Abstract
Organic solvents and apolar media are used in the studies of nucleic acids to modify the conformation and function of nucleic acids, to improve solubility of hydrophobic ligands, to construct molecular scaffolds for organic synthesis, and to study molecular crowding effects. Understanding how organic solvents affect nucleic acid interactions and identifying the factors that dominate solvent effects are important for the creation of oligonucleotide-based technologies. This review describes the structural and catalytic properties of DNA and RNA oligonucleotides in organic solutions and in aqueous solutions with organic cosolvents. There are several possible mechanisms underlying the effects of organic solvents on nucleic acid interactions. The reported results emphasize the significance of the osmotic pressure effect and the dielectric constant effect in addition to specific interactions with nucleic acid strands. This review will serve as a guide for the selection of solvent systems based on the purpose of the nucleic acid-based experiments.
Keywords: Oligonucleotide, Ribozyme, DNAzyme, Organic solvent, Molecular crowding, Dielectric constant
Introduction
Synthetic DNA and RNA oligonucleotides, now available in high purity at relatively low cost, are used in many medical and technological applications. Oligonucleotides that are designed for specific recognition of target sequences by complementary base pairing are employed for the regulation and analysis of cellular gene expression in basic science experiments and for gene regulation and diagnostics in clinical applications (Schubert and Kurreck 2004; Pan and Clawson 2006; Ni et al. 2011). Due to the specificity of base pairing interactions, nucleic acids are also used for the preparation of defined assemblies of DNA and RNA strands in nanotechnology applications (Smith et al. 2013). Thermodynamic stability of nucleic acid structures has been investigated in aqueous solutions containing salts, and the quantitative studies have enabled prediction of thermodynamic stabilities of hybridization and of the folded conformations of DNA and RNA.
Nucleotides have binding sites for water and cations, and the stabilities of these interactions depend on the solution composition such as the water content and the concentration and species of salt. DNA and RNA strands are highly hydrated in water solution: the groove of a DNA duplex has an ordered water molecule network (Westhof 1988; Marky and Kupke 2000), and the double-helical conformation is changed in response to the relative humidity (Saenger 1983). Moreover, the negative charge of nucleotide phosphates must be screened by bound cations (Record et al. 1998; Draper et al. 2005). Cation binding is essential for the base-pair formation and the folding and catalytic activity of deoxyribozymes (DNAzymes) and ribozymes. These nucleic acid-based enzymes have been shown to inhibit gene expression in vivo (Yen et al. 2004; Citti and Rainaldi 2005; Li et al. 2008; Liu et al. 2012), have been used to study RNA folding in cells (Mahen et al. 2010), and can serve as probes in biosensors (Sekella et al. 2002; Liu et al. 2009).
In medical and technological applications and under certain conditions in cells, oligonucleotides are required to function in non-aqueous media. The intracellular environment contains high concentrations of macromolecules, small metabolites, and osmolyte compounds. These components directly and indirectly affect nucleic acid interactions in cells: macromolecules sterically exclude other molecules and increase the solution viscosity, and organic components solvated by water reduce the amount of free water and change the dielectric permittivity (Asami et al. 1976; Srere 1981; Parsegian and Rau 1984; Garlid 2000; Luby-Phelps 2000; Tanizaki et al. 2008; Cuervo et al. 2014). The environments near and within the cell membrane, inside amphiphilic drug delivery polymer vesicles, and proximal to the surface of biosensors are the conditions in which there are short-range ordering or disordering of water molecules with unusual solvent properties. In addition, organic solvents are used in many experiments involving nucleic acids such as precipitation with ethanol, 2-propanol, and high-molecular-weight polyethylene glycol (PEG); conformational denaturation with formamide and dimethylsulfoxide (DMSO); solubilization of nucleic acid-interacting hydrophobic compounds with N,N-dimethylformamide (DMF), DMSO, and methanol (Dong et al. 2002; Lee et al. 2013); and characterization of the effects of molecular crowding environment (Nakano et al. 2014a).
When an organic compound is used instead of water as a solvent for nucleic acids, many factors are altered. Because organic solvents have distinct properties of the hydrogen bonding, polarity, and hydrophobicity, the solvation of nucleic acids depends on the solvent used. It is notable that, when organic solvents are macroscopically dissolved in water, the solutions are not always microscopically homogeneous. In these mixed solutions, water–water interactions are disrupted, and cosolvent–cosolvent interactions and water–cosolvent interactions are formed. Thus, the solvent properties such as water content and dielectric constant are different from those of water solutions (Fig. 1b). In particular, many organic solutions have a low dielectric constant, which is also characteristic of regions of the intracellular environment of viral particles, the surfaces and interiors of certain proteins, and the surfaces of nucleic acids (Fig. 1a). While quantitative research on the thermodynamic stabilities of DNA and RNA structures has been widely performed using polar and hydrophilic solutions of water, there are increasing numbers of reports on the thermodynamic and kinetic characteristics of nucleic acids under conditions created by the use of organic solvents. The following sections describe the applications of organic solutions for oligonucleotide-based technologies and the studies of thermodynamic stability and catalytic activity of oligonucleotides in organic solutions with little or no water (at most a few percent) and in aqueous solutions with organic cosolvents (from a few to several tens of percent by volume or weight per unit volume of solution, represented by vol% or wt%, respectively). When the definition of the percentage amount of cosolvents was not clearly stated in literature reports, the amount is referred to as percent (%) in this review.
Fig. 1.
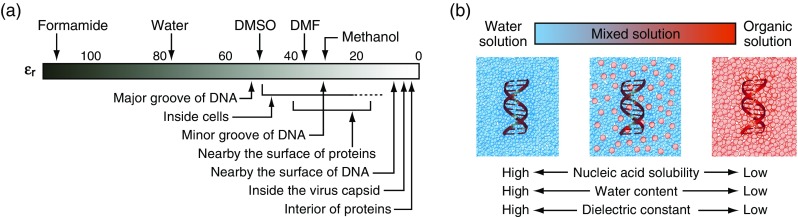
a Relative dielectric constants ε r of organic solutions and those of cells, DNAs, and proteins (Asami et al. 1976; Lamm and Pack 1997; Young et al. 1998; Pitera et al. 2001; Cuervo et al. 2014). b Solvent properties of water, organic solvents, and mixed aqueous solutions with organic cosolvents
Organic solutions with a small amount of water
Organic solutions are poor solvents for DNA and RNA. Addition of organic solvent to solutions of polynucleotides results in the formation of compact conformations and precipitation. For example, methanol, ethanol, 1-propanol, 2-propanol, t-butanol, acetone, and ethylene glycol at 40–70 wt% induce the phase transition of plasmids and phage DNAs from an elongated coil to a compact globular conformation. The dielectric constant of these solutions is suggested to be the key factor in determining the conformational behavior of the polynucleotides (Arscott et al. 1995; Mel’niov et al. 1999). Short oligonucleotides are relatively soluble in organic solutions in comparison with polynucleotides. Single-stranded DNAs are soluble in 97 % methanol and in acetone: A 45-mer DNA is more soluble than a 108-mer DNA, although the solubility is strongly dependent on the species and concentration of the salt used (Stanlis and McIntosh 2003). A short DNA oligonucleotide (21-mer) is soluble in nearly anhydrous solutions of glycerol, ethylene glycol, formamide, methanol, and DMSO in the presence of a small amount of water (e.g., 99 % organic solvent and 1 % water) (Bonner and Klibanov 2000). In these same solutions, the structure of the 21-base-pair duplex is unstable, and no duplex structure is formed in formamide, methanol, or DMSO. On the other hand, it has also been reported that a 21-base-pair DNA duplex is not denatured in 95 vol% DMF (Ke et al. 2010). Molecular dynamics simulations and atomic force microscopy have shown that a 12-base-pair DNA duplex dissociates into single strands in diethylbenzene, 1-propanol, and octane, and the DNA strands separate at the interface of the low-polarity medium and water (Cui et al. 2007). Thus, water is not unique as a medium for solubilization of nucleic acids, although the base-pairing interactions tend to be disrupted in non-aqueous solutions. Mixtures of organic solvents and water could be better solvating agents than either of the individual solvents because the water and organic molecules can orient to minimize conformational stress around a nucleotide chain (Hammouda and Worcester 2006). A study using molecular dynamics simulations showed that a small number of highly stable water molecules and neutralization of phosphate charges are crucial requirements for the transfer of a 7-base-pair DNA duplex from water to an apolar medium such as carbon tetrachloride (Arcella et al. 2014). Solvation of nucleotides in organic solutions in the presence of a small amount of water is a complex phenomenon that depends on the nature of the environment to which the nucleic acids are initially exposed (Ababneh et al. 2003).
