Abstract
BACKGROUND
Differences in fast beta (20–28 Hz) electroencephalogram (EEG) oscillatory activity distinguish some individuals with psychiatric and substance use disorders, suggesting that it may be a useful endophenotype for studying the genetics of disorders characterized by neural hyper-excitability. Despite the high heritability estimates provided by twin and family studies, there have been relatively few genetic studies of beta EEG, and to date only one genetic association finding has replicated (i.e., GABRA2).
METHOD
In a sample of 1,564 individuals from 117 families of European Ancestry (EA) drawn from the Collaborative Study on the Genetics of Alcoholism (COGA), we performed a Genome-Wide Association Study (GWAS) on resting-state fronto-central fast beta EEG power, adjusting regression models for family relatedness, age, sex, and ancestry. To further characterize genetic findings, we examined the functional and behavioral significance of GWAS findings.
RESULTS
Three intronic variants located within DSE (dermatan sulfate epimerase) on 6q22 were associated with fast beta EEG at a genome wide significant level (p<5×10−8). The most significant SNP was rs2252790 (p<2.6×10−8; MAF= 0.36; β= 0.135). rs2252790 is an eQTL for ROS1 expressed most robustly in the temporal cortex (p= 1.2×10−6) and for DSE/TSPYL4 expressed most robustly in the hippocampus (p=7.3×10−4; β= 0.29). Previous studies have indicated that DSE is involved in a network of genes integral to membrane organization; gene-based tests indicated that several variants within this network (i.e., DSE, ZEB2, RND3, MCTP1, and CTBP2) were also associated with beta EEG (empirical p<0.05), and of these genes, ZEB2 and CTBP2 were associated with DSM-V Alcohol Use Disorder (AUD; empirical p<0.05).
DISCUSSION
In this sample of EA families enriched for AUDs, fast beta EEG is associated with variants within DSE on 6q22; the most significant SNP influences the mRNA expression of DSE and ROS1 in hippocampus and temporal cortex, brain regions important for beta EEG activity. Gene-based tests suggest evidence of association with related genes, ZEB2, RND3, MCTP1, CTBP2, and beta EEG. Converging data from GWAS, gene expression, and gene-networks presented in this study provide support for the role of genetic variants within DSE and related genes in neural hyperexcitability, and has highlighted two potential candidate genes for AUD and/or related neurological conditions: ZEB2 and CTBP2. However, results must be replicated in large, independent samples.
Keywords: Genome-wide Association Study (GWAS), Endophenotype, Electrophysiology, Resting EEG
Introduction
The resting-state human electroencephalography (EEG) represents the ongoing oscillations of spontaneous and continuous brain electrical activity, typically recorded while the participant is in a relaxed state (Niedermeyer, 1999). EEG is traditionally decomposed into the following frequency bands: delta (0–3 Hz), theta (4–7 Hz), alpha (8–12 Hz), beta (13–28 Hz), and gamma (>29 Hz), with each band reflecting different topography and brain activity (Niedermeyer, 1999). For example, alpha rhythm reflects a relaxed state and has a posterior occipital topography, while beta rhythm reflects an active brain state and is present all over the scalp but predominantly at fronto-central loci (Rangaswamy and Porjesz, 2014). Dynamic coordination of lower frequencies (theta or alpha rhythms from subcortical region) and higher frequencies (beta or gamma rhythms from cortical sites) through a mechanism of phase-amplitude coupling modulates thalamo-cortical and cortico-cortical activity (Canolty and Knight, 2010; Malekmohammadi et al., 2015). Further, coherence at the beta frequency may serve to establish transient physiological connections among neurons in the hippocampus and related brain structures (Leung, 1992a; Vecchio et al., 2016a). While local excitatory-inhibitory interactions underlying sensory, motor and perceptual functions involve local gamma-band oscillations, more integrative cognitive functions mediated by long-range cortical interactions often involve the beta range (Donner and Siegel, 2011). Resting-state brain activity in the beta range (herein referred to as beta EEG) is associated with several behavioral traits, including alcohol use disorders (Bauer, 2001; Begleiter and Porjesz, 1999; Choi et al., 2013; Gilmore et al., 2010a; Lee et al., 2014; Rangaswamy et al., 2002). Given these associations, and the high degree of genetic influence observed (Malone et al., 2014; van Beijsterveldt et al., 1996), beta EEG has been proposed as a useful endo-phenotype (Gottesman and Gould, 2003) for identifying genetic risk factors for disorders characterized by disinhibitory traits (Edenberg et al., 2004; Porjesz et al., 2002). Despite the promise of the endo-phenotype concept however, the genetic complexity of resting-state EEG (Malone et al., 2014), coupled with the scant number of replicable and/or clinically useful genetic variants uncovered by this approach (Iacono et al., 2016), has necessitated large scale genetic association studies of beta EEG, utilizing best-practices in genetic epidemiology.
