Abstract
Plant development is predominantly postembryonic and tuned in to respond to environmental cues. All living plant cells can be triggered to de-differentiate, assume different cell identities, or form a new organism. This developmental plasticity is thought to be an adaptation to the sessile lifestyle of plants. Recent discoveries have advanced our understanding of the orchestration of plant developmental switches by transcriptional master regulators, chromatin state changes, and hormone response pathways. Here, we review these recent advances with emphasis on the earliest stages of plant development and on the switch from pluripotency to differentiation in different plant organ systems.
Introduction
Recent studies in both animals and plants have revealed that the epigenome contributes to cell identity and function [1, 2]. The epigenome comprises alternative chromatin states that can impact gene activity; they are not accompanied by alterations in nucleotide sequence but can nevertheless be passed on to daughter cells. It is now clear that a number of attributes of the chromatin impact the accessibility of the genome for transcription, including: the three-dimensional organization of the chromatin in the nucleus; chromatin condensation by linker histones and non-histone proteins; histone modifications or the presence of alternative histones (i.e., histone variants); the position and occupancy of the nucleosomes; and covalent modification of the DNA by methylation [3, 4]. Upon perceiving a relevant cue, enzyme complexes (Box 1) can alter the existing chromatin state, making new genomic regions accessible while closing others off, thus generating a “legible genome” that is specific to cell type, developmental stage, or environmental condition. In this review, we discuss some of the major developmentally or environmentally triggered transcriptional reprogramming events in plants, with special emphasis on the role of chromatin and the epigenome.
Early stages in plant development and response to environmental cues
From fertilization to embryo development
In angiosperms, seed development is initiated by a double-fertilization event, during which the egg cell and the central cell each fuse with a male sperm cell, resulting in the formation of the embryo and the endosperm, respectively (Fig. 1). The embryo and the endosperm are surrounded by maternal tissues such as the seed coat, which derives from the integuments [5]. Proper seed formation is achieved by the coordinated development of these three different tissue types [6]. The embryo initiates a shoot and a root apical meristem, two leaf-like structures called cotyledons, and a short stem termed the hypocotyl [7].
Fig. 1.

Function of chromatin regulators in seed development. a Prior to fertilization, DNA hypomethylation by DME enables maternal expression of PRC2, which deposits H3K27me3 at auxin response genes to inhibit the development of non-embryonic tissues. PRC2 acts in concert with LHP1 and histone H2A deubiquitinases (UBPs). b Post fertilization, paternally expressed genes raise the auxin hormone level and activate non-embryonic tissue development; this requires the endosperm expressed TF AGL62. The increased auxin levels reduce PRC2 expression. c CHD and SWI/SNF chromatin remodelers contribute to embryo pattering after fertilization. Black, chromatin regulators; blue, transcription factors; green, hormones
The endosperm is a nourishing tissue that supports embryo growth [8]. Its initiation and its correct development are necessary for the establishment of a viable seed [9]. Endosperm development prior to fertilization is inhibited by FIS (FERTILIZATION-INDEPENDENT SEED)-PRC2 (POLYCOMB REPRESSIVE COMPLEX2), which acts in the female gametophyte and during endosperm development. Loss of FIS-PRC2 function causes autonomous endosperm development without fertilization, resulting in seeds that develop an endosperm but no embryo [10]. More recently, histone ubiquitination and the hormone auxin were linked to endosperm formation. Two H2A deubiquitinases, Ubiquitin-Specific Protease 12 (UBP12) and UBP13, are partners of the H3K27me3-binding protein Like Heterochromatin Protein 1 (LHP1) [11]. These proteins are expressed in the central cell of the mature female gametophyte and are recruited to several Polycomb targets, where they are required for elevated H3K27me3 levels and for the repression of transcription. In the absence of LHP1 or UBP12/UBP13, autonomous endosperm develops, suggesting that LHP1 or UBP12/UBP13 may repress FIS-PRC2 targets [12] (Fig. 1a). Elevating auxin levels, either genetically or pharmacologically, induces replication of the central cell in the absence of fertilization [13], suggesting that auxin may promote endosperm formation. Indeed, FIS-PRC2 directly silences two auxin biosynthesis pathway genes, YUCCA10 (YUC10) and TRYPTOPHAN AMINOTRANSFERASE RELATED 1 (TAR1) in the maternal gametophyte; this lowers auxin levels in the central cell prior to fertilization. After fertilization, paternal expression of auxin biosynthesis genes enables an auxin increase in the fertilized central cell, which triggers the initiation of endosperm formation, bypassing the block by the maternal FIS-PRC2 [13] (Fig. 1b).
The endosperm is also the main site of genomic imprinting in flowering plants, an epigenetic phenomenon that results in the expression of a gene from just one of the two available alleles in a parent-of-origin-dependent manner [14]. Imprinting has evolved independently in mammals and flowering plants [15]. Differential DNA methylation underlies most imprinted gene expression [16]. Global removal of methylation from cytosines found in CG dinucleotides by the DEMETER (DME) DNA glycosylase occurs in the maternal genome of the endosperm in Arabidopsis thaliana, leading to hypomethylation [17, 18]. DME is expressed in the companion cells of the gametes, including the central cell of the female gametophyte before fertilization [19] (Fig. 1a, b). Genes that are exclusively maternally expressed (MEGs) are characterized by loss of repressive DNA methylation. Paternally expressed genes (PEGs) arise when reduced maternal DNA methylation enables an alternative epigenetic silencing mechanism—polycomb repression—to silence the maternal alleles [20, 21]. In some cases, parent-of-origin-specific H3K27me3 is not dependent on differential DNA methylation [22].
Among the numerous MEGs that have been identified are the FIS-PRC2 components MEDEA (MEA) and FIS2 [22]. As discussed above, mutation of the MEA or FIS2 components of FIS-PRC2 causes the formation of endosperm prior to fertilization and embryo abortion. This is due, at least in part, to de-repression of the maternal alleles of the PEGs YUC10 and TAR1 and to increased auxin levels [13, 23]. Interestingly, while some of the same genes (including the auxin biosynthesis genes) are imprinted in many different flowering plants, the majority of the imprinted genes are species-specific [24–26]. Moreover, imprinting at the same gene may be achieved by different mechanisms in different plant species. In Arabidopsis lyrata, an outcrossing plant species closely related to A. thaliana, many PEGs arise due to CHG methylation and repression of the maternal alleles, and the maternal endosperm genome is not hypomethylated [27]. One biological role of gene dosage or of imprinting in the endosperm may be as a hybridization barrier that underlies speciation [28, 29]. The maternal FIS-PRC2 may also buffer paternal genetic variation to prevent its influence on seed development [30]. Finally, imprinted gene expression may transmit environmental cues that are perceived by the mother plant to modulate seed germination [31].
A clever genetic trick has been used to enable egg cell fertilization in a prc2 null mutant background [32]. This gave rise to viable embryos that became abnormal only after germination, pinpointing the developmental window during which PRC2 function is first required in plant development [33]. Thus, unlike in animals [34], PRC2 is not strictly essential for embryo formation in plants. Other chromatin regulators are important for the development of the embryo proper (Fig. 1c); for example, double mutants in the redundantly acting SWI/SNF (SWItch/Sucrose Non-Fermentable) subfamily chromatin remodelers MINUSCULE1 (MINU1) and MINU2 cause embryo lethality, with abnormal cell divisions apparent by the globular stage [35]. Double mutants in the BRAHMA (BRM) and SPLAYED (SYD) SWI/SNF subfamily chromatin remodelers, which have overlapping roles, also cause embryo lethality, as do mutations in the SWI/SNF chromatin remodeling complex components SWI3A or SWI3B [36–40]. In the case of brm mutants, the embryo defect may result from reduced auxin response; double mutants in brm and the auxin response factor monopteros (mp) are embryo lethal [40].
Unlike the egg cell and the central cell, which are fertilized and give rise to the embryo and the endosperm, the maternal tissue of the ovule does not participate in the fertilization process, yet it also undergoes drastic changes in response to fertilization. The integuments undergo rapid cell division and expansion to form the seed coat [41], while the proximal region of the nucellus undergoes programmed cell death (PCD) [42]. Sporophyte PRC2 (EMBRYONIC FLOWER2 (EMF2)/VERNALIZATION2 (VRN2)-PRC2) exerts a block on seed coat development before fertilization, and lack of the core PRC2 subunits VRN2 and EMF2 results in dosage-dependent autonomous seed coat development [43]. Auxin and gibberellin (GA) signaling are activated in the seed coat post-fertilization, and exogenous application of GA3 or 2,4-D (auxin) or overproduction of these hormones promotes fertilization-independent seed coat development [23]. The production of auxin in the unfertilized central cell is sufficient to drive seed coat development [23, 43], and the endosperm-specific transcription factor (TF) AGAMOUS-like MADS box protein 62 (AGL62) [44] promotes the transport of auxin from the endosperm to the integuments via the transcriptional upregulation of a PGP-type auxin transporter [23, 45]. Genetically, auxin and PRC2 act in the same pathway, with auxin acting upstream of PRC2 and downregulating PRC2 accumulation, whereas GA is activated when PRC2 is removed from the integuments (Fig. 1a, b). These findings uncover a precisely tuned developmental switch, operating at the intersection of hormones and chromatin regulators, that provides coordinated development of the embryo, endosperm, and seed coat. It also balances the maternal and paternal genomes, thereby impacting survival and speciation.