Nucleic acids are made more compatible with organic solutions by conjugation with hydrophobic groups such as long alkyl chains, lipids, and amphiphilic polymers or by the introduction of non-natural nucleotides modified with hydrophobic groups. A hydrophobic 6-mer DNA with dodecyl phosphotriester linkages self-assembles into a G-quadruplex in chloroform when metal ions are extracted from water solution (Shibata et al. 2013). It is remarkable that the G-quadruplex DNAzyme formed by the association of 6-mer DNA strands modified at the 5′-end with PEG catalyzes the oxidative reaction of 3-aminophthalic hydrazine in methanol; the active DNAzyme was prepared by mixing the DNA and the substrate in water containing K+ followed by lyophilization and redissolvation in methanol (Abe et al. 2012). Organic solvent-soluble oligonucleotides can also be prepared by using quaternary ammonium surfactants as counterions; the complex of DNA and didodecyldimethylammonium is soluble in organic solutions such as DMF and tetrahydrofuran, and this method has been used for coupling of a DNA strand with highly hydrophobic compounds (Liu et al. 2014). These studies indicate that oligonucleotides are soluble and that their base-paired structures are maintained in various organic solutions when there are metal ions carried over from water solution or when amphiphilic cations are added to screen the charges of the nucleotide phosphates.
Organic solvents have been utilized to drive DNA-mediated organic reactions that produce compounds having desired optical, electrical, or magnetic properties (Gartner and Liu 2001). In the DNA-guided organic synthesis, the association of complementary DNA strands in the presence of organic solvents promotes the coupling reaction between the hydrophobic molecules covalently linked to the 5′- or 3′-ends of oligonucleotides. Hydrophobic reactants linked through association of DNA oligonucleotides (10- to 30-mers) prehybridized in water containing the cationic surfactant cethyltrimethylammonium results in amine acylation in the solutions of 95 % or higher amounts of DMF, dichloromethane, tetrahydrofuran, and acetonitrile (Rozenman and Liu 2006; Rozenman et al. 2007). More structurally complex double-crossover molecules based on DNA self-assembly are promising molecular scaffolds for DNA-based template synthesis of hydrophobic compounds at the water–chloroform interface (Lin et al. 2008). In this study, amphiphilic DNA double-crossover molecules were constructed using hydrophobically modified thymidines with benzylation at the 2′ position or with replacement of one of the phosphate oxygens by a methyl group. Organic cosolvents are also effective for DNA-based asymmetric catalyses: For example, a Henry reaction was shown to be mediated by a double-stranded DNA in the presence of methanol, ether, toluene, and DMSO at about 14 vol% (Fan et al. 2008). Further, a Diels-Alder reaction, a Michael addition, and a Friedel-Crafts reaction were mediated by a copper complex bound to the chiral DNA duplex in the presence of DMF, tetrahydrofuran, methanol, DMSO, 1,4-dioxane at 10–33 vol% (Megens and Roelfes 2010). When oligonucleotides are used as molecular scaffolds for organic synthesis, optimization of the solvent system is important because organic solvents generally destabilize nucleic acid base pairing.
Aqueous solutions containing organic cosolvents
Nucleic acids are soluble in aqueous solutions mixed with water-soluble organic cosolvents. Studies using circular dichroism (CD) and electron paramagnetic resonance (EPR) spectroscopy have shown that the addition of various cosolvents at 20 wt% or higher does not substantially change the overall conformation of short duplexes or hairpins (2–16-mers) but does alter the conformations of single-stranded DNA and RNA (Nakano et al. 2008, 2014b). Nuclear magnetic resonance (NMR) analyses indicate that a relatively low concentration of cosolvent, such as 1,4-dioxane at 10 vol% or DMSO at 5 vol%, disrupts weak noncanonical interactions in the group II intron ribozyme (Furler et al. 2009) and in the transactivation response element RNA from human immunodeficiency virus type 1 (HIV-1) (Lee et al. 2013). Thus, organic cosolvents in amounts of up to a few tens of percent by volume or weight do not significantly affect base pairing but do disrupt weak interactions of flexible residues.
There are many reports on the stability of oligonucleotide structures in mixed solutions. In most cases, the thermal stability of base pairing decreases in the presence of cosolvents (discussed further below). This destabilization effect under reduced water content is proposed to be relevant to the function of DNA unwinding enzymes such as helicases (Cui et al. 2007). Moreover, the ability of organic cosolvents to decrease the thermodynamic stability of base pairs has the practical benefit of regulating oligonucleotide structure formation in various applications: For example, the efficiencies of polymerase chain reaction (PCR) and DNA sequencing are improved by the addition of 10 % DMSO, which inhibits reassociation of template DNA strands (Winship 1989; Jensen et al. 2010). Detection of target sequences using molecular beacons is also improved by addition of cosolvents (methanol, ethanol, isopropanol, acetonitrile, formamide, DMF, DMSO, ethylene glycol, and glycerol), showing a 70-fold rate enhancement in 56 vol% ethanol compared with the rate in the absence of cosolvents, as a result of decreasing the activation energy for the hybridization (Dave and Liu 2010). Likewise, the rate of strand replacement in a 30-base-pair DNA duplex is enhanced by the addition of cosolvents such as isopropanol, ethanol, and DMSO; e.g., a 15-fold enhancement was observed in 20 vol% isopropanol (Zhang et al. 2015). It is worth noting that, in contrast to the case of oligonucleotide duplexes, the stability of 9- and 10-mer duplexes of peptide nucleic acids (PNAs) having a non-ionic backbone are not disrupted by the addition of DMF or dioxane up to 70 % (Sen and Nielsen 2006, 2007) or of methanol, ethylene glycol, or low-molecular-weight PEGs at 20 wt% (Nakano et al. 2012b). These observations suggest an important role of the nucleotide backbone in the destabilization of oligonucleotide duplexes.
There are several possible explanations for the effects of cosolvents on the stability of nucleic acid structures. When the molarity of cosolvent is high (e.g., about 2 M for the 20 wt% solution of a compound with the molecular weight of 100), binding interactions with nucleotides might be significant. The single-stranded conformation may have greater capability to interact with cosolvents than the base-paired duplex in which bases are stacked within the helix. Particularly, formamide, DMSO, and diethylsuloxide (e.g., at 40 vol%) destabilize DNA base pairing because of the formation of multiple hydrogen bonds and hydrophobic interactions with the nucleotide bases (Escara and Hutton 1980; Blake and Delcourt 1996; Markarian et al. 2006). On the other hand, the enhancement of the renaturation rate of base pairs formed by E. coli and bacteriophage λ DNAs in the solutions with DMSO and formamide has been attributed to the effect of the dielectric constant (Escara and Hutton 1980). To clarify how the solvent properties affect the stability of nucleic acid structures, it is preferable to employ cosolvents that have no strong specific interactions with nucleotides. In addition, although experiments using varied concentrations of a single species of cosolvent have been used to correlate particular solvent properties with base-pair stability or hybridization kinetics, such experiments can be misleading when multiple solvent properties are simultaneously changed. Systematic comparisons of the effects of different types of cosolvents such as ethylene glycol derivatives, small primary alcohols, and aprotic solvents are useful as certain cosolvents have similar dielectric constant but different water activities and vice versa (Fig. 2). Many studies have emphasized the importance of the osmotic pressure effect arising from the reduced water activity and of the dielectric constant effect on nucleic acids. Several examples are given in the following sections.
Fig. 2.
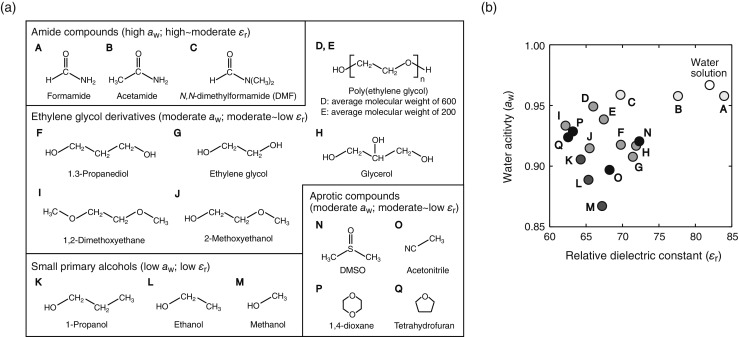
a Organic solvents used for the preparation of mixed aqueous solutions. The properties of water activity a w and relative dielectric constant ε r of 20 wt% solutions are indicated. b Values of the water activity and the relative dielectric constant determined for the 20 wt% solutions of organic cosolvents containing 1 M NaCl (Nakano et al. 2004, 2012a)
Effects of reduced water activity
The stabilities of DNA and RNA duplexes with 6 or 7 base pairs in a 1 M NaCl solution are decreased in the presence not only of formamide, DMSO, and DMF, but also of methanol, ethanol, ethylene glycol, 1-propanol, 2-propanol, and dioxane in the amount of 10 mol% (Albergo and Turner 1981; Hickey and Turner 1985). The magnitudes of the destabilization depend on the cosolvent used, and there is not a good correlation between duplex stability and viscosity, dipole moment, surface tension, or the solubility of adenine. A study using solutions of cosolvents such as ethylene glycol, methanol, 1,2-dimethoxyethane, and low-molecular-weight PEGs also demonstrated that thermodynamic stabilities of DNA duplexes and hairpins (8- to 18-mer) decreased as the amount of cosolvents was increased up to 40–50 wt% and the effect of cosolvents on the stabilities reduced as the amount of NaCl concentration was increased up to 1 M (Nakano et al. 2004, 2012b). Organic compounds dissolved in water act as osmolytes, interacting with water molecules and decreasing the activity of water to less than 1 (Parsegian et al. 2000; Blandamer et al. 2005). For example, ethylene glycol derivatives, small primary alcohols, and aprotic compounds in the amount of 20 wt% create solutions with water activities of 0.87–0.95 (Fig. 2b). The reduction in the water activity affects the equilibrium of reactions that are accompanied by the association or dissociation of water molecules, which holds true for many nucleic acid reactions. Consistent with this interpretation, there is a correlation between the water activity and the stability of DNA duplexes or hairpins (8- to 20-mer), suggesting that the osmotic pressure dominates the effect on the base-pair stability under the experimental salt condition of 1 M NaCl (Nakano et al. 2004, 2008, 2012b; Karimata et al. 2007). The degree of destabilization in the presence of amide compounds such as formamide, DMF, and acetamide is greater than expected strictly from an effect of osmotic pressure (Nakano et al. 2014b); these cosolvents have specific interactions with nucleotides. It has been argued that PEG directly interacts with DNA strands due to its hydrophobic nature (Knowles et al. 2011; Buscaglia et al. 2013b); however, this effect of PEG is likely not significant at high salt concentrations, because the degree of duplex destabilization in PEG-containing solutions at 1 M NaCl was found to be similar to those observed in the solutions with 1,2-dimethoxyethane and 1-propanol, which have water activity similar to that of the PEG solution (Nakano et al. 2004, 2012b). Accordingly, the choice of solvent system is important for investigating the effect of solvent properties because all organic compounds have the potential to interact with nucleic acids.
