Abstract
Intrinsic flexibility is closely related to protein function, and a plethora of important regulatory proteins have been found to be flexible, multi-domain or even intrinsically disordered. On the one hand, understanding such systems depends on how these proteins behave in solution. On the other, small-angle X-ray scattering (SAXS) is a technique that fulfills the requirements to study protein structure and dynamics relatively quickly with few experimental limitations. Molecular chaperones from Hsp70 and Hsp90 families are multi-domain proteins containing flexible and/or disordered regions that play central roles in cellular proteostasis. Here, we review the structure and function of these proteins by SAXS. Our general approach includes the use of SAXS data to determine size and shape parameters, as well as protein shape reconstruction and their validation by using accessory biophysical tools. Some remarkable examples are presented that exemplify the potential of the SAXS technique. Protein structure can be determined in solution even at limiting protein concentrations (for example, human mortalin, a mitochondrial Hsp70 chaperone). The protein organization, flexibility and function (for example, the J-protein co-chaperones), oligomeric status, domain organization, and flexibility (for the Hsp90 chaperone and the Hip and Hep1 co-chaperones) may also be determined. Lastly, the shape, structural conservation, and protein dynamics (for the Hsp90 chaperone and both p23 and Aha1 co-chaperones) may be studied by SAXS. We believe this review will enhance the application of the SAXS technique to the study of the molecular chaperones.
Keywords: SAXS, Multi-domain proteins, Protein dynamics, Molecular chaperones, Mortalin Hsp70
Introduction
The comprehension of biological systems depends on an understanding of highly dynamic macromolecules and processes. Multi-domain, flexible or intrinsically disordered proteins cannot be neglected in the genome (Fukuchi et al. 2011). In fact, 75 % of signaling proteins have more than 30 amino acid-long segments that are predicted to be disordered and/or flexible (Dunker et al. 2008). Flexible regions can work as linkers connecting folded domains in a multi-domain protein, specifying some preferred conformation. They can also work in inter-domain signal transduction (Ma et al. 2011). They serve as binding sites for interacting partners, helping in molecular recognition, working as activation/inhibition modules, exhibiting post-translational modification sites (Berlow et al. 2015; Dunker et al. 2002; Tompa 2005, 2011; Tompa and Csermely 2004; Trudeau et al. 2013; Uversky et al. 2000; Wang et al. 2011), among others. Hence, protein dynamics is closely related to protein function.
A direct consequence of flexible regions is an increase in protein conformational dynamics in solution, resulting in conformational polydispersity (Kikhney and Svergun 2015). Therefore, more than one biophysical technique is usually required to study flexible proteins (Putnam et al. 2007). Indeed, structural studies of such proteins by protein crystallization are limited by several factors including high conformational dynamics, protein degradation, and a low propensity to crystallize. Nuclear magnetic resonance (NMR) spectroscopy is the method of choice to study protein structure and dynamics (Berlow et al. 2015; Lemak et al. 2014), but it also has its own limitations like protein size and concentration, as well as its high cost (Putnam et al. 2007).
Usually, eukaryotic multi-domain proteins are difficult to purify due to their instability and propensity to aggregate and degrade (Berlow et al. 2015). Alternatively, folded domains can be isolated and their structures studied by crystallization. However, despite the invaluable contribution of this approach, information about full-length protein dynamics is needed to understand protein structure-function-dynamic relationships. Thus, full-length protein behavior must be checked by some coherent method once is unknown how disordered regions participate in protein function in vivo.
In this context, small-angle X-ray scattering (SAXS) studies can provide structural information of flexible/disordered proteins, and contribute to knowledge on hydrodynamic behavior and shape/topology of macromolecules (Kikhney and Svergun 2015; Tompa 2011; Rambo and Tainer 2011; Barbosa et al. 2013).
Molecular chaperones from the Hsp70 and Hsp90 families and many of their co-chaperones are multi-domain and/or flexible proteins involved in protein folding and degradation, translocation through membranes, avoidance of aggregation–disaggregation, and other processes (Batista et al. 2015a; Borges and Ramos 2005; da Silva and Borges 2011; Kampinga and Craig 2010; Nillegoda et al. 2015). Therefore, these proteins exert critical control over cellular protein quality (Batista et al. 2015a). They are formed by one or more folded domains connected by flexible linkers, and they can present intrinsically disordered regions.
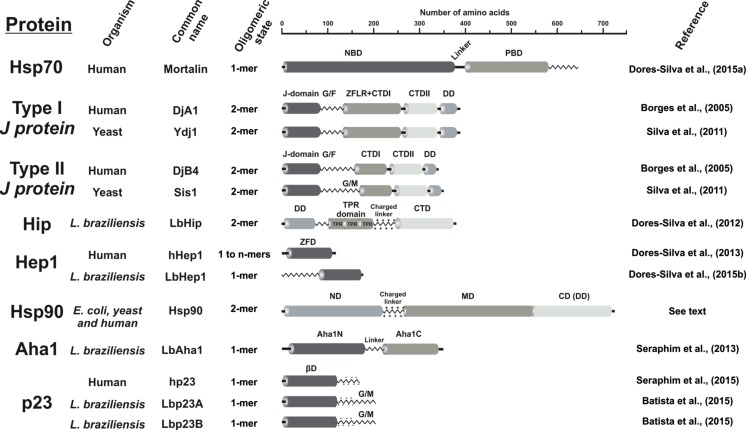
Figure 1 depicts the primary structure organization of the Hsp70 and Hsp90 families and some of their co-chaperones, which are the focus of this review. The presence of autonomous domains/regions as well as flexible/disordered regions is indicated by zigzag lines.
Fig. 1.
Domain architecture of Hsp70 and Hsp90 molecular chaperones families and some of their co-chaperones that form the focus in this review. The abbreviated names of the domains are placed above the domain (see text for identification). The size of the domains and regions corresponds to their peptide lengths. Protein domains are described in the text. Zigzag lines represent flexible regions or intrinsic unfolded sequences
Here, we review the application of SAXS for studying multi-domain molecular chaperones containing flexible linkers and/or disordered regions. Briefly, our approach after SAXS data collection involves: (1) application of ab initio modeling strategies, such as DAMMIN or DAMMIF; (2) validation of the generated model by correlating hydrodynamic properties predicted for the ab initio model with the experimental ones determined by SAXS, analytical ultracentrifugation (AUC) and analytical size exclusion chromatography (aSEC); and (3) use of available three-dimensional structures for the proteins or their domains to infer about protein organization and dynamics, programs such as GENFIT, CRYSOL and EOM, to name a few, were used in the latter approach. In a general way, this approach provides an understanding of the domain organization of the target protein and insights into their mechanisms of action. However, some systems studied by SAXS do not conform to this general strategy, so here we show additional examples of how to use this remarkable technique to study proteins in solution.
SAXS: principles and applications
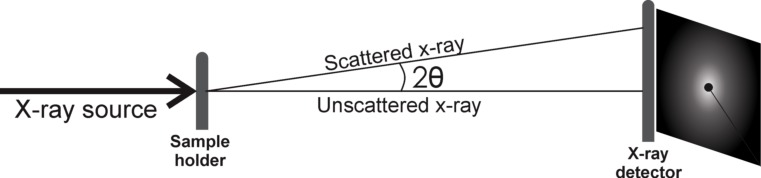
SAXS is an important tool in the analysis of biologically relevant macromolecules like proteins, micelles and liposomes (from surfactants or lipids) and nucleic acids in solution (Barbosa et al. 2013; Blanchet and Svergun 2013). The technique is based on the scattering of X-rays as they interact with the electrons from the scattering particle. Figure 2 illustrates the experimental setup. First, a collimated beam of X-rays can be produced in a conventional X-ray tube, or in a synchrotron ring or even in a free electron laser source (FELS) like the one under construction in Hamburg, Germany. The main difference between the X-ray sources is the beam brilliance.
Fig. 2.
Small-angle X-ray scattering experimental setup
After the X-ray beam exits a monochromator, it passes through two or three slits to collimate the beam. It then hits the sample holder and is scattered to the detector positioned tens of centimeters to a few meters from the sample.
The photons that cross the sample, but do not scatter, are called the direct beam, and the scattering angle is defined as the angle between a certain photon and the direct beam (Fig. 2). The scattering vector is related to the scattering angle, and is used to express the scattered intensity as follows:
| 1 |
where λ is the X-ray wavelength and 2θ the scattering angle (Fig. 2).
SAXS can be applied to a large number of systems and in this review we show the main features of the technique and its application to the study of modular proteins in solution, particularly molecular chaperones.