Seed maturation and dormancy
In higher plants, seed development can be divided into two phases, morphogenesis (embryo and endosperm development) and maturation. Maturation ensures that the fully developed embryo accumulates sufficient storage compounds, while water content decreases and abscisic acid (ABA) levels increase. Finally, the seed acquires desiccation tolerance and enters a metabolically quiescent state [46]. The initiation of seed maturation is mainly controlled by three B3 domain TFs: LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID (ABA) INSENTITIVE3 (ABI3) and FUSCA3 (FUS3). These factors work in concert with the CCAAT-box binding complex component LEC1 to regulate ABA, auxin, GA, and sugar responses [47]. They form a complex gene regulatory network that activates largely overlapping downstream genes that are involved in starch and lipid biosynthesis. They also regulate the biosynthesis and/or catabolism of the hormones ABA and GA to balance their ratio during seed dormancy and germination [47, 48] (Fig. 2).
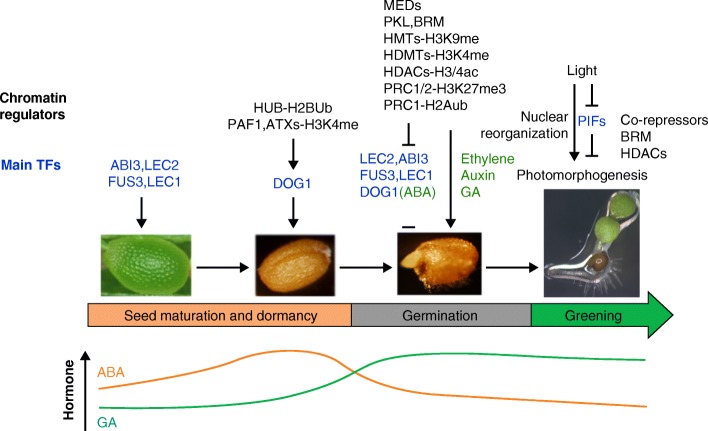
Fig. 2.
Involvement of chromatin regulators during seed maturation, dormancy, and germination. Master transcription factors (TFs), such as ABI3, LEC2, FUS3, LEC1, and DOG1, promote seed maturation and dormancy and trigger ABA hormone accumulation. The H2B ubiquitinase HUB, the PAF1 complex, and H3K4 methyltransferases (ATXs) promote DOG1 transcription. Subsequently, during germination, a large number of different chromatin regulators jointly repress expression of the LEC2, ABI3, FUS3, LEC1, and DOG1 TF and increase the GA/ABA hormone ratio. These chromatin regulators include MEDIATOR (MED) components, H3K9 methyltransferases (HMTs), H3K4 demethylases (HDMTs), HDACs, and the PRC1 and PRC2 Polycomb complexes. Finally, light signaling promotes photomorphogenesis, activates the seedling program, and triggers large scale nuclear reorganization, which is mediated by the PIF TFs and chromatin regulators such as BRM and HDACs
Seed dormancy is initiated during early seed maturation and continues after the seed completes its development, but then is gradually overcome during dry storage (after-ripening). The plant hormone ABA and the DELAY OF GERMINATION 1 (DOG1) protein are essential regulators of seed dormancy. ABA is produced during seed maturation and is necessary for the induction of seed dormancy and for maintenance of the dormant state following seed shedding. Factors that modulate ABA levels or signal transduction alter the seed dormancy level [49, 50]. DOG1, which was isolated by quantitative trait locus analysis, has been identified as a major and “specific” regulator of seed dormancy in A. thaliana and other plants, and DOG1 levels and activity are intricately regulated [51–53]. This regulation includes positive autoregulation at sites that are subject to natural variation [54]. A study of dormancy cycling in the soil implicated seed bank factors involved in the dynamics of chromatin remodeling in changing dormancy status by altering the expression of key regulators such as DOG1 [55]. The H2B ubiquitinases HISTONE MONOUBIQUITINATION 1 (HUB1) and HUB2 and the ARABIDOPSIS TRITHORAX-RELATED 7 (ATXR7) H3K4 methyltransferase promote seed dormancy by upregulating expression of DOG1 and other genes, presumably by influencing their H2Bub and H3K4 methylation statuses [56–58]. By contrast, SIN3-LIKE (SNL) co-repressor proteins promote seed dormancy by preventing the acetylation of H3K9/18 or H3K14 at genes linked to germination [59]. In addition, the HDA9 HDAC promotes seed dormancy by repressing genes that are related to photosynthesis and photoautotrophic growth [60, 61] (Fig. 2). During seed maturation, the expression of master transcriptional activators of seed dormancy is therefore upregulated by chromatin modifications that are potentially linked to transcriptional elongation, while genes that promote germination and photosynthesis are repressed by histone deacetylation.
Seed germination and greening (photomorphogenesis)
After the release of dormancy by environmental signals such as stratification [62], seed germination commences with protrusion of the radicle through the seed coat [47]. This process is facilitated by GA, which is newly synthesized in the imbibed embryo, and is inhibited by ABA [47]. Additional hormones such as ethylene and auxin also play roles in germination [63]. Low doses of auxin promote germination, whereas high doses inhibit this process [63]. Upon germination, the seed maturation program is silenced and seedling identity genes are activated. The broad changes in transcriptional programs that accompany this key developmental transition are orchestrated by a myriad of events that remodel and modify chromatin state (Fig. 2).
Repression of the seed maturation/dormancy program involves both the EMF2-PRC2 complex and PRC1, which silence the expression of seed maturation loci such as ABI3, LEC2, DOG1, and CHOTTO1 (CHO1)/AINTEGUMENTA-LIKE 5 (AIL5) during germination [33, 64–69]. The PRC1 complex is recruited by VP1/ABI3-like (VAL) proteins and PHD domain-containing ALFIN1-like (AL) proteins. AL protein can bind to H3K4me3. Two homologs of ZUOTIN-RELATED FACTOR1 (ZRF1), possible readers of H2Aub, contribute to Polycomb-mediated silencing of ABI3, CRUCIFERIN 3 (CRU3), and CHO1/AIL5 [70]. Histone deacetylases (HDACs) such as HDA19 and HDA6 also repress seed maturation genes [71–74]; these enzymes are recruited by diverse TFs, including VAL2, SCARECROW-LIKE15 (SCL15), and BRI1-EMS-SUPPRESSOR1 (BES1), and by the TOPLESS (TPL) co-repressor [73, 74]. Finally, H3K9 methylation by SU(VAR)3-9 HOMOLOG 4 (SUVH4) and SUVH5 and chromatin remodeling by the chromodomain (CHD) family member PICKLE (PKL) and by the SWI/SNF chromatin remodeler BRM also contribute to the silencing of dormancy and of embryonic genes [75–79]. The histone H3K4me2/3 demethylases LYSINE SPECIFIC DEMETHYLASE LIKE 1 (LDL1) and LDL2, by contrast, assist in the process by removing activating histone modifications from the seed dormancy genes [80].
To promote germination, the histone arginine demethylases JUMONJI DOMAIN-CONTAINING PROTEIN 20 (JMJ20) and JMJ22 remove repressive histone arginine methylation from two GA biosynthesis genes, GIBBERELLIN 3 BETA-HYDROXYLASE1 (GA3OX1) and GA3OX2 [81]. In addition, SNL co-repressors slow the speed of seed germination by inhibiting auxin synthesis and directly repress the expression of auxin transporters such as AUXIN RESISTANT1 (AUX1) [82]. Increased H3 lysine 9 or 18 acetylation (H3K9/18 ac) at AUX1 was observed in snl1 snl2 mutants. AUX1 enhances radicle emergence by promoting CYCLIN D expression [82].
