Abstract
To study the physiological roles of NADH and NADPH homeostasis in cancer, we studied the effect of NNT knockdown on physiology of SK-Hep1 cells. NNT knockdown cells show limited abilities to maintain NAD+ and NADPH levels and have reduced proliferation and tumorigenicity. There is an increased dependence of energy production on oxidative phosphorylation. Studies with stable isotope tracers have revealed that under the new steady-state metabolic condition, the fluxes of TCA and glycolysis decrease while that of reductive carboxylation increases. Increased [α-ketoglutarate]/[succinate] ratio in NNT-deficient cells results in decrease in HIF-1α level and expression of HIF-1α regulated genes. Reduction in NADPH level leads to repression of HDAC1 activity and an increase in p53 acetylation. These findings suggest that NNT is essential to homeostasis of NADH and NADPH pools, anomalies of which affect HIF-1α- and HDAC1-dependent pathways, and hence retrograde response of mitochondria.
Abbreviations: 3-BrPA, 3-bromopyruvate; ΔΨm, mitochondrial membrane potential; DMS, dimethyl succinate; HDAC, histone deacetylase; ETC, electron transport chain; HIF-1, hypoxia-inducible factor 1; IGF-1R, insulin-like growth factor 1 receptor; IDH, isocitrate dehydrogenase; LC-TOF-MS, liquid-chromatography time-of-flight mass spectrometry; NADH, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; NNT, nicotinamide nucleotide transhydrogenase; OCR, oxygen consumption rate; ROS, reactive oxygen species; SK-Sc, SK-Hep1 cells expressing scrambled control shRNA; SK-shNNT, SK-Hep1 cells expressing NNT-specific shRNA; STC2, stanniocalcin 2; TCA, tricarboxylic acid; VEGFA, vascular endothelial growth factor A
Keywords: Transhydrogenase, Mitochondria, Metabolomics, HIF-1α, HDAC1
Graphical abstract
Highlights
-
•
NNT is essential to maintenance of NAD(H) and NADP(H) homeostasis.
-
•
Changes in NAD(H) and NADP(H) pools constitute important mitonuclear signal.
-
•
NADH feedback controls TCA cycle and regulates HIF-1α signaling.
-
•
NADPH, essential to mitochondrial antioxidative system, regulates deacetylases.
1. Introduction
Mitochondria are at the hub of metabolism, energy production, redox homeostasis. A number of anabolic and catabolic reactions, such as tricarboxylic acid (TCA) cycle, electron transport, steroidogenesis, are compartmentalized in mitochondria [1]. Mitochondria send signals to nucleus through retrograde signaling, which triggers expression of specific genes; maintains mitochondrial and cellular homeostasis; and enables adaptation to various stimuli and stresses.
Nicotinamide adenine dinucleotide (NADH) and nicotinamide adenine dinucleotide phosphate (NADPH) are important co-enzymes of metabolism, and act as electron carriers in the processes of energy transduction, biosynthesis and redox homeostasis [2], [3], [4], [5]. These pyridine nucleotides and their derivatives are involved in signal transduction and cell regulatory pathways [6], like NAD+ acts as substrate for deacetylases [7]. NADPH can allosterically activate histone deacetylase 1 (HDAC1) in vitro [5]. Moreover, protein acetylation/deacetylation are important in energetic stress response [8].
Nicotinamide nucleotide transhydrogenase (NNT) is a nuclear gene-encoded protein located in the mitochondrial inner membrane [9]. It couples the proton flow down electrochemical proton gradient to hydride transfer from NADH to NADP+ [10]. Under pathophysiological conditions, NNT catalyzes the reverse reaction to generate NADH and maintain mitochondrial membrane potential (ΔΨm) via proton pumping.
The transhydrogenase of E. coli provides about 40% of NADPH during bacterial growth on glucose; and its expression is essential to growth of bacteria on carbon source that cannot be directly metabolized to generate NADPH [11], [12]. C57BL/6 J mice, which has a multi-exon deletion of NNT gene, show impaired insulin secretion and glucose homeostasis [13]. Paradoxically, C57BL/6J mice are resistant to heart failure and death. Their cardiac mitochondria generate less superoxide and hydrogen peroxide than those of C57BL/6N mice that express NNT protein [14], suggesting NNT can function in a reverse mode to generate more reactive oxygen species (ROS). The physiological roles of NNT are more complicated than previously thought. C57BL/6J mice are protected from chemical carcinogenesis than other mouse strains, in terms of initiation and growth of preneoplastic and neoplastic lesions [15], [16], [17]. It is speculative that NNT may play a growth-regulatory role in cancer.
Hypoxia-inducible factor 1 (HIF-1) plays an important role in cellular adaptive responses to hypoxia through multiple genes transactivation involved in survival, proliferation, metabolism, migration and angiogenesis [18]. HIF-1, a heterodimeric transcription factor, consists of HIF-1α and HIF-1β subunits [19]. HIF-1α can be hydroxylated at proline residues by prolyl hydroxylases, which require oxygen and α-ketoglutarate as co-substrates, and ferrous ion and ascorbate as cofactors [20]. The hydroxylated HIF-1 preferably binds VHL protein, and undergoes subsequent ubiquitination and degradation [21]. HIF-1α regulates an intricate network of hypoxic, metabolic and redox signaling pathways [22].
Here we report that NNT deficiency increases NADH, electron transfer via electron transport chain (ETC), and ATP generation in hepatoma cells. The redox homeostasis is disturbed, with reduction in NADPH pool and increased ROS generation. The fluxes of TCA cycle and reverse carboxylation are altered. These changes lead to decreases in HIF-1α level and HDAC1 activity, to changes in gene expression and protein acetylation, and eventually to a slowdown in proliferation. Our findings suggest that NNT serves to fine-tune [NADH]/[NAD+] and [NADPH]/[NADP+] ratios, and regulates mitochondrial retrograde signaling response.
2. Materials and methods
2.1. Cell culture, expression vectors encoding shNNT and tdTomato
SK-Hep-1 cells (ATCC catalog number: HTB-52) were maintained in DMEM, supplemented with 10% (v/v) FCS, 25 mM glucose, 2 mM pyruvate, 6 mM glutamine, and 100 units/ml penicillin 0.1 mg/ml streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The detail of shRNA sequence, tdTomato expressing construct and experiments were shown in Supplemental materials and methods.
2.2. In vivo tumor growth
All animal study was approved by the Institutional Animal Care and Use Committee of Chang Gung University (IACUC No. CGU10-035). Please refer to the Supplemental materials and methods for additional details.
2.3. [U-13C6] glucose and [U-13C5] glutamine labeling and isotopologue analysis
About 2×105 cells were seeded in complete medium, and cultured at 37 °C for 72 h. They were washed twice with serum-free DMEM (without glucose and glutamine). The medium was replaced with the [U-13C6] glucose labeling medium supplemented with 10% dialyzed FCS, 25 mM [U-13C6] glucose, 6 mM glutamine and 2 mM pyruvate, or with [U-13C5] glutamine labeling medium supplemented with 10% dialyzed FCS, 25 mM glucose, 6 mM [U-13C5] glutamine and 2 mM pyruvate. The labeled cells were rinsed twice with ice-cold PBS, and scraped in 1 ml of 80% ice-cold methanol. The sample was then dried under nitrogen gas, and dissolved in water prior to analysis. The dissolved sample was subject to liquid-chromatography time-of-flight mass spectroscopic (LC-TOF-MS) analysis as depicted in Supplemental Experimental Procedures.
2.4. Statistical methods
Data are presented as mean±SD. The mass spectrometry data were analyzed as depicted in Supplemental materials and methods. Other data were analyzed by one-way analysis of variance (ANOVA) and Student's t-test where appropriate. A p value of less than 0.05 is considered significant.
