Abstract
The overproduction of reactive oxygen species (ROS) generates oxidative stress in cells. Oxidative stress results in various pathophysiological conditions, especially cancers and neurodegenerative diseases (NDD). The Keap1–Nrf2 [Kelch-like ECH-associated protein 1–nuclear factor (erythroid-derived 2)-like 2] regulatory pathway plays a central role in protecting cells against oxidative and xenobiotic stresses. The Nrf2 transcription factor activates the transcription of several cytoprotective genes that have been implicated in protection from cancer and NDD. The Keap1–Nrf2 system acts as a double-edged sword: Nrf2 activity protects cells and makes the cell resistant to oxidative and electrophilic stresses, whereas elevated Nrf2 activity helps in cancer cell survival and proliferation. Several groups in the recent past, from both academics and industry, have reported the potential role of Nrf2-mediated transcription to protect from cancer and NDD, resulting from mechanisms involving xenobiotic and oxidative stress. It suggests that the Keap1–Nrf2 system is a potential therapeutic target to combat cancer and NDD by designing and developing modulators (inhibitors/activators) for Nrf2 activation. Herein, we review and discuss the recent advancement in the regulation of the Keap1–Nrf2 system, its role under physiological and pathophysiological conditions including cancer and NDD, and modulators design strategies for Nrf2 activation.
Keywords: Oxidative stress, Keap1, Nrf2, Inhibitors, Cancer, Neurodegenerative disorders
Introduction
Reactive oxygen species (ROS) are biological molecules produced naturally as a by-product of oxygen metabolism by aerobic organisms. Under physiological conditions, the ROS level will be in equilibrium in the system. ROS plays a vital role in the physiology of cell signalling. The production of ROS is intracellularly contributed by several enzymes, which include NA(D)PH oxidase, cytochrome P450-dependent oxygenases and xanthine oxidase. Moreover, ROS is also produced non-enzymatically in mitochondrial complex I and III of the electron transport chain (Turrens 2003).
Oxidative stress is the overproduction or imbalanced production of ROS, which disturbs the normal antioxidant mechanism in the system. Oxidative stress and ROS production are involved in the pathogenesis of numerous neurodegenerative diseases (NDD), including Alzheimer’s, Parkinson’s and Huntington’s diseases (Behl 2005; Browne and Beal 2006), atherosclerosis (Sugamura and Keaney 2011), rheumatoid arthritis (Filippin et al. 2008), ischaemia and stress (Jiang and Duong 2016). The age factor introduces several common pathogenic mechanisms which have been identified as sources of oxidative stress in neurodegenerative disorders. Some of the phenomena involved are changes in tissue antioxidant status, mitochondrial dysfunction and comprised energy status, excitotoxicity, defects in the homeostasis in redox-active trace metals, formation of advanced glycation end products (AGEs), aberrant protein metabolism and proteasome, dysfunction and several environmental or genetic risk factors.
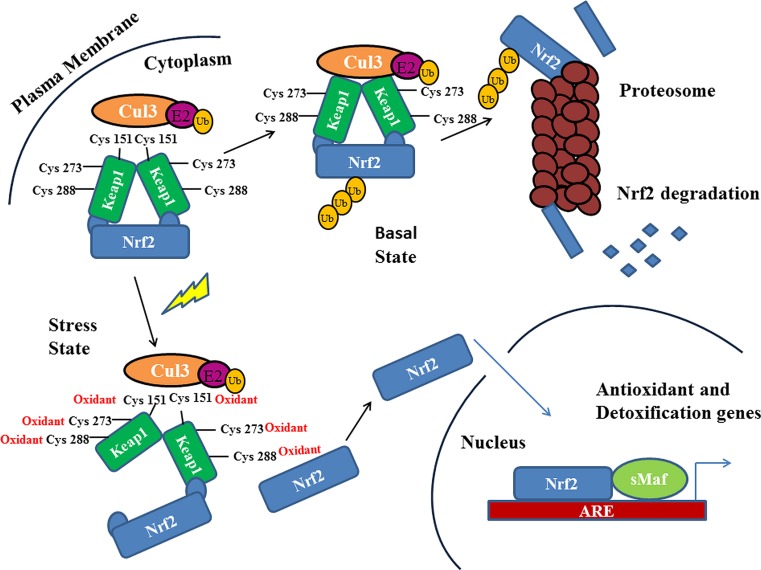
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a basic leucine zipper (bZIP) transcription factor with a cap ‘n’ collar (CNC) structure (Chan et al. 1995). The Nrf2 transcription factor is ubiquitously expressed and present in various organs and tissues, including the kidney, muscle, lung, heart, liver and brain. The Nrf2 transcription factor is tightly regulated by the repressor protein, Keap1 (Kelch-like ECH-associated protein 1), in the cytoplasm, which subsequently plays an essential role in Nrf2 degradation by the ubiquitin–proteasome pathway (Furukawa and Xiong 2005). Under oxidative stress, Nrf2 dissociates from Keap1, translocates to the nucleus and transactivates several cytoprotective genes to combat the oxidative stress (Taguchi et al. 2011) (Fig. 1).
Fig. 1.
Under homeostatic conditions, Nrf2 is negatively regulated and ubiquitinated through Keap1, and degraded by the proteasomal degradation pathway. Under stressed conditions, Nrf2 dissociates from Keap1, translocates into the nucleus and activates cytoprotective genes
As Nrf2 plays a central role in protecting cells from damage, it has been implicated in preventing major diseases. Recent reports have shown the importance of the Keap1–Nrf2 system as a therapeutic target for cancer and NDD (Bryan et al. 2013). In recent years, both academia and industry have shown immense interest in the Keap1–Nrf2 system targets to discover and develop inhibitors to activate Nrf2 transactivation function by inhibiting the Keap1–Nrf2 interaction. Here, we discuss the recent advancement in the regulation of the Keap1–Nrf2 pathway and their importance as potential therapeutic targets for diseases such as cancer and NDD.
Structural insights of Keap1–Nrf2 transcription factor
The cytoplasmic repressor protein, Keap1
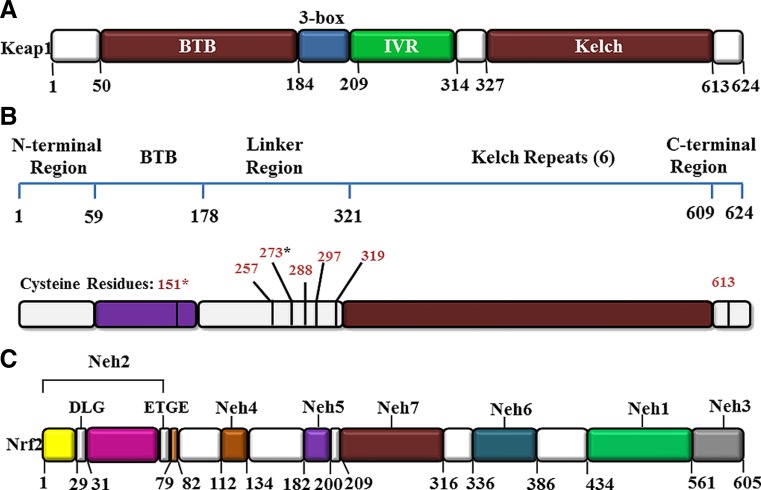
Keap1 is a dimeric protein consisting of 624 amino acid residues. It is the nearest homologue of the kelch protein of Drosophila, which binds to actin (Itoh et al. 1999; Zipper and Mulcahy 2002). Keap1 acts as a substrate adapter protein for the E3 ubiquitin ligase complex formed by Cul3 and Rbx1 and targets Nf-E2/Nrf2 for ubiquitination and degradation by the proteasome (McMahon et al. 2004; Bryan et al. 2013) (Fig. 1). The Keap1 protein is mainly located in the cytoplasm; however, it also shuttles between cytoplasm and nucleus (Sun et al. 2011). The amino acid sequence of Keap1 is highly conserved among mice, rats and humans. Structurally, Keap1 can be sub-divided into five different domains, namely, the N-terminal region (NTR), the broad-complex, tramtrack and bric-à-brac (BTB) domain, the intervening region (IVR) or the BACK domain, double glycine repeats (DGR) or β-propeller domain and the C-terminal region (Stogios and Privé 2004) (Fig. 2). The β-propeller domain and the C-terminal region together is called Keap1–DC (Keap1–DC, hereafter).
Fig. 2.
Domain structures of Keap1 and Nrf2. a Keap1 consists of three major functional domains: the BTB, IVR, and the Kelch/β-propeller domains. b The reactive cysteine residues in Keap1 are indicated with asterisks. c The Nrf2 protein contains six domains, Neh1–Neh6. The ETGE and DLG motifs in the Neh2 domain are essential for the direct interaction with the Kelch domain of Keap1
The BTB domain is essential for homodimerisation of the Keap1 protein. The BTB domain along with the IVR domain play an essential role for Nrf2 polyubiquitination and 26S proteasomal mediated degradation under basal conditions. The N-terminal of the BTB domain interacts with the Cullin-3 (Canning et al. 2013; Chauhan et al. 2013). The BTB domain forms a dimer and consists of three β-sheets flanked by six α-helices. The β1 helix is essential for the formation of the dimeric interface. The N-terminal residues form the domain-swapped β-sheet, which also plays a key role in the homodimerisation interface formation (Cleasby et al. 2014).
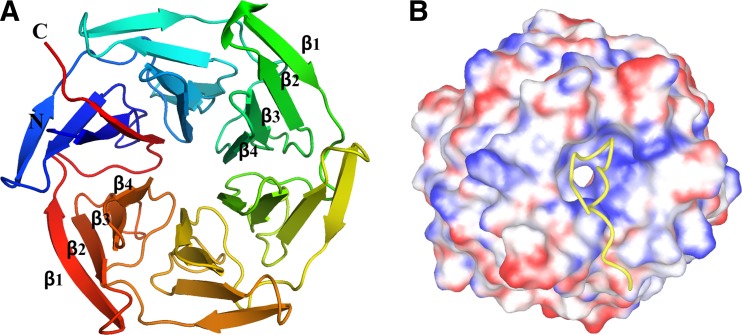
The overall tertiary structure of Keap1–DC belongs to the β-propeller domain with a size of approximately 49 Å × 36 Å (Padmanabhan et al. 2006) (Fig. 3). It consists of a tandem of six twisted four-stranded (β1–β4) antiparallel β-sheets resembling a six-bladed β-propeller structure with pseudo six-fold symmetry. Like other β-propeller proteins, β-strands of both termini meet and, in this case, the disc-like conformation is tightened by a ‘3 + 1’ arrangement of β-strands (‘Velcro’ closure or ‘molecular clasp’).
