Abstract
Traditional DNA-based typing focuses primarily on interrogating the exons of HLA genes that form the antigen recognition domain (ARD). The relevance of mismatching donor and recipient for HLA variation outside the ARD on hematopoietic stem cell transplantation (HCT) outcomes is unknown. This study was designed to evaluate the frequency of variation outside the ARD in 10/10 (HLA-A, -B, -C, -DRB1, -DQB1) matched unrelated donor transplant pairs (n=360). Next generation DNA sequencing was used to characterize both HLA exons and introns for HLA-A, -B, -C alleles; exons 2, 3 and the intervening intron for HLA-DRB1; and exons only for HLA-DQA1 and -DQB1. Over 97% of alleles at each locus were matched for their nucleotide sequence outside of the ARD exons. Of the 4320 allele comparisons overall, only 17 allele pairs were mismatched for non-ARD exons, 41 for noncoding regions and 9 for ARD exons. The observed variation between donor and recipient usually involved a single nucleotide difference (88% of mismatches); 88% of the non-ARD exon variants impacted the amino acid sequence. The impact of amino acid sequence variation caused by substitutions in exons outside ARD regions in D-R pairs will be difficult to assess in HCT outcome studies since these mismatches do not occur very frequently.
Keywords: Hematopoietic stem cell transplantation, Histocompatibility testing, Human leukocyte antigens, Polymorphism, genetic, Sequence analysis, DNA
Introduction
In hematopoietic stem cell transplantation (HCT), the optimal match for unrelated donor and recipient is based on HLA-A, -B, -C, -DRB1, and sometimes –DQB1, since these loci strongly impact survival [1–4]. More recently, selection of a permissive HLA-DPB1 T cell epitope mismatch has been added to the optimal matching criteria [2,5]. Matching uses high resolution typing assignments based on the sequences of exons encoding the HLA molecule’s antigen recognition domain (ARD). The ARD is the region of the HLA protein that binds an antigenic peptide and interacts with the antigen receptors of T lymphocytes and the natural killer cell immunoglobulin-like receptors. It includes the α1 and α2 domains of class I proteins, specified by exons 2 and 3, and the α1 and β1 domains of the class II proteins, specified by exon 2 of the class II HLA genes. Alleles sharing the nucleotide sequence in the ARD exons form the G allele groups; alleles encoding HLA proteins sharing their polypeptide sequences in this region form P groups [6].
While the majority of HLA allelic polymorphism alters the DNA sequence in the ARD-encoding exons, additional polymorphisms have been observed outside of this region in random individuals. For example, based on IPD-IMGT/HLA 3.23.0, the HLA assignment, A*02:01:01G, includes 52 alleles that share the nucleotide sequence of exons 2 and 3 but that differ in the DNA sequence of exon 1, exons 4–8 or the noncoding regions (introns, 5′ and 3′ untranslated regions (UTR)). The majority of these alleles (63%) encode different amino acid sequences (e.g., A*02:09, A*02:66) from that encoded by the allele giving the G group its name (A*02:01:01:01) or cause variation in the expression of the protein (e.g., A*02:01:01:02L, A*02:43N). The remaining alleles included in A*02:01:01G encode synonymous substitutions in exons other than exons 2 and 3 (e.g., A*02:01:15) or in noncoding regions (e.g., A*02:01:01:03). Since typing has focused on the ARD exons, little is known about the frequency of alleles within a G group.
While variation within a G group outside of the ARD exons resulting in the loss of HLA expression clearly will affect allorecognition, the impact of amino acid substitutions outside the ARD on the ability of T lymphocytes to recognize this variation is poorly understood. For example, DRB1*14:01 and DRB1*14:54 differ by a single amino acid substitution (Y112H) in the second extracellular domain of the beta chain. Based on a crystal structure of HLA-DR, substitution of Y112 (DRB1*14:01) may alter an intra-molecular salt bridge between H112 and E162 near the cell surface but does not appear to be in a position to impact the conformation of the antigen-binding site or the interaction with the T-cell receptor [7]. A retrospective analysis of the impact of mismatches between DRB1*14:01 and DRB1*14:54 in bone marrow transplant pairs found no evidence that outcome was impacted although the sample size was small [8]. In vitro studies of stimulation between DRB1*14:01 and DRB1*14:54 expressing cells showed weak alloreactivity but in only a single direction (Roelen et al., manuscript in preparation). For class I, the peptide binding motifs of B*44:02 and B*44:27 which differ in the α3 domain (V199A) and 3 other amino acids in transmembrane and cytoplasmic regions, are indistinguishable [9] and cellular assays found no CTL stimulation in a mixed lymphocyte reaction between cells carrying the two alleles [10]. Other allele pairs that differ only in exons outside ARD-encoding exons vary in the transmembrane or cytoplasmic regions, hidden from direct interaction with the TCR, while variation in the HLA leader peptide is not present in the mature protein. Thus the consequences of mismatches from this non-ARD variation are likely to be minimal but the data supporting this conclusion are very limited.