3. Results
3.1. NNT deficiency causes growth slowdown and reduces tumorigenicity
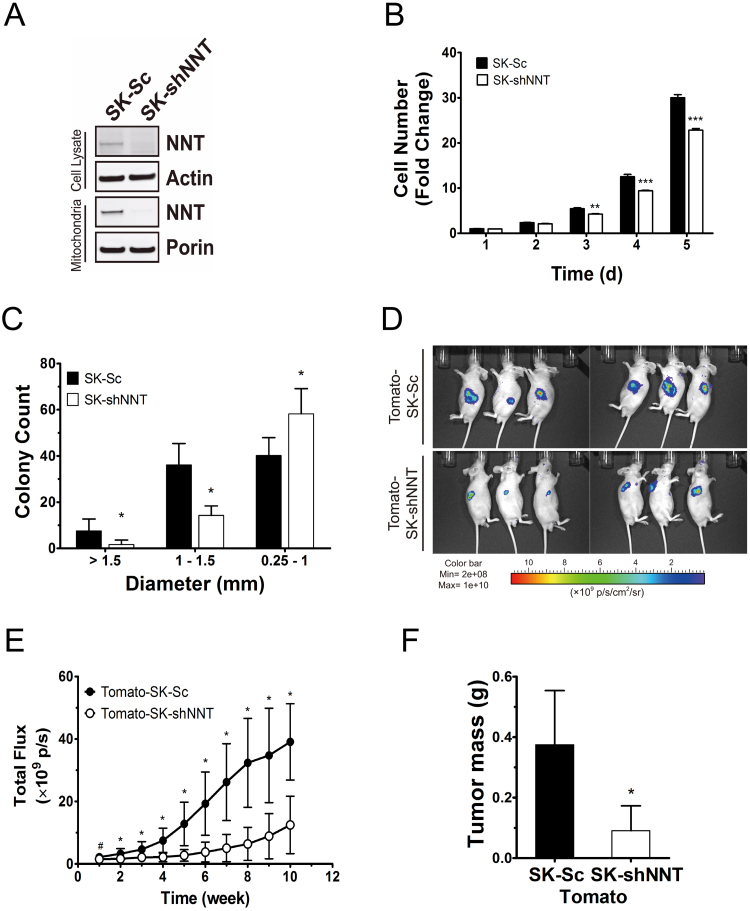
To study the effect of NNT deficiency on cell growth, we introduced NNT shRNA into SK-Hep1 cells using retroviral expression vector. The expression level of NNT in either whole cell lysate or mitochondrial fraction was reduced to a negligible level in NNT knockdown cells (SK-shNNT), as compared with that of scrambled control cells (SK-Sc) (Fig. 1A). Knockdown of NNT expression caused reduction in monolayer growth, growth potential and tumorigenicity. The number of SK-shNNT cells was 25% lower than that of SK-Sc cells after 4–5 days of growth (Fig. 1B). SK-shNNT cells formed smaller colonies than SK-Sc cells did (Fig. 1C). Both SK-shNNT and SK-Sc cells were tagged with fluorescent protein tdTomato to derive Tomato-SK-shNNT and Tomato-SK-Sc cells, and injected subcutaneously on the dorsum of athymic mice. The fluorescence signal emitted, indicative of tumor growth, was monitored (Fig. 1D). The fluorescence of cells increased over time as tumors formed and grew. The fluorescence of tumors formed by Tomato-SK-shNNT cells was significantly lower than that of Tomato-SK-Sc cells (Fig. 1E). At the 10th week, the average mass of Tomato-SK-shNNT tumors excised from host mice was 75% lower than that of Tomato-SK-Sc tumors (Fig. 1F).
Fig. 1.
NNT deficiency reduces proliferation and tumorigenesis of hepatoma. (A) Expression of NNT, porin and actin of cell lysate and mitochondria fraction in SK-shNNT and SK-Sc cells analyzed by immunoblotting. (B) Growth curves of SK-shNNT and SK-Sc cells. Data are mean±SD, n=6. ** p<0.01 *** p<0.005 vs. SK-Sc cells at indicated time. (C) Colonies formed by SK-shNNT and SK-Sc cells were rated according to their diameters. Data are mean±SD, n=9. * p<0.05, vs. SK-Sc cells. (D) The tdTomato fluorescent signal of tumors formed by Tomato-SK-shNNT and Tomato-SK-Sc cells at 10th week after injection. (E) Temporal change in tumor fluorescence during tumor growth. Data are mean±SD, n=34. # p<0.05, * p<0.005, vs. Tomato-SK-Sc cells. (F) The tumors were retrieved from cell-injected mice, and weighed at 10th week after implantation. Data are mean±SD, n=34. * p<0.05, vs. Tomato-SK-Sc cells.
3.2. NNT serves to maintain the redox homeostasis in mitochondria
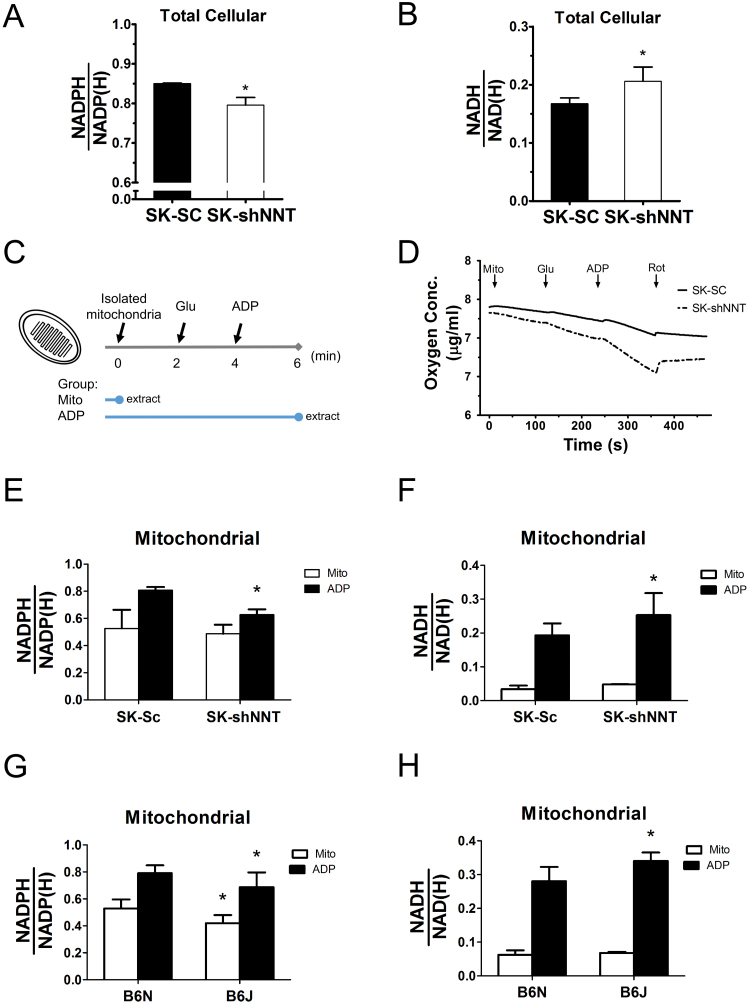
To study if NNT deficiency impairs the redox homeostasis, we quantified the levels of NADH and NADPH in the respective pools. Cellular NADPH level of SK-shNNT cells was lower than that of SK-Sc cells (Fig. 2A), while the cellular NADH level of the former was higher than that of the latter (Fig. 2B). To address the importance of NNT in regulation of the mitochondrial redox state, we measured the levels of NAD(P)+ and NAD(P)H in mitochondria under active respiratory state. We isolated mitochondria from cells; stimulated respiration with glutamate and ADP; and quantified the levels of reduced and oxidized pyridine dinucleotides. The strategy and functionality of mitochondria was shown by the ability of glutamate and ADP to stimulate oxygen consumption and thus respiratory activity (Fig. 2C and D). The basal NADPH levels of SK-shNNT and SK-Sc cells were 0.487±0.066 and 0.526±0.138, while the basal NADH levels of in these cells were 0.048±0.001 and 0.034±0.011, respectively. Upon glutamate and ADP addition, the NADPH level of SK-Sc cells increased by 53.6%, while that of SK-shNNT cells increased by 28.7% (Fig. 2E). The relative NADH level of SK-shNNT cells was higher than that of SK-Sc cells (Fig. 2F). A similar conclusion is reached for mitochondria isolated from livers of C57BL/6 N (designated B6N) and C57BL/6 J (designated B6J). The B6J had mutation in NNT gene, and did not express a functional product. Though ADP treatment increased NADPH in mitochondria of both strains, the relative NADPH level in mitochondria of B6N increased to a significantly higher extent than that of B6J (Fig. 2G). It was accompanied by a higher relative NADH level in mitochondria of B6J, as compared to that of B6N (Fig. 2H). These findings suggest that NNT plays an important role in maintenance of mitochondrial redox homeostasis.
Fig. 2.
NNT deficiency causes disturbance in pyridine adenine dinucleotide pools. SK-shNNT and SK-Sc cells were seeded, and after attachment, cultured for 1 day prior to extraction and analysis of pyridine adenine dinucleotides as described in Supplemental materials and methods. Cellular (abbreviated as ) and (abbreviated as ) are shown. Data are mean±SD, n=6. * p<0.05 vs. SK-Sc cells. (C) Diagram showing sequential addition of Glutamate (Glu) and ATP, and extraction of untreated (mito), and Glu- and ADP-treated (ADP) mitochondria. (D) Mitochondria isolated from SK-shNNT and SK-Sc cells were successively treated with glutamate and ADP for 2 min. The electron transfer was terminated with rotenone (Rot). The oxygen concentration (μg/ml) was continuously monitored amperometrically. The (E) and ) (F), of mito and ADP treatment groups for mitochondria from SK-shNNT and SK-Sc cells are shown. Data are mean±SD, n=5. * p<0.05 vs. SK-Sc cells. The ratios, (G) and (H), of mito and ADP treatment groups for mitochondria from B6J and B6N cells are shown. Data are mean±SD, n=5. * p<0.05 vs. B6N.
