Abstract
Tetrabromobisphenol A (TBBPA) is a brominated flame retardant used globally at high volumes, primarily in the epoxy resin of circuit boards. It has been detected in the environment and in humans. The National Toxicology Program found that chronic oral TBBPA treatment of 250 mg/kg and higher caused an increased incidence of uterine lesions in female Wistar Han rats. The present laboratory has previously reported changes in gene expression associated with estrogen homeostasis in liver and uterine tissue of adult female Wistar Han rats after five days of gavage with 250 mg/kg of TBBPA. Microarray analysis of tissue from these same TBBPA-treated rats was performed to detect additional pathways perturbed by TBBPA. Microarray analysis of uterine tissue detected downregulation of genes in pathways of the immune response following TBBPA treatment. These results, along with validation of associated gene expression changes using droplet digital PCR, are reported here. Our findings suggest mechanisms that may be related to estrogen-mediated immunosuppression.
Keywords: tetrabromobisphenol A, uterine toxicity, rats, gene expression, immunosuppression
1. Introduction
Tetrabromobisphenol A (TBBPA; CAS No. 79-94-7) is a brominated flame retardant (BFR) used to retard the start and spread of fire. TBBPA is the most widely used BFR, with an estimated global market volume of between 150,000 and 210,000 tons per year (Alaee et al., 2003; de Wit et al., 2010). The primary application of TBBPA is as a reactive component in the epoxy resins for printed circuit boards, though it is also used additively in acrylonitrile-butadiene-styrene (ABS) resin (Alaee et al., 2003).
TBBPA has been detected in humans and the environment. A study by Dodson et al. (2012) detected TBBPA in household dust at concentrations ranging from <10 to 3400 ng/g. TBBPA has been detected in human milk samples, indicating risk of exposure through breastfeeding (Cariou et al., 2008). As other BFRs such as polybrominated diphenyl ethers and hexabromocyclododecanes are phased out, additive use of TBBPA and concomitant exposure may increase (Covaci et al., 2009).
Previous studies have observed decreased thyroxine (T4) levels and other endocrine effects after TBBPA exposures (Cope et al., 2015; Hamers et al., 2006; Sanders et al., 2016; Van der Ven et al., 2008). TBBPA has some structural similarity to T4 and may compete with the hormone for binding to transthyretin (Meerts et al., 2000). It may also compete for binding to human estrogen sulfotransferase (Gosavi et al., 2013). Chronic TBBPA treatment by gavage to female Wistar Han rats in a National Toxicology Program (NTP) 2-year cancer bioassay resulted in increased incidence of epithelial atypical hyperplasia at 250 mg/kg, the lowest dose studied, and higher and uterine tumors (adenomas, adenocarcinomas, and malignant mixed Müllerian cell tumors) at higher doses (Dunnick et al., 2015; NTP, 2013).
The present laboratory has demonstrated altered expression of genes involved in cell proliferation and biosynthesis and metabolism of estrogen in liver and uterine tissue of female Wistar Han rats treated with five daily doses of 250 mg/kg of TBBPA by gavage (Sanders et al., 2016). Changes in expression of target genes such as Cyp1b1, Comt, Esr1, Hsd17b2, Igf1, Ppara, Sult2a1, and Ugt1a1 support the hypothesis that disruption of estrogen homeostasis is a major mode-of-action for the increased incidence of uterine lesions in the TBBPA-treated rats of the NTP bioassay. In this study global gene expression changes were investigated by microarray analysis to search for additional affected biological pathways in liver and uterine tissue from the same five-day TBBPA-exposed rats previously studied in Sanders et al. (2016).
2. Methods
2.1 Animals and treatments
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the National Institute of Environmental Health Sciences (NIEHS). Animals were maintained in an AAALAC-approved animal care facility. The animals used in this study are the same animals used in Sanders et al. (2016), which describes the procedures in greater detail. Briefly, female Wistar Han rats were obtained from Charles River, Raleigh NC (age: 12 weeks at dosing; weight: 181–239 g). Each rat (n=10 rats/treatment group) received 5 consecutive daily doses by gavage of either TBBPA (250 mg/kg) or vehicle only (1:1:3 ratio of ethanol, Cremophore EL®, and water). The TBBPA (3,3′,5,5′-tetrabromobisphenol A) used in this study was obtained from Sigma-Aldrich (St. Louis, MO) with a chemical purity of 97%. Doses were administered by gavage at a volume of 4 mL/kg with a #16 ball-tipped feeding needle attached to a syringe. Rats of the same treatment group were housed two per cage and received NIH#31 food (Zeigler Bros., 2010) and water ad libitum. The estrous cycles of the rats were staged/synchronized and monitored by methods described by Whitten (1958) and Hubscher et al. (2005). At initial dosing, 18 of 20 rats were in diestrus (one TBBPA-treated rat was in metestrus and one vehicle control rat was in proestrus). Each rat was euthanized by CO2 asphyxiation 24 hours following the final dose. At the terminal timepoint, 17 of 20 rats were in diestrus (two vehicle controls and one TBBPA-treated rat were in metestrus).