Previous studies report differences in the magnitude of beta EEG among individuals with alcohol use disorders (AUD) and related problems (Bauer, 2001; Begleiter and Porjesz, 1999; Gilmore et al., 2010b; Propping et al., 1981; Rangaswamy et al., 2002; Winterer et al., 1998), gambling addiction (Choi et al., 2013), and Attention Deficit Hyperactive Disorder (ADHD; (Lee et al., 2014)). Researchers have consistently reported that individuals affected with DSM (III-R and IV) Alcohol Dependence (AD) show higher beta EEG (Bauer, 2001; Propping et al., 1981; Rangaswamy et al., 2002; Winterer et al., 1998). Further decomposition of the beta frequency band demonstrates that increased fast beta power (>19 Hz) is of key importance in the association of beta EEG and AUD. For example, multiple studies have reported that fast beta EEG is superior to severity of illness, depression level, and childhood conduct problems in predicting relapse in abstinent individuals with AUD (Bauer, 2001; Saletu-Zyhlarz et al.). Since elevated beta EEG is present in the offspring of alcoholics prior to the onset of risky drinking (Begleiter and Porjesz, 1999; Deckel et al., 1996; Rangaswamy et al., 2002), it has been suggested that excess beta power precedes the development of AUDs and is likely related to an underlying genetic predisposition for developing AUD, rather than a consequence of heavy alcohol use. Begleiter and colleagues have suggested that this may be an electrophysiological index of an imbalance in the excitation–inhibition homeostasis in the cortex, which underlies a predisposition to develop AUD and related disorders (Begleiter and Porjesz, 1999; Porjesz et al., 2005). Further supporting this hypothesis is the association of beta EEG and other disorders characterized by behavioral disinhibition such as behavior problems and hyperactivity in children (Deckel et al., 1996), externalizing psychopathology (e.g., substance abuse symptoms) in a community sample of adolescents (Gilmore et al., 2010b), ADHD (Choi et al., 2013), and internet addiction with comorbid depression (Lee et al., 2014). Although the precise role of increased beta EEG in these behaviors and disorders remains unclear, this literature suggests that there is variation in fast beta EEG among individuals with AUD and related psychopathology.
Resting state EEG is highly heritable (Malone et al., 2014; van Beijsterveldt et al., 1996), with studies reporting heritability (h2) estimates of monopolar resting state EEG power ranging from 0.49 to 0.85 (Malone et al., 2014; Smit et al., 2005; van Beijsterveldt et al., 1996). Bipolar EEG derivations offer an improvement over monopolar EEG derivations in the spatial resolution of the electrical sources, and reduce volume conduction effects (Ingber and Nunez, 1995; Nunez et al., 1997). In addition, the stability of EEG signals is excellent and under standardized conditions, there are high test-retest correlations. Studies that have examined the heritability of bipolar eyes-closed resting EEG power have shown comparable estimates to monopolar derivations (Tang et al., 2007b) and indicate that bipolar derivations are in greater accord with genetic findings in brain anatomy (Tang et al., 2007a). Despite the high heritability estimates provided by twin and family studies, there have been relatively few large (i.e., adequately powered) genetic studies of beta EEG (Iacono et al., 2016), and to date only one finding has replicated. An early analysis found linkage between beta EEG and a region of chromosome 4 (Porjesz et al., 2002) harboring variants in the gene that encodes the GABA α2 receptor subunit (GABRA2), which were subsequently associated with both beta EEG and AD (Edenberg et al., 2004). More recently, a study of 586 individuals of European ancestry (EA) with DSM-IV AD, and 603 ancestrally matched individuals without AD, replicated the association between beta activity and several GABRA2 variants (Lydall et al., 2011). To date, only two genome-wide association studies (GWAS) of beta EEG have been conducted (Hodgkinson et al., 2010; Malone et al., 2014). In a study of 322 Native-American individuals, there were no genome-wide significant associations reported for beta EEG (Hodgkinson et al., 2010). We note that there were genome-wide significant findings for other EEG parameters; an association was observed among theta power (and AD) and several variants in SGIP1 (Hodgkinson et al., 2010). A recent GWAS of several EEG measures (including monopolar beta EEG, assessed at the central electrode) in 4,026 adolescent twins and their parents (Malone et al., 2014) did not report any genome-wide significant variants, but replicated the previous associations observed between beta EEG and GABRA2 and expanded our understanding of the genetic epidemiology of other EEG parameters (i.e., resting-state theta EEG).
Given that beta EEG is highly heritable and has been found to be related to several externalizing traits including AUD (Bauer, 2001; Begleiter and Porjesz, 1999; Choi et al., 2013; Gilmore et al., 2010b; Lee et al., 2014; Rangaswamy et al., 2002), genetic analysis of beta EEG may aid in our understanding basic brain functioning, and potentially differences and similarities among individuals with a range of behavioral and psychiatric disorders. As the elevation of beta power reported in individuals with AUD has a largely anterior topography, particularly in the higher frequency fast beta band (20–28 Hz; (Rangaswamy et al., 2002)), the primary aim of this study was to conduct a GWAS of fast beta (20–28 Hz) EEG power (bipolar derivation at fronto-central loci) in families from the Collaborative Study on the Genetics of Alcoholism (COGA); many of these families were densely affected with AD. In an effort to move beyond genotype-phenotype association and further characterize genetic association findings, the secondary aims of this study were to examine the functional and behavioral significance of GWAS findings. To this end, we explore the functional significance of GWAS variants using publically available gene expression data. In addition, we explore biological networks using publically available prediction programs.
SAMPLE & METHODS
Sample
COGA recruited DSM-III-R and DSM-IV AD probands from inpatient and outpatient treatment facilities through six participating sites: State University of New York Downstate Medical Center, University of Connecticut Health Science Center, Indiana University School of Medicine, University of Iowa College of Medicine, University of California School of Medicine, and Washington University School of Medicine. Recruitment and assessment procedures, including a clinical interview, neurophysiological assessments and DNA collection have been described previously (Begleiter et al., 1995; Foroud et al., 2000). Probands and family members were administered the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA), a poly-diagnostic interview (Bucholz et al. 1994; Hesselbrock et al. 1999). Individuals below the age of 18 were administered an adolescent version of the SSAGA. The laboratory and data-collection procedures were identical at each of the sites (Begleiter et al., 1998). Institutional review boards at all sites approved the study.