Fig. 3.
The tertiary structure of the Kelch domain of Keap1. a A cartoon diagram of the Kelch/β-propeller domain (PDB Id: 2DYH). It contains six blades; each blade is formed by four β-strands, β1–β4. b Electrostatic surface potential of the Kelch/β-propeller domain of Keap1 in complex with the Nrf2 peptide containing the ETGE motif (shown as a yellow ribbon). The Nrf2 peptide binds at the bottom region of the Kelch domain
The human Keap1 consists of 27 cysteines acting as ROS sensors in the regulation of cellular homeostasis. Among the cysteine residues, Cys151, Cys171, Cys273 and Cys288 are highly reactive, which are present in the BTB–IVR domains of Keap1 (Cleasby et al. 2014) (Fig. 2b).
Nrf2 transcription factor
Nrf2 (or NFE2L2) is a ubiquitously expressed antioxidant transcription factor, which tightly regulates the expression of cytoprotective genes in response to both exogenous and in-situ signals (Marzec et al. 2007). It belongs to the bZIP factors of the CNC family. The CNC family proteins regulate gene expression, tissue differentiation and development in a variety of organisms. The CNC domain, comprising a sequence of 43 conserved amino acids, is located N-terminally to the basic DNA-binding domain. The family members of the CNC domain heterodimerise with small Maf proteins (Derjuga et al. 2004) and exhibit high homology in their DNA binding and leucine zipper domains; however, their biological functions are different. Most of the CNC family members are transcription activators, but Bach1 and Bach2 are transcription repressors. Nrf2 is the most studied CNC family member and is responsible for the expression of constitutive and inducible levels of phase II enzymes and endogenous antioxidants. An Nrf2 null mouse was found to exhibit decreased expression of phase II enzymes (Chan et al. 1993). Nrf3 is another important factor which mainly finds a population in the placental tissue.
The Nrf2 protein is comprised of six highly conserved Neh (Nrf2–ECH homology) domains, Neh1–Neh6 (Fig. 2c). The Neh1 domain contains the CNC-type bZIP region which is crucial for DNA binding and dimerisation with other transcription factors (Nioi et al. 2005). The Neh1 domain is required for homo- or heterodimerisation with Maf proteins (MafF, MafG and MafK) and also with leucine zipper containing protein domains (Motohashi et al. 2002). The Neh3 domain lies at the C-terminal region of Nrf2, acts as a transactivation domain to promote the transcription of antioxidant response element (ARE)-dependent genes by means of interacting with the chromo-ATPase/helicase DNA binding protein family member CHD6 (Nioi et al. 2005). The Neh4 and Neh5 domains of Nrf2 coordinate with co-activators CBP (CREB/ATF4) and BRG1 (brahma-related gene 1), respectively (Moi et al. 1994). The Neh6 domain plays a key role in the Keap1-independent degradation pathway of Nrf2. The degradation of Nrf2 in stressed cells is predominantly mediated by the redox-insensitive Neh6 domain (McMahon et al. 2004). The Neh2 domain is present at the N-terminal region of Nrf2. It possesses two motifs, namely, DLG and ETGE motifs. These two motifs of Neh2 are mainly responsible for the direct interaction with the negative regulator, Keap1, which subsequently guide the degradation of an excess of Nrf2 factor to maintain homeostatic conditions (McMahon et al. 2004).
Molecular mechanism of the Keap1–Nrf2 pathway
The Keap1–Nrf2 system is a vital member of regulating cells under a homeostatic environment. This system counters the xenobiotic and oxidative responses which defend the cells from external and internal toxicity. At basal homeostatic conditions, Keap1 maintains a consistent generation of Nrf2 and retains its low levels in the cytoplasm. A specific cysteine residue modification of Keap1 is responsible for the conformational change of the protein (Fig. 2b) in cells under oxidative stress conditions. This causes the initial detachment of DLG motif from the Keap1–Nrf2 complex, leaving the ETGE motif still in association with the protein, according to the hinge and latch model (Tong et al. 2007). Therefore, Keap1-bound Nrf2 is released and translocated inside the nucleus of the cell. Post-translocation, Nrf2 associates with other transcription factors and binds to the ARE region of phase II enzymes expressing genes. The Nrf2 transcriptional activity proceeds with the aid of other co-transcription factors in complex with the former. The stress conditions lead to the suspension of Keap1–Nrf2 interactions and causes transcription of cytoprotective genes like NQO1, GSTs and GCL, which in turn, scavenges the cellular oxidative stress (Kansanen et al. 2013) (Fig. 1). According to another model, Cul3 gets dissociated from the Keap1–Cul3 complex in the presence of ROS. Hence, ubiquitination of Nrf2 is halted, which leads Nrf2 to escape from the proteasomal degradation and results in its subsequent nuclear translocation (Niture and Jaiswal 2010).
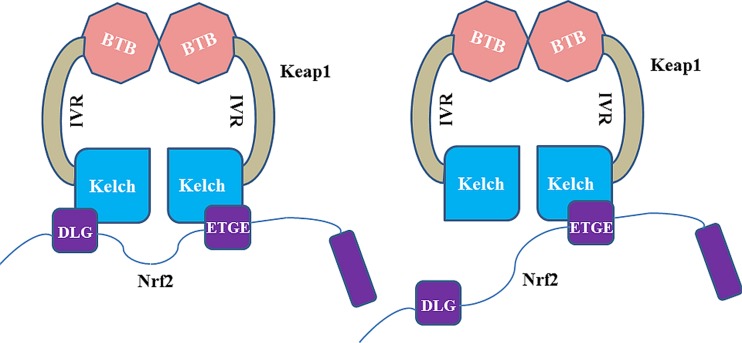
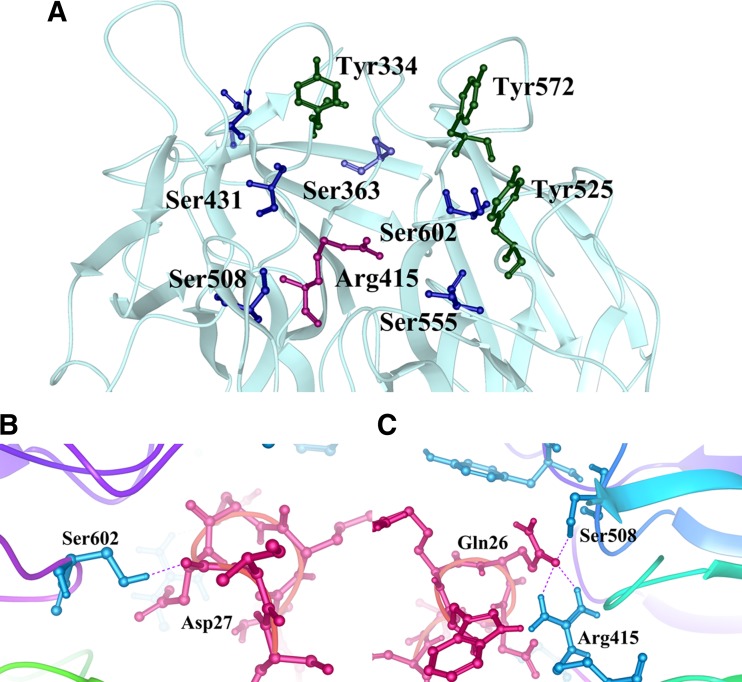
The ETGE and DLG motifs of the Neh2 domain binds to the two Keap1–DC domains of the Keap1 homodimer, in a hinge and latch fashion (Padmanabhan et al. 2006; Tong et al. 2007) (Fig. 4). The ETGE motif has stronger binding affinity than the DLG motif with Keap1–DC. The connecting loops that protrude from the central core of the β-propeller form a binding cavity with abundant ionic residues in the cavity surface exposed to the solvent region and hydrophobic residues towards the internal cavity surface (Fig. 5). The Keap1–DC sequence contains highly conserved glycine, tyrosine and tryptophan residues. These conserved residues are vital for repressor activity of the kelch domain. Mutation of these residues leads to abrogation of the repression activity (Singh et al. 2006).
Fig. 4.
Two-site binding model (referred to as the hinge and latch model) of Keap1 and Nrf2. Left Under homeostatic conditions, the Keap1 homodimer binds to a single Nrf2 chain at two sites of the Kelch domain monomers and recognises the DLG and ETGE motifs in Nrf2 simultaneously. Right Under stress conditions, conformational changes occur in the Keap1 homodimer, which subsequently disrupt the weakly binding DLG motif interaction with the Kelch domain
Fig. 5.
Crystal structure of the Kelch domain of Keap1 in complex with the Nrf2 peptide (PDB Id: 2DYH). a Near the Nrf2 peptide binding region. The key residues are indicated by sticks. The tyrosines, serines and arginine residues are coloured in green, blue and pink, respectively. b and c Showing the Nrf2–peptide (red sticks) interaction with Keap1 residues (blue sticks). The hydrogen bonds are shown by dotted lines
Regulation of the Nrf2 degradation pathway
The Keap1–Nrf2 complex triggers the degradation of Nrf2 factor via CUL3-dependent E3-ubiquitin ligase-mediated ubiquitination and its successive degradation through proteasome (Fig. 1) (Kobayashi et al. 2004). Cullin family proteins are hydrophobic in nature and have a high substrate specificity towards a multimeric complex of E3 ligases. Cullin proteins play an essential role by providing solid scaffolds to these E3 ligases, which polyubiquinates the substrate with the help of E2 ligases (Saha and Deshaies 2008). The removal of the Rbx1 RING domain from the C-terminal of Cullin is a primary reason for Nedd8-mediated enhancement in CRL (Cullin RING ubiquitin ligases) E3 activity, which results in the polyubiquitination of Nrf2 (Saifee and Zheng 2008). The Neh2 domain of Nrf2 is important for Keap1-dependent degradation of Nrf2 in cells under basal homeostatic conditions. Especially, the Neh2 domain, containing lysine residues at 44, 50, 52, 53, 56, 64 and 68, are ubiquitinated by CUL3-dependent E3-ubiquitin ligase (Sekhar et al. 2002).