When the seedling emerges from the soil, photomorphogenesis commences; this is characterized by reduced hypocotyl elongation, by cotyledon opening and expansion, and by chlorophyll biosynthesis [83]. The switch from heterotrophic to autotrophic growth is accompanied by large-scale transcriptional reprogramming in the context of chromatin (Fig. 2). Light exposure triggers nuclear architecture reorganization, which involves events such as nuclear size expansion, heterochromatin condensation and globally increased RNA Pol II activity [84]. This nuclear architectural change is induced mainly by blue light and is independent of local DNA methylation changes [84]. Germination is coupled to red/far-red light sensing by the light labile PHYTOCHROME-INTERACTING FACTORs (PIFs). For example, PIF1 inhibits seed germination in the dark by increasing ABA and by decreasing GA levels and response, as well as by repressing genes that are required for cell wall loosening [85]. PIF1 recruits the LEUNIG HOMOLOG (LUH) of the Groucho family transcriptional co-repressor to a subset of its targets [86]. PIF1 also inhibits chlorophyll biosynthesis by recruiting the SWI/SNF chromatin remodeling ATPase BRM to the chlorophyll biosynthesis gene PROTOCHLOROPHYLLIDE OXIDOREDUCTASE C (PORC) to repress its expression [87]. The CHD chromatin remodeling ATPase PKL is required for 80% of the gene expression changes triggered by GA [88]. Finally, CCAAT-box binding factors redundantly repress light-controlled hypocotyl elongation, interact with HDA15, and bind to the promoters of hypocotyl elongation genes such as IAA10 and XTH17 [89]. Germination and establishment of autotrophic seedling growth thus not only rely on chromatin modification and remodeling in response to environmental cues that trigger repression of the embryonic and dormancy programs, but are also accompanied by altered hormone environments and large scale nuclear reorganization.
Pluripotency and differentiation in plant development
De-differentiation and callus formation
All living plant cells can de-differentiate (i.e., form callus) when exposed to a combination of auxin and cytokinin (CK) hormones, and it has been proposed that de-differentiation occurs through a root developmental pathway [90]. Asexual propagation via induced de-differentiation and subsequent regeneration of a new plant is of economical importance for diverse species from oil palms to orchids [91]. Callus formation in plants—like induced pluripotency in animals —requires epigenetic reprogramming [92]. In agreement with these findings, callus formation is accompanied by rapid loss of H3K27me3 from many genes, including several that are linked to the auxin pathway [93]. However, induced differentiation from leaves also requires PRC2 activity, presumably to silence the foliar gene-expression program [93]. PKL opposes callus formation and mutants in which this chromatin remodeling ATPase is defective are hypersensitive to CK and show enhanced callus greening [94]. By contrast, several HDACs promote callus formation and are transcriptionally upregulated during callus induction [95]. For example, mutation of HDA9 or HD-TUINS PROTEIN 1 (HDT1) causes reduced callus formation that is correlated with a lack of meristematic gene activity (Fig. 3) [95].
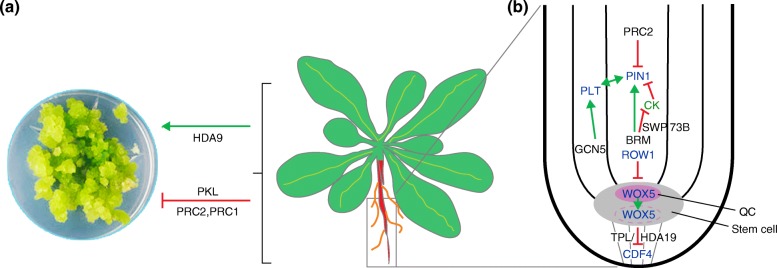
Fig. 3.
Role of chromatin remodelers in callus formation and root development. a Spontaneous de-differentiation of meristematic plant tissues is prevented by Polycomb repression (PRC2, PRC1), while the CHD chromatin remodeler PKL prevents induced callus formation. Histone deacetylation (HDA9) promotes de-differentiation in induced callus. b Stem cell maintenance is promoted by upregulation of auxin transport (via PIN1) through BRM/SWP73B and histone acetylation (GCN5) to promote expression of the PLT TFs. It also requires repression of CDF4 by WOX5 and TPL/HDA19. Differentiation is promoted by PRC2 (which represses PIN1 expression) and by ROW1, which prevents expansion of WOX5 expression. Black, chromatin regulators; blue, transcription factors; green, hormones
A recent genome-wide transcriptome comparison between wild-type leaves and leaf explant-derived calli identified 10,405 differentially expressed genes [96]. Not surprisingly, key TFs involved in leaf development were downregulated in the calli. In addition, 115 genes that are involved in chromatin remodeling were differentially expressed in calli. Notably, the expression of chromatin regulators that act in opposition to Polycomb repression (H3K4 methyltransferases of the Trithorax family of proteins) was elevated; these chromatin regulators may promote the expression of meristematic genes (Fig. 3). Reprogramming of cell identity through de-differentiation is not perfect and frequently results in DNA hypomethylation [97]. In the case of the oil palm, hypomethylation of the retrotransposon Karma in the B class floral homeotic gene results in flower-patterning defects and failure to form seeds [97]. On the other hand meristematic cells in plants need to be protected from de-differentiation. Polycomb repression prevents spontaneous de-differentiation and the repressive marks set by PRC2 are crucial for maintaining the identity of differentiation programs [98]. In particular, loss of PRC2 function leads to loss of cell identity and to callus formation from meristems in the shoot and root [33, 98]. The spontaneous callus formed from meristematic tissues in polycomb mutants differs from the induced callus in that it frequently produces somatic embryos [33, 98, 99]. In conclusion, de-differentiation of mature plant tissues is accompanied by large-scale epigenetic reprogramming in response to hormonal cues; this can result in epigenome defects in asexually produced plants. At the same time, plant meristematic tissues require Polycomb repression to block spontaneous de-differentiation.
Root formation and the root stem cell niche
Chromatin regulators have been implicated in the establishment and maintenance of the primary and lateral root meristems. The EMF2-PRC2 complex directly represses the expression of the auxin transport protein PIN-FORMED1 (PIN1), which is important for rootward auxin flux, and thus reduces auxin accumulation and meristematic activity in both the primary and lateral root [100]. As a consequence, the rate of lateral root initiation is increased in prc2 mutants [100]. PRC2 is expressed in the meristem and in the vasculature, and upstream regulators that control the spatiotemporal accumulation of PRC2 at the transcriptional level have recently been identified [101]. By contrast, the SWI/SNF chromatin remodeler BRM directly activates the expression of PIN1 in the root [102]. Knockdown of SWI/SNF Associated Protein 73 (SWP73) causes defective roots with short meristems that have increased CK levels [103]. SWP73 represses the expression of ATP/ADP isopentenyltransferase (IPT) enzymes that regulate the rate-limiting step in CK biosynthesis. SWP73 binds to the IPT3 and IPT7 loci and destabilizes a positive gene regulatory loop (Fig. 3) [103]. The histone acetyltransferase GENERAL CONTROL NONDEREPRESSIBLE 5 (GCN5) promotes expression of PLETHORA (PLT) genes, which act in a positive feedback with the auxin pathway to promote maintenance of the root stem cell niche [104]. Finally, deacetylation also plays a role in the root meristem. The WUSCHEL HOMEOBOX5 (WOX5) gene is expressed in the quiescent center (QC) and promotes stem cell fate in the surrounding initial cells (Fig. 3b). WOX5 directly represses expression of the TF CYCLING DOF FACTOR 4 (CDF4), which promotes differentiation, in the QC and in the columella stem cells [105]. WOX5 protein moves into the columella stem cells and recruits the TPL/HDA19 repressor complex to lower H3 acetylation at the CDF4 locus regulatory region. WOX5 expression in turn is confined to the QC by the PHD domain-containing protein REPRESSOR OF WUSCHEL1 (ROW1) [106]. When ROW1 binds to the activating H3K4me3 marks on the WOX5 promoter, it silences WOX5 expression by an as yet uncharacterized mechanism, restricting shootward expansion of the WOX5 expression domain (Fig. 3). The transition from cell proliferation to differentiation in the root is preceded by eviction of the canonical histone H3.1 and its replacement with the H3.3 histone variant [107]. Thus, a multilayered chromatin regulatory and hormonal network controls root meristem maintenance and size.
SAM initiation and maintenance
Maintenance of the shoot apical meristem (SAM) is tightly controlled by opposite-acting pathways. WUSCHEL (WUS) and CLAVATA3 (CLV3) are two key stem cell regulators, with WUS being expressed specifically in the organizing center (OC) located below the stem cell pool (Fig. 4c). WUS non-cell-autonomously maintains stem cell identity by upregulating CLV3 expression [108]. CLV3 is processed into a small peptide that limits WUS expression and prevents uncontrolled SAM proliferation [109]. Recently, the bHLH TF HECATE1 (HEC1) was shown to repress WUS and CLV3 expression by integrating CK and auxin signals [110]. The TF FAR-RED ELONGATED HYPOCOTYL3 (FHY3) acts as a direct repressor of CLV3, thus maintaining the stem cell pool [111]. The GRAS family TF HAIRY MERISTEM (HAM) physically interacts with WUS/WOX proteins in various stem cell niches, and HAM and WUS regulate similar sets of genes [112]. WUS also represses the expression of the differentiation-related gene KANADI1 (KAN1; Fig. 4a) [113]. Recently, the interaction between TPL/HDAC and WUS, which is required for stem cell fate promotion, was mapped to the WUS box and not to the EAR motif frequently implicated in transcriptional repression [114]. A separate pathway for SAM initiation and maintenance acts through the homeodomain TF SHOOTMERISTEM-LESS (STM), which induces CK biosynthesis [115]. CK acts as a positive regulator of WUS expression, mainly through the perception of CK by two CK receptors, ARABIDOPSIS HIS KINASE 2 (AHK2) and AHK4 [116]. A long-distance SAM-promoting pathway that involves the bypass (bps) signal has recently been uncovered [117]. bps1 mutants fail to maintain meristem identity and WUS expression because of defects in CK response.