All COGA DNA samples were genotyped for a 96 SNP (single nucleotide polymorphism) array (Fluidigm SNPtrace, Rutgers University Cell and DNA Repository) that included 64 ancestry informative markers. The principal components derived from these SNPs were used to assign ancestry, and were the basis for the selection of the European Ancestry (EA) families. Prioritization of families was based on the most informative families, defined as those with the largest number of alcohol dependent family members with DNA and electrophysiological measurements. The analytic sample consisted of all family members with both resting state EEG and GWAS data available: 1,564 individuals (824 females and 740 males; average age: 31.6) from 117 multi-generational families affected with AD. Family sizes ranged from 4 to 39 individuals with an average of 13.4 individuals (with EEG data) per family.
EEG Recording & Processing
Prior to neurophysiological assessments, participants were required to have abstained from alcohol for a minimum of 3 weeks. Individual were excluded from neurophysiological assessment if they had any of the following: (1) recent substance or alcohol use (i.e., positive breath-analyzer test); (2) hepatic encephalopathy/cirrhosis of the liver; (3) significant history of head injury, seizures or neurosurgery; (4) uncorrected sensory deficits; (5) taking medication known to influence brain functioning; and (6) other acute/chronic medical illnesses that affect brain function.
Participants were seated comfortably in a dimly lit sound-attenuated temperature-regulated booth (Industrial Acoustics, Bronx, NY). They were instructed to keep their eyes closed and remain relaxed, but to not fall asleep. EEG data were collected in the awake, eyes-closed condition for 4.25 minutes. Each participant wore a fitted electrode cap (Electro-Cap International, Eaton, OH) using the 19-channel montage as specified according to the 10–20 international system (Supplementary Figure 1). The nose was used as a reference, and a forehead electrode served as the ground electrode. Both vertical and horizontal eye movements were monitored with electrodes that were placed supraorbitally and at the outer canthus of the left eye to perform ocular artifact correction. Electrode impedances were maintained below 5 kΩ. Electrical activity was amplified 100,000 times by Sensorium (Charlotte, VT) EPA-2 electrophysiology amplifiers with either a bandpass between 0.02 and 50 Hz and digitized on a Concurrent (Atlanta, GA) 5550 computer at a sampling rate of 256 Hz or a band pass between 0.02 Hz and 100.0 Hz on a Neuroscan system (Version 4.1 to 4.5) (Neurosoft, Inc., El Paso, TX) at sampling rates of 500 Hz or 512 Hz. All six collection sites used identical experimental procedures and EEG acquisition hardware and software programs.
A continuous interval comprising 256 seconds of eyes-closed resting EEG data was analyzed. The raw data were subjected to wavelet filtering and reconstruction to eliminate very high and low frequencies (Bruce & Gao, 1994; Strang & Nguyen, 1996). The s12 wavelet was used to perform a six-level analysis, and the output signal was reconstructed with levels d6–d3, roughly equivalent to applying a bandpass filter with a range of 2–64 Hz to the data. Subsequently, eye movements were removed by using the method developed by Gasser et al. (Gasser and Laemmli, 1987; Gasser et al., 1986). The filtered artifact-free data were transformed into bipolar derivations. Bipolar derivations were used in preference to monopolar derivations to improve the spatial resolution of the electrical sources (Ingber and Nunez, 1995; Nunez et al., 1997). Bipolar derivations were analyzed in 254 overlapping 2-second epochs by use of a Fourier transform. After windowing effects were minimized by application of a Hamming function (Hamming, 1983), the resulting spectral densities, sampled at 0.5 Hz intervals, were aggregated into bands, divided by the bandwidth, and then averaged across epochs. As the elevation of beta power reported in individuals with alcohol use problems has a largely anterior topography, particularly in the higher frequency fast beta band (20–28 Hz; (Rangaswamy et al., 2002)), the current study examines fast beta EEG (20–28 Hz) at fronto-central pairs: Fz-Cz, F3-C3, and F4-C4. Given the high degree of correlation observed among these phenotypes, GWAS results are presented for F3-C3, for which the most robust effects were observed.
Genotyping, Imputation and Quality Review
Genotyping of 1,564 individuals from 117 EA families was performed at the Center for Inherited Disease Research (CIDR) using the Illumina 2.5M array (Illlumina, San Diego, CA, USA). COGA’s quality control (QC) approach has been previously reported (Wetherill et al., 2015). Briefly, individuals with a genotype rate <98% were excluded from analysis, and SNPs with a genotyping rate <98% were excluded from analysis. The 795 genotyped founders were used to remove SNPs which violated Hardy-Weinberg equilibrium (HWE; p<10−6). SNPs with minor allele frequency (MAF) less than 3% in the founders were also removed from further analysis. The reported pedigree structure was assessed using a pruned set of 1,519,440 SNPs. Pairwise identity by descent estimates were computed in PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) to detect pairs of individuals whose allele sharing was not consistent with the reported family relationship. Family structures were altered as needed, and then SNP genotypes were tested for Mendelian inconsistencies (Pedcheck; O’Connell & Weeks, 1998) with the revised family structure. The cleaned genotype data were imputed to 1000 genomes (EUR and AFR, Phase 3, b37, October 2014) with build hg19 using SHAPEIT (https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html) and IMPUTE2 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html). To avoid ambiguities in strand designation, SNPs with A/T or C/G alleles were removed. After imputation, genotype probabilities ≥ 0.90 were changed to genotypes. Mendelian errors in the imputed SNPs were reviewed and resolved as described in Wetherill et al., 2015 (Wetherill et al., 2015). All SNPs with imputation genotyping rate < 98% and MAF < 0.03 were excluded from association analyses.