Variation in the noncoding regions between donor and recipient would not be expected to contribute to an alloreactive mismatch unless the variation altered the expression of the HLA protein. Intronic regions controlling HLA expression include sequences at the 5′ and 3′ ends of introns involved in mRNA splicing; for example, in DRB4*01:03:01:02N, this splicing motif is altered eliminating RNA expression [11]. HLA expression is also impacted by 5′ and 3′ untranslated regions containing, for example, promoter and microRNA binding sites [12–14]. Variation in these sites has been observed, for example, among HLA-A [15] and among HLA-C alleles [16], altering expression levels. However, limited information exists on variation in noncoding regions among alleles that share the sequence of their exons. For example, A*02:01:01:02L, A*02:01:01:04, and A*02:01:01:06 differ from A*02:01:01:01 by only 1 or 2 nucleotide substitutions in their noncoding regions, one impacting a 5′ regulatory region enhancer resulting in reduced expression [17]. Next generation DNA sequencing provides a tool to characterize the frequency and potential impact of intronic variation within the HLA gene system; however, evaluation of the impact of variation that does not lie in known regulatory regions will require additional assays of gene expression or protein function.
In order to determine the feasibility of studies to measure the impact of mismatches outside the ARD on the clinical outcome of HCT, it is essential to assess the frequency with which these non-ARD mismatches occur. The purpose of this study was to characterize the HLA diversity found in regions outside the ARD-encoding exons in donors (D) and recipients (R) matched at high resolution for five loci, HLA-A, -B, -C, -DRB1, and -DQB1 (i.e., 10/10 match).
Materials and Methods
HLA-A, -B, -C, -DRB1, and -DQB1 high resolution matched D - R pairs (n=360) were selected based on availability of retrospective HLA typing performed by Sanger-based sequencing and DNA from the CIBMTR Repository. These transplantations, carried out by 91 transplant centers, were facilitated by the National Marrow Donor Program (NMDP)/Be The Match® between the years 2000 and 2015. The majority of this population self-identified as Caucasian (82%) (Table 1). Each pair was originally HLA typed by an NMDP-affiliated transplant center during unrelated donor selection using their local typing strategy and matching criteria. Cells from donor and recipient were submitted to the NMDP repository and the HLA assignments and match status were confirmed by retrospective Sanger sequencing of the exons encoding the antigen recognition domains of HLA-A, -B, -C, -DRB1, -DQB1 and –DPB1 [18].
Table 1.
Race/ethnicity of donors and recipients (n=720)
| Race/Ethnicity | Donor Percent | Recipient Percent |
|---|---|---|
| African American | 3 | 4 |
| Asian Pacific Islander | 3 | 3 |
| Caucasian | 76 | 88 |
| Hispanic | 4 | 4 |
| Native American | 0 | 0 |
| Decline/Unknown | 14 | 1 |
Long-range amplification of HLA loci was performed in separate polymerase chain reactions using DNA from EBV-transformed cell lines. PCR primers are described in Supplemental Table 1. Amplicons from one individual were then pooled and sheared to an average size of 400 bp by sonication (Covaris LE220 focused ultrasonicator). A library was constructed using Illumina’s TruSeq Nano kit and the DNA fragments tagged with one unique index combination. Libraries from 96 individuals were combined and sequenced simultaneously in a single 500 cycle (V2 or V3) paired-end run using an Illumina MiSeq. Data analysis used the Conexio Genomics Assign™ MPS software (version 1.0.0.792; IPD-IMGT/HLA database 3.17.0.1). Although several thousand paired reads are generated for each locus, for Assign alignment, reads are selected based on fragment size, diversity and broad quality segregation (reads are placed in bins with a mean quality score of 1–10, 11–21, 21–30, 31–40 and 41–50). Reads are accumulated until there are at least 100 forward reads and 100 reverse reads covering each position in the analysis. The minimal cut-off used for average read depth used for allele assignment in this study was 100 for each locus although most loci had an average read depth between 200–300. The Assign software generates the initial consensus assembly without reference to the list of known alleles so there is no inherent bias against calling novel mutations in the calling algorithm. Class I sequence analysis evaluated nucleotides from the first nucleotide of start codon to the last nucleotide of the termination codon with the exception of 52 nucleotides at the 3′ end of intron 2 for HLA-B and –C which were not evaluated. Exons 2 and 3 and intron 2 (excluding the 5′ 150 intron nucleotides) were evaluated for DRB1. Exons 1–3 and the first half of exon 4 (exon containing the termination codon) were evaluated for DQA1 and exons 2 – 6 (exon containing the termination codon) for DQB1. Regions not evaluated contain repeated sequence motifs which made alignment of reads difficult and/or characteristics that challenge the sequencing chemistry producing high nonspecific backgrounds. These regions were excluded because their sequences were not reliable. Next generation sequencing (NGS) assignments for all loci were compared to prior HLA assignments obtained by retrospective Sanger DNA-based sequencing with the exception of DQA1 which was not previously characterized. DQ introns were not analyzed because they contain many repeated motifs and the two alleles in an individual may differ dramatically by large insertions or deletions, sometimes involving these repeated motifs, making accurate assessment of the sequence difficult without further assays. Differences between D and R were flagged. Exon and intron variants between D and R were confirmed by repeating the PCR amplification and sequencing the locus in isolation using NGS and/or by Sanger-based sequencing [20]. New alleles have been submitted to GenBank and IPD-IMGT/HLA for allele assignment. Supplemental Table 2 lists novel alleles that were shared by donor and recipient and that are not listed in other tables.
