Abstract
First described about 15 years ago, BLUF (Blue Light Using Flavin) domains are light-triggered switches that control enzyme activity or gene expression in response to blue light, remaining activated for seconds or even minutes after stimulation. The conserved, ferredoxin-like fold holds a flavin chromophore that captures the light and somehow triggers downstream events. BLUF proteins are found in both prokaryotes and eukaryotes and have a variety of architectures and oligomeric forms, but the BLUF domain itself seems to have a well-preserved structure and mechanism that have been the focus of intense study for a number of years. Crystallographic and NMR structures of BLUF domains have been solved, but the conflicting models have led to considerable debate about the atomic details of photo-activation. Advanced spectroscopic and computational methods have been used to analyse the early events after photon absorption, but these too have led to widely differing conclusions. New structural models are improving our understanding of the details of the mechanism and may lead to novel tailor-made tools for optogenetics.
Keywords: Photo-activation, Optogenetics, Allostery, Flavin
Introduction
All living organisms need to be able to sense their environment in order to respond appropriately to the prevailing conditions around them, and the ability to adapt to different levels of sunlight is particularly important, even for many non-photosynthetic organisms. Higher plants have at least five types of sensory photoreceptors to optimise the energy yield from photosynthesis while minimising UV-mediated damage (Heijde and Ulm 2012). Various photoreceptor types using different chromophores have evolved for different purposes and are receptive to different wavelengths of light. Among these are four families of blue-light photoreceptors with flavin chromophores: photoactive yellow protein (PYP), light-oxygen-voltage (LOV), cryptochromes and BLUF (Blue Light Using Flavin) proteins. Members of each family share a common mechanism but are responsible for controlling a diverse range of cellular responses (Zoltowski and Gardner 2011). BLUF proteins are unique in being the only family of photoreceptors known to show photo-induced proton-coupled electron transfer. BLUF domains were independently discovered by different research groups in and around 2002, in the unicellular flagellate Euglena gracilis (Iseki et al. 2002) and in the purple bacterium Rhodobacter sphaeroides (Masuda and Bauer 2002). Euglena gracilis shows a photophobic response that depends on a photo-activated adenyl cyclase (PAC) found near the base of the flagellum that increases the level of cAMP when illuminated (Iseki et al. 2002). Rhodobacter sphaeroides controls the expression of genes related to photosynthesis through a light-sensitive protein called AppA, which interacts with a DNA-binding protein called PpsR (Masuda and Bauer 2002). AppA and PAC are just two examples of many photo-sensitive proteins carrying the BLUF domain, about 100 amino acid residues long, that is responsible for the detection of light (Gomelsky and Klug 2002). Genome sequencing has since revealed the presence of BLUF domains in a wide variety of organisms, including unicellular eukaryotes and about 10% of prokaryotes (Losi and Gartner 2008). Many BLUF proteins, including AppA and PAC, carry an extra domain downstream from the BLUF domain, with enzymatic or other properties, and the majority of these “group I” proteins appear to be homodimers. BlrP1, for example, is a dimeric cyclic nucleotide phosphodiesterase from Klebsiella pneumonia that shows a fourfold increase in enzyme activity under light conditions (Barends et al. 2009; Tyagi et al. 2008). PAC is unusual in having α and β subunits, each with a BLUF domain, that come together to form an α2β2 tetramer (Iseki et al. 2002). Many other BLUF proteins have fewer than 200 amino acid residues and are designated “group II” proteins. These proteins have little more than the BLUF domain in each subunit, but may carry secondary structural elements in the C-terminal region that are required for stability.
AppA is the most studied BLUF protein. First identified in the 1990s (Gomelsky and Kaplan 1995, 1998), it was only realised to be a light-sensing protein some years later. It carries a C-terminal SCHIC (Sensor Containing Heme Instead of Cobalamin) domain that senses redox conditions. AppA interacts with the transcription repressor PpsR in the dark. Masuda and Bauer (2002) suggested that AppA could convert PpsR from an active DNA-binding tetramer to an inactive dimer by reducing a disulfide bond in the PpsR tetramer. The interaction between AppA and PpsR was reported to be blocked by blue light, relieving the repression of transcription (Kraft et al. 2003), so that gene expression occurs only under suitable conditions of light and redox potential (Braatsch et al. 2002). Later work, including crystal structures and hydrogen/deuterium exchange of AppA complexed with PpsR, suggested a quite different mechanism, in which blue light dissociated the AppA/PpsR complex from DNA but did not appreciably alter the affinity of the two protein components (Winkler et al. 2013). A complete model of the full-length 450-residue protein has yet to be published, but crystal structures have been determined for residues 3 to 399, missing only the cysteine-rich region at the C-terminus which is involved in promoting the reduction of PpsR (Winkler et al. 2013). Regardless of the true interactions between AppA and PpsR, it is striking that the BLUF domain from a eukaryotic adenylate cyclase can substitute for the N-terminal region of AppA, as demonstrated before the protein structure had been determined by replacing the N-terminal domain of AppA with the equivalent region of PAC to give a functional chimeric protein despite the sequence identity of the exchanged domains being only about 30% (Han et al. 2004). The sequences of known BLUF models are compared in Fig. 1.