Role of the Nrf2–Keap1 pathway in cancer
Nrf2 activation upregulates the various set of enzymes for the detoxification of chemical carcinogens and confers protection against carcinogenicity, mutagenicity and other types of toxicity (Yu and Kensler 2005). Several studies have shown that Nrf2 protects against oxidative stress, chemotherapeutic agents and radiotherapy (Lau et al. 2008; Kensler and Wakabayashi 2010; Takahashi et al. 2015). However, Nrf2 disruption has enabled the cells towards carcinogens, which lead to the progression of inflammation and, finally, cancer formation (Slocum and Kensler 2011; Takahashi et al. 2015). This dual action of Nrf2 has been termed as a ‘double-edged sword’ with respect to the benefits or risks of the Keap1–Nrf2 pathway in cells (Lau et al. 2008). The Nrf2 transcription factor is associated with the phase II enzymes gene regulation, as it maintains the appropriate level of these enzymes inside the cell. The excessive Nrf2 expression leads to the survival of both normal as well as cancerous cells. Hence, the Nrf2 downstream gene expression balance is required to obtain the clinical benefits and with less side effects. In this context, the development of Nrf2 inhibitors is challenging for cancer treatment. Discovery of the dual role of Nrf2 enabled scientists to understand the Nrf2 signalling in cancer and development of pharmacological compounds targeting Nrf2 for the prevention of cancer and treatment (reviewed in Jaramillo and Zhang 2013). The maintenance of proper homeostatic conditions by the development of Nrf2 inhibitors/activators is a vital therapeutic strategy.
Several studies have shown, by using Nrf2 knockout mice, that Nrf2 protects against chemical carcinogens induced tumour formation in the stomach and skin (Ramos-Gomez et al. 2001), intestines (Osburn et al. 2007) and bladder (Fahey et al. 2002). Nrf2 has the ability to reduce the ROS and DNA damage in chemical-induced carcinogen cells (Morito et al. 2003). Other studies showed the protective role of Nrf2 in mice, harbouring a single nucleotide polymorphism (SNP) in the promoter region of the mouse Nrf2 gene. Reduced expression of Nrf2 has been observed in mice with SNPs. An antineoplastic compound, brusatol, is an Nrf2 inhibitor that increases the chemotherapeutic efficacy of cisplatin, a common chemotherapeutic (Ren et al. 2011). PI3K inhibitors (Mitsuishi et al. 2012) and Nrf2 siRNA inhibit Nrf2 in cancer cells. Cancer suicide gene therapy, which is an alternative approach, has been utilised to target the cancer cells with high Nrf2 levels. Cancer cells with high ARE activity have been transferred with Nrf2-driven lentiviral vectors containing thymidine kinase (TK) and treated with a pro drug, ganciclovir (GCV). This enabled the killing of TK-containing tumour cells as well as neighbouring cells by the bystander effect (Leinonen et al. 2012).
It has been observed by identifying several genes in the pentose phosphate pathway including glucose-6-phosphate dehydrogenase (G6PD), phosphogluconate dehydrogenase (PGD), transketolase (TKT) and transaldolase 1 (TALDO1), which are responsible for the regeneration of nicotinamide adenine dinucleotide phosphate (NADPH). Also, other metabolic genes, including malic enzyme 1 (ME1), pyrophosphate phosphoribosyl amidotransferase (PPAT), methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) and isocitrate dehydrogenase 1 (IDH1), were also identified as transcription targets of Nrf2. These proteins are responsible for purines synthesis, which are the building blocks of DNA and RNA, which in turn, leads to the proliferation of cancer cells (Mitsuishi et al. 2012). Several studies have shown that elevated levels of Nrf2 in cancer cells are less sensitive to chemotherapeutic treatments and renders more resistant to cancer cells against a variety of anti-cancer agents (Shibata et al. 2008a, b; Wang et al. 2008).
Several mechanisms are shown in behaviour following elevated levels of Nrf2 in cancer cells, which includes: (a) somatic mutation: gain-of-function mutations in Nrf2 and loss-of-function mutations in Keap1 and CUL3 have been identified in several human cancers. Mutations in the Keap1 gene have been identified in human lung adenocarcinoma cell lines, which involve a glycine to cysteine substitution in the kelch domain of Keap1. It exhibited reduced affinity of Keap1 to the Nrf2 and, thereby, activation of Nrf2 in the cancerous cells (Padmanabhan et al. 2006). Mutations also occur more frequently in Nrf2 than in Keap1. Mutations in the ETGE and DLG motifs impair two-site substrate recognition of Keap1, which leads to the stabilisation of Nrf2 and, subsequently, activation of target genes of Nrf2 (Shibata et al. 2011; Kim et al. 2010a, b). Somatic mutations in CUL3 were identified in hereditary type 2 papillary renal cell carcinoma (Ooi et al. 2013). (b) Epigenetic silencing of Keap1 by hypermethylation: epigenetic modification of Keap1 promotes the activation of Nrf2. Methylation in the promoter region of Keap1 affects its expression and hinders the ability to bind to the Nrf2. Therefore, this leads to the expression of Nrf2 (Wang et al. 2008; Muscarella et al. 2011). Conversely, DNA methylation by DNA methyltransferases (DNMTs) appears to downregulate the Nrf2 expression indirectly (Khor et al. 2014; Yu et al. 2010; Rajabi et al. 2016; Tagde et al. 2016b). (c) Accumulation of p21 and p62 disrupts the Nrf2–Keap1 complex: it has been demonstrated that p53 negatively regulates Nrf2 and specifically suppresses the transcription of target genes of Nrf2 (Faraonio et al. 2006). p21 (a direct downstream target of p53) associates with the DLG motif of Nrf2, which leads to the disruption of Keap1 binding with Nrf2. As a result, Nrf2 is stabilised in response to p21 upregulation. Another protein, p62 (sequestosome 1 protein), which modulates the activity of Nrf2, is a scaffold protein that binds to the polyubiquitinated proteins and targets aggregated proteins and damaged organelles for degradation. p62 directly interacts with the kelch domain of Keap1 via its STGE motif that is similar to the Nrf2 ETGE motif, thereby disrupting the Keap1–Nrf2 complex (Komatsu et al. 2010; Lau et al. 2010). It causes a decrease in the ubiquitination of Nrf2, an increase in Nrf2 stability and, ultimately, leads to the enhanced expression of ARE-bearing genes. (d) Transcriptional upregulation of Nrf2 by oncogenes: oncogenes like KRAS, BRAF and C-MYC increased the mRNA level of Nrf2 and target genes of Nrf2 (DeNicola et al. 2011). The C-MYC oncogene is involved in both increased and decreased expression of phase II antioxidant genes, depending on the pleiotropic effect (Levy and Forman 2010). The oncogenic transmembrane protein MUC1-C transcribes the C-MYC mRNA and proteins, which in turn, upregulates the C-MYC gene expression. Upregulation of C-MYC genes leads to decrease of the Nrf2 stability (Tagde et al. 2016a; Bouillez et al. 2016). (e) Metabolic activation of Nrf2 by Kreb cycle intermediates: in Kreb’s cycle, fumarate modifies cysteine residues within Keap1, which disrupt the ability to ubiquitinate Nrf2. This leads to the prolonged activation of Nrf2 (Adam et al. 2011).
Role of Nrf2–Keap1 in neurodegenerative disorders
The protective effect of Nrf2 against neurodegeneration due to oxidative stress has been well studied. The Nrf2 transcription factor induces the expression of a variety of cytopreventive and detoxification enzymes, which will confer protection in neurodegenerative disorders. The target genes of Nrf2 have been involved in the regulation of glutathione (GSH), antioxidant proteins/enzymes, drug-metabolising enzymes or drug transporters, proteasome subunits, pentose phosphate pathway enzymes and enzymes involved in nucleotide synthesis (Yamazaki et al. 2015; Tagde et al. 2014; Hasegawa et al. 2016). The central nervous system is sensitive to oxidative stress, which confers pathological features, including the accumulation of aberrant protein aggregates, microglial activation and mitochondrial dysfunction. These pathological processes will generate ROS, which in turn, cause oxidative stress and damage to lipids, proteins and DNA. These pathophysiological events encompass a wide variety of neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease, and ischaemia and stroke.
Alzheimer’s disease
Alzheimer’s disease (AD) is a progressive neurodegenerative disease characterised by loss of neuronal integrity, memory impairment and cognitive decline. AD is characterised by the extracellular deposition of amyloid-β (Aβ) in diffuse and neuritic senile plaques and the intra-nuclear accumulation of paired helical filaments (PHFs; consisting of the hyperphosphorylated microtubule-associated tau protein) in neurofibrillary tangles (NFTs), dystrophic neurites and neuropil threads (Cummings 2004). Studies have shown that the expression of target genes of Nrf2 were increased in AD patients. It has been demonstrated that the expression levels of HO-1, GCLM and p62 are increased in the temporal cortices of AD patients compared with that of controls (Marcus et al. 1998). In the normal hippocampus, Nrf2 is expressed in neurons and predominantly localised in the nucleus. In the AD brain, Nrf2 predominantly localises in the cytoplasm of hippocampal neurons and not a component of Aβ plaques or NFTs. Immunoblotting studies suggested that the Nrf2 levels were reduced in AD cases (Ramsey et al. 2007). Another study has shown that the expression of astroglial HO-1 is elevated in the temporal and hippocampus in mild cognitive impairment in AD as compared with controls (Schipper et al. 2006). Nrf2 expression was increased due to pharmacological or lentivirus, which enables Nrf2 to increase the expression of antioxidant genes. This ameliorates the ROS accumulation of Aβ peptides (Eftekharzadeh et al. 2010). Nrf2 has been stabilised by the building up of dysfunctional DJ-1, which prevents the association of Keap1 with Nrf2 and leads to the proteasomal degradation (Clements et al. 2006). Collectively, the activation of Nrf2 could be a therapeutic target for AD, which will ameliorate this disease.
