| ACE507 | S248N/K250S | 560 (40) | 0.76 | 42 | 66 |
| ACE506 | K250E | 540 (38) | 0.11 | 300 | 170 |
| BKM232 | K216S/H223T | 490 (34) | >50 | 61 | 110 |
| BKM225 | K220N | 490 (34) | >50 | 67 | 130 |
| BKM226 | K220E | 490 (33) | >50 | 51 | 130 |
| BKM233 | K216S/K220S | 450 (31) | >50 | 65 | 82 |
| ACE504 | K240E/K243E | 440 (29) | >50 | 140 | 57 |
| ACE505 | K216E/K250E | 370 (22) | >50 | 1000 | 190 |
| BKM228 | K216E/H223E | 370 (22) | >50 | 300 | 130 |
| BKM229 | K216E/K220E | 300 (16) | >50 | 800 | 150 |
| ACE503 | K202E/K220E | <150 (13) | >50 | 100 000 | 410 |
| BKM195 | H198A/K205A | NA | 0.21 | 35 000 | NA |
| BKM199 | R230A | NA | 0.64 | 75 000 | NA |
aNaCl concentration (conductivity) at elution during Heparin-Sepharose chromatography.
bIC50 values for monomeric biotin–heparin binding measured by ELISA.
cIC50 values for LRP6 binding measured by FACS assay.
dIC50 values for Wnt3a-stimulated canonical Wnt signaling inhibition measured by STF.
Mutants are organized in the table based on binding characteristics to Heparin-Sepharose from tightest to weakest binders.
NA - not tested.
Design of DKK2C2 variants to reduce HS binding and clearance
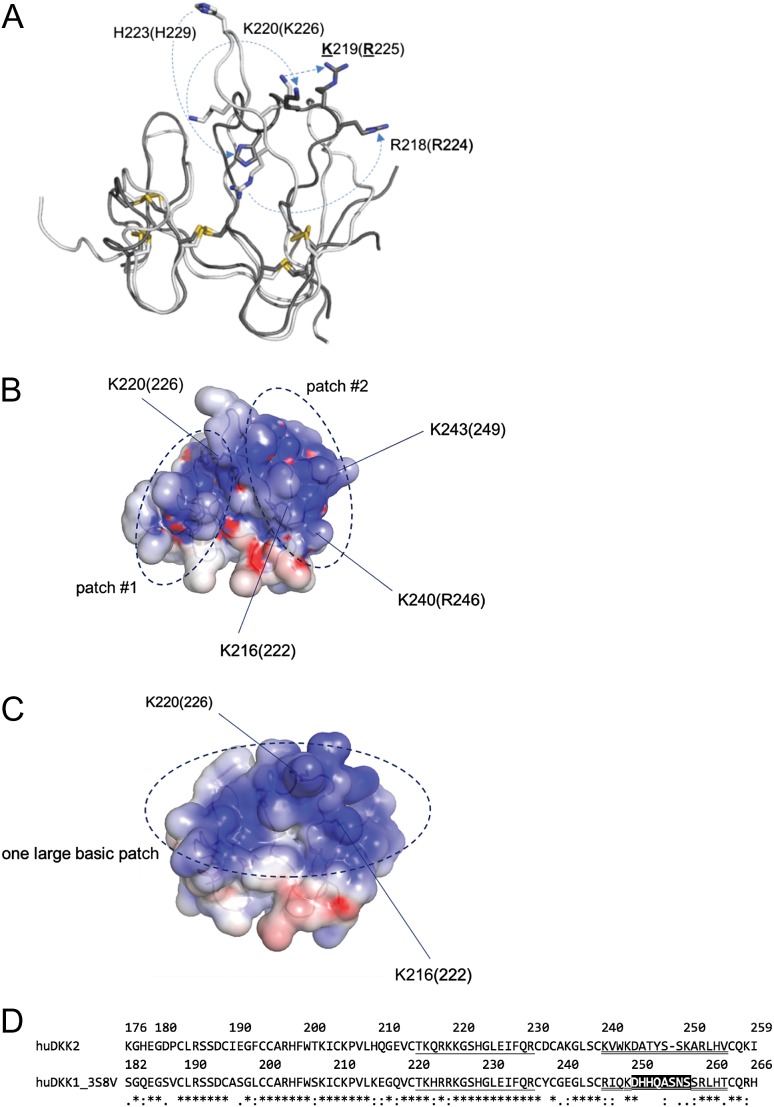
With the goal of creating DKK2C2 mutations that would reduce or eliminate HS binding, while retaining its affinity for LRP6, we relied on the structures of unbound DKK2C2 (nuclear magnetic resonance spectroscopy (NMR) structure, 2JTK.pdb: (Chen et al., 2008)) and its close homolog DKK1C2 bound to LRP6 (X-ray structures: 3S8V.pdb and 3S2K.pdb: (Ahn et al., 2011; Cheng et al., 2011)) to guide our design. Inspection of the structures revealed major differences in the conformation of basic residues we deemed would contribute to HS binding (Fig. 3A). Consequently, we decided to design mutated variants based on patches of strong positive charge density for both the unbound and bound conformations, which we calculated using the Adaptive Poisson–Boltzmann Solver (APBS, Baker et al., 2001) as accessed through Pymol (The PyMOL Molecular Graphics System, Version 1.7.4.0 Schrödinger, LLC). APBS allows for the more accurate modeling of the delocalized charge density than the simple assignment of Coulomb point charges to the functional groups of basic residues, which in turn allowed us to prioritize which residues to target for mutations.
Fig. 3.