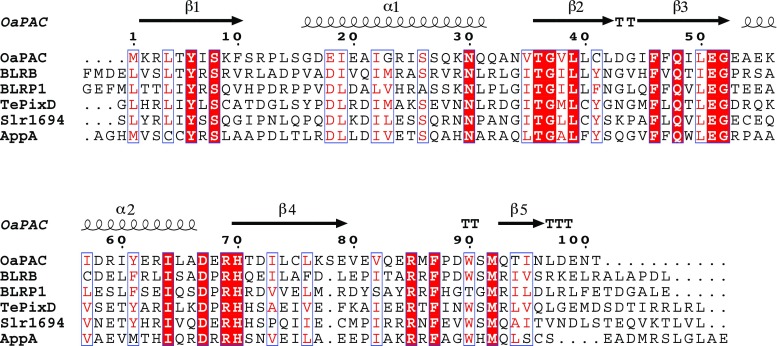
Fig. 1.
A sequence alignment of the six BLUF (Blue Light Using Flavin) domains for which independent experimental models have been deposited in the Protein Data Bank. The conserved residues, shown on a red background, are clustered around the flavin chromophore. Secondary structure elements from the crystal structure of OaPAC, a photo-activated adenyl cyclase (PAC) from Oscillatoria acuminata, are indicated with coils and arrows to indicate α-helices and β-strands, respectively, and turns are indicated by T. Tyr 8, Gln 48, Met 92 and Trp 90 of OaPAC form the quartet of residues that attract the most interest in functional studies. Asn 30 forms hydrogen bonds directly with the flavin. This figure was created with the program Espript (Robert and Gouet 2014)
Currently there is huge interest in optogenetics and synthetic biology, and BLUF domains appear to be an excellent plug-and-play unit for making biomolecular machines or gene circuitry light sensitive (Masuda 2013). However, a deep understanding of the structure and mechanism of the BLUF domain will be required to exploit it to maximum effect, while the extensive literature describing BLUF proteins reveals conflicting results and conclusions derived from an array of biophysical techniques. In particular, the conformational changes to the protein upon photo-stimulation have proved controversial. There have been some recent developments indicating a move towards a more concordant view of the common mechanistic features of BLUF proteins, and this may herald the development of artificial proteins with the desired, light-dependent properties.
Photo-activation
All BLUF proteins show conserved tyrosine, glutamine and methionine residues intimately related to function (Fig. 1). The photocycle is initiated by light causing an electron and then a proton to transfer from the conserved tyrosine to the flavin, yielding a bi-radical (Dragnea et al. 2005; Gauden et al. 2005, 2006). The reaction is not photo-reversible, and within 10 ns the photo-excited state falls back to the signalling state with recombination of the bi-radical (Gauden et al. 2007; Laptenok et al. 2015). Replacing the conserved glutamine with alanine causes the bi-radical to decay orders of magnitude more slowly (Fudim et al. 2015; Toh et al. 2008), with the signalling state possibly surviving from seconds to minutes depending on the nature of the BLUF protein (Toh et al. 2008). Photo-activation brings about a red-shift of about 10 nm in the main visible absorption peak of the flavin (Masuda and Bauer 2002), a feature that distinguishes BLUF proteins from other flavin-based photoreceptors. Upon photo-activation, the carbonyl bond at O4 of the flavin is weakened as its hydrogen bonds with neighbouring residues become stronger; larger changes are inferred in the protein itself, but with no dramatic change of fold (Bonetti et al. 2009; Gauden et al. 2006; Masuda et al. 2004; Unno et al. 2005). There is little consensus on how changes at the flavin are signalled by BLUF domains, but the conserved glutamine residue is intimately involved (Domratcheva et al. 2016; Fudim et al. 2015; Grinstead et al. 2006; Udvarhelyi and Domratcheva 2013; Unno et al. 2006). Replacing the conserved tyrosine, glutamine or methionine residues may block the photocycle and yield a protein in a pseudo-lit state that is active in the dark (Masuda et al. 2008; Yuan et al. 2011).
BLUF proteins remain locked in the dark state if the flavin chromophore is replaced with roseoflavin, a natural analogue with a dimethylamino group at C8. Quantum mechanical calculations have suggested energetic reasons for the photocycle not to be triggered (Merz et al. 2011), but it also seems likely that protonation of the flavin at the dimethylamino group could have an effect. The added positive charge may prevent electron density building up at the opposite end of the flavin, around the O4 atom, so that stronger hydrogen bonding to the glutamine side-chain cannot occur. More problematic issues to be answered are how the native protein changes upon absorbing light, how this change may trigger subsequent events and how the protein returns to its resting state.
Structural models
Crystal structures of three different BLUF domains were published almost simultaneously in 2005 (Anderson et al. 2005; Jung et al. 2005; Kita et al. 2005). Two of these, BlrB and TePxD (also known as Tll0078), are group II proteins. Like AppA, BlrB is a BLUF protein from Rhodobacter sphaeroides but is less than one-third of its length per polypeptide chain and much less well characterised, either biologically or in vitro. TePxD regulates phototaxis of the cyanobacterium Thermocynecoccus elongates (Okajima et al. 2005), forming a decamer in the dark and responding to light by changing its oligomeric state (Tanaka et al. 2009). Full-length AppA has 450 residues per monomer, but truncation allowed the N-terminal 117 residues to crystallise (Anderson et al. 2005). These models show that BLUF domains bind the chromophoric flavin between two α-helices, which themselves lie against a five-stranded β-sheet, but raise a number of questions regarding the photo-sensing mechanism. Unravelling the mechanistic details has however been complicated by the sequence and functional diversity of the BLUF family.