The stabilities of the left-handed Z-form, branched junction motifs, and triple- and quadruple-helical structures of DNA oligonucleotides are increased in many mixed solutions (Spink and Chaires 1999; Miyoshi et al. 2006; Nakano et al. 2008; Muhuri et al. 2009). The increase in stabilities of these non-canonical structures is due to the release of water during the structure formation, which is favored under the reduced water activity. The addition of methanol at 8 and 16 vol% also increases the stability of tertiary structure formed by a 58-mer fragment of E. coli rRNA (Shiman and Draper 2000). The GAAA tetraloop-receptor motif is stabilized by low-molecular-weight PEG up to around 15 wt% (Downey et al. 2007), and the structures of yeast tRNAs are stabilized by 20 wt% methanol and low-molecular-weight PEG (Strulson et al. 2014). In addition, the folding of the 71-mer adenine-specific riboswitch and the affinity for the ligand analog 2-aminopurine are enhanced by the addition of low-molecular-weight PEG up to 30 wt% (Kumar et al. 2012). Increases in osmotic pressure enhance cooperativity of tertiary folding because of the destabilizing effect on base-paired intermediates (Shiman and Draper 2000; Nakano et al. 2014b; Strulson et al. 2014). These studies highlight the importance of the osmotic pressure on tertiary folding and function of RNA. It is remarkable that the effects of cosolvents on the tRNA and the adenine-specific riboswitch are pronounced at Mg2+ concentrations of 0.5 mM and lower, but the magnitude of the effect is smaller at higher Mg2+ concentrations (Kumar et al. 2012; Strulson et al. 2014). Thus, the effect appears to become less significant at high ionic strengths.
Effects of reduced dielectric constant
The dielectric constant determines the efficiency of cation binding that screens the electronegative potential of nucleotide phosphates. A low dielectric constant medium enhances the electrostatic attraction between the phosphate group and cations, overcoming the electrostatic repulsion between the backbone phosphate groups. The interaction energy for cation binding can be estimated using the simple electrostatic interaction model based on the Coulomb interaction that is inversely proportional to the dielectric constant. For example, 20 wt% solutions of primary alcohols, ethylene glycol derivatives, and aprotic compounds have relative dielectric constants ε r of between 62 and 72, whereas the value of pure water is about 80 at 25 °C (Fig. 2b). The reduction in the ε r from 80 to 60 results in about a 1.3-fold stronger electrostatic interaction, suggesting the increased efficiency of cation binding to nucleotide phosphates in low dielectric constant solutions.
There are several studies of the effect of dielectric constant on the melting temperature (T m) of DNA duplexes. A study using mixed solutions with ethylene glycol and 1,4-dioxane at varied concentrations suggested a dominant effect of the dielectric constant on the stability of DNA base pairing when a relatively low amount of cosolvent (not less than about 0.69 partial volume fraction of water) is used (de Xammar Oro and Grigera 1995). This study showed that the T m values of calf thymus DNA scales with the dielectric constant in the range above 58. Another study suggested that the interaction between DNA and Na+ during base-pair formation is enhanced in the presence of glycerol, which is demonstrated by the increase in T m of calf thymus DNA as glycerol is added up to 40 vol% at low ionic strength of Na+; but this effect is not apparent at high Na+ concentrations (Sorokin et al. 1997). In contrast, in experiments using mixed solutions of glycerol, ethylene glycol, and acetamide, the water activity of solutions rather than the dielectric constant is supposed to impact the base-pair stability of E coli DNA (Spink and Chaires 1999).
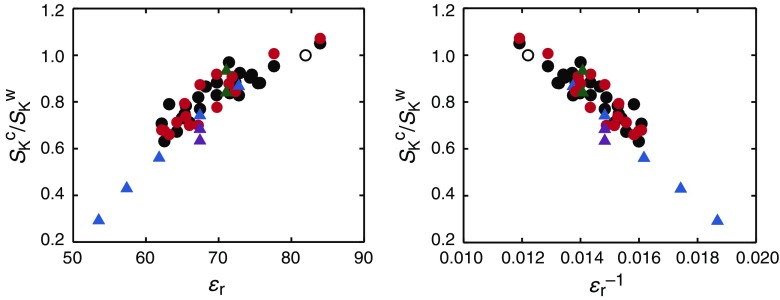
Systematic investigations using an 11-base-pair RNA duplex showed that duplex stability in 1 M NaCl does not correlate with the dielectric constant, water activity, or viscosity of aqueous solutions with 5–20 wt% cosolvents such as methanol, ethylene glycol, 1,2-dimethoxyethane, and low-molecular-weight PEGs (Nakano et al. 2014b). It is probable that base-pair stability is determined by multiple factors, including specific interactions with cosolvents, and consequently the investigations using different kinds of cosolvents would have provided no clear correlation between a single solvent property parameter and duplex stability. This study showed linear dependences of the free energy change (∆G) or the equilibrium constant (K) of the duplex formation on NaCl concentration of mixed solutions, analyzed by S K (= ∂(–∆G)/∂ln [NaCl] = RT ∂ln K/∂ln [NaCl], where R is the gas constant and T is the absolute temperature). Interestingly, there is a correlation between the dependence on NaCl concentration and the dielectric constant of solutions. In addition to the above study, there are several reports of the NaCl concentration dependence of the thermodynamic stability of duplex and hairpin structures of DNA and RNA oligonucleotides in 5–50 wt% solutions of cosolvents such as those shown in Fig. 2a. Based on these reports, effects of the dielectric constant of mixed solutions, either ε r or ε r −1, on the NaCl concentration dependence were analyzed using the ratio to the dependence in water, S K c/S K w (the superscripts c and w represent the parameter for the mixed solution with cosolvents and water solution, respectively), as indicated in Fig. 3. The calculation of the ratio cancels the terms arising from the differences in the length and sequence of oligonucleotides examined. Although values of the dielectric constant used are those of bulk solvent, a good correlation was obtained regardless of the nucleotide sequence used in the studies. In addition, the analysis of T m of E. coli DNA in the range of 10–50 vol% DMSO has revealed a relationship between ∂T m/∂log [Na+] and the reciprocal of the dielectric constant (Escara and Hutton 1980). The reported ratios of the dependence of T m in low dielectric constant media to the dependence in water are in good agreement with the degree of the reductions given in Fig. 3. It is probable that the strength of electrostatic interactions mainly determines the degree of salt concentration dependence.
Fig. 3.