Rational design of HS-binding mutations in DKK2C2. (A) Schematic diagram showing conformational shifts between DKK2C2 (2JTK.pdb, white), based on an NMR structure at pH 5, and DKK1C2 (3S8V.pdb, dark gray), based on an X-ray structure at pH 8.8. Residue numbers are for DKK2C2 (open) and DKK1C2 (in parentheses). A number of basic residues undergo large conformational shifts between the two structures, such as H223 (229), K220 (226), and R218 (224). (B) Schematic representation showing the locations of basic patches #1 and #2 on the surface of DKK2C2 (2JTK.pdb). (C) Schematic representation showing the location of the large basic patch on the surface of DKK1C2 (3S8V.pdb) (all images in A–C were created in Pymol [The PyMOL Molecular Graphics System, Version 1.7.4.0 Schrödinger, LLC], electrostatic surfaces were created using APBS (Baker et al., 2001)). (D) Alignment of human DKK2C2 (as numbered in 2JTK.pdb) and DKK1C2 (as numbered in 3S8V.pdb); both 3S8V and a nearly identical structure of DKK1C2 bound to LRP6 (3S2K.pdb) have missing coordinates for residues D250–S257, which are indicated through a black background; the loop between conserved disulfide-bound cysteines C214 (220) and C231 (237), which undergoes the largest conformational shift between DKK2C2 and DKK1C2–LRP6 structures is single underlined; the C-terminal loop between conserved disulfide-bound cysteines C239 (245) and C256 (263) is double-underlined.
The two possible conformations of DKK2C2 resulted in two different sets of charged patches (Fig. 3B and C). Accordingly, two sets of variants were designed to cover either conformation; note DKK2 numbering throughout. The first set of mutations was introduced based on the NMR structure of unbound DKK2C2 (Chen et al., 2008). In this conformation, two basic patches were identified on the surface of the protein (Fig. 3B). Alteration of one patch was brought about by mutation R185N (ACE502 in Table I) to generate a glycosylation motif 185–187 NSS, and the double charge reversal mutations K202E/K220E (ACE503 in Table I). Alteration of the second patch was achieved by the introduction of double charge reversal mutations K240E/K243E (ACE504 in Table I) and K216E/K250E (ACE505 in Table I), the single charge reversal mutation K250E (ACE506 in Table I), and double mutations S248N/K250S (ACE507 in Table I) to obtain the glycosylation motif 248–250 NSS. The second set of mutations was introduced based on the X-ray structures of DKK1C2, bound to LRP6 (Ahn et al., 2011, Cheng et al., 2011). In this structure, a conserved loop (87% identity between DKK1C2 and DKK2C2) between the sixth and the seventh conserved cysteine folds into a helical conformation, which relocates residues H223 and K220, connecting the two basic patches in DKK2C2 to form one extended patch (Fig. 3C). Residues K202, R197, H223, K220 and K216 form this extended basic patch, which lies distal to the LRP6 binding interface, as discerned in mutational binding studies of DKK1 and LRP6 (Ahn et al., 2011; Cheng et al., 2011). We have confidence these surfaces exist for bound DKK2C2 given the high degree of sequence similarity between DKK1C2 and DKK2C2 preceding the C-terminal loop (Fig. 3D). Mutations introduced from inspection of the DKK1C2 bound structure were the glycosylation mutation K220N (BKM225 in Table I) to obtain the glycosylation motif 220–222 NGS, the single charge reversals K220E (BKM226 in Table I), H223E (BKM227 in Table I), R197E (BKM230 in Table I), K202E (BKM231 in Table I), and the double charge reversals K216E/H223E (BKM228 in Table I). The placement of K220 on DKK1C2 is juxtaposed to K216 and prompted double charge reversal mutations K216E/K220E (BKM229 in Table I). The double charge reversals K216E/H223E and K216E/K220E were also attempted as double charge neutralization mutations K216S/H223T and K216S/K220S, respectively (BKM232 and BKM233 in Table I).
Purification and characterization of HSA-DKK2C2 HS-binding mutants
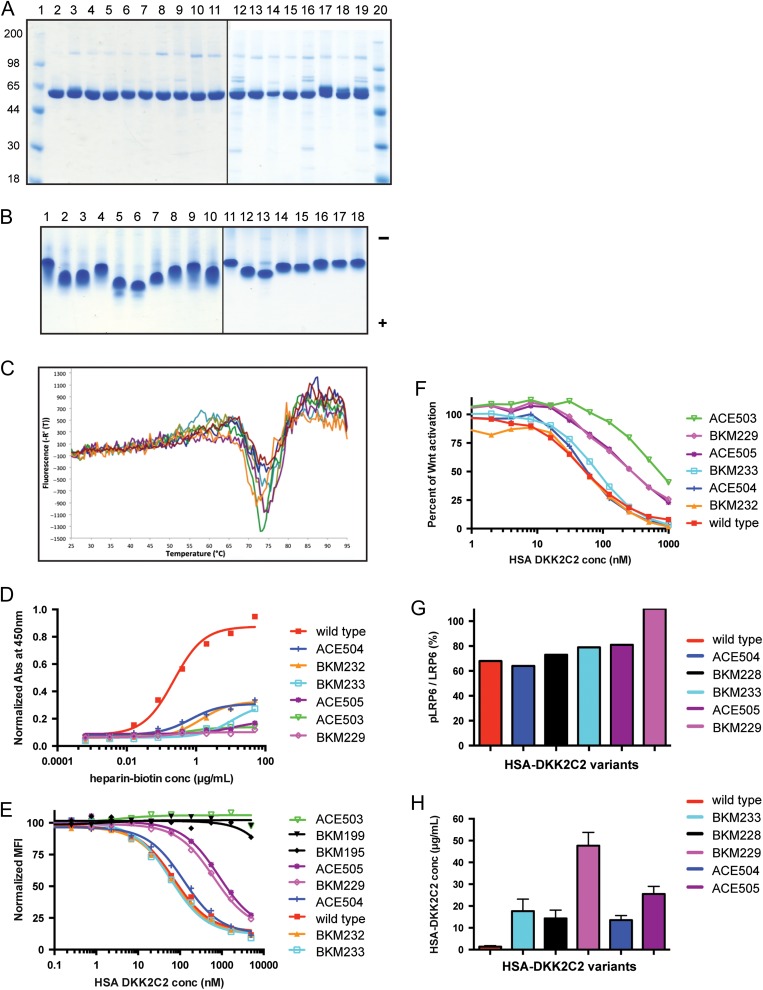
In total, 15 variants of HSA-DKK2C2 were expressed transiently by CHO cells and purified by either strong cation or anion exchange chromatography (see Materials and Methods). All molecules were primarily monomeric, stable and the average yield following ion exchange chromatography was greater than 100 mg/l. All of the purified mutants presented as a single prominent band of ~60 kDa under reducing conditions, migrating at the same position as purified HSA-DKK2C2 with the wild-type DKK2C2 sequence (Fig. 4A and selected mutants Fig. 1A). ACE507 was designed to encode a novel glycosylation site into the protein. As seen in Lanes 17–19, a doublet of bands at 60 kDa was observed, consistent with partial glycosylation at this site. Preparations enriched in glycosylated (Fig. 4A, Lane 17) and nonglycosylated (Fig. 4A, Lane 18) forms were generated by ion exchange chromatography. The percent purity of all mutants was above 86%, as measured by analytical SEC (Supplementary Table SIV and selected mutants Fig. 1B). SEC revealed that all the mutants migrate largely as monomer, eluting from the column as a single prominent peak at 9.5 min with an apparent molecular weight of ~60 kDa (Fig. 4A).