Parkinson’s disease
Parkinson’s disease (PD) is characterised by loss of dopaminergic neurons in a brain region known as the substantia nigra (SN) and the presence of α-synuclein-containing inclusion bodies (Lewy bodies) in the cytoplasm of neurons in various regions of the brain, including the SN, olfactory bulb and neocortex (Mosley et al. 2006). Activation of NOQ1 and HO-1 by the nuclear localisation of Nrf2 was induced in the SN of PD patients (Ramsey et al. 2007). The investigators have also proposed that Nrf2 might regulate different gene products in various neuronal subpopulations (i.e. hippocampal neurons vs. nigral neurons for AD and PD brains, respectively). Activation of the Nrf2–ARE system in PD has been demonstrated by the upregulation of nigra immunoreactivity for regulated proteins, including NOQ1 and HO-1 (Riedl et al. 1999). Other studies found the localisation of Nrf2 in the SN of PD brain in addition to cytoplasm, observed in neurons (Ramsey et al. 2007). Collectively, the data suggest that the Nrf2–ARE signal has been activated in PD, which will counteract Nrf2-activated gene transcription.
The oncogene DJ-1 is an oxidative stress sensor gene as well as transcription regulator. Under oxidative stress conditions, the expression of DJ-1 increases and helps in reducing oxidative stress via stress-induced cell death. Excessive oxidation of DJ-1 leads to loss of function, which results in the oxidative stress in PD (Ariga et al. 2013). Increased oxidative stress was observed due to the reduced expression of Nrf2-dependent genes in patients with deficient DJ-1 genes. This shows that levels of dysfunctional Nrf2 are related to the pathogenesis of the onset of familial PD (Zhang et al. 2013). DJ-1 is required for optimal induction of NOQ1, and the loss of DJ-1 is accompanied by reduced expression in NOQ1. DJ-1 prevents Keap1 from interacting with Nrf2, thereby preventing the ubiquitination of Nrf2. In the absence of DJ-1, Nrf2 is unstable and transcriptional response will be blunt in basal conditions as well as induced conditions (Clements et al. 2006). The protective role of Nrf2 in PD has also been studied using in vivo and in vitro models. The dopamine analogue 6-hydroxyldopamine (6-HAD), which is highly neurotoxic, is found to activate the Nrf2–ARE system, which in turn, activates the cellular defence mechanism to protect against oxidative stress (Jakel et al. 2005).
Huntington’s disease
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder characterised by a cognitive decline in chorea form movements and behavioural difficulties. HD is caused by a CAG trinucleotide expansion (≥35 repeats) in exon 1 of the huntingtin (htt) gene (Walker 2007). Polyglutamine expansion leads to a conformational change in the huntingtin protein, causing the formation of large aggregations in the nucleus and cytoplasm. These htt and mutant htt (mhtt) genes are expressed ubiquitously in human tissues, spiny neurons of the putamen and caudate nucleus in HD (Arrasate et al. 2004; Albin et al. 1992). The recent evidence suggested that the involvement of ROS in HD pathogenesis is by dysfunctioning of the mitochondrial complex II (Calkins et al. 2005). In the primary phase of HD, the focus is on the activation of the astrocyte and microglia by an overexpression of the vital cytoprotective genes via the Keap1–Nrf2–ARE system by Nrf2 agonist; this might protect the brain injury caused by the ROS (Magesh et al. 2012). Activation of the Keap1–Nrf2–ARE pathway by small molecules in astrocytes accelerates the resistance of neurons to the non-excitotoxic glutamate toxicity (Kraft et al. 2004). Protection against the ROS in co-culture neuron culture is conferred by the precise astrocyte stimulation of Nrf2 (Shih et al. 2003). In the animal model of HD, astrocyte-specific activation of Nrf2 reduces the disease pathogenesis (Calkins et al. 2005). Thus, the activation and overexpression Nrf2 is an encouraging therapeutic target in the treatment of HD.
Ischaemia and stroke
Recent studies have also reported the role of Nrf2 in ischaemic and haemorrhagic stroke (Jiang et al. 2016). As the cerebrum has high lipid content and high oxygen consumption, it is susceptible to oxidative damage (Adibhatla and Hatcher 2010). The high ROS level during reperfusion leads to the pathophysiology of cerebral ischaemia–reperfusion (IR) injury and haemorrhage (Kontos 1989; Flamm et al. 1978; Aronowski and Zhao 2011). Mitochondrial dysfunction and endoplasmic reticulum stress within brains are the main factors causing IR injury. It has been reported that GST and NQO1 are decreased significantly in Nrf2-deficient mice (Ramos-Gomez et al. 2001).
The literature studies suggest that Nrf2 activation may protect neurons, astrocytes, oligodendrocytes and microglia against oxidative stress and, hence, the Keap1–Nrf2 pathway is one of the therapeutic approaches to the neurovascular system. The activation of Nrf2 is shown to inhibit the effect of the mitochondrial uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) on cytoplasmic Ca2+ ions, which in turn, enhances the mitochondrial function, inhibited the production of ROS and increased the levels of antioxidant enzymes (Leirós et al. 2013).
The ER stress may trigger the Nrf2-dependent transcriptional regulation of phase II detoxifying enzymes by means of forming misfolded and/or unfolded proteins in the ER lumen due to ROS (Xu et al. 2010). As mentioned before, under oxidative stress, p62 dissociates Nrf2 from Keap1 by direct competition, which subsequently activates Nrf2 and mediates the autophagic degradation of Keap1 (Faraonio et al. 2006). The cross-talk between p62 and the Keap1–Nrf2 pathway in the regulation of autophagy may play an important role in the removal of ROS, prevention of oxidative damage and alleviation of ER stress during cerebral IR injury (Wang et al. 2013).
Ischaemic stroke occurs due to a decrease in blood supply to part of the brain, which causes brain tissue damage in the corresponding area. Recent studies showed that the Nrf2 expression level is significantly increased in the ischaemic penumbra, whereas the Nrf2 level is not detected in the core ischemic zone. These findings indicate that Nrf2 activation may be important and may contribute to cell protection and survival against ischaemia (Dang et al. 2012).
Haemorrhagic stroke occurs due to the bleeding of blood vessels in the brain, either directly into the brain parenchyma or into the subarachnoid space surrounding the brain tissue. The former is known as intracerebral haemorrhage (ICH) and the latter condition is known as subarachnoid haemorrhage (SAH) (Feigin et al. 2005). Reports using ICH models have shown that Nrf2 subjected to both pre-treatment and post-treatment are significantly effective in expressing Nrf2-dependent cytoprotective proteins, which subsequently cause a reduction in oxidative burden to brain tissue and, thus, increase the recovery of neurological function (Zhao et al. 2007). It has been reported that, using SAH rat models, the Nrf2–ARE pathway is activated in the cortex during an early stage of SAH. It suggests that Nrf2 signalling may involve in the pathogenesis of early brain injury (EBI) induced by SAH (Chen et al. 2011).
Key structural features of Keap1 for drug design
Keap1 is a β-propeller structure that consists only of β-sheets and loops. These β-sheets are diverse in composition and are connected with each other through loops. The loops are rather stable according to computational simulations of crystallographic structures and constitute an important part of the functionality of the protein (Fig. 5). The loops enclose into a central cavity, which forms the binding cavity of the protein. The conserved tyrosines and tryptophans form pi-pi stacking interactions in the hydrophobic site of the binding cavity. The important tyrosines Tyr334, Tyr572, and Tyr525 form hydrophobic interactions with ligands and help in maintaining a stable sequestration (Fig. 5a). The arginine residues are responsible for the electrostatic interactions and the serine residues are mainly involved in both inter-/intra-molecular hydrogen bond interactions. Mutagenesis studies revealed that the glycine residues at the positions 364 and 430 are important for the Keap1–Nrf2 association. The serine-mutated proteins were found to stabilise in a delayed manner or possess minor fluctuations throughout compared to glycine-mutated ones. But, eventually, the hydrogen bonding with the residues at these positions was not affected, even after mutation. This was reasoned based upon the involvement of the backbone carbonyl atoms to link to their partners. This involvement was sustained even after mutation (Cheng et al. 2015).
Chemical library synthesis studies revealed that the polar compounds like triazole substitution at the side groups are an important potency enhancing feature in the designed inhibitors. The increase in the potency is because of the increase in the electrostatic interactions between the protein–ligand complexes (Zhuang et al. 2014). But the double-ring structure in the inhibitors needs to be maintained throughout and carefully installed with R groups for favourable interactions with the residues surrounding the tyrosine residues because the latter engage the double-ring structure. Also, halogen substitution has not been proven to be a successful idea while designing inhibitors.
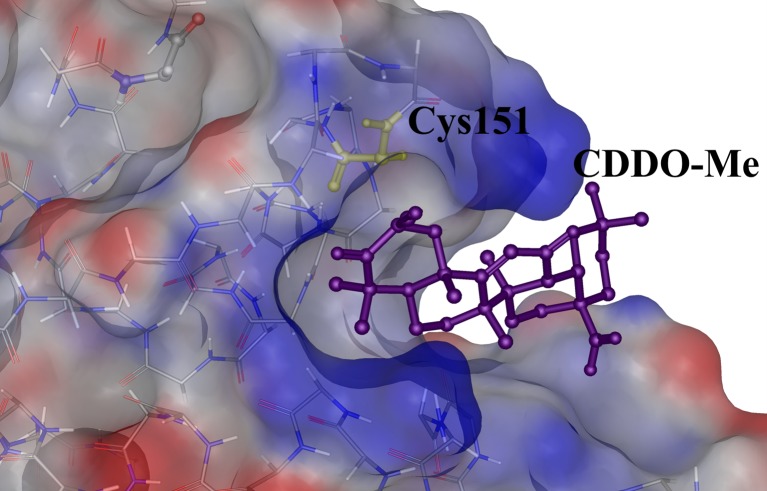
The modification of any of the 27 highly reactive cysteines (Fig. 2b) on all three domains of Keap1 may provide an alternative solution to the inhibition of Keap1. The cysteine residues (C151, C273 and C288) on the BTB-IVR domain decrease the binding of CUL3 to the former and, thus, the Nrf2 degradation pathway is affected (Cleasby et al. 2014). However, Nrf2 is not released substantially for nuclear import. In such cases, overexpression of Nrf2 helps in the induction of the ARE pathway. The triterpenoid CDDO adducts covalently bonded with C151 of the BTB domain and ensures that there is no interaction of the CUL3 with the Keap1 (Cleasby et al. 2014) (Fig. 6). The intervening region (IVR) of Keap1 consists of the cysteine residues C273 and C288 that attract electrophilic binding and, hence, cause distortion of the structural fold that leads to improper positioning of the protein domains and, hence, disrupt the binding of the degradation complex required for its regular functioning (Cleasby et al. 2014).