The NaCl concentration dependence of the stability of DNA and RNA structures relative to the dependence in the absence of cosolvents (S K c/S K w) against the dielectric constant (ε r or ε r −1) of mixed solutions with organic cosolvents at 5–50 wt%. The data are derived from the reports for a 20-mer DNA that forms a hairpin (blue) (Karimata et al. 2007), 18-mer DNA duplex and hairpin (purple) (Nakano et al. 2008), 13-mer DNA duplexes (green) (Nakano and Sugimoto 2014), an 11-mer RNA duplex (black) (Nakano et al. 2014b), and a 9-mer RNA duplex (red; our unpublished results). The data obtained in water solution are indicated by open circles
The dependence of the structural stability on salt concentration (the S K value) is related to the degree of cation binding increased during the structure formation (Record et al. 1998; Bloomfield et al. 2000). Thus, the data in Fig. 3 indicate smaller increases in the number of bound Na+ during structure formation in lower dielectric constant solutions compared to those with higher dielectric constants: For example, 60–70 % fewer Na+ ions are bound in medium with ε r of 60 than in water solution. It may appear contradictory that the cation binding is enhanced in low dielectric constant media. It should be noted that a low dielectric constant enhances the efficiency of cation binding both to less condensed and more condensed nucleic acid structures, and the data in Fig. 3 provide the differences between the numbers of thermodynamically associated cations before and after the structure formation. Consistent with the experimental results, a computer simulation demonstrated that low dielectric constant medium leads to stronger charge neutralization of a DNA helix (12-mer) relative to that in high dielectric constant conditions and consequently enhances the electrostatic helix–helix attraction with a smaller increase in the number of bound cations (Tan and Chen 2006).
The polyelectrolyte theory applied to nucleic acids explains that the charge spacing distance of a nucleotide chain b determines, or is determined by, the thermodynamic binding parameter Ψ (Record et al. 1998; Bloomfield et al. 2000). The Ψ value represents the degree of binding interaction of a single phosphate group with counterions in the condensation layer and with those in the surrounding ion atmosphere mediated by weak nonspecific interactions in a rapid equilibrium between association and dissociation. When nucleic acid strands are assumed to be ideal polyelectrolytes having uniformly distributed charges along a cylinder axis, the value of Ψ is calculated by the equation, Ψ = 1 – 1/(2ξ), where ξ is the charge density parameter representing the degree of monovalent cation condensation (ξ = e 2/4πε 0 ε r k B Tb, where e is the magnitude of the elementary charge, ε 0 is the vacuum permittivity, k B is the Boltzmann constant, and T is the absolute temperature). The value of Ψ depends on the charge spacing distance along the helix (Fig. 4a) and on the dielectric constant (Fig. 4b). For a typical B-form helical conformation of a DNA duplex in water (ε r = 80), the b ds (the subscript ds represents the parameter for a duplex strand) is 0.17 nm (Record et al. 1998; Bloomfield et al. 2000) and the Ψ for monovalent cation binding is 0.88. Assuming that the duplex conformation is unchanged in the medium of ε r = 60 (hence, the b ds remains 0.17 nm), the Ψ is 0.91. Thus, this calculation shows a greater degree of cation binding to the duplex in the lower dielectric constant medium. In the case of a single-stranded DNA in the medium with ε r of 80, the b ss (the subscript ss represents the parameter for a single strand) is 0.43 nm and the Ψ is 0.69, although the values depend to some extent on the nucleotide sequence (Record et al. 1998; Bloomfield et al. 2000). If the helical conformation is unchanged in medium with ε r of 60 (the b ss remains 0.43 nm), the Ψ is 0.77. Based on these Ψ values, the reaction cycle for the transition from a single strand to a duplex in water solution and in a mixed solution having a low dielectric constant shown in Fig. 4c is considered. The dielectric constant affects cation binding to both the single strand and the duplex, and the degree of cation binding, which increases upon duplex formation (∆Ψ = Ψ ds – Ψ ss), decreases linearly as the dielectric constant decreases (Fig. 4d). Thus, the ∆Ψ c in mixed solutions is smaller than ∆Ψ w in water. For example, the ∆Ψ c value in medium with ε r of 60 is 0.14 (=0.91–0.77), which is 75 % of the ∆Ψ w value of 0.19 (=0.88–0.69). In the case of RNA strands, the b ds for a typical RNA duplex (A-form conformation) is 0.13 nm (Record et al. 1998; Bloomfield et al. 2000), and the Ψ for monovalent cation binding is 0.91 in medium with ε r of 80 and 0.93 in the medium with ε r of 60. If the calculation is performed with the b ss values used above, the ∆Ψ c for the RNA duplex formation in the medium with ε r of 60 is 0.16 (=0.93–0.77), which is about 73 % of the ∆Ψ w value of 0.22 (=0.91–0.69) in water.
Fig. 4.
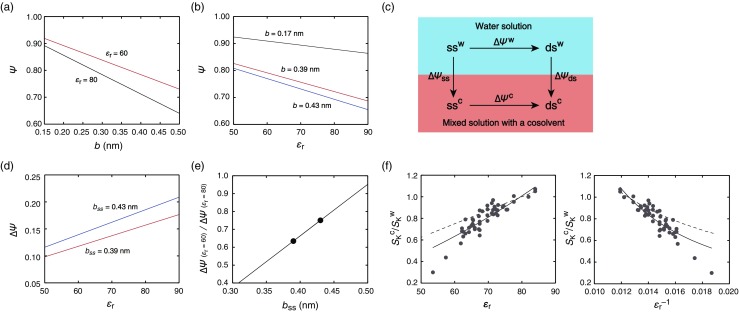
a Effect of the charge spacing distance b on the degree of interaction with monovalent cations Ψ in the medium of ε r = 80 (black) or 60 (red) at 25 °C. b Effect of ε r on Ψ for the charge spacing distance 0.17 nm (black), 0.43 nm (blue), or 0.39 nm (red). c The reaction cycle for the formation of a duplex (ds) from a single strand (ss) in water solution (superscript w) or mixed solution with a low dielectric constant cosolvent (superscript c). d Effect of the ε r on the change in Ψ during the duplex formation (b ds = 0.17 nm) from a single strand of b ss = 0.43 nm (blue) or 0.39 nm (red). e Effect of the b ss on the ∆Ψ in the medium of ε r = 60 relative to that in ε r = 80 with an assumption that the value of b ds = 0.17 nm is invariant. The data points for b ss = 0.43 and 0.39 nm described in the text are marked by closed circles. f The curve fits to the data in Fig. 3 using a linear approximation of b ss as a function of ε r. The broken lines represent the theoretical curve generated using a constant value 0.39 nm for b ss
However, experimentally, the conformations of single strands but not duplexes were found to be changed in mixed solutions relative to the conformations in water (Nakano et al. 2008, 2014b). Furthermore, a study by EPR spectroscopy reports shortening of the helical length of a single-stranded DNA from 0.48 nm in water to 0.33 nm in 50 wt% PEG with the average molecular weight of 200, but no alteration in the length of a DNA duplex (Nakano et al. 2008). This phenomenon is consistent with a report of the collapse of long linear polyelectrolyte chains of poly(2-vinylpyridine) quaternized with ethyl bromide in poor solvents consisting of 1-propanol and 2-pentanone (Loh et al. 2008). As demonstrated in Fig. 4b, d, only a slight change in b affects the values of Ψ and ∆Ψ. Assuming that b ss is changed to 0.39 nm (about 10 % shortening of the distance), the Ψ in the medium with ε r of 60 is calculated to be 0.79. Consequently, the ∆Ψ c value when ε r is 60 is 0.12 (= 0.91 – 0.79) for DNA and 0.14 (= 0.93 – 0.79) for RNA, 63 % and 64 % of their ∆Ψ w values, respectively. The reductions are consistent with the data given in Fig. 3. The theoretical curves, using a linear approximation of b ss as a function of ε r, fitted to the data in Fig. 3 are shown in Fig. 4f. The curve fits the data better than the use of a constant value for b ss. The deviation of the data for a low dielectric constant (ε r < 60) from the fit might be due to a less significant electrostatic effect under high cosolvent concentrations (de Xammar Oro and Grigera 1995) or to the inappropriate use of the first-order approximation. In addition, the analysis using the reaction cycle shown in Fig. 4c suggests that the degree of ion binding increased by the transfer of a single strand from water to a mixed solution (∆Ψ ss) is greater than the case of the transfer of a duplex (∆Ψ ds). It is possible that the flexibility of a single strand allows shortening of the helical length. This scenario implies the role of intrinsically disordered residues to enhance the efficiency of cation binding to nucleic acids in low dielectric constant media.