Fig. 4.
Chromatin remodelers regulate differentiation and proliferation. a In the organizing center of the shoot apical meristem, WUS expression is promoted by chromatin remodeling (via MINU1 and MINU2) and possibly chromatin assembly (involving FAS1). WUS in turn represses differentiation genes such as KANADI1 (KAN) via histone deacetylation (involving TPL and HDA). In the leaf primordium, founder cell driver transcription factors (AS1, AS2, and JLO) repress KNOX gene expression via Polycomb repression, histone deacetylation, and histone variant incorporation (involving PRC2, LHP1, HDA6, and HirA). b In the inflorescence meristem center, stem cell maintenance is promoted by chromatin remodeling (SYD), histone arginine methylation (PRMT5), and a chromatin reader (OBE). SYD directly promotes expression of WUS, a positive regulator of stem cell fate. PRMT5, on the other hand, upregulates CLV3. CLV3 counteracts SAM overproliferation by inhibiting WUS expression. Flower primordium initiation at the flanks of the inflorescence meristem requires an auxin-triggered switch from a compacted chromatin state (TPL/HDA19) to an open chromatin state (BRM/SYD) at MP-bound target genes such as LFY and FIL that promote flower primordium fate. Finally, flower patterning requires the removal of Polycomb repression at floral homeotic genes such as AP3 and AG. This is enabled by the concerted action of the chromatin remodelers SYD and BRM that are recruited by SEP3 and LFY. PKL and histone H3K4 methylation (ATX1, ULT1) also contribute to overcoming Polycomb repression at AP3 and AG. c In different regions of the leaf, the chromatin remodeler BRM promotes either differentiation or cell division. Towards the tip of the leaf, BRM and TCP4 activate the ARR16 TF gene. ARR16 inhibits CK response, which promotes differentiation. At the base of the leaf, BRM and AN3 induce expression of genes encoding the GRF and HEC1 TFs. GRF and HEC1 have been implicated in promoting cell proliferation in the leaf. d After floral organ initiation, the flower meristem terminates. The floral homeotic TF AG represses WUS expression directly and indirectly by promoting Polycomb repression. KNU, a direct target of AG, also represses WUS. The expression of STM, a KNOX gene, is silenced by PRC1. The class A floral homeotic gene AP1 lowers CK hormone levels to promote differentiation. Black, chromatin regulators; blue, transcription factors; green, hormones
Only a handful of chromatin regulators have been implicated in SAM establishment and maintenance. FASCIATA1 (FAS1), a subunit of the A. thaliana homolog of CHROMATIN ASSEMBLY FACTOR-1 (CAF-1), which is responsible for replication-coupled incorporation of the canonical histone H3.1, is linked to stem cell maintenance [118]. fas1 mutants enhance the SAM defects of weak wus mutants, yet fas single mutants have an expanded WUS-expressing domain and an enlarged SAM [118]. One explanation for this apparent discrepancy may be that fas1 mutants fail to initiate a functional organizing center upon germination; this defect may result in the establishment of new WUS-expressing cell populations that subsequently fuse to form one large fasciated meristem (Fig. 4a). A similar sequence of events was observed in double mutants of the redundantly acting MINU1 and MINU2 SWI/SNF chromatin remodeling ATPases; hypomorph minu1 minu2 mutants form multiple primary meristems [35]. PROTEIN ARGININE METHYLTRANSFERASE 5 (PRMT5), a member of the type II arginine methyltransferase family, directly binds to the regulatory regions of the gene encoding the CLV3 peptide receptor CORYNE (CRN) (Fig. 4b). The resulting H3R4me2 methylation represses CRN expression and this promotes meristem homeostasis [119]. Similarly, OBERON3 (OBE3), which encodes a PHD finger-containing protein, acts as a positive regulator of WUS expression in a mutual positive feedback loop (Fig. 4b) [120]. Finally, the SWI/SNF chromatin remodeler SYD directly promotes the maintenance of WUS expression [121].
Leaf development
To enable leaf initiation at the flanks of the shoot apex, the MYB TF ASYMMETRIC LEAVES 1 (AS1) and its partner the LATERAL ORGAN BOUNDARY (LBD) domain TF AS2 repress the expression of pluripotency genes. AS1 and AS2 directly recruit the HirA histone H3.3 chaperone and PRC2 to the regulatory regions of the Class-I KNOTTED1-like homeobox (KNOX) family genes BREVIPEDICELLUS (BP) and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 2 (KNAT2) to silence them [122, 123]. The LBD protein JAGGED LATERAL ORGAN (JLO) contributes to AS2-mediated KNOX repression by forming a trimeric complex with AS1. Loss of JLO function leads to the ectopic expression of STM and BP [124]. The HDAC HDA6 also interacts with AS1 and directly represses KNOX gene expression [125]. More recently, LHP1, also known as TERMINAL FLOWER2 (TFL2), was shown to contribute to KNOX gene repression via direct physical interaction with AS1 and AS2 (Fig. 4a) [126]. LHP1 may promote the spread of H3K27me3 [127]. LHP1 and AS1/AS2 have many additional direct targets that have roles in leaf development and maturation [126, 128].
A. thaliana leaf cells also face a choice between proliferation and differentiation. Leaf differentiation is promoted by recruitment of the chromatin remodeler BRM and the dedicated BRM complex component SWI3C by the CINCINNATA-like TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (CIN-TCP) TF TCP4 [129]. BRM, together with TCP4, reduces CK responsiveness by promoting the expression of an inhibitor of CK response, ARABIDOPSIS RESPONSE REGULATOR 16 (ARR16). The transcription co-activator ANGUSTIFOLIA3 (AN3), on the other hand, promotes cell proliferation in leaves [130]. AN3 directly induces the expression of GROWTH REGULATING FACTORS (GRFs) and HEC1. These genes are also direct targets of the SWI/SNF complex components SWP73B and BRM, with which AN3 physically interacts (Fig. 4c). A subsequent study additionally implicated SWP73B in leaf polarity [131]. Similar interactions and roles for AN3 and SWI/SNF were also observed in maize leaf development [132]. Thus, AS1 is at the center of a chromatin repressor hub that promotes leaf initiation, whereas opposite roles of the SWI/SNF complex in leaf maturation are distinguished by the presence of the AN3 co-activator (Fig. 4b, c).
Flower development
Organogenesis (flower primordium initiation) from stem cell descendants at the periphery of the inflorescence meristem requires an auxin maximum that activates the AUXIN RESPONSE FACTOR 5 (ARF5) or MONOPTEROS (MP) [133]. When auxin levels are low, negative regulators of auxin response, the auxin-labile AUX/IAA proteins, bind to ARFs such as MP and generate a repressive chromatin environment [133]. AUX/IAA proteins physically interact with and recruit the TPL/HDA19 co-repressor and additionally prevent MP from interacting with the SWI/SNF ATPases SYD and BRM [40, 134]. The histone deacetylation generates a repressive chromatin environment near MP-binding sites that prevents the activation of auxin response genes in the absence of the hormonal cue. Increased auxin levels in the primordium founder cells lead to AUX/IAA protein degradation, loss of TPL/HDA19, and physical interaction of SWI/SNF complexes with MP. SYD and BRM open up the chromatin at MP target loci such as LEAFY (LFY) and FILAMENTOUS FLOWERS (FIL) [40]. Histone acetylation might also contribute to this process. For example, the bZIP11 TF recruits the GCN5 histone acetyltransferase to promote the expression of auxin biosynthesis genes [135].
Floral meristems (FMs) arise from subapical stem cells in the center of the inflorescence [136] and give rise to the primordia of the floral organs [137]. In stage 2 flowers, the FM is fully formed and floral organ primordium patterning is initiated by the activation of the floral homeotic genes. Prior to this developmental time point, floral homeotic genes are silenced by Polycomb repression [137]. ARABIDOPSIS TRITHORAX 1 (ATX1) promotes upregulation of the floral homeotic genes through H3K4 tri-methylation [138]. In addition, the master regulator of floral cell fate, LFY, together with the MADS-domain TF SEPALLATA3 (SEP3) recruits the SWI/SNF chromatin remodelers BRM and SYD to the class B and class C floral homeotic genes [39]. The activity of the remodelers is absolutely required to upregulate the floral homeotic genes, and the combined LFY and SEP3 expression domains in the FM overlap with the sites where these floral homeotic genes are induced. The SAND domain-containing protein ULTRAPETALA1 (ULT1) acts in parallel with LFY to activate the C class floral homeotic gene AGAMOUS (AG) [139]. Finally, the CHD chromatin remodeler PKL also promotes flower patterning and upregulation of the floral homeotic genes [140].
