Abstract
Bacteria organize DNA into self-adherent conglomerates called nucleoids that are replicated, transcribed, and partitioned within the cytoplasm during growth and cell division. Three classes of proteins help condense nucleoids: (1) DNA gyrase generates diffusible negative supercoils that help compact DNA into a dynamic interwound and multiply branched structure; (2) RNA polymerase and abundant small basic nucleoid-associated proteins (NAPs) create constrained supercoils by binding, bending, and forming cooperative protein–DNA complexes; (3) a multi-protein DNA condensin organizes chromosome structure to assist sister chromosome segregation after replication. Most bacteria have four topoisomerases that participate in DNA dynamics during replication and transcription. Gyrase and topoisomerase I (Topo I) are intimately involved in transcription; Topo III and Topo IV play critical roles in decatenating and unknotting DNA during and immediately after replication. RNA polymerase generates positive (+) supercoils downstream and negative (−) supercoils upstream of highly transcribed operons. Supercoil levels vary under fast versus slow growth conditions, but what surprises many investigators is that it also varies significantly between different bacterial species. The MukFEB condensin is dispensable in the high supercoil density (σ) organism Escherichia coli but is essential in Salmonella spp. which has 15 % fewer supercoils. These observations raise two questions: (1) How do different species regulate supercoil density? (2) Why do closely related species evolve different optimal supercoil levels? Control of supercoil density in E. coli and Salmonella is largely determined by differences encoded within the gyrase subunits. Supercoil differences may arise to minimalize toxicity of mobile DNA elements in the genome.
Keywords: Gyrase, Topoisomerase, Nucleoid-associated proteins, Supercoil density, MukFEB condensin
Introduction
Progress in understanding the structure/function mechanics of the bacterial chromosome greatly improved with development of techniques that allow real time visualization of bacterial nucleoids inside living cells and the ability to follow specific chromosomal loci using fluorescent labels (Kleckner et al. 2014; Wang et al. 2008). Nucleoids are self-adherent filaments stochastically organized locally into approximately 10-kb domains in growing cells (Cunha et al. 2005; Espeli et al. 2008; Hadizadeh Yazdi et al. 2012; Postow et al. 2004; Stein et al. 2005; Wiggins et al. 2010). The 4-Mb circular Escherichia coli chromosome is the best studied nucleoid in prokaryotic biology. In this review I focus on DNA condensation that exploits negative supercoiling to produce a highly interwound plectonemic network of DNA. This network exhibits two forms of movement that will juxtapose two points along the network over distances of 10 to 100 kb, depending on cell growth conditions. These movements are slithering, which is a reptilian-like sliding of the interwound network, and branching, which is a rotational movement that causes segments to extrude and be resorbed along a fiber axis (Higgins et al. 1996) (Fig. 1). Supercoil diffusion is critical for many biochemical reactions of the chromosome, including site-specific recombination, transcription, transposition of mobile elements, and initiation of DNA replication (Higgins and Vologodskii 2015). Yet this DNA movement is not discernable in real time, even with the highest resolution techniques currently available. I discuss the control of supercoil density in two closely related species, E. coli and Salmonella Typhimurium. These organisms have essentially identical tool kits of proteins that generate diffusible and constrained supercoiling, but they maintain significantly different values of supercoil density (σ = − 0.070 and – 0.060, respectively). Multiple studies show that negative supercoils present in E. coli are partitioned between a diffusible supercoiled state that moves rapidly throughout thousands of base pairs of chromosomal DNA and constrained supercoils that behave locally in a manner reminiscent of nucleosomes.
Fig. 1.
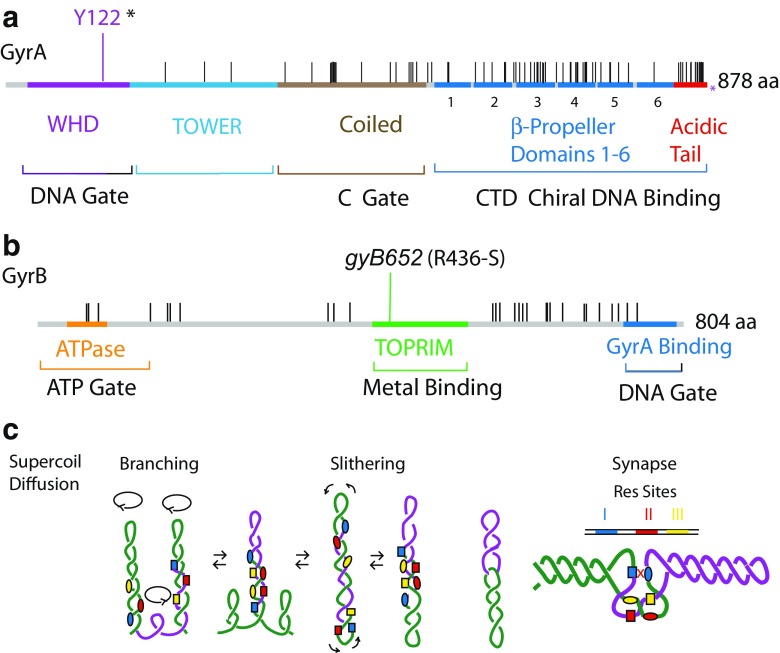