The dielectric constant also affects the equilibrium between a unimolecular hairpin and a dimer duplex of self-complementary DNA and RNA oligonucleotides (Nakano et al. 2012a). The equilibrium is shifted toward the dimer duplex by the addition of ethylene glycol derivatives and primary alcohols at 20 wt%, caused by enhanced binding of Na+ and Mg2+ to the loop nucleotides of a hairpin structure. There was an increase by a factor of 3 in the affinity of Na+ binding and by a factor of 6 in the affinity of Mg2+ binding in the medium of 20 wt% 1,2-dimethoxyethane (ε r = 62) relative to water. In the case of formation of the G-quadruplexes driven by monovalent ion binding to guanine bases, the structural stability is increased in the mixed solutions of low-molecular-weight PEG, ethanol, and acetonitrile relative to that in water; and the stabilization is thought to result from changes in the osmotic pressure or from specific interactions of cosolvents (Miyoshi et al. 2006; Vorlíčková et al. 2006, 2007; Buscaglia et al. 2013a). In contrast to the reports, the dielectric constant effect is suggested to underlie effects on the stability of the G-quadruplex formed by the thrombin binding aptamer (15-mer) in the mixed solutions of methanol, ethanol, and propanol up to 40 vol% (ε r = ~50), as there is a linear relationship between T m values and the reciprocal of the dielectric constant of the solutions (Smirnov and Shafer 2007). Another study using various 20 wt% solutions of cosolvents showed that G-quadruplexes (formed by 15- and 21-mers) are stabilized more in a lower dielectric constant solution due to a greater than 10-fold increase in the affinity of Na+ binding in the medium of ε r close to 60 relative to that in water (our unpublished results). The reduced dielectric constant of 10 vol% 1,4-dioxane (ε r = ~70) also causes the 1.5-fold increase in the affinity of specific Mg2+ binding to the catalytic core domain of the group II intron ribozyme relative to water solution (Furler et al. 2009). These studies support the hypothesis that effects of the dielectric constant are dominant in low concentrations of cations, and other factors such as water activity and specific interactions with nucleotides could dominate in saturating concentrations of cations. It has been argued that increasing ionic strength attenuates local accumulation of organic compounds at the surface of DNA (Nordstrom et al. 2006) and that low ionic strength induces hydrophobic interactions within single-stranded DNA (Markarian and Schlenoff 2010). Therefore, it is important to consider the effect of cosolvents on nucleic acids in terms of the salt conditions.
Solvent effects on the catalytic activities of DNAzymes and ribozymes
The use of organic solvents can expand the function of DNAzymes and ribozymes by increasing the solubility of hydrophobic substrates. A relatively low amount of cosolvents (e.g., DMSO at less than 10 vol%) have been used during in vitro selection of catalytic DNA and RNA and in Diels-Alder reactions (Seelig and Jäschke 1999; Chandra and Silverman 2008) and aldol reactions (Fusz et al. 2005) catalyzed by these molecules. The RNA ligation reaction catalyzed by DNA aptazymes in the presence of herbicides (alachlor and atrazine) proceeds more rapidly and in higher yields upon the addition of 10 % methanol, ethanol, DMSO, acetone, or DMF. The cosolvents might facilitate product release, stabilize certain structures, prevent the formation of inactive conformations, and/or improve the association between the DNA and substrates (Behera et al. 2013). Because the catalytically active structures of the DNAzymes and ribozymes are maintained by weak interactions compared with base-paired secondary structures, solvent conditions that significantly disrupt RNA interactions are not suitable for use.
Formation of the structures of natural ribozymes that catalyze site-specific phosphodiester bond cleavage of RNA is accompanied by water release and metal ion binding (Rangan and Woodson 2003; Bevilacqua et al. 2004; Doudna and Lorsch 2005; Lilley 2005; Toor et al. 2008). The effects of osmotic pressure effect on the catalytic activities of a lead-dependent ribozyme and a hairpin ribozyme have been reported. The lead-dependent ribozyme, called the leadzyme, is comprised of a short stem-loop structure (Fig. 5a) and cleaves RNA in the presence of Pb2+. This ribozyme was initially generated by in vitro selection from random sequence libraries (Pan and Uhlenbeck 1992), and the same sequence motifs are found in yeast phenylalanine-specific tRNA and in human mRNAs (Barciszewska et al. 2005). The addition of low-molecular-weight PEG (2.5–7.5 vol%) enhances the catalytic activity of an 11-mer leadzyme in the presence of 25 mM Pb2+ by about a factor of 2 (Giel-Pietraszuk and Barciszewski 2012). This enhancement is attributed to a reduction in water activity that facilitates the release of water during catalysis. Hairpin ribozymes are found in the satellite RNA of plant viruses such as tobacco ringspot virus (Buzayan et al. 1986; Hampel and Tritz 1989). These ribozymes are comprised of a substrate-binding domain loop and a catalytic loop domain (Fig. 5a), and cleave RNA in the presence of divalent metal ions. As the amount of low-molecular-weight PEG is increased to 10 %, the cleavage rate of an 85-mer hairpin ribozyme in the presence of 1 mM Mg2+ increases by several fold; PEG likely facilitates tertiary folding, which is accompanied by the release of water molecules, while there is no evidence for the dielectric constant effect in this system (Herve et al. 2006).
Fig. 5.
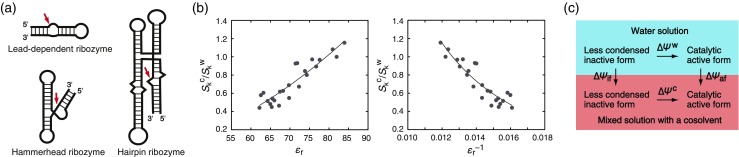
a The ribozyme structures described in the text and their cleavage sites (red arrows). b Plots of the NaCl concentration dependence of the cleavage rate constant of a hammerhead ribozyme relative to the dependence in the absence of cosolvents (S k c/S k w) against the dielectric constant (ε r or ε r −1) of mixed solutions in the amount of 10 or 20 wt% cosolvents (Nakano et al. 2014b). The curve fits are drawn based on the calculation performed in Fig. 4f. c The reaction cycle for the formation of the catalytically active form of ribozymes in water solution and a mixed solution with a low dielectric constant cosolvent
The hammerhead ribozymes are found in RNA satellites of plant viruses and in genomes including those of humans (Bourdeau et al. 1999; Martick et al. 2008; de la Pena and Garcia-Robles 2010; Hammann et al. 2012). These ribozymes are comprised of three stems (Fig. 5a), and cleave RNA in the presence of divalent metal ions (Forster and Symons 1987; Uhlenbeck 1987; Haseloff and Gerlach 1988; Martick and Scott 2006; Lee et al. 2008). Cosolvents such as acetonitrile, methanol, DMF, DMSO, ethylene glycol, and glycerol have minimal effects on the rate and yield of the intermolecular cleavage of a 17-mer substrate by a 38-mer ribozyme in 10 mM Mg2+; but at concentrations higher than 60 vol%, these cosolvents completely inhibit the cleavage reaction (Feig et al. 1998; Mikulecky and Feig 2002). A study using a similar ribozyme motif (intermolecular cleavage of a 15-mer substrate by a 43-mer ribozyme) showed that 20 wt% solutions of ethylene glycol derivatives, small primary alcohols, and aprotic compounds enhance the RNA cleavage rate by several fold in the presence of 10 mM Mg2+ and by greater than 10 fold at lower Mg2+ concentrations (Nakano et al. 2009, 2015a). Because the tertiary structure of the ribozyme used in the study is not very stable, the efficiency of Mg2+ binding is important for the formation of the catalytically active structure and the cleavage rate. The MgCl2 concentration required for rapid catalysis, in which the binding of Mg2+ is mediated by specific interactions, is changed in the mixed solutions. The concentration of MgCl2 necessary decreases as the dielectric constant of solutions is decreased; e.g., the concentration decreases greater than by a factor of 10 in 20 wt% 1,2-dimethoxyethane (ε r = 62) than in the absence of cosolvents (Nakano et al. 2015a).
In the reaction catalyzed by the hammerhead ribozyme, divalent metal ions can be replaced by monovalent cations if concentrations are sufficiently high (Murray et al. 1998). Catalysis in the presence of Na+ and no divalent cation is mediated by highly cooperative and nonspecific weak interactions with diffusive cations that contribute to folding into the catalytic active form. The NaCl concentration dependence of the cleavage rate constant k (S k = ∂ln k/∂ln [NaCl], in a linear dependence) is changed in the presence of cosolvents (Nakano et al. 2014b). The dependence decreases with the dielectric constant of the solution (Fig. 5b), and about 40 % less Na+ is required in 20 wt% 1,2-dimethoxyethane (ε r = 62) than in the absence of cosolvents. Assuming that the substrate cleavage rate is determined by the amount of the catalytic active structure formed, the degree of Na+ binding upon formation of the active structure (∆Ψ, which is proportional to S k) is decreased in low dielectric constant solutions (S k c/S k w < 1 and hence ∆Ψ c/∆Ψ w < 1). Thus, the analysis using the reaction cycle in Fig. 5c suggests greater Na+ binding to less condensed inactive forms (∆Ψ if) than to the catalytic active form (∆Ψ af), analogous to the case of a duplex formation. About a 40 % decrease in Na+ binding in the medium with ε r of about 60 is greater than the decreases observed for duplex formation (60–70 %) given in Fig. 3, possibly because of greater Na+-induced collapse upon formation of the active structure of the ribozyme than occurs during duplex formation.