Fig. 4.
Characterization of HSA-DKK2C2 variants. (A) Reducing SDS-PAGE stained with SimplyBlue SafeStain examining purified HSA-DKK2C2 variants. Lanes 1 and 20: molecular weight standards; Lane 2: wild type HSA-DKK2C2 ACE464; Lane 3: BKM225; Lane 4: BKM226; Lane 5: BKM227; Lane 6: BKM228; Lane 7: BKM229; Lane 8: BKM230; Lane 9: BKM231; Lane 10: BKM232; Lane 11: BKM233; Lane 12: ACE502; Lane 13: ACE504; Lane 14: ACE505; Lane 15: ACE506—pH 5.5 purification; Lane 16: ACE506—pH 6.5 purification; Lane 17: ACE507—pH 5.5 purification, glycosylated form; Lane 18: ACE507—pH 5.5 purification, hypo-glycosylated form; Lane 19: ACE507—pH 6.5 purification. (B) Approximately 4 μg of wild-type HSA-DKK2C2 and each of the variants were analyzed by native PAGE under non-reducing conditions and stained with SimplyBlue SafeStain. Lane 1: wild type HSA-DKK2C2 ACE464; Lane 2: BKM225; Lane 3: BKM226; Lane 4: BKM227; Lane 5: BKM228; Lane 6: BKM229; Lane 7: BKM230; Lane 8: BKM231; Lane 9: BKM232; Lane 10: BKM233; Lane 11: ACE502; Lane 12: ACE504; Lane 13: ACE505; Lane 14: ACE506—pH 5.5 purification; Lane 15: ACE506—pH 6.5 purification; Lane 16: ACE507—pH 5.5 purification, glycosylated form; Lane 17: ACE507—pH 5.5 purification, hypo-glycosylated form; Lane 18: ACE507—pH 6.5 purification. Direction of anode (−) and cathode (+) is indicated. (C) A graphical depiction of the DSF profiles of selected HSA-DKK2C2 mutants. Thermal denaturation profiles for the six mutants BKM229 (brown); ACE505 (magenta); BKM228 (cyan); ACE504 (green); ACE506 (orange); and BKM233 (blue), and wild type HSA-DKK2C2 ACE464 (red), from 25°C to 95°C. (D) A graphical depiction of the results of heparin–biotin ELISA for selected HSA-DKK2C2 mutants. Titrations curve for biotin–heparin binding to HSA-DKK2C2 mutants plated at 15 µg/ml. Detection was at 450 nm with streptavidin-horseradish peroxidase. Illustrated is a representative of duplicate experiments for these variants. (E) A graphical depiction of the results of HSA-DKK2C2 mutant competition with anti-LRP6 antibody for binding to LRP6. LRP6 binding curves for HSA-DKK2C2 molecules impaired for either heparin or LRP6 binding (BKM195 and BKM199), following competition with anti-LRP6 monoclonal antibody. (F) A graphical depiction of percent canonical Wnt pathway activation in STF assay by Wnt3a in response to varying concentrations of wild type HSA-DKK2C2 ACE464 (red) and selected HSA-DKK2C2 variants. Plotted are the average values from triplicate experiments. (G) Bar graph depicting the ratio of phosphorylated LRP6 to total LRP6 detected by Western blotting for cells treated with each of the six HSA-DKK2C2 variants. This experiment was performed at three concentrations of DKK2C2 molecule (250, 500 or 1000 nM). Plotted values are depicting values generated from incubation with 1000 nM. (H) PK analysis of HSA-DKK2C2 variants. A bar graph depicting average serum levels from 2 or 3 mice intravenously dosed with 10 mg/kg of HSA-DKK2C2 molecules 24 h postinjection. Variants were detected by quantitative western blotting and quantified against a standard curve of wild type HSA-DKK2C2 ACE464 in serum. In D–H wild type refers to ACE464.