Association between allele mismatch and ancestry match was evaluated using the Chi-square test. A log-rank test was used to determine the sample size needed to assess the impact on outcome of non-ARD variation.
Results
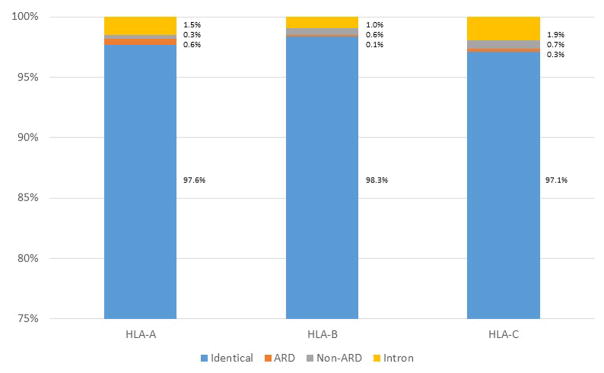
The 360 10/10 high resolution matched D - R pairs allowed the comparison of 720 allele pairs for each of six loci, HLA-A, -B, -C, -DRB1, -DQA1 and -DQB1. Since most (78%) pairs were mismatched for HLA-DP as identified by the prior Sanger sequencing, the DP assignments were not evaluated. NGS sequencing determined that the majority of the HLA-A, -B, -C allele pairs were matched for sequences outside the ARD exons including both exons and introns: 97.6% of the 720 allele pairs were matched for HLA-A; 98.3%, HLA-B; 97.1%, HLA-C (Table 2, Figure 1). Of the approximately one third of the large DRB1 gene that was sequenced, 98.5% of the 720 allele pairs were matched. DQ matching was only evaluated for the exon sequences; these loci were also well matched in the 720 comparisons: DQA1, 99.3% and DQB1, 99.9%.
Table 2.
Summary of mismatches between D – R alleles
| HLA Locus | Identical No. Allele Pairs |
D – R Differ No. Allele Pairs |
||||
|---|---|---|---|---|---|---|
| ARD exon | Non-ARD exon | Noncoding region | ||||
| Nonsynonymous | Synonymous | Nonsynonymous | Synonymous | |||
| A | 703 | 0 | 4 | 2 | 0 | 11 |
| B | 708 | 1 | 0 | 3 | 1 | 7 |
| C | 699 | 0 | 2 | 4 | 1 | 14 |
| DRB1 | 709 | 0 | 0 | 2 | 0 | 9 |
| DQA1 | 715 | 1 | 0 | 4 | 0 | Not evaluated |
| DQB1 | 719 | 1 | 0 | 0 | 0 | Not evaluated |
| Total | 4253 | 3 | 6 | 15 | 2 | 41 |
Figure 1.
Summary of HLA class I matching between unrelated donor and recipient by locus. The four categories include: (1) donor and recipient carry identical alleles (exons and introns); (2) donor and recipient exhibit a difference in the exons encoding the ARD; (3) donor and recipient exhibit a difference in the non-ARD encoding exons; and (4) donor and recipient exhibit a difference in an intron. Each bar chart represents 720 allele comparisons.
Of the 67 cases where D and R were mismatched for a specific allele pair, there was no association with pairs that were mismatched for self-identified race/ethnicity (p=0.87). Four allele mismatches had known differences (multiple race vs Caucasian or Caucasian vs Hispanic), 15 involved Caucasian and unknown race and 48 involved D and R matched for race/ethnicity. Three individuals (two Caucasian vs unknown race; one both Caucasian) had two allele pairs mismatched (A+C and B+DRB1).