2.2 Tissue collection and RNA and cDNA preparation
Liver and uterus were excised at necropsy. The central portion of the left lobe of the liver was collected and cubed. The uterus was collected in three sections by locating the midpoint of the branched uterine horns and cutting one to two mm to either side. This uterine collection technique followed a protocol used in the NTP chronic study (NTP, 2013). The proximal section (nearest the cervix) and distal section (nearest the ovaries) of the uterus were processed separately. The small portion of tissue between the proximal and distal sections was not analyzed. Uterine and liver tissues were flash-frozen in liquid nitrogen upon collection and were stored at −80°C.
RNA for use in the microarray analysis was prepared from liver and uterus tissues as described previously (Sanders et al., 2016). In brief, frozen tissue samples (50–60 mg) were weighed and minced in Qiagen (Germantown, MD) RLT buffer containing β-mercaptoethanol (1:100). Sample volume was brought to 1.4 mL (liver) or 1 mL (uterus) and homogenized with an Omni μH handheld homogenizer (Omni International, Kennesaw, GA). Homogenates were centrifuged at maximum speed in an Eppendorf 5430R microfuge for three minutes and 3 × 350-μL aliquots of lysate were transferred into 2-mL tubes (uterus was divided equally and brought up to 350 μL with additional RLT). Total RNA from the lysate was isolated in a QIAcube (Qiagen) following the QIAcube protocol for animal tissues and cells using the RNeasy Mini Kit with DNase treatment. Elution volume was 50 μL (liver) or 30 μL (uterus), and a second elution of 30 μL was performed manually for the uterus samples to maximize recovery. The RNA concentration for each eluate was determined using a Nanodrop 2000c (Thermo Scientific, Wilmington, DE). Eluates with similar concentrations from the same sample were pooled prior to further analysis. RNA integrity was verified with high-resolution capillary gel electrophoresis in a QIAxcel Advanced instrument (Qiagen); RNA integrity scores (RIS) for our samples ranged from 7.5 to 9.6, with a median RIS of 8.9. RNA samples were stored at −80°C.
Droplet digital PCR (ddPCR) was used in this study to confirm the microarray results. For ddPCR analysis, RNA was prepared nearly identically to the RNA from microarray analysis with a few changes noted below. Minced tissue samples were homogenized in individual tubes with a FastPrep-24 5G Sample Preparation System (MP Biomedicals, Santa Ana, CA) using 1.4 mm ceramic spheres (MP Lysing Matrix D) for one cycle of 40 seconds at 6.0 m/s.
cDNA for ddPCR was prepared using 2.5 U/μL Moloney Murine Leukemia Virus (MuLV) Reverse Transcriptase (Applied Biosystems, Life Technologies, Grand Island, NY), 0.25 mM dNTP mix, 5 mM MgCl2, PCR Buffer II (Applied Biosystems), 2.5 μM Random Hexamers, 0.25 U/μL RNase inhibitor, and 20 ng/μL of RNA. Reactions were incubated in a Bio-Rad T100 thermal cycler (Bio-Rad Laboratories, Hercules, CA) for one cycle of 10 min at 25°C, 60 min at 42°C, 5 min at 95°C, then held at 4°C until removal to storage at −20°C.
2.3 Microarray analysis
Microarray analysis was conducted using total RNA isolated from liver and uterine tissue from three rats from each of the two treatment groups using Affymetrix (Santa Clara, CA) Rat Genome 230 2.0 GeneChip® arrays, which are comprised of more than 31,000 probe sets over the transcribed rat genome. The sample size of 3 animals per group was used as recommended by the NIEHS Microarray Core Facility, which performed the procedure. In brief, 100 ng total RNA was amplified as directed in the Affymetrix 3′ IVT Express kit protocol. 12.5 μg of amplified biotin-aRNAs was fragmented and 10 μg was hybridized to each array at 45°C for 16 hours in a rotating hybridization oven using Affymetrix Eukaryotic Target Hybridization Controls and protocol. The array slides were stained with streptavidin/phycoerythrin utilizing a double-antibody staining procedure, then washed for antibody amplification according to the GeneChip Hybridization, Wash, and Stain Kit user manual. The arrays were scanned in an Affymetrix Scanner 3000 and data were obtained using the GeneChip® Command Console Software (AGCC; Version 3.2) utilizing the MAS5 algorithm to generate .CHP files. Data preprocessing, normalization, and error modeling was performed with Omicsoft (Version 7.0). Subsequent data analysis of CEL files is described below.
2.4 Sample analysis using ddPCR
To validate the microarray analysis, treatment-related changes in gene expression of select genes were measured using ddPCR in a Bio-Rad QX200 system. The target genes are identified in Results. Probe-based assays were purchased from Bio-Rad and Applied Biosystems. Droplets were generated from 20 μL reactions containing ddPCR Supermix for Probes (Bio-Rad), gene-specific assay, RNase-free water, and cDNA. The ddPCR reactions were performed in a T100 thermal cycler (Bio-Rad) in 40 μL reactions containing generated droplets and oil (Droplet Generation Oil for Probes, Bio-Rad).