Association Analysis
Primary analyses were conducted in GWAF (Genome-Wide Association analyses with Family) on 12,972,748 SNPs, of which 1,519,440 were genotyped directly, using a generalized estimating equation (GEE) framework to control for the relatedness in the family sample (Chen and Yang, 2010). Sex and log-transformed age (at the time of EEG recording) were included as covariates in the model, as each of these variables were associated with fast beta EEG (p<0.0001). The first three principal components (PC1-PC3) computed from SNPRelate (Zheng et al, 2012) were also included as covariates to reduce the risk of false-positive associations owing to population stratification. Established thresholds for genome-wide significance (p<5×10−8) were utilized. In an effort to assess the influence of alcohol use problems on the genetic associations observed for beta EEG, given the association of AUD and beta EEG in this and previous studies, we conducted a secondary analysis in which we repeated the initial GWAS as described above, with the addition of DSM-V AUD severity as a covariate.
Functional Analyses
We utilized publicly available data from the UK Brain Expression Consortium (BRAINEAC; http://www.braineac.org/) to examine whether the most significant GWAS variant for fast beta EEG was an expression quantitative trait locus (eQTL) for any known gene. BRAINEAC draws on data from 134 neuropathologically normal individuals of EA and assesses 10 different regions of the brain, including: cerebellar cortex, frontal cortex, hippocampus, medulla (inferior olivary nucleus), occipital cortex (primary visual cortex), putamen, substantia nigra, thalamus, temporal cortex, and intralobular white matter (Trabzuni et al., 2011). All p-values presented are Bonferonni corrected for multiple-testing, based on the ten brain regions examined. Due to the large number of brain regions examined, only the SNPs genome-wide associated with fast beta EEG were examined in BRAINEAC to minimize multiple-testing. All associations that withstood multiple testing were examined in the Genotype-Tissue Expression Project (GTex) database (www.gtexportal.org) to confirm eQTL findings (For brain eQTLs, sample sizes ranged from 70–127). GeneMANIA Cytoscape 3.0.0 plugin (Mostafavi et al., 2008), a multiple association network integration algorithm for predicting gene function, was employed to identify genes in related gene networks; physical, co-expression, co-localization and pathway gene-gene interactions were evaluated. Once a gene network was identified (via GeneMANIA as described above), DAVID (Database for Annotation, Visualization, and Integrated Discovery; (Dennis et al., 2003) bioinformatics resource was used to assess gene functions common to this network. Functional Categories and Gene Ontologies were evaluated based on enrichment scores (Fisher Exact Probability Value or “EASE Score”) and tests of statistical significance, including p-values adjusted for multiple testing (Bonferonni correction).
Post-Hoc Analyses
Following the identification of gene networks of relevance to GWAS findings (as described above via GeneMANIA), gene-based tests of association with fast beta EEG were conducted in PLINK (Purcell et al., 2007), using set-based analyses (“--set-test”). Gene-based tests included all available SNPs in a given gene, corrected for the number of independent signals (i.e., linkage disequilibrium blocks) within that gene set. In addition, association models were adjusted for the relatedness in the family sample, sex, log-transformed age, and ancestry. Tests of association were accepted as significant if the 100,000 permutations of the set-based regression analysis (Bonferroni corrected for the number of independent signals within the set) produced an empirical p-value <0.05. Subsequently, these procedures were repeated for tests of gene-based association with DSM-V AUD severity only among genes that were associated with beta EEG. In addition, GABRA2 variants previously shown to be associated with aspects of beta EEG (Edenberg et al., 2004; Lydall et al., 2011; Malone et al., 2014) were tested for association with fast beta EEG (20–28 Hz) at fronto-central pairs: Fz-Cz, F3-C3, and F4-C4.
Results
Association Analysis
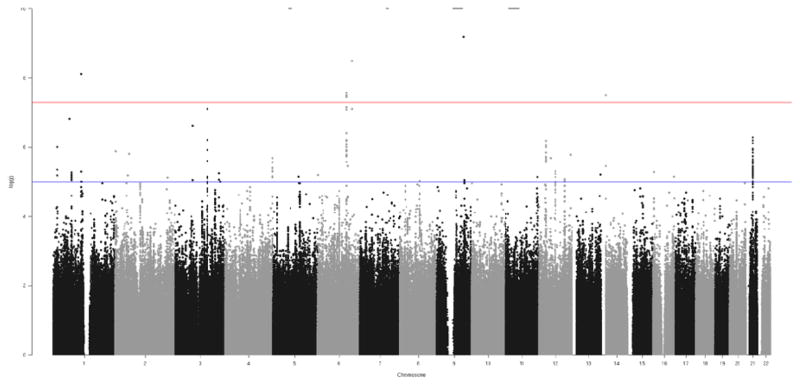
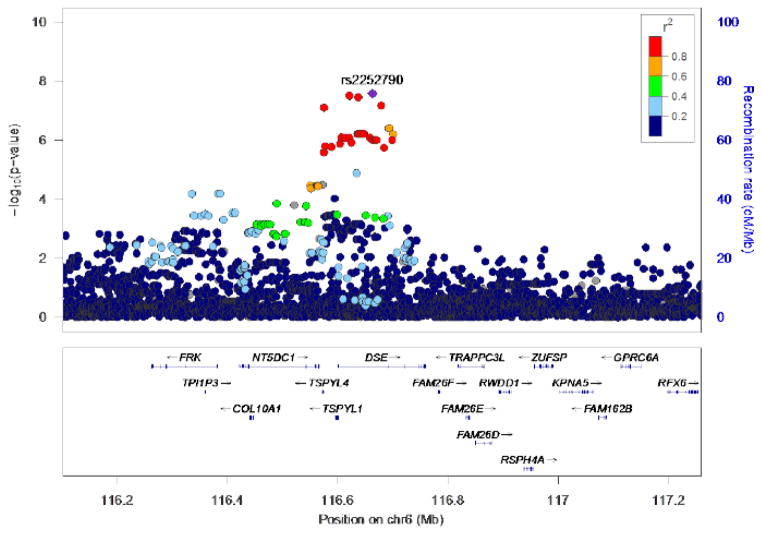
Three highly correlated (i.e., in high linkage disequilibrium; r2 =1.0; D-prime=1.0 based on hg19 1000 Genomes from the CEU sample) intronic SNPs (rs10456907, rs13214667, rs2252790) located within DSE (dermatan sulfate epimerase) on 6q22 were associated with fast beta EEG at a genome wide significant level (p<5×10−8). All variants associated with beta EEG at p <5×10−7 are detailed in Table 1 and depicted in Figures 1 and 2. Conditional analyses, and the high degree of linkage disequilibrium observed among the most significant SNPs (Figure 2), suggest that a single genome-wide signal is implicated. The most significant SNP was rs2252790 (p<2.6×10−8; MAF: 0.36; β: 0.135; Table 1). GWAS results adjusted for DSM-V AUD severity (i.e., GWAS model included DSM-V AUD severity as a covariate) yielded similar results as the primary analyses: intronic DSE variant rs2252790 remained the most significantly associated SNP. However, p-values were slightly less robust (Supplemental Table 1) with only one of three DSE variants remaining genome-wide significant (rs2252790). Three additional sub-threshold (5×10−7>p>5×10−8; Table 1) signals were also detected, including a signal on the long arm of Chromosome 3 (3q11.2; UROC1, FRMD4B; Supplemental Figure 2), an intergenic signal on the long arm of Chromosome 12 (12q14; Supplemental Figure 3), and an intergenic signal on the long arm of Chromosome 21 (21q21; Supplemental Figure 4).
