Abstract
Here we report functional characterization of the Streptomyces coelicolor M145 gene SCO1678, which encodes a GntR-like regulator of the FadR subfamily. Bioinformatic analysis suggested that SCO1678 is part of putative operon (gnt) involved in gluconate metabolism. Combining the results of SCO1678 knockout, transcriptional analysis of gnt operon, and Sco1678 protein-DNA electromobility shift assays, we established that Sco1678 protein controls the gluconate operon. It does so via repression of its transcription from a single promoter located between genes SCO1678 and SCO1679. The knockout also influenced, in a medium-dependent manner, the production of secondary metabolites by S. coelicolor. In comparison to the wild type, on gluconate-containing minimal medium, the SCO1678 mutant produced much less actinorhodin and accumulated a yellow-colored pigment, likely to be the cryptic polyketide coelimycin. Possible links between gluconate metabolism and antibiotic production are discussed.
1. Introduction
Bacteria of genus Streptomyces are abundant soil dwellers having unparalleled capacity to produce bioactive small molecules and to assimilate complex plant (e.g., lignins and cellulose) and animal polymers [1]. These properties fuel interest in Streptomyces as a source of novel drug candidates [2], valuable hydrolytic enzymes [3], and ecofriendly plant protection strategies [4]. Streptomycetes have evolved a complicated regulatory network that coordinates their primary metabolism with biosynthetic pathways responsible for the production of specialized secondary metabolites and breakdown of polymers [5, 6]. Core regulators of primary metabolism of streptomycetes are intimately linked to antibiotic production [7]. Streptomyces genomes are also very large, between 6 and 11 Mbp, and harbor a lot of regulatory genes. The overall understanding of how these regulatory genes control transcription is still unclear [8]. This limits our current ability to take full advantage of genomic potential of Streptomyces for its biomedical and industrial applications. Therefore, it is important to continue functional characterization of various regulatory genes in well-known model species, like the best studied Streptomyces coelicolor A3(2) (or its derivative M145).
Recently we have carried out extensive in silico analysis of GntR family transcriptional factors, one of the biggest and yet poorly understood groups of regulators in Streptomyces [9]. As a result, twelve GntR regulators have been described that are conserved across the Streptomycetaceae family. We refer to them as “core GntRs” to highlight their potential important function in Streptomyces biology. Two of them, WhiH and DasR, have been extensively studied in the past. WhiH controls early steps of sporulation [10, 11], while DasR is a pleiotropic regulator of multiple carbohydrate transporters, chitin metabolism and antibiotic production genes [12]. A third regulator, Sco0823, was recently proposed by us to participate in ferric ion uptake [9]. Functions of the nine other core GntRs are yet to be explored. Here we report functional characterization of S. coelicolor M145 gene SCO1678, which encodes a GntR type regulator from the FadR subfamily. We demonstrate that SCO1678 encodes a repressor of the gluconate operon and that the promoter of gluconate kinase gene SCO1679 is the main target of Sco1678 regulatory action. Interestingly, a knockout of SCO1678 also altered the secondary metabolite profile of S. coelicolor. Our work reveals one more regulatory checkpoint that links primary metabolism and antibiotic production in actinomycetes.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
Bacterial strains used in this work are listed in Table 1. E. coli DH5α, ET12567 (pUZ8002), and BW25113 (carrying pIJ790) were used for routine cloning, to perform intergeneric conjugation with Streptomyces species and to carry out RedET-mediated gene replacement, respectively [13]. E. coli DH5α and ET12567 (pUZ8002) were grown at 37°C in Luria-Bertani (LB) medium [14]; strain BW25113 was grown at 28°C in 2x YT medium (Tryptone: 20 g, Yeast extract: 10 g, and sodium chloride: 5 g per 1 l of distilled water). All Streptomyces strains were grown at 28°C. Solid SFM medium (soya flour, mannitol, agar—20 g/liter each) was used to harvest Streptomyces spores and to plate Streptomyces-E. coli matings. To analyze the gene transcription profile, precultures of Streptomyces strains were grown in TSB medium for 24 h, and then mycelium was harvested by centrifugation, washed three times with water, and inoculated into SMM medium with 1% glucose or gluconate for 36 h [15]. To analyze antibiotic (actinorhodin (ACT), undecylprodigiosin (RED), and coelimycin (CPK)) production, liquid YMPG, R2YE, SMM, and Oxoid agar media, respectively, were used [15, 16]. Where needed, media were supplemented with respective antibiotics.
Table 1.
Bacterial strains and plasmids used in this work.
| Strains or plasmids | Description | Source or reference |
|---|---|---|
| S. coelicolor M145 | SCP1−, SCP2− derivative of A3(2); ACT and RED producer | [15] |
| S. coelicolor ΔgntR | SCO1678 knockout in M145 | This work |
| S. coelicolor pKC-SCO1678 | SCO1678 overexpression in M145 | This work |
| S. coelicolor pKC1139 | M145 with pKC1139 empty vector | This work |
| S. coelicolor pGUS | M145 carrying pGUS | This work |
| S. coelicolor pGUS-gntRp | M145 carrying pGUS-gntRp | This work |
| Escherichia coli DH5α | Routine cloning host | Life Technologies |
| E. coli ET12567 (pUZ8002) | Host for conjugative DNA transfer | [13] |
| E. coli BW25113 (pIJ790) | Host for recombineering experiments | [13] |
| E. coli BL21 (DE3) GOLD | Strain for recombinant protein production | Stratagene |
| pIJ790 | ts-plasmid carrying genes for λ-RED recombination, CmlR | [13] |
| pLeere | Carrying acc(3)IV flanked by loxP-sites, apramycin and ampicillin resistances | Luzhetskyy |
| pKC1139 | pSG5 ts-replicon; apramycin-resistant (AmR) shuttle vector | [15] |
| pKC-SCO1678 | pKC1139 harboring SCO1678 with promoter region; SCO1678 overexpression, AmR | This work |
| pKC0702-SCO1678 | pKC0702 harboring SCO1678 with 3-kb flanking region, HygR | This work |
| pKC0702-SCO1678::Am | SCO1678 knockout construct; Δsco1678::hyg (HygR and AmR) | This work |
| pGUS | Promoterless gusA-containing plasmid, apramycin resistance | [17] |
| pGUS-gntRp | pGUS with gntRp-gusA fusion, AmR | This work |
| pET28a | Protein expression vector, pET-system | Novagen |
| pET28a-SCO1678 | Sco1678-6His protein expression | This work |
2.2. DNA Techniques
Isolation of plasmid DNA from E. coli and chromosomal DNA from Streptomyces, DNA digestion by restriction endonucleases, agarose gel electrophoresis, and DNA ligation were performed using standard protocols [14, 15]. E. coli transformation and intergenetic E. coli-Streptomyces matings were performed as described in [15]. DNA amplification by PCR was generated with Taq (NEB) and Phusion (NEB) DNA polymerases. All plasmids were verified by DNA sequencing.