X-ray crystallographic models of six different BLUF domains, with a little less than 20% sequence identity, have now been deposited in the Protein Data Bank (Table 1). Overall the models share a very similar environment around the flavin chromophore, including both polar and apolar amino acids, except for one marked conformational difference discussed below. Either flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN) is found bound to the protein, but the position of the isoalloxazine ring is independent of its side-groups. Among the absolutely conserved residues near the chromophore, the trio of tyrosine, glutamine and methionine residues has attracted the most interest. Unfortunately, the crystal structures of BLUF proteins are not generally of sufficient resolution to enable reliable determination of the orientation of the side-chains around the flavin, so that, for example, it has been proposed that the glutamine oxygen atom receives a hydrogen bond from the tyrosine side-chain in the dark (Anderson et al. 2005), or in the signalling state (Yuan et al. 2006).
Table 1.
A list of current experimental models of BLUF domains in the Protein Data Bank (www.rcsb.org/pdb)
| Protein Data Bank entry | Protein | Resolutiona | Annotation | Group | Number of residues per model subunit |
|---|---|---|---|---|---|
| 1X0P | TePixDb | 2.0 | Electron transport | II | 143 |
| 1YRX | APPA | 2.3 | Novel (truncated) | I | 117 |
| 2IYG | APPA | 2.3 | Dark state | I | 108 |
| 2IYI | APPA | 2.95 | Light-induced intermediate | I | 108 |
| 4HH0 | APPA | 2.6 | Dark-state C20S mutant | I | 385 |
| 4HH1 | APPA | 3.5 | Wild-type dark state | I | 385 |
| 2BUN | APPA | NMR | I | 120 | |
| 2BYC | BLRB | 1.9 | Dark state | I | 136 |
| 3GFX | BLRP1 | 2.4 | Hydrolase | II | 400 |
| 3GFY | BLRP1 | 2.6 | No metal ions | II | 400 |
| 3GFZ | BLRP1 | 2.05 | pH 6 | II | 400 |
| 3GG0 | BLRP1 | 2.55 | pH 9 | II | 400 |
| 3GG1 | BLRP1 | 2.3 | pH 8 | II | 400 |
| 2KB2 | BLRP1 | NMR | II | 148 | |
| 3MZI | SLR1694 | 2.3 | Y8F mutant | I | 139 |
| 2HFN | SLR1694 | 1.8 | I | 140 | |
| 2HFO | SLR1694 | 2.1 | I | 140 | |
| 4YUS | OAPAC | 1.8 | Dark state, full-length. | II | 350 |
aResolution, where appropriate, is given in Ångstrom
bThe structure of TePixD was solved before the role of the protein in light detection was realised
The recently solved structure of dark-state OaPAC, a PAC from Oscillatoria acuminata, has the highest resolution of the models in Table 1 and is used for illustration (Ohki et al. 2016). At 1.8 Å resolution, the data allow the side-chain conformations around the flavin to be modelled with confidence, even if the hydrogen atoms are invisible, showing the tyrosine donates a hydrogen bond to the glutamine (Fig. 2). bPAC from the soil bacterium Beggiatoa has a very similar sequence to that of OaPAC (Stierl et al. 2011). OaPAC and bPAC are homodimers carrying an N-terminal BLUF domain and a C-terminal class III adenylate cyclase (AC) domain. They are both small but show a large increase in activity upon light exposure. The arrangement of Tyr 6, Gln 48 and Met 92 in OaPAC is similar to that of the equivalent residues in other BLUF models, although some models show a very different position of the methionine with its displacement by a partly conserved tryptophan residue (Trp 90 in OaPAC) found near the start of the fifth β-strand (Fig. 3). Of the known BLUF models, only BlrP1 is missing this tryptophan, with threonine instead (Table 1). The majority of independent BLUF domains in crystal structures show the conserved methionine close to the flavin and the conserved tryptophan on the protein surface. A dramatic difference is found in some models, such as AppA (Jung et al. 2006), with the conserved methionine having flipped to the protein surface and been replaced by the tryptophan. These two forms are called the Metout and Metin conformations, and their relevance to protein function remains highly contentious. Although support for the Metout conformation has been obtained using spectroscopy (Wu and Gardner 2009), crystallography (Barends et al. 2009) and computational methods (Sadeghian et al. 2008), it is not seen for all BLUF domains, and its relevance to physiological function has been questioned (Khrenova et al. 2013). The suggestion that N-terminal truncation of AppA is responsible for an artefactual conformation has been rejected (Unno et al. 2012), but this rejection leaves open the question of C-terminal truncation, and does not explain the structure of BlrP1 models, which also show both Metout and Metin conformations (Barends et al. 2009).
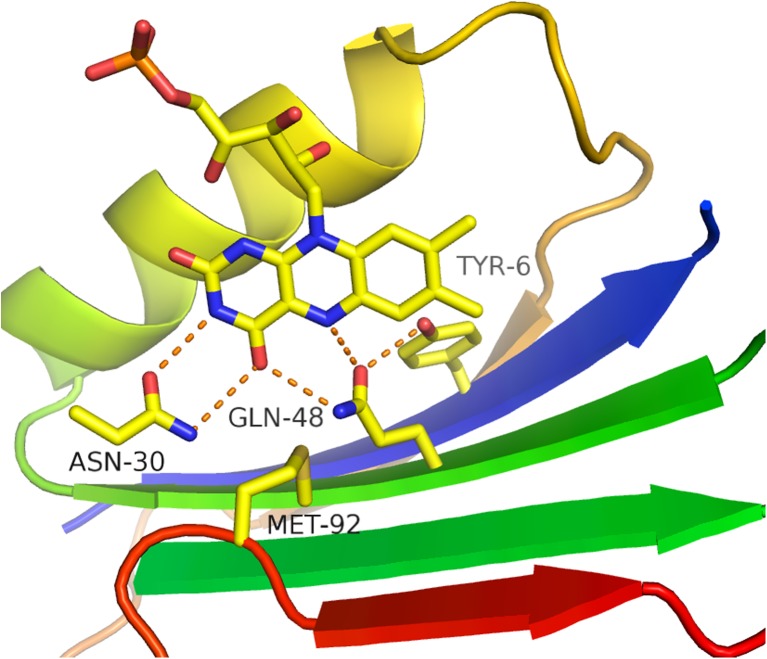
Fig. 2.