Guinier’s law
One of the first interesting physical quantities that can be easily extracted from a SAXS curve is the protein Radius of Gyration (Rg). It is related to the way that the protein mass is distributed with respect to its center of mass. Andre Guinier (1911–2000) discovered the relationship between the scattered intensity in the small angle region (q → 0) and the scattering particle Rg (Guinier 1939). Guinier showed that the scattered intensity in this region can be expressed as (Guinier 1939; Guinier and Fournet 1955):
| 2 |
Thus, a plot of Ln (I(q)) versus q 2 should be a straight line and the modulus of the angular coefficient equal to Rg 2/3, whereas the linear coefficient is the forward scattered intensity, I(0), which is related to the protein molecular weight. Such an approximation is an interesting tool for studying proteins in solution, particularly when only the main structural features are of interest.
Kratky plot
The Kratky plot is another interesting tool that can be used to characterize globular or (partially) unfolded proteins (Kratky and Porod 1949; Glatter and Kratky 1982). It was first proposed by Kratky to study polymers in solution (Kratky and Porod 1949), but is now also widely used in the analysis of protein SAXS data. This plot is sensitive to the degree of compactness of a protein, thus it is particularly interesting for the study of folding/unfolding processes and of natively unfolded, intrinsically disordered proteins. The Kratky plot is obtained by simply representing the experimental data as I(q) q 2 versus q where a globular protein is represented as a bell-like shaped Kratky plot. For globular proteins, intensity is proportional to q −4 for large q values. For unfolded proteins, however, the Kratky plot should present a plateau, since scattering intensity is proportional to q − 2 (Kratky and Porod 1949; Barbosa et al. 2013; Glatter and Kratky 1982).
The Kratky plot is indeed a useful tool in the study of proteins in solution, in particular because it does not require any pre-treatment of the experimental data except for the proper subtraction of the buffer from the protein scattering curve. In fact, if there are problems such as an uncorrected buffer subtraction, the Kratky plot should be avoided, since it can lead to a misinterpretation of the SAXS data.
The pair distance distribution function
This is probably the most used tool for the SAXS data analysis of proteins in solution. The pair distance distribution function (PDDF, p(r)), can be obtained from the experimental scattering curve without any geometrical model assumption.
For diluted systems, e.g., where the interference function does not take place over the SAXS curves or where the scattering system is monodisperse, an indirect Fourier transform connects the scattering intensity, I(q), to the PDDF, p(r). The PDDF function is related to the scattering particle inner structure such that it gives information on the protein size (maximum dimension, Dmax) and shape (Glatter and Kratky 1982; Guinier and Fournet 1955; Svergun et al. 1987). The PDDF is of particular interest in the study of molecular chaperones since knowledge of their envelope is often relevant. Furthermore, SAXS allows the application of several different analyses. It is commonly used for the reconstruction of protein shape (ab initio methods), and can be combined with different methods such as molecular dynamics simulation (Avila et al. 2014). We usually use GNOM software to calculate the p(r) curves (Svergun 1992).
Ab initio low resolution shape reconstruction
The protein envelope is a key challenge in the study of molecular chaperones. They undergo conformational changes to perform their cellular activities, and thus the knowledge of protein shape under a variety of environments is of great interest. A few software algorithms can reconstruct the protein envelope using the PDDF. The most common are the DAMMIN (Svergun 1999) and DAMMIF (Franke and Svergun 2009), both developed by the research group of Prof. D. I. Svergun at the EMBL, Hamburg, Germany (Svergun 2015). Briefly, both search for an ensemble of a few thousand spheres (“dummy atoms”) in certain positions in such a way that the theoretical SAXS curve calculated with this configuration is similar to the experimental one.
This procedure has several advantages, but it also has some disadvantages such as the number of possible mathematical solutions that can be achieved without physical meaning. In order to overcome this problem, the software applies penalties to ensure the protein envelope compactness and inter-connectivity (Svergun 1999). Moreover, to ensure that the final envelope is the most probable one, analyses should be repeated several (few dozen) times and averaged using DAMAVER software (Volkov and Svergun 2003) to get the final protein envelope. One should bear in mind that these methods require the monodispersity of the system, since the software looks for a mean structure that describes the scattering profile and the presence of more than one scattering particle in the system will disturb the final result.
High-resolution structures
There are some interesting methodologies in the SAXS data analysis that can be applied to study molecular chaperones in solution if their three-dimensional structures are known. Software is available to calculate theoretical scattering patterns, like CRYSOL (Svergun et al. 1995), SASMOL (Ortore et al. 2009) and GENFIT (Spinozzi et al. 2014). These programs rely on knowledge of the protein structure to calculate the theoretical scattering pattern. One should bear in mind that in order to apply these methods the protein structures in the crystals and in solution must be alike. This must be checked before any further analysis. Moreover, it is also important to check if the available protein three-dimensional structure (the PDB file) is complete (i.e., with all the residues). This is important because the absence of some specific regions in the crystallographic structure (like flexible regions that are generally missing in the PDB file) can affect the theoretical calculation of the SAXS profile.
Ensemble optimization
EOM is generally applied in the study of multi-domains proteins, with partially known three-dimensional structures (Bernado et al. 2007; Tria et al. 2015). This is of particular interest for proteins with compact domains linked by flexible regions, such that they could adopt several possible conformations in solution. EOM creates a pool of random conformations (around 10,000 structures), or conformers using the protein domain structures and the primary structure to account for flexible regions. An ensemble of possible conformations (around 50 structures) is then chosen based on the adjustment of the average theoretical scattering pattern of the ensemble on the experimental curve. Hence, the software chooses the best ensemble to produce the theoretical profile.
Several different methods can be applied to the study of proteins in solution. This review does not aim to cover all the SAXS methods and a more detailed description of these models can be found elsewhere (Barbosa et al. 2013; Hura et al. 2009; Jacques and Trewhella 2010; Koch et al. 2003; Mertens and Svergun 2010; Putnam et al. 2007; Rambo and Tainer 2010, 2011). In the next section, we show some examples based on our experience using SAXS to understand molecular chaperones.
Understanding molecular chaperones by SAXS
Mortalin: human mitochondrial Hsp70
Molecular chaperones belonging to the Hsp70 family perform a central role in proteostasis, defined as the concept of competing and integrated biological pathways within cells that control the biogenesis, folding, trafficking, and degradation of proteins, among others. It plays a pivotal role together with other molecular chaperone families (Batista et al. 2015a; Borges and Ramos 2005; da Silva and Borges 2011). Several activities have been ascribed to Hsp70, which can be executed by multiple induced or non-induced isoforms. Several co-chaperones control its mechanical cycle, different regulatory mechanisms, and organelle-specific isoforms (Arndt et al. 2007; Bukau et al. 2006; Daugaard et al. 2007; Hartl and Hayer-Hartl 2002; Mayer and Bukau 2005; Meimaridou et al. 2009).
Human mortalin is the mitochondrial Hsp70 that acts mainly in importing and folding cytosolic proteins to the mitochondrial matrix (Bohnert et al. 2007; Dolezal et al. 2006; Fan and Young 2011; Mokranjac and Neupert 2009). It is also present in other subcellular locations related to its action in p53 “kidnapping” and cancer (Kaul et al. 2007; Ran et al. 2000; Wadhwa et al. 1993). As observed for all Hsp70 isoforms, mortalin is a modular protein formed by two conserved domains (Fig. 1). The N-terminal nucleotide-binding domain (NBD) has ATPase activity critical for their cycle mechanism, while the C-terminal peptide-binding domain (PBD) is involved in the interaction with client proteins (Batista et al. 2015a; da Silva and Borges 2011). A highly conserved hydrophobic linker works as a bidirectional allosteric signaling component connecting both domains (da Silva and Borges 2011; Kampinga and Craig 2010). Nevertheless, the study of the recombinant human mortalin, as with other mitochondrial Hsp70 proteins, is limited by its tendency to aggregate at higher concentrations (Dores-Silva et al. 2013; Sichting et al. 2005; Zhai et al. 2008). Thus, to investigate the structure and function of this protein, a special co-chaperone and dilute concentrations, which are usually appropriate for some spectroscopic techniques such as circular dichroism and fluorescence, are necessary (see below).