2.3. Construction of pGUS-gntRp Plasmid
Approximately 230-bp of the promoter region of gntR was amplified with primers SCO1678gusXbaI and SCO1678gusKpnI, digested, and cloned into XbaI and KpnI sites of pGUS-vector giving pGUS-gntRp. Oligonucleotides used throughout this work are listed in Table 2.
Table 2.
Primers used in this work.
| Primer name | Sequence | Purpose/PCR product |
|---|---|---|
| SCO1678-f | AATAAAAGCTTCGTGACTGAAGAAGAGCGAA | To amplify SCO1678 with 3-kb flanks |
| SCO1678-r | AATAATCTAGAAGGGAGTAATGAGGCTACGA | |
| SCO1678_acc_f | GACTTTTTGTTCCCGTGACATGCCCCGTACGCTGAGTGCATGGATATCTCTAGATACCG | To amplify aac(3)IV for SCO1678 replacement |
| SCO1678_acc_r | TCCTCGACTGACGACGTCTCCCCCTGCCGCCCCGGCGCTCAAACAAAAGCTGGAGCTC | |
| SCO1678NcoI-f | AATTAACCATGGGCAGCACACCGGGCCGGGGGCT | SCO1678 ORF for protein production |
| SCO1678HindIII-r | AATTAAAAGCTTGGGCGCCAGGATGTCCAGCT | |
| SCO1678gusXbaI | AATAATCTAGACAGGGGACCGATGGTGGTCT | Promoter region of SCO1678 to clone into pGUS |
| SCO1678gusKpnI | AATAAGGTACCGCACTCAGCGTACGGGGCAT | |
| SCO1678 cmpl-f | AATAAGGATCCAGGGGACCGATGGTGGTCTT | SCO1678 with 500 bp promoter |
| SCO1678 cmpl-r | AATAATCTAGATTCCTCGACTGACGACGTCT | |
| SCO1679Cy5-f | AGCCAGTGGCGATAAGCTGTGATCGCGGGGCCGAGG | Cy5-labeled gntR-K intergeneric region |
| SCO1679Cy5-r | AGCCAGTGGCGATAAGGTTGCTGCATCGCACTCTCG | |
| SCO1680Cy5-f | AGCCAGTGGCGATAAGGCAGCCGGACGAGGCGGGAG | Cy5-labeled gntP promoter region |
| SCO1680Cy5-r | AGCCAGTGGCGATAAGGGTGGTTCCCTTGCGGTGAT | |
| SCO1681Cy5-f | AGCCAGTGGCGATAAGCTGGCTGGTGAAGGAGTACT | Cy5-labeled gntZ promoter region |
| SCO1681Cy5-r | AGCCAGTGGCGATAAGCGTCGTACTCCTGTTCCTGC | |
| SCO1263Cy5-f | AGCCAGTGGCGATAAGCAGGGCCAGCAGACCGCCCA | Cy5-labeled SCO1263 promoter |
| SCO1263Cy5-r | AGCCAGTGGCGATAAGCGCTCCATCCCAGCGGACGC | |
| RTSCO1678-f | TGCGAGTGGAACGTCTACGA | RT-PCR analysis of SCO1678 |
| RTSCO1678-r | GTCCTCGAACATCACGTCGT | |
| RTSCO1679-f | AACATCGCCAAGATGACGGC | RT-PCR analysis of SCO1679 |
| RTSCO1679-r | GCCCGTTCGGTGATCTCCTC | |
| RTSCO1680-f | TGTTCTTCGAGGTCGGCATC | RT-PCR analysis of SCO1680 |
| RTSCO1680-r | CTTGAGCAGCATCAGCACGA | |
| RTSCO1681-f | TCGACATCCTGGTCAACAAC | RT-PCR analysis of SCO1681 |
| RTSCO1681-r | GGTGCTGAACTCCTCGTCCT | |
| RTSCO4991-f | ACCGGTCTGATCTTCGGCAT | RT-PCR analysis of SCO4991 |
| RTSCO4991-r | ATCCAGGCGGAGAACACCCA | |
| RTgntR-K_f | ACTCGACGAGGTGCATGGAC | RT-PCR of gntR-K intergeneric region |
| RTgntR-K_r | GCCGTCATCTTGGCGATGTT | |
| RTgntK-P_f | TGATCGAGGACCGGATGTCG | RT-PCR of gntK-P intergeneric region |
| RTgntK-P_r | TGGTGATGACCTTGTCCAGC | |
| RTgntP-Z_f | ACATGTCGACCACGCACGC | RT-PCR of gntP-Z intergeneric region |
| RTgntP-Z_r | ACCGTCGGTCACGTCGAACA | |
| RThrdB_f | CGAGGACGAGGCGACCGAGGAG | RT-PCR analysis of hrdB gene |
| RThrdB_r | CAGCTTGTCCTCGGCGAACAGA |
2.4. Construction of pKC-gntR Overexpression Plasmid and pSET-gntR for Complementation
SCO1678 (=gntR) coding sequence with 230-bp promoter region was amplified with SCO1678 cmpl-f/SCO1678 cmpl-r primer pair. PCR product was digested with BamHI and XbaI and cloned into respective sites of moderate copy number vector pKC1139 and integrative plasmid pSET152, giving pKC-gntR and pSET-gntR, respectively.