Seventeen allele pairs of the 4320 evaluated (0.4%) differed in non-ARD encoding exons; 15 of these differed by nonsynonymous substitutions (Table 3). These nonsynonymous substitutions included three pairs that differed for non-expressed alleles: B*51:11N and C*04:09N (observed twice). Eleven of the alternative alleles, including the two non-expressed alleles, were common or well-documented [21]; three were novel. The remaining three alleles, A*01:37, B*27:13 and DQA1*04:02, are not considered common or well documented. [These latter alleles are not detected by many high resolution typing methods in current use and so may be found more frequently when allele level resolution testing is applied.] One of the common and well documented null alleles was identified as a mismatch by the transplant center so this 9/10 matched pair should not have been included in the sample set. Review of the transplant center typing of the remaining two null alleles showed that one was in an unresolved ambiguous typing (C*04:01 or C*04:09N) and one (B*51:11N) was mis-assigned as the expressed allele (Supplemental Table 3).
Table 3.
Mismatches between donor and recipient observed in the non-ARD-encoding exons
| Allele 1 | Allele 2 | No. Observed | Location of Variation Exon/Codon AA Substitution/Location in Proteina |
CWD Status of Allele 2b |
|---|---|---|---|---|
| A*01:01:01:01 | A*01:37 | 1 | Exon 4/T228M/α3 | Not CWD, confirmed by Sanger |
| A*03:01:01:01 | A*03:26 | 1 | Exon 4/K268E/α3 | WD, confirmed by Sanger |
| B*27:05:02 | B*27:13 | 1 | Exon 1/A-20E/leader | Not CWD; GenBank: KX686520 (*27:13) |
| B*40:01:02 | B*40:01var GenBank: KX686514 |
1 | Exon 4/265 synonymous | Novel, confirmed by Sanger |
| B*44:02:01:03 | B*44:27:01 | 1 | Exons 4,5,7/V199A, V282I, A305T, C325S/α3, TM, Cyt | Common |
| B*51:01:01 | B*51:11N | 1 | Exon 4/185 insertion/not expressed | WD |
| C*03:03:01 | C*03:03var GenBank: KX686519 |
1 | Exon 5/P276L/TM | Novel |
| C*04:01:01 | C*04:09N | 2 | Exon 7/341 deletion/not expressed | Common |
| C*04:01:01 | C*04:01var GenBank: KX686516 |
1 | Exon 1/−5 Synonymous | Novel |
| C*07:01:01:01 | C*07:18 | 1 | Exon 6/A324V/cyt | Common |
| DRB1*14:01:01 | DRB1*14:54:01 | 2 | Exon 3/Y112H/β2 | Common |
| DQA1*03:01:01 | DQA1*03:03:01 | 2 | Exon 3/A160D/α2 | Common |
| DQA1*04:01:01 | DQA1*04:02 | 1 | Exon 3/T138I/α2 | Not CWD |
| DQA1*05:05:01 | DQA1*05:09 | 1 | Exon 1/E1K/α1 | WD |
TM, transmembrane; Cyt, cytoplasmic tail
[19]; CWD, common or well documented
Forty one (0.9%) allele pairs differed only in intronic regions; DQ was not evaluated. The majority (85%) of the allele pairs differed for only a single apparently random nucleotide substitution in one of the several introns of the HLA gene (Table 4).
Table 4.
Mismatches between donor and recipient observed only in the noncoding regionsa
| Allele 1 | Allele 2 | No. Observed | Variationb |
|---|---|---|---|
| A*01:01:01:01 | A*01:01:01:04 GenBank: KX707637 |
1 | Intron 3 1329 G>C |
| A*01:01:01:01 | A*01:01:01var GenBank: KX707636 |
1 | Intron 7 2769 T>C |
| A*02:01:01:01 | A*02:01:01:10 GenBank: KX707632 |
1 | Intron 2 542 A>C |
| A*02:01:01:01 | A*02:01:01:05 | 1 | Intron 3 with insertion at 1407 nucleotide 9 T>C |
| A*03:01:01:01 | A*03:01:01:10 GenBank: KX707635 |
1 | Intron 2 655 C>T |
| A*03:01:01:01 | A*03:01:01:09 GenBank: KX707638 |
1 | Intron 5 2150 T>A |
| A*03:01:01:01 | A*03:01:01:05 | 1 | Intron 6 2606 C>T |
| A*24:02:01:01 | 24:02:01:05 | 2 | Intron 3 1384 A>G |