Optimal concentration of cDNA for subsequent assays was determined by running ddPCR with pooled untreated uterus cDNA at 1, 5, 10, and 20 ng, along with no-template controls (Table S1). All assays were optimized for annealing temperature from 55°C to 65°C, with 56°C being optimal for all tested assays. Subsequent cycling conditions for assays were: one cycle of 10 min at 95°C, 40 cycles of 30 sec at 94°C and 1 min at 56°C, and one cycle of 10 min at 98°C before holding at 4°C until transfer to the droplet reader. The ramp rate was 2.5°C/sec.
For target gene expression assays in uterine samples, treatment groups of either 9 (control distal and treated proximal) or 10 (control proximal and treated distal) rats were assayed. Liver samples were not analyzed by ddPCR due to negative liver pathway data. All samples were run in triplicate. QuantaSoft version 1.5.38.1118 (Bio-Rad) was used to analyze droplet readings. Thresholds for droplets were manually set immediately beneath the positive droplet band. Data were then exported into Microsoft Excel.
2.5 Data analysis
CEL files of data from microarray hybridization were imported into Partek Flow software (Partek, St. Louis, MO) and processed to create a list of differentially expressed genes with false discovery rate of <0.3 and p-value of <0.05. The false discovery rate was set at <0.3 on consultation with a statistician because outbred stocks of rats exhibit more interindividual variability. The data were evaluated using Ingenuity Pathway Analysis (IPA) (Qiagen, Redwood City, CA). Pathway analysis of the proximal uterus and liver returned few pathways. The top ten pathways that were perturbed in the distal uterus were selected for further validation by ddPCR.
The ddPCR data are presented as mean ± SD and are from n = 9–10 rats/treatment group and 3–6 replicates/rat. Samples with low droplet generation (<3000 droplets) were excluded from the analysis. Concentration data (represented as copies per μL) were imported into GraphPad Prism 7.01 for statistical analysis. The D’Agostino-Pearson omnibus normality test was performed, and data were subsequently analyzed using the two-tailed unpaired t-test. For gene expression data that was not normally distributed, the Mann-Whitney Rank Sum test was used instead with exact p-values reported. Data were considered to be statistically significant at p<0.05.
3. Results
Microarray analysis with Partek Flow indicated that 275 genes were differentially expressed between TBBPA-treated and control distal uterus (Table S2). For the proximal uterus and liver, 22 and 64 genes were differentially expressed, respectively (Tables S3 and S4). IPA analysis of the differentially expressed genes indicated that the top canonical pathways perturbed by TBBPA treatment in the distal uterus involved the immune system (Table 1). Pathway analysis of the liver and proximal uterus did not suggest that any specific pathways were perturbed between TBBPA-treated and control samples (Tables S5 and S6). To further validate the pathway response in the distal uterus, thirteen genes were chosen from the list of differentially expressed genes for analysis by ddPCR (Table 2).
Table 1.
Top 10 Canonical Pathways in TBBPA-treated distal uterus determined from microarray data using Ingenuity Pathway Analysis
| Ingenuity Canonical Pathways | -log(p-value) |
|---|---|
| Graft-versus-Host Disease Signaling | 6.16 |
| Communication between Innate and Adaptive Immune Cells | 5.65 |
| Dendritic Cell Maturation | 5.41 |
| Allograft Rejection Signaling | 4.67 |
| Altered T Cell and B Cell Signaling in Rheumatoid Arthritis | 4.17 |
| OX40 Signaling Pathway | 4.03 |
| Autoimmune Thyroid Disease Signaling | 3.86 |
| Antigen Presentation Pathway | 3.38 |
| Phagosome Formation | 3.36 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 2.83 |
Table 2.