2.5. Construction and Verification of SCO1678 Knockout Strain
An in-frame deletion mutant S. coelicolor ΔgntR was constructed using REDIRECT technology [13]. For this purpose, gene SCO1678 with 3-kb flanking regions was amplified from chromosomal DNA of S. coelicolor with the following primers: SCO1678-f and SCO1678-r. The PCR product was digested with HindIII and XbaI restriction endonucleases and subsequently cloned into respective sites of pKC0702. The obtained plasmid pKC0702-SCO1678 was transformed into E. coli BW25113 where replacement of SCO1678 by apramycin resistance cassette aac(3)IV was accomplished. The latter was amplified from plasmid pLeere using SCO1678_acc_f and SCO1678_acc_r primer pair. The knockout plasmid pKC0702-SCO1678::Am was introduced into the wild type S. coelicolor M145 followed by screening of apramycin-resistant and hygromycin-sensitive colonies. Positive clones were indicative that a double crossover had occurred between the homologous regions of the M145 genome and on the knockout construct. Markerless mutant S. coelicolor ΔgntR was generated by implication of site-specific recombinase Cre as described in [18]. The SCO1678 disruption and marker eviction were confirmed via PCR (primers SCO1678 cmpl-f/SCO1678 cmpl-r).
2.6. RT-PCR
RNA for semiquantitative RT-PCR was isolated using RNeasy mini kit (Qiagen) according to recommendations of the supplier. RNA samples were checked for DNA contamination by PCR. For cDNA synthesis 3 μg of total RNA was incubated with random primers for five minutes at 72°C. The remaining components (RNase inhibitor, dNTPs, reverse transcriptase buffer, DTT, and ProtoScript II reverse transcriptase (NEB)) were subsequently added and reverse transcription (RT) was carried out at 42°C for 60 min. 200 ng of synthesized RT products was used as a template for subsequent PCR analysis with primers listed in Table 2. Obtained PCR products were separated on a 1.5% agarose gel to analyze the transcription profile of genes of interest.
2.7. Production and Purification of His-Tagged SCO1678 Protein
To produce C-terminally hexahistidine-tagged Sco1678 protein (GntR-His), its ORF was cloned into pET28a expression vector using PCR and primers SCO1678NcoI-f and SCO1678HindIII-r. Resulting plasmid was labeled as pET28a-SCO1678. For GntR-His production, E. coli BL21 (DE3) GOLD carrying pET28a-SCO1678 was grown in LB supplemented with tetracycline and kanamycin until OD600 reached 0.5; then the culture was induced with 1 mM IPTG (isopropylthiogalactoside) and incubated for six hours at 22°C. Cells were collected by centrifugation and resuspended in a lysis buffer (50 mM Na2HPO4, 300 mM NaCl, and 20 mM imidazole, pH 7) containing proteinase inhibitor (Roche). Cells lysis was achieved by two consecutive passages through a French press (American Instrument Corporation) at 1000 psi. The cell lysate was centrifuged at 18000 rpm for 30 minutes and soluble fraction was applied to Ni-NTA agarose resin (Qiagen), washed two times with wash buffer (50 mM Na2HPO4, 300 mM NaCl, and 40 mM imidazole, pH 7). The protein was eluted with 200 mM imidazole and dialyzed against storage buffer (50 mM Na2HPO4, 300 mM NaCl, and 5% glycerol, pH 7). Protein concentration was determined by Bradford assay.
2.8. Electromobility Shift Assay (EMSA) of DNA-Protein Complexes
Putative promoter regions of SCO1263, gntP, gntZ, and gntR-K were amplified from chromosomal DNA of S. coelicolor with primers listed in Table 2 and subsequently labeled with indocarbocyanine (Cy5) as described in [19]. Cy5-labeled probes (0.2 pmol) were incubated with purified recombinant GntR-His (see above) in concentrations of 0.93 to 4.18 pmol in binding buffer (20 mM Tris/HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 5% (v/v) glycerol, and 0.5 mM EDTA) for 25 min at 25°C. Electrophoresis was carried out in 8% native polyacrylamide gel in 1x TBE buffer at 150 V for 60 min. DNA bands were visualized by fluorescence imaging using a Typhoon Trio variable mode imager (GE Healthcare). EMSA was used to test whether gluconate, glucono-1,5-lactone, and glucose are potential effector molecules for Sco1678 (at final concentration of 5 mM in reaction mixture).
2.9. Analysis of β-Glucuronidase Activity
Strains carrying pGUS-gntR or pGUS plasmids were grown for 36 h in SMM supplemented with certain carbon source. Transcription level of β-glucuronidase from gntR promoter was examined according to [17].
2.10. Analysis of Antibiotic Production
ACT and RED production levels were quantified as described in [15], in YMPG and R2YE media, respectively. ACT production was analyzed in SMM medium with either glucose or gluconate as the sole carbon source.
3. Results
3.1. Analysis of gnt Operon in S. coelicolor
Gene SCO1678 encodes a 233 aa transcriptional factor from the GntR family of regulators. As a member of this family Sco1678 protein consists of a N-terminal DNA-binding domain with GntR-like helix-turn-helix motif followed by a C-terminal effector binding/oligomerization domain. Based on the secondary structure of the C-terminal domain, Sco1678 was proposed to fall into the FadR subfamily of GntRs [9]. Divergently to SCO1678, genes of putative gluconate (gnt) operon are located (Figure 1(a)). Gene SCO1679 (gntK) encodes gluconokinase that phosphorylates gluconate to glucono-5-phosphate which is then metabolized in the pentose phosphate pathway. Gene SCO1680 (gntP) encodes gluconate permease that transports the molecule into a cell. The next two genes, SCO1681-SCO1682 (gntZ-gntZ2), overlap by 4 nucleotides and encode gluconate dehydrogenase and zinc-binding alcohol dehydrogenase, respectively. RT-PCR confirmed transcriptional coupling of genes SCO1679-1680-1681 (Figure 1(b)).