Recently, we reported on structural analyses of the recombinant human mortalin by SAXS and other biophysical tools (Dores-Silva et al. 2015a). The mortalin SAXS curves were obtained at dilute and aggregation-free concentrations (~0.6 mg.mL−1) allowing the determination of structural parameters like MM (Molecular Mass), Rg and Dmax as well as the construction of an ab initio model. MM and Rg were ~70 kDa and 36 Å, respectively, indicating protein was both homogeneous and monomeric. The p(r) curves generated by GNOM software (Svergun 1992) showed mortalin has a Dmax of around 130 Å. Thus it was possible to create a reliable ab initio model for this protein using DAMMIN and DAMAVER (Svergun 1999; Volkov and Svergun 2003). The first program generated 20 independent models that were merged using the latter. The final mortalin ab initio model had an elongated shape that presented hydrodynamic properties predicted by the HydroPro program (de la Torre et al. 2000), and similar to those experimentally observed by AUC and aSEC. Both NBD and PBD crystallographic structures were well accommodated into the mortalin ab initio model required by the above assumptions. Moreover, GENFIT (Spinozzi et al. 2014) and the crystallographic structures of DnaK in open (PDB ID: 4B9Q) and closed (PDB ID: 2KHO) conformations were used to show that apomortalin behaves as an equilibrium of these conformations that shifts to the closed state in the test conditions (Dores-Silva et al. 2015a). Therefore, the SAXS technique is a powerful tool for unveiling the structure of limited amounts of recombinant human mortalin at spectroscopic concentrations (Dores-Silva et al. 2015a).
J-proteins: elongated and flexible proteins
J-proteins form a large group of proteins considering the domain organization and cellular localization (Kampinga and Craig 2010; Mayer and Bukau 2005). They are involved in protein folding, since they interact with client proteins through hydrophobic surfaces and avoid protein aggregation (Borges and Ramos 2005; Cyr and Ramos 2015; Fan et al. 2003; Kampinga and Craig 2010; Mayer and Bukau 2005; Summers et al. 2009). J-proteins are Hsp70 co-chaperones that stimulate ATPase and act as client protein scanning factors (Kampinga and Craig 2010; Karzai and McMacken 1996; Laufen et al. 1999; Rudiger et al. 2001; Summers et al. 2009).
From the structural point of view, J-proteins are characterized by the presence of an α-helical domain (the J-domain), which binds to Hsp70 and stimulates its ATPase activity (Greene et al. 1998; Kampinga and Craig 2010; Szabo et al. 1994). Based on domain organization, the J-protein family can be divided into four types of which the best studied are types I and II (Cyr and Ramos 2015; Kampinga and Craig 2010). They have the J-domain at their N-terminal regions (Fig. 1) followed by a flexible and disordered G/F-rich region (Szyperski et al. 1994), which works as a spacer between the J-domain and the rest of the protein (Borges et al. 2012; Kampinga and Craig 2010; Silva et al. 2011; Summers et al. 2009).
The central and C-terminal regions of type I and II J-proteins are formed by the C-terminal domain (CTD), which can be shared into two similar β-sheet folded subdomains (CTDI and CTDII) (Li et al. 2003; Sha et al. 2000). The CTDI is the main client protein binding site (Li et al. 2003; Sha et al. 2000) and delivers client protein to Hsp70 (Borges et al. 2012; Kampinga and Craig 2010; Rudiger et al. 2001; Silva et al. 2011; Summers et al. 2009). In type I J-proteins, there is a Cys-rich region forming a zinc-finger-like region (ZFLR; Fig. 1) between the G/F-rich region and CTDI. Some type II J-proteins have a Gly/Met (G/M)-rich region instead of a ZFLR. Type III J-proteins only have the conserved J-domain in a part of the protein, not necessarily in the N-terminal region (Mayer and Bukau 2005), and select client proteins for Hsp70 (Kampinga and Craig 2010). Type IV J-proteins have the J-domain with a degenerated HPD motif responsible for interaction with Hsp70 and activation of its ATPase activity (Botha et al. 2007).
Types I and II J-proteins are structurally the most similar and form homodimers through the DD located in the C-terminus. Except for some type II J-proteins, the homodimers are critical for their intrinsic chaperone activity (Cyr and Ramos 2015; Kampinga and Craig 2010; Li et al. 2003; Sha et al. 2000). The main differences lie in the central portion of these proteins (Fig. 1).
Recently, the importance of type I and II J-proteins in metazoa was emphasized by their synergic effect in the disaggregation action of Hsp70 plus Hsp110, a Hsp70 nucleotide exchange factor (Nillegoda et al. 2015). Indeed, these proteins interact with each other through a mechanism that depends on J-domain coupling in the CTD (Nillegoda et al. 2015).
Interestingly, human J-proteins type I (DjA1) and II (DjB4) were studied by SAXS and hydrodynamic approaches, and the results indicate they are modular/multi-domain, highly elongated proteins with different structures (Borges et al. 2005). AUC and SAXS data indicate that DjA1 and DjB4 are both elongated homodimers in solution and long enough (>150 Å) to simultaneously interact with both Hsp70 domains (Borges et al. 2005). In spite of the higher MM, DjA1 had a smaller Dmax than DjB4. Analyzing the ab initio models developed for these proteins and considering the dimerization site lies at the C-terminal region, the J-domains of DjB4 were positioned at opposite ends of the ab initio model, while they were side-by-side in the DjA1 (Borges et al. 2005). Therefore, the central portion of these proteins might be responsible for their divergent structures. Based on additional interaction sites between type I and II J-proteins with Hsp70, the SAXS results suggested different interaction mechanisms for these proteins (Borges et al. 2005).
The yeast J-proteins belonging to types I and II, Ydj1 and Sis1, respectively, diverge in terms of selectivity to client proteins (Cyr 1995; Lu and Cyr 1998), which was related to their central portion, since Sis1 has a G/M-rich region while Ydj1 has a ZFLR. In 2004, Cyr and colleagues (Fan et al. 2004) exchanged the central part of Sis1 and Ydj1, and vice-versa, yielding two chimeras (called SYS and YSY). Briefly, the chimeras exhibited exchangeble specificity for client proteins (Fan et al. 2004). Through SAXS, it was showed that Ydj1 and Sis1 have a similar domain organization to the human ortologues, i.e. Sis1 is more elongated than Ydj1 (Ramos et al. 2008). Indeed, the chimeras had their shapes changed. YSY became similar to Sis1 and was more elongated than SYS, which resembled Ydj1. Therefore, the central portions of these proteins are responsible for their structure and function (Ramos et al. 2008; Summers et al. 2009).
The crystallographic structure of the Sis1 CTD indicated its dimeric structure resembled a bent horseshoe with a hydrophobic cleft capable of binding client proteins. It lies on the surface of the CTDI (Sha et al. 2000). The CTD of the Ydj1 protomer has an L-shape where the ZFLR is the small leg and they might be “looking to each other” in the dimeric structure. Like Sis1, the Ydj1 CTDI has a hydrophobic cleft that binds client proteins (Li et al. 2003). In both cases, the flexible G/F-regions guarantee independence to the J-domain from the CTD and are also close to the hydrophobic cleft and can influence its interaction with client proteins (Johnson and Craig 2001; Sondheimer et al. 2001; Yan and Craig 1999).
In order to study the central portion of Sis1 and Ydj1, deletion mutants of these regions were prepared and purified in the folded state. They revealed elongated homodimers, as evidenced by AUC experiments (Silva et al. 2011). SAXS experiments indicated that all proteins studied are elongated and flexible, probably due to the presence of disordered and flexible regions. Even the truncation of the G/M+CTDI regions from Sis1 did not alter the mutant flexibility as evaluated by the Kratky plot (Silva et al. 2011). NMR experiments confirmed that the G/M region of Sis1 is indeed disordered (Borges et al. 2012). Furthermore, the modular behavior of J-proteins is critical for client protein transfer to Hsp70 (Borges et al. 2012; Silva et al. 2011) allowing the Hsp70 ATPase activation to be executed by the independent J-domain (Summers et al. 2009) in an “anchoring and docking” mechanism (Cyr and Ramos 2015; Hu et al. 2008). The deletion of the central region of Ydj1 resulted in a protein with similar shape and Dmax to that observed in the Sis1 equivalent deletion, confirming this is the region responsible for the Type I J-protein characteristic shape (Silva et al. 2011).
Using SAXS data obtained for yeast J-proteins, truncated mutants as well as crystallographic structures available for Sis1 and Ydj1 domains, different modeling routines and rigid body simulations were performed to understand their organization (Silva et al. 2011). Similar aproaches were used for human J-proteins (Borges et al. 2005), yeast J-proteins and chimeras (Ramos et al. 2008). All cases took into account the dimeric structure of the dimerization site located at the C-terminal region (Borges et al. 2005; Sha et al. 2000). Therefore, the CTD of Sis1 or Ydj1 were positioned at the center of the ab initio model and the J-domain of each protomer were placed at opposite ends. In this position, the J-domain is fully available to stimulate the ATPase activity of the yeast Hsp70 as experimentally observed, despite a reduction of the protein Dmax (Silva et al. 2011).