Fig. 6.
Crystal structure of the Keap1–BTB domain in complex with a ligand, CDDO-Me (PDB Id: 4CXT). The reactive cysteine residue Cys151 is shown as yellow sticks and the CDDO-Me molecule is shown as purple sticks
Keap1 inhibition for therapy
The role of Keap1 in both cancer and NDD is equally important considering its contradictory function in the pathology of both diseases. The function is performed with the assistance of other associated transcription factors and adaptors like p65, p62, Cul3 E3 ligase and so on. The various therapeutic mechanisms for the achievement of the expression of cytoprotective genes involves:
Gene activation of Nrf2
Provision of a ketogenic diet and/or administration of phyto-antioxidants like genistein or antioxidant enzymes like peroxiredoxin to the cells
Deactivation of Keap1: covalent or non-covalent inhibitors (direct inhibition)
Stimulation for the p62 directed autophagy of Keap1, which leads to lowering Keap1 populations and, hence, increased Nrf2 concentration relatively in the cytoplasm
Physical methods like acupuncture have also been found to activate Nrf2-dependent ARE expression
The increase in Nrf2 concentration in the nucleus ensuring that free Nrf2 nuclear translocation has taken place confirms the activation of the Nrf2–ARE pathway biochemically. Also, other probing techniques involve the evaluation of fold change in antioxidant expression. The antioxidants analysed are HO-1, NQO-1, GST, SOD and GGT.
The maintenance of redox balance is mandatory for the regulation of cell homeostasis. Similarly to Nrf2, Keap1 is involved in some other cellular pathways as well. IK-κβ is a substrate analogue of Keap1 that is involved in the activation of the NF-κB signalling pathway, causing deleterious effects in the cell (Kim et al. 2010a, b). Interaction studies reveal that the ETGE motif of Nrf2 is also present in IK-κβ, which produces a similar contact fingerprint between the two molecules (Jiang et al. 2013). This contradictory regulation of antioxidant-mediated cytoprotection and tumour progression by the same molecule, Keap1, exerts a functional burden on the protein. Hence, designing inhibitor compounds considering interaction patterns that do not disturb the regulation of the NF-κB signalling pathway can be a better strategy for use in overcoming any undesirable side effects.
The function of Nrf2 in neuronal cells especially in in vivo studies has been proven in ROS scavenging and depletion of oxidative stress effects. Various pharmacological agents like curcumin (Yang et al. 2009), resveratrol (Ren et al. 2011), MMF (Cho et al. 2015), DMF (Albrecht et al. 2012) and other FAEs (Linker et al. 2011) have been found to exert Nrf2-dependent reduction of oxidative stress. The other agents that can be used for the treatment of neurodegenerative disorders through Nrf2-dependent strategies are sulphoraphane, VEDA-1209 and synthetic triterpenoids. However, these molecules are filtered out in either clinical trials or toxicological reviews in humans. FAEs can be used for neuroprotection related to retinal injuries (Cho et al. 2015). Nrf2 could be activated indirectly through the inhibition of GSH synthesis as in cancer cells, by the addition of buthionine sulphoximine (BSO) (Furfaro et al. 2012; Tagde et al. 2014). Based on the building blocks of constituents, the inhibitory agents have been classified into peptide and chemical inhibitors of Keap1.
Peptide inhibitors
Extensive studies have been carried out to distinguish the structural implications of Keap1–peptide interactions. Based on the information, highly potent peptides targeted to inhibiting Keap1–Nrf2 interactions (KNI) were designed. As already known, the interaction pattern of Nrf2 with Keap1 has been elucidated through mutagenesis to distinguish an important fragmentation of binding in the nanomolar binding of Nrf2 peptide (5nM).
The ETGE motif forms a β-turn and binds to the bottom region of the kelch domain of Keap1. It establishes significant electrostatic interactions with the important residues like Ser508, Arg415 and Ser602 with its own glutamate residues (Gln26, Asp27), along with van der Waals interactions (Fig. 5b, c). Previous studies have indicated the possibilities of other proteins like prothymosin-α and sequestosome to compete with Nrf2 binding and act as KNI disruptors (Padmanabhan et al. 2008). Sequestosome-1, also called p62, has a Keap1 interacting region that helps in the binding of Nrf2. Nrf2, unlike sequestosome, has two binding sites to Keap1. These double binding sites help in the formation of its degradation complex with CUL-E3 ligase adaptors. One of the initial efforts to study KNI was done using prothymosin-α, a nuclear oncoprotein found in proliferating mammalian cells and helps in the prevention of apoptosis.
Mutant Marburg viral proteins determined the importance of a homologous motif present in the solvent-exposed K-loop of the protein domain. The residues Gly211 and Glu212 in the motif are very important for the Keap1 interaction. The Arg415 residue of Keap1 is as important for the binding of the viral protein MVP24 as much as it is important for the binding of Nrf2 (Edwards et al. 2014).
The in vivo studies showed that the proteins like sestrins that could provoke the Nrf2–ARE pathway through the autophagic degradation of Keap1 mediated by p53, thereby protecting the liver from oxidative damage. Sestrin (Sesn2) induction invokes p62, a competitive antagonist of Keap1, which mimics the binding of the ETGE motif of Nrf2 to the kelch domain, but only under overexpressed conditions of Sesn2. Under normal conditions, p62 does not replace Nrf2 binding antagonistically because of the weak binding it projects on Keap1 as compared to the ETGE motif (Bae et al. 2013).
Many peptides have been designed through mutagenesis experiments to detect the important residues involved in binding to the Keap1 protein. Basic structural compositions in the design of peptide inhibitors are summarised as follows (Hancock et al. 2013):
Mutagenesis studies determine the presence of similar seven-residue stretches in both ETGE and DLG motifs for the mandatory binding of the PPi, with aspartate and phenylalanine at the two ends.
The first aspartate acid is irreplaceable, as it helps in the stabilisation of the secondary structure of the peptide. The hairpin conformation of the peptide is important for the proper orientation of the binding residues into the central cylindrical cavity of Keap1.
The presence of glutamate residues in the third and sixth positions of the peptide is important for tethering to the arginine residues through salt bridge interactions.
Glutamate residue on the second position may be replaced with a simpler amino acid that may ensure a more intense bonding of the peptide inhibitor.
Competitive inhibition of the DLG motif has been proposed to be threshold strategy to disrupt the KNI.
Replacement of the N-termini of the peptides with lipid-conjugated amino acids either retained or increased the activity of the peptide. It was hypothesised that the fatty acids formed micelles that provided a larger effective molecular volume that helps in bonding with the aromatics inside the protein cavity.
The presence of acetyl substitution does not give any binding results with the Kelch domain of the protein.
Chemical inhibitors of Keap1
Research has been carried out in extreme zeal towards the discovery of various chemical moieties that mimic the binding of Nrf2 to Keap1 and serve as a therapeutic drug inhibitor. As discussed earlier, the binding of the lead molecules to the Nrf2 binding site or the oxidation of reactive cysteines in the BTB binding pocket of Keap1 is mandatory to disrupt the protein–protein interactions.
Researchers have recently identified various natural KNI disruptors like baicalein, caffeic acid, falcarindiol, sulphoraphane, resveratrol, curcumin and lipoic acid (Table 1). Synthetic compounds and their derivatives have been studied for the protein–peptide disruption activity, many of which crystal structures have been reported. Crystal structure information of protein–ligand complexes provides a real-time insight into the behavioural fingerprint of the KNI disruptors in general. Pharmacophore mapping and quantitative structure–activity relationship (QSAR)-based screening of existing databases could provide potential leads for molecular docking studies. Based on the success in proposed interaction patterns, a real-time study of the complex behaviour could conceptually prepare the lead for biochemical experimentation.
Table 1.
Tabular representation of major inhibitor discoveries for Keap1 domains
| Inhibitor | Domain affected | Reference |
|---|---|---|
| Quercetin | Kelch domain | Tanigawa et al. (2007) |
| Falcarindiol | Cysteine-rich areas of Keap1 (BTB and IVR), especially Cys151 | Ohnuma et al. (2010) |
| Mono- and dimethyl fumarate | Cysteine modification | Linker et al. (2011) |
| WTX (Wilms tumour gene on X chromosome) | Kelch domain; competitive binding | Camp et al. (2012) |
| Sestrins | Rbx1-mediated autophagic degradation of Keap1 | Bae et al. (2013) |
| ML334 | Kelch domain | Hu et al. (2013) |
| Cpd16 | Kelch domain | Marcotte et al. (2013) |
| Synthetic peptide inhibitors | Kelch domain | Hancock et al. (2013) |
| SKI-II, sphingosine kinase inhibitor | Formation of Keap1 dimers | Mercado et al. (2014) |
| Baicalein | Keap1 dependent/independent manner | Qin et al. (2014) |
| Monocyclic, bicyclic and tricyclic ethynylcyanodienones | Undetermined | Li et al. (2015) |
| PF-4708671 (S6K1-specific inhibitor) | p62-induced autophagic degradation of Keap1 | Park et al. (2015) |
The Keap1-targeted inhibition mechanism has been mainly carried out by inhibitors that could interrupt the cross-talk between the former and Nrf2 (Magesh et al. 2012). One of the methodologies of reduction in the Keap1 population, hampering the redox balance, involves the dimerisation of Keap1 to prevent Nrf2 binding. Sulphoraphane and CDDO–imidazole compounds were able to decrease the levels of Keap1 by its homodimerisation (Fig. 5). However, poor selectivity and increased toxicity problems set back the former compounds. Other CDDO analogues that have been tested for conditions other than neurodegeneration involve CDDO-Me for diabetics with chronic kidney disease. Tecfidera, a dimethyl fumarate drug, was recently approved by the U.S. Food and Drug Administration (FDA) for the treatment of multiple sclerosis (Venci and Gandhi 2013). SK-II, a sphingosine kinase inhibitor, also carried out the same process but with a relatively stronger covalent bonding between the homodimers (Mercado et al. 2014). The mechanism through which dimerisation takes place has been hypothesised at the release of endogenous electrophilic compounds resulting in the reduction of the cysteine residues on Keap1 that act as cellular stress sensors. The induction of the arachidonic acid pathway by the inhibitor has been hypothesised in the release of electrophilic fatty acids (EFA), inactivating Keap1.