Figure 1.
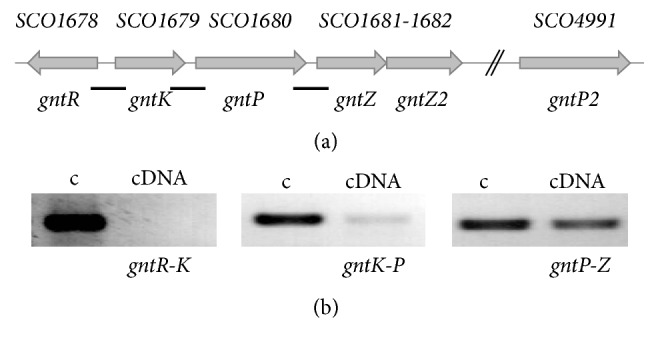
Genes for gluconate operon in S. coelicolor (a) and their transcriptional organization (b). Three black rectangles beneath the SCO genes indicate fragments amplified during RT-PCR analysis of intergenic regions (see (b)).
Another carbohydrate transport membrane protein Sco4991 shows 39% identity to GntP permease of Bacillus subtilis and 38% to S. coelicolor gluconate permease Sco1680. The former, therefore, might be involved in gluconate uptake as well. This agrees with recent global analysis of the entire array of transport proteins in S. coelicolor, where Sco1680 and Sco4991 were annotated as high-affinity gluconate permease and gluconate permease, respectively [20].
3.2. Expression and Knockout of SCO1678
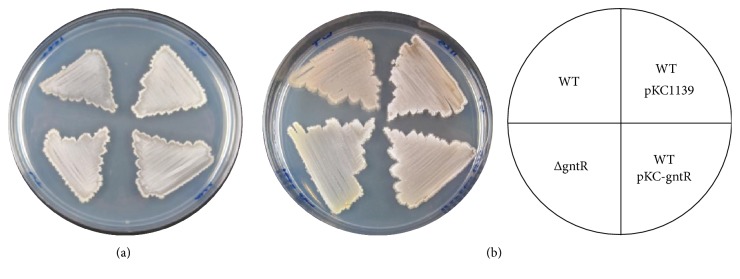
Organization and regulation of gnt operons in Escherichia coli and Bacillus subtilis are well studied [21, 22]. In both cases transcription of gnt genes is repressed by GntR, a protein that served as a prototype for the entire GntR family of regulators. From available in silico data we propose that SCO1678 also encodes GntR. If, contrary to our assumption, SCO1678 encodes an activator of gnt genes, then its deletion would lead to arrested or significantly reduced growth in presence of gluconate as the sole carbon source. To probe function of Sco1678, the SCO1678 gene was overexpressed on a moderate copy number plasmid pKC1139 and in-frame deletion mutant S. coelicolor ΔgntR was generated. Both strains as well as the wild type were grown in liquid SMM or on MM-agar plates supplemented with either glucose or gluconate (1%, w/v). No differences in growth rate and sporulation were detected among the strains (Figure 2). Complementation of ΔgntR with SCO1678 (plasmid pSET-gntR) also had no recognizable effects on growth. Our results agree with the assumption that SCO1678 encodes a repressor of the gnt genes. We noted, though, that ΔgntR on gluconate-containing medium had different coloration, probably because of changes in secondary metabolism, as discussed below (see Section 3.7).
Figure 2.
S. coelicolor strains grown on MM with glucose (a) or gluconate (b). WT: S. coelicolor M145; ΔgntR: S. coelicolor ΔgntR; WT pKC-gntR: S. coelicolor harboring pKC-gntR; WT pKC1139: S. coelicolor plus empty vector pKC1139.
3.3. Transcriptional Profile of gnt Genes
To further elucidate SCO1678 function, expression of the gnt operon of S. coelicolor was investigated by RT-PCR. For this purpose, a wild type strain and S. coelicolor ΔgntR were grown in liquid SMM medium supplemented with either glucose or gluconate as a sole carbon source. Data are summarized in Figure 3. We note here that our analysis was not quantitative, and transcription level can only be roughly compared within one strain. Transcription of gntK, gntP, gntZ, and SCO4991 in wild type strain M145 when cultured in gluconate-containing medium indicates their involvement in gluconate metabolism and transport. However, transcription of gntK and SCO4991 was also detected in M145 in presence of glucose, although their level was lower comparing to that in presence of gluconate. Further analysis of expression of gnt gene in S. coelicolor ΔgntR revealed that in presence of either tested sugar, all four gene transcriptions were detectable.
Figure 3.
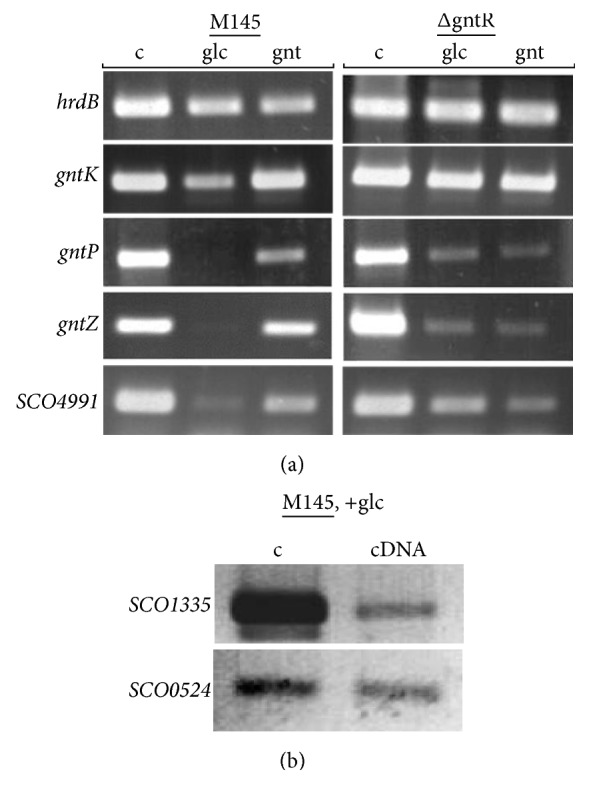
Transcriptional profile of (a) gnt genes in S. coelicolor M145 (M145) and ΔgntR (ΔgntR), and (b) SCO1335 and SCO0524 genes possibly related to pentose phosphate pathway. As a template for RT-PCR chromosomal DNA (c) and cDNA obtained from strains grown in either glucose (glc) or gluconate- (gnt-) containing SMM were used.