3.3. NNT deficiency alters mitochondrial physiology and increases oxidative phosphorylation
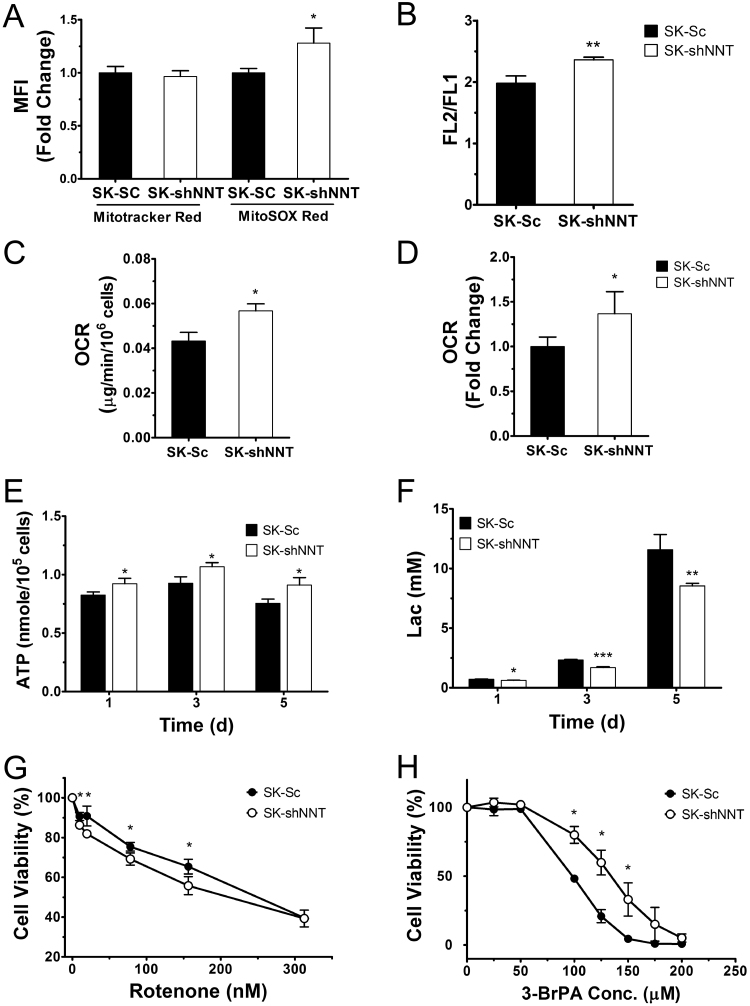
To test whether NNT deficiency alters mitochondrial physiology, we stained SK-NNT and SK-Sc cells with MitoSOX Red and Mitotracker Red, respectively, to detect mitochondrial ROS or mass. SK-shNNT cells had higher ROS generation but similar mitochondrial mass when compared to those of SK-Sc cells (Fig. 3A). As revealed by JC-1 staining, SK-shNNT cells maintained a higher ΔΨm than SK-Sc cells (Fig. 3B). It was consistent with the western analysis of mitochondrial proteins (Supplemental Fig. 1). Expression of mitochondrial proteins did not differ significantly between these cells. The changes in mitochondrial redox states of NADH and NADPH in NNT-deficient cells may be associated with changes in respiratory functions. We measured oxygen consumption rates (OCRs) of SK-Sc and SK-shNNT cells. The cellular OCR of SK-shNNT cells was 30% higher than that of SK-Sc cells (Fig. 3C). Consistent with this, the OCR of mitochondria in response to malate/pyruvate/ADP treatment was more than 35% higher in SK-shNNT cells than SK-Sc cells (Fig. 3D), suggesting that the flow of electrons from NADH to ETC is enhanced in NNT-deficient cells. Increase in oxygen consumption in SK-shNNT was accompanied by increase in ATP synthesis. SK-shNNT cells had 11.9%, 15.1% and 20.7% higher ATP level than SK-Sc cells after 1, 3 and 5 days of cultivation, respectively (Fig. 3E). The lactate in the corresponding culture medium from SK-shNNT cells on day 1, 3 and 5 were 11.4%, 27.3% and 26.3% lower than those of SK-Sc cells, respectively (Fig. 3F).
Fig. 3.
NNT deficient cells show distinct metabolic activities. SK-shNNT and SK-Sc cells were cultured for 2 days, stained with Mitotracker Red 580FM (A), MitoSOX Red (A) and JC-1 (B) dyes for cytometric analysis of mitochondrial mass, ROS generation and ΔΨm. Data are mean±SD, n=6 or 3. * p<0.05, ** p<0.01 vs. SK-Sc cells. (C) Cells were harvested for measurement of their OCRs. Data are mean±SD, n=9. * p<0.05 vs. SK-Sc cells. (D) Mitochondria isolated from SK-shNNT and SK-Sc cells were assayed for OCR associated with complex I activities. Data are mean±SD, n=9. * p<0.05 vs. SK-Sc cells. (E and F) SK-shNNT and SK-Sc cells were incubated in complete medium for 1, 3 and 5 day. Cellular ATP content (E) and lactate in medium (F) were quantified. Data are mean±SD, n=3, 4. * p<0.05, ** p<0.01, *** p<0.005 vs. SK-Sc cells at indicated time. (G and H) SK-shNNT and SK-Sc cells were treated with indicated concentrations of rotenone (G) or 3-BrPA (H) for 48 h, and their cell viabilities were determined. Data are mean±SD, n=3, 6. * p<0.05 vs. SK-Sc cells.
The shift in energy metabolism from glycolysis to TCA was accompanied by an increased sensitivity of these cells to rotenone. SK-Sc and SK-shNNT cells displayed a dose-dependent reduction in viability upon rotenone treatment (Fig. 3G). The viability of SK-shNNT cells was lower than that of SK-Sc cells at rotenone concentrations ranging from 10 to 80 nM. Moreover, SK-shNNT cells showed an increased resistance to cytotoxic effect of glycolytic inhibitor 3-bromopyruvate (3-BrPA). The 50% cytotoxic concentration (CC50) of 3-BrPA for SK-shNNT cells was 138.5±1.12 μM, while that for SK-Sc cells was 100.1±1.02 μM (Fig. 3H). These findings suggest that SK-shNNT cells are more dependent on mitochondrial metabolism for energy than the control counterparts.
3.4. Nutritional requirement differs between NNT-deficient and control cells
SK-shNNT and SK-Sc cells differed in their dependence of cell growth on glutamine and glucose. Proliferation of these cells decreased significantly, as concentrations of glucose and glutamine in medium decreased. In particular, SK-shNNT cells grew slower than SK-Sc cells did. Glutamine deprivation had a significant effect on growth (Supplemental Fig. 2A), suggesting that glutaminolysis and/or complete oxidation of glutamine may play an important role in energy metabolism and biosynthesis in hepatoma cells. Glucose deprivation reduced the growth rate of SK-Sc cells to a greater extent than that of SK-shNNT cells, implying that SK-shNNT cells are less dependent on glucose for growth (Supplemental Fig. 2B). In contrast to SK-Sc cells, the growth of SK-shNNT cells was significantly reduced in the absence of pyruvate (Supplemental Fig. 2C). This may be attributed to changes in steady-state of metabolism involved in energy metabolism and anaplerosis in the absence of NNT.