Polyphenols like curcumin, resveratrol and caffeic acid have been shown to induce Nrf2-dependent antioxidant expression in the body. Curcumin, a yellow compound bioactive in turmeric, and its derivatives were found to possess anti-oxidative and anti-inflammatory properties that could cure cerebral oedema and is mainly helpful in cases of kidney pathologies through activation of the Nrf2–ARE pathway (Yang et al. 2009; Son et al. 2008). Caffeic acid, a phenolic acid, consists of both nucleophilic and an electrophilic moiety in its structure. The electrophilic moiety (Michael acceptor) is the supporting structural advantage that enforces the disruption of KNI (Sirota et al. 2015; Lee et al. 2010). Caffeic acid has also been found to bind potentially with Keap1 at the Nrf2 binding site (Pang et al. 2016). A recent study on structural activity and structure–property relationships of a proposed inhibitor recognised the presence of a symmetric structure with an aromatic ring in the centre that could tether to the hydrophobic binding segment inside the central core of the protein. The symmetric modification of benzene rings at the ends of the structure could increase the potency of the drug (Jiang et al. 2015).
Another natural Nrf2 activator is a carnosic acid which promotes the expression of the antioxidant cocktail in neuronal cells by the disruption of KNI through the induction of electrophilic compounds that is sensed by the reactive cysteines on Keap1. Oxidation of any of the cysteines lying in the BTB–IVR domain region alters the conformation of the protein layout, pulling apart the dimer structure. Molecular in vitro studies have proven that carnosic acid binds with the BTB domain of Keap1, with weak binding to the IVR region as well. Synthetically available compound CDDO/bardoxolone is an important inhibitor of the BTB domain with which the first crystal of the complex was reported (Cleasby et al. 2014) (Fig. 6). Pterisolic acid B, a naturally occurring diterpenoid, was recently discovered and that could inactivate Keap1 by modifying the Cys171 residue of the BTB domain (Dong et al. 2016); the Nrf2 degradation pathway is, thus, affected by this activator and its anhydrous analogue.
A search in the US Clinical Trials website (https://clinicaltrials.gov/) related to Nrf2 and Keap1 showed a few molecules, including sulphoraphane (SFN) (in broccoli sprouts), resveratrol, CXA-10, RTA-408, dimethyl fumarate (DMF) and monomethyl fumarate (MMF). Though the drug molecule bardoxolone methyl was shown to be a promising candidate, the drug was recently withdrawn from phase III clinical trials in end-stage renal disease patients with type II diabetes due to serious adverse effects and mortality among the patients receiving the active drug (Tayek and Kalantar-Zadeh 2013; Zhang 2013; de Zeeuw et al. 2013). The side effects of other synthetic compounds selected for clinical trials are still unknown.
Conclusion
The maintenance of cell redox balance is critical for appropriate functioning. The development of novel peptide and chemical inhibitors of the Keap1–Nrf2 complex would be an effective therapeutic strategy for NDD and cancers. Past research works indicate that interaction-based designing of inhibitors can induce specificity and selectivity of the inhibitor compounds on the binding region on Keap1. Concentration on the physical aspect of the protein–protein interaction may assist in designing a better equipped competitive inhibitor for Keap1. Also, the research focus needs to be shifted to find potential drug binding sites in other domains as well which can trigger Nrf2 activation through the oxidation of reactive cysteines.
Acknowledgments
BP is grateful to the Indian Council of Medical Research (ICMR) (no. 001/101/2015/00787) and Department of Science and Technology, SERB (no. SR/SO/BB-0108/2012), Government of India, India for the financial support.
Abbreviations
- ARE
Antioxidant response element
- AD
Alzheimer’s disease
- Aβ
Amyloid-β
- AGEs
Advanced glycation end products
- ATF4
Activating transcription factor 4
- BRG1
Brahma-related gene 1
- BTB
Broad-complex, tramtrack and bric-à-brac
- CBP
CREB-binding protein
- CDDO
1-[2-Cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]
- CDDO-Me
2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid methyl ester or bardoxolone methyl
- CHD
Chromodomain helicase DNA binding protein 6
- CNC
Cap ‘n’ collar
- CREB
cAMP response element binding protein
- CRL
Cullin-RING ubiquitin ligases
- DJ1
Protein deglycase DJ-1
- DMF
Dimethyl fumarate
- EFA
Electrophilic fatty acids
- FAE
Fumaric acid esters
- FCCP
Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- GCL
Gamma-glutamylcysteine ligase
- GCLM
Glutamate cysteine ligase modifier
- GGT
Gamma-glutamyl transferase
- GCV
Ganciclovir
- G6PD
Glucose-6-phosphate dehydrogenase
- GSH
Glutathione
- GST
Glutathione S-transferase
- HO-1
Heme oxygenase-1
- 6-HAD
6-Hydroxyldopamine
- HD
Huntington’s disease
- ICH
Intracerebral haemorrhage
- IDH1
Isocitrate dehydrogenase 1
- IKKB
Inhibitor of nuclear factor kappa-B kinase subunit beta
- IR
Ischaemia–reperfusion
- IVR
Intervening region
- Keap1
Kelch-like ECH-associated protein 1
- Maf
Musculoaponeurotic fibrosarcoma
- ME1
Malic enzyme 1
- MMF
Monomethyl fumarate
- MTHFD2
Methylenetetrahydrofolate dehydrogenase 2
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NDD
Neurodegenerative diseases
- NEDD8
Neural precursor cell expressed, developmentally downregulated 8
- Neh
Nrf2–ECH homology
- NFT
Neurofibrillary tangles
- NQO1
NAD(P)H:quinone oxidoreductase 1
- Nrf2
Nuclear factor (erythroid-derived 2)-like 2
- PD
Parkinson’s disease
- PGD
Phosphogluconate dehydrogenase
- PHF
Paired helical filaments
- PI3K
Phosphoinositide 3-kinase
- PPAT
Phosphoribosyl pyrophosphate amidotransferase
- QSAR
Quantitative structure–activity relationship
- RBX1
Ring box protein 1
- ROS
Reactive oxygen species
- SAH
Subarachnoid haemorrhage
- SK-II
Sphingosine kinase inhibitor 2
- SN
Substantia nigra
- SNP
Single nucleotide polymorphism
- SOD
Superoxide dismutase
- TALDO1
Transaldolase 1
- TK
Thymidine kinase
- TKT
Transketolase
Compliance with ethical standards
Conflict of interest
Prashant Deshmukh declares that he has no conflicts of interest. Sruthi Unni declares that she has no conflicts of interest. Gopinath Krishnappa declares that he has no conflicts of interest. Balasundaram Padmanabhan declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Prashant Deshmukh and Sruthi Unni contributed equally to this work.
References
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, Stevens M. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in Keap1 succination and Nrf2 signaling. Cancer Cell. 2011;20(4):524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibhatla RM, Hatcher JF. Lipid oxidation and peroxidation in CNS health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2010;12(1):125–169. doi: 10.1089/ars.2009.2668. [DOI] [PubMed] [Google Scholar]
- Albin RL, Reiner A, Anderson KD, Dure LS, Handelin B, Balfour R, Whetsell WO, Penney JB, Young AB. Preferential loss of striato-external pallidal projection neurons in presymptomatic Huntington’s disease. Ann Neurol. 1992;31(4):425–430. doi: 10.1002/ana.410310412. [DOI] [PubMed] [Google Scholar]
- Albrecht P, Bouchachia I, Goebels N, Henke N, Hofstetter HH, Issberner A, Kovacs Z, Lewerenz J, Lisak D, Maher P, Mausberg AK. Effects of dimethyl fumarate on neuroprotection and immunomodulation. J Neuroinflammation. 2012;9(163):2094–2099. doi: 10.1186/1742-2094-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga H, Takahashi-Niki K, Kato I, Maita H, Niki T, Iguchi-Ariga SM. Neuroprotective function of DJ-1 in Parkinson’s disease. Oxidative Med Cell Longev. 2013 doi: 10.1155/2013/683920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronowski J, Zhao X. Molecular pathophysiology of cerebral hemorrhage: secondary brain injury. Stroke. 2011;42(6):1781–1786. doi: 10.1161/STROKEAHA.110.596718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, Rhee SG. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17(1):73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Behl C. Oxidative stress in Alzheimer’s disease: implications for prevention and therapy. New York: Springer; 2005. pp. 65–78. [DOI] [PubMed] [Google Scholar]
- Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, Tagde A, Wong KK, Kufe D. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76(6):1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006;8(11–12):2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- Bryan HK, Olayanju A, Goldring CE, Park BK. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol. 2013;85(6):705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Calkins MJ, Jakel RJ, Johnson DA, Chan K, Kan YW, Johnson JA. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc Natl Acad Sci U S A. 2005;102(1):244–249. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, Tang X, Zheng N, Major MB, Moon RT. Wilms tumor gene on X chromosome (WTX) inhibits degradation of Nrf2 protein through competitive binding to Keap1 protein. J Biol Chem. 2012;287(9):6539–6550. doi: 10.1074/jbc.M111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Marsden BD, Gileadi O. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem. 2013;288(11):7803–7814. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A. 1993;90(23):11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Cheung MC, Moi P, Chan K, Kan YW. Chromosomal localization of the human NF-E2 family of bZIP transcription factors by fluorescence in situ hybridization. Hum Genet. 1995;95(3):265–269. doi: 10.1007/BF00225191. [DOI] [PubMed] [Google Scholar]
- Chauhan N, Chaunsali L, Deshmukh P, Padmanabhan B. Analysis of dimerization of BTB-IVR domains of Keap1 and its interaction with Cul3, by molecular modeling. Bioinformation. 2013;9(9):450. doi: 10.6026/97320630009450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Fang Q, Zhang J, Zhou D, Wang Z. Role of the Nrf2–ARE pathway in early brain injury after experimental subarachnoid hemorrhage. J Neurosci Res. 2011;89(4):515–523. doi: 10.1002/jnr.22577. [DOI] [PubMed] [Google Scholar]
- Cheng IC, Chen YJ, Ku CW, Huang YW, Yang CN. Structural and dynamic characterization of mutated Keap1 for varied affinity toward Nrf2: a molecular dynamics simulation study. J Chem Inf Model. 2015;55(10):2178–2186. doi: 10.1021/acs.jcim.5b00300. [DOI] [PubMed] [Google Scholar]