Genes assayed for droplet digital PCR validation with microarray fold change (treated vs. control) in the distal uterus and Entrez Gene database information
| Gene | Fold change | Gene name and associated function |
|---|---|---|
| Ccl6 | −2.74094242 | Chemokine (C-C motif) ligand 6 / cytokine activity, apoptosis |
| Clca4 | −24.398129 | Chloride channel accessory 4 / chloride transport |
| Clec4a3 | −2.82542789 | C-type lectin domain family 4, member A3 / may play a role in immune response, phagocytosis |
| Clec7a | −2.18258692 | C-type lectin domain family 7, member A / pathogen recognition, regulated by Esr1 |
| Ctse | −3.81489471 | Cathepsin E / antigen processing, regulates Il18 |
| Dmbt1 | −7.61495061 | Deleted in malignant brain tumors 1 / associated with endometrioid cancer, role in interaction of tumor cells and immune system |
| Ecm1 | −1.9850104 | Extracellular matrix protein 1 / angiogenesis, inflammatory response |
| Fcgr1a | −2.22506708 | Fc fragment of IgG, high affinity Ia, receptor (CD64) / immune response, antigen presentation |
| Fcgr3a | −3.81127061 | Fc fragment of IgG, low affinity IIIa, receptor (CD16) / immune response regulation |
| Iapp | −4.37469083 | Islet amyloid polypeptide / apoptosis |
| Il18 | −2.14066052 | Interleukin 18 / cytokine activity, T cell activation |
| RT1-DMb | −2.03443864 | RT1 class II, locus DMb / may play a role in antigen presentation |
| Tlr4 | −2.04418908 | Toll-like receptor 4 / innate immunity activation, pathogen recognition |
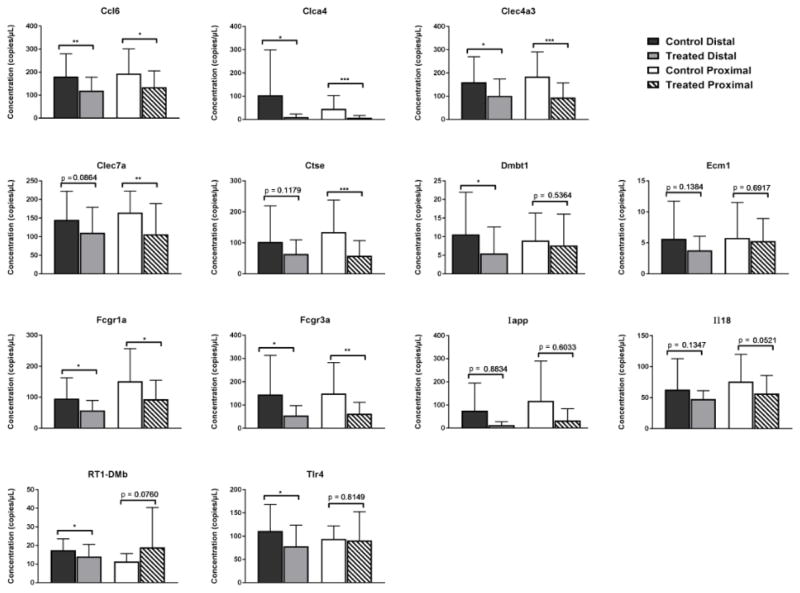
The microarray results were confirmed using ddPCR, a digital PCR technology that uses nanoliter-sized droplets for precise absolute quantification of nucleic acids in a sample (Hindson et al., 2011; Pinheiro et al., 2012). Massive sample partitioning into water-in-oil droplets enables ddPCR to reliably measure small fold differences among samples (Berman et al., 2012). The ddPCR data (Figure 1) validated that several genes involved in immune system responsiveness were significantly downregulated in one or both uterine tissue sections. Significant changes in expression were observed in the uterine tissues of TBBPA-treated female Wistar Han rats in the following genes: Ccl6, Clca4, Clec4a3, Clec7a, Ctse, Dmbt1, Fcgr1a, Fcgr3a, RT1-DMb, and Tlr4. For Clec7a and Ctse, the change was limited to the proximal uterus, while for Dmbt1, RT1-DMb, and Tlr4 the change only occurred in the distal uterus.
Figure 1.
TBBPA-dependent changes in assayed gene expression in proximal or distal uterus. Data are presented as mean ± SD and are from n = 9–10 rats/treatment group and 3–6 replicates/rat. The mean is significantly different at *p<0.05, **p<0.01, and ***p<0.001.
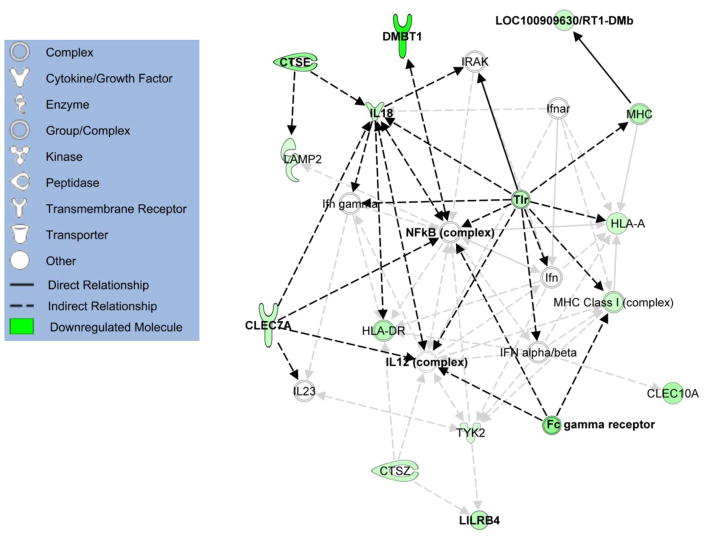
A network generated from IPA (Figure 2) depicts the relationships and interactions between eight of the thirteen genes that were differentially expressed in the distal uterus. Several of these relationships center around toll-like receptors (TLR), interleukin 18 (IL18), and nuclear factor kappa-B (NF-κB).