Water-soluble organic polymers are used as macromolecular crowding agents to mimic the crowded environment in cells (Minton 1998). Addition of high-molecular-weight PEG or dextran in the amount of 5–25 wt% enhances the RNA cleavage activity of group I ribozyme (195-mer RNA), human delta virus (HDV)-like ribozyme (76-mer RNA), and hairpin ribozyme (intermolecular cleavage of a 48-mer substrate by a 35-mer ribozyme) by several fold in a low concentration of Mg2+, around 1 mM (Kilburn et al. 2010; Strulson et al. 2013; Desai et al. 2014; Paudel and Rueda 2014). The rate enhancements result from increased compactness of the RNA structures through the excluded volume effect of the polymer additives. In the case of a hammerhead ribozyme motif (43-mer ribozyme and 15-mer substrate), the addition of high-molecular-weight PEG increases the cleavage rate at unsaturated concentrations but not at saturated concentrations of Mg2+ (Nakano et al. 2009). This study showed that the Mg2+ concentration required for fast catalysis is decreased in the presence of PEG with an average molecular weight of 8000, as is also the case for smaller cosolvents (Nakano et al. 2015a). Because polymer crowding substantially modifies the water activity and the dielectric constant (Nakano et al. 2004, 2012a), the effects of polymer additives may, at least partly, arise from changes in these solvent properties. Consistent with this possibility, values of ∆Ψ for the hybridization of 15, 25, 33, and 35 base pairs of DNA decrease linearly with the concentration of PEG with an average molecular weight of 3000 (0–30 wt%) (Markarian and Schlenoff 2010). It has also been reported that PEG with an average molecular weight of 8000 at 20 wt% shortens the length of a single-stranded oligonucleotide but not the length of a duplex (Nakano et al. 2008). In addition, the magnitude of the effect of polymer additives is sensitive to the ionic strength because the cation concentration affects the size of unfolded RNAs (Denesyuk and Thirumalai 2013). These results indicate that polymer solutions have effects on cation binding to nucleic acids in addition to their excluded volume effect.
Concluding remarks
This review described the importance of the use of organic solvents in nucleic acid research. Use of organic solvents or cosolvents has applications in oligonucleotide-based technologies, such as PCR, construction of self-assembled DNA nanostructures, and DNA-guided organic synthesis, through the modulation of the stability and kinetics of hybridization. Cosolvents alter water activity and dielectric constant and thus impact base paring and tertiary structure formations. Finally, the presence of organic solvents influences the activities of nucleic acid catalysts. Although organic solvents affect nucleic acid interactions by a combination of mechanisms, only a few factors appear to dominate the cosolvent effect at extremes of salt concentrations. Considerations of these dominant factors are helpful during selection of the solvent system. For example, based on the solvent properties provided in Fig. 2, it is plausible that solvents that reduce the dielectric constant, like 1,2-dimethoxyethane, are beneficial to promote nucleic acid folding at low ionic strengths, whereas solvents that reduce the water activity, such as methanol, are beneficial to disrupt base pairing at high ionic strengths. More comprehensive studies will provide valuable information that will guide optimization of solvent systems for nucleic acid experiments.
Experimental systems using organic solvents facilitate the study of how the molecular environment affects the interactions of nucleic acids. The intracellular medium consists of a large number of organic molecules in a confined space (Luby-Phelps 2000), and the water activity and the dielectric constant are reduced relative to water. The free Mg2+ concentration in mammalian cells is relatively low, 0.25–1 mM (Grubbs 2002). The experiment systems using organic solvents that mimic low water activity, low dielectric constant, and low Mg2+ concentration of the intracellular environment suggest that characteristics of nucleic acid hybridization, tertiary folding, and expression of the catalytic activity of ribozymes in cells differ significantly from those in aqueous buffer (Nakano et al. 2015b). Further studies using organic solvents will advance our understanding of structures and functions of DNA and RNA molecules under non-aqueous conditions including intracellular environments.
Acknowledgments
This work was partly supported by Grants-in-Aid for Scientific Research from JSPS and MEXT (Ministry of Education, Culture, Sports, Science and Technology)-Supported Program for the Strategic Research Foundation at Private Universities (2014–2019), Japan, and the Hirao Taro Foundation of KONAN GAKUEN for Academic Research.
Compliance with Ethical Standards
Conflict of interest
Shu-ichi Nakano declares that he has no conflict of interest.
Naoki Sugimoto declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human or animal subjects performed by the author.
Contributor Information
Shu-ichi Nakano, Phone: +81-78-303-1429, Email: shuichi@center.konan-u.ac.jp.
Naoki Sugimoto, Phone: +81-78-303-1416, Email: sugimoto@konan-u.ac.jp.
References
- Ababneh AM, Large CC, Georghiou S. Solvation of nucleosides in aqueous mixtures of organic solvents: relevance to DNA open basepairs. Biophys J. 2003;85:1111–1127. doi: 10.1016/S0006-3495(03)74548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, et al. Structure formation and catalytic activity of DNA dissolved in organic solvents. Angew Chem Int Ed Engl. 2012;51:6475–6479. doi: 10.1002/anie.201201111. [DOI] [PubMed] [Google Scholar]
- Albergo DD, Turner DH. Solvent effects on thermodynamics of double-helix formation in (dG-dC)3. Biochemistry. 1981;20:1413–1418. doi: 10.1021/bi00509a002. [DOI] [PubMed] [Google Scholar]
- Arcella A, Portella G, Collepardo-Guevara R, Chakraborty D, Wales DJ, Orozco M. Structure and properties of DNA in apolar solvents. J Phys Chem B. 2014;118:8540–8548. doi: 10.1021/jp503816r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arscott PG, Ma C, Wenner JR, Bloomfield VA. DNA condensation by cobalt hexaammine (III) in alcohol-water mixtures: dielectric constant and other solvent effects. Biopolymers. 1995;36:345–364. doi: 10.1002/bip.360360309. [DOI] [PubMed] [Google Scholar]
- Asami K, Hanai T, Koizumi N. Dielectric properties of yeast cells. J Membr Biol. 1976;28:169–180. doi: 10.1007/BF01869695. [DOI] [PubMed] [Google Scholar]
- Barciszewska MZ, Szymanski M, Wyszko E, Pas J, Rychlewski L, Barciszewski J. Lead toxicity through the leadzyme. Mutat Res. 2005;589:103–110. doi: 10.1016/j.mrrev.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Behera AK, Schlund KJ, Mason AJ, Alila KO, Han M, Grout RL, Baum DA. Enhanced deoxyribozyme-catalyzed RNA ligation in the presence of organic cosolvents. Biopolymers. 2013;99:382–391. doi: 10.1002/bip.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Brown TS, Nakano S, Yajima R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Blake RD, Delcourt SG. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 1996;24:2095–2103. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandamer MJ, Engberts JB, Gleeson PT, Reis JC. Activity of water in aqueous systems; a frequently neglected property. Chem Soc Rev. 2005;34:440–458. doi: 10.1039/b400473f. [DOI] [PubMed] [Google Scholar]
- Bloomfield VA, Crothers DM, Tinoco I Jr (2000) Interaction of nucleic acids and water and ions. Nucleic acids: structures, properties and functions, Chap.11. University Science Books, Sausalito, pp 475–534
- Bonner G, Klibanov AM. Structural stability of DNA in nonaqueous solvents. Biotechnol Bioeng. 2000;68:339–344. doi: 10.1002/(SICI)1097-0290(20000505)68:3<339::AID-BIT12>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Ferbeyre G, Pageau M, Paquin B, Cedergren R. The distribution of RNA motifs in natural sequences. Nucleic Acids Res. 1999;27:4457–4467. doi: 10.1093/nar/27.22.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia R, Gray RD, Chaires JB. Thermodynamic characterization of human telomere quadruplex unfolding. Biopolymers. 2013;99:1006–1018. doi: 10.1002/bip.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia R, Miller MC, Dean WL, Gray RD, Lane AN, Trent JO, Chaires JB. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan JM, Gerlach WL, Bruening G. Satellite tobacco ringspot virus RNA: A subset of the RNA sequence is sufficient for autolytic processing. Proc Natl Acad Sci U S A. 1986;83:8859–8862. doi: 10.1073/pnas.83.23.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra M, Silverman SK. DNA and RNA can be equally efficient catalysts for carbon-carbon bond formation. J Am Chem Soc. 2008;130:2936–2937. doi: 10.1021/ja7111965. [DOI] [PubMed] [Google Scholar]