3.5. NNT deficiency induces anomalous cellular metabolism
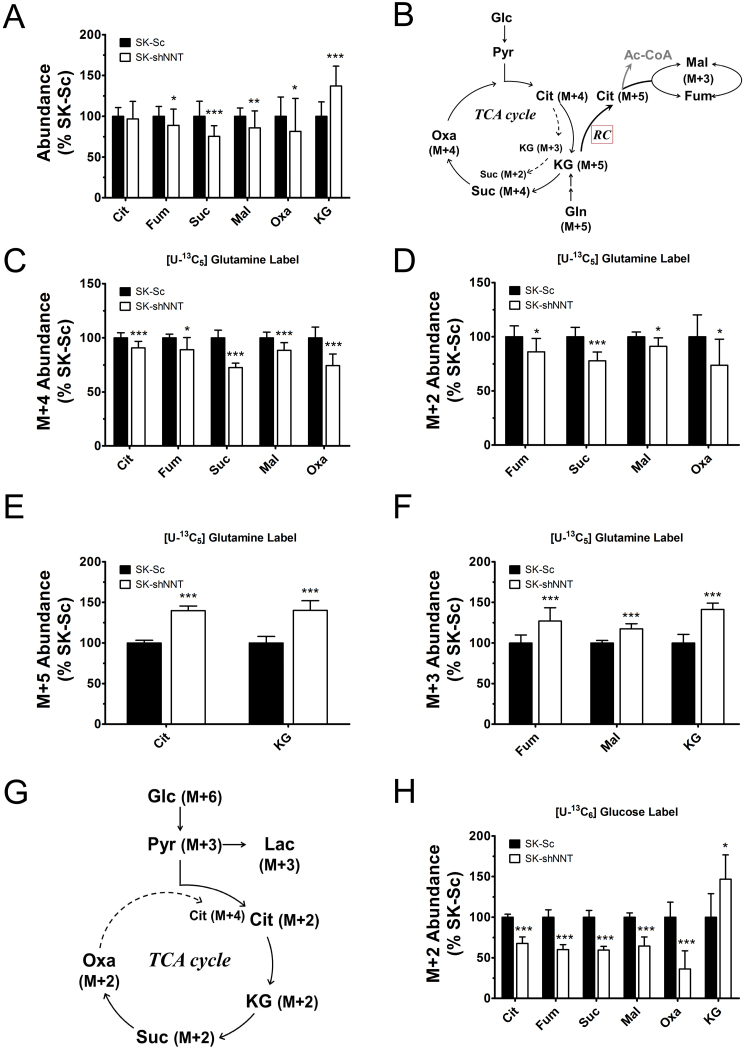
The anomalies of mitochondrial functions may be attributed to changes in steady-state metabolism in NNT-deficient cells. To test such hypothesis, we studied the effect of NNT knockdown on glucose and glutamine metabolism. The control and knockdown cells were cultured in the presence of [U-13C6] glucose or [U-13C5] glutamine, and extracted for metabolite analysis. The levels of fumarate, succinate, malate and oxaloacetate, which include all isotopologue forms, were lower in SK-shNNT cells than in SK-Sc cells (Fig. 4A; Supplemental Fig. 3A). The levels of citrate in these cells were nearly the same. In contrast, the level of α-ketoglutarate was significantly elevated in SK-shNNT cells.
Fig. 4.
NNT deficiency reduces the flux of TCA cycle. SK-shNNT and SK-Sc cells were cultured in complete medium for 3 days, and subsequently cultured in normal medium, [U-13C6] glucose-, or [U-13C5] glutamine-labeling medium for 4 additional hours. The relative abundances of metabolites are expressed as the percentage of those of SK-Sc. (A) The levels of total citrate (Cit), fumarate (Fum), succinate (Suc), malate (Mal), oxaloacetate (Oxa), and α-ketoglutarate (KG) in labeled cells are shown. Data are mean±SD, n=18. * p<0.05, ** p<0.01, *** p<0.005 vs. SK-Sc cells. (B) A schematic diagram depicts oxidation and reductive carboxylation of [U-13C5] glutamine and formation of M+N isotopologues of various metabolites, where N denotes the number of 13C atoms. The levels of M+4 (C), M+2 (D), M+5 (E) and M+3 (F) isotopologues of indicated metabolites from [U-13C5] glutamine tracer are shown. Data are mean±SD, n=9. * p<0.05, *** p<0.005 vs. SK-Sc cells. (G) A schematic diagram depicts oxidation of [U-13C6] glucose and formation of M+N isotopologues of various metabolites. The levels of M+2 (H) isotopologues of indicated metabolites in [U-13C6] glucose-labeled cells are shown. Data are mean±SD, n=9. * p<0.05, *** p<0.005 vs. SK-Sc cells.
As glutaminolysis plays an important role in metabolism, we examined the contribution of [U-13C5] glutamine tracer to TCA cycle metabolites. The labeling pattern of metabolites derived from [U-13C5] glutamine was shown (Fig. 4B; Supplemental Fig. 3B and C). The levels of M+4 and M+2 isotopologue forms of succinate, fumarate, malate, oxaloacetate and citrate decreased, indicating the reduced flux through the forward reactions of TCA cycle (Fig. 4C and D; Supplemental Fig. 3D). Increased levels of M+5 and M+3 isotopologues of α-ketoglutarate suggest a reduction in activity of the α-ketoglutarate dehydrogenase reaction (Fig. 4E and F; Supplemental Fig. 3E). In contrast, the levels of M+5 citrate, and M+3 malate and fumarate were elevated in SK-shNNT cells (Fig. 4E and F). It is likely that the rate of reductive carboxylation is enhanced in NNT-deficient cells. Similarly, the labeling pattern of metabolites derived from [U-13C6] glucose tracer revealed that the rates of glycolysis and forward reactions of TCA cycle diminish in NNT-deficient cells (Fig. 4G; Supplemental Fig. 4A and B). The level of M+3 pyruvate in SK-shNNT cells significantly decreased, reducing the supply of acetyl-CoA to TCA cycle (Supplemental Fig. 4D). It was accompanied by decreases in their levels of M+3 lactate and M+2 isotopologue forms of citrate, fumarate, succinate, malate and oxaloacetate (Fig. 4H; Supplemental Fig. 4C and D). Consistent with the results obtained in [U-13C5] glutamine labeling study, M+2 and M+3 α-ketoglutarate (i.e. arising from the first and second round of TCA cycle, respectively) accumulated in SK-shNNT cells than SK-Sc cells (Fig. 4H; Supplemental Fig. 4D), implying an inhibition of the downstream α-ketoglutarate dehydrogenase reaction. The M+4 isotopologues of fumarate, succinate, malate and oxaloacetate, which are generated by combined pyruvate carboxylation and TCA cycle, decreased in abundance (Supplemental Fig. 4D). It is possible that the contribution to pools of 4 carbon intermediates by pyruvate carboxylase or malic enzyme is lowered in NNT-deficient cells.
The changes in the intermediates of glycolysis and TCA cycle are associated with metabolism of certain amino acids in NNT-deficient cells. There was no significant difference in the level of glutamine, which is taken up from medium, between SK-shNNT and SK-Sc cells (Supplemental Fig. 5A). Glutamate was decreased by 15% in abundance in SK-shNNT cells (Supplemental Fig. 5B). The levels of aspartate and alanine, which can be derived from oxaloacetate and pyruvate via transamination, in SK-shNNT cells decreased by 21.4% and 16.5%, respectively, as compared to those of SK-Sc cells (Supplemental Fig. 5D and E). The levels of phenylalanine in SK-shNNT and SK-Sc cells were the same (Supplemental Fig. 5F).
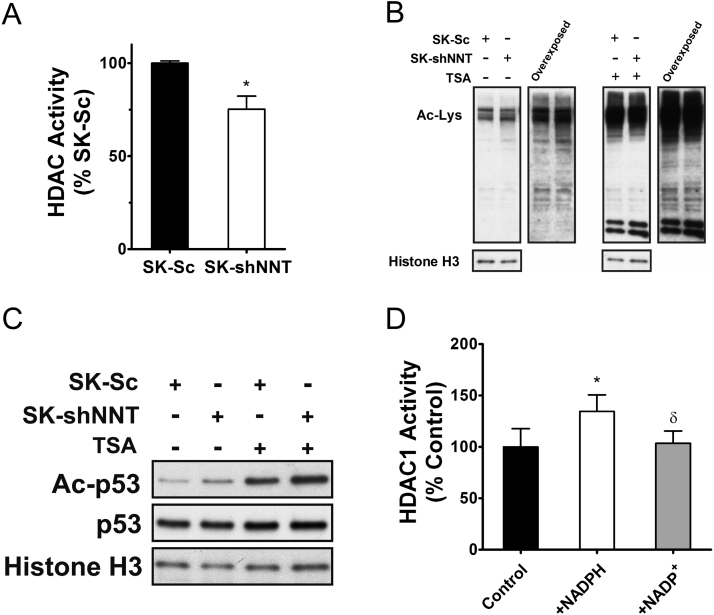
3.6. Reduction in cellular NADPH content downregulates the HDAC activity
Previous study has shown that NADPH acts as an allosteric activator of HDAC [5]. We hypothesized that the reduced NADPH level in NNT-deficient cells lowers their HDAC activity. To test such hypothesis, we assayed the total HDAC activities of SK-Sc and SK-shNNT cells. Total HDAC activity of SK-shNNT cells was 25% lower than that of SK-Sc cell (Fig. 5A). There was no difference in expression of a major form of deacetylase HDAC1 between SK-Sc and SK-shNNT cells (Supplemental Fig. 6). The reduction in HDAC activity was accompanied by increased acetylation of nuclear proteins in SK-shNNT cells, as compared to that of SK-Sc cells (Fig. 5B). Of proteins that undergo acetylation, p53 is an important regulator of growth [23]. Acetylation of p53 increased in SK-shNNT cells (Fig. 5C). To study the effect of NADPH on HDAC activity, we used recombinant HDAC1 protein in an in vitro assay. NADPH but not NADP+ activated HDAC1 activity in vitro (Fig. 5D). These findings imply that the reduction in cellular NADPH level in NNT-deficient leads to increased protein acetylation.
Fig. 5.