To further assess product quality, samples were analyzed by native gel electrophoresis. Unlike SDS-PAGE in which separation is based primarily on size, native gel electrophoresis is sensitive to charge and can readily detect single charge differences in proteins, that result from mutagenesis or post translational modifications such as glycation, deamindation, oxidation, desialyation or exopeptidase action that impact a protein's charge where the spread of isoforms during native gel electrophoresis can reveal differences in molecular charge distribution (Danwen and Code, 2016). ACE464 and ACE486 without mutations in DKK2C2 have a calculated pI of 6.75 (predicted using the Biotools program). Mutants that resulted in addition of a single negative charge (BKM225, ACE502, ACE507) and (BKM227, BKM227) had calculated pI values of 6.68 and 6.65, respectively. Mutants that resulted in addition of two (BKM230, BKM231, BKM226, BKM233 and ACE506), three (BKM228) and four negative charges (ACE505, BKM229 and ACE504) had pI values of 6.61, 6.52 and 6.48, respectively. All of the samples migrated as predicted on native gels (Fig. 4B); for instance, mutants gaining negative charge as a result of double glutamic acid substitutions migrated faster toward the cathode than single those bearing single glutamic acid substitutions.
HSA-DKK2C2 mutant stability and self-association
Thermal stability can be used to assess protein quality and solubility, where a change in the temperature at which a protein denatures is indicative of changes in structure or associative forces. For such studies, wild-type HSA-DKK2C2 (ACE464) and each of the engineered HS-binding mutants was subjected to differential scanning fluorimetry (DSF), a method of ramping temperature and monitoring the binding of a non-specific thermofluor to exposed hydrophobic surfaces (selected mutants, Fig. 4C). Upon protein unfolding, exposed hydrophobic surfaces bind the dye, resulting in an increase in fluorescence due to the exclusion of water. All of the mutants were stable and behaved essentially the same as wild-type HSA-DKK2C2 with observed melting temperatures (Tm) values of 75 ± 2°C. The single transition titrations observed primarily reflect the dye binding properties of HSA. These data indicate that the mutations in DKK2C2 did not impact the thermal stability of the fusion proteins. Additionally, the mutations resulted in relatively little effect on the propensity of HSA-DKK2C2 to self-associate, as measured by DLS (selected mutants, Fig. 1C). DLS is often used to measure the concentration dependence of protein–protein interactions in solution, and to obtain a diffusion interaction parameter (KD) where a positive value indicates net repulsive forces and a negative KD value indicates net attractive forces between molecules. All HS-binding mutants exhibited a weak tendency to self-associate in PBS at pH 7.5, which was slightly weaker but comparable to that of wild-type HSA-DKK2C2 (ACE464). These assays indicated that a head-to-head comparison of these proteins for HS binding would be reasonable.
HSA-DKK2C2 mutant characterization by heparin-sepharose chromatography
Heparin, a soluble member of the HS family, is frequently used as a model compound in experimental and theoretical studies of protein–HS interactions (Mottarella et al., 2014). The HSA-DKK2C2 variants were tested for ability to interact with heparin by measuring binding to a heparin-sepharose resin and assessing the salt concentration required for elution from the resin. Wild-type HSA-DKK2C2 (ACE464) and each of the HS-binding variants were individually subjected to heparin-sepharose chromatography under the same conditions. Based on elution using a linear salt gradient, all mutants exhibited reduced heparin binding compared to wild type (Table I). Wild-type HSA-DKK2C2 (ACE464) bound tightest to the heparin-sepharose column, eluting from the column with 650 mM salt. In contrast, ACE503 showed weakest binding and failed to bind the resin in binding buffer (150mM salt). Double charge reversals had the largest impact, with these molecules eluting with generally half the salt concentration required for wild-type HSA-DKK2C2 (ACE464) elution. All of the mutants exhibited some degree of reduced heparin binding relative to wild-type HSA-DKK2C2 (ACE464), eluting at lower salt concentrations with affinities that were dependent on the mutation.
HSA-DKK2C2 mutant characterization examining monomeric heparin–biotin binding by ELISA
The reduced binding affinity of the mutants for heparin observed with heparin-sepharose chromatography was confirmed using an ELISA based heparin-binding assay. Wild-type HSA-DKK2C2 (ACE464) and each of the HS-binding variants were examined in dilution series for binding to monomeric heparin–biotin in a plate-based format using ELISA. As observed with heparin sepharose, the binding of the mutants to monomeric heparin was similarly impacted, where the mutants that exhibited the lowest affinity for heparin sepharose showed the lowest affinity for monomeric heparin by ELISA (Table I and selected mutants Fig. 4D). Again all double charge reversals (BKM228, BKM229, BKM233, ACE504 and ACE505) practically eliminated heparin binding, whereas the other mutations caused varying degrees of reduced heparin binding. To evaluate whether alterations in the binding of LRP6, the cell-surface receptor for DKK2, might similarly impact heparin binding, two DKK variants deficient for LRP6 binding, BKM195 and BKM199 (Wang et al., 2008; Cheng et al., 2011), were tested for their ability to bind heparin–biotin in the ELISA format. Heparin binding was not impacted by these particular mutations, indicating that the LRP6 interaction site is distinct from the heparin-binding sites of DKK2 (Table I).
HSA-DKK2C2 mutant characterization examining LRP6 binding
To assess whether our HS-binding mutations influenced binding of DKK2C2 to its receptor LRP6, we developed a FACS binding assay. Affinities were quantified by competition of binding by a high affinity anti-LRP6 antibody for the same domain of LRP6 (propeller 3) bound by DKK2C2. In a reporter format, LRP6-expressing cells were first incubated with the HSA-DKK2C2 variants in a dilution series and free LRP6 that was not bound to DKK2 was measured with the anti-LRP6 antibody. In the FACS assay, wild-type HSA-DKK2C2 (ACE464) bound with an IC50 of 60 nM. The IC50 values for HS-binding mutants ranged between 20 and 1000 nM (Table I and selected mutants Fig. 4E). As expected, published mutants shown to abolish LRP6 binding, BKM195 and BKM199 (Wang et al., 2008; Cheng et al., 2011), had IC50 values of greater than 30 000 nM in this assay. The weakest HS-binding variant, ACE503, was also dead in terms of LRP6 binding in this assay. The impact on free LRP6 binding of the other double charge reversal variants, hampered for heparin binding, was either substantial (ACE505 and BKM229: 17- and 13-fold reduced IC50 relative to wild type, respectively) or moderate (BKM228 and ACE504: 5 and 2.3-fold reduced IC50 relative to wild type, respectively). These particular mutants are appealing in terms of their suitability for systemic delivery, exhibiting reduced heparin binding and therefore potential for extended half-life, and yet retention of LRP binding and presumable inhibition of canonical Wnt signaling.