Absence of gntP, gntZ transcription, and reduced level of gntK transcription in presence of glucose and transcription of all four genes in the presence of gluconate implied that Sco1678 is a gluconate-dependent repressor for the transcription of gnt genes. Transcription of gntK and SCO4991 in the presence of glucose was puzzling. However, glucose can be converted into gluconate and gluconate-6-phosphate and further metabolized through the pentose phosphate pathway. In this case two enzymes are involved: glucose-1-dehydrogenase (Sco1335) converts glucose to glucono-1,5-lactone which is further metabolized to gluconate by gluconolactonase (Sco0524). These same molecules may induce gnt operon to some extent. Both aforementioned genes were expressed during wild type growth in glucose-containing SMM (Figure 3(b)). Transcription of SCO4991 in presence of either carbohydrate implies that this transporter can be involved in uptake of not only gluconate but other sugars as well.
3.4. Binding of Recombinant Sco1678 Protein to Promoter Regions of Putative Target Genes
Pure hexahistidine-tagged Sco1678 (GntR-His; 0.93 to 4.14 pmol) was incubated with Cy5-labeled DNA fragments encompassing promoters of gntR-gntK, gntP, and gntZ, and reaction products were separated in native acrylamide gel. As shown in Figure 4, GntR-His shifts gntR-gntK intergeneric region starting from a concentration of 0.93 pmol. No band shifts were observed with promoters of gntP and gntZ even when higher concentrations of the protein were applied. This agrees with our RT-PCR analysis of gnt operon (see Figure 1(b)), showing that gntK, gntP, and gntZ are transcribed as a polycistronic mRNA. Therefore, Sco1678 binds only to gntR-gntK region.
Figure 4.
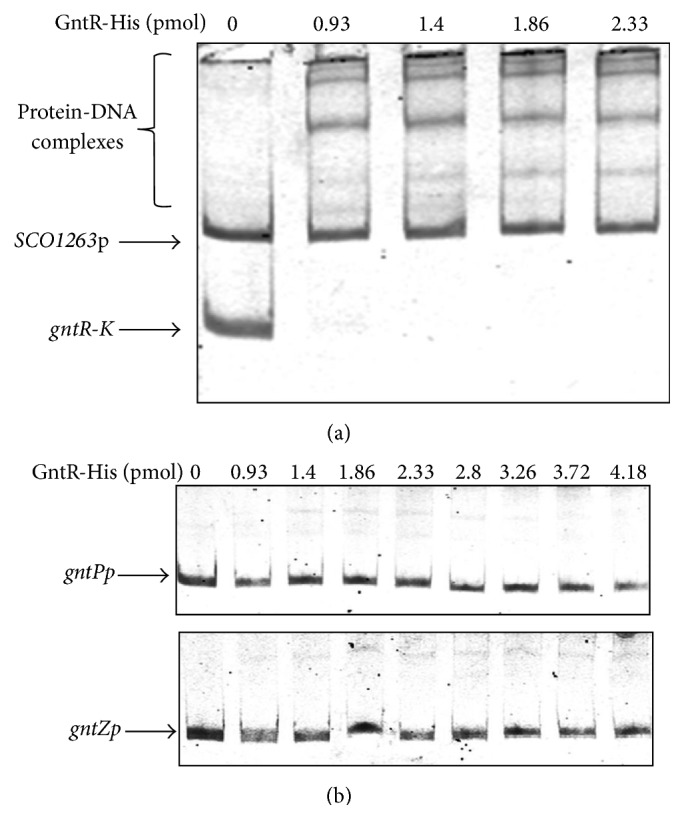
Binding of Sco1678 (GntR) to gntR-K region (a) and to promoters of gntP and gntZ (b). 0.2 pmol DNA fragment was incubated with indicated GntR-His concentration. Promoter region of SCO1263 was used in reaction as a negative control to check GntR-His specificity to its target (gnt) genes.
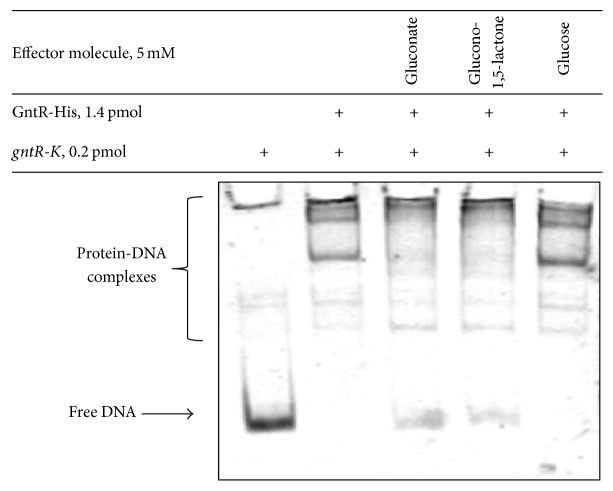
3.5. Identification of Sco1678 Effector Molecules
GntR type transcriptional factors act as repressors of gene transcription. Upon binding the appropriate effector molecule, the GntR repressors are no longer able to recognize promoters [23]. In most cases, such an effector molecule is a metabolite from the pathway where products of target genes are involved. To identify the putative effector molecule of GntR the DNA-binding shift assay was performed in the presence of gluconate and gluconolactone as well as glucose (the latter was used as a negative control). For this purpose, purified GntR-His was incubated with gntR-K intergeneric region in binding buffer that contains putative ligand (5 mM) and subsequently separated in a native acrylamide gel. As shown in Figure 5 gluconate and gluconolactone interfered with binding of GntR-His to the gntR-K region but could not release the DNA completely. The presence of glucose did not affect protein-DNA interaction.