Hydrogen bonds formed by conserved residues with the flavin chromophore in the BLUF domain of OaPAC. The figure shows a ribbon diagram of the N-terminal 100 residues of the protein, with helix 1 omitted for clarity. The ribbon is coloured from blue to red (N to C terminal end). The flavin mononucleotide (FMN) is shown as a stick model, with carbon atoms coloured yellow, oxygen red and nitrogen blue. The O4 of the flavin receives hydrogen bonds from both Asn 30 and Gln 48, two absolutely conserved side-chains in the BLUF family
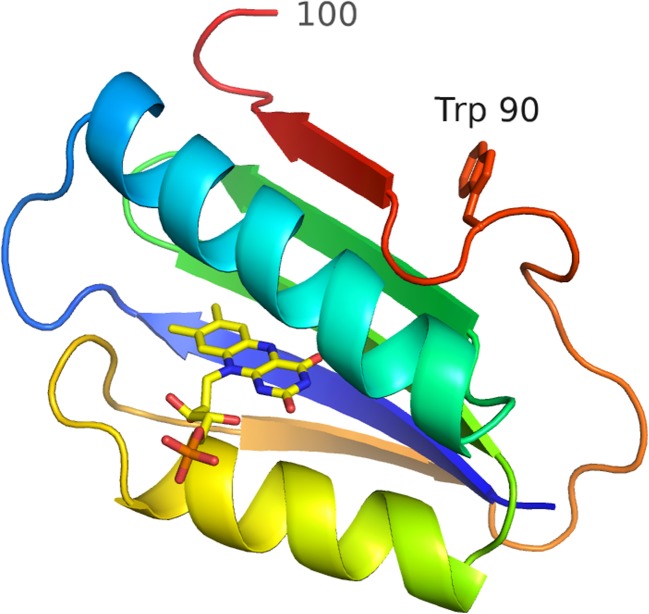
Fig. 3.
Ribbon diagram of the BLUF domain of OaPAC, coloured as in Fig. 2. The side-chain of Trp 90 is shown as a stick model, and the prominent kink in the backbone between this residue and the β5-strand is clearly visible. Residues at this point play an important role in signalling at the domain surface the changes that occur around the buried flavin upon photo-excitation
Structural changes in the BLUF domain
To add to the confusion regarding the question of whether the Metin or Metout model represents the dark state, it has been suggested that a flip between these structures may occur upon photo-activation of the protein. Such a large change in the tertiary structure involving the β5-strand could explain the stability of the signalling state, which may persist for minutes in the case of AppA. Direct evidence for such a change has proved hard to obtain from crystallography studies, since crystals of the AppA N-terminal domain do not show a complete red-shift upon light exposure, apparently being blocked from complete activation (Jung et al. 2006). More recent isotope-edited vibrational spectroscopic results obtained with the BLUF photoreceptor SyPixD have been interpreted in terms of register shifts of the β2- and β5-strands, and a change in backbone rigidity being the crucial difference in the signalling state (Mehlhorn et al. 2015).
Several studies have suggested that the conserved glutamine side-chain flips upon photo-activation so that its nitrogen and oxygen atoms exchange places. Anderson and colleagues (Anderson et al. 2005) noted that such a flip might explain the spectroscopically detected changes in hydrogen bonding to the flavin (Masuda et al. 2005; Unno et al. 2006). However, Fourier transform infrared (FTIR) spectroscopy studies of the blue-light photoreceptor TePixD (Takahashi et al. 2007) and AppA (Iwata et al. 2011) indicate that the hydrogen bond donated by the tyrosine to the glutamine oxygen becomes stronger upon photo-activation, which is consistent with the proposal that the glutamine residue forms an imidic tautomer as the hydrogen bond pattern around the flavin changes (Collette et al. 2014; Domratcheva et al. 2016; Khrenova et al. 2013). It remains unclear how the imidic tautomer is maintained for any length of time, given the roughly 40 kJ/mol energy cost over the amidic form, but the stronger hydrogen bonds formed around the flavin must provide some compensation (Fig. 4). Until high-resolution crystal structures are determined for a BLUF protein in both the resting and signalling states, it will be impossible to resolve the questions of structure changes upon activation by crystallography.