Table 1.
Variants associated with Fast beta EEG (20–28 Hz; bipolar derivation at the fronto-central electrode pair (F3-C3)) at p-value<5×10−7
| SNP | CHR | BASE PAIR | AL1 | AL2 | MAF | BETA | SE | P-VALUE | GENE |
|---|---|---|---|---|---|---|---|---|---|
| rs112196420 | 3 | 69499803 | T | G | 0.03 | −0.339 | 0.066 | 2.30E-07 | FRMD4B |
| rs16837279 | 3 | 125932261 | G | T | 0.216 | −0.131 | 0.026 | 6.00E-07 | intergenic |
| rs1687482 | 3 | 126215694 | T | G | 0.151 | −0.162 | 0.03 | 7.60E-08 | UROC1 |
| rs910391 | 6 | 116575344 | T | G | 0.362 | 0.128 | 0.024 | 7.90E-08 | TSPYL4, DSE |
| rs2982842 | 6 | 116606807 | G | A | 0.359 | 0.114 | 0.023 | 8.00E-07 | DSE |
| rs12212556 | 6 | 116613907 | G | A | 0.359 | 0.114 | 0.023 | 8.30E-07 | DSE |
| rs10456906 | 6 | 116621035 | A | C | 0.359 | 0.114 | 0.023 | 8.30E-07 | DSE |
| rs10456907 | 6 | 116621036 | C | T | 0.362 | 0.133 | 0.024 | 3.10E-08 | DSE |
| rs13209267 | 6 | 116635819 | C | A | 0.358 | 0.116 | 0.023 | 6.10E-07 | DSE |
| rs12213442 | 6 | 116636907 | A | G | 0.358 | 0.116 | 0.023 | 6.10E-07 | DSE |
| rs13214667 | 6 | 116637440 | A | G | 0.364 | 0.134 | 0.024 | 3.50E-08 | DSE |
| rs12203516 | 6 | 116638619 | G | A | 0.357 | 0.116 | 0.023 | 6.10E-07 | DSE |
| rs12214926 | 6 | 116643292 | T | C | 0.357 | 0.116 | 0.023 | 6.00E-07 | DSE |
| rs12204234 | 6 | 116647587 | C | T | 0.358 | 0.116 | 0.023 | 6.00E-07 | DSE |
| rs2501053 | 6 | 116658732 | A | G | 0.359 | 0.114 | 0.023 | 8.20E-07 | DSE |
| rs2501052 | 6 | 116658955 | A | G | 0.358 | 0.114 | 0.023 | 8.20E-07 | DSE |
| rs2501050 | 6 | 116663206 | G | A | 0.358 | 0.114 | 0.023 | 9.80E-07 | DSE |
| rs2252790 | 6 | 116663323 | G | A | 0.36 | 0.135 | 0.024 | 2.60E-08 | DSE |
| rs4946160 | 6 | 116667472 | C | A | 0.358 | 0.114 | 0.023 | 9.80E-07 | DSE |
| rs3021301 | 6 | 116670364 | A | G | 0.358 | 0.114 | 0.023 | 9.80E-07 | DSE |
| rs2498710 | 6 | 116670636 | G | A | 0.358 | 0.114 | 0.023 | 9.80E-07 | DSE |
| rs2858848 | 6 | 116679275 | A | G | 0.364 | 0.131 | 0.024 | 6.70E-08 | DSE |
| rs2640873 | 6 | 116692795 | A | G | 0.403 | 0.115 | 0.023 | 3.90E-07 | DSE |
| rs2640872 | 6 | 116693384 | T | C | 0.403 | 0.115 | 0.023 | 3.90E-07 | DSE |
| rs2213563 | 6 | 116698931 | A | G | 0.374 | 0.112 | 0.023 | 9.90E-07 | DSE |
| rs574871 | 6 | 116700809 | C | T | 0.403 | 0.113 | 0.023 | 6.20E-07 | DSE |
| rs258430 | 12 | 28033502 | G | A | 0.573 | −0.111 | 0.022 | 6.30E-07 | intergenic |
| rs258442 | 12 | 28045607 | T | C | 0.567 | −0.109 | 0.022 | 9.90E-07 | intergenic |
| rs258444 | 12 | 28046102 | G | A | 0.574 | −0.109 | 0.022 | 8.60E-07 | intergenic |
| rs10843021 | 12 | 28050947 | C | T | 0.568 | −0.109 | 0.022 | 9.90E-07 | intergenic |
| rs399384 | 21 | 27968714 | T | C | 0.365 | −0.111 | 0.023 | 7.40E-07 | intergenic |
| rs384901 | 21 | 27970165 | G | T | 0.385 | −0.11 | 0.022 | 7.40E-07 | intergenic |
| rs222957 | 21 | 27977823 | A | G | 0.376 | −0.111 | 0.022 | 6.10E-07 | intergenic |
| rs376583780 | 21 | 28003040 | T | C | 0.328 | −0.118 | 0.023 | 5.10E-07 | intergenic |
| rs370690781 | 21 | 28003042 | T | C | 0.328 | −0.118 | 0.023 | 5.10E-07 | intergenic |
| rs372255102 | 21 | 28003047 | T | C | 0.328 | −0.118 | 0.023 | 5.10E-07 | intergenic |
| rs376311445 | 21 | 28003049 | A | C | 0.328 | −0.118 | 0.023 | 5.10E-07 | intergenic |
| rs56387300 | 21 | 28003051 | T | C | 0.328 | −0.118 | 0.023 | 5.10E-07 | intergenic |
Note: Genome-wide significant (p-value<5×10−8) variants are bolded.