Canonical Wnt signaling inhibition by HSA-DKK2C2 mutants
To assess whether LRP binding retained by the HSA-DKK2C2 mutants was sufficient to inhibit LRP-Wnt-Frizzled ternary complex formation and canonical Wnt signaling, HS-binding mutants were tested in the STF assay. Altering heparin binding generally impacted the ability of HSA-DKK2C2 to inhibit canonical Wnt signaling (Table I and selected mutants Fig. 4F) as the majority of DKK2C2 variants exhibited a reduction in dose-dependent inhibition of Wnt3a-stimulated signaling, relative to the wild-type ACE464 molecule. ACE486, bearing wild-type DKK2 sequence, had an apparent IC50 of 92 nM while IC50 values for DKK2C2 mutants ranged from 34 to 410 nM. In general, those mutations with the greatest effect on heparin binding (double charge reversal variants ACE503, ACE505, BKM229 and BKM228) exhibited the largest reductions in their ability to inhibit Wnt3a signaling. The one exception was the double charge reversal variant ACE504 which behaved essentially as wild type in the STF assay. This variant emerged as the best candidate in terms of activity and potential for extended half-life in serum, given its reduction in heparin binding. The effects of mutations on Wnt signaling were not as pronounced as was seen with LRP6 binding, which could reflect differences in the binding of DKK2C2 to LRP6 in the absence of other components of the LRP6 signaling complex.
Phospho-LRP6 inhibition by HSA-DKK2C2 mutants
As part of its inhibitory function, DKK proteins prevent the Wnt3a-dependent phosphorylation of LRP6 (pLRP6), thereby reducing overall pLRP6 levels (Binnerts et al., 2009). Since LRP6 phosphorylation is a conserved mechanism required for activation of the canonical Wnt pathway and heparin-binding mutations impact canonical signaling as measured by STF, we assessed whether incubation with HSA-DKK2C2 variants defective for heparin binding impacted cellular pLRP6 status (Supplementary Fig. S4). HSA-DKK2C2 variants impacted cellular pLRP6 consistent with observed activities of these molecules in the STF assay (Fig. 4G). This assay was performed with HSA-DKK2C2 variants at 250, 500 and 1000 nM. The rank order was generally consistent between concentrations. Ratios decreased an average of 8% with each increasing step in concentration. As expected pLRP6 status was unaffected by incubation with the double charge reversal mutant ACE504, consistent with its wild-type activity in the STF assay. While BKM228, BKM229 and ACE505 were the weakest in inhibiting Wnt3a signaling as measured by the STF readout, only incubation with ACE505 and BKM229 had a significant effect on LRP phosphorylation status. This likely reflects the weaker affinities of these two variants for LRP6. Altogether three distinct assays (LRP6 binding, STF transcriptional reporter activation and LRP6 phosphorylation) indicated that BKM229 and ACE505 had the largest impact on Wnt pathway activity. At the same time, these assays indicated that ACE504, with practically undetectable heparin binding, had relatively no functional impact on either LRP6 binding and phosphorylation, or Wnt pathway inhibition. This molecule displayed the desired attributes of an exogenously delivered Wnt pathway inhibitor, pending a sustained serum residence time.
PKs measurements of HSA-DKK2C2 mutants
Five of the weakest heparin-binding variants of HSA-DKK2C2 (BKM233, BKM228, BKM229, ACE504 and ACE505) were selected for PK assessment in mice. We anticipated that a reduction in HS binding would decrease non-specific cell interactions and thereby increase HSA-DKK2C2 serum exposure. Mice (3 per group, 2 for ACE504) were injected intravenously with 10 mg/kg of the HSA-DKK2C2 variants. Blood was drawn and serum prepared after 24 h. Levels of DKK2 in the serum were measured using a quantitative western blot protocol against a standard curve of HSA-DKK2C2 in serum, using independent measurements with HSA and DKK2C2 antibodies as conformation of serum levels. All five of the mutants tested showed much higher levels of DKK2 in the serum at the 24 h time point, as compared to wild type ACE464 (Fig. 4H and Supplementary Fig. S5). Wild-type HSA-DKK2C2 (ACE464) levels were observed below the LOQ of 1 µg/ml at 24 h, whereas mutant levels ranged from 10 to 50 µg/ml. One mutant, BKM229, was detected in serum at circulating levels of ~15 µg/ml after 48 h (Supplementary Fig. S6). The clear improvement in serum PK of the modified HSA-DKK2C2 molecules implied that non-specific cell interactions due to HS binding were hampering circulating levels of the wild-type molecule.