These results agreed with the literature since the J-domain can stimulate Hsp70 ATPase activity (Laufen et al. 1999; Pierpaoli et al. 1998). Therefore, this activity depends on the availability of the J-domain to interact with Hsp70. Nevertheless, the truncated mutants could not interact with model client proteins nor cooperate with Hsp70 to refold luciferase (Borges et al. 2012; Silva et al. 2011). These results indicated that the main client-protein binding site in CTDI was eliminated, and the reduction in the Sis1 and Ydj1 maximum dimensions, harmed the client protein transfer to Hsp70 (Borges et al. 2012; Silva et al. 2011).
HIP: a highly elongated Hsp70 co-chaperone
Hsc70-Interacting Protein (HIP) is a co-chaperone to enable cognate Hsp70 to stabilize two chaperone molecules bound to the same client protein (da Silva and Borges 2011; Hohfeld et al. 1995). As depicted in Fig. 1, HIP is a dimeric multi-domain protein composed by three conserved regions consisting of an N-terminal DD, a tetratricopeptide repeat (TPR) domain, and a C-terminal region, the latter two being involved in Hsc70 interaction (Lamb et al. 1995).
Using SAXS and other biophysical tools, our research group reported the structure of HIP from the protozoan Leishmania braziliensis (LbHIP) (Dores-Silva et al. 2012). It behaves like a highly elongated dimer in solution with MM and Rg values of 83 kDa and 54 Å, calculated using the Guinier analysis. Moreover, the Kratky plot indicated that LbHIP is slightly flexible in solution. Such behavior is probably due to the presence of several linkers connecting the α-helical folded domains and some disordered regions. The p(r) curve showed that LbHIP is a prolate particle in solution with Dmax of around 200 Å. The protein envelope was consistent with the average ab initio model that displayed hydrodynamic properties (predicted by HydroPro) in agreement with the experimental data determined by AUC and aSEC. Furthermore, the ab initio model allowed the prediction of the relative position of the LbHIP conserved domains.
We concluded that both mammalian HIP and LbHIP have N-terminal DDs located at the center of the ab initio model. The C-terminal region of each protomer was found at opposite ends of this model (Dores-Silva et al. 2012). The TPR domains of each protomer were supposed to be located between the N- and C-terminal regions. In addition, chemical-induced unfolding followed by SAXS and AUC indicated that LbHIP dimer is quite stable and the unfolding transition observed at 2.6 M urea should be related to a part of the protein other than the DD, e.g., the TPR and the C-terminal portion (Dores-Silva et al. 2012). In summary, SAXS studies were critical for understanding the LbHIP structural features, validating it as a scaffold protein for stabilizing two Hsc70 molecules and interacting with the same client protein, since it has two independent binding sites and it is of sufficient size to cooperate with two Hsp70.
Hep1: polydisperse or partially unstructured?
Hsp70-escort protein 1 (Hep1) is an essential protein for mitochondrial biogenesis since it acts by solubilizing and keeping the mtHsp70 protein, like mortalin, in a functional state (Burri et al. 2004; Sichting et al. 2005; Szklarz et al. 2005; Yamamoto et al. 2005). This small protein has a zinc-finger motif formed by four cysteines in a conserved zinc-finger domain (ZFD) almost 100 amino acids long (Fig. 1) (Momose et al. 2007; Vu et al. 2012; Yamamoto et al. 2005).
We reported the structure–function relationship of the human Hep1 (hHep1) and, surprisingly, it was a mixture of at least four species in which the monomer was the main species as shown by AUC data (Dores-Silva et al. 2013). Therefore, information regarding hHep1 shape and envelope were not obtained by SAXS, since it was not possible to evaluate the p(r) function. One should bear in mind that this requires monodispersity of the proteins. It is well known that the Guinier law can be used for calculating apparent MM for the scattering particles (Glatter 1977; Guinier and Fournet 1955). The SAXS data obtained indicated a MM for hHep1 different from that expected for a monomer or any other monodisperse oligomer, confirming that hHep1 behaves as a polydisperse system. Thus, we analyzed the SAXS data by simple visual inspection using the I(q→0)/cprot value (Eq. 2) to obtain information on the protein oligomerization process. The increment of hHep1 concentration led to a gradual increase in I(q→0)/cprot, suggesting that hHep1 oligomerizes in a concentration-dependent manner. In addition, the presence of EDTA modified the increment effect in the I(q→0)/cprot value as a function of hHep1 concentration, suggesting that some hHep1 oligomers were destabilized by EDTA (Dores-Silva et al. 2013). Hence, the SAXS technique provides not only structural aspects of a protein but also information on the system itself, like eventual inter-particle interactions.
LbHep1 is the Hep1 orthologue found in the genome of the L. braziliensis protozoan. It has a conserved ZFD in its C-terminal region, but it has a 70 amino acid N-terminal sequence of unknown function that shares similarities with no known protein. SAXS experiments showed that, unlike hHep1, LbHep1 behaves as a highly elongated monomer in solution but in a rather different way from hHep1, at least at the concentrations tested. SAXS data were collected at several LbHep1 concentrations and the Guinier approximation was used to determine the MM and Rg values. The data revealed that LbHep1 behaves as a monodisperse system with MM and Rg of around 20 kDa and 33 Å, respectively, which agrees with the MM value for the monomeric species (18 kDa) obtained from the primary structure. Furthermore, the p(r) curve suggested that LbHep1 has Dmax of about 130 Å, and had a maximum value of p(r) for r ~30 Å, suggesting a quite elongated shape consistent with hydrodynamic data for LbHep1.
These results were explained by the presence of an unstructured region in the N-terminus (Fig. 1) as observed from circular dichroism data (Dores-Silva et al. 2015b). Therefore, the LbHep1 model consists of a disordered N-terminal region of still unknown function connected to the ZFD domain located in the C-terminal region, which in turn retains the solubility action over L. braziliensis mtHsp70 (Dores-Silva et al. 2015b). LbHep1 has dimensions compatible with that observed for L. braziliensis mtHsp70 (unpublished data) allowing us to hypothesize that LbHep1 might interact with both domains of that protein.
Hsp90: a multi-domain, dimeric and elongated protein
Hsp90 is a 80- to 90-kDa highly-conserved chaperone that plays a central role in the cell physiology of eukaryotes. It participates in protein folding, degradation, protein maturation, signaling, membrane translocation, and others functions (Batista et al. 2015a; Eckl and Richter 2013; Li and Buchner 2013; Silva et al. 2013). A plethora of proteins are known “clients” of Hsp90 including kinases, glucocorticoid hormone receptors, transcription factors, and proteins involved in intracellular traffic, genome maintenance and RNA metabolism (Eckl and Richter 2013; Flynn et al. 2015; Li and Buchner 2013; Mayer and Le Breton 2015; Zhao and Houry 2005). Since the function of Hsp90 is closely related to critical signaling pathways, it emerged as a potential therapeutic target against cancer and other diseases such as those caused by protozoan parasites (Pallavi et al. 2010; Patki and Pawar 2013; Seraphim et al. 2014; Shonhai et al. 2011; Young et al. 2001).
Hsp90 is a homodimer in solution where each monomer has three domains (Fig. 1) (Krukenberg et al. 2011; Nemoto et al. 1995). At the N-terminus, the N-domain (ND) contains an ATP-binding site with a low ATPase activity. It dimerizes transiently with the ND of the adjacent Hsp90 protomer during the Hsp90 functional cycle (Chadli et al. 2000; Obermann et al. 1998; Panaretou et al. 1998; Pullen and Bolon 2011). In Fig. 1, the ND is a site for Hsp90 interaction with some co-chaperones (Ali et al. 2006; Olesen et al. 2015). The M-domain (MD) is involved in ATP hydrolysis and forms a platform for client proteins and co-chaperones interaction (Cunningham et al. 2012; Lotz et al. 2003; Meyer et al. 2003, 2004). The C-domain (CD) is the Hsp90 DD and interacts with client proteins and TPR-based co-chaperones (Nemoto et al. 1995; Onuoha et al. 2008; Worrall et al. 2008).
Hsp90 proteins are remarkably flexible macromolecules undergoing conformational changes related to ATP binding and hydrolysis during its conformational cycle (Krukenberg et al. 2011; Richter et al. 2008). Hsp90 exists in multiple conformational equilibria subtly influenced by ATP/ADP-binding (Mickler et al. 2009; Ratzke et al. 2012). At least three conformational states have been observed for Hsp90 proteins in which apo-Hsp90 fluctuates between open and closed conformations. ATP-binding Hsp90 tends to favor the closed state (Southworth and Agard 2008). The third discrete and compact state corresponds to Hsp90 ADP-bound (Mickler et al. 2009; Southworth and Agard 2008). Hsp90 conformational changes upon nucleotide binding are species-specific (Southworth and Agard 2008).