- Cho H, Hartsock MJ, Xu Z, He M, Duh EJ. Monomethyl fumarate promotes Nrf2-dependent neuroprotection in retinal ischemia–reperfusion. J Neuroinflammation. 2015;12(1):239. doi: 10.1186/s12974-015-0452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleasby A, Yon J, Day PJ, Richardson C, Tickle IJ, Williams PA, Callahan JF, Carr R, Concha N, Kerns JK, Qi H. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6):e98896. doi: 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements CM, McNally RS, Conti BJ, Mak TW, Ting JPY. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JL. Dementia with Lewy bodies: molecular pathogenesis and implications for classification. J Geriatr Psychiatry Neurol. 2004;17:112–119. doi: 10.1177/0891988704267473. [DOI] [PubMed] [Google Scholar]
- Dang J, Brandenburg LO, Rosen C, Fragoulis A, Kipp M, Pufe T, Beyer C, Wruck CJ. Nrf2 expression by neurons, astroglia, and microglia in the cerebral cortical penumbra of ischemic rats. J Mol Neurosci. 2012;46(3):578–584. doi: 10.1007/s12031-011-9645-9. [DOI] [PubMed] [Google Scholar]
- de Zeeuw D, Akizawa T, Audhya P, Bakris GL, Chin M, Christ-Schmidt H, Goldsberry A, Houser M, Krauth M, Lambers Heerspink HJ, McMurray JJ, BECON Trial Investigators Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N Engl J Med. 2013;369:2492–2503. doi: 10.1056/NEJMoa1306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Kenneth HY, Yeo CJ, Calhoun ES, Scrimieri F. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derjuga A, Gourley TS, Holm TM, Heng HH, Shivdasani RA, Ahmed R, Andrews NC, Blank V. Complexity of CNC transcription factors as revealed by gene targeting of the Nrf3 locus. Mol Cell Biol. 2004;24(8):3286–3294. doi: 10.1128/MCB.24.8.3286-3294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Liu W, Shen Z, Li L, Chen S, Lei X. Pterisolic acid B is a Nrf2 activator by targeting C171 within Keap1–BTB domain. Sci Rep. 2016;6:19231. doi: 10.1038/srep19231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep. 2014;6(6):1017–1025. doi: 10.1016/j.celrep.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekharzadeh B, Maghsoudi N, Khodagholi F. Stabilization of transcription factor Nrf2 by tBHQ prevents oxidative stress-induced amyloid beta formation in NT2N neurons. Biochimie. 2010;92(3):245–253. doi: 10.1016/j.biochi.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99(11):7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281(52):39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- Feigin VL, Rinkel GJ, Lawes CM, Algra A, Bennett DA, van Gijn J, Anderson CS. Risk factors for subarachnoid hemorrhage: an updated systematic review of epidemiological studies. Stroke. 2005;36(12):2773–2780. doi: 10.1161/01.STR.0000190838.02954.e8. [DOI] [PubMed] [Google Scholar]
- Filippin LI, Vercelino R, Marroni NP, Xavier RM. Redox signalling and the inflammatory response in rheumatoid arthritis. Clin Exp Immunol. 2008;152(3):415–422. doi: 10.1111/j.1365-2249.2008.03634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm ES, Demopoulos HB, Seligman ML, Poser RG, Ransohoff JO. Free radicals in cerebral ischemia. Stroke. 1978;9(5):445–447. doi: 10.1161/01.STR.9.5.445. [DOI] [PubMed] [Google Scholar]
- Furfaro AL, Macay JRZ, Marengo B, Nitti M, Parodi A, Fenoglio D, Marinari UM, Pronzato MA, Domenicotti C, Traverso N. Resistance of neuroblastoma GI-ME-N cell line to glutathione depletion involves Nrf2 and heme oxygenase-1. Free Radic Biol Med. 2012;52(2):488–496. doi: 10.1016/j.freeradbiomed.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25(1):162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R, Schaap M, Pfister H, Wells G. Peptide inhibitors of the Keap1–Nrf2 protein–protein interaction with improved binding and cellular activity. Org Biomol Chem. 2013;11(21):3553–3557. doi: 10.1039/c3ob40249e. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Takahashi H, Rajabi H, Alam M, Suzuki Y, Yin L, Tagde A, Maeda T, Hiraki M, Sukhatme VP, Kufe D. Functional interactions of the cystine/glutamate antiporter, CD44v and MUC1-C oncoprotein in triple-negative breast cancer cells. Oncotarget. 2016;7(11):11756–11769. doi: 10.18632/oncotarget.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Magesh S, Chen L, Wang L, Lewis TA, Chen Y, Khodier C, Inoyama D, Beamer LJ, Emge TJ, Shen J. Discovery of a small-molecule inhibitor and cellular probe of Keap1–Nrf2 protein–protein interaction. Bioorg Med Chem Lett. 2013;23(10):3039–3043. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel RJ, Kern JT, Johnson DA, Johnson JA. Induction of the protective antioxidant response element pathway by 6-hydroxydopamine in vivo and in vitro. Toxicol Sci. 2005;87(1):176–186. doi: 10.1093/toxsci/kfi241. [DOI] [PubMed] [Google Scholar]
- Jaramillo MC, Zhang DD. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Duong TQ. Methylene blue treatment in experimental ischemic stroke: a mini review. Brain Circ. 2016;2(1):48–53. doi: 10.4103/2394-8108.178548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZY, Chu HX, Xi MY, Yang TT, Jia JM, Huang JJ, Guo XK, Zhang XJ, You QD, Sun HP. Insight into the intermolecular recognition mechanism between Keap1 and IKKβ combining homology modelling, protein–protein docking, molecular dynamics simulations and virtual alanine mutation. PLoS One. 2013;8(9):e75076. doi: 10.1371/journal.pone.0075076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZY, Xu LL, Lu MC, Chen ZY, Yuan ZW, Xu XL, Guo XK, Zhang XJ, Sun HP, You QD. Structure–activity and structure–property relationship and exploratory in vivo evaluation of the nanomolar Keap1–Nrf2 protein–protein interaction inhibitor. J Med Chem. 2015;58(16):6410–6421. doi: 10.1021/acs.jmedchem.5b00185. [DOI] [PubMed] [Google Scholar]
- Jiang S, Deng C, Lv J, Fan C, Hu W, Di S, Yan X, Ma Z, Liang Z, Yang Y (2016) Nrf2 weaves an elaborate network of neuroprotection against stroke. Mol Neurobiol 1–16 [DOI] [PubMed]
- Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1–Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1(1):45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31(1):90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor TO, Fuentes F, Shu L, Paredes-Gonzalez X, Yang AY, Liu Y, Smiraglia DJ, Yegnasubramanian S, Nelson WG, Kong ANT. Epigenetic DNA methylation of antioxidative stress regulator Nrf2 in human prostate cancer. Cancer Prev Res. 2014;7(12):1186–1197. doi: 10.1158/1940-6207.CAPR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI. Suppression of NF-κB signaling by KEAP1 regulation of IKKβ activity through autophagic degradation and inhibition of phosphorylation. Cell Signal. 2010;22(11):1645–1654. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220(4):446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kontos HA. Oxygen radicals in CNS damage. Chem Biol Interact. 1989;72(3):229–255. doi: 10.1016/0009-2797(89)90001-X. [DOI] [PubMed] [Google Scholar]
- Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24(5):1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58(5):262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30(13):3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shin DH, Kim JH, Hong S, Choi D, Kim YJ, Kwak MK, Jung Y. Caffeic acid phenethyl ester-mediated Nrf2 activation and IκB kinase inhibition are involved in NFκB inhibitory effect: structural analysis for NFκB inhibition. Eur J Pharmacol. 2010;643(1):21–28. doi: 10.1016/j.ejphar.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Leinonen HM, Ruotsalainen AK, Määttä AM, Laitinen HM, Kuosmanen SM, Kansanen E, Pikkarainen JT, Lappalainen JP, Samaranayake H, Lesch HP, Kaikkonen MU. Oxidative stress-regulated lentiviral TK/GCV gene therapy for lung cancer treatment. Cancer Res. 2012;72(23):6227–6235. doi: 10.1158/0008-5472.CAN-12-1166. [DOI] [PubMed] [Google Scholar]
- Leirós M, Alonso E, Sanchez JA, Rateb ME, Ebel R, Houssen WE, Jaspars M, Alfonso A, Botana LM. Mitigation of ROS insults by streptomyces secondary metabolites in primary cortical neurons. ACS Chem Neurosci. 2013;5(1):71–80. doi: 10.1021/cn4001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Forman HJ. C‐Myc is a Nrf2‐interacting protein that negatively regulates phase II genes through their electrophile responsive elements. IUBMB Life. 2010;62(3):237–246. doi: 10.1002/iub.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zheng S, Higgins M, Morra RP, Jr, Mendis AT, Chien CW, Ojima I, Mierke DF, Dinkova-Kostova AT, Honda T. New monocyclic, bicyclic, and tricyclic ethynylcyanodienones as activators of the Keap1/Nrf2/ARE pathway and inhibitors of inducible nitric oxide synthase. J Med Chem. 2015;58(11):4738–4748. doi: 10.1021/acs.jmedchem.5b00393. [DOI] [PubMed] [Google Scholar]
- Linker RA, Lee DH, Ryan S, van Dam AM, Conrad R, Bista P, Zeng W, Hronowsky X, Buko A, Chollate S, Ellrichmann G. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134(3):678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1–Nrf2–ARE pathway as potential preventive and therapeutic agents. Med Res Rev. 2012;32(4):687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte D, Zeng W, Hus JC, McKenzie A, Hession C, Jin P, Bergeron C, Lugovskoy A, Enyedy I, Cuervo H, Wang D. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorg Med Chem. 2013;21(14):4011–4019. doi: 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Marcus DL, Thomas C, Rodriguez C, Simberkoff K, Tsai JS, Strafaci JA, Freedman ML. Increased peroxidation and reduced antioxidant enzyme activity in Alzheimer’s disease. Exp Neurol. 1998;150(1):40–44. doi: 10.1006/exnr.1997.6750. [DOI] [PubMed] [Google Scholar]
- Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, Kleeberger SR. Functional polymorphisms in the transcription factor Nrf2 in humans increase the risk of acute lung injury. FASEB J. 2007;21(9):2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem. 2004;279(30):31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- Mercado N, Kizawa Y, Ueda K, Xiong Y, Kimura G, Moses A, Curtis JM, Ito K, Barnes PJ. Activation of transcription factor Nrf2 signalling by the sphingosine kinase inhibitor SKI-II is mediated by the formation of Keap1 dimers. PLoS One. 2014;9(2):e88168. doi: 10.1371/journal.pone.0088168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morito N, Yoh K, Itoh K, Hirayama A, Koyama A, Yamamoto M, Takahashi S. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22(58):9275–9281. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- Mosley RL, Benner EJ, Kadiu I, Thomas M, Boska MD, Hasan K, Laurie C, Gendelman HE. Neuroinflammation, oxidative stress, and the pathogenesis of Parkinson’s disease. Clin Neurosci Res. 2006;6(5):261–281. doi: 10.1016/j.cnr.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294(1):1–12. doi: 10.1016/S0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- Muscarella LA, Parrella P, D’Alessandro V, la Torre A, Barbano R, Fontana A, Tancredi A, Guarnieri V, Balsamo T, Coco M, Copetti M. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics. 2011;6(6):710–719. doi: 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]
- Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25(24):10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niture SK, Jaiswal AK. Hsp90 interaction with INrf2 (Keap1) mediates stress-induced Nrf2 activation. J Biol Chem. 2010;285(47):36865–36875. doi: 10.1074/jbc.M110.175802. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ohnuma T, Nakayama S, Anan E, Nishiyama T, Ogura K, Hiratsuka A. Activation of the Nrf2/ARE pathway via S-alkylation of cysteine 151 in the chemopreventive agent-sensor Keap1 protein by falcarindiol, a conjugated diacetylene compound. Toxicol Appl Pharmacol. 2010;244(1):27–36. doi: 10.1016/j.taap.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Ooi A, Dykema K, Ansari A, Petillo D, Snider J, Kahnoski R, Anema J, Craig D, Carpten J, Teh BT, Furge KA. CUL3 and Nrf2 mutations confer an Nrf2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013;73(7):2044–2051. doi: 10.1158/0008-5472.CAN-12-3227. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer. 2007;121(9):1883–1891. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21(5):689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Nakamura Y, Yokoyama S. Structural analysis of the complex of Keap1 with a prothymosin α peptide. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2008;64(4):233–238. doi: 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang C, Zheng Z, Shi L, Sheng Y, Wei H, Wang Z, Ji L. Caffeic acid prevents acetaminophen-induced liver injury by activating the Keap1–Nrf2 antioxidative defense system. Free Radic Biol Med. 2016;91:236–246. doi: 10.1016/j.freeradbiomed.2015.12.024. [DOI] [PubMed] [Google Scholar]
- Park JS, Kang DH, Bae SH. PF-4708671, a specific inhibitor of p70 ribosomal S6 kinase 1, activates Nrf2 by promoting p62-dependent autophagic degradation of Keap1. Biochem Biophys Res Commun. 2015;466(3):499–504. doi: 10.1016/j.bbrc.2015.09.059. [DOI] [PubMed] [Google Scholar]
- Qin S, Deng F, Wu W, Jiang L, Yamashiro T, Yano S, Hou DX. Baicalein modulates Nrf2/Keap1 system in both Keap1-dependent and Keap1-independent mechanisms. Arch Biochem Biophys. 2014;559:53–61. doi: 10.1016/j.abb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Rajabi H, Tagde A, Alam M, Bouillez A, Pitroda S, Suzuki Y, Kufe D. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016 doi: 10.1038/onc.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in Nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98(6):3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey CP, Glass CA, Montgomery MB, Lindl KA, Ritson GP, Chia LA, Hamilton RL, Chu CT, Jordan-Sciutto KL. Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol. 2007;66(1):75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Fan C, Chen N, Huang J, Yang Q. Resveratrol pretreatment attenuates cerebral ischemic injury by upregulating expression of transcription factor Nrf2 and HO-1 in rats. Neurochem Res. 2011;36(12):2352–2362. doi: 10.1007/s11064-011-0561-8. [DOI] [PubMed] [Google Scholar]
- Riedl AG, Watts PM, Brown CT, Jenner P. P450 and heme oxygenase enzymes in the basal ganglia and their roles in Parkinson’s disease. Adv Neurol. 1999;80:271–286. [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32(1):21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee NH, Zheng N. A ubiquitin-like protein unleashes ubiquitin ligases. Cell. 2008;135(2):209–211. doi: 10.1016/j.cell.2008.09.049. [DOI] [PubMed] [Google Scholar]
- Schipper HM, Bennett DA, Liberman A, Bienias JL, Schneider JA, Kelly J, Arvanitakis Z. Glial heme oxygenase-1 expression in Alzheimer disease and mild cognitive impairment. Neurobiol Aging. 2006;27(2):252–261. doi: 10.1016/j.neurobiolaging.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Sekhar KR, Yan XX, Freeman ML. Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene. 2002;21(44):6829–6834. doi: 10.1038/sj.onc.1205905. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135(4):1358–1368. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1–Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K, Yoshimatsu Y, Tachimori Y, Kushima R, Kiyono T, Yamamoto M. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia. 2011;13(9):864–873. doi: 10.1593/neo.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23(8):3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3(10):e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota R, Gibson D, Kohen R. The role of the catecholic and the electrophilic moieties of caffeic acid in Nrf2/Keap1 pathway activation in ovarian carcinoma cell lines. Redox Biol. 2015;4:48–59. doi: 10.1016/j.redox.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum SL, Kensler TW. Nrf2: control of sensitivity to carcinogens. Arch Toxicol. 2011;85(4):273–284. doi: 10.1007/s00204-011-0675-4. [DOI] [PubMed] [Google Scholar]
- Son TG, Camandola S, Mattson MP. Hormetic dietary phytochemicals. Neuromol Med. 2008;10(4):236–246. doi: 10.1007/s12017-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Privé GG. The BACK domain in BTB-kelch proteins. Trends Biochem Sci. 2004;29(12):634–637. doi: 10.1016/j.tibs.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Sugamura K, Keaney JF., Jr Reactive oxygen species in cardiovascular disease. Free Radic Biol Med. 2011;51(5):978–992. doi: 10.1016/j.freeradbiomed.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Wu T, Zhao F, Lau A, Birch CM, Zhang DD. KPNA6 (Importin α7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response. Mol Cell Biol. 2011;31(9):1800–1811. doi: 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagde A, Singh H, Kang MH, Reynolds CP. The glutathione synthesis inhibitor buthionine sulfoximine synergistically enhanced melphalan activity against preclinical models of multiple myeloma. Blood Cancer J. 2014;4:e229. doi: 10.1038/bcj.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, Tai YT, Hideshima T, Anderson K, Avigan D, Kufe D. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127(21):2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagde A, Rajabi H, Stroopinsky D, Gali R, Alam M, Bouillez A, Kharbanda S, Stone R, Avigan D, Kufe D (2016b) MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget. doi:10.18632/oncotarget.9777 [DOI] [PMC free article] [PubMed]
- Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, Raina D, Hasegawa M, Suzuki Y, Tagde A, Bronson RT. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34(40):5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigawa S, Fujii M, Hou DX. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med. 2007;42(11):1690–1703. doi: 10.1016/j.freeradbiomed.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Tayek JA, Kalantar-Zadeh K. The extinguished BEACON of bardoxolone: not a Monday morning quarterback story. Am J Nephrol. 2013;37(3):208–211. doi: 10.1159/000346950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27(21):7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552(2):335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venci JV, Gandhi MA. Dimethyl fumarate (Tecfidera): a new oral agent for multiple sclerosis. Ann Pharmacother. 2013;47(12):1697–1702. doi: 10.1177/1060028013509232. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington’s disease. Lancet. 2007;369(9557):218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373(1):151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wang W, Kang J, Li H, Su J, Wu J, Xu Y, Yu H, Xiang X, Yi H, Lu Y, Sun L. Regulation of endoplasmic reticulum stress in rat cortex by p62/ZIP through the Keap1–Nrf2–ARE signalling pathway after transient focal cerebral ischaemia. Brain Inj. 2013;27(7–8):924–933. doi: 10.3109/02699052.2013.793397. [DOI] [PubMed] [Google Scholar]
- Xu H, Zhou YL, Zhang XY, Lu P, Li GS. Activation of PERK signaling through fluoride-mediated endoplasmic reticulum stress in OS732 cells. Toxicology. 2010;277(1):1–5. doi: 10.1016/j.tox.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tanji K, Wakabayashi K, Matsuura S, Itoh K. Role of the Keap1/Nrf2 pathway in neurodegenerative diseases. Pathol Int. 2015;65(5):210–219. doi: 10.1111/pin.12261. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhang X, Fan H, Liu Y. Curcumin upregulates transcription factor Nrf2, HO-1 expression and protects rat brains against focal ischemia. Brain Res. 2009;1282:133–141. doi: 10.1016/j.brainres.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Yu X, Kensler T. Nrf2 as a target for cancer chemoprevention. Mutat Res. 2005;591(1–2):93–102. doi: 10.1016/j.mrfmmm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, Foster BA, Kan YW, Kong AN. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One. 2010;5(1):e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD. Bardoxolone brings Nrf2-based therapies to light. Antioxid Redox Signal. 2013;19(5):517–518. doi: 10.1089/ars.2012.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, An C, Gao Y, Leak RK, Chen J, Zhang F. Emerging roles of Nrf2 and phase II antioxidant enzymes in neuroprotection. Prog Neurobiol. 2013;100:30–47. doi: 10.1016/j.pneurobio.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sun G, Zhang J, Strong R, Dash PK, Kan YW, Grotta JC, Aronowski J. Transcription factor Nrf2 protects the brain from damage produced by intracerebral haemorrhage. Stroke. 2007;38(12):3280–3286. doi: 10.1161/STROKEAHA.107.486506. [DOI] [PubMed] [Google Scholar]
- Zhuang C, Narayanapillai S, Zhang W, Sham YY, Xing C. Rapid identification of Keap1–Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J Med Chem. 2014;57(3):1121–1126. doi: 10.1021/jm4017174. [DOI] [PubMed] [Google Scholar]
- Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277(39):36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]