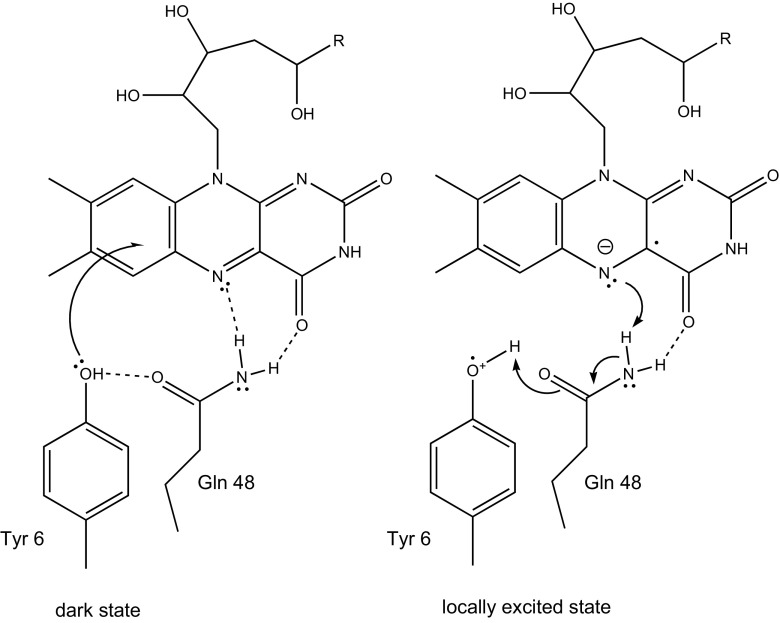
Fig. 4.
A schematic diagram showing the movement of an electron from the conserved tyrosine upon photo-excitation, giving a bi-radical form. Rearrangement of the hydrogen bonding between the protein and prosthetic group allows the tyrosine to lose a hydrogen atom and the flavin to gain one, while at the same time Gln 48 isomerises to the imidic form
Both the activation and deactivation processes of OaPAC are each accurately described by a single exponential function that decays in seconds. This life-time of the activated state is far greater than expected for a simple charge transfer, suggesting that concerted bond breaking is required to switch back to the dark, resting state. Simple transition state theory suggests that a first order reaction rate of 0.2 per second involves an activation energy of about 76 kJ/mol, which implies the concerted breaking of several hydrogen bonds. The activation energy for reversion to the resting, dark state has been estimated from temperature dependence studies to be 89 kJ/mol for the blue-light photoreceptor YcgF (Schroeder et al. 2008) and 81 kJ/mol for TePixD (Fukushima et al. 2005), but AppA has the most stable signalling state of any BLUF domain, remaining active for tens of minutes and relaxing via a bi-phasic process (Kraft et al. 2003; Laan et al. 2003). The significantly more stable signalling state of AppA and its non-exponential relaxation are consistent with a more dramatic conformational change than in the case of OaPAC, so that truncated versions of AppA may not in fact prove ideal model systems for the study of BLUF proteins in general. However, further experimental evidence is required to demonstrate any fundamental differences in the motions or conformations of particular BLUF proteins.
Signalling mechanism
The group II BLUF proteins simply change oligomeric state upon exposure to light and do not trigger enzyme activity changes directly (Kuroi et al. 2014; Nakajima et al. 2016). Group I proteins show various quaternary structures, in which the BLUF domains may or may not form subunit interfaces and which may remain unchanged on photoactivation. OaPAC is dimeric at all times, and both the BLUF domain and adenylate cyclase domain form the dimer interface (Fig. 5). The major issue to be resolved is therefore the nature of the signal transmitted over a substantial distance, without major conformational switching or change in oligomeric state. A coiled-coil between the BLUF domains plays an essential role in the photo-activation mechanism, carrying a signal from the flavin chromophores, which are themselves about 35 Å apart, to the two active sites over 45 Å away (Ohki et al. 2016). However, the coiled-coil appears to be rigid in the crystal structure, and it is hard to imagine any substantial mechanical mechanism like that observed in haemoglobin. Light-minus-dark differences in the FTIR spectra of bPAC have been interpreted to indicate a significant tertiary change from random coil to helical (Stierl et al. 2014). On the basis of the dark-state OaPAC model, the FTIR spectra of bPAC can be interpreted as a stiffening of the coiled-coil at the N-terminal end, which is sensed at the AC domain. Mutations within the coiled-coil of OaPAC are known to produce a pseudo-lit state, and the simplest explanation is that these buried residues, distant from the active site, are able to mimic the vibrational characteristics of the native, photo-activated protein (Ohki et al. 2016).
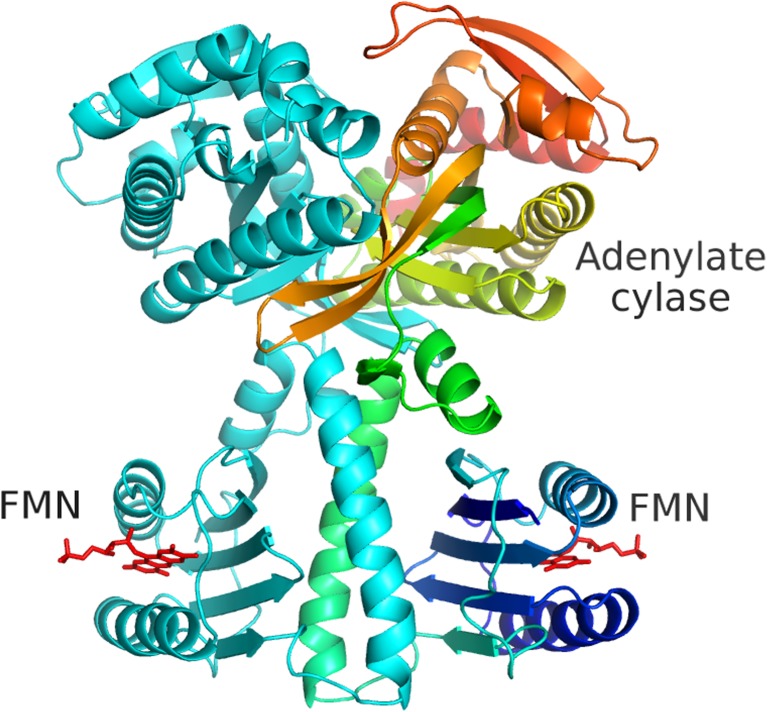
Fig. 5.