In the recent years, SAXS has been widely used to understand the structure and dynamics of full-length Hsp90 in solution (Bron et al. 2008; Cunningham et al. 2012; Krukenberg et al. 2008, 2009a, b; Partridge et al. 2014; Southworth and Agard 2008; Street et al. 2010, 2011). Using ab initio modeling strategies, as discussed above, Bron et al (2008) pointed out that Hsp90 has an elongated V-shaped solution structure, resembling the “flying seagull” conformation seen in the open state (Bron et al. 2008). The effects of nucleotides, osmolytes, temperature and pH on the Hsp90 conformation have already been monitored by SAXS (Street et al. 2010; Cunningham et al. 2012; Partridge et al. 2014; Krukenberg et al. 2009a). Due to the multi-domain and flexible nature of the Hsp90, it adopts several conformations in solution and changes in the Hsp90 structure are easily detected by inspecting the p(r) function. The Hsp90 presents a wide p(r) distribution when in its open state, whereas a closed state shows a sharper p(r) function (Cunningham et al. 2012). In spite of these large changes in the p(r), subtle shifts in the conformational equilibrium of the Hsp90 might also be detected and even different Hsp90 conformations might be estimated (Street et al. 2010; Partridge et al. 2014; Krukenberg et al. 2009a). Lastly, SAXS has been used to compare species-specific Hsp90 structures and conformational changes (Krukenberg et al. 2009b), highlighting the use of this technique to understand the features of orthologous Hsp90 proteins.
p23: structured conserved proteins
The p23 co-chaperone is a small protein formed by a folded N-terminal β-sheet domain (βD) and an acidic C-terminal disordered region (Weaver et al. 2000; Weikl et al. 1999). It is involved in a plethora of cellular processes like proteostasis, protein transport, ribosome biogenesis, transcriptional, and genome stability (Echtenkamp and Freeman 2014; Echtenkamp et al. 2011). The p23 interacts with the Hsp90 ND and MD, and the ND dimerization upon ATP binding stabilizes the interaction between the two proteins (Ali et al. 2006; Karagöz et al. 2011; Martinez-Yamout et al. 2006). This interaction allows p23 to regulate Hsp90 function by inhibiting its ATPase activity (McLaughlin et al. 2006); however, the exact role of this inhibition remains controversial. Some authors described p23 as a stabilizer of Hsp90–client-protein interaction (Dittmar et al. 1997; McLaughlin et al. 2006). On the other hand, p23 may be a release factor for Hsp90 (Young and Hartl 2000). p23 βD is involved in preventing aggregation by holding partially or fully unfolded client proteins (Forafonov et al. 2008; Seraphim et al. 2015; Weikl et al. 1999). The function of the C-terminal tail is unclear. Martinez-Yamout and colleagues reported that the C-terminal region of hp23 is disordered and proposed that it works like “fly casting” to increase hp23 solubility when bound to a client protein. Moreover, it could work as a flexible binding site for partners (Martinez-Yamout et al. 2006). A role in p23 regulation in apoptotic cells and telomerase complexes has been suggested for this region (Mollerup and Berchtold 2005; Woo et al. 2009).
Recently, we used SAXS to study the structural properties of p23 proteins from human (hp23) (Seraphim et al. 2015) and L. braziliensis (Lbp23A and Lbp23B) (Batista et al. 2015b). To understand how the C-terminal segment of hp23 affects its structure/function, four hp23 constructs were prepared: full-length hp23; hp231-142; hp231-131; and hp231-117. Circular dichroism spectra of hp23, hp231-142 and hp231-117 were typically of β-sheet-rich proteins, whereas hp231-131 presented some α-helix content. In addition, hp23 and hp231-142 had a higher content of disordered structure compared to hp231-131 and hp231-117 (Seraphim et al. 2015). These results pointed to the disordered C-terminal tail of hp23 and suggested that the region between the amino acids 118–131 could be prone to α-helical structure (Seraphim et al. 2015).
Thermal-induced unfolding experiments revealed that the thermal stability of hp23 decreased as the length of the deletion increases. Thus, we hypothesized that the hp23 C-terminus stabilizes the βD structure. SAXS experiments showed that all hp23 constructs were elongated monomers in solution with hp231-160 presenting higher Rg and Dmax than hp231-142. hp231-131 and hp231-117 were smaller than the two other constructs and had similar dimensions despite their different sequence lengths. This was observed by AUC and aSEC experiments which showed that the C-terminal region is responsible for the elongated shape of the hp23 and suggests 118–131 α-helix region likely interacts with the βD (Seraphim et al. 2015). Thus, we proposed a mechanism where the C-terminal segment works as an auto-inhibitory module (Trudeau et al. 2013) regulating the structure and function of the hp23 (Seraphim et al. 2015).
In contrast to human, L. braziliensis protozoan possesses two p23 proteins named Lbp23A and Lbp23B that share 21 % of identity. Both structures were obtained folded with the same characteristic p23 signatures as their human and yeast counterparts (Batista et al. 2015b). Notwithstanding the Lbp23A and Lbp23B similar secondary structures, they were significantly different. Lbp23A was more stable than Lbp23B as shown by thermally and chemically induced unfolding (Batista et al. 2015b). In addition, Lbp23A had different substrate specificity compared to the Lbp23B in chaperone activity assays. SAXS profiles of Lbp23A and Lbp23B revealed both are elongated monomers in solution with a similar size and dimensions. The p(r) curves also revealed similar overall structures, confirmed by generating ab initio models using DAMMIN (Svergun 1999) and DAMAVER (Volkov and Svergun 2003). Despite the low conservation, slightly different secondary structures, stability and substrate specificity, Lbp23A and Lbp23B had a highly conserved solution structure (Batista et al. 2015b).
The same comparison was done between Lbp23A and Lbp23B comparing human p23 with the L. braziliensis proteins. All three had highly conserved solution structures, despite their differences in sequence and functional features. This strongly suggests that SAXS is valuable for studying the solution structure of evolutionarily distant proteins.
Aha1: a multi-domain flexible protein
Aha1 is a 38-kDa protein formed by two independent regions, the N-terminal (Aha1N) and the C-terminal domains (Aha1C) joined by a flexible disordered linker (Koulov et al. 2010; Meyer et al. 2004; Panaretou et al. 2002; Seraphim et al. 2013). Aha1 stimulates Hsp90 ATPase activity, participating in processes such as protein kinase (Lotz et al. 2003; Sun et al. 2012) and glucocorticoid receptor maturation (Harst et al. 2005). In addition, Aha1 acts on metabolic enzymes, mitochondrial proteins, membrane transporters, vesicle trafficking, and protein degradation-related proteins. However, it is not known if this occurs in an Hsp90-dependent or -independent manner (Sun et al. 2015).
Aha1 stimulates Hsp90 through a mechanism where its Aha1N domain interacts primarily with the Hsp90 MD, followed by an interaction between the Aha1C domain and the Hsp90 ND (Koulov et al. 2010; Lotz et al. 2003; Meyer et al. 2004; Retzlaff et al. 2010). The crystal structure of the yeast Aha1N domain complexed to Hsp90 MD (Meyer et al. 2004) and the NMR structure of the human Aha1C domain (PDB ID: 1X53) have been solved. In spite of this, the overall structure of Aha1 and its behavior in solution remained unknown. In order to fill this gap, we produced and characterized the recombinant L. braziliensis Aha1 (LbAha1) protein (Seraphim et al. 2013). LbAha1 is formed by two domains with different chemical stabilities that behave as a highly elongated monomer in solution, as attested by circular dichroism, fluorescence, aSEC and AUC techniques (Seraphim et al. 2013).
Using SAXS, we confirmed the protein behaves as an elongated monomer in solution with Rg of 36 ± 2 Å and Dmax of 140 ± 10 Å (Seraphim et al. 2013). This dimension was compatible with that observed for the L. braziliensis Hsp90NM construct (unpublished data) allowing LbAha1 to interact with both L. braziliensis Hsp90, ND and MD. The LbAha1 ab initio model was reconstructed using the DAMMIN and DAMAVER programs (Svergun 1999; Volkov and Svergun 2003). The final model emphasized the LbAha1 elongated feature and presented hydrodynamic properties in agreement with the experimental ones (Seraphim et al. 2013).
To go further into the LbAha1 dynamics, we wondered if LbAha1 could adopt a preferential organization in solution. To answer this question, we applied the EOM (Bernado et al. 2007; Tria et al. 2015). This strategy assumes that the scattering curve of a protein is consequence of a mixture of conformational species due to flexibility. Based on available Aha1N and Aha1C domain three-dimensional structures, the EOM generated 10,000 random conformers that could be adopted by LbAha1. From this pool, the EOM selected the best ensemble (composed of 20–50 structures) that had an averaged theoretical SAXS curve similar to the experimental one. It is important to mention that with this method all the structures in the ensemble contribute the same weight to the final theoretical SAXS curve (Seraphim et al. 2013).