| A*68:01:02:01 | A*68:01:02:02 | 2 | Intron 7 2770 G>A |
| B*08:01:01:01 | B*08:01:01:03 GenBank: KX707641 |
1 | Intron 3 1037 C>T |
| B*15:01:01:01 | B*15:01:01:06 GenBank: KX707642 |
2 | Intron 5 2324 G>T |
| B*15:01:01:01 | B*15:01:01:05 GenBank: KX707644 |
1 | Intron 6 2534 G>A, 2541 T>C |
| B*35:03:01 | B*35:03:01:02 GenBank: KX707640 |
1 | Intron 4 1915 C>T |
| B*44:02:01:03 | B*44:02:01:05 GenBank: KX707643 |
1 | Intron 3 1126 T>G |
| B*44:02:01:01 | B*44:02:01:04 GenBank: KX707639 |
1 | Intron 6 2560 A>G |
| C*02:02:02:01 | C*02:02:02:04 GenBank: KX711699 |
1 | Intron 2 639 A>G |
| C*03:04:01:01 | C*03:04:01:04 GenBank: KX711698 |
1 | Intron 6 2675 C>G |
| C*04:01:01:01 | C*04:01:01:06 | 1 | Intron 5 2206 A>G |
| C*04:01:01:01 | C*04:01:01:05 GenBank: KX711700 |
6 | Intron 1 152 G>A |
| C*04:01:01:06 | C*04:01:01:05 | 1 | Intron 1 152 G>A, intron 5 2206 G>A |
| C*05:01:01:02 | C*05:01:01:04 GenBank: KX711701 |
1 | Intron 3 1179 A>C, 1183 C>T, 1184 T>C |
| C*05:01:01:02 | C*05:01:01:05 GenBank: KX711703 |
1 | Intron 3 1310 G>A, 1321 T>C, 1323 A>G, 1353 T>G, 1355 G>A |
| C*07:01:01:01 | C*07:01:01:06 GenBank: KX711697 |
1 | Intron 5 2275 A>G |
| C*07:01:01:01 | C*07:01:01:07 GenBank: KX711702 |
1 | Intron 5 2322 G>A |
| DRB1*03:01:01 | DRB1*03:01:01var GenBank: KX756065 |
1 | Intron 2 5881–5883 GAC>ACT |
| DRB1*03:01:01 | DRB1*03:01:01var GenBank: KX756064 |
3 | Intron 2 6386 C>A |
| DRB1*07:01:01 | DRB1*07:01:01var GenBank: KX756066 |
1 | Intron 2 7298 T>C |
| DRB1*13:02:01 | DRB1*13:02:01var GenBank: KX756061 |
1 | Intron 1 5186 C>T |
| DRB1*14:54:01var1 GenBank: KX756060 |
DRB1*14:54:01var2 GenBank: KX756062 |
1 | Intron 2 6250 G>A; both alleles have 7172 intron 2 G>A variationc |
| DRB1*15:01:01:01 (02/03/04) | DRB1*15:01:01:01 (02/03/04)var1 GenBank: KX756063 |
1 | Intron 2 7380 A>C (var 1) |
| DRB1*15:01:01:01 (02/03/04)var1 | DRB1*15:01:01:01 (02/03/04)var2 GenBank: KX774264 |
1 | Intron 2 6237 A>G |
All variation has been confirmed by NGS sequencing of two independent PCR amplifications with the second sequence of the variant locus obtained in the absence of other HLA amplicons from that individual. It is still possible, however, that some variants result from artifacts such as the presence of reads from other co-amplified loci or alleles. For example, C*05:01:01var with 5 nucleotide changes shares this region with the second allele in the heterozygote, C*07:01:01:01, but the reads include C*05-specific residues at 5′ and 3′ ends.
Numbering is based on IMGT genomic sequence alignments with the reference allele (e.g., B*07:02:01 for HLA-B alignments) [26].
All DRB1*14:54 alleles sequenced in this study (22 alleles) carry intron 2 7172 G>A variation defining DRB1*14:54var1.
Although presumably matched at high resolution using retrospective Sanger sequencing, nine allele pairs (0.2%) differed within ARD exons (Table 5, Supplemental Table 3). Three of these pairs differed for a nonsynonymous substitution: (1) A DQA1 allele pair that was not typed previously; (2) The inadvertent inclusion of a 9/10 matched D – R pair in the sample set (HLA-B mismatch); and (3) A mis-assignment by the transplant center and retrospective typing laboratory (DQB1*06:79:01). It was not possible to determine if the transplant center was aware of the synonymous substitutions that differed between donor and recipient because the transplant center assignments were reported using only two fields of HLA nomenclature.
Table 5.