- Citti L, Rainaldi G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther. 2005;5:11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Dans PD, Carrascosa JL, Orozco M, Gomila G, Fumagalli L. Direct measurement of the dielectric polarization properties of DNA. Proc Natl Acad Sci U S A. 2014;111:E3624–E3630. doi: 10.1073/pnas.1405702111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Yu J, Kuhner F, Schulten K, Gaub HE. Double-stranded DNA dissociates into single strands when dragged into a poor solvent. J Am Chem Soc. 2007;129:14710–14716. doi: 10.1021/ja074776c. [DOI] [PubMed] [Google Scholar]
- Dave N, Liu J. Fast molecular beacon hybridization in organic solvents with improved target specificity. J Phys Chem B. 2010;114:15694–15699. doi: 10.1021/jp106754k. [DOI] [PubMed] [Google Scholar]
- de la Pena M, Garcia-Robles I. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 2010;11:711–716. doi: 10.1038/embor.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Xammar Oro JR, Grigera JR. On the thermal stability of DNA in solution of mixed solvents. J Biol Phys. 1995;21:151–154. doi: 10.1007/BF00712343. [DOI] [Google Scholar]
- Denesyuk NA, Thirumalai D. Entropic stabilization of the folded states of RNA due to macromolecular crowding. Biophys Rev. 2013;5:225–232. doi: 10.1007/s12551-013-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Kilburn D, Lee HT, Woodson SA. Increased ribozyme activity in crowded solutions. J Biol Chem. 2014;289:2972–2977. doi: 10.1074/jbc.M113.527861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Wang S, Stewar G, Hwang H-M, Fu PP, Yu H. Effect of organic solvents and biologically relevant ions on the light-induced DNA cleavage by pyrene and its amino and hydroxy derivatives. Int J Mol Sci. 2002;3:937–947. doi: 10.3390/i3090937. [DOI] [Google Scholar]
- Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat Struct Mol Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- Downey CD, Crisman RL, Randolph TW, Pardi A. Influence of hydrostatic pressure and cosolutes on RNA tertiary structure. J Am Chem Soc. 2007;129:9290–9291. doi: 10.1021/ja072179k. [DOI] [PubMed] [Google Scholar]
- Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- Escara JF, Hutton JR. Thermal stability and renaturation of DNA in dimethyl sulfoxide solutions: acceleration of the renaturation rate. Biopolymers. 1980;19:1315–1327. doi: 10.1002/bip.1980.360190708. [DOI] [PubMed] [Google Scholar]
- Fan J, Sun G, Wan C, Wang Z, Li Y (2008) Investigation of DNA as a catalyst for Henry reaction in water. Chem Commun 3792–3794 [DOI] [PubMed]
- Feig AL, Ammons GE, Uhlenbeck OC. Cryoenzymology of the hammerhead ribozyme. RNA. 1998;4:1251–1258. doi: 10.1017/S1355838298980943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Symons RH. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-X. [DOI] [PubMed] [Google Scholar]
- Furler M, Knobloch B, Sigel RKO. Influence of decreased solvent permittivity on the structure and magnesium (II)-binding properties of the catalytic domain 5 of a group II intron ribozyme. Inorg Chim Acta. 2009;362:771–776. doi: 10.1016/j.ica.2008.03.095. [DOI] [Google Scholar]
- Fusz S, Eisenfuhr A, Srivatsan SG, Heckel A, Famulok M. A ribozyme for the aldol reaction. Chem Biol. 2005;12:941–950. doi: 10.1016/j.chembiol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Garlid KD. The state of water in biological systems. Int Rev Cytol. 2000;192:281–302. doi: 10.1016/S0074-7696(08)60530-6. [DOI] [PubMed] [Google Scholar]
- Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J Am Chem Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giel-Pietraszuk M, Barciszewski J. Hydrostatic and osmotic pressure study of the RNA hydration. Mol Biol Rep. 2012;39:6309–6318. doi: 10.1007/s11033-012-1452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs RD. Intracellular magnesium and magnesium buffering. Biometals. 2002;15:251–259. doi: 10.1023/A:1016026831789. [DOI] [PubMed] [Google Scholar]
- Hammann C, Luptak A, Perreault J, de la Pena M. The ubiquitous hammerhead ribozyme. RNA. 2012;18:871–885. doi: 10.1261/rna.031401.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammouda B, Worcester D. The denaturation transition of DNA in mixed solvents. Biophys J. 2006;91:2237–2242. doi: 10.1529/biophysj.106.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel A, Tritz R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Herve G, Tobe S, Heams T, Vergne J, Maurel MC. Hydrostatic and osmotic pressure study of the hairpin ribozyme. Biochim Biophys Acta. 2006;1764:573–577. doi: 10.1016/j.bbapap.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Hickey DR, Turner DH. Solvent effects on the stability of A7U7p. Biochemistry. 1985;24:2086–2094. doi: 10.1021/bi00329a042. [DOI] [PubMed] [Google Scholar]
- Jensen MA, Fukushima M, Davis RW. DMSO and betaine greatly improve amplification of GC-rich constructs in de novo synthesis. PLoS ONE. 2010;5:e11024. doi: 10.1371/journal.pone.0011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimata H, Nakano S, Sugimoto N. Effects of polyethylene glycol on DNA duplex stability at different NaCl concentrations. Bull Chem Soc Jpn. 2007;80:1987–1994. doi: 10.1246/bcsj.80.1987. [DOI] [Google Scholar]
- Ke F, Luu YK, Hadjiargyrou M, Liang D. Characterizing DNA condensation and conformational changes in organic solvents. PLoS ONE. 2010;5:e13308. doi: 10.1371/journal.pone.0013308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT., Jr Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc Natl Acad Sci U S A. 2011;108:12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Endoh T, Murakami K, Sugimoto N. Dehydration from conserved stem regions is fundamental for ligand-dependent conformational transition of the adenine-specific riboswitch. Chem Commun. 2012;48:9693–9695. doi: 10.1039/c2cc34506d. [DOI] [PubMed] [Google Scholar]
- Lamm G, Pack GR. Calculation of dielectric constants near polyelectrolytes in solution. J Phys Chem B. 1997;101:959–965. doi: 10.1021/jp9623453. [DOI] [Google Scholar]
- Lee TS, Lopez CS, Giambasu GM, Martick M, Scott WG, York DM. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J Am Chem Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Vogt CE, McBrairty M, Al-Hashimi HM. Influence of dimethylsulfoxide on RNA structure and ligand binding. Anal Chem. 2013;85:9692–9698. doi: 10.1021/ac402038t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Nosrati M, Kashani-Sabet M. Knockdown of telomerase RNA using hammerhead ribozymes and RNA interference. Methods Mol Biol. 2008;405:113–131. doi: 10.1007/978-1-60327-070-0_10. [DOI] [PubMed] [Google Scholar]
- Lilley DM. Structure, folding and mechanisms of ribozymes. Curr Opin Struct Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Lin J, Seeman NC, Vaidehi N. Molecular-dynamics simulations of insertion of chemically modified DNA nanostructures into a water-chloroform interface. Biophys J. 2008;95:1099–1107. doi: 10.1529/biophysj.107.119230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lou X, Du J, Guan M, Wang J, Ding X, Zhao J. DNAzyme-based fluorescent microarray for highly selective and sensitive detection of lead(II) Analyst. 2012;137:70–72. doi: 10.1039/C1AN15633K. [DOI] [PubMed] [Google Scholar]
- Liu K, Zheng L, Liu Q, de Vries JW, Gerasimov JY, Herrmann A. Nucleic acid chemistry in the organic phase: from functionalized oligonucleotides to DNA side chain polymers. J Am Chem Soc. 2014;136:14255–14262. doi: 10.1021/ja5080486. [DOI] [PubMed] [Google Scholar]
- Loh P, Deen GR, Vollmer D, Fischer K, Schmidt M, Kundagrami A, Muthukumar M. Collapse of linear polyelectrolyte chains in a poor solvent: when does a collapsing polyelectrolyte collect its counterions? Macromolecules. 2008;41:9352–9358. doi: 10.1021/ma8014239. [DOI] [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/S0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markarian MZ, Schlenoff JB. Effect of molecular crowding and ionic strength on the isothermal hybridization of oligonucleotides. J Phys Chem B. 2010;114:10620–10627. doi: 10.1021/jp103213w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markarian SA, Asatryan AM, Grigoryan KR, Sargsyan HR. Effect of diethylsulfoxide on the thermal denaturation of DNA. Biopolymers. 2006;82:1–5. doi: 10.1002/bip.20454. [DOI] [PubMed] [Google Scholar]
- Marky LA, Kupke DW. Enthalpy-entropy compensations in nucleic acids: contribution of electrostriction and structural hydration. Methods Enzymol. 2000;323:419–441. doi: 10.1016/S0076-6879(00)23376-4. [DOI] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens RP, Roelfes G. Organic co-solvents in aqueous DNA-based asymmetric catalysis. Org Biomol Chem. 2010;8:1387–1393. doi: 10.1039/b921385f. [DOI] [PubMed] [Google Scholar]
- Mel’niov SM, Khan MO, Lindman B, Jönsson B. Phase behavior of single DNA in mixed solvents. J Am Chem Soc. 1999;121:1130–1136. doi: 10.1021/ja981491e. [DOI] [Google Scholar]