Such structures were also analyzed by the Hydropro program and showed average hydrodynamic properties in agreement with the experimental ones. The Rg and Dmax-values observed for the selected LbAha1 conformers displayed the same behavior as the 10,000 initial conformers. We concluded that LbAha1 has a high flexibility in solution with a relative independence between Aha1N and Aha1C domains (Seraphim et al. 2013). Lastly, we found that LbAha1 domains likely possess some distention-contraction behavior once the hydrated volume of LbAha1 was similar to that of a hydrated particle of the same MM, but it is slightly smaller than those estimated for the ab initio model and EOM selected conformers (Seraphim et al. 2013). In summary, our SAXS approach revealed that LbAha1 has dimension, flexibility and domain independence in agreement with the proposed general mechanism for Aha1–Hsp90 interaction.
Conclusions
Comprehension of biological systems relies on a knowledge of how macromolecules behave in aqueous environments. A network of dynamic interactions between dynamic macromolecules, such as proteins, drives processes that are essential for cell developmental and cell physiology. A plethora of proteins are formed by more than one domain connected through unstructured or flexible regions, or they present disordered regions, or they can be intrinsically unstructured. These regions are important sites for regulation and usually mediate protein–protein interactions. Thus, the study of these highly flexible, multi-domain and dynamic proteins is needed to comprehend how their structures are intrinsically linked to their cellular function.
Few techniques allow the study of the behavior of an overall protein in solution without regard for molecular size, dynamics or sample preparation. Here, we presented an overview of an old, but remarkably up-to-date, technique to understand the behavior of macromolecules in solution. This review is the result of our understanding of structures and functions of molecular chaperones of the Hsp70 and Hsp90 families by applying the SAXS technique and other biophysical tools. These proteins belong to a multi-domain class of proteins and they exemplify the potential of SAXS to study proteins in solution, well beyond the generation of ab initio models. SAXS can predict and interpret domain organization in a large and flexible protein. It can also determine macromolecular behavior such as oligomeric state, conformational changes, dynamics, and even structure conservation. We show that the combination of SAXS, NMR and X-ray crystallography provide an invaluable understanding of the structural features that drive protein function (Berlow et al. 2015; Lemak et al. 2014; Putnam et al. 2007). Moreover, a comparison of SAXS-generated models with hydrodynamic measurements (AUC, aSEC) can validate the SAXS theoretical analyses.
Acknowledgments
J.C. Borges thanks FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and CNPq (Conselho Nacional de Pesquisa e Desenvolvimento) for financial support (grants #2007/05001-4, #2011/23110-0, #2012/50161-8, #2014/07206-6 and #471415/2013-8) and a Research Fellowship (#306125/2012-9). L.R.S. Barbosa also thanks FAPESP and CNPq for financial support (#2012/50161-8, #2015/07572-5, #2015/15822-1 and #303048/2012-3). P.R. Dores-Silva thanks FAPESP for grant (#2014/16646-0). We also thank the Brazilian Synchrotron Light Laboratory (LNLS/CNPEM-ABTLuS, Campinas, Brazil) for the use of the SAXS beamline.
Compliance with Ethical Standards
Conflict of interest
Júlio C. Borges declares that he has no conflict of interest.
Thiago V. Seraphim declares that he has no conflict of interest.
Paulo R. Dores-Silva declares that he has no conflict of interest.
Leandro R. S. Barbosa declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Júlio C. Borges, Email: borgesjc@iqsc.usp.br
Leandro R. S. Barbosa, Email: lbarbosa@if.usp.br
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Rogon C, Hohfeld J. To be, or not to be—molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64:2525–2541. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila CL, Torres-Bugeau CM, Barbosa LR, Sales EM, Ouidja MO, Socías SB, Celej MS, Raisman-Vozari R, Papy-Garcia D, Itri R, Chehín RN. Structural characterization of heparin-induced glyceraldehyde-3-phosphate dehydrogenase protofibrils preventing alpha-synuclein oligomeric species toxicity. J Biol Chem. 2014;289:13838–13850. doi: 10.1074/jbc.M113.544288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa LRS, Spinozzi F, Mariani P, Itri R (2013) Small-angle X-ray scattering applied to proteins in solution. In: Ruso JM, Piñeiro Á (eds) Proteins in solution and at interfaces: methods and applications in biotechnology and materials science. Wiley, New York, pp 49–72
- Batista FA, Gava LM, Pinheiro GM, Ramos CH, Borges JC. From conformation to interaction: techniques to explore the Hsp70/ Hsp90 network. Curr Protein Pept Sci. 2015;16:735–753. doi: 10.2174/1389203716666150505225744. [DOI] [PubMed] [Google Scholar]
- Batista FAH, Almeida GS, Seraphim TV, Silva KP, Murta SMF, Barbosa LRS, Borges JC. Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities. FEBS J. 2015;282:388–406. doi: 10.1111/febs.13141. [DOI] [PubMed] [Google Scholar]
- Berlow RB, Dyson HJ, Wright PE. Functional advantages of dynamic protein disorder. FEBS Lett. 2015;589:2433–2440. doi: 10.1016/j.febslet.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle x-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Blanchet CE, Svergun DI. Small-angle x-ray scattering on biological macromolecules and nanocomposites. Annu Rev Phys Chem. 2013;64:37–54. doi: 10.1146/annurev-physchem-040412-110132. [DOI] [PubMed] [Google Scholar]
- Bohnert M, Pfanner N, van der Laan M. A dynamic machinery for import of mitochondrial precursor proteins. FEBS Lett. 2007;581:2802–2810. doi: 10.1016/j.febslet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Borges JC, Ramos CHI. Protein folding assisted by chaperones. Protein Pept Lett. 2005;12:257–261. doi: 10.2174/0929866053587165. [DOI] [PubMed] [Google Scholar]
- Borges JC, Fischer H, Craievich AF, Ramos CH. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J Biol Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- Borges JC, Seraphim TV, Mokry DZ, Almeida FCL, Cyr DM, Ramos CHI. Identification of regions involved in substrate binding and dimer stabilization within the central domains of yeast Hsp40 Sis1. PLoS ONE. 2012;7:e50927. doi: 10.1371/journal.pone.0050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha M, Pesce ER, Blatch GL. The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: regulating chaperone power in the parasite and the host. Int J Biochem Cell Biol. 2007;39:1781–1803. doi: 10.1016/j.biocel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Bron P, Giudice E, Rolland JP, Buey RM, Barbier P, Díaz JF, Peyrot V, Thomas D, Garnier C. Apo-Hsp90 coexists in two open conformational states in solution. Biol Cell. 2008;100:413–425. doi: 10.1042/BC20070149. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Burri L, Vascotto K, Fredersdorf S, Tiedt R, Hall MN, Lithgow T. Zim17, a novel zinc finger protein essential for protein import into mitochondria. J Biol Chem. 2004;279:50243–50249. doi: 10.1074/jbc.M409194200. [DOI] [PubMed] [Google Scholar]
- Chadli A, Bouhouche I, Sullivan W, Stensgard B, McMahon N, Catelli MG, Toft DO. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci U S A. 2000;97:12524–12529. doi: 10.1073/pnas.220430297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CN, Southworth DR, Krukenberg KA, Agard DA. The conserved arginine 380 of Hsp90 is not a catalytic residue, but stabilizes the closed conformation required for ATP hydrolysis. Protein Sci. 2012;21:1162–1171. doi: 10.1002/pro.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Ramos CH (2015) Specification of Hsp70 function by type I and type II Hsp40. In: The networking of chaperones by co-chaperones. Subcellular Biochemistry. Springer, Switzerland, pp 91–102
- da Silva KP, Borges JC. The molecular chaperone Hsp70 family members function by a bidirectional heterotrophic allosteric mechanism. Protein Pept Lett. 2011;18:132–142. doi: 10.2174/092986611794475057. [DOI] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- de la Torre JG, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KD, Demady DR, Stancato LF, Krishna P, Pratt WB. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dores-Silva PR, Silva ER, Gomes FER, Silva KP, Barbosa LRS, Borges JC. Low resolution structural characterization of the Hsp70-interacting protein—hip - from Leishmania braziliensis emphasizes its high asymmetry. Arch Biochem Biophys. 2012;520:88–98. doi: 10.1016/j.abb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Dores-Silva PR, Minari K, Ramos CHI, Barbosa LRS, Borges JC. Structural and stability studies of the human mtHsp70-escort protein 1: an essential mortalin co-chaperone. Int J Biol Macromol. 2013;56:140–148. doi: 10.1016/j.ijbiomac.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Dores-Silva PR, Barbosa LRS, Ramos CHI, Borges JC. Human mitochondrial Hsp70 (mortalin): shedding light on ATPase activity, interaction with adenosine nucleotides, solution structure and domain organization. PLoS ONE. 2015;10:e0117170. doi: 10.1371/journal.pone.0117170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores-Silva PR, Beloti LL, Minari K, Silva SMO, Barbosa LRS, Borges JC. Structural and functional studies of Hsp70-escort protein—Hep1 - of Leishmania braziliensis. Int J Biol Macromol. 2015;79:903–912. doi: 10.1016/j.ijbiomac.2015.05.042. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Echtenkamp FJ, Freeman BC. The molecular chaperones interaction networks in protein folding and degradation. New York: Springer; 2014. Emergence and characterization of the p23 molecular chaperone; pp. 207–232. [Google Scholar]