Discussion
HS-binding proteins such as DKK2 present challenges in drug development because of their short serum half-lives, resulting in the need for frequent injections and larger doses (Gardell et al., 2003; Lin et al., 2010; Okkerse et al., 2016). In this study, we sought to engineer an efficacious and long-lived variant of the Wnt inhibitor that could augment the activity of endogenous DKK2. Poor expression, aggregation, disulfide scrambling and proteolytic lability affected the generation of a suitable DKK2 polypeptide. While active in cell-based assays, DKK2C2 produced in E. coli was found to have very low exposure in rodents with no measurable titer after 30 min following intravenous administration. Notably, this construct expressed poorly in either CHO or HEK293 cells. In contrast, N-terminal HSA fusions of DKK2C2 were active, showed high levels of expression and exhibited reduced proteolytic lability and aggregation. HSA was chosen as the fusion platform for DKK2C2 because of its desirable pharmacological attributes such as its long-lived nature, high solubility and stability in serum, and its lack of immune reactivity. The effectiveness of HSA to improve the aforementioned properties of its fusion partner has been previously demonstrated with the glucagon-like peptide-1 agonist albiglutide, the first albumin fusion protein drug approved by the FDA in 2014, and other albumin fusion proteins currently in clinical trials (Rogers et al., 2015). As part of the optimization of the HSA-DKK2C2 construct, we varied the length of the linker sequence from within DKK2 connecting HSA with the C2 domain; HSA-DKK2C2 (H174-I259) was selected for all subsequent studies. We further improved the homogeneity of HSA-DKK2C2 polypeptides by deletion of the pro-peptide sequence of HSA (residues 1–25). We recommend this modification for subsequent studies, in order to reduce the heterogeneity of HSA fusion proteins.
Our selection of HSA as the preferred fusion partner for DKK2C2 arose from a detailed evaluation of HSA, Fc and XTEN as fusion partners. While all three fusions led to increased expression relative to DKK2C2 alone, the resulting Fc and XTEN-DKK2C2 fusion products contained high levels of aggregation and clipping (data not shown). Expression levels of XTEN-DKK2C2 (molecular weight 22 970 Da) were ~100 mg/l with recoverable titers of 10 mg/l. SDS-PAGE analysis of XTEN-DKK2C2 under non-reducing conditions revealed that most of the product had formed extensive disulfide-linked complexes and we were unable to purify the monomeric protein. For Fc-DKK2C2 fusion designs (molecular weight 73 045 Da), we achieved expression levels of 30–100 mg/l. Mass spectrometry analysis of the Fc-DKK2C2 fusion revealed extensive cleavages within the DKK2 linker sequence at residues M172 and S173 as well as internal cleavages within DKK2C2 at residues R218, K220, E226 and S249. Recoverable titers of an Fc product with intact DKK2C2 attached accounted for only ~10% of the expressed product. The purified protein ran as a dimer by SDS-PAGE analysis under non-reducing conditions as expected, but the material eluted in the excluded volume of the column with apparent molecular weight of >700 kDa (based on a high molecular weight reference standard) when evaluated by SEC, indicating aggregation. Due to aggregation as well as low recovery of intact protein, efforts toward DKK2C2 Fc fusions were stopped. In contrast, observed titers for HSA-DKK2C2 fusion proteins (average molecular weights of 76 400 Da) were 100–300 mg/l with high purification yields and no issues of aggregation or clipping. Observed cleavages within the DKK2 linker sequences in the Fc fusions prompted our evaluation of different linker lengths in the early designs of the HSA fusion proteins. The results with the Fc fusion were somewhat surprising based on published studies using an Fc-fusion protein containing full-length DKK2 instead of the DKK2C2 version we studied (Min et al., 2011). In the paper by Min et al. (2011), there was no characterization of the DKK2-Fc protein they produced and in animal studies the protein was administered locally at the injury site and therefore there was no PK analysis of the administered protein. Fc fusion proteins in general are an excellent platform for extending serum half-life (Levin et al., 2015; Strohl, 2015); however, prior experience engineering an Fc fusion with sonic hedgehog, a different low molecular weight HS-binding protein of similar size and charge, revealed reduced exposure of the fusion protein (Pepinsky et al., 2002, Shapiro et al., 2003). In these studies, AUC of a 1 mg/kg dose decreased from 9 µg × h/ml for the unmodified protein to 4.3 µg × h/ml for the Fc fusion. Thus, simply designing carriers to increase half-life without addressing the inherent issue of HS binding may not improve exposure. Furthermore, two copies of the target protein in the Fc fusion designs can exacerbate these issues. In the studies with sonic hedgehog, we were able to increase AUC of a 1 mg/kg dose from 9 µg × h/ml to 390 µg × h/ml by engineering sites for targeted PEGylation, using a design-based mutagenesis approach that complements the strategy we applied for DKK2C2 in this study.
Similar issues were encountered in the recent development of long-acting recombinant forms of Factor VIII and IX as Fc fusion proteins for treatment of individuals with hemophilia A and B. While the approved monovalent-Fc fusion proteins of Factor VIII (Eloctate®) and Factor IX (Alprolix®) circumvent potential liabilities of bivalent Fc designs, they did not address clearance mechanisms inherit to the blood factors, which led to only modest 1.5–3-fold increases in half-life (Strohl, 2015). Notably, Fc engineering of Factor VIII only increased the half-life in humans from 12 h without Fc to 19 h. Although significant in that it translated into patient benefit by shifting dosing regimens from daily infusions to 2–3 times per week, these findings further highlight the need for directed engineering efforts that address clearance mechanisms in order to achieve desired improvements in serum half-life. HSA fusions of Factor IX currently under development show similar half-lives as the monovalent-Fc product (Strohl, 2015). While we did not perform in vivo PK testing with the Fc-DKK2C2 construct due to aggregation, based on data published for Factor IX, we anticipate that the Fc-DKK2C2 would perform similar to the HSA product and would not overcome the effect of HS binding.
While the HSA-DKK2C2 fusion protein we produced with the wild-type DKK2C2 sequence exhibited favorable attributes in terms of stability and solubility, it displayed poor, albeit improved, PK in rodents; following administration at 10 mg/kg, DKK2 was detectable in tissue only up to 3 h, and in serum at the LOQ of 1 µg/ml at 24 h. We surmised that the reduced serum half-life of the molecule could be attributed to non-specific cell interactions resulting from the binding of DKK2C2 to HS given the surface density of positive charge of DKK2C2 (pI of 9.1). By manipulating positively charged amino acids predicted to participate in this interaction, we engineered multiple DKK2C2 variants that dramatically reduced heparin binding. We demonstrate that most of these molecules are monomeric, stable and retain function, i.e. LRP6 binding and phosphorylation, and canonical Wnt pathway inhibition.