Structure and sequence alignment of DNA gyrase A and B genes (GyrA, GyrB) from Salmonella and Escherichia coli. a For GyrA the catalytic tyrosine (Y122) is shown in purple. There are 77 positions with amino acid substitutions between the species (black lines). The reference sequence is Salmonella GyrA, which is 3 amino acids longer than E. coli GyrA and terminates at 875 amino acids. b GyrB is more highly conserved than GyrA, with the two proteins differing at only 28 positions in this 804-amino acid protein. Domains for GyrA include the DNA Gate, the C Gate, and the CTD Chiral DNA Binding elements. For GyrB domains include the ATP Gate, Metal Binding site, and GyrA Gate interaction region. c. Long-range diffusion mechanisms of negative supercoiling is illustrated for a segment of DNA that includes two directly repeated recombination sites for the γδ resolvase (Res). One Res dimer binds to three DNA sites, numbered I (blue), II (red), and III (yellow). Branch extrusion (c, left) or reptilian slithering (c, center) move the two Res sites into a three-node tangle. (c, far right). The products of recombination are two catenated circles (c right). In vivo-catenated links are separated by topoisomerase IV (Topo IV)
Constrained supercoils
The E. coli chromosome contains about 30,000 supercoils (σ = − 0.07). Approximately half of these supercoils can diffuse freely (σD) and the other half are constrained (σC) so that DNA writhe persists when a chromosomal region is relaxed, resulting in the relationship σ = σD + σC (Higgins and Vologodskii 2015). RNA polymerase (RNAP) is one protein that constrains supercoils. The enzyme has five proteins (α2 ββ’ω) with a molecular mass of about 400 kDa. About 3,000 molecules of RNAP are present in E. coli cells grown exponentially in rich medium, of which two-thirds are actively engaged in transcription. One consequence of transcription is that each polymerase unwinds a short segment of the DNA template that moves with the enzyme. This creates a linking number change (ΔLk) of −1.7 supercoils per RNA polymerase (Gamper and Hearst 1982) so that RNAP accounts for about 3400 supercoils, or roughly 10 % of σC. A group of abundant small basic proteins that are often called histone-like or nucleic-associated proteins (NAPs) bind and bend DNA; these proteins are thought to account for the rest of constrained supercoiling. The leading member is HU protein, a hetero-dimer composed of HupA and HupB protomers in E. coli (Dillon and Dorman 2010). HU can produce supercoiled DNA when incubated with a relaxed substrate and an enzyme such as calf thymus topoisomerase I (Topo I), and it accounts for 6000 supercoils or roughly 40 % of the constrained DNA supercoils. About 50 % of σC is presumed to associate with the NAPs IHF, H-NS, STPA, FIS, and DPS (Johnson et al. 2005; Pul and Wagner 2010).
Diffusible supercoils
DNA gyrase is primarily responsible for maintaining approximately 15,000 diffusible supercoils in E. coli. A tetramer of two proteins, GyrA and GyrB (Fig. 1), gyrase was the first type II topoisomerase discovered. The enzyme introduces two (-) supercoils in each enzymatic cycle (Fig. 2). By first breaking both strands of DNA and then forming a transient covalent phospho-tyrosine (Y122) complex with both of the DNA strands, the enzyme passes one double-strand segment through the gap and reseals the bond before releasing the DNA product. Gyrase is a factor in many DNA transactions and a fundamental force in compacting chromosomal DNA; consequently, it has an impact on much of the chemistry of DNA metabolism (Higgins et al. 2005). The structure and sequence of the gyrase subunits are highly conserved. After millions of years of evolutionary divergence separating E. coli and Salmonella there are only 28 amino acid changes in GyrB and 77 amino acid differences in GyrA (Fig. 1). All four subunits have been purified and cross-complement each other in in vitro supercoiling reactions (Pang et al. 2005). Nonetheless, Salmonella has a supercoil density that is 15 % lower than that of E. coli (Champion and Higgins 2007; Higgins et al. 2005), and E. coli is not viable at the wild-type (WT) Salmonella supercoil density. The mystery of how these differences arose and why they have been stably maintained is discussed in a following section.
Fig. 2.