- Echtenkamp FJ, Zelin E, Oxelmark E, Woo JI, Andrews BJ, Garabedian M, Freeman BC. Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol Cell. 2011;43:229–241. doi: 10.1016/j.molcel.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckl JM, Richter K. Functions of the Hsp90 chaperone system: lifting client proteins to new heights. Int J Biochem Mol Biol. 2013;4:157–165. [PMC free article] [PubMed] [Google Scholar]
- Fan ACY, Young JC. Function of cytosolic chaperones in Tom70-mediated mitochondrial import. Prot Pept Lett. 2011;18:122–131. doi: 10.2174/092986611794475020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:MFROHF>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Mishra P, Bolon DN. Mechanistic asymmetry in Hsp90 dimers. J Mol Biol. 2015;427:2904–2911. doi: 10.1016/j.jmb.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forafonov F, Toogun OA, Grad I, Suslova E, Freeman BC, Picard D. p23/Sba1p protects against Hsp90 inhibitors independently of its intrinsic chaperone activity. Mol Cell Biol. 2008;28:3446–3456. doi: 10.1128/MCB.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi S, Hosoda K, Homma K, Gojobori T, Nishikawa K. Binary classification of protein molecules into intrinsically disordered and ordered segments. BMC Struct Biol. 2011;11:29. doi: 10.1186/1472-6807-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter O. A new method for the evaluation of small-angle scattering data. J Appl Cryst. 1977;10:415–421. doi: 10.1107/S0021889877013879. [DOI] [Google Scholar]
- Glatter O, Kratky O. Small angle x-ray scattering. New York: Academic; 1982. [DOI] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinier A. X-ray diffraction at very low angles: applications to study of ultra-microscopic phenomena. Ann Phys. 1939;12:161–236. [Google Scholar]
- Guinier A, Fournet G. Small angle scattering of X-rays. New York: Wiley; 1955. [Google Scholar]
- Harst A, Lin H, Obermann WM. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem J. 2005;387:789–796. doi: 10.1042/BJ20041283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Protein folding—molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Minami Y, Hartl FU. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- Hu JB, Wu YK, Li JZ, Qian XG, Fu ZQ, Sha BD. The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1. BMC Struct Biol. 2008;8:3. doi: 10.1186/1472-6807-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Trewhella J. Small-angle scattering for structural biology-expanding the frontier while avoiding the pitfalls. Protein Sci. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Craig EA. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J Cell Biol. 2001;152:851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Bio. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rüdiger SG. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci U S A. 2011;108:580–585. doi: 10.1073/pnas.1011867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Deocaris CC, Wadhwa R. Three faces of mortalin: a housekeeper, guardian and killer. Exp Gerontol. 2007;42:263–274. doi: 10.1016/j.exger.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Kikhney AG, Svergun DI. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015;589:2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- Koch MHJ, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/S0033583503003871. [DOI] [PubMed] [Google Scholar]
- Koulov AV, LaPointe P, Lu B, Razvi A, Coppinger J, Dong MQ, Matteson J, Laister R, Arrowsmith C, Yates JR, Balch WE. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21:871–884. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratky O, Porod G. Roentgenuntersuchung Geloester Fadenmolekuele. Rec Trav Chim Pays-Bas. 1949;68:1106–1122. doi: 10.1002/recl.19490681203. [DOI] [Google Scholar]
- Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Southworth DR, Street TO, Agard DA. pH-dependent conformational changes in bacterial Hsp90 reveal a Grp94-like conformation at pH 6 that is highly active in suppression of citrate synthase aggregation. J Mol Biol. 2009;390:278–291. doi: 10.1016/j.jmb.2009.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Bottcher UM, Southworth DR, Agard DA. Grp94, the endoplasmic reticulum Hsp90, has a similar solution conformation to cytosolic Hsp90 in the absence of nucleotide. Protein Sci. 2009;18:1815–1827. doi: 10.1002/pro.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions—to Tpr or not to Tpr. Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/S0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak A, Wu B, Yee A, Houliston S, Lee HW, Gutmanas A, Fang X, Garcia M, Semesi A, Wang YX, Prestegard JH, Arrowsmith CH. Structural characterization of a flexible two-domain protein in solution using small angle X-ray scattering and NMR data. Structure. 2014;22:1862–1874. doi: 10.1016/j.str.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Buchner J. Structure, function and regulation of the Hsp90 machinery. Biomed J. 2013;36:106–117. doi: 10.4103/2319-4170.113230. [DOI] [PubMed] [Google Scholar]
- Li JZ, Oian XG, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Ma B, Tsai CJ, Haliloglu T, Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure. 2011;19:907–917. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Yamout MA, Venkitakrishnan RP, Preece NE, Kroon G, Wright PE, Dyson HJ. Localization of sites of interaction between p23 and Hsp90 in solution. J Biol Chem. 2006;281:14457–14464. doi: 10.1074/jbc.M601759200. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Le Breton L. Hsp90: breaking the symmetry. Mol Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Sobott F, Yao ZP, Zhang W, Nielsen PR, Grossmann JG, Laue ED, Robinson CV, Jackson SE. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- Mertens HDT, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/S1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Liao C, Hu B, Roe SM, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:1402–1410. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim Biophys Acta. 2009;1793:33–41. doi: 10.1016/j.bbamcr.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Mollerup J, Berchtold MW. The co-chaperone p23 is degraded by caspases and the proteasome during apoptosis. FEBS Lett. 2005;579:4187–4192. doi: 10.1016/j.febslet.2005.06.045. [DOI] [PubMed] [Google Scholar]
- Momose T, Ohshima C, Maeda M, Endo T. Structural basis of functional cooperation of Tim15/Zim17 with yeast mitochondrial Hsp70. EMBO Rep. 2007;8:664–670. doi: 10.1038/sj.embor.7400990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Ohara-Nemoto Y, Ota M, Takagi T, Yokoyama K. Mechanism of dimer formation of the 90-kDa heat-shock protein. Eur J Biochem. 1995;233:1–8. doi: 10.1111/j.1432-1033.1995.001_1.x. [DOI] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, Guilbride DL, Wade RC, Morimoto RI, Mayer MP, Bukau B. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen SH, Ingles DJ, Zhu JY, Martin MP, Betzi S, Georg GI, Tash JS, Schönbrunn E. Stability of the human Hsp90-p50Cdc37 chaperone complex against nucleotides and Hsp90 inhibitors, and the influence of phosphorylation by casein kinase 2. Molecules. 2015;20:1643–1660. doi: 10.3390/molecules20011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuoha SC, Couistock ET, Grossmann JG, Jackson SE. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J Mol Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Ortore MG, Spinozzi F, Mariani P, Paciaroni A, Barbosa LR, Amenitsch H, Steinhart M, Ollivier J, Russo D. Combining structure and dynamics: non-denaturing high-pressure effect on lysozyme in solution. J R Soc Interface. 2009;6(Suppl 5):S619–634. doi: 10.1098/rsif.2009.0163.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavi R, Roy N, Nageshan RK, Talukdar P, Pavithra SR, Reddy R, Venketesh S, Kumar R, Gupta AK, Singh RK, Yadav SC, Tatu U. Heat shock protein 90 as a drug target against protozoan infections: biochemical characterization of HSP90 from Plasmodium falciparum and Trypanosoma evansi and evaluation of its inhibitor as a candidate drug. J Biol Chem. 2010;285:37964–37975. doi: 10.1074/jbc.M110.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/S1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Partridge JR, Lavery LA, Elnatan D, Naber N, Cooke R, Agard DA (2014) A novel N-terminal extension in mitochondrial TRAP1 serves as a thermal regulator of chaperone activity. eLife 3 [DOI] [PMC free article] [PubMed]