Interestingly, flower patterning and the activation of the floral homeotic genes is linked to flower meristem termination [137]. Like the vegetative SAM and the reproductive inflorescence meristem, FMs express the pluripotency factors WUS and STM [135] (Fig. 4d). The class C floral homeotic gene AG directly represses the expression of the stem cell-promoting gene WUS in the center of the flower meristem with the help of PRC2 [141]. In addition, AG activates the zinc finger protein KNUCKLES (KNU), which in turn directly represses WUS and thus terminates meristem identity (Fig. 4d) [142]. ARF3 binds to the chromatin of WUS in an AG-dependent manner and directly represses WUS expression to promote FM determinacy [143]. In addition, the SAND domain protein ULT1 represses WUS expression, working together with its partner ULT1 INTERACTING FACTOR 1 (UIF1), a MYB and EAR domain-containing TF that can bind to WUS regulatory regions [144]. In parallel, AtRING1a and AtRING1b (core components of the PRC1 complex) contribute to the termination of floral stem cell fate through repression of KNOX genes [145]. Finally, the class A floral homeotic gene AP1 suppresses meristematic activity in the axils of the outermost floral organs, the sepals, by lowering CK levels (Fig. 4d). AP1 directly represses the expression of the CK biosynthetic gene LONELY GUY1 (LOG1) and directly upregulates the CK degradation gene CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3) (Fig. 4d) [146]. AP1 can physically interact with transcriptional co-repressors linked to histone deactylation and with SWI/SNF group chromatin remodelers [147–149]. Thus, in flowers, tightly regulated chromatin state switches promote organ initiation, flower patterning, and meristem termination.
Discussion
The picture that emerges from the recent investigations is that developmental transitions in plants are orchestrated by the combined activities of transcription factors, hormone response pathways, and regulators of chromatin state. There is crosstalk between these three regulatory layers. For example, transcription factors recruit chromatin enzymes but are also dependent on chromatin remodeling for the ability to bind target genes. The hormonal pathways trigger chromatin state changes, and chromatin modification and remodeling alter hormone accumulation, signaling, and response. Finally, hormone environments alter transcription factor activity and transcription factors modulate hormone levels and response. In addition, the large-scale transcriptional reprogramming that occurs during major developmental switches relies on many diverse chromatin regulators; this enhances both the robustness of the underlying chromatin state changes and the plant's ability to fine-tune the response to diverse cues. Other conclusions are less universal. For example, while Polycomb repressive complexes and SWI/SNF chromatin remodeling ATPases frequently act in opposition, they can also jointly promote a specific reprogramming event by acting on the same or on different targets.
A longstanding question has been whether the writers, erasers, and readers of the chromatin state changes that accompany major reprogramming events are permissive—working by allowing master transcriptional regulators to exert their roles in transcriptional reprogramming or by preventing them from doing so—or whether they can also be decisive, that is to say they can interpret intrinsic and extrinsic cues to trigger the reprogramming events. While the jury is still out on this question, what has become clear is that the boundaries between TFs and the chromatin regulators are becoming more and more blurred. Some TFs in plants are more promiscuous in their genome occupancy than chromatin regulators [150]. In addition, as outlined above, it has become apparent that many developmental or environmental cues are directly interpreted by chromatin regulators and modulate their spatial, temporal, and condition-dependent accumulation or activity [101, 151–154].
To better understand developmental transition in the context of chromatin in plants, the future presents a number of challenges:
To elucidate the cell-, tissue- and condition-dependent roles of chromatin regulators using spatially restricted loss- and gain-of-function mutants in these regulators combined with cell- and tissue-specific epigenome analyses.
To identify in temporal resolution the order and logic of the series of chromatin state changes that lead to the repression and activation of new gene expression programs.
To define the composition of the individual or multifunctional complexes that trigger chromatin state changes and to determine how their formation and activity are controlled by extrinsic or intrinsic cues.
To uncover the biological roles in plant development or stress responses of the large number of predicted chromatin regulators present in plant genomes whose biological roles are not yet understood.
Box 1. Chromatin regulators
Genomic DNA that is wrapped around the histone octamer in nucleosomes is much less accessible than DNA that is not in contact with histones. Nucleosome occupancy (the fraction of a specific genomic DNA fragment that is wrapped around a histone octamer in a population of cells/nuclei) or nucleosome positioning (the identity of the specific DNA fragment wrapped around the histone octamer in a larger region of interest) can be altered by chromatin remodeling using the energy derived from ATP hydrolysis to break the histone–DNA contacts [155]. Plants have a large number of chromatin remodelers, but the SWItch/ Sucrose Non-Fermentable (SWI/SNF) complexes formed around BRAHMA (BRM) and SPLAYED (SYD) and the chromodomain (CHD) family chromatin remodeling ATPase PICKLE (PKL) are the most studied [156].
Histone variants are predominantly incorporated into nucleosomes outside of DNA replication and differ in primary sequence from “canonical” histones. These sequence differences impact the properties of the histone variants and those of the nucleosome particles that contain them [157]. In this review, the histone variants H2A.Z, H3.3, and H1.3 are discussed.
Covalent modification of histones is executed by “writers”—enzymes that covalently alter amino acids in the histones through acetylation, methylation, ubiquitylation, or phosphorylation, for example [158]. Many of these enzymes act in complexes. Histone acetyltransferases (HATs) generally cause increased genome accessibility (less compaction), whereas the effects of lysine methylation are strongly context dependent. Polycomb Repressive Complex 2 (PRC2) generates tri-methylation on lysine 27 of histone H3 (H3K27me3), a transcription-repressive mark, whereas tri-methylation on lysine 4 of histone H3 (H3K4me3) is associated with open chromatin and active transcription. Histone arginine methylation is frequently repressive. Ubiquitination on lysine 121 of histone H2A (H2AK121ub) is generated by PRC1, an enzyme complex that also contains non-histone proteins that strongly compact chromatin. Ubiquitination on lysine 143 of histone H2B (H2BK143ub) promotes transcriptional elongation.
Additional non-histone proteins have specific protein domains (such as PHD domains) that can recognize histone modifications; these downstream effectors are called “readers”. Readers interpret the chromatin state and contribute to the final chromatin compaction and transcription outcome.
Finally, all covalent histone marks are reversible, their removal being executed by so-called “erasers”. There are a myriad of erasers in plants; in this review, histone deacetylases (HDACs), which remove histone lysine acetylation, feature most prominently. HDACs, on their own or together with Polycomb repression, compact chromatin to silence unnecessary or detrimental gene expression programs.
In plants, cytosine DNA methylation occurs in three sequence contexts: CG, CHG, and CHH (where H equals A, T, or C) [159]. Specialized complexes have been linked to the initiation, maintenance, and removal of cytosine methylation. Cytosine methylation is frequently associated with transposable elements, and some of these transposable elements have been co-opted for the transcriptional regulation of nearby genes, generally silencing gene expression when methylated. Removal of CGme is executed by the DEMETER (DME) DNA glycosylase, which has prominent roles in imprinting in the endosperm.
Acknowledgements
We apologize to those whose work was not cited because of space limitations. We thank Samantha Klasfeld for assistance in preparing figures. Work in the Wagner lab is supported by funding from National Science Foundation (NSF MCB-1614355 and IOS-1557529 to DW).