Fig. 1.
Manhattan plot of GWAS results for the fast beta EEG (20–28 Hz; bipolar derivation at the fronto-central electrode pair (F3-C3)).
Negative log-transformed p-values for SNPs are plotted against base-pair position for each chromosome. Three intronic DSE variants on chromosome 6 exceeded the genome-wide significance threshold of 5 × 10−8, indicated by the red line.
Figure 2. Association results for fast beta EEG on Chromosome 6.
Y-axis denotes the −log10(p-value) for association. X-axis is the physical position on the chromosome (Mb). The most significantly associated SNP (rs2252790) is shown in purple. The extent of linkage disequilibrium (as measured by r2) between each SNP and the most significantly associated SNP is indicated by the color scale at top left. Larger values of r2 indicate greater linkage disequilibrium (LD). LD is based on hg19 1000 Genomes from the CEU sample.
Functional Analyses
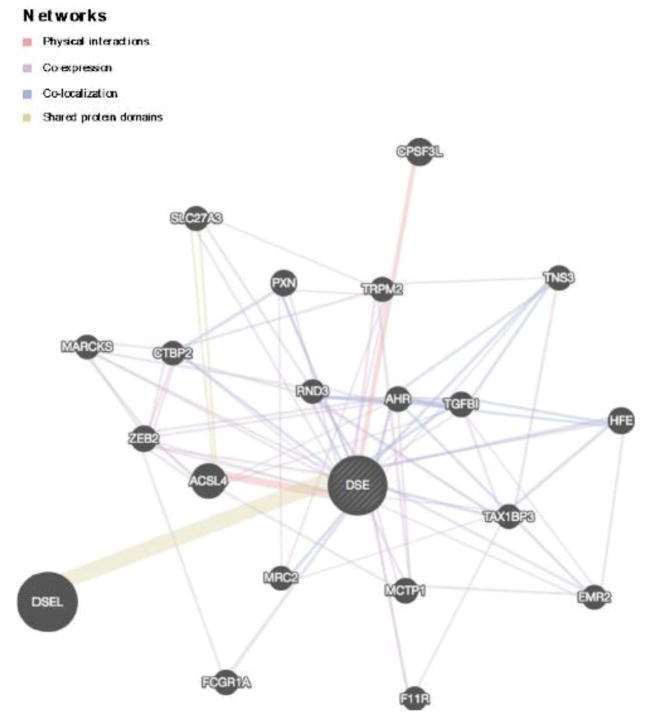
Braineac indicates that rs2252790 is nominally associated with the mRNA expression of DSE/TSPYL1 (TSPY like 4), and ROS1 (ROS proto-oncogene 1, receptor tyrosine kinase) in several brain regions. Two of these findings survived a Bonferroni multiple test correction (p<5×10−3): rs2252790 is an eQTL for DSE/TSPYL1 expression in hippocampus tissue (p=1.26×10−4) and for ROS1 expression in temporal cortex tissue (p=1.20×10−6). In the GTEx database, rs2252790 is associated with the expression of DSE/TSPYL1, ROS1, NT5DC1, and FRK, with the most robust effects observed for DSE expression (p=8.0×10−20). GeneMANIA indicated that DSE is involved in the following network of genes: DSEL, ACSL4, CPSF3L, HFE, ZEB2, PXN, AHR, MCTP1, TGFBI, FCGR1A, TNS3, TRPM2, SLC27A3, EMR2, TAX1BP3, MRC2, F11R, RND3, MARCKS, CTBP2. These genes are detailed in Table 2 and Figure 3. DAVID indicated that 11/21 of these genes were integral to membrane organization (enrichment score: 1.33, p-value=0.03); however, this enrichment score did not survive a Bonferonni correction (p-value= 0.09).
Table 2.