Mismatches between donor and recipient observed in the ARD-encoding exons
| Mismatch in Pair | No. Observed | Location of Variation Exon/Codon AA Substitution/Location in Protein |
Comments |
|---|---|---|---|
| A*03:01:02 vs A*03:01:01:01 | 1 | Exon 3/156 synonymous | Confirmed by Sanger; GenBank: KX686515 (*03:01:02) |
| A*68:01:02:01 vs A*68:01:01:02 A*68:01:02:02 vs A*68:01:01:02 A*68:01:02:02var vs A*68:01:01:02 |
3 | Exon 2/10 synonymous | Var: intron variation in allele |
| B*35:12:01 vs B*35:03:01 | 1 | Exon 3/V99L, N114D, Y116F/α2 | 9/10 match mistakenly in sample set; prior typing consistent with NGS |
| C*03:04:53 vs C*03:04:01 | 1 | Exon 2/2 synonymous | Confirmed by Sanger; GenBank: KX686518 |
| C*04:01:10 vs C*04:01:01:06 | 1 | Exon 2/42 synonymous | Confirmed by Sanger |
| DQA1*01:14 vs DQA1*01:03:01 | 1 | Exon 2/F15L/α1 | Not typed previously; confirmed by Sanger; GenBank:KX686517 |
| DQB1*06:79:01 vs. DQB1*06:02:01 | 1 | Exon 2/V38A/β1 | Missed in prior typing; confirmed by Sanger; GenBank: KX686513 (*06:79) |
Discussion
This is the first study to evaluate the genetic variation and characterize mismatching outside of the ARD in a cohort of HLA-matched unrelated D - R pairs. The paucity of exonic mismatches outside of the ARD is striking. Over 98% of the allele comparisons were matched for all exon sequences. Limited variation between D and R was observed in all the loci except DQB1. The frequency of non-ARD exon variants will be determined as more individuals are tested with sequencing methods that characterize the entire gene.
The majority (15/17) of the nucleotide variation in the non-ARD exons resulted in single amino acid substitutions or loss of protein expression. The largest protein alteration was B*44:02:01:03 vs B*44:27:01 which altered four amino acids. This variation has already been shown to not impact the peptide binding repertoire or stimulate alloreactive T cells [9,10]. Based on this observation and on the location of some variants in transmembrane, cytoplasmic or leader sequences, it is anticipated that the impact of the remaining amino acid variation on allorecognition will be negligible although further functional studies are needed to confirm this.
Most variation observed between allele pairs was intronic variation consisting primarily of single nucleotide substitutions located in various introns. HLA-C exhibited the most variation but all loci showed similar levels (DQA1 and DQB1 not evaluated). While most of the intron variation seems to be randomly located, the substitution in C*03:04:01:04 at the 3′ end of intron 6 is near the mRNA splice junction but does not impact the motif [22]. The remaining variation does not appear to contribute to an alloreactive mismatch. Introns might be expected to experience less selective pressure compared to protein-encoding exons and therefore might exhibit more sequence variation; however, selective pressure may also conserve noncoding regions that control gene expression or impact sites in the vicinity of selected exon variants [22–24]. The data presented here suggest that the introns will be highly conserved; further data on noncoding sequences will be needed to support this observation.
Although DQA1 was not typed at the time of transplant, the tight linkage between DQA1 and DQB1 predicts that, if DQB1 is matched, DQA1 will also be matched [25]. This was found to be true in the majority of pairs where 99.2% of the DQA1 alleles were matched.
One explanation for the high degree of matching outside the ARD-encoding exons is that the transplant centers were using typing methods that characterized additional exons and selecting donors matched at this higher than G-level resolution. Unfortunately, the typing strategies of each of the 91 transplant centers are not available and the assignments provided by the centers to NMDP may have been truncated or incomplete (e.g., only the most “common” assignment listed) so it is not possible to address this possibility directly. Indirect evidence supporting a majority of typing at ARD resolution is the transplant center typing of DRB1*14:01 vs. DRB1*14:54 where the high frequency of the latter allele differing in exon 3 generated an early interest in matching outside of exon 2 [7,8]. Of the 12 D-R pairs that included DRB1*14:01 or DRB1*14:54, only 1 pair showed transplant center typing for both D and R at the level of DRB1*14:54 (data not shown). The remainder included assignments that either did not resolve the two alleles or listed only DRB1*14:01 even though the individuals actually carried DRB1*14:54. This modest probe into typing resolution coupled with the fact that 60% of the pairs were typed between 2000 and 2010, suggests that the majority of assignments were made based on ARD-encoding exons only.
The impact of amino acid sequence variation caused by substitutions in exons outside ARD regions in D-R pairs will be difficult to assess in HCT outcome studies since these mismatches do not occur very frequently. Assuming that exonic variation outside the ARD has the same impact as variation inside the ARD, a mismatch at HLA-A, -B, -C or -DRB1 would lead to approximately a 10% decrease in 5 year overall survival (50% vs. 60%) [1]. A total of 11 non-ARD exonic mismatches involving HLA-A, -B, -C or -DRB1 were observed in the current cohort for a frequency of 3%. Based on a log-rank test for 80% power at a significance level of p=0.05 or 0.01, the required sample size to detect a difference would be approximately 5,916 and 8,806, respectively.