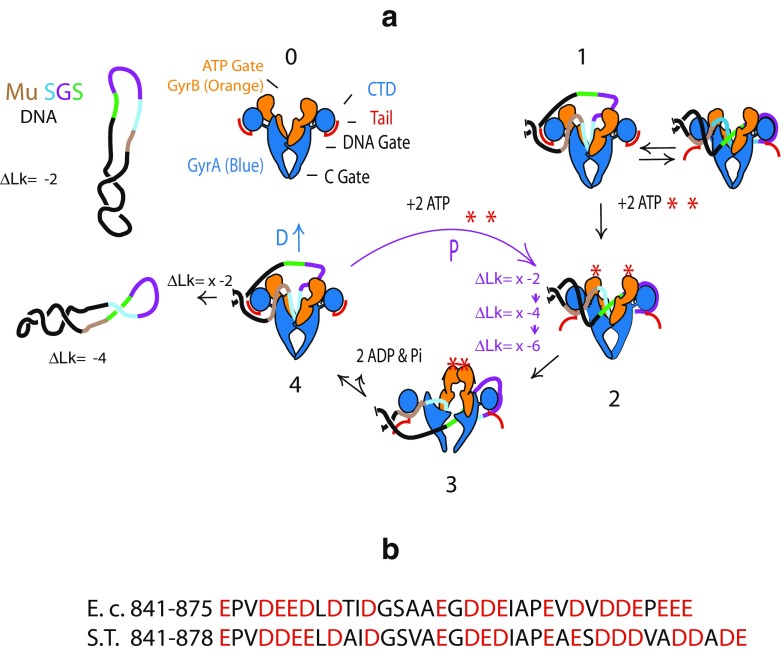
a Model for supercoil density control based a branch point leading to DNA dissociation (D) or processive reactions (P) where the enzyme remains bound to the same site. ΔLk Linking number change, Pi inorganic phosphorus, Mu SGS strong DNA gyrase cleavage site at the center of the Mu genome, CTD C-terminal domain. b Comparison of the C-terminal acidic tails of E. coli (E.c.) and Salmonella Typhimurium (S.T.) Acidic tail residues are shown in red
Gyrase mutations with different phenotypes in two species
In biochemistry and molecular genetics it is often assumed that metabolic pathways and enzymes that catalyze the critical reactions all work the same way. If you understand the mechanism of an essential E. coli enzyme (DNA gyrase or RNA polymerase for example), you expect the phenotype of specific mutations to apply in most types of bacteria. This is the assumption in the emerging fields of “metabolomics” and systems biology. However, the assumption is often wrong. Zenhua Pang studied a temperature-sensitive (TS) mutation of gyrB in Salmonella (Pang et al. 2005). The gyrB-652 mutation changes R436-S in the TOWPRIM domain of GyrB (Fig. 1), and it makes cells TS for growth at 42 °C and resistant to low (5 μg/ml) levels of nalidixic acid. Normally, nalidixic acid resistance mutations map near the catalytic tyrosine present in the gyrA gene. To study the catalytic mechanism of the mutant, Pang cloned, expressed, and purified GyrA, GyrB, and GyrB652 proteins to assemble and study Salmonella WT gyrase and GyrB652 enzyme activity in vitro (Pang et al. 2005). WT gyrase and GyrB652 were both proficient in supercoiling relaxed circular DNA substrates at 30 °C. Surprisingly, the GyrB652 enzyme supercoiled DNA faster at 42 °C than at 30 °C. GyrB652 supercoiling reactions carried out at 42 °C worked twice as fast as those at 30 °C. However, GyrB652 gyrase had a slower catalytic rate compared to the WT gyrase at all temperatures tested, and it is this slow enzyme mechanism in Salmonella that causes problems at high temperature.
GyrB652 was the first topoisomerase exhibiting growth rate toxicity (Champion and Higgins 2007). To determine whether the R436-S GyrB protein would also cause TS growth in E. coli, Champion and Higgins (2007) used the lambda RED “recombineering” (recombination-mediated genetic engineering) method (Datta et al. 2006) to introduce this mutation into the E. coli gyrB chromosomal gene and found that this single base pair change was lethal in E. coli at all growth temperatures. This experiment provided the first example of a gyrase mutation that caused strikingly different phenotypes in two closely related species of bacteria. A subsequent study carried out by the Higgins’ group showed that many TS mutations in GyrA and Topo IV decrease catalytic efficiency rather than causing protein dysfunction at the high temperature (Rovinskiy et al. 2012). In all of these cases, chromosomal supercoiling losses are observed even at the permissive growth temperatures.
Species-specific phenotypes of other homologous genes
To better understand the GyrB phenotype, Keith Champion studied other phenotypes linked to negative supercoiling (Champion and Higgins 2007). In this study, he first measured the supercoil density of pUC18 plasmid DNA isolated from Salmonella and found to be 15 % lower than that in the same plasmid isolated from E. coli. Experiments with supercoil-dependent Z-DNA-forming plasmids confirmed that E. coli creates a significantly higher DNA torsional strain than does Salmonella. Second, he introduced either a Salmonella GyrB652 protein or a WT Salmonella GyrB protein into WT E. coli and observed that both proteins were toxic, even at very low expression levels, suggesting that WT and mutant gyrase subunits from Salmonella cause problems in vivo for E. coli. Third, species differences in supercoil density changed many phenotypes linked to replication and chromosome segregation. For example, a null mutation in the mukB subunit of condensin is viable in E. coli, but lethal in Salmonella. The explanation is that at a low supercoil set point, the Salmonella chromosome relies more heavily on condensin to compact and organize the chromosome than does E. coli with higher supercoil compaction forces (Holmes and Cozzarelli 2000; Sawitzke and Austin 2000). Fourth, strains with null mutations in seqA were healthy in Salmonella, whereas E. coli seqA deletions were sick and exhibited RecA-induced SOS phenotypes. The high supercoil density of E. coli promotes premature initiation at the origin of replication (OriC) because a replisome can auto-assemble at unpaired regions near the OriC. SeqA suppresses this reaction by binding to hemi-methylated sites within the OriC and sequestering it from interacting with the replication initiation protein DnaA for many minutes. At the lower supercoil density of Salmonella, there is no over-initiation and SeqA sequestration is not needed.