Decreased NADPH in NNT-deficient cells leads to reduction in HDAC activity. (A) The HDAC activities in nuclear extracts of SK-shNNT and SK-Sc cells were assayed for HDAC activity. Data are mean±SD, n=6. * p<0.05 vs. SK-Sc cells. (B) SK-shNNT and SK-Sc cells were treated without or with 1 μM Trichostatin A (TSA) for 5 h, and their nuclear extracts were subject to western blotting with antibodies to acetylated-lysine (Ac-Lys) and histone H3 (internal control). Immunoblot was exposed to film for 1 min or 5 min (overexposed). A representative experiment out of three is shown. (C) Acetylated p53 (Ac-p53), total p53 and histone H3 (internal control) were analyzed by western blotting with respective antibodies. A representative experiment out of three is shown. (D) Recombinant HDAC1 was treated without (Control) or with 1 mM NADPH or NADP+, and assayed for its activity. Data are expressed as the percentage of HDAC1 activity of control group. Data are mean±SD, n=6. * p<0.05 vs. control; δ p<0.05 vs. NADPH treatment group.
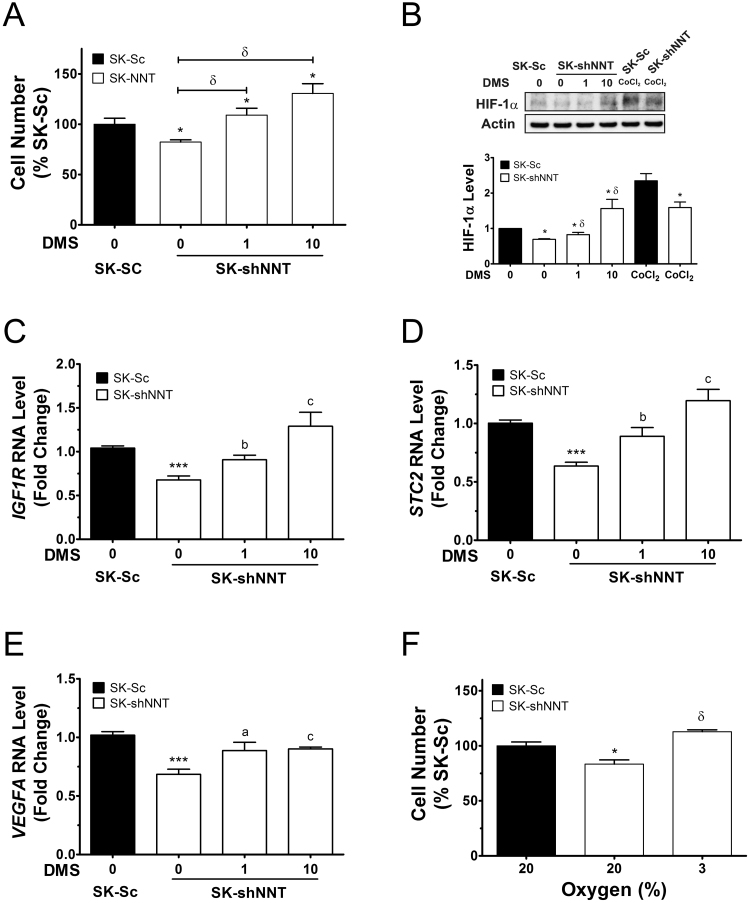
3.7. NNT deficiency causes reducing HIF-1α expression and its transcription of downstream genes
HIF-1α, involved in regulation of adaptive responses to hypoxia, is itself post-translationally regulated by proline hydroxylation and ubiquitin-mediated degradation [24]. The increase in α-ketoglutarate level relative to succinate level may alter the HIF prolyl hydroxylase activity and HIF-1α degradation [25]. The ratio between levels of α-ketoglutarate and succinate for SK-shNNT cells was higher than that of SK-Sc cells. Consistent with this, the level of HIF-1α in SK-shNNT cells was lower than that of SK-Sc cells (Fig. 6B). Treatment with dimethyl succinate (DMS), a cell-permeable form of succinate, stabilized cellular HIF-1α in SK-shNNT cells (Fig. 6B). The HIF-1α level increased 2.25-fold in these cells after treatment with 10 mM DMS. Control treatment with hypoxia-mimetic compound CoCl2 increased HIF-1α protein. HIF-1α stabilization was paralleled by enhanced proliferation. Treatment with 1 and 10 mM DMS caused over 30% and 55% increase in growth of SK-shNNT cells (Fig. 6A). Consistent with this, cultivation of SK-shNNT cells under 3% oxygen enhanced their growth (Fig. 6F).
Fig. 6.
Reduction in HIF-1α signaling in NNT-deficient cells. (A) SK-shNNT cells were cultured in the absence or presence of 1 or 10 mM DMS for 3 days. Untreated SK-Sc cells served as control. Data are expressed as the percentage of untreated SK-Sc cells. Data are mean±SD, n=6. * p<0.05 vs. SK-Sc cells; δ p<0.05 vs. untreated SK-shNNT cells. (B) Western blotting with antibodies to HIF-1α and actin (internal control). As positive control, SK-shNNT and SK-Sc cells were treated with 100 μM cobalt chloride (CoCl2). Untreated SK-Sc cells served as control. A representative experiment out of three is shown. The relative intensity was normalized to that of actin, and is expressed as fold change of untreated SK-Sc cells. Data are mean±SD, n=3. * p<0.05 vs. SK-Sc cells; δ p<0.05 vs. untreated SK-shNNT cells. (C–E), SK-shNNT cells were similarly treated, while the untreated SK-Sc cells served as control. RNA was extracted for RT-qPCR analysis of expression of IGF1R, STC2 and VEGFA genes. Data are expressed as fold change relative to that of untreated SK-Sc cells. Data are mean±SD, n=7. * p<0.05 vs. SK-Sc cells; δ p<0.05, a p<0.05, b p<0.01, c p<0.005 vs. untreated SK-shNNT cells. (F) SK-shNNT cells were cultured under oxygen tension of 3 or 20%. SK-Sc cells cultured under oxygen tension of 20% served as control. Data are expressed as the percentage of SK-Sc cells. Data are mean±SD, n=6. * p<0.005 vs. SK-Sc cells cultured under oxygen tension of 20%; δ p<0.005 vs. untreated SK-shNNT cells oxygen tension of 3%.
Stabilization of HIF-1α leads to upregulated transcription of its target genes. Using RT-qPCR, we studied the expression of the transcripts of HIF-1α-regulated genes insulin-like growth factor 1 receptor (IGF-1R), stanniocalcin 2 (STC2) and vascular endothelial growth factor A (VEGFA). The level of IGF-1R transcript in SK-NNT cells increased by more than 30% and 90%, respectively, after treatment with 1 and 10 mM DMS (Fig. 6C); the level of STC2 transcript increased by 40% and 88% respectively in treated cells (Fig. 6D); and the level of VEGFA transcript increased by 30% and 32% respectively in treated cells (Fig. 6E). These findings imply that downregulation of HIF-1α and its target genes may contribute to growth slowdown of NNT-deficient cells.
4. Discussion
We have derived hepatoma cells expressing NNT-specific shRNA or scrambled control shRNA, and studied the effect of NNT deficiency on metabolism and proliferation. Our findings have shown that NNT deficiency impairs in vitro and in vivo growth of hepatoma cells. We further demonstrate that the growth slowdown is associated with changes in metabolism and pyridine adenine dinucleotide pools, which may affect protein acetylation and HIF-1α-dependent transcription.
It was previously shown that C57BL/6 J mice harboring deletion of NNT gene are less susceptible to chemical-induced carcinogenesis than other strains [16], [26], probably due to lower rate of proliferation [26]. It is not certain if deletion NNT gene is the sole factor affecting proliferation and tumor growth. Our finding that knockdown of NNT expression significantly reduces proliferation in vitro and in vivo confirms such notion.
NNT is the proton-translocating inner membrane protein that catalyzes hydride transfer between NAD+ and NADP+ [27]. The proton flow down the electrochemical gradient (i.e. from intermembrane space to matrix) drives reduction of NADP+ at the expense of NADH. NNT, unlike other NADPH-generating enzymes, is believed to couple NADPH generation to mitochondrial respiration. The NNT contributes to at least 14–23% of the NADPH pool of energetic mitochondria (Fig. 2E and G). This is in the same range as the amount of NADPH supposedly generated by NNT in cardiomyocytes [14]. The decrease in NADPH level is accompanied by increased ROS generation in NNT-deficient cells (Fig. 3A). Notably, cellular NADPH abundance is around 7% lower in NNT-deficient cells than that in control cells. The difference in NADPH level between NNT-deficient and control cells at cellular level is relatively mild as compared with that at mitochondrial level. It is probably ascribed to the presence of other NADPH-generating enzymes, such as malic enzyme, isocitrate dehydrogenase (IDH), glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase, which compensate for the deficit [6], [28].