- Patki JM, Pawar SS. HSP90: chaperone-me-not. Pathol Oncol Res. 2013;19:631–640. doi: 10.1007/s12253-013-9675-4. [DOI] [PubMed] [Google Scholar]
- Pierpaoli EV, Sandmeier E, Schonfeld HJ, Christen P. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J Biol Chem. 1998;273:6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- Pullen L, Bolon DN. Enforced N-domain proximity stimulates Hsp90 ATPase activity and is compatible with function in vivo. J Biol Chem. 2011;286:11091–11098. doi: 10.1074/jbc.M111.223131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95(8):559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos CHI, Oliveira CLP, Fan CY, Torriani IL, Cyr DM. Conserved central domains control the quaternary structure of type I and type II Hsp40 molecular chaperones. J Mol Biol. 2008;383:155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran QT, Wadhwa R, Kawai R, Kaul SC, Sifers RN, Bick RJ, Smith JR, Pereira-Smith OM. Extramitochondrial localization of mortalin/mthsp70/PBP74/GRP75. Biochem Biophys Res Comm. 2000;275:174–179. doi: 10.1006/bbrc.2000.3237. [DOI] [PubMed] [Google Scholar]
- Ratzke C, Nguyen MN, Mayer MP, Hugel T. From a ratchet mechanism to random fluctuations evolution of Hsp90’s mechanochemical cycle. J Mol Biol. 2012;423:462–471. doi: 10.1016/j.jmb.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, Richter K, Buchner J. Asymmetric activation of the Hsp90 dimer by its cochaperone Aha1. Mol Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Richter K, Soroka J, Skalniak L, Leskovar A, Hessling M, Reinstein J, Buchner J. Conserved conformational changes in the ATPase cycle of human Hsp90. J Biol Chem. 2008;283:17757–17765. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphim TV, Alves MM, Silva IM, Gomes FE, Silva KP, Murta SM, Barbosa LR, Borges JC. Low resolution structural studies indicate that the activator of Hsp90 ATPase 1 (Aha1) of Leishmania braziliensis has an elongated shape which allows its interaction with both N- and M-domains of Hsp90. PLoS ONE. 2013;8:e66822. doi: 10.1371/journal.pone.0066822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphim TV, Ramos CHI, Borges JC. The molecular chaperones interaction networks in protein folding and degradation. New York: Springer; 2014. The interaction networks of Hsp70 and Hsp90 in the Plasmodium and Leishmania parasites; pp. 445–481. [Google Scholar]
- Seraphim TV, Gava LM, Mokry DZ, Cagliari TC, Barbosa LRS, Ramos CHI, Borges JC. The C-terminal region of the human p23 chaperone modulates its structure and function. Arch Biochem Biophys. 2015;565:57–67. doi: 10.1016/j.abb.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Sha BD, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure. 2000;8:799–807. doi: 10.1016/S0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Shonhai A, Maier AG, Przyborski JM, Blatch GL. Intracellular protozoan parasites of humans: the role of molecular chaperones in development and pathogenesis. Protein Pept Lett. 2011;18:143–157. doi: 10.2174/092986611794475002. [DOI] [PubMed] [Google Scholar]
- Sichting M, Mokranjac D, Azem A, Neupert W, Hell K. Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 2005;24:1046–1056. doi: 10.1038/sj.emboj.7600580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Borges JC, Cyr DM, Ramos CHI, Torriani IL. Central domain deletions affect the SAXS solution structure and function of Yeast Hsp40 proteins Sis1 and Ydj1. BMC Struct Biol. 2011;11:40. doi: 10.1186/1472-6807-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva KP, Seraphim TV, Borges JC. Structural and functional studies of Leishmania braziliensis Hsp90. Biochim Biophys Acta. 2013;1834:351–361. doi: 10.1016/j.bbapap.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ(+)] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth DR, Agard DA (2008) Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell 32(5):631–640 [DOI] [PMC free article] [PubMed]
- Spinozzi F, Ferrero C, Ortore MG, De Maria Antolinos A, Mariani P. GENFIT: software for the analysis of small-angle X-ray and neutron scattering data of macromolecules in solution. J Appl Cryst. 2014;47:1132–1139. doi: 10.1107/S1600576714005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Krukenberg KA, Rosgen J, Bolen DW, Agard DA. Osmolyte-induced conformational changes in the Hsp90 molecular chaperone. Protein Sci. 2010;19:57–65. doi: 10.1002/pro.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, Douglas PM, Ramos CHI, Cyr DM. Polypeptide transfer from Hsp40 to Hsp70 molecular chaperones. Trends Biochem Sci. 2009;34:230–233. doi: 10.1016/j.tibs.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Prince T, Manjarrez JR, Scroggins BT, Matts RL. Characterization of the interaction of Aha1 with components of the Hsp90 chaperone machine and client proteins. Biochim Biophys Acta. 2012;1823:1092–1101. doi: 10.1016/j.bbamcr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Sun L, Hartson SD, Matts RL. Identification of proteins associated with Aha1 in HeLa cells by quantitative proteomics. Biochim Biophys Acta. 2015;1854:365–380. doi: 10.1016/j.bbapap.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI (2015) Svergun’s group web page. http://www.embl-hamburg.de/research/unit/svergun/index.html
- Svergun DI, Feĭgin LA, Taylor GW. Structure analysis by small-angle X-ray and neutron scattering. New York: Plenum; 1987. [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. doi: 10.1107/S0021889895007047. [DOI] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system—DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarz LKS, Guiard B, Rissler M, Wiedemann N, Kozjak V, van der Laan M, Lohaus C, Marcus K, Meyer HE, Chacinska A, Pfanner N, Meisinger C. Inactivation of the mitochondrial heat shock protein Zim17 leads to aggregation of matrix Hsp70s followed by plelotropic effects on morphology and protein biogenesis. J Mol Biol. 2005;351:206–218. doi: 10.1016/j.jmb.2005.05.068. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wuthrich K. NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues-2-108) containing the highly conserved J domain. Proc Natl Acad Sci U S A. 1994;91:11343–11347. doi: 10.1073/pnas.91.24.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21:419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Tria G, Mertens HDT, Kachala M, Svergun DI. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau T, Nassar R, Cumberworth A, Wong ETC, Woollard G, Gsponer J. Structure and intrinsic disorder in protein autoinhibition. Structure. 2013;21:332–341. doi: 10.1016/j.str.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu MT, Zhai P, Lee J, Guerra C, Liu S, Gustin MC, Silberg JJ. The DNLZ/HEP zinc-binding subdomain is critical for regulation of the mitochondrial chaperone HSPA9. Protein Sci. 2012;21:258–267. doi: 10.1002/pro.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Mitsui Y, Sugimoto Y. Differential subcellular-distribution of mortalin in mortal and immortal mouse and human fibroblasts. Exp Cell Res. 1993;207:442–448. doi: 10.1006/excr.1993.1213. [DOI] [PubMed] [Google Scholar]
- Wang ML, Kurland CG, Caetano-Anolles G. Reductive evolution of proteomes and protein structures. Proc Natl Acad Sci U S A. 2011;108:11954–11958. doi: 10.1073/pnas.1017361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AJ, Sullivan WP, Felts SJ, Owen BAL, Toft DO. Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J Biol Chem. 2000;275:23045–23052. doi: 10.1074/jbc.M003410200. [DOI] [PubMed] [Google Scholar]
- Weikl T, Abelmann K, Buchner J. An unstructured C-terminal region of the Hsp90 co-chaperone p23 is important for its chaperone function. J Mol Biol. 1999;293:685–691. doi: 10.1006/jmbi.1999.3172. [DOI] [PubMed] [Google Scholar]
- Woo SH, An S, Lee HC, Jin HO, Seo SK, Yoo DH, Lee KH, Rhee CH, Choi EJ, Hong SI, Park IC. A truncated form of p23 down-regulates telomerase activity via disruption of Hsp90 function. J Biol Chem. 2009;284:30871–30880. doi: 10.1074/jbc.M109.052720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall LJ, Wear MA, Page AP, Walkinshaw MD. Cloning, purification and characterization of the Caenorhabditis elegans small glutamine-rich tetratricopeptide repeat-containing protein. Biochim Biophys Acta. 2008;1784:496–503. doi: 10.1016/j.bbapap.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Momose T, Yatsukawa Y, Ohshima C, Ishikawa D, Sato T, Tamura Y, Ohwa Y, Endo T. Identification of a novel member of yeast mitochondrial Hsp70-associated motor and chaperone proteins that facilitates protein translocation across the inner membrane. FEBS Lett. 2005;579:507–511. doi: 10.1016/j.febslet.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/MCB.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai P, Stanworth C, Liu S, Silberg JJ. The human escort protein hep binds to the ATPase domain of mitochondrial Hsp70 and regulates ATP hydrolysis. J Biol Chem. 2008;283:26098–26106. doi: 10.1074/jbc.M803475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Houry WA. Hsp90: a chaperone for protein folding and gene regulation. Biochem Cell Biol. 2005;83:703–710. doi: 10.1139/o05-158. [DOI] [PubMed] [Google Scholar]