Life on a flat chromosome
Many reports dealing with gyrase and supercoiling start with a litany of the cellular functions reported to be intimately connected to gyrase-driven negative supercoiling. The list includes initiation of replication, recombination, transcription, transposition, homologous and site-specific recombination, among others. However, bacterial cells can survive, grow, and carry out well-organized cell divisions even while their chromosomes have lost all detectable diffusible negative supercoils (Higgins 2014; Rovinskiy et al. 2012). Such growth is not possible for E. coli, which is the paradigm case, so this result in Salmonella is not widely understood or appreciated, although it may be more characteristic of most bacterial species.
Discovery of viable flat chromosomes started with experiments by Booker, who set out to study DNA topology of the highly transcribed rrnG operon of Salmonella (Booker et al. 2010). Booker introduced supercoil sensors upstream and downstream of the 5-kb rrnG operon and measured supercoil-dependent resolution efficiency. The results revealed that in Salmonella with a WT complement of genes for DNA topology and transcription, there was a significant excess of negative supercoiling upstream from the promoter, and a supercoil deficit downstream from the transcription terminator. These findings confirmed the Liu and Wang hypothesis of twin domains of opposite handed supercoiling up- and downstream of a transcribing RNA polymerase (Liu and Wang 1987) and proved that twin domain supercoiling exists even in WT cells. Booker extended the experiment to test how mutations in TopA (E. coli strains with a Topo I enzyme of low activity) and TS mutations in gyrase altered the result. TopA mutants significantly increased (−) supercoiling upstream of the promoter, thereby confirming its role in preventing hyper-supercoiling on the chromosome (Drolet et al. 2003). A TS GyrB mutant growing at permissive temperature (30 °C) caused a significant loss of (−) supercoiling downstream of the terminator. However, the GyrB652 mutation caused a complete loss of negative supercoiling in a 9-kb domain downstream from the rrnG terminator, even though this strain grows at the permissive temperature (30 °C).
The complete loss of supercoiling downstream of rrnG in strains with GyrB652 raised an intriguing possibility. A flat chromosome lacking nearly all negative supercoiling might be produced in cells with a sluggish GyrB subunit. This hypothesis was tested and confirmed in several ways (Rovinskiy et al. 2012). First, supercoil sensors at eight positions were evaluated for chromosome supercoiling. Analysis of these positions revealed that 95 % of all diffusible chromosomal supercoiling disappeared in GyrB652 cells growing exponentially at 30 °C. Second, if transcription can run the chromosome flat, can supercoiling be restored by blocking transcription?. Rifampicin was added for 10 min after cells had been induced for resolvase expression, and this treatment restored supercoil density to 70 % of the WT level. Third, the possibility of a slow GyrB652 influencing the normal rate of transcription elongation was tested by measuring the rates of RNA polymerase elongation in the Lac operon at the same positions used to analyze recombination. The average RNA polymerase elongation rate fell from 50 nucleotides/s in WT cells growing at 30 °C to roughly half that in the GyrB652 mutants. This result reaffirms the close linkage between transcription and gyrase catalytic rates. Fourth, if rates of transcription elongation and gyrase supercoiling are tuned to match in WT cells, what happens to supercoiling if one slows down RNAP? When an RNAP mutant with a slow elongation rate was introduced into Salmonella cells, the global supercoil density of the chromosome increased, confirming again the intimate connection between gyrase catalytic activity and the transcription elongation properties of RNAP. This result confirms that RNAP is critical for establishing the mean supercoil density of chromosomes in collaboration with gyrase and Topo I.
What controls supercoil density?
To examine critical elements that participate in regulating supercoil levels, Nikolay Rovinskiy made transgenic swaps of GyrA between E. coli and Salmonella. As Champion had found earlier for GyrB, GyrA proteins caused supercoiling disruptions as single-copy transgenes. For his thesis Rovinskiy constructed chimeras to see if one GyrA region might control proper supercoiling. He found that in both cases when a transgenic GyrA from the wrong host was fused to the correct species GyrA sequence containing the last two pinwheel elements and acidic tail, proper supercoiling was restored. One model proposed for species-specific supercoiling involves the pinwheel elements of the C-terminal domain (CTD) and the C-terminal 35–38 acidic tail (Fig. 1a red tail on GyrA in Fig. 2b). Purified E. coli and Salmonella GyrA dimers do not bind to DNA, but GyrA dimers lacking the tail do bind to DNA because the positively charged residues of the CTD inhibit DNA binding. As shown in Fig. 2a (Stage 0), gyrase forms two chambers, with the top chamber open and the lower C chamber closed. When DNA enters the upper chamber (Stage 1), it is drawn to the floor, which bends DNA into a V shape caused by two 75° bends on opposing sides of the DNA cleavage site (Wendorff et al. 2012). DNA bending orients the DNA arms so that they compete with GyrA CTD for DNA binding and formation of a chiral (+) loop on one pinwheel element. Looping of the DNA over the pinwheels places the DNA (green) transfer segment over the DNA cleavage site. In solution, gyrase–DNA complexes are stable with a half-life of > > 24 h. (Higgins and Cozzarelli 1982). DNase footprinting (Oram et al. 2006) and topology mapping (Kirkegaard and Wang 1981) indicate that in solution the equilibrium strongly favors a (+) wrapped conformation. However, stretching tension, which is applied in both single molecule rotorbead studies (Basu et al. 2012) and magnetic tweezers experiments (Fernandez-Sierra et al. 2015), exerts tension that appears to shift the equilibrium to favor the unwrapped condition. In Stage 2, binding of ATP to both GyrB subunits stabilizes the + chiral loop (Basu et al. 2012) and the enzyme closes the upper chamber by dimerization of ATP–GyrB subunits (Stage 3). At Stage 3, cleaved complexes are efficiently formed with both GyrA subunits attached to opposing DNA strands via a phospho-tyrosine linkage to Y122. In the cleaved state, solid body rotation of the GyrA Tower and GyrB TOPRIM domains opens both the DNA gate for passage of the transfer DNA segment (green) down to the lower chamber and the C gate at the bottom. ATP hydrolysis promotes re-ligation of the covalent intermediate complex and opening of the ATP gate, which places the enzyme in position to follow one of two tracks—release of DNA and return to Stage 0, or retain the bend and proceed to Stage 2 for another rapid reaction. In rotor bead experiments, processive supercoil cycles rapidly introduce >100 supercoils in continuous bursts (Basu et al. 2012).