Authors’ contributions
All authors wrote, read, and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ABA
Abscisic acid
- ABI3
ABA INSENTITIVE3
- AG
AGAMOUS
- AHK2
ARABIDOPSIS HIS KINASE 2
- AIL5
AINTEGUMENTA-LIKE 5
- AN3
ANGUSTIFOLIA3
- ARF5
AUXIN RESPONSE FACTOR 5
- AS1
ASYMMETRIC LEAVES 1
- ATX1
ARABIDOPSIS TRITHORAX 1
- AUX1
AUXIN RESISTANT1
- BP
BREVIPEDICELLUS
- bps
bypass
- BRM
BRAHMA
- CDF4
CYCLING DOF FACTOR 4
- CHD
Chromodomain
- CHO1
CHOTTO1
- CK
Cytokinin
- CLV3
CLAVATA3
- CRN
CORYNE
- DME
DEMETER
- DOG1
DELAY OF GERMINATION 1
- EMF2
EMBRYONIC FLOWER2
- FAS1
FASCIATA1
- FIS
FERTILIZATION-INDEPENDENT SEED
- FM
Floral meristem
- GA
Gibberellin
- GA3OX1
GIBBERELLIN 3 BETA-HYDROXYLASE1
- GCN5
GENERAL CONTROL NONDEREPRESSIBLE 5
- HAM
HAIRY MERISTEM
- HDAC
Histone deacetylase
- HEC1
HECATE1
- HUB1
HISTONE MONOUBIQUITINATION 1
- IPT
Isopentenyltransferase
- JLO
JAGGED LATERAL ORGAN
- JMJ20
JUMONJI DOMAIN-CONTAINING PROTEIN 20
- KNOX
KNOTTED1-like homeobox
- KNU
KNUCKLES
- LBD
LATERAL ORGAN BOUNDARY
- LDL1
LYSINE SPECIFIC DEMETHYLASE LIKE 1
- LEC2
LEAFY COTYLEDON2
- LFY
LEAFY
- LHP1
Like Heterochromatin Protein 1
- MEA
MEDEA
- MEG
Maternally expressed gene
- MINU1
MINUSCULE1
- mp
monopteros
- PEG
Paternally expressed gene
- PIF
PHYTOCHROME-INTERACTING FACTOR
- PIN1
PIN-FORMED1
- PRC2
POLYCOMB REPRESSIVE COMPLEX2
- QC
Quiescent center
- ROW1
REPRESSOR OF WUSCHEL1
- SAM
Shoot apical meristem
- SEP3
SEPALLATA3
- SNL
SIN3-LIKE
- STM
SHOOTMERISTEM-LESS
- SUVH4
SU(VAR)3-9 HOMOLOG 4
- SWI/SNF complex
SWItch/Sucrose Non-Fermentable chromatin remodeling complex
- SWP73
SWI/SNF Associated Protein 73
- SYD
SPLAYED
- TAR1
TRYPTOPHAN AMINOTRANSFERASE RELATED 1
- TF
Transcription factor
- TPL
TOPLESS
- UBP12
Ubiquitin-Specific Protease 12
- ULT1
ULTRAPETALA1
- VAL
VP1/ABI3-like
- VRN2
VERNALIZATION2
- WOX5
WUSCHEL HOMEOBOX5
- WUS
WUSCHEL
- YUC10
YUCCA10
References
- 1.Pikaard CS, Mittelsten SO. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol. 2014;6:a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moris N, Pina C, Arias AM. Transition states and cell fate decisions in epigenetic landscapes. Nat Rev Genet. 2016;17:693–703. doi: 10.1038/nrg.2016.98. [DOI] [PubMed] [Google Scholar]
- 3.He G, Elling AA, Deng XW. The epigenome and plant development. Annu Rev Plant Biol. 2011;62:411–35. doi: 10.1146/annurev-arplant-042110-103806. [DOI] [PubMed] [Google Scholar]
- 4.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 5.Berger F, Hamamura Y, Ingouff M, Higashiyama T. Double fertilization—caught in the act. Trends Plant Sci. 2008;13:437–43. doi: 10.1016/j.tplants.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Sreenivasulu N, Wobus U. Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol. 2013;64:189–217. doi: 10.1146/annurev-arplant-050312-120215. [DOI] [PubMed] [Google Scholar]
- 7.Wendrich JR, Weijers D. The Arabidopsis embryo as a miniature morphogenesis model. New Phytol. 2013;199:14–25. doi: 10.1111/nph.12267. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Berger F. Endosperm: food for humankind and fodder for scientific discoveries. New Phytol. 2012;195:290–305. doi: 10.1111/j.1469-8137.2012.04182.x. [DOI] [PubMed] [Google Scholar]
- 9.Hehenberger E, Kradolfer D, Kohler C. Endosperm cellularization defines an important developmental transition for embryo development. Development. 2012;139:2031–9. doi: 10.1242/dev.077057. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1997;94:4223–28. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:855–66. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derkacheva M, Liu S, Figueiredo DD, Gentry M, Mozgova I, Nanni P, et al. H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat Plants. 2016;2:16126. doi: 10.1038/nplants.2016.126. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo DD, Batista RA, Roszak PJ, Köhler C. Auxin production couples endosperm development to fertilization. Nat Plants. 2015;1:15184. doi: 10.1038/nplants.2015.184. [DOI] [PubMed] [Google Scholar]
- 14.Gehring M. Genomic imprinting: insights from plants. Annu Rev Genet. 2013;47:187–208. doi: 10.1146/annurev-genet-110711-155527. [DOI] [PubMed] [Google Scholar]
- 15.Pires ND, Grossniklaus U. Different yet similar: evolution of imprinting in flowering plants and mammals. F1000Prime Rep. 2014;6:63. doi: 10.12703/P6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife. 2014;3:1–24. doi: 10.7554/eLife.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibarra CA, Feng XQ, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–4. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park K, Kim MY, Vickers M, Park J-S, Hyun Y, Okamoto T, et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci U S A. 2016;113:201619047. doi: 10.1073/pnas.1619047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J-S, Frost JM, Park K, Ohr H, Park GT, Kim S, et al. Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2017;114:2078–83. [DOI] [PMC free article] [PubMed]
- 20.Moreno-Romero J, Jiang H, Santos-Gonzalez J, Köhler C. Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 2016;1:1–14. doi: 10.15252/embj.201593534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Köhler C. Epigenetic processes in flowering plant reproduction. J Exp Bot. 2017;68(4):797–807. doi: 10.1093/jxb/erw486. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues JA, Zilberman D. Evolution and function of genomic imprinting in plants. Genes Dev. 2015;29:2517–31. doi: 10.1101/gad.269902.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler C. Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife. 2016;5:1–23. doi: 10.7554/eLife.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatorangan MR, Laenen B, Steige K, Slotte T, Köhler C. Rapid evolution of genomic imprinting in two species of the Brassicaceae. Plant Cell. 2016;28:1815–27. doi: 10.1105/tpc.16.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, et al. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, et al. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci U S A. 2013;110:19639–44. doi: 10.1073/pnas.1309182110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klosinska M, Picard CL, Gehring M. Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat Plants. 2016;2:16145. doi: 10.1038/nplants.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff P, Jiang H, Wang G, Santos-Gonzalez J, Kohler C. Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. Elife. 2015;4:1–14. doi: 10.7554/eLife.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gehring M, Satyaki PR. Endosperm and imprinting, inextricably linked. Plant Physiol. 2017;173:143–54. doi: 10.1104/pp.16.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pires ND, Bemer M, Müller LM, Baroux C, Spillane C, Grossniklaus U. Quantitative genetics identifies cryptic genetic variation involved in the paternal regulation of seed development. PLoS Genet. 2016;12:1–19. doi: 10.1371/journal.pgen.1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piskurewicz U, Iwasaki M, Susaki D, Megies C, Kinoshita T, Lopez-Molina L. Dormancy-specific imprinting underlies maternal inheritance of seed dormancy in Arabidopsis thaliana. Elife. 2016;5:1–23. doi: 10.7554/eLife.19573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowack MK, Shirzadi R, Dissmeyer N, Dolf A, Endl E, Grini PE, et al. Bypassing genomic imprinting allows seed development. Nature. 2007;447:312–5. doi: 10.1038/nature05770. [DOI] [PubMed] [Google Scholar]
- 33.Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang Y, Silva-Ortega CO, Wu S, Yamaguchi N, Wu MF, Pfluger J, et al. Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J. 2012;72:1000–14. doi: 10.1111/tpj.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarnowski TJ, Ríos G, Jásik J, Swiezewski S, Kaczanowski S, Li Y, et al. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell. 2005;17:2454–72. doi: 10.1105/tpc.105.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, et al. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development. 2006;133:3223–30. doi: 10.1242/dev.02508. [DOI] [PubMed] [Google Scholar]
- 38.Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, et al. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007;19:403–16. doi: 10.1105/tpc.106.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, Sang Y, Bezhani S, Yamaguchi N, Han S, Li Z, et al. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci U S A. 2012;109:3576–81. doi: 10.1073/pnas.1113409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife. 2015;4 doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haughn G, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 2005;10:472–7. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Domínguez F, Cejudo FJ. Programmed cell death (PCD): an essential process of cereal seed development and germination. Front Plant Sci. 2014;5:366. doi: 10.3389/fpls.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roszak P, Köhler C. Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc Natl Acad Sci U S A. 2011;108:20826–31. doi: 10.1073/pnas.1117111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang I-H, Steffen JG, Portereiko MF, Lloyd A, Drews GN. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell. 2008;20:635–47. doi: 10.1105/tpc.107.055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44:179–94. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 46.Raz V, Bergervoet JHW, Koornneef M. Sequential steps for developmental arrest in Arabidopsis seeds. Development. 2001;128:243–52. doi: 10.1242/dev.128.2.243. [DOI] [PubMed] [Google Scholar]
- 47.Holdsworth MJ, Bentsink L, Soppe WJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki M, McCarty DR. Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol. 2008;11:548–53. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Gacio MDC, Matilla-Vázquez MA, Matilla AJ. Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal Behav. 2009;4:1035–49. doi: 10.4161/psb.4.11.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nonogaki H. Seed dormancy and germination-emerging mechanisms and new hypotheses. Front Plant Sci. 2014;5:233. doi: 10.3389/fpls.2014.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164:711–29. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chahtane H, Kim W, Lopez-Molina L. Primary seed dormancy: a temporally multilayered riddle waiting to be unlocked. J Exp Bot. 2017;68(4):857–869. doi: 10.1093/jxb/erw377. [DOI] [PubMed] [Google Scholar]