Figure 5.
EMSA-mediated identification of putative effector molecules for recombinant Sco1678 protein. Binding of Sco1678 (1.4 pM) to gntR-K intergenic region was tested in presence of effectors mentioned in the figure. See Materials and Methods for workup conditions.
3.6. Autoregulatory Function of SCO1678
Most GntR regulators either repress or activate the transcription of their own genes. To explore this further, promoter of SCO1678 was fused with gusA reporter gene encoding β-glucuronidase, and the resulting construct was introduced into both S. coelicolor ΔgntR and wild type strains. Data are summarized in Figure 6. Fivefold increased SCO1678 transcription was observed in the wild type grown in SMM-glucose medium as compared to those grown in SMM-gluconate medium. No differences in transcription were observed for SCO1678 deletion mutant grown in presence of either glucose or gluconate, and it was equal to the transcription level observed for the wild type grown on gluconate. Our data showed that Sco1678 upregulates its own gene transcription and that the SCO1678 promoter is not activated in the presence of the effector molecule gluconate.
Figure 6.
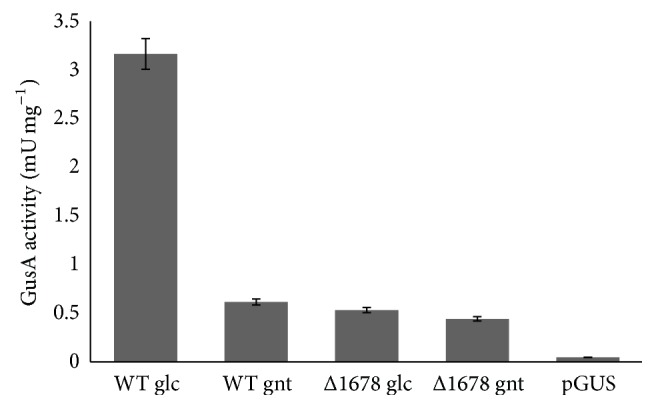
Transcriptional activity of SCO1678 (gntR) promoter in S. coelicolor WT (M145) and ΔgntR. The β-glucuronidase activity was measured from both strains carrying pGUS-gntR grown for 36 hours in SMM medium supplemented with glucose (WT glc, Δ1678 glc) or gluconate (WT gnt, Δ1678 gnt). S. coelicolor pGUS was grown in SMM with glucose and used as a control strain. Data represent mean values of three independent replicates. Error bars, ±2SD.
3.7. S. coelicolor ΔgntR Produces Yellow Polyketide Coelimycin
S. coelicolor produces prolifically at least five natural compounds: ACT, RED, methylenomycin, calcium-dependent antibiotic, and coelimycin [24]. Secondary metabolite production by Streptomyces is tightly linked to physiological and nutritional status. To investigate putative influence of SCO1678 deletion on ACT and RED production, strains were grown in YMPG, R2YE, and SMM media supplemented with either glucose or gluconate. No differences in antibiotics production were observed in YMPG and R2YE media in comparison to the wild type. In glucose-containing SMM medium both strains produced ACT (Figure 7(a)). However, the level of its production was decreased in the presence of gluconate. Moreover, gluconate triggered the production of a yellow compound, most likely coelimycin (CPK), in S. coelicolor ΔgntR. Yellow pigment accumulation was also observed when strains were grown on gluconate-containing Oxoid agar plates (Figures 7(b) and 7(c)). Secondary metabolism profile (coloration of agar plates) reverted to wild type when SCO1678 was introduced (on integrative plasmid pSET-gntR) into ΔgntR (data not shown). The identity of yellow pigment to CPK was established via analytical HPLC-MS analysis. The new peak (absent in extracts of M145) showed identical retention time (16.5 min) with standard Coelimycin P1, and based on the high resolution and high accurate MS data (349.1214 Da; (M+H)+), identical molecular formulae are generated as coelimycin P1; UV absorbance spectra of our peak and CPK were also the same.
Figure 7.