DSE -related gene network identified via GeneMANIA, a multiple association network integration algorithm utilizing physical, co-expression, co-localization, and pathway gene-gene interactions observed in previous studies.
| GENE SYMBOL | CHROMOSOME LOCATION | OFFICIAL GENE NAME |
|---|---|---|
| ACSL4 | X | acyl-CoA synthetase long-chain family member 4 |
| CPSF3L | 1 | cleavage and polyadenylation specific factor 3-like |
| FCGR1A | 1 | Fc fragment of IgG receptor Ia |
| SLC27A3 | 1 | solute carrier family 27 member 3 |
| F11R | 1 | F11 receptor |
| ZEB2 | 2 | zinc finger E-box binding homeobox 2 |
| RND3 | 2 | Rho family GTPase 3 |
| MCTP1 | 5 | multiple C2 and transmembrane domain containing 1 |
| TGFBI | 5 | transforming growth factor beta induced |
| DSE | 6 | dermatan sulfate epimerase |
| HFE | 6 | hemochromatosis |
| MARCKS | 6 | myristoylated alanine rich protein kinase C substrate |
| AHR | 7 | aryl hydrocarbon receptor |
| TNS3 | 7 | tensin 3 |
| CTBP2 | 10 | C-terminal binding protein 2 |
| PXN | 12 | paxillin |
| TAX1BP3 | 17 | Tax1 binding protein 3 |
| MRC2 | 17 | mannose receptor C type 2 |
| DSEL | 18 | dermatan sulfate epimerase-like |
| ADGRE2 | 19 | adhesion G protein-coupled receptor E2 |
| TRPM2 | 21 | transient receptor potential cation channel subfamily M member 2 |
Note: ACSL4 was not assessed in the current study
Figure 3.
DSE related gene network identified via GeneMANIA, a multiple association network integration algorithm utiilizing physical, co-expression, and co-localization gene-gene interactions as well as genes with shared protein domains observed in previous studies.
Post-Hoc Analyses
Following the identification of gene networks of relevance to GWAS findings (i.e., genes shown to interact with DSE via physical, co-expression, co-localization and/or pathway analyses in previous studies, curated using GeneMANIA as described above), gene-based analyses indicated that when considered as a set (all available SNPs in a given gene, corrected for the number of independent signals within that gene set), variants within the following genes are associated with fast beta EEG (empirical p-value<0.05): DSE, ZEB2, MCTP1, RND3 and CTBP2 (Table 3). In addition, gene-based analyses indicated that ZEB2 and CTBP2 were also associated with DSM-V AUD (empirical p-value<0.05; Table 3). Table 3 details the number of variants examined in each gene-set, the number of variants nominally associated with beta EEG and/or AUD (p<0.05), the number of independent signals represented among each of the SNPs tested, and the empirical p-value based on 100,000 permutations. In addition, 20/26 GABRA2 variants previously shown to be associated with aspects of beta EEG were modestly (p<0.05) associated with fast beta EEG at specific fronto-central pairs (Table 4). Of these 20 variants, 5 survive a multiple-test correction (0.05/3 LD Blocks: p<0.017).
Table 3.
Gene-based association of DSE-related genes, fast beta EEG, and DSM-V AUD
| GENE SYMBOL | # OF SNPS TESTED | # OF SIGNIFICANT SNPS | # OF INDEPENDENT SIGNALS | EMPIRICAL P-VALUE | ||
|---|---|---|---|---|---|---|
| Beta EEG | DSM-V AUD | Beta EEG | DSM-V AUD | |||
| DSE | 602 | 163 | 89 | 5 | <0.000 | 0.095 |
| CTBP2 | 780 | 53 | 64 | 5 | 0.019 | 0.044 |
| ZEB2 | 218 | 23 | 43 | 5 | 0.032 | 0.022 |
| MCTP1 | 1651 | 140 | 110 | 5 | 0.044 | 0.068 |
| RND3 | 90 | 4 | 3 | 1 | 0.046 | 0.210 |
| TNS3 | 936 | 52 | Not tested | 5 | 0.065 | Not tested |
| MRC2 | 167 | 15 | Not tested | 5 | 0.082 | Not tested |
| F11R | 180 | 16 | Not tested | 5 | 0.125 | Not tested |
| MARCKS | 61 | 20 | Not tested | 5 | 0.176 | Not tested |
| CPSF3L | 103 | 10 | Not tested | 5 | 0.204 | Not tested |
| TRPM2 | 270 | 7 | Not tested | 5 | 0.267 | Not tested |
| ADGRE2 | 317 | 19 | Not tested | 5 | 0.319 | Not tested |
| TGFBI | 142 | 4 | Not tested | 4 | 0.345 | Not tested |
| DSEL | 77 | 3 | Not tested | 5 | 0.374 | Not tested |
| PXN | 143 | 6 | Not tested | 5 | 0.404 | Not tested |
| HFE | 115 | 22 | Not tested | 5 | 0.583 | Not tested |
| SLC27A3 | 46 | 4 | Not tested | 3 | 0.591 | Not tested |
| AHR | 117 | 11 | Not tested | 5 | 0.608 | Not tested |
| FCGR1A | 4 | 0 | Not tested | 0 | 1.000 | Not tested |
| TAX1BP3 | 115 | 0 | Not tested | 1 | 1.000 | Not tested |
Note: Following the identification of DSE related genes identified via GeneMANIA, gene-based tests of association with fast beta EEG were conducted in PLINK (Purcell et al., 2003), using set-based analyses (“--set-test”). Gene-based tests included all available SNPs in a given gene, corrected for the number of independent signals (i.e., linkage disequilibrium blocks) within that gene set. In addition, all association models were adjusted for the relatedness in the family sample, sex, log-transformed age, and ancestry. Tests of association were accepted as significant if the 100,000 permutations of the set-based regression analysis produced an empirical p-value <0.05. Subsequently, these procedures were repeated for tests of gene-based association with DSM-V AUD severity only among genes that were associated with beta EEG. Table 3 details the number of SNPs examined in each gene-set, the number of SNPs within each gene-set associated with beta EEG and AUD (p<0.05), the number of independent signals represented among each of the SNPs tested, and the empirical p-value based on 100,000 permutations.
Table 4.