We have proposed that the Salmonella/E. coli supercoil density difference is controlled by the energetics of the branch point of distributive versus processive reaction. The E. coli tail has 35 residues with 19 acidic side chains, whereas Salmonella’s tail has 38 residues and 21 acidic amino acids. This difference means that the Salmonella tail is a better DNA competitor than E. coli, and consequently, a less processive enzyme. When bound to the identical sites, E. coli gyrase is predicted to be more processive and to achieve high supercoiled levels faster than Salmonella gyrase.
Critical partners with gyrase in chromosome supercoiling reactions are high-affinity binding sites that promote processive bursts and contribute to the processive/distributive equilibrium (Chong et al. 2014). The strongest known gyrase DNA binding site is the strong DNA gyrase cleavage site (SGS) at the center of the Mu genome, which is stronger than all other gyrase binding sites in E. coli and Salmonella (Pato and Banerjee 2000). Because of its stability and processive properties, the Mu SGS has been used in both rotorbead and molecular tweezers experiment to study supercoiling cycles. Unfortunately, very little is known about processivity and gyrase dynamics when gyrase is bound to sites like the BIMES of E. coli and Salmonella (Espeli and Boccard 1997; Stern et al. 1984; Yang and Ames 1988). We note that when a site is too strong, it can interfere with competing DNA processes. In dilute solution the off-rate of a DNA with the SGS from gyrase is longer than 1–2 days (Scheirer and Higgins 1997), and collisions between gyrase and RNA polymerase, DNA polymerase, and other helicases can lead to traffic jams during transcription, replication, and DNA repair in vivo. Therefore, gyrase and its optimum binding sites are most likely under strong evolutionary pressure for optimal genetic fitness. Recent results also show that even synonymous codons can have remarkable selective phenotypes in E. coli (Brandis and Hughes 2016).
Why do bacteria evolve significantly different supercoil set points?
Work on gyrase from species like Borrelia burgdorferi and Mycobacterium tuberculosis (M tb) show that gyrase from different bacterial species varies significantly in terms of mechanism and the ability to supercoil DNA in vitro. The DNA binding and wrapping mechanism of M tb gyrase differs dramatically from that of E. coli, resulting in the in vitro set point of the former being lower than that of the E. coli gyrase (Tretter and Berger 2012a, b). The GyrA of Borrelia burgdorferi and E. coli differs in terms of C-terminal β-pinwheel structure, and the Borrelia GyrA DNA binding domain is actually expressed from an independent promoter inside the GyrA gene such that the C-terminal region functions as an independent DNA-bending protein. This protein will substitute for the E. coli HU protein in biochemical assays for in vitro Mu transposition (Knight and Samuels 1999; Ruthenburg et al. 2005). These differences between gyrases from very distantly related bacteria are not too surprising. However, why do supercoil differences arise between closely related species like E. coli and Salmonella that have nearly identical genetic maps and enzymes?
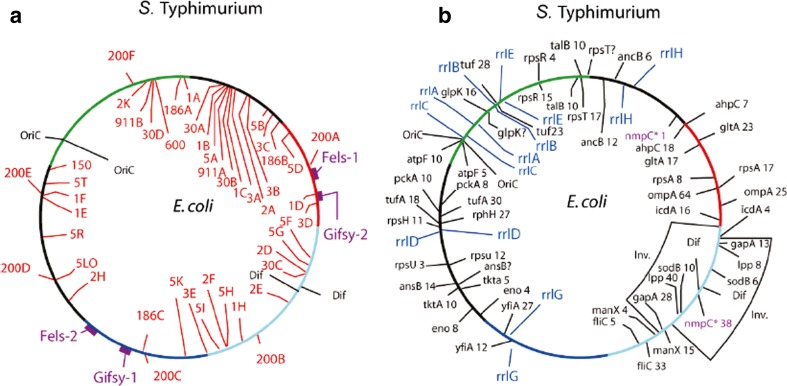
One possible answer to this question is the presence of multiple mobile DNA elements in the two organisms (Fig. 3a). The transcriptional landscape and location of highly expressed genes is remarkably conserved between E. coli and Salmonella (Fig. 3b). The sequential order of the most highly transcribed genes is preserved along with the relative transcript abundance measured with microarray technology. However, the types and number of mobile elements are dramatically different between the species (Fig. 3a) Escherichia coli K12 encodes a large cast of insertion sequence (IS) elements, which includes multiple copies of IS1, IS3, IS4, IS5, IS30, IS150, IS186, and IS911. These IS sequences are active and move frequently (Manna et al. 2001). Escherichia coli K12 has no inducible plaque-forming prophage.