Figure 2.
Ingenuity Pathway Analysis network showing the relationships between several assayed genes (emphasized with black arrows). Molecules shown in green were downregulated in TBBPA-treated distal uterus according to microarray data. Intensity of green indicates strength of downregulation.
4. Discussion
Downregulation of pathways and genes linked to immune system responses following TBBPA exposure highlights a possible link between immunosuppression and the observed TBBPA-induced uterine epithelial tumors in female Wistar Han rats (NTP, 2013). Studies conducted in vitro have demonstrated effects of TBBPA on human natural killer cells and murine splenocytes and macrophages (Han et al., 2009; Hurd & Whalen, 2011; Kibakaya et al., 2009; Pullen et al., 2003). Han et al. (2009) found that TBBPA in vitro induced proinflammatory cytokine expression through activation of Akt/MAPK/NF-κB/AP-1 signaling while Kibakaya et al. (2009) found that TBBPA had immunosuppressive effects by decreasing the lytic function of natural killer cells. In human natural killer cells, TBBPA causes decreases in cell-surface proteins; these proteins are necessary for the tumor-destroying function of natural killer cells (Hurd & Whalen, 2011). If these direct effects are applicable to in vivo TBBPA exposures, then it could be suggested that immunological defenses against infection and tumors could be reduced, thereby possibly enhancing susceptibility to infectious disease or cancer.
Dunnick et al. (2015) postulated that competition between TBBPA and endogenous estrogen for conjugating enzymes may result in elevated levels of the hormone in the uterus, leading to tumor formation following chronic administration of high doses of TBBPA. This hypothesis was supported by observed expression changes in genes involved in estrogen biosynthesis and metabolism (such as Cyp1b1, Comt, Hsd17b2, Sult2a1, and Ugt1a1) in the TBBPA-treated rats described in the present work (Sanders et al., 2016). Sanders et al. (2016) found that Esr1, Ppara, and Igf1 were upregulated in both proximal and distal uterus in the TBBPA-treated rats of this study while Esr2 was upregulated in distal uterus and downregulated in proximal uterus. Estrogen modulation of the immune system responses is well established in mammals and necessary for protection of the fetus during pregnancy (Jansson & Holmdahl, 1998). Therefore, it is plausible that immunosuppressive responses could occur at the site-of-action as a result of elevated endogenous estrogen levels in the tissue following repeated TBBPA exposure, especially at doses of 250 mg/kg or higher.
In mice TBBPA exposure through diet for four weeks resulted in alterations in cytokines, including decreases in Th2 cytokines, following an induced respiratory syncytial virus infection (Watanabe et al., 2010). Dunnick et al. (2016) further found that 13 weeks of repeat TBBPA exposure (1000 mg/kg) in female Wistar Han rats activated the interferon pathway in the liver, adding evidence to the hypothesis that long-term exposure to TBBPA could result in immunomodulatory changes that promote cancer.
Following the uterine collection protocol of the NTP chronic study (NTP, 2013), distal and proximal uteri were analyzed separately. Differences between proximal and distal uterus have been seen in rats, such as higher myometrial oxytocin receptor (MOR) levels in the proximal uterus in late pregnancy, which could be experimentally altered by administration of estradiol (Gorodeski et al., 1990). We hypothesize that estrogen signaling from the ovary may impact the changes seen in the distal uterus as that uterus section is closest to the ovaries.
The current study revealed perturbations in several immune system pathways in the uterus in response to TBBPA exposure. It was hypothesized that genes previously found to be altered by the present laboratory using a targeted approach in Sanders et al. (2016) would also show changes in an untargeted approach such as microarray analysis. However, none of the genes previously found to be altered following TBBPA treatment (which were involved in estrogen and TBBPA metabolism, cell proliferation, and cell cycle regulation) were found to be differentially expressed in these microarray results. Subsequent ddPCR analysis of the genes studied in Sanders et al. (2016) confirmed these gene changes were real, even though the microarray analysis did not detect them (Figure S7). The difference between the microarray and ddPCR analyses could be attributed to the smaller sample size for the microarray analysis; however, it may also have occurred because the differentially expressed genes from Sanders et al. (2016) had subtle changes in expression that the microarray was not sensitive enough to detect.
Analysis of specific immune-related genes with ddPCR highlighted a few genes of interest with differential expression between the uteri of vehicle- and TBBPA-treated rats. Ctse, with decreased expression in the TBBPA-treated rats in this work, encodes the peptidase cathepsin E. Cathepsin E has been found to have a role in host defense against tumor cells, possibly through tumor-associated macrophage-mediated cytotoxicity (Kawakubo et al., 2007). Female cathepsin E-deficient mice have previously been found to develop swollen uteri and tumors in the uterine horn as well as increased incidence of mammary tumors (Goto et al., 2014; Kawakubo et al., 2014). Cathepsin-E has been found to be influenced by ovarian hormones in rats as ovariectomized Wistar rats demonstrate increased cathepsin-E activity that can be slightly decreased through administration of estradiol (Gladson et al., 1998). Given that Ctse was downregulated in the proximal uterus of TBBPA-treated rats in this study, it is plausible that Ctse may have a role in the uterine carcinogenesis found in the NTP study.