- Mikulecky PJ, Feig AL. Cold denaturation of the hammerhead ribozyme. J Am Chem Soc. 2002;124:890–891. doi: 10.1021/ja016878n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 1998;295:127–149. doi: 10.1016/S0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Karimata H, Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J Am Chem Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- Muhuri S, Mimura K, Miyoshi D, Sugimoto N. Stabilization of three-way junctions of DNA under molecular crowding conditions. J Am Chem Soc. 2009;131:9268–9280. doi: 10.1021/ja900744e. [DOI] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/S1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Nakano S, Sugimoto N. Roles of the amino group of purine bases in the thermodynamic stability of DNA base pairing. Molecules. 2014;19:11613–11627. doi: 10.3390/molecules190811613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Karimata H, Ohmichi T, Kawakami J, Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J Am Chem Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- Nakano S, et al. Conformation and the sodium ion condensation on DNA and RNA structures in the presence of a neutral cosolute as a mimic of the intracellular media. Mol BioSyst. 2008;4:579–588. doi: 10.1039/b718806d. [DOI] [PubMed] [Google Scholar]
- Nakano S, Karimata HT, Kitagawa Y, Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J Am Chem Soc. 2009;131:16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- Nakano S, Hirayama H, Miyoshi D, Sugimoto N. Dimerization of nucleic acid hairpins in the conditions caused by neutral cosolutes. J Phys Chem B. 2012;116:7406–7415. doi: 10.1021/jp302170f. [DOI] [PubMed] [Google Scholar]
- Nakano S, Yamaguchi D, Tateishi-Karimata H, Miyoshi D, Sugimoto N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys J. 2012;102:2808–2817. doi: 10.1016/j.bpj.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev. 2014;114:2733–2758. doi: 10.1021/cr400113m. [DOI] [PubMed] [Google Scholar]
- Nakano S, Kitagawa Y, Miyoshi D, Sugimoto N. Hammerhead ribozyme activity and oligonucleotide duplex stability in mixed solutions of water and organic compounds. FEBS Open Bio. 2014;4:643–650. doi: 10.1016/j.fob.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Kitagawa Y, Yamashita H, Miyoshi D, Sugimoto N. Effects of cosolvents on the folding and catalytic activities of the hammerhead ribozyme. ChemBioChem. 2015;16:1803–1810. doi: 10.1002/cbic.201500139. [DOI] [PubMed] [Google Scholar]
- Nakano S, Kitagawa Y, Miyoshi D, Sugimoto N. Effects of background anionic compounds on the activity of the hammerhead ribozyme in Mg2+-unsaturated solutions. J Biol Inorg Chem. 2015;20:1049–1058. doi: 10.1007/s00775-015-1286-y. [DOI] [PubMed] [Google Scholar]
- Ni X, Castanares M, Mukherjee A, Lupold SE. Nucleic acid aptamers: clinical applications and promising new horizons. Curr Med Chem. 2011;18:4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom LJ, Clark CA, Andersen B, Champlin SM, Schwinefus JJ. Effect of ethylene glycol, urea, and N-methylated glycines on DNA thermal stability: the role of DNA base pair composition and hydration. Biochemistry. 2006;45:9604–9614. doi: 10.1021/bi052469i. [DOI] [PubMed] [Google Scholar]
- Pan WH, Clawson GA. Antisense applications for biological control. J Cell Biochem. 2006;98:14–35. doi: 10.1002/jcb.20790. [DOI] [PubMed] [Google Scholar]
- Pan T, Uhlenbeck OC. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+ Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Parsegian VA, Rau DC. Water near intracellular surfaces. J Cell Biol. 1984;99:196–200. doi: 10.1083/jcb.99.1.196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc Natl Acad Sci U S A. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel BP, Rueda D. Molecular crowding accelerates ribozyme docking and catalysis. J Am Chem Soc. 2014;136:16700–16703. doi: 10.1021/ja5073146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitera JW, Falta M, van Gunsteren WF. Dielectric properties of proteins from simulation: the effects of solvent, ligands, pH, and temperature. Biophys J. 2001;80:2546–2555. doi: 10.1016/S0006-3495(01)76226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Woodson SA. Structural requirement for Mg2+ binding in the group I intron core. J Mol Biol. 2003;329:229–238. doi: 10.1016/S0022-2836(03)00430-3. [DOI] [PubMed] [Google Scholar]
- Record MT, Jr, Zhang W, Anderson CF. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv Protein Chem. 1998;51:281–353. doi: 10.1016/S0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- Rozenman MM, Liu DR. DNA-templated synthesis in organic solvents. ChemBioChem. 2006;7:253–256. doi: 10.1002/cbic.200500413. [DOI] [PubMed] [Google Scholar]
- Rozenman MM, Kanan MW, Liu DR. Development and initial application of a hybridization-independent, DNA-encoded reaction discovery system compatible with organic solvents. J Am Chem Soc. 2007;129:14933–14938. doi: 10.1021/ja074155j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger W. Principles of nucleic acid structure. New York: Springer; 1983. [Google Scholar]
- Schubert S, Kurreck J. Ribozyme- and deoxyribozyme-strategies for medical applications. Curr Drug Targets. 2004;5:667–681. doi: 10.2174/1389450043345092. [DOI] [PubMed] [Google Scholar]
- Seelig B, Jäschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- Sekella PT, Rueda D, Walter NG. A biosensor for theophylline based on fluorescence detection of ligand-induced hammerhead ribozyme cleavage. RNA. 2002;8:1242–1252. doi: 10.1017/S1355838202028066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Nielsen PE. Unique properties of purine/pyrimidine asymmetric PNA•DNA duplexes: differential stabilization of PNA•DNA duplexes by purines in the PNA strand. Biophys J. 2006;90:1329–1337. doi: 10.1529/biophysj.105.073213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Nielsen PE. On the stability of peptide nucleic acid duplexes in the presence of organic solvents. Nucleic Acids Res. 2007;35:3367–3374. doi: 10.1093/nar/gkm210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Dohno C, Nakatani K. G-quadruplex formation of entirely hydrophobic DNA in organic solvents. Chem Commun. 2013;49:5501–5503. doi: 10.1039/c3cc42221f. [DOI] [PubMed] [Google Scholar]
- Shiman R, Draper DE. Stabilization of RNA tertiary structure by monovalent cations. J Mol Biol. 2000;302:79–91. doi: 10.1006/jmbi.2000.4031. [DOI] [PubMed] [Google Scholar]
- Smirnov IV, Shafer RH. Electrostatics dominate quadruplex stability. Biopolymers. 2007;85:91–101. doi: 10.1002/bip.20609. [DOI] [PubMed] [Google Scholar]
- Smith D, Schuller V, Engst C, Radler J, Liedl T. Nucleic acid nanostructures for biomedical applications. Nanomedicine. 2013;8:105–121. doi: 10.2217/nnm.12.184. [DOI] [PubMed] [Google Scholar]
- Sorokin VA, Gladchenko GO, Valeev VA, Sysa IV, Petrova LG, Blagoi YP. Effect of salt and organic solvents on DNA thermal stability and structure. J Mol Struct. 1997;1:237–240. doi: 10.1016/S0022-2860(96)09711-6. [DOI] [Google Scholar]
- Spink CH, Chaires JB. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- Srere PA. Protein crystals as a model for mitochondrial matrix proteins. Trends Biochem Sci. 1981;6:4–7. doi: 10.1016/0968-0004(81)90003-7. [DOI] [Google Scholar]
- Stanlis KK, McIntosh JR. Single-strand DNA aptamers as probes for protein localization in cells. J Histochem Cytochem. 2003;51:797–808. doi: 10.1177/002215540305100611. [DOI] [PubMed] [Google Scholar]
- Strulson CA, Yennawar NH, Rambo RP, Bevilacqua PC. Molecular crowding favors reactivity of a human ribozyme under physiological ionic conditions. Biochemistry. 2013;52:8187–8197. doi: 10.1021/bi400816s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strulson CA, Boyer JA, Whitman EE, Bevilacqua PC. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA. 2014;20:331–347. doi: 10.1261/rna.042747.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ZJ, Chen SJ. Ion-mediated nucleic acid helix-helix interactions. Biophys J. 2006;91:518–536. doi: 10.1529/biophysj.106.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki S, Clifford J, Connelly BD, Feig M. Conformational sampling of peptides in cellular environments. Biophys J. 2008;94:747–759. doi: 10.1529/biophysj.107.116236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Vorlíčková M, Bednářová K, Kypr J. Ethanol is a better inducer of DNA guanine tetraplexes than potassium cations. Biopolymers. 2006;82:253–260. doi: 10.1002/bip.20488. [DOI] [PubMed] [Google Scholar]
- Vorlíčková M, Bednářová K, Kejnovská I, Kypr J. Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions. Biopolymers. 2007;86:1–10. doi: 10.1002/bip.20672. [DOI] [PubMed] [Google Scholar]
- Westhof E. Water: an integral part of nucleic acid structure. Annu Rev Biophys Biophys Chem. 1988;17:125–144. doi: 10.1146/annurev.bb.17.060188.001013. [DOI] [PubMed] [Google Scholar]
- Winship PR. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989;17:1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L, et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Young MA, Jayaram B, Beveridge DL. Local dielectric environment of B-DNA in solution: Results from a 14 ns molecular dynamics trajectory. J Phys Chem B. 1998;102:7666–7669. doi: 10.1021/jp9823188. [DOI] [Google Scholar]
- Zhang T, Shang C, Duan R, Hakeem A, Zhang Z, Lou X, Xia F. Polar organic solvents accelerate the rate of DNA strand replacement reaction. Analyst. 2015;140:2023–2028. doi: 10.1039/C4AN02302A. [DOI] [PubMed] [Google Scholar]