- 53.Cyrek M, Fedak H, Ciesielski A, Guo Y, Sliwa A, Brzezniak L, et al. Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 2016;170:947–55. doi: 10.1104/pp.15.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakabayashi K, Bartsch M, Ding J, Soppe WJJ. Seed dormancy in Arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet. 2015;11:1–20. doi: 10.1371/journal.pgen.1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Footitt S, Muller K, Kermode AR, Finch-Savage WE. Seed dormancy cycling in Arabidopsis: chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals. Plant J. 2015;81:413–25. doi: 10.1111/tpj.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19:433–44. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Geyer R, van Zanten M, Carles A, Li Y, Hörold A, et al. Identification of the Arabidopsis reduced dormancy 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One. 2011;6:1–8. doi: 10.1371/journal.pone.0022241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell. 2013;25:149–66. doi: 10.1105/tpc.112.108191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146:149–61. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Zanten M, Zöll C, Wang Z, Philipp C, Carles A, Li Y, et al. HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 2014;80:475–88. doi: 10.1111/tpj.12646. [DOI] [PubMed] [Google Scholar]
- 62.Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012;35:1769–86. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 63.Bentsink L, Koornneef M. Seed dormancy and germination. Arab Book. 2008;6:e0119. [DOI] [PMC free article] [PubMed]
- 64.Tang X, Lim MH, Pelletier J, Tang M, Nguyen V, Keller WA, et al. Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J Exp Bot. 2012;63:1391–404. doi: 10.1093/jxb/err383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen D, Molitor A, Liu C, Shen W-H. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 2010;20:1332–44. doi: 10.1038/cr.2010.151. [DOI] [PubMed] [Google Scholar]
- 66.Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol. 2010;20:1853–9. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 67.Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M. VAL-and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in arabidopsis. Curr Biol. 2013;23:1324–9. doi: 10.1016/j.cub.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 68.Molitor AM, Bu Z, Yu Y, Shen WH. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014;10. [DOI] [PMC free article] [PubMed]
- 69.Merini W, Romero-Campero FJ, Gomez-Zambrano A, Zhou Y, Turck F, Calonje M. The Arabidopsis Polycomb Repressive Complex 1 (PRC1) components AtBMI1A, B and C impact gene networks throughout all stages of plant development. Plant Physiol. 2016;173:627–41. [DOI] [PMC free article] [PubMed]
- 70.Feng J, Chen D, Berr A, Shen W-H. ZRF1 chromatin regulators have polycomb silencing and independent roles in development. Plant Physiol. 2016;172:1746–59. doi: 10.1104/pp.16.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, et al. HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell. 2013;25:134–48. doi: 10.1105/tpc.112.096313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chhun T, Chong SY, Park BS, Wong ECC, Yin JL, Kim M, et al. HSI2 repressor recruits MED13 and HDA6 to down-regulate seed maturation gene expression directly during arabidopsis early seedling growth. Plant Cell Physiol. 2016;57:1689–706. doi: 10.1093/pcp/pcw095. [DOI] [PubMed] [Google Scholar]
- 73.Gao M-J, Li X, Huang J, Gropp GM, Gjetvaj B, Lindsay DL, et al. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat Commun. 2015;6:7243. doi: 10.1038/ncomms8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ryu H, Cho H, Bae W, Hwang I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun. 2014;5:4138. doi: 10.1038/ncomms5138. [DOI] [PubMed] [Google Scholar]
- 75.Zheng J, Chen F, Wang Z, Cao H, Li X, Deng X, et al. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012;193:605–16. doi: 10.1111/j.1469-8137.2011.03969.x. [DOI] [PubMed] [Google Scholar]
- 76.Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:13839–44. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J. The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 2012;159:418–32. doi: 10.1104/pp.112.194878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008;147:1143–57. doi: 10.1104/pp.108.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han S-K, Sang Y, Rodrigues A, Wu M-F, Rodriguez PL, Wagner D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell. 2012;24:4892–906. doi: 10.1105/tpc.112.105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao M, Yang S, Liu X, Wu K. Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front Plant Sci. 2015;6:159. doi: 10.3389/fpls.2015.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, et al. Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell. 2012;22:736–48. doi: 10.1016/j.devcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z, Chen F, Li X, Cao H, Ding M, Zhang C, et al. Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat Commun. 2016;7:13412. doi: 10.1038/ncomms13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Wit M, Galvão VC, Fankhauser C. Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol. 2016;67:513–37. doi: 10.1146/annurev-arplant-043015-112252. [DOI] [PubMed] [Google Scholar]
- 84.Bourbousse C, Mestiri I, Zabulon G, Bourge M, Formiggini F, Koini MA, et al. Light signaling controls nuclear architecture reorganization during seedling establishment. Proc Natl Acad Sci U S A. 2015;112:E2836–44. doi: 10.1073/pnas.1503512112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee N, Park J, Kim K, Choi G. The transcriptional coregulator LEUNIG_HOMOLOG inhibits light-dependent seed germination in Arabidopsis. Plant Cell. 2015;27:2301–13. doi: 10.1105/tpc.15.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang D, Li Y, Zhang X, Zha P, Lin R. The SWI2/SNF2 chromatin-remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Mol Plant. 2016;10:155–67. doi: 10.1016/j.molp.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 88.Park J, Oh D-H, Dassanayake M, Nguyen KT, Ogas J, Choi G, et al. Gibberellin signaling requires chromatin remodeler PICKLE to promote vegetative growth and phase transitions. Plant Physiol. 2017;173:1463–74. [DOI] [PMC free article] [PubMed]
- 89.Tang Y, Liu X, Liu X, Li Y, Wu K, Hou X. Arabidopsis NF-YCs mediate the light control of hypocotyl elongation via modulating histone acetylation. Mol Plant. 2017;10:260–73. doi: 10.1016/j.molp.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 90.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell. 2010;18:463–71. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 91.Tedder A, Helling M, Pannell JR, Shimizu-Inatsugi R, Kawagoe T, Van Campen J, et al. Female sterility associated with increased clonal propagation suggests a unique combination of androdioecy and asexual reproduction in populations of Cardamine amara (Brassicaceae) Ann Bot. 2015;115:763–76. doi: 10.1093/aob/mcv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–68. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He C, Chen X, Huang H, Xu L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 2012;8:1–13. doi: 10.1371/journal.pgen.1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu YG, et al. The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in arabidopsis calli. Plant Cell Physiol. 2011;52:618–28. doi: 10.1093/pcp/pcr022. [DOI] [PubMed] [Google Scholar]
- 95.Lee K, Park OS, Jung SJ, Seo PJ. Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J Plant Physiol. 2016;191:95–100. doi: 10.1016/j.jplph.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 96.Lee K, Park OS, Seo PJ. RNA-Seq analysis of the Arabidopsis transcriptome in pluripotent calli. Mol Cells. 2016;39:484–94. doi: 10.14348/molcells.2016.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ong-Abdullah M, Ordway JM, Jiang N, Ooi SE, Kok S, Sarpan N, et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature. 2015;525:533–7. doi: 10.1038/nature15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ikeuchi M, Iwase A, Rymen B, Harashima H, Shibata M, Ohnuma M, et al. PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants. 2015;1:15089. doi: 10.1038/nplants.2015.89. [DOI] [PubMed] [Google Scholar]
- 99.Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, et al. Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell. 2011;23:3204–14. doi: 10.1105/tpc.111.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu X, Xu T, He Y. A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol Plant. 2014;7:977–88. doi: 10.1093/mp/ssu035. [DOI] [PubMed] [Google Scholar]