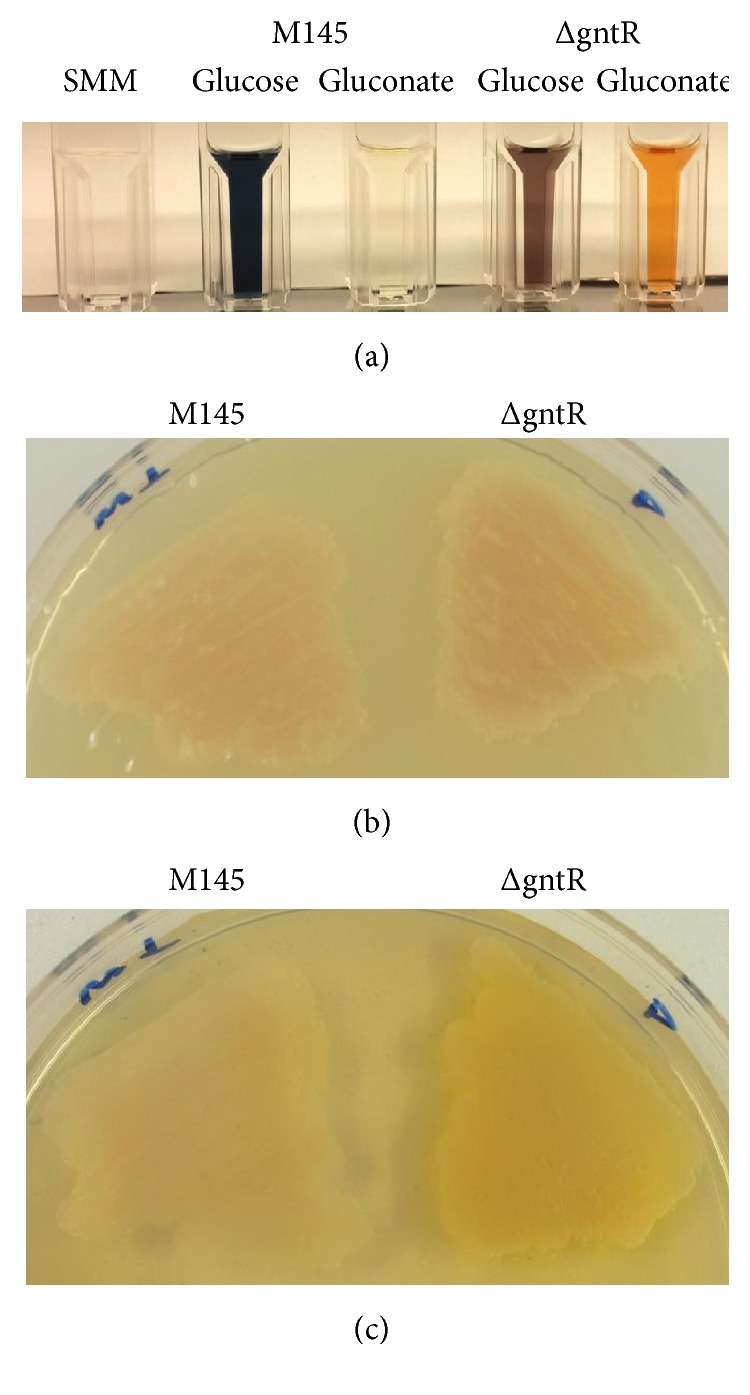
Gluconate inhibits ACT production in S. coelicolor M145 and triggers production of yellow-pigmented compound, CPK, in S. coelicolor ΔgntR. Spent media in cuvettes are shown (a) where respective strains were grown. The production levels correspond to equal amounts of biomass, as judged by Bradford protein assay. On glucose-containing SMM agar both M145 and ΔgntR produce ACT, and yellow pigment was not obvious (b), while on gluconate-containing Oxoid agar (c) these strains differ in secondary metabolism profile.
4. Discussion
Streptomycetes can metabolize a variety of carbohydrates including gluconic acid. In this work, we address for the first time the genetic basis of the ability of model strain, S. coelicolor, to utilize gluconate. First, it is imported into the cell by gluconate permease (GntP = Sco1680, maybe Sco4991), after phosphorylation to gluconate-6-phosphate by gluconokinase (GntK = Sco1679) and it then enters the pentose phosphate pathway. Typically, genes responsible for gluconate uptake are organized into an operon under negative control of the GntR repressor [25] (see Figure 1). Transcription level of key gnt genes of S. coelicolor were high when grown in gluconate-containing medium and were absent or severely repressed when grown in the presence of glucose. These observations point to involvement of respective proteins in gluconate metabolism. By combining the results of SCO1678 knockout, RT-PCR of gnt operon, and EMSA of Sco1678 protein, we can safely conclude that Sco1678 is repressor of gnt operon.
Sco1678 binds to promoter region of gluconokinase, but not permease or dehydrogenase, suggesting that these genes are transcribed as polycistronic mRNA. This hypothesis was confirmed by RT-PCR because we could amplify gntK-P and gntP-Z intergenic regions from cDNA (Figure 1(b)).
In SMM medium gluconate inhibits ACT production in both strains. This is not the first case where gluconate has abolished antibiotic production. For instance, cocultivation of S. coelicolor and Pseudomonas fluorescens BBc6R8, a producer of gluconic acid, stops ACT production [26]. Gluconate inhibits prodigiosin biosynthesis in Serratia sp. ATCC39006. In this case PigT regulator activates transcription of the biosynthetic operon pigA–O, but addition of gluconate decreases transcription of the latter. PigT shows high level homology to E. coli GntR protein [27]. Our working hypothesis is that gluconate, in the absence of the other carbon sources and regulatory function of Sco1678, serves as a metabolic signal that switches secondary metabolism from production of typical metabolites (ACT and RED) to minor or cryptic ones, such as coelimycin. We speculate that this secondary metabolic switch is mediated by a regulatory protein not yet known. Work is currently underway in our laboratories to experimentally explore this hypothesis.
5. Conclusions
Protein Sco1678, encoded within Streptomyces coelicolor M145 genome, belongs to twelve of the most conserved regulators of GntR family across class of Actinobacteria. Here we show for the first time that SCO1678 gene encodes GntR, or repressor of gluconate utilization operon. Its repressor function is exerted via binding to single promoter upstream of gluconokinase gene SCO1679 and is responsive to gluconate. We revealed that GntR in S. coelicolor constitutes a regulatory checkpoint for secondary metabolism, because under certain growth conditions the SCO1678 knockout decreases actinorhodin titers and induces the production of otherwise cryptic polyketide coelimycin. Our data suggest that further studies of SCO1678 (gntR) are worth pursuing. They will lead to further insight into coordination of primary and secondary metabolic pathways and help devise novel approaches towards the induction of silent gene clusters in actinomycetes.
Acknowledgments
The authors thank Kaitlyn Pinkett for proofreading their manuscript. Olga Tsypik was supported by DAAD fellowship. Research groups of Bohdan Ostash and Victor Fedorenko are supported by Grants BG-41Nr and BG-46F, respectively, from the Ministry of Education and Science of Ukraine.