Fig. 3.
a Map of E. coli mobile DNA elements (inside) and S. Typhimurium mobile elements (outside). E. coli includes multiple copies of insertion sequence 1 (IS1; 1A–H), IS2 (2A–H), IS3 (3A–F), IS5 (5A–R), IS30 (30A–C), IS150 (150), IS186 (186A–C), IS600 (600), and IS911 (911A–B). In Salmonella Typhimurium LT2 there are six copies of IS200 (200A–F) in addition to four lysogenic plaque-forming prophages, namely, Fels-1, Gifsy-2, Gyfsy-1, and Fels-2, shown in purple in clockwise order. b The positions of the 27 most highly transcribed protein operons are shown in black and those of the seven ribosomal RNA operons are shown in blue. Steady-state RNA/DNA level for the protein-encoding genes from microarray experiments in exponential phase on LB medium are listed following the gene name
In contrast, Salmonella has none of the IS elements present in E. coli. It has six copies of the IS200 element that appear to be inactive since they have not been seen to move in almost 50 years of study in Salmonella. More significant are the four plaque-forming prophage, Gifsy-1, Gifsy-2, Fels-1, and Fels-2. These viruses are under selection because they include multiple genes and operons that increase the ability of Salmonella to infect and reside in mammalian hosts (Bacciu et al. 2004; Coombes et al. 2005; Figueroa-Bossi and Bossi 1999; Ho et al. 2002). These elements are also potentially lethal if and when spontaneous induction leads to viral replication and cell lysis. A low supercoil density could provide insulation from spontaneous induction and lytic phage development.
Comparing the phage lytic/lysogenic equilibriums in E. coli and Salmonella
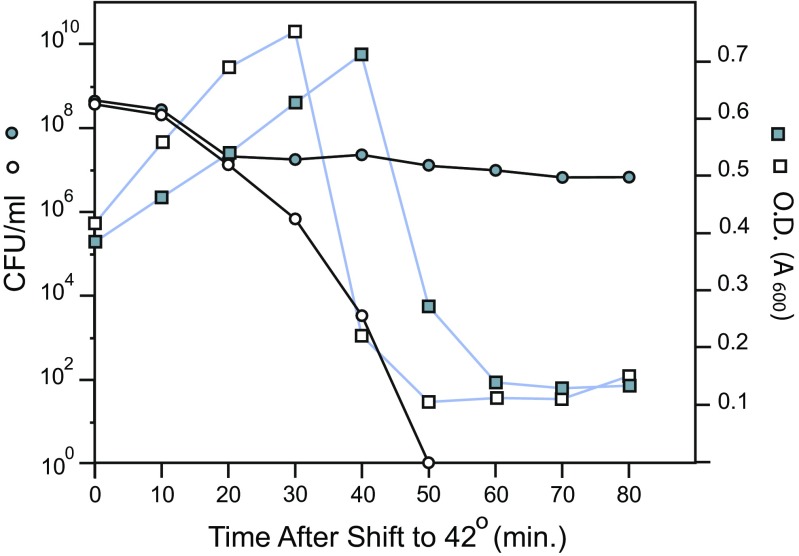
To prove that a lower supercoil density in Salmonella can reduce the lytic burden of four lysogenic viruses, we needed to test the lytic induction potential of both species. An efficient induction method to study Salmonella prophages was not available; therefore, Champion carried out experiments that compared the lethality of the bacteriophage Mu (Champion and Higgins 2007) in E. coli and Salmonella (Fig. 4). Lysogenic WT strains of E. coli and Salmonella were isolated by infecting cells with the thermal-inducible plaque-forming MupAp1 phage. Multiple cultures of both species were grown at 30 °C, then shifted to a 42 °C shaking water bath, and the OD600 as well as the number of viable cells that plated on LB agar at 30 °C were sampled at 10-min intervals for the next 80 min. Escherichia coli cultures began to lyse at 30 min after the temperature shift, while Salmonella cultures began lysis 10 min later. Thus, Mu phage development took longer in Salmonella than it did in E. coli. More dramatically, the number of cells surviving the shift to 42 °C were vastly different. In E. coli, viable cell numbers fell immediately after the shift and rapidly declined; by 50 min, viability had fallen by 8 log units and no survivors were detected. In Salmonella, viable cell numbers also dropped immediately after the temperature shift, but they then leveled off at 20 min and the viable cell count remained at 107/ml for the next hour. Whereas both species support the Mu lytic cycle, Salmonella survived the strong thermo-induction 10,000,000-fold better than E. coli. The results of this experiment strongly support the hypothesis that the low supercoil density in Salmonella is an important factor in tolerating four prophage genomes in one bacterial chromosome.
Fig. 4.