Ribbon diagram of the complete OaPAC dimer. Each subunit contains 366 residues, consisting of a BLUF domain, helical region (coiled-coil) and C-terminal adenylate cyclase domain. One dimer is coloured light blue, and the other is shown in dark blue (N-terminus) to red (C-terminus). The flavin molecules are shown as red stick models. Activation of the adenylate cyclase domains by light involves signal transduction from the flavin through the central coiled-coil to the active sites
A pattern is then emerging of signalling by changes in rigidity near the start of the β5 strand, rather than moving levers or domain shifts. The conserved tryptophan at this position plays an unknown role in sensing the movement of the flavin and in transmitting this information to other domains or proteins downstream. BlrP1 shows that tryptophan is not absolutely required for this process and that BLUF domains can operate without a shared interface. Different BLUF proteins show different behaviour if the conserved tryptophan is replaced with phenylalanine or alanine, with recovery of the dark state being accelerated in some cases and retarded in others, but dark-state activity is increased (Bonetti et al. 2009; Laan et al. 2006; Stierl et al. 2014). A similar pseudo-lit state is found if the key methionine is replaced with alanine, which can also be explained by a change in the relative stability of different orientations of the tryptophan side-chain at the protein surface (Fudim et al. 2015; Stierl et al. 2014). It may be that different BLUF domains have rather different signal transduction mechanisms, but at present the evidence is strongly pointing towards the type of model proposed by Kotani almost 50 years ago (Kotani 1968), whereby vibrational changes elicit effects at a distance with no change in average structure. These vibrational properties are becoming more widely recognised as a general property of allosteric proteins (Cooper and Dryden 1984; Townsend et al. 2015).
Discussion
Since the first models of BLUF domains were determined, the central issues to resolve have been the roles of individual residues at the flavin binding site and how their conformational changes are transmitted (Bonetti et al. 2009). Numerous studies of other BLUF domains using advanced spectroscopic techniques show that the principal visible flavin absorption bands shift to longer wavelengths upon light exposure, and that the carbonyl oxygen O4 becomes a better hydrogen bond acceptor; however, the structural basis for these changes has remained obscure. The relative importance of the Metin and Metout conformations is still actively debated (Goyal and Hammes-Schiffer 2017), although a number of studies offer no support for any role for the Trpin/Metout conformation (Dragnea et al. 2009; Mehlhorn et al. 2015; Toh et al. 2008). The field remains highly active and attractive to specialists in a variety of biophysical techniques so that further progress towards an inclusive model of BLUF domain function is eagerly expected.
Acknowledgements
JRHT would like to thank MEXT and OpenEye Scientific Software Inc. for financial support.
Compliance with ethical standards
Conflict of interest
Sam-Yong Park declares that he has no conflicts of interest.
Jeremy R. H. Tame declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Anderson S, Dragnea V, Masuda S, Ybe J, Moffat K, Bauer C. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44:7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends TRM, Hartmann E, Griese JJ et al (2009) Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018 [DOI] [PubMed]
- Bonetti C, Stierl M, Mathes T et al (2009) The role of key amino acids in the photoactivation pathway of the Synechocystis Slr 1694 BLUF domain. Biochemistry 48:11458–11469 [DOI] [PubMed]
- Braatsch S, Gomelsky M, Kuphal S, Klug G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 2002;45:827–836. doi: 10.1046/j.1365-2958.2002.03058.x. [DOI] [PubMed] [Google Scholar]
- Collette F, Renger T, Schmidt am Busch M. Revealing the functional states in the active site of BLUF photoreceptors from electrochromic shift calculations. J Phys Chem B. 2014;118:11109–11119. doi: 10.1021/jp506400y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- Domratcheva T, Hartmann E, Schlichting I, Kottke T. Evidence for tautomerisation of glutamine in BLUF blue light receptors by vibrational spectroscopy and computational chemistry. Sci Rep. 2016;6:22669. doi: 10.1038/srep22669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragnea V, Waegele M, Balascuta S, Bauer C, Dragnea B. Time-resolved spectroscopic studies of the AppA blue-light receptor BLUF domain from Rhodobacter sphaeroides. Biochemistry. 2005;44:15978–15985. doi: 10.1021/bi050839x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragnea V, Arunkumar AI, Yuan H, Giedroc DP, Bauer CE. Spectroscopic studies of the AppA BLUF domain from Rhodobacter sphaeroides: addressing movement of tryptophan 104 in the signaling state. Biochemistry. 2009;48:9969–9979. doi: 10.1021/bi9009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudim R, Mehlhorn J, Berthold T, Weber S, Schleicher E, Kennis JT, Mathes T. Photoinduced formation of flavin radicals in BLUF domains lacking the central glutamine. FEBS J. 2015;282:3161–3174. doi: 10.1111/febs.13297. [DOI] [PubMed] [Google Scholar]