C-type lectin receptors, such as Clec4a3 and Clec7a, are involved in antigen presentation; both Clec4a3 and Clec7a were downregulated in proximal uterus following TBBPA treatment. The interaction between C-type lectin receptors and tumor glycoproteins indicates potential for immune surveillance as cells with aberrant glycosylation are detected and removed (Aarnoudse et al., 2006). CLEC7A, or Dectin-1, recognizes N-glycan structures found on the surface of some tumor cells and directs natural killer cells to aid in killing the tumor cell; Dectin-1-deficient mice have been found to have increased rates of tumor growth (Chiba et al., 2014).
Cytokines regulate various immunological responses, so downregulation in cytokine activity as implicated through decreased expression of Ccl6 in both TBBPA-treated proximal and distal uterus may be evidence of an immunotoxic insult mediated by TBBPA exposure. The cytokine CCL6 is a chemoattractant for helper T cells, B cells, and macrophages (Bizargity & Bonney, 2009). Additionally, CCL6 may play an important role in early brain development by attracting primitive macrophages and microglia into the developing brain (Bilbo & Schwarz, 2012). Another gene downregulated in both uterine sections in this study, Clca4, has been shown to be downregulated in breast tumors, and CLCA4 expression inhibits breast cancer cell proliferation (Yu et al., 2013).
TBBPA has recently been classified as a Group 2A carcinogen by the International Agency for Research on Cancer (IARC), indicating TBBPA is probably carcinogenic to humans based on sufficient evidence of carcinogenicity in experimental animals (Grosse et al., 2016). One of the findings from the IARC Monograph Working Group influencing this classification was the immunosuppressive effects of TBBPA (Grosse et al., 2016).
The current study was done using a dose of 250 mg/kg of TBBPA, a dose at which increased incidence of uterine epithelial atypical hyperplasia and neoplasia was seen in the NTP chronic bioassay (Dunnick et al., 2015; NTP, 2013). Given the results of this hypothesis-generating study and other in vivo and in vitro studies, it is important that the effects of TBBPA on the immune system be further characterized as one of the mechanisms by which chronic TBBPA exposure may lead to uterine tumors.
5. Conclusions
This work further characterizes the mechanism(s) by which TBBPA may contribute to the increased incidence of uterine tumors observed in female Wistar Han rats from the NTP chronic bioassay. The observed changes in gene expression shown here indicate that TBBPA treatment could lead to downregulation of the immune response. Previous data derived from the same animals in Sanders et al. (2016) supported a hypothesis that disruption of estrogen homeostasis is a major mechanism for the increased incidence of uterine lesions observed in TBBPA-treated rats at doses of 250 mg/kg and higher in the NTP study. With evidence from this work, it is further hypothesized that TBBPA exposure may trigger estrogen-mediated immunosuppression in these rats. Work is ongoing to characterize TBBPA-mediated effects on these pathways at lower dose exposures.
Supplementary Material
Highlights.
Perturbation of biological pathways in TBBPA-exposed female rats was studied
Gene expression changes were observed with microarray and validated with ddPCR
Genes in immune response pathways in uterus were downregulated following TBBPA
Immunosuppression may play a role in uterine tumor formation after TBBPA exposure
Acknowledgments
This work was supported by the Intramural Research Program of the National Cancer Institute of the National Institutes of Health [Project: ZIA BC 011476]. Microarray hybridizations were performed by the NIEHS Molecular Genomics Core.