Lytic response of a Mu lysogen in E. coli and Salmonella. Fresh overnight cultures of lysogens of E. coli (open squares) and Salmonella Typhimurium (filled squares) were diluted 1:100 in fresh LB and grown at 30 °C to the log phase (OD600 = 0.4). Part of each culture was shifted to a shaking water bath maintained at 42 °C, and the OD600 and colony-forming units (CFU) were measured at 10-min intervals through the lysis period. E. coli cultures began to lyse at 30 min, whereas lysis in Salmonella cultures was delayed to 40 min. The number of viable Salmonella cells (filled circles) dropped by 50-fold, while the number of viable E. coli cells (open circles) dropped by seven orders of magnituce —i.e., no survivors after 40 min at 42 °C
In the final analysis, the critical factor for gene expression systems for all organisms is that they must be highly efficient at matching gene expression output with growth rate demands of the most common niches occupied by the species. In bacteria, transcription and translation are intimately connected by temporal synchrony. Ribosomes follow the emerging transcript in most genes, and there is a large collection of proteins dedicated to delivering specific proteins to their proper cell location. The genetic investment is huge, and examples of proteins, RNAs, and small molecules that collaborate include gyrase, DksA, NusA, NusG, MFD, Rho, RfhA, GreA, GreB, RNAP, ppGpp, tmRNA, Topo I, cAMP, and cyclic GMP. Other results indicate that transcription is coupled with DNA repair and replication machinery so that conflicts between RNA and DNA polymerases are quickly and efficiently resolved (Cagliero et al. 2014; Hanawalt and Spivak 2008; Tehranchi et al. 2010).
In the post-genomic era, there has been a shift in focus from funding research designed to uncover biochemical mechanisms and test genetic relationships to funding correlation science. New fields of proteomics, metabolomics, and translational science are based on the notion that the current level of understanding of biological systems is sufficient to enable epidemics to be predicted, fundamental problems to be solved with massive computational power and cures can be developed from designer drugs that target almost any protein. Daily media predictions hint that new work in mouse genetics will lead to cures for diabetes, cancer, and Parkinson’s disease. However, there has been little interest in testing the limits of our knowledge. Work described above shows that unexpected responses of E. coli and Salmonella to highly homologous proteins from the other specie proves that we lack information about important connections between gyrase, condensins, and the transcription machinery. In addition, the Ventner group in La Jolla published a recent paper in Science in which they describe the smallest free-living organism, which they isolated by whittling down a larger bacterial genome to only 531,560 bp and 473 genes (Hutchinson et al. 2016). Shockingly, 147 essential genes in this bacterium have no known structure or function. Correlations have little meaning without an understanding of cause and effect. Shakespeare said it well. “There are more things under heaven and earth, Horatio, than are dreamt of in your philosophy” (Hamlet I.5:159–167).
Acknowledgments
Work in the Higgins lab has been supported by the United States National Institutes of Health grant GM-33143.
Compliance with ethical standards
Conflict of interest statement
The author declares that he has no conflict of interest regarding this article.
Ethical approval statement
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘DNA supercoiling, protein interactions and genetic function’ edited by Laura Finzi and Wilma Olson.
References
- Bacciu D, Falchi G, Spazziani A, Bossi L, Marogna G, Leori GS, Rubino S, Uzzau S (2004) Transposition of the heat-stable toxin astA gene into a gifsy-2-related prophage of Salmonella enterica serovar Abortusovis. J Bacteriol 186:4568–4574 [DOI] [PMC free article] [PubMed]
- Basu A, Schoeffler AJ, Berger JM, Bryant Z (2012) ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat Struct Mol Biol 19(538–546):S531 [DOI] [PMC free article] [PubMed]
- Booker BM, Deng S, Higgins NP (2010) DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol Microbiol 78:1348–1364 [DOI] [PubMed]
- Brandis G, Hughes D. The selective advantage of synonymous codon usage bias in Salmonella. PLoS Genet. 2016;12:e1005926. doi: 10.1371/journal.pgen.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliero C, Zhou YN, Jin DJ. Spatial organization of transcription machinery and its segregation from the replisome in fast-growing bacterial cells. Nucleic Acids Res. 2014;42:13696–13705. doi: 10.1093/nar/gku1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion K, Higgins NP (2007) Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J Bacteriol 189:5839–5849 [DOI] [PMC free article] [PubMed]
- Chong S, Chen C, Ge H, Xie XS. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Wickham ME, Brown NF, Lemire S, Bossi L, Hsiao WW, Brinkman FS, Finlay BB (2005) Genetic and molecular analysis of GogB, a phage-encoded type III-secreted substrate in Salmonella enterica serovar typhimurium with autonomous expression from its associated phage. J Mol Biol 348:817–830 [DOI] [PubMed]
- Cunha S, Woldringh CL, Odijk T (2005) Restricted diffusion of DNA segments within the isolated Escherichia coli nucleoid. J Struct Biol 150:226–232 [DOI] [PubMed]
- Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci. 2003;8:D210–D221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- Espeli O, Boccard F (1997) In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol Microbiol 26:767–777 [DOI] [PubMed]
- Espeli O, Mercier R, Boccard F (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68:1418–1427 [DOI] [PubMed]