- Fukushima Y, Okajima K, Shibata Y, Ikeuchi M, Itoh S. Primary intermediate in the photocycle of a blue-light sensory BLUF FAD-protein, Tll0078, of Thermosynechococcus elongatus BP-1. Biochemistry. 2005;44:5149–5158. doi: 10.1021/bi048044y. [DOI] [PubMed] [Google Scholar]
- Gauden M, Yeremenko S, Laan W et al (2005) Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry 44:3653–3662 [DOI] [PubMed]
- Gauden M, van Stokkum IH, Key JM, Luhrs D, van Grondelle R, Hegemann P, Kennis JT. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc Natl Acad Sci USA. 2006;103:10895–10900. doi: 10.1073/pnas.0600720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauden M, Grinstead J, Laan W et al (2007) On the role of aromatic side chains in the photoactivation of BLUF domains. Biochemistry 46:7405–7415 [DOI] [PubMed]
- Gomelsky M, Kaplan S. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1995;177:4609–4618. doi: 10.1128/jb.177.16.4609-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M, Kaplan S. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel fad binding domain. J Biol Chem. 1998;273:35319–35325. doi: 10.1074/jbc.273.52.35319. [DOI] [PubMed] [Google Scholar]
- Gomelsky M, Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/S0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- Goyal P, Hammes-Schiffer S. Role of active site conformational changes in photocycle activation of the AppA BLUF photoreceptor. Proc Natl Acad Sci USA. 2017;30:1621393114. doi: 10.1073/pnas.1621393114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstead JS, Avila-Perez M, Hellingwerf KJ, Boelens R, Kaptein R. Light-induced flipping of a conserved glutamine sidechain and its orientation in the AppA BLUF domain. J Am Chem Soc. 2006;128:15066–15067. doi: 10.1021/ja0660103. [DOI] [PubMed] [Google Scholar]
- Han Y, Braatsch S, Osterloh L, Klug G. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci USA. 2004;101:12306–12311. doi: 10.1073/pnas.0403547101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012;17:230–237. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Iseki M, Matsunaga S, Murakami A et al (2002) A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415:1047–1051 [DOI] [PubMed]
- Iwata T, Watanabe A, Iseki M, Watanabe M, Kandori H. Strong donation of the hydrogen bond of tyrosine during photoactivation of the BLUF domain. J Phys Chem Lett. 2011;2:1015–1019. doi: 10.1021/jz2003974. [DOI] [Google Scholar]
- Jung A, Domratcheva T, Tarutina M et al (2005) Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc Natl Acad Sci USA 102:12350–12355 [DOI] [PMC free article] [PubMed]
- Jung A, Reinstein J, Domratcheva T, Shoeman RL, Schlichting I. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J Mol Biol. 2006;362:717–732. doi: 10.1016/j.jmb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Khrenova MG, Nemukhin AV, Domratcheva T. Photoinduced electron transfer facilitates tautomerization of the conserved signaling glutamine side chain in BLUF protein light sensors. J Phys Chem B. 2013;117:2369–2377. doi: 10.1021/jp312775x. [DOI] [PubMed] [Google Scholar]
- Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005;349:1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- Kotani M (1968) Fluctuation in quaternary structure of proteins and cooperative ligand binding I. Suppl Prog Theor Phys (Extra Number) 233–241
- Kraft BJ, Masuda S, Kikuchi J, Dragnea V, Tollin G, Zaleski JM, Bauer CE. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: a novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 2003;42:6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- Kuroi K, Okajima K, Ikeuchi M, Tokutomi S, Terazima M. Transient conformational fluctuation of TePixD during a reaction. Proc Natl Acad Sci USA. 2014;111:14764–14769. doi: 10.1073/pnas.1413222111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan W, van der Horst MA, van Stokkum IH, Hellingwerf KJ. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: a key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem Photobiol. 2003;78:290–297. doi: 10.1562/0031-8655(2003)078<0290:ICOTPP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Laan W, Gauden M, Yeremenko S, van Grondelle R, Kennis JT, Hellingwerf KJ. On the mechanism of activation of the BLUF domain of AppA. Biochemistry. 2006;45:51–60. doi: 10.1021/bi051367p. [DOI] [PubMed] [Google Scholar]
- Laptenok SP, Lukacs, A, Brust R et al (2015) Electron transfer quenching in light adapted and mutant forms of the AppA BLUF domain. Faraday Discuss 177:293–311 [DOI] [PubMed]
- Losi A, Gartner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- Masuda S. Light detection and signal transduction in the BLUF photoreceptors. Plant Cell Physiol. 2013;54:171–179. doi: 10.1093/pcp/pcs173. [DOI] [PubMed] [Google Scholar]
- Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/S0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Masuda S, Hasegawa K, Ishii A, Ono TA. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr 1694 of Synechocystis sp. PCC6803. Biochemistry. 2004;43:5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- Masuda S, Hasegawa K, Ono TA. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44:1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- Masuda S, Hasegawa K, Ohta H, Ono TA. Crucial role in light signal transduction for the conserved Met93 of the BLUF protein PixD/Slr1694. Plant Cell Physiol. 2008;49:1600–1606. doi: 10.1093/pcp/pcn132. [DOI] [PubMed] [Google Scholar]