It can be envisaged that the loss of NNT protein spares NADH, which is re-oxidized to NAD+ via ETC. We observed an increase in NADH levels in mitochondria of NNT-deficient cells and those isolated from liver of C57BL/6J mice. It is in agreement with the previous finding that silencing of NNT expression increases NADH/NAD+ ratio in osteosarcoma cells [29]. Contradictory result on NNT silencing has been reported. The NADH/NAD+ ratio decreases in NNT-knockdown renal carcinoma cells [30]. A number of factors may cause such discrepancy. Yin et al. reported that the NADH/NAD+ ratio increased in NNT-specific siRNA-transfected pheochromocytoma cells which were induced to differentiate for 1 day, but the ratio decreased after 4 more days of differentiation [2]. It is conceivable that factors, like cell type, differentiation state, and the compensatory effect by other redox-metabolizing enzymes, may affect the outcome of NNT silencing.
The increase in mitochondrial NADH/NAD+ ratio in NNT-deficient cells is linked to changes in activities of TCA cycle and glycolysis. The rate of glycolysis in SK-shNNT cells, as measured by production of M+3 pyruvate by [U-13C6] glucose-labeled cells, decreased drastically by over 30%. A number of factors may account for reduction in glycolytic flux in these cells. Phosphofructose kinase 1 and pyruvate kinase, regulatory enzymes, are allosterically inhibited by ATP surplus [31]. Glycolysis can be negatively regulated through activation p53-dependent pathway and/or inactivation of HIF-1α-dependent pathway [32]. The flux of the entry of glutamine into TCA, as estimated from M+4 citrate in [U-13C5] glutamine-labeled cells, was 10–15% lower in SK-shNNT cells than in SK-Sc cells. Specifically, M+3 α-ketoglutarate accumulated while the level of M+2 succinate decreased, implying an inhibition of α-ketoglutarate dehydrogenase step and/or enhancement of glutamate dehydrogenase step. The increase in the flux of glutamate dehydrogenation is indicated by a significant drop in intracellular glutamate level. Consistent with this, M+2 and M+3 α-ketoglutarate increased in abundance in [U-13C6] glucose-labeled SK-shNNT cells. The flux of the entry of glucose into TCA cycle, as estimated from M+2 and M+4 citrate in [U-13C6] glucose-labeled cells, also decreased substantially in SK-shNNT cells. It is likely that the fluxes of glycolysis and TCA cycle are lowered in NNT-deficient cells. Slow-down of TCA cycle flux can be attributed to inhibition at several steps. α-Ketoglutarate dehydrogenase may be inhibited by an increase in allosteric regulator NADH [33], and/or by a loss of activating succinylation [34]; the activity of isocitrate dehydrogenase can be reduced by an increase in ATP [35]. Despite a moderate decline in the flux of TCA cycle, a higher proportion of NADH feed electrons into ETC in mitochondria of NNT-deficient cells, generating more ATP. There is a shift in energy metabolism from glycolysis to oxidative phosphorylation.
Apart from energy metabolism, cells depend on TCA cycle to meet their anabolic needs. Oxaloacetate can be converted to aspartate and asparagine; citrate can be cleaved by citrate lyase to release acetyl-CoA for lipid biosynthesis. It has been shown that cancer cells can use reductive carboxylation for formation of citrate [36], [37]. NADP+-dependent IDHs, namely IDH1 and IDH2, are involved in the process, and catalyze the conversion of α-ketoglutarate to isocitrate. Isocitrate is then isomerized to citrate by aconitase. The citrate derived from reductive carboxylation (i.e. M+5 citrate in [U-13C5] glutamine-labeled cells) increased in abundance in SK-shNNT cells. This may represent an attempt of these cells to compensate for the decline in the level of citrate which is contributed by glucose and glutamine in forward reactions of TCA cycle. It is, at first sight, rather unexpected that the reductive carboxyation is enhanced in NNT-deficient cells, which have less NAPDH. The rise in α-ketoglutarate level, and thus lower [citrate]/[α-ketoglutarate] ratio, in these cells may drive the reductive carboxylation in the direction of citrate formation. It is supported by the finding that reductive carboxylation in von Hippel-Lindau (VHL) tumor suppressor gene-deficient renal carcinoma cells is causally related to low [citrate]/[α-ketoglutarate] ratio [38]. In spite of an increased flux of reverse carboxylation, the overall levels of 4-carbon metabolites, such as oxaloacetate, malate and fumarate, decrease. Changes in intermediates of TCA cycle and NADPH also affect amino acid pools in NNT-deficient cells. The pool of aspartate abates as a result of a decline in intracellular oxaloacetate and alanine levels. These findings suggest that anabolism may be adversely affected in NNT-deficient cells.
Protein acetylation represents an important post-translational modification that regulates transcription and metabolism [39], [40]. Consistent with the previous finding [5], we found that NADPH but not NADP+ is an activator of HDAC1. Additionally, NADPH maintains cellular antioxidative defense, and blocks the oxidative modification of HDAC1 that inactivates deacetylase activity [41]. Decreased NADPH level, and thus decreased HDAC activity, in NNT-deficient cells increase acetylation of various nuclear proteins. Of acetylated proteins, p53 is transcriptional regulator, modulating such cellular processes as cell cycle arrest, senescence, apoptosis, differentiation, development, tissue homeostasis, and regeneration [23], [42], [43]. Acetylation of p53 enhances its transcriptional activity [44]. HDAC1, 2 and 3 can specifically downregulate the transactivating activity of p53 [45]. Acetylation p53 increases in NNT-deficient cells, and the resultant p53 activation is likely to cause growth slowdown. Changes in transcription of several p53-dependent genes have been observed in our microarray study (unpublished data). Transcription of cyclin dependent kinase inhibitor CDKN1A, which halts cell cycle progression through G1/S and S/G2 checkpoints [46], is elevated in these cells. Another cell cycle regulator GADD45B is transcriptionally upregulated too [47]. What's more, p53 may regulate metabolism. It can inhibit glycolysis by increasing transcription of TP53-induced glycolysis and apoptosis regulator (TIGAR), which dephosphorylates fructose-2,6-bisphosphate and reduces the activity of phosphofructokinase 1 [48]. Indeed, the level of TIGAR transcript increases in NNT-deficient cells (unpublished data). In addition, p53 upregulates expression of miR-34a, which represses expression of glycolytic enzymes, such as hexokinase 1, hexokinase 2, glucose-6-phosphate isomerase and lactate dehydrogenase A, and pyruvate dehydrogenase kinase 1 [49], [50]. miR-34a-mediated repression of these enzymes leads to inhibition of glycolysis and enhancement of mitochondrial respiration [49]. These findings suggest that redox imbalance in NNT-deficient cells alters HDAC activity and nuclear protein acetylation.
HIF-1α-mediated signaling may be altered in the absence of NNT. A high [α-ketoglutarate]/[succinate] ratio may promote prolyl hydroxylase-mediated degradation of HIF-1α, and causes a reduction in basal level of HIF-1α. It is probable that a decrease in HIF-1α adversely affects cellular proliferation. It is not unprecedented that HIF-1α expression is associated with proliferation. Suppression of HIF-1α activity impairs growth of gastric tumors [51]. HIF-1α expression enhances proliferation of hepatoma cells [52]. The proliferation slowdown of NNT-deficient cells may be due to reduced expression of HIF-1α-dependent genes, such as IGF1R and STC2. These genes are known to promote growth [53], [54]. DMS treatment that stabilized HIF-1α significantly upregulated expression of these genes. It is likely that changes in metabolites associated with NNT-deficiency lead to growth retardation via HIF-1α signaling.