As part of the design of mutants of DKK2C2, we engineered a construct in which a glycosylation site was introduced at a site that could sterically impact HS binding through addition of bulky glycan structures. Introduction of glycosylation has been successfully used to promote the serum half-life of proteins, reduce their proteolysis, and impact self-association and solubility by reducing aggregation (Baloh et al., 1998; van Veen et al., 2004; Shental-Bechor and Levy, 2008, Hanson et al., 2009, Danwen et al., 2016). While insertion of the glycan indeed decreased heparin binding, the efficiency of glycosylation was low, resulting in lower yield of the glycosylated product. Because we were unable to completely remove unglycosylated protein, activity measurements of the construct were compromised by nonglycosylated protein in the sample, and do not accurately represent the loss in activity due to the glycan modification.
Most pertinent to our intention, we find that five pairs of mutations extend serum half-life of HSA-DKK2C2; at 24 h, these molecules were detected at 10–50-fold higher levels than that of the wild-type molecule. Most significant, mutant BKM229 persisted in serum at levels of ~15 µg/ml after 48 h. Four out of five of the HSA-DKK2C2 variants bear a charge reversal of K216, which contributes to basic patches both in the unbound DKK2C2 and LRP6-bound DKK1C2 structures. Altered residues in BKM228 and BKM229, which are based on the DKK1C2 bound structure, are located in a conserved loop (residues C214–C231) that is stabilized through the formation of a near complete helix, close to hydrophobic residues I227 and F228 which contact LRP6 (Ahn et al., 2011; Cheng et al., 2011). Our LRP6 binding data hint at the possibility that these mutations affect signaling more than mutations in ACE504, which are located in a flexible loop that is less directly connected to the binding site. In summary, we found a subset of mutants based on both the unbound DKK2C2 and bound DKK1C1 structures that had weak affinity for heparin and extended serum half-life, highlighting the merit of both design strategies.
We present these molecules, with acceptable drug-like properties, as lead development candidates. From our engineering efforts to reduce HS binding, we effectively generated a panel of HSA-DKK2C2 mutants with altered affinity for LRP6 and a range of activity with respect to Wnt pathway inhibition. In terms of therapeutic applicability, this roster of mutants could be advantageous in that complete inhibition of canonical Wnt signaling could in fact be deleterious, while dialing down activity of the pathway to an optimal level over time (influenced by molecule half-life) may evoke the appropriate response. Furthermore, since HSA contains a single free cysteine, cross-linking agents can be targeted to this site to generate hetero-bifunctionality, enabling therapeutic diversification of these HSA-DKK2C2 molecules. This cysteine additionally allows for the generation of labeled probes such as through biotinylation or fluorescent conjugates that can be used for assay development.
Given the role of the Wnt pathway in tissue injury and repair (Zhou et al., 2016), the HSA-DKK2C2 polypeptides described herein can be used for treatment and prevention of both ischemic and nephrotoxic kidney injury, as well as lung injury. Additionally, inhibition of canonical Wnt signaling has been indicated for the treatment of numerous cancers including non–small-cell carcinomas, melanomas, hepatocellular and colorectal cancers, breast and prostate (Paul and Dey, 2008; Polakis, 2012; Xue et al., 2016). The potent, long-acting HSA-DKK2C2 molecules generated in this study should be applicable for treatment of these and other indications that involve aberrant Wnt signaling.
Supplementary Material
Acknowledgements
The authors thank Alan Buckler and Werner Meier for leadership and support, Jeremy Duffield, Michael Crackower for helpful discussions, Judy George for conditioned media, Monika Vecchi and Susan Foley for mass spectrometry analysis, Allan Capili and Janine Ferrant-Orgettas for overseeing aspects of the work, and Qin Wang for assisting in the design of PK studies and for calculating PK parameters. The PEDS board member responsible for editing this manuscript was Dr Karyn O'Neil.
References
- Ahn V.E., Chu M.L., Choi H.J., Tran D., Abo A. and Weis W.I. (2011) Dev. Cell, 21, 862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A., Liu G., Yaniv A., Gazit A. and Aaronson S.A. (2001) Nat. Cell. Biol., 3, 683–686. [DOI] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J. and McCammon J.A. (2001) Proc. Natl. Acad. Sci. USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh R.H., Tansey M.G., Lampe P.A. et al. (1998) Neuron, 21, 1291–1302. [DOI] [PubMed] [Google Scholar]
- Binnerts M.E., Tomasevic N., Bright J.M. et al. (2009) Mol. Biol. Cell., 20, 3552–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E., Carano R., Cochran A.G., et al. (2014). ANTI-LRP6 ANTIBODIES. U.S., GENENTECH, INC., South San Francisco, CA (US). U.S. Patent No. 8846041.