- Fernandez-Sierra M, Shao Q, Fountain C, Finzi L, Dunlap D (2015) E. coli gyrase fails to negatively supercoil diaminopurine-substituted DNA. J Mol Biol 427:2305–2318 [DOI] [PMC free article] [PubMed]
- Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- Gamper HB, Hearst JE. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982;29:81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF (2012) Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol 86:1318–1333 [DOI] [PMC free article] [PubMed]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Higgins NP. RNA polymerase: chromosome domain boundary maker and regulator of supercoil density. Curr Opin Microbiol. 2014;22C:138–143. doi: 10.1016/j.mib.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NP, Cozzarelli NR. The binding of gyrase to DNA: analysis by retention to nitrocellulose filters. Nucleic Acids Res. 1982;10:6833–6847. doi: 10.1093/nar/10.21.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NP, Vologodskii AV (2015) Topological behavior of plasmid DNA. Microbiol Spectr 3(2). doi: 10.1128/microbiolspec.PLAS-0036-2014 [DOI] [PMC free article] [PubMed]
- Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the GD resolution system in Salmonella typhimurium. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NP, Deng S, Pang Z, Stein R, Champion K, Manna D. Domain behavior and supercoil dynamics in bacterial chromosomes. In: Higgins NP, editor. The bacterial chromosome. Washington: ASM Press; 2005. pp. 133–153. [Google Scholar]
- Ho TD, Figueroa-Bossi N, Wang M, Uzzau S, Bossi L, Slauch JM. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:5234–5239. doi: 10.1128/JB.184.19.5234-5239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes VF, Cozzarelli NR. Closing the ring: Links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson C, Chuang R-Y, Noskov VN et al (2016) Design and synthesis of a minimal bacterial genome. Science. doi: 10.1126/science.aad6253 [DOI] [PubMed]
- Johnson RC, Johnson LM, Schmidt JW, Gardner JF. The major nucleoid proteins in the structure and function of the E. coli chromosome. In: Higgins NP, editor. The bacterial chromosome. Washington: ASM Press; 2005. pp. 65–132. [Google Scholar]
- Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Fisher JK, Stouf M, White MA, Bates D, Witz G. The bacterial nucleoid: nature, dynamics and sister segregation. Curr Opin Microbiol. 2014;22:127–137. doi: 10.1016/j.mib.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Samuels DS (1999) Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J 18:4875–4881 [DOI] [PMC free article] [PubMed]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Wang X, Higgins NP. Mu and Is1 transposition exhibits strong orientation bias at the E. coli bgl locus. J Bacteriol. 2001;183:3328–3335. doi: 10.1128/JB.183.11.3328-3335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M, Travers AA, Howells AJ, Maxwell A, Pato ML. Dissection of the bacteriophage Mu strong gyrase site (SGS): significance of the SGS right arm in Mu biology and DNA gyrase mechanism. J Bacteriol. 2006;188:619–632. doi: 10.1128/JB.188.2.619-632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Chen R, Manna D, Higgins NP. A gyrase mutant with low activity disrupts supercoiling at the replication terminus. J Bacteriol. 2005;187:7773–7783. doi: 10.1128/JB.187.22.7773-7783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato ML, Banerjee M. Genetic analysis of the strong gyrase site (SGS) of bacteriophage Mu: localization of determinants required for promoting Mu replication. Mol Microbiol. 2000;37:800–810. doi: 10.1046/j.1365-2958.2000.02042.x. [DOI] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pul U, Wagner R. Nucleoid-associated proteins: structural properties. In: Dorman CJ, Dame RT, editors. Bacterial chromatin. New York: Springer; 2010. pp. 175–204. [Google Scholar]
- Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet. 2012;8:e1002845. doi: 10.1371/journal.pgen.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL (2005) A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem 280:26177–26184 [DOI] [PubMed]
- Sawitzke JA, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci USA. 2000;97:1671–1676. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheirer KE, Higgins NP. The DNA cleavage reaction of DNA gyrase. Comparison of stable ternary complexes formed with enoxacin and CcdB protein. J Biol Chem. 1997;272:27202–27209. doi: 10.1074/jbc.272.43.27202. [DOI] [PubMed] [Google Scholar]
- Stein R, Deng S, Higgins NP. Measuring chromosome dynamics on different timescales using resolvases with varying half-lives. Mol Microbiol. 2005;56:1049–1061. doi: 10.1111/j.1365-2958.2005.04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MJ, Ames GF-L, Smith M, Robinson EC, Higgins CF. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter EM, Berger JM (2012a) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: I. A nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J Biol Chem 287:18636–18644 [DOI] [PMC free article] [PubMed]
- Tretter EM, Berger JM. Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II. The shape of the GyrA subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency. J Biol Chem. 2012;287:18645–18654. doi: 10.1074/jbc.M112.345736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reyes-Lamothe R, Sherratt DJ (2008) Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev 22:2426–2433 [DOI] [PMC free article] [PubMed]
- Wendorff TJ, Schmidt BH, Heslop P, Austin CA, Berger JM. The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J Mol Biol. 2012;424:109–124. doi: 10.1016/j.jmb.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J (2010) Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci USA 107:4991–4995 [DOI] [PMC free article] [PubMed]
- Yang Y, Ames GF (1988) DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci USA 85:8850–8854 [DOI] [PMC free article] [PubMed]