- Mehlhorn J, Lindtner T, Richter F et al (2015) Light-induced rearrangement of the b5 strand in the BLUF photoreceptor SyPixD (Slr1694). J Phys Chem Lett 6:4749–4753 [DOI] [PubMed]
- Merz T, Sadeghian K, Schutz M. Why BLUF photoreceptors with roseoflavin cofactors lose their biological functionality. Phys Chem Chem Phys. 2011;13:14775–14783. doi: 10.1039/c1cp21386e. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Kuroi K, Nakasone Y, Okajima K, Ikeuchi M, Tokutomi S, Terazima M. Anomalous pressure effects on the photoreaction of a light-sensor protein from Synechocystis, PixD (Slr1694), and the compressibility change of its intermediates. Phys Chem Chem Phys. 2016;18:25915–25925. doi: 10.1039/C6CP05091C. [DOI] [PubMed] [Google Scholar]
- Ohki M, Sugiyama K, Kawai F et al (2016) Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium. Proc Natl Acad Sci USA 113:6659–6664 [DOI] [PMC free article] [PubMed]
- Okajima K, Yoshihara S, Fukushima Y et al (2005) Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J Biochem 137:741–750 [DOI] [PubMed]
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324 [DOI] [PMC free article] [PubMed]
- Sadeghian K, Bocola M, Schütz M. A conclusive mechanism of the photoinduced reaction cascade in blue light using flavin photoreceptors. J Am Chem Soc. 2008;130:12501–12513. doi: 10.1021/ja803726a. [DOI] [PubMed] [Google Scholar]
- Schroeder C, Werner K, Otten H, Kratzig S, Schwalbe H, Essen LO. Influence of a joining helix on the BLUF domain of the YcgF photoreceptor from Escherichia coli. Chembiochem. 2008;9:2463–2473. doi: 10.1002/cbic.200800280. [DOI] [PubMed] [Google Scholar]
- Stierl M, Stumpf P, Udwari D et al (2011) Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem 286:1181–1188 [DOI] [PMC free article] [PubMed]
- Stierl M, Penzkofer A, Kennis JT, Hegemann P, Mathes T. Key residues for the light regulation of the blue light-activated adenylyl cyclase from Beggiatoa sp. Biochemistry. 2014;53:5121–5130. doi: 10.1021/bi500479v. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Okajima K, Suzuki H, Nakamura H, Ikeuchi M, Noguchi T. FTIR study on the hydrogen bond structure of a key tyrosine residue in the flavin-binding blue light sensor TePixD from Thermosynechococcus elongatus. Biochemistry. 2007;46:6459–6467. doi: 10.1021/bi7004653. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nakasone Y, Okajima K, Ikeuchi M, Tokutomi S, Terazima M. Oligomeric-state-dependent conformational change of the BLUF protein TePixD (Tll0078) J Mol Biol. 2009;386:1290–1300. doi: 10.1016/j.jmb.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Toh KC, van Stokkum IH, Hendriks J et al (2008) On the signaling mechanism and the absence of photoreversibility in the AppA BLUF domain. Biophys J 95:312–321 [DOI] [PMC free article] [PubMed]
- Townsend PD, Rodgers TL, Pohl E, Wilson MR, McLeish TCB, Cann MJ (2015) Global low-frequency motions in protein allostery: CAP as a model system. Biophys Rev 7:175–182 [DOI] [PMC free article] [PubMed]
- Tyagi A, Penzkofer A, Griese J, Schlichting I, Kirienko NV, Gomelsky M. Photodynamics of blue-light-regulated phosphodiesterase BlrP1 protein from Klebsiella pneumoniae and its photoreceptor BLUF domain. Chem Phys. 2008;354:130–141. doi: 10.1016/j.chemphys.2008.10.003. [DOI] [Google Scholar]
- Udvarhelyi A, Domratcheva T. Glutamine rotamers in BLUF photoreceptors: a mechanistic reappraisal. J Phys Chem B. 2013;117:2888–2897. doi: 10.1021/jp400437x. [DOI] [PubMed] [Google Scholar]
- Unno M, Sano R, Masuda S, Ono TA, Yamauchi S. Light-induced structural changes in the active site of the BLUF domain in AppA by Raman spectroscopy. J Phys Chem B. 2005;109:12620–12626. doi: 10.1021/jp0522664. [DOI] [PubMed] [Google Scholar]
- Unno M, Masuda S, Ono TA, Yamauchi S. Orientation of a key glutamine residue in the BLUF domain from AppA revealed by mutagenesis, spectroscopy, and quantum chemical calculations. J Am Chem Soc. 2006;128:5638–5639. doi: 10.1021/ja060633z. [DOI] [PubMed] [Google Scholar]
- Unno M, Tsukiji Y, Kubota K, Masuda S. N-terminal truncation does not affect the location of a conserved tryptophan in the BLUF domain of AppA from Rhodobacter sphaeroides. J Phys Chem B. 2012;116:8974–8980. doi: 10.1021/jp305873z. [DOI] [PubMed] [Google Scholar]
- Winkler A, Heintz U, Lindner R, Reinstein J, Shoeman RL, Schlichting I (2013) A ternary AppA–PpsR–DNA complex mediates light-regulation of photosynthesis-related gene expression. Nat Struct Mol Biol 20:859–867 [DOI] [PMC free article] [PubMed]
- Wu Q, Gardner KH (2009) Structure and insight into blue light-induced changes in the BlrP1 BLUF domain. Biochemistry 48:2620–2629 [DOI] [PubMed]
- Yuan H, Anderson S, Masuda S, Dragnea V, Moffat K, Bauer C (2006) Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry 45:12687–12694 [DOI] [PMC free article] [PubMed]
- Yuan H, Dragnea V, Wu Q, Gardner KH, Bauer CE (2011) Mutational and structural studies of the PixD BLUF output signal that affects light-regulated interactions with PixE. Biochemistry 50:6365–6375 [DOI] [PMC free article] [PubMed]
- Zoltowski BD, Gardner KH. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]