It is encouraging that the high resolution HLA typing carried out to date was able to identify a match over all exons in over 98% of the donor recipient pairs. At present, it does not appear to be necessary to increase the resolution of HLA typing beyond the ARD in selecting a matched unrelated donor except in cases of common non-expressed variants, like C*04:09N, within G-group assignments. The advantage that next generation sequencing does offer, however, is an unambiguous allele assignment for the majority of samples. Because these D – R pairs were matched for 10/10, this likely explains the very conserved nature of their gene sequences. Mismatched pairs that carry haplotypes that have been separated by past recombinational events may exhibit more variation. Further study is warranted to confirm these findings in larger and more diverse cohorts.
Supplementary Material
Acknowledgments
This study was supported by Office of Naval Research grants N00014-14-1-0209 and N00014-15-1-0052 to Georgetown University. The Office of Naval Research did not participate in the design or conduct of the study. The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement 5U24-CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U10HL069294 from NHLBI and NCI; a contract HHSH250201200016C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-14-1-0028 and N00014-15-1-0848 from the Office of Naval Research. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
Footnotes
Author contributions:
Lihua Hou: Developed the NGS typing method, performed the DNA sequencing
Cynthia Vierra-Green: Selected the donor and recipient samples, analyzed the data
Ana Lazaro: Submitted the sequences to IPD-IMGT/HLA for allele assignment
Colleen Brady: Prepared data, reviewed the manuscript
Michael Haagenson: Prepared demographic data and reviewed the manuscript
Stephen Spellman: Designed study, wrote the manuscript
Carolyn Katovich Hurley: Developed the NGS typing method, analyzed the data, wrote the manuscript
Conflicts of interest: Georgetown University has filed a patent application on which coauthor Hurley is an inventor of the HLA typing and Sanger-based sequencing technology.
References
- 1.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, Fernandez-Vina M, Flomenberg N, Horowitz M, Hurley CK, Noreen H, Oudshoorn M, Petersdorf E, Setterholm M, Spellman S, Weisdorf D, Williams TM, Anasetti C. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 2.Pidala J, Lee SJ, Ahn KW, Spellman S, Wang HL, Aljurf M, Askar M, Dehn J, Fernandez VM, Gratwohl A, Gupta V, Hanna R, Horowitz MM, Hurley CK, Inamoto Y, Kassim AA, Nishihori T, Mueller C, Oudshoorn M, Petersdorf EW, Prasad V, Robinson J, Saber W, Schultz KR, Shaw B, Storek J, Wood WA, Woolfrey AE, Anasetti C. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124:2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furst D, Muller C, Vucinic V, Bunjes D, Herr W, Gramatzki M, Schwerdtfeger R, Arnold R, Einsele H, Wulf G, Pfreundschuh M, Glass B, Schrezenmeier H, Schwarz K, Mytilineos J. High-resolution HLA matching in hematopoietic stem cell transplantation: a retrospective collaborative analysis. Blood. 2013;122:3220–3229. doi: 10.1182/blood-2013-02-482547. [DOI] [PubMed] [Google Scholar]
- 4.Morishima Y, Kashiwase K, Matsuo K, Azuma F, Morishima S, Onizuka M, Yabe T, Murata M, Doki N, Eto T, Mori T, Miyamura K, Sao H, Ichinohe T, Saji H, Kato S, Atsuta Y, Kawa K, Kodera Y, Sasazuki T. Biological significance of HLA locus matching in unrelated donor bone marrow transplantation. Blood. 2015;125:1189–1197. doi: 10.1182/blood-2014-10-604785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD, Dubois V, Horowitz MM, Madrigal JA, Morishima Y, Oudshoorn M, Ringden O, Spellman S, Velardi A, Zino E, Petersdorf EW. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Muller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard A, Terasaki PI, Tiercy JM, Trowsdale J. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Lazaro AM, Masaberg C, Haagenson M, Vierra-Green C, Spellman S, Dakshanamurthy S, Ng J, Hurley CK. Evaluating the potential impact of mismatches outside the antigen recognition site in unrelated hematopoietic stem cell transplantation: HLA-DRB1*1454 and DRB1*140101. Tissue Antigens. 2009;73:595–598. doi: 10.1111/j.1399-0039.2009.01245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasi A, Crocchiolo R, Bontempelli M, Carcassi C, Carella G, Crespiatico L, Garbarino L, Mascaretti L, Mazzi B, Mazzola G, Miotti V, Porfirio B, Tagliaferri C, Valentini T, Vecchiato C, Fleischhauer K, Sacchi N, Bosi A, Martinetti M. The conundrum of HLA-DRB1*14:01/*14:54 and HLA-DRB3*02:01/*02:02 mismatches in unrelated hematopoietic SCT. Bone Marrow Transplant. 2011;46:916–922. doi: 10.1038/bmt.2010.246. [DOI] [PubMed] [Google Scholar]