Footnotes
Conflicts of interest
The authors declare that they have no actual or potential competing financial interests associated with this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarnoudse CA, Garcia Vallejo JJ, Saeland E, van Kooyk Y. Recognition of tumor glycans by antigen-presenting cells. Curr Opin Immunol. 2006;18(1):105–111. doi: 10.1016/j.coi.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Alaee M, Arias P, Sjodin A, Bergman A. An overview of commercially used brominated flame retardants, their applications, their use patterns in different countries/regions and possible modes of release. Environ Int. 2003;29(6):683–689. doi: 10.1016/S0160-4120(03)00121-1. [DOI] [PubMed] [Google Scholar]
- Berman J, Heredia N, Regan J, Cooper S, Tzonev S, Hefner E, Karlin-Neumann G. Droplet Digital™ PCR: High-Resolution Copy Number Variation Analysis. Bio-Rad Bulletin. 2012;6475:1–2. [Google Scholar]
- Bilbo SD, Schwarz JM. The immune system and developmental programming of brain and behavior. Front Neuroendocrinol. 2012;33(3):267–286. doi: 10.1016/j.yfrne.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizargity P, Bonney EA. Dendritic cells: a family portrait at mid-gestation. Immunology. 2009;126(4):565–578. doi: 10.1111/j.1365-2567.2008.02918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou R, Antignac JP, Zalko D, Berrebi A, Cravedi JP, Maume D, Marchand P, Monteau F, Riu A, Andre F, Le Bizec B. Exposure assessment of French women and their newborns to tetrabromobisphenol-A: occurrence measurements in maternal adipose tissue, serum, breast milk and cord serum. Chemosphere. 2008;73(7):1036–1041. doi: 10.1016/j.chemosphere.2008.07.084. [DOI] [PubMed] [Google Scholar]
- Chiba S, Ikushima H, Ueki H, Yanai H, Kimura Y, Hangai S, Nishio J, Negishi H, Tamura T, Saijo S, Iwakura Y, Taniguchi T. Recognition of tumor cells by Dectin-1 orchestrates innate immune cells for anti-tumor responses. Elife. 2014;3:e04177. doi: 10.7554/eLife.04177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope RB, Kacew S, Dourson M. A reproductive, developmental and neurobehavioral study following oral exposure of tetrabromobisphenol A on Sprague-Dawley rats. Toxicology. 2015;329:49–59. doi: 10.1016/j.tox.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Covaci A, Voorspoels S, Abdallah MA, Geens T, Harrad S, Law RJ. Analytical and environmental aspects of the flame retardant tetrabromobisphenol-A and its derivatives. J Chromatogr A. 2009;1216(3):346–363. doi: 10.1016/j.chroma.2008.08.035. [DOI] [PubMed] [Google Scholar]
- de Wit CA, Herzke D, Vorkamp K. Brominated flame retardants in the Arctic environment--trends and new candidates. Sci Total Environ. 2010;408(15):2885–2918. doi: 10.1016/j.scitotenv.2009.08.037. [DOI] [PubMed] [Google Scholar]
- Dunnick JK, Morgan DL, Elmore SA, Gerrish K, Pandiri A, Ton TV, Shockley KR, Merrick BA. Tetrabromobisphenol A activates the hepatic interferon pathway in rats. Toxicol Lett. 2016;266:32–41. doi: 10.1016/j.toxlet.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick JK, Sanders JM, Kissling GE, Johnson CL, Boyle MH, Elmore SA. Environmental chemical exposure may contribute to uterine cancer development: studies with tetrabromobisphenol A. Toxicol Pathol. 2015;43(4):464–473. doi: 10.1177/0192623314557335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladson M, Srinivasan N, Malini T, Arunakaran J, Aruldhas MM, Govindarajulu P. Interaction of estradiol, progesterone and corticosterone on uterine connective tissue degrading enzymes. Endocr Res. 1998;24(1):89–103. doi: 10.3109/07435809809031871. [DOI] [PubMed] [Google Scholar]
- Gorodeski GI, Sheean LA, Utian WH. Myometrial Oxytocin Receptors Levels in the Pregnant Rat Are Higher in Distal Than in Proximal Portions of the Horn and Correlate with Disparate Oxytocin Responsive Myometrial Contractility in These Segments. Endocrinology. 1990;127(3):1136–1143. doi: 10.1210/endo-127-3-1136. [DOI] [PubMed] [Google Scholar]
- Gosavi RA, Knudsen GA, Birnbaum LS, Pedersen LC. Mimicking of estradiol binding by flame retardants and their metabolites: a crystallographic analysis. Environ Health Perspect. 2013;121(10):1194–1199. doi: 10.1289/ehp.1306902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Ozaki Y, Suzumori N, Yasukochi A, Kawakubo T, Furuno T, Nakanishi M, Yamamoto K, Sugiura-Ogasawara M. Role of cathepsin E in decidual macrophage of patients with recurrent miscarriage. Mol Hum Reprod. 2014;20(5):454–462. doi: 10.1093/molehr/gau008. [DOI] [PubMed] [Google Scholar]
- Grosse Y, Loomis D, Guyton KZ, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Mattock H, Straif K. Carcinogenicity of some industrial chemicals. The Lancet Oncology. 2016;17(4):419–420. doi: 10.1016/S1470-2045(16)00137-6. [DOI] [PubMed] [Google Scholar]