Disclosure
Present address of Olga Tsypik and Roman Makitrynskyy is Albert-Ludwigs-University of Freiburg, Pharmazeutische Biologie, 79104 Freiburg, Germany.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Barka E. A., Vatsa P., Sanchez L., et al. Taxonomy, physiology, and natural products of actinobacteria. Microbiology and Molecular Biology Reviews. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones M. B., Nierman W. C., Shan Y., et al. Reducing the bottleneck in discovery of novel antibiotics. Microbial Ecology. 2017;73(3):658–667. doi: 10.1007/s00248-016-0889-3. [DOI] [PubMed] [Google Scholar]
- 3.Lewin G. R., Carlos C., Chevrette M. G., et al. Evolution and ecology of actinobacteria and their bioenergy applications. Annual Review of Microbiology. 2016;70(1):235–254. doi: 10.1146/annurev-micro-102215-095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rey T., Dumas B. Plenty is no plague: Streptomyces symbiosis with crops. Trends in Plant Science. 2017;22(1):30–37. doi: 10.1016/j.tplants.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Liu G., Chater K. F., Chandra G., Niu G., Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiology and Molecular Biology Reviews. 2013;77(1):112–143. doi: 10.1128/mmbr.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Book A. J., Lewin G. R., McDonald B. R., et al. Evolution of high cellulolytic activity in symbiotic Streptomyces through selection of expanded gene content and coordinated gene expression. PLoS Biology. 2016;14(6) doi: 10.1371/journal.pbio.1002475.e1002475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He J. M., Zhu H., Zheng G. S., et al. Direct involvement of the master nitrogen metabolism regulator GlnR in antibiotic biosynthesis in Streptomyces. The Journal of Biological Chemistry. 2016;291:26443–26454. doi: 10.1074/jbc.m116.762476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín J. F., Liras P. Cascades and networks of regulatory genes that control antibiotic biosynthesis. Sub-Cellular Biochemistry. 2012;64:115–138. doi: 10.1007/978-94-007-5055-5_6. [DOI] [PubMed] [Google Scholar]
- 9.Tsypik O., Yushchuk O., Zaburannyi N., et al. Transcriptional regulators of GntR family in Streptomyces coelicolor A3(2): analysis in silico and in vivo of YtrA subfamily. Folia Microbiologica. 2016;61(3):209–220. doi: 10.1007/s12223-015-0426-7. [DOI] [PubMed] [Google Scholar]
- 10.Persson J., Chater K. F., Flärdh K. Molecular and cytological analysis of the expression of Streptomyces sporulation regulatory gene whiH. FEMS Microbiology Letters. 2013;341(2):96–105. doi: 10.1111/1574-6968.12099. [DOI] [PubMed] [Google Scholar]
- 11.Bush M. J., Tschowri N., Schlimpert S., Flärdh K., Buttner M. J. C-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nature Reviews Microbiology. 2015;13(12):749–760. doi: 10.1038/nrmicro3546. [DOI] [PubMed] [Google Scholar]
- 12.Urem M., Światek-Połatyńska M. A., Rigali S., van Wezel G. P. Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Molecular Microbiology. 2016;102(2):183–195. doi: 10.1111/mmi.13464. [DOI] [PubMed] [Google Scholar]
- 13.Gust B., Chandra G., Jakimowicz D., Yuqing T., Bruton C. J., Chater K. F. λ red-mediated genetic manipulation of antibiotic-producing Streptomyces. Advances in Applied Microbiology. 2004;54:107–128. doi: 10.1016/s0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- 16.Pawlik K., Kotowska M., Kolesiński P. Streptomyces coelicolor A3(2) produces a new yellow pigment associated with the polyketide synthase Cpk. Journal of Molecular Microbiology and Biotechnology. 2010;19(3):147–151. doi: 10.1159/000321501. [DOI] [PubMed] [Google Scholar]
- 17.Makitrynskyy R., Ostash B., Tsypik O., et al. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biology. 2013;3(10) doi: 10.1098/rsob.130121.130121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedoryshyn M., Welle E., Bechthold A., Luzhetskyy A. Functional expression of the Cre recombinase in actinomycetes. Applied Microbiology and Biotechnology. 2008;78(6):1065–1070. doi: 10.1007/s00253-008-1382-9. [DOI] [PubMed] [Google Scholar]
- 19.Tiffert Y., Supra P., Wurm R., Wohlleben W., Wagner R., Reuther J. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Molecular Microbiology. 2008;67(4):861–880. doi: 10.1111/j.1365-2958.2007.06092.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z., Sun N., Wu S., Li Y.-Q., Wang Y. Genomic data mining reveals a rich repertoire of transport proteins in Streptomyces. BMC Genomics. 2016;17, article 510 doi: 10.1186/s12864-016-2899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong S., Porco A., Isturiz T., Conway T. Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. Journal of Bacteriology. 1996;178(11):3260–3269. doi: 10.1128/jb.178.11.3260-3269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita Y., Miwa Y. Identification of an operator sequence for the Bacillus subtilis gnt operon. Journal of Biological Chemistry. 1989;264(7):4201–4206. [PubMed] [Google Scholar]
- 23.Hoskisson P. A., Rigali S. Advances in Applied Microbiology. chapter 1. Vol. 69. Elsevier; 2009. Variation in form and function the helix-turn-helix regulators of the GntR superfamily; pp. 1–22. [DOI] [PubMed] [Google Scholar]
- 24.Jeong Y., Kim J.-N., Kim M. W., et al. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nature Communications. 2016;7 doi: 10.1038/ncomms11605.11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haydon D. J., Guest J. R. A new family of bacterial regulatory proteins. FEMS Microbiology Letters. 1991;79(2-3):291–295. doi: 10.1111/j.1574-6968.1991.tb04544.x. [DOI] [PubMed] [Google Scholar]
- 26.Galet J., Deveau A., Hôtel L., Leblond P., Frey-Klett P., Aigle B. Gluconic acid-producing Pseudomonas sp. prevent γ-actinorhodin biosynthesis by Streptomyces coelicolor A3(2) Archives of Microbiology. 2014;196(9):619–627. doi: 10.1007/s00203-014-1000-4. [DOI] [PubMed] [Google Scholar]
- 27.Fineran P. C., Everson L., Slater H., Salmond G. P. C. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology. 2005;151(12):3833–3845. doi: 10.1099/mic.0.28251-0. [DOI] [PubMed] [Google Scholar]