- 9.Bade-Doeding C, Cano P, Huyton T, Badrinath S, Eiz-Vesper B, Hiller O, Blasczyk R. Mismatches outside exons 2 and 3 do not alter the peptide motif of the allele group B*44:02P. Hum Immunol. 2011;72:1039–1044. doi: 10.1016/j.humimm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Bettens F, Schanz U, Tiercy JM. Lack of recognition of HLA class I mismatches outside alpha1/alpha2 domains by CD8+ alloreactive T lymphocytes: the HLA-B44 paradigm. Tissue Antigens. 2013;81:414–418. doi: 10.1111/tan.12102. [DOI] [PubMed] [Google Scholar]
- 11.Sutton VR, Kienzle BK, Knowles RW. An altered splice site is found in the DRB4 gene that is not expressed in HLA-DR7, Dw11 individuals. Immunogenetics. 1989;29:317–322. doi: 10.1007/BF00352841. [DOI] [PubMed] [Google Scholar]
- 12.Rene C, Lozano C, Eliaou JF. Expression of classical HLA class I molecules: regulation and clinical impacts: Julia Bodmer Award Review 2015. HLA. 2016;87:338–349. doi: 10.1111/tan.12787. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rene C, Lozano C, Villalba M, Eliaou JF. 5′ and 3′ untranslated regions contribute to the differential expression of specific HLA-A alleles. Eur J Immunol. 2015;45:3454–3463. doi: 10.1002/eji.201545927. [DOI] [PubMed] [Google Scholar]
- 16.O’hUigin C, Kulkarni S, Xu Y, Deng Z, Kidd J, Kidd K, Gao X, Carrington M. The molecular origin and consequences of escape from miRNA regulation by HLA-C alleles. Am J Hum Genet. 2011;89:424–431. doi: 10.1016/j.ajhg.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balas A, García-Sánchez F, Gómez-Reino F, Vicario JL. HLA class I allele (HLA-A2) expression defect associated with a mutation in its enhancer B inverted cat box in two families. Hum Immunol. 1994;41:69–73. doi: 10.1016/0198-8859(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 18.Spellman S, Setterholm M, Maiers M, Noreen H, Oudshoorn M, Fernandez-Vina M, Petersdorf E, Bray R, Hartzman RJ, Ng J, Hurley CK. Advances in the selection of HLA-compatible donors: refinements in HLA typing and matching over the first 20 years of the National Marrow Donor Program Registry. Biol Blood Marrow Transplant. 2008;14:37–44. doi: 10.1016/j.bbmt.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu B, Mack SJ, Lazaro A, Lancaster A, Thomson G, Cao K, Chen M, Ling G, Hartzman R, Ng J, Hurley CK. HLA-A, -B, -C, -DRB1 allele and haplotype frequencies in an African American population. Tissue Antigens. 2007;69:73–85. doi: 10.1111/j.1399-0039.2006.00728.x. [DOI] [PubMed] [Google Scholar]
- 21.Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, Moraes ME, Pereira SE, Kempenich JH, Reed EF, Setterholm M, Smith AG, Tilanus MG, Torres M, Varney MD, Voorter CE, Fischer GF, Fleischhauer K, Goodridge D, Klitz W, Little AM, Maiers M, Marsh SG, Muller CR, Noreen H, Rozemuller EH, Sanchez-Mazas A, Senitzer D, Trachtenberg E, Fernandez-Vina M. Common and well-documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens. 2013;81:194–203. doi: 10.1111/tan.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson S, Starkey A, MacKenzie A. Evidence of uneven selective pressure on different subsets of the conserved human genome; implications for the significance of intronic and intergenic DNA. BMC Genomics. 2009;10:614. doi: 10.1186/1471-2164-10-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabbagh A, Luisi P, Castelli EC, Gineau L, Courtin D, Milet J, Massaro JD, Laayouni H, Moreau P, Donadi EA, Garcia A. Worldwide genetic variation at the 3′ untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. Genes Immun. 2014;15:95–106. doi: 10.1038/gene.2013.67. [DOI] [PubMed] [Google Scholar]
- 25.Klitz W, Maiers M, Spellman S, Baxter-Lowe LA, Schmeckpeper B, Williams TM, Fernandez-Vina M. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens. 2003;62:296–307. doi: 10.1034/j.1399-0039.2003.00103.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.