- Hamers T, Kamstra JH, Sonneveld E, Murk AJ, Kester MH, Andersson PL, Legler J, Brouwer A. In vitro profiling of the endocrine-disrupting potency of brominated flame retardants. Toxicol Sci. 2006;92(1):157–173. doi: 10.1093/toxsci/kfj187. [DOI] [PubMed] [Google Scholar]
- Han EH, Park JH, Kang KW, Jeong TC, Kim HS, Jeong HG. Risk assessment of tetrabromobisphenol A on cyclooxygenase-2 expression via MAP kinase/NF-kappaB/AP-1 signaling pathways in murine macrophages. J Toxicol Environ Health A. 2009;72(21–22):1431–1438. doi: 10.1080/15287390903212873. [DOI] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher CH, Brooks DL, Johnson JR. A quantitative method for assessing stages of the rat estrous cycle. Biotech Histochem. 2005;80(2):79–87. doi: 10.1080/10520290500138422. [DOI] [PubMed] [Google Scholar]
- Hurd T, Whalen MM. Tetrabromobisphenol A decreases cell-surface proteins involved in human natural killer (NK) cell-dependent target cell lysis. J Immunotoxicol. 2011;8(3):219–227. doi: 10.3109/1547691X.2011.580437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson L, Holmdahl R. Estrogen-mediated immunosuppression in autoimmune diseases. Inflamm Res. 1998;47(7):290–301. doi: 10.1007/s000110050332. [DOI] [PubMed] [Google Scholar]
- Kawakubo T, Okamoto K, Iwata J, Shin M, Okamoto Y, Yasukochi A, Nakayama KI, Kadowaki T, Tsukuba T, Yamamoto K. Cathepsin E prevents tumor growth and metastasis by catalyzing the proteolytic release of soluble TRAIL from tumor cell surface. Cancer Res. 2007;67(22):10869–10878. doi: 10.1158/0008-5472.CAN-07-2048. [DOI] [PubMed] [Google Scholar]
- Kawakubo T, Yasukochi A, Toyama T, Takahashi S, Okamoto K, Tsukuba T, Nakamura S, Ozaki Y, Nishigaki K, Yamashita H, Yamamoto K. Repression of cathepsin E expression increases the risk of mammary carcinogenesis and links to poor prognosis in breast cancer. Carcinogenesis. 2014;35(3):714–726. doi: 10.1093/carcin/bgt373. [DOI] [PubMed] [Google Scholar]
- Kibakaya EC, Stephen K, Whalen MM. Tetrabromobisphenol A has immunosuppressive effects on human natural killer cells. J Immunotoxicol. 2009;6(4):285–292. doi: 10.3109/15476910903258260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerts IATM, van Zanden JJ, Luijks EAC, van Leeuwen-Bol I, Marsh G, Jakobsson E, Bergman A, Brouwer A. Potent competitive interactions of some brominated flame retardants and related compounds with human transthyretin in vitro. Toxicol Sci. 2000;56(1):95–104. doi: 10.1093/toxsci/56.1.95. [DOI] [PubMed] [Google Scholar]
- NTP. TR-587: Technical Report Pathology Tables and Curves for TR-587: Tetrabromobisphenol A (TBBPA) National Toxicology Program, Health and Human Services; 2013. Found at: http://ntp.niehs.nih.gov/go/38602. Retrieved from http://ntp.niehs.nih.gov/go/38602. [Google Scholar]
- Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen S, Boecker R, Tiegs G. The flame retardants tetrabromobisphenol A and tetrabromobisphenol A-bisallylether suppress the induction of interleukin-2 receptor alpha chain (CD25) in murine splenocytes. Toxicology. 2003;184(1):11–22. doi: 10.1016/s0300-483x(02)00442-0. [DOI] [PubMed] [Google Scholar]
- Sanders JM, Coulter SJ, Knudsen GA, Dunnick JK, Kissling GE, Birnbaum LS. Disruption of estrogen homeostasis as a mechanism for uterine toxicity in Wistar Han rats treated with tetrabromobisphenol A. Toxicol Appl Pharmacol. 2016;298:31–39. doi: 10.1016/j.taap.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ven LT, Van de Kuil T, Verhoef A, Verwer CM, Lilienthal H, Leonards PE, Schauer UM, Canton RF, Litens S, De Jong FH, Visser TJ, Dekant W, Stern N, Hakansson H, Slob W, Van den Berg M, Vos JG, Piersma AH. Endocrine effects of tetrabromobisphenol-A (TBBPA) in Wistar rats as tested in a one-generation reproduction study and a subacute toxicity study. Toxicology. 2008;245(1–2):76–89. doi: 10.1016/j.tox.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Watanabe W, Shimizu T, Sawamura R, Hino A, Konno K, Hirose A, Kurokawa M. Effects of tetrabromobisphenol A, a brominated flame retardant, on the immune response to respiratory syncytial virus infection in mice. Int Immunopharmacol. 2010;10(4):393–397. doi: 10.1016/j.intimp.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Whitten WK. Modification of the oestrous cycle of the mouse by external stimuli associated with the male; changes in the oestrous cycle determined by vaginal smears. J Endocrinol. 1958;17(3):307–313. doi: 10.1677/joe.0.0170307. [DOI] [PubMed] [Google Scholar]
- Yu Y, Walia V, Elble RC. Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells. PLoS One. 2013;8(12):e83943. doi: 10.1371/journal.pone.0083943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler Bros., Inc. Rodent NIH-31 Open Formula Auto. 2010 from http://www.zeiglerfeed.com/Literature/Rodent%20NIH-31%20Open%20Formula%20Auto.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.