- 101.De Lucas M, Pu L, Turco G, Gaudinier A, Morao AK, Harashima H, et al. Transcriptional regulation of Arabidopsis Polycomb Repressive Complex 2 coordinates cell type proliferation and differentiation. Plant Cell. 2016;28:2616–31. doi: 10.1105/tpc.15.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang S, Li C, Zhao L, Gao S, Lu J, Zhao M, et al. The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell. 2015;27:1670–80. doi: 10.1105/tpc.15.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jégu T, Domenichini S, Blein T, Ariel F, Christ A, Kim SK, et al. A SWI/SNF chromatin remodelling protein controls cytokinin production through the regulation of chromatin architecture. PLoS One. 2015;10:1–18. doi: 10.1371/journal.pone.0138276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kornet N, Scheres B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell. 2009;21:1070–9. doi: 10.1105/tpc.108.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, et al. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell. 2015;33:576–88. doi: 10.1016/j.devcel.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y, Jiao Y, Liu Z, Zhu Y-X. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat Commun. 2015;6:1–8. doi: 10.1038/ncomms7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Otero S, Desvoyes B, Peiró R, Gutierrez C. Histone H3 dynamics reveal domains with distinct proliferation potential in the Arabidopsis root. Plant Cell. 2016;28:1361–71. doi: 10.1105/tpc.15.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perales M, Reddy GV. Stem cell maintenance in shoot apical meristems. Curr Opin Plant Biol. 2012;15:10–6. doi: 10.1016/j.pbi.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 109.Galli M, Gallavotti A. Expanding the regulatory network for meristem size in plants. Trends Genet. 2016;32:372–83. doi: 10.1016/j.tig.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 110.Schuster C, Gaillochet C, Medzihradszky A, Busch W, Daum G, Krebs M, et al. A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev Cell. 2014;28:438–49. doi: 10.1016/j.devcel.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 111.Li D, Fu X, Guo L, Huang Z, Li Y, Liu Y, et al. FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc Natl Acad Sci U S A. 2016;113:9375–80. doi: 10.1073/pnas.1602960113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, et al. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 2015;517:377–80. doi: 10.1038/nature13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gruel J, Landrein B, Tarr P, Schuster C, Refahi Y, Sampathkumar A, et al. An epidermis-driven mechanism positions and scales stem cell niches in plants. Sci Adv. 2016;2 doi: 10.1126/sciadv.1500989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dolzblasz A, Nardmann J, Clerici E, Causier B, van der Graaff E, Chen J, et al. Stem cell regulation by Arabidopsis WOX genes. Mol Plant. 2016;9:1028–39. doi: 10.1016/j.molp.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 115.El-Showk S, Ruonala R, Helariutta Y. Crossing paths: cytokinin signalling and crosstalk. Development. 2013;140:1373–83. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- 116.Adibi M, Yoshida S, Weijers D, Fleck C. Centering the organizing center in the Arabidopsis thaliana shoot apical meristem by a combination of cytokinin signaling and self-organization. PLoS One. 2016;11:1–28. doi: 10.1371/journal.pone.0147830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee D-K, Parrott DL, Adhikari E, Fraser N, Sieburth LE. The mobile bypass signal arrests shoot growth by disrupting SAM maintenance, cytokinin signaling, and WUS expression. Plant Physiol. 2016;171:2178–90. doi: 10.1104/pp.16.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–42. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 119.Yue M, Li Q, Zhang Y, Zhao Y, Zhang Z, Bao S. Histone H4R3 methylation catalyzed by SKB1/PRMT5 is required for maintaining shoot apical meristem. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0083258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin TF, Saiga S, Abe M, Laux T. OBE3 and WUS interaction in shoot meristem stem cell regulation. PLoS One. 2016;11:1–13. doi: 10.1371/journal.pone.0155657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kwon CS, Kwon CS, Chen C, Chen C, Wagner D, Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005;19:992–1003. doi: 10.1101/gad.1276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guo M, Thomas J, Collins G, Timmermans MCP. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell. 2008;20:48–58. doi: 10.1105/tpc.107.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lodha M, Marco CF, Timmermans MCP. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 2013;27:596–601. doi: 10.1101/gad.211425.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rast MI, Simon R. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell. 2012;24:2917–33. doi: 10.1105/tpc.112.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo M, Yu CW, Chen FF, Zhao L, Tian G, Liu X, et al. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 2012;8:4–13. doi: 10.1371/journal.pgen.1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li Z, Li B, Liu J, Guo Z, Liu Y, Li Y, et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J Integr Plant Biol. 2016;58:959–70. doi: 10.1111/jipb.12485. [DOI] [PubMed] [Google Scholar]
- 127.Veluchamy A, Jegu T, Ariel F, Latrasse D, Mariappan KG, Kim SK, et al. LHP1 regulates H3K27me3 spreading and shapes the three-dimensional conformation of the Arabidopsis genome. PLoS One. 2016;11:1–25. doi: 10.1371/journal.pone.0158936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Husbands AY, Benkovics AH, Nogueira FTS, Lodha M, Timmermans MCP. The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell. 2015;27:3321–35. doi: 10.1105/tpc.15.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, et al. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell. 2013;24:438–45. doi: 10.1016/j.devcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014;26:210–29. doi: 10.1105/tpc.113.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sacharowski SP, Gratkowska DM, Sarnowska EA, Kondrak P, Jancewicz I, Porri A, et al. SWP73 subunits of Arabidopsis SWI/SNF chromatin remodeling complexes play distinct roles in leaf and flower development. Plant Cell. 2015;27:1889–906. doi: 10.1105/tpc.15.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nelissen H, Eeckhout D, Demuynck K, Persiau G, Walton A, van Bel M, et al. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize Leaf. Plant Cell. 2015;27:1605–19. doi: 10.1105/tpc.15.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weijers D, Wagner D. Transcriptional responses to the auxin hormone. Annu Rev Plant Biol. 2016;67:539–74. doi: 10.1146/annurev-arplant-043015-112122. [DOI] [PubMed] [Google Scholar]
- 134.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–6. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 135.Weiste C, Dröge-Laser W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat Commun. 2014;5:3883. doi: 10.1038/ncomms4883. [DOI] [PubMed] [Google Scholar]
- 136.Burian A, Barbier de Reuille P, Kuhlemeier C. Patterns of stem cell divisions contribute to plant longevity. Curr Biol. 2016;26:1385–94. doi: 10.1016/j.cub.2016.03.067. [DOI] [PubMed] [Google Scholar]
- 137.Wils CR, Kaufmann K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim Biophys Acta. 1860;2016:95–105. doi: 10.1016/j.bbagrm.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 138.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2004;13:627–37. doi: 10.1016/S0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 139.Engelhorn J, Moreau F, Fletcher JC, Carles CC. ULTRAPETALA1 and LEAFY pathways function independently in specifying identity and determinacy at the Arabidopsis floral meristem. Ann Bot. 2014;114:1497–505. doi: 10.1093/aob/mcu185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aichinger E, Villar CBR, Di Mambro R, Sabatini S, Köhler C. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell. 2011;23:1047–60. doi: 10.1105/tpc.111.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu X, Kim YJ, Muller R, Yumul RE, Liu C, Pan Y, et al. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell. 2011;23:3654–70. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sun B, Looi L-S, Guo S, He Z, Gan E-S, Huang J, et al. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science. 2014;343:1248559. doi: 10.1126/science.1248559. [DOI] [PubMed] [Google Scholar]
- 143.Liu X, Dinh TT, Li D, Shi B, Li Y, Cao X, et al. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2004;80:629–41. doi: 10.1111/tpj.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moreau F, Thévenon E, Blanvillain R, Lopez-Vidriero I, Franco-Zorrilla JM, Dumas R, et al. The Myb-domain protein ULTRAPETALA1 INTERACTING FACTOR 1 controls floral meristem activities in Arabidopsis. Development. 2016;143:1108–19. doi: 10.1242/dev.127365. [DOI] [PubMed] [Google Scholar]
- 145.Chen D, Molitor AM, Xu L, Shen WH. Arabidopsis PRC1 core component AtRING1 regulates stem cell-determining carpel development mainly through repression of class I KNOX genes. BMC Biol. 2016;14:112. doi: 10.1186/s12915-016-0336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Han Y, Zhang C, Yang H, Jiao Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111:6840–5. doi: 10.1073/pnas.1318532111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sridhar VV, Surendrarao A, Liu Z. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development. 2006;133:3159–66. doi: 10.1242/dev.02498. [DOI] [PubMed] [Google Scholar]
- 148.Gregis V, Sessa A, Dorca-Fornell C, Kater MM. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009;60:626–37. doi: 10.1111/j.1365-313X.2009.03985.x. [DOI] [PubMed] [Google Scholar]
- 149.Smaczniak C, Immink RGH, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci U S A. 2012;109:1560–5. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, et al. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science. 2014;345:1605–9. doi: 10.1126/science.1256888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci U S A. 2000;97:10637–42. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Su Y, Kwon CS, Bezhani S, Huvermann B, Chen C, Peragine A, et al. The N-terminal ATPase AT-hook-containing region of the Arabidopsis chromatin-remodeling protein SPLAYED is sufficient for biological activity. Plant J. 2006;46:685–99. doi: 10.1111/j.1365-313X.2006.02734.x. [DOI] [PubMed] [Google Scholar]
- 153.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 154.Jeong CW, Roh H, Dang TV, Choi YDY, Fischer RL, Lee JS, et al. An E3 ligase complex regulates SET-domain polycomb group protein activity in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2011;108:8036–41. doi: 10.1073/pnas.1104232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bell O, Tiwari VK, Thomä NH, Schübeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–64. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 156.Han S-K, Wu M-F, Cui S, Wagner D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015;83:62–77. doi: 10.1111/tpj.12877. [DOI] [PubMed] [Google Scholar]
- 157.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–82. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]