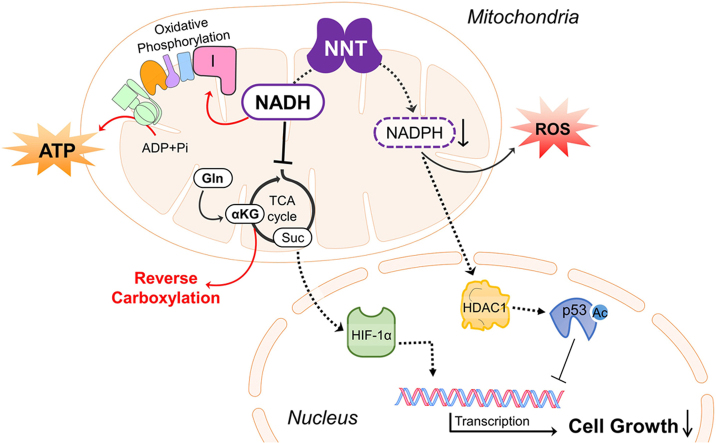
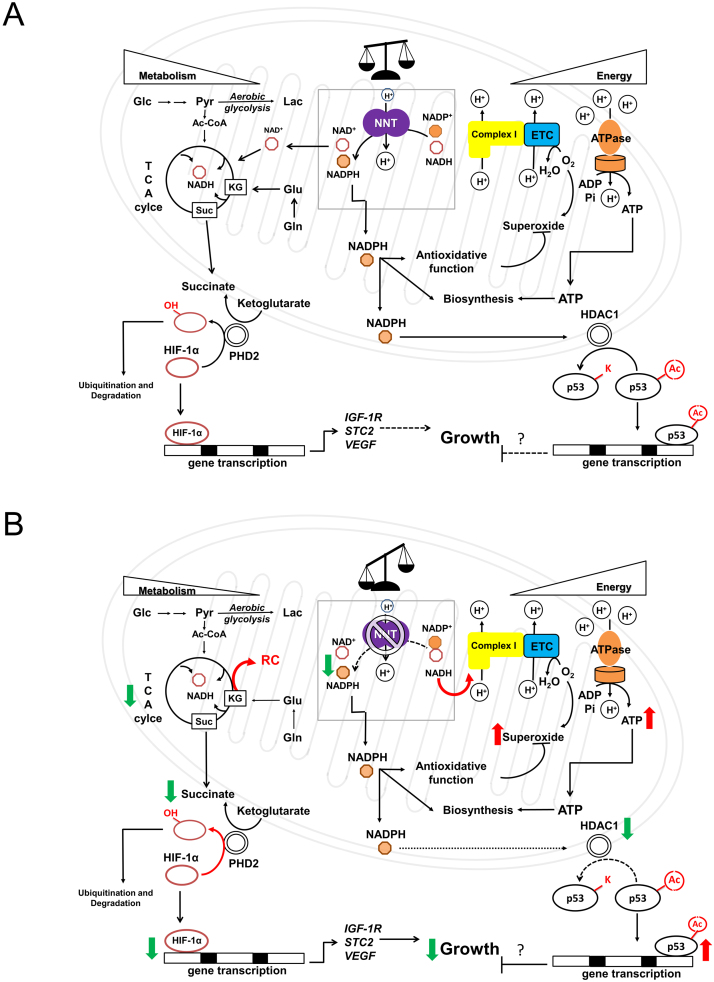
Our findings suggest that redox homeostasis and metabolism are intimately linked. NADPH plays indispensable roles in antioxidative defense and biosynthesis of molecules essential to cells. NADH acts as both a coenzyme in transfer of reducing equivalent and a regulator of energy metabolism (e.g. TCA cycle). NNT plays an important role in maintaining a finely-adjusted balance between NADPH and NADH. As proton flow is required for the reaction of NNT, it also provides a means of sensing whether the mitochondria are energetic or not. If any excess in NADH occurs in energetic mitochondria, NNT acts to transfer hydride ion to generate more NADPH. This helps to remove any ROS generated within actively respiring mitochondria. When the protonmotive force is lowered in less active mitochondria, any NADH left is preserved for oxidative phosphorylation. There is less need for antioxidative defense at this moment. Under pathological condition in which protonmotive force is dissipated in de-energized mitochondria, NNT may operate in reverse mode to catalyze hydride transfer from NADPH to NAD+, and produce NADH. It has been proposed that NNT may work in reverse mode under transient ischemia [9], [55]. Furthermore, a recent study points out that NNT may function in reverse mode in failing heart to generate oxidative stress [14]. The state of mitochondria can be communicated to nucleus via signaling pathways. Under normal circumstances, HIF-1α level is maintained at a proper level. After dimerization with HIF-1β, it binds to promoters of HIF-1-regulated genes and maintains their transcription. At the same time, activities of HDACs (e.g. HDAC1) and acetylation of cellular proteins (e.g. p53) are kept at certain steady levels. All these molecular events are critical to cellular proliferation (Fig. 7). In the absence of NNT, a higher steady state level of NADH is reached, leading to increased ETC activity and ATP generation. These molecular changes are accompanied by decreases in the fluxes of glycolysis and TCA cycle, particularly at α-ketoglutarate dehydrogenase step. Thus, the steady-state NADH level represents a compromise between decreased NADH production and its virtually-ceased NNT-mediated consumption. The increase in [α-ketoglutarate]/[succinate] ratio promotes prolyl hydroxylase-mediated degradation of HIF-1α. Reduction in transcription of HIF-1-regulated genes ensues. NNT deficiency reduces cellular NADPH content, leading to decreased HDAC activity and increased nuclear protein acetylation. Changes in protein acetylation result in alteration of their functions. All these molecular changes precipitate growth slowdown. It is likely that NNT participates in fine adjustment of pyridine adenine dinucleotide pools, which are involved in retrograde signaling for metabolic and redox control of cell growth.
Fig. 7.
A schematic diagram showing the proposed model of the physiological roles of NNT in hepatoma cells. (A) Normally, NNT is involved in sensing of mitochondrial energetic state and fine adjustment of NADPH and NADH pools. NADPH affects HDACs and protein acetylation; NADH and ATP can regulate metabolic pathways, such as glycolysis and TCA cycle. The [α-ketoglutarate]/[succinate] ratio can regulate HIF-1α ubiquitination and proteolysis. (B) In the absence of NNT, NADH is shunted to ETC for its-re-oxidation to NAD+. A steady-state condition is attained, and is characterized by a higher steady-state level of NADH and a lower steady-state flux of TCA cycle. The NADPH level decreases, which lead to a decrease in HDAC activity and an increase in protein acetylation. The acetylated p53 accumulates, and transactivates the growth-suppressive genes. Additionally, as a consequence to metabolic changes, an increase in [α-ketoglutarate]/[succinate] ratio activates prolyl hydroxylase, and increases HIF-1α degradation. Transcription of HIF-1α-regulated growth-promoting genes is reduced. The red upward or green downward arrow indicates an increase or a decrease in the entity alongside it. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Author contributions
H.-Y. H. and M.-L. C. designed the study. H.-Y. H., Y.-T. L. and P.-R. W. performed the in vitro and animal experiments, and analyzed the data. Y.-T. L., G.G. L. and M.-L. C. contributed to stable isotope tracer study. H.-Y. H. supervised the work and wrote the manuscript. G.G. L. and M.-L. C. reviewed the first draft.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The authors are grateful to Prof. Arnold Stern of New York University for his critical review of the article. We appreciate the help of Chang Gung University Core Instrument Center in flow cytometric and IN Cell Analysis. The research work was supported, in whole or in part, by grants from Chang Gung Memorial Hospital (BMRP819, BMRP564, CMRPD1C0752, CMRPD1C0753, CMRPD1E0421, CMRPD1E0422, CMRPD3D0192, CMRPD1C0442, CMRPD1C0443, CMRPD1C0763, CMRPD3D0191, CMRPD3D0193, CMRPD1F0471, CMRPD1F0511 and CMRPD1C0762) and Ministry of Science and Technology (MOST 104-2320-B-182-022-MY3 and MOST 104-2320-B-182-017-MY3).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.04.035.