- Bourhis E., Tam C., Franke Y., Bazan J.F., Ernst J., Hwang J., Costa M., Cochran A.G. and Hannoush R.N. (2010) J. Biol. Chem., 285, 9172–9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang K., Shao Y., Huang J., Li X., Shan J., Wu D. and Zheng J.J. (2008) J. Biol. Chem., 283, 23364–23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Biechele T., Wei Z., Morrone S., Moon R.T., Wang L. and Xu W. (2011) Nat. Struct. Mol. Biol., 18, 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. and Nusse R. (2012) Cell, 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- Danwen Q., Code C., Quan C., Gong B.J., Arndt J., Pepinsky B., Rand K.D. and Houde D. (2016) Pharm. Res., 33, 1383–1398. [DOI] [PubMed] [Google Scholar]
- Datta-Mannan A., Yaden B., Krishnan V., Jones B.E. and Croy J.E. (2013) J. Pharmacol. Exp. Ther., 344, 616–623. [DOI] [PubMed] [Google Scholar]
- Gardell L.R., Wang R., Ehrenfels C. et al. (2003) Nat. Med., 9, 1383–1389. [DOI] [PubMed] [Google Scholar]
- Glinka A., Wu W., Delius H., Monaghan A.P., Blumenstock C. and Niehrs C. (1998) Nature, 391, 357–362. [DOI] [PubMed] [Google Scholar]
- Hanlon A.D., Larkin M.I. and Reddick R.M. (2010) Biophys. J., 98, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S.R., Culyba E.K., Hsu T.L., Wong C.H., Kelly J.W. and Powers E.T. (2009) Proc. Natl. Acad. Sci. USA, 106, 3131–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann G., Prospero T., Brinkmann V. et al. (1998) Curr. Biol., 8, 125–134. [DOI] [PubMed] [Google Scholar]
- He X.M. and Carter D.C. (1992) Nature, 358, 209–215. [DOI] [PubMed] [Google Scholar]
- Kawakita A., Yanamoto S., Yamada S., Naruse T., Takahashi H., Kawasaki G. and Umeda M. (2014) Pathol. Oncol. Res., 20, 253–261. [DOI] [PubMed] [Google Scholar]
- Kontermann R.E. (2016) Expert. Opin. Biol. Ther., 16, 903–915. [DOI] [PubMed] [Google Scholar]
- Lehermayr C., Mahler H.C., Mader K. and Fischer S. (2011) J. Pharm. Sci., 100, 2551–2562. [DOI] [PubMed] [Google Scholar]
- Levin D., Golding B., Strome S.E. and Sauna Z.E. (2015) Trends Biotechnol., 33, 27–34. [DOI] [PubMed] [Google Scholar]
- Li L., Mao J., Sun L., Liu W. and Wu D. (2002) J. Biol. Chem., 277, 5977–5981. [DOI] [PubMed] [Google Scholar]
- Li Q., Shen K., Zhao Y., He X., Ma C., Wang L., Wang B., Liu J. and Ma J. (2013) FEBS Lett., 587, 1742–1748. [DOI] [PubMed] [Google Scholar]
- Lin S.L., Li B., Rao S. et al. (2010) Proc. Natl. Acad. Sci. USA, 107, 4194–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., Tamai K. and He X. (2009) Dev. Cell., 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B. and Niehrs C. (2003) Gene, 302, 179–183. [DOI] [PubMed] [Google Scholar]
- Mao B., Wu W., Davidson G. et al. (2002) Nature, 417, 664–667. [DOI] [PubMed] [Google Scholar]
- Mao B., Wu W., Li Y., Hoppe D., Stannek P., Glinka A. and Niehrs C. (2001) Nature, 411, 321–325. [DOI] [PubMed] [Google Scholar]
- Min J.K., Park H., Choi H.J. et al. (2011) J. Clin. Invest., 121, 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottarella S.E., Beglov D., Beglova N., Nugent M.A., Kozakov D. and Vajda S. (2014) J. Chem. Inf. Model., 54, 2068–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkerse P., Hay J.L., Versage E. et al. (2016) Br. J. Clin. Pharmacol., 82, 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S. and Dey A. (2008) Neoplasma, 55, 165–176. [PubMed] [Google Scholar]
- Pepinsky R.B., Shapiro R.I., Wang S. et al. (2002) J. Pharm. Sci., 91, 371–387. [DOI] [PubMed] [Google Scholar]
- Polakis P. (2012) Cold Spring Harb. Perspect. Biol., 4, a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B., Dong D., Li Z. and Li Z. (2015) Curr. Pharm. Des., 21, 1899–1907. [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos P., Piccinini E., Penttinen A.M. et al. (2016) Neurobiol Dis., 96, 335–345. [DOI] [PubMed] [Google Scholar]
- Sarrazin S., Lamanna W.C. and Esko J.D. (2011) Cold Spring Harb. Perspect. Biol., 3, a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenov M.V., Tamai K., Brott B.K., Kuhl M., Sokol S. and He X. (2001) Curr. Biol., 11, 951–961. [DOI] [PubMed] [Google Scholar]
- Shapiro R.I., Wen D., Levesque M., Hronowski X., Gill A., Garber E.A., Galdes A., Strauch K.L. and Taylor F.R. (2003) Protein Expr. Purif., 29, 272–283. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D. and Levy Y. (2008) Proc. Natl. Acad. Sci. USA, 105, 8256–8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvian L., Jin P., Carmillo P. et al. (2006) Biochemistry, 45, 6801–6812. [DOI] [PubMed] [Google Scholar]
- Sinclair A.M. and Elliott S. (2005) J. Pharm. Sci., 94, 1626–1635. [DOI] [PubMed] [Google Scholar]
- Strohl W.R. (2015) BioDrugs, 29, 215–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio S., Kashima A., Mochizuki S., Noda M. and Kobayashi K. (1999) Protein Eng., 12, 439–446. [DOI] [PubMed] [Google Scholar]
- van Amerongen R. (2012) Cold Spring Harb. Perspect. Biol., 4, a007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen H.A., Geerts M.E., van Berkel P.H. and Nuijens J.H. (2004) Eur. J. Biochem., 271, 678–684. [DOI] [PubMed] [Google Scholar]
- Wang K., Zhang Y., Li X., Chen L., Wang H., Wu J., Zheng J. and Wu D. (2008) J. Biol. Chem., 283, 23371–23375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Wang Y., Dabdoub A. et al. (2004) Cell, 116, 883–895. [DOI] [PubMed] [Google Scholar]
- Xue G., Romano E., Massi D. and Mandala M. (2016) Cancer Treat. Rev., 49, 1–12. [DOI] [PubMed] [Google Scholar]
- Zhou D., Tan R.J., Fu H. and Liu Y. (2016) Lab. Invest., 96, 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.