Appendix A. Supplementary material
Supplementary material
References
- 1.Schaffer S.W., Suleiman M.S. Springer; New York, NY: 2007. Mitochondria. The Dynamic Organelle. [Google Scholar]
- 2.Yin F., Sancheti H., Cadenas E. Silencing of nicotinamide nucleotide transhydrogenase impairs cellular redox homeostasis and energy metabolism in PC12 cells. Biochim. Biophys. Acta. 2012;1817(3):401–409. doi: 10.1016/j.bbabio.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 2008;10(2):179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., Sauve A.A. NAD+ metabolism: bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta. 2016;1864(12):1787–1800. doi: 10.1016/j.bbapap.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelauer M., Krall A.S., McBrian M.A., Li J.Y., Kurdistani S.K. Stimulation of histone deacetylase activity by metabolites of intermediary metabolism. J. Biol. Chem. 2012;287(38):32006–32016. doi: 10.1074/jbc.M112.362467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollak N., Dolle C., Ziegler M. The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. Biochem. J. 2007;402(2):205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13(4):225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menzies K., Auwerx J. An acetylation rheostat for the control of muscle energy homeostasis. J. Mol. Endocrinol. 2013;51(3):T101–T113. doi: 10.1530/JME-13-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoek J.B., Rydstrom J. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem. J. 1988;254(1):1–10. doi: 10.1042/bj2540001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson J.B., Leung J.H., Stout C.D., Schurig-Briccio L.A., Gennis R.B. Review and Hypothesis. New insights into the reaction mechanism of transhydrogenase: swivelling the dIII component may gate the proton channel. FEBS Lett. 2015;589(16):2027–2033. doi: 10.1016/j.febslet.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Sauer U., Canonaco F., Heri S., Perrenoud A., Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 2004;279(8):6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- 12.Imam S., Noguera D.R., Donohue T.J. Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst. Biol. 2013;7:89. doi: 10.1186/1752-0509-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toye A.A., Lippiat J.D., Proks P., Shimomura K., Bentley L., Hugill A., Mijat V., Goldsworthy M., Moir L., Haynes A., Quarterman J., Freeman H.C., Ashcroft F.M., Cox R.D. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48(4):675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- 14.Nickel A.G., von Hardenberg A., Hohl M., Loffler J.R., Kohlhaas M., Becker J., Reil J.C., Kazakov A., Bonnekoh J., Stadelmaier M., Puhl S.L., Wagner M., Bogeski I., Cortassa S., Kappl R., Pasieka B., Lafontaine M., Lancaster C.R., Blacker T.S., Hall A.R., Duchen M.R., Kastner L., Lipp P., Zeller T., Muller C., Knopp A., Laufs U., Bohm M., Hoth M., Maack C. Reversal of mitochondrial transhydrogenase causes oxidative stress in heart failure. Cell Metab. 2015;22(3):472–484. doi: 10.1016/j.cmet.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Kushida M., Kamendulis L.M., Peat T.J., Klaunig J.E. Dose-related induction of hepatic preneoplastic lesions by diethylnitrosamine in C57BL/6 mice. Toxicol. Pathol. 2011;39(5):776–786. doi: 10.1177/0192623311409596. [DOI] [PubMed] [Google Scholar]
- 16.Hanigan M.H., Kemp C.J., Ginsler J.J., Drinkwater N.R. Rapid growth of preneoplastic lesions in hepatocarcinogen-sensitive C3H/HeJ male mice relative to C57BL/6J male mice. Carcinogenesis. 1988;9(6):885–890. doi: 10.1093/carcin/9.6.885. [DOI] [PubMed] [Google Scholar]
- 17.Goldsworthy T.L., Fransson-Steen R. Quantitation of the cancer process in C57BL/6J, B6C3F1 and C3H/HeJ mice. Toxicol. Pathol. 2002;30(1):97–105. doi: 10.1080/01926230252824770. [DOI] [PubMed] [Google Scholar]
- 18.Semenza G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001;13(2):167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsila M., Koivunen P., Gunzler V., Kivirikko K.I., Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 2003;278(33):30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 22.Masson N., Ratcliffe P.J. Hypoxia signaling pathways in cancer metabolism: the importance of co-selecting interconnected physiological pathways. Cancer Metab. 2014;2(1):3. doi: 10.1186/2049-3002-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 24.Weidemann A., Johnson R.S. Biology of HIF-1alpha. Cell Death Differ. 2008;15(4):621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y., Mansfield K.D., Bertozzi C.C., Rudenko V., Chan D.A., Giaccia A.J., Simon M.C. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol. Cell. Biol. 2007;27(3):912–925. doi: 10.1128/MCB.01223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bursch W., Chabicovsky M., Wastl U., Grasl-Kraupp B., Bukowska K., Taper H., Schulte-Hermann R. Apoptosis in stages of mouse hepatocarcinogenesis: failure to counterbalance cell proliferation and to account for strain differences in tumor susceptibility. Toxicol. Sci. 2005;85(1):515–529. doi: 10.1093/toxsci/kfi129. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen A., Karlsson G.B., Rydstrom J. Proton-translocating transhydrogenase: an update of unsolved and controversial issues. J. Bioenerg. Biomembr. 2008;40(5):463–473. doi: 10.1007/s10863-008-9170-x. [DOI] [PubMed] [Google Scholar]
- 28.Agledal L., Niere M., Ziegler M. The phosphate makes a difference: cellular functions of NADP. Redox Rep. 2010;15(1):2–10. doi: 10.1179/174329210X12650506623122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullen A.R., Hu Z., Shi X., Jiang L., Boroughs L.K., Kovacs Z., Boriack R., Rakheja D., Sullivan L.B., Linehan W.M., Chandel N.S., DeBerardinis R.J. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7(5):1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gameiro P.A., Laviolette L.A., Kelleher J.K., Iliopoulos O., Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J. Biol. Chem. 2013;288(18):12967–12977. doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han H.S., Kang G., Kim J.S., Choi B.H., Koo S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016;48:e218. doi: 10.1038/emm.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray L.R., Tompkins S.C., Taylor E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014;71(14):2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tretter L., Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360(1464):2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson G.E., Xu H., Chen H.L., Chen W., Denton T.T., Zhang S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J. Neurochem. 2015;134(1):86–96. doi: 10.1111/jnc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi F., Chen X., Beard D.A. Detailed kinetics and regulation of mammalian NAD-linked isocitrate dehydrogenase. Biochim. Biophys. Acta. 2008;1784(11):1641–1651. doi: 10.1016/j.bbapap.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., DeBerardinis R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., Kelleher J.K., Vander Heiden M.G., Iliopoulos O., Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gameiro P.A., Yang J., Metelo A.M., Perez-Carro R., Baker R., Wang Z., Arreola A., Rathmell W.K., Olumi A., Lopez-Larrubia P., Stephanopoulos G., Iliopoulos O. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17(3):372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Martile M., Del Bufalo D., Trisciuoglio D. The multifaceted role of lysine acetylation in cancer: prognostic biomarker and therapeutic target. Oncotarget. 2016 doi: 10.18632/oncotarget.10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drazic A., Myklebust L.M., Ree R., Arnesen T. The world of protein acetylation. Biochim. Biophys. Acta. 2016;1864(10):1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Doyle K., Fitzpatrick F.A. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J. Biol. Chem. 2010;285(23):17417–17424. doi: 10.1074/jbc.M109.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charni M., Aloni-Grinstein R., Molchadsky A., Rotter V. p53 on the crossroad between regeneration and cancer. Cell Death Differ. 2016 doi: 10.1038/cdd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 44.Terui T., Murakami K., Takimoto R., Takahashi M., Takada K., Murakami T., Minami S., Matsunaga T., Takayama T., Kato J., Niitsu Y. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer Res. 2003;63(24):8948–8954. [PubMed] [Google Scholar]
- 45.Juan L.J., Shia W.J., Chen M.H., Yang W.M., Seto E., Lin Y.S., Wu C.W. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 2000;275(27):20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 46.Michael D., Oren M. The p53 and Mdm2 families in cancer. Curr. Opin. Genet. Dev. 2002;12(1):53–59. doi: 10.1016/s0959-437x(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y.A., Kim M.Y., Yu H.Y., Mishra S.K., Lee J.H., Choi K.S., Kim J.H., Xiang Y.K., Jung Y.S. Gadd45beta is transcriptionally activated by p53 via p38alpha-mediated phosphorylation during myocardial ischemic injury. J. Mol. Med. 2013;91(11):1303–1313. doi: 10.1007/s00109-013-1070-9. [DOI] [PubMed] [Google Scholar]
- 48.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 49.Kim H.R., Roe J.S., Lee J.E., Cho E.J., Youn H.D. p53 regulates glucose metabolism by miR-34a. Biochem. Biophys. Res. Commun. 2013;437(2):225–231. doi: 10.1016/j.bbrc.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 50.Xiao X., Huang X., Ye F., Chen B., Song C., Wen J., Zhang Z., Zheng G., Tang H., Xie X. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci. Rep. 2016;6:21735. doi: 10.1038/srep21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoeltzing O., McCarty M.F., Wey J.S., Fan F., Liu W., Belcheva A., Bucana C.D., Semenza G.L., Ellis L.M. Role of hypoxia-inducible factor 1alpha in gastric cancer cell growth, angiogenesis, and vessel maturation. J. Natl. Cancer Inst. 2004;96(12):946–956. doi: 10.1093/jnci/djh168. [DOI] [PubMed] [Google Scholar]
- 52.Xu Z., Liu E., Peng C., Li Y., He Z., Zhao C., Niu J. Role of hypoxia-inducible-1alpha in hepatocellular carcinoma cells using a Tet-on inducible system to regulate its expression in vitro. Oncol. Rep. 2012;27(2):573–578. doi: 10.3892/or.2011.1533. [DOI] [PubMed] [Google Scholar]
- 53.Liu Q., Xu Z., Mao S., Chen W., Zeng R., Zhou S., Liu J. Effect of hypoxia on hypoxia inducible factor-1alpha, insulin-like growth factor I and vascular endothelial growth factor expression in hepatocellular carcinoma HepG2 cells. Oncol. Lett. 2015;9(3):1142–1148. doi: 10.3892/ol.2015.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Law A.Y., Wong C.K. Stanniocalcin-2 is a HIF-1 target gene that promotes cell proliferation in hypoxia. Exp. Cell Res. 2010;316(3):466–476. doi: 10.1016/j.yexcr.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 55.Sheeran F.L., Rydstrom J., Shakhparonov M.I., Pestov N.B., Pepe S. Diminished NADPH transhydrogenase activity and mitochondrial redox regulation in human failing myocardium. Biochim. Biophys. Acta. 2010;1797(6–7):1138–1148. doi: 10.1016/j.bbabio.2010.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material