Associations of previously reported GABRA2 variants and fast beta EEG (20–28 Hz; bipolar derivation at the fronto-central electrode pairs (FZ-CZ, F3-C3, F4-C4))
| SNP | Base Pair Location | Major Allele | Minor Allele | MAF | β | p-value | β | p-value | β | p-value |
|---|---|---|---|---|---|---|---|---|---|---|
| FZ-CZ | F3-C3 | F4-C4 | ||||||||
| rs561779 | 46238778 | G | A | 0.59 | −0.04 | 0.046 | 0.01 | 0.680 | 0.00 | 0.980 |
| rs572227 | 46251393 | C | T | 0.59 | −0.05 | 0.014* | 0.00 | 0.940 | −0.01 | 0.620 |
| rs573400 | 46252066 | T | C | 0.59 | −0.04 | 0.047 | 0.01 | 0.690 | 0.00 | 0.980 |
| rs532780 | 46261366 | T | C | 0.58 | −0.04 | 0.035 | 0.01 | 0.560 | 0.00 | 0.920 |
| rs548583 | 46263344 | G | A | 0.58 | −0.06 | 0.011* | 0.01 | 0.820 | −0.01 | 0.720 |
| rs496650 | 46264385 | A | C | 0.58 | −0.04 | 0.050 | 0.01 | 0.600 | 0.00 | 0.940 |
| rs540363 | 46274246 | A | G | 0.58 | −0.04 | 0.039 | 0.01 | 0.610 | 0.00 | 0.950 |
| rs526752 | 46276629 | A | G | 0.58 | −0.05 | 0.031 | 0.01 | 0.580 | 0.00 | 0.950 |
| rs530329 | 46281119 | T | C | 0.58 | −0.05 | 0.031 | 0.01 | 0.580 | 0.00 | 0.950 |
| rs483160 | 46287075 | G | T | 0.58 | −0.05 | 0.025 | 0.01 | 0.640 | 0.00 | 0.950 |
| rs279871 | 46305733 | T | C | 0.57 | −0.06 | 0.009* | 0.00 | 0.860 | −0.01 | 0.640 |
| rs279867 | 46308303 | A | C | 0.58 | −0.05 | 0.029 | 0.01 | 0.590 | 0.00 | 0.990 |
| rs279866 | 46309764 | A | G | 0.58 | −0.04 | 0.036 | 0.01 | 0.550 | 0.00 | 0.960 |
| rs279863 | 46313022 | C | A | 0.58 | −0.04 | 0.032 | 0.01 | 0.580 | 0.00 | 0.950 |
| rs279858 | 46314593 | T | C | 0.57 | −0.06 | 0.011* | 0.01 | 0.830 | −0.01 | 0.700 |
| rs279843 | 46325204 | C | T | 0.57 | −0.05 | 0.028 | 0.00 | 0.980 | −0.01 | 0.680 |
| rs279846 | 46329886 | C | T | 0.55 | −0.04 | 0.066 | 0.01 | 0.590 | 0.01 | 0.700 |
| rs279826 | 46334209 | A | G | 0.55 | −0.04 | 0.059 | 0.01 | 0.590 | 0.01 | 0.770 |
| rs279828 | 46334810 | A | C | 0.55 | −0.04 | 0.045 | 0.01 | 0.660 | 0.01 | 0.770 |
| rs279837 | 46339323 | A | G | 0.57 | −0.04 | 0.031 | 0.01 | 0.770 | 0.00 | 0.830 |
| rs279841 | 46340763 | G | A | 0.57 | −0.05 | 0.017* | 0.00 | 0.980 | −0.01 | 0.830 |
| rs189957 | 46346679 | A | G | 0.55 | −0.04 | 0.051 | 0.01 | 0.600 | 0.01 | 0.790 |
| rs1442062 | 46377076 | A | G | 0.25 | 0.03 | 0.280 | 0.02 | 0.460 | 0.03 | 0.280 |
| rs3756007 | 46391064 | C | T | 0.04 | −0.02 | 0.770 | −0.02 | 0.710 | −0.03 | 0.620 |
| rs894269 | 46393612 | C | T | 0.78 | −0.01 | 0.720 | 0.03 | 0.200 | 0.01 | 0.580 |
| rs1545234 | 46404413 | A | G | 0.70 | 0.01 | 0.630 | 0.03 | 0.230 | 0.02 | 0.340 |
Withstands a multiple-test correction
Discussion
Although previous studies have reported variation in beta EEG among individuals diagnosed with AD and related conditions, there have been relatively few studies examining genetic variants in relation to beta EEG and only one finding that has been replicated to date (GABRA2 (Porjesz et al. 2002b; Edenberg et al. 2004; Lydall et al. 2011; Ittiwut et al. 2012; Malone et al. 2014)). Subsequently, associations between GABRA2, AD (Edenberg et al. 2004; Lappalainen et al. 2005; Covault et al, 2004; Drgon et al., 2006; Fehr et al., 2006; Soyka et al. 2008; Enoch et al. 2006, 2010; Roh et al. 2011), drug dependence (Agrawal et al. 2006; Enoch et al. 2010; Ehlers & Gizer 2013), and externalizing behavior (Dick et al., 2013; Salvatore et al., 2015; Trucco et al., 2016; Wang et al., 2016) have been observed, indicating the potential of genetic studies of beta EEG to facilitate discovery of genes underlying disinhibitory behavior. However, the number of replicable genetic variants uncovered by this approach has been limited (Iacono et al., 2016), necessitating large scale genetic association studies of beta EEG utilizing the best-practices of genetic epidemiology.
In a GWAS of fronto-central fast beta EEG in families of EA, we report a genome-wide significant signal in an intronic region of DSE (dermatin sulfate) on 6q22. The most significant SNP, rs2252790 (p<2.6×10−8; MAF: 0.36), was positively associated with fast beta EEG (β: 0.135). Taken together, data from Brainiac and GTEx suggest that rs2252790 is associated with the expression of DSE/TSPYL1 (note, that DSE and TSPYL1 have overlapping regions) and ROS1 in several brain tissues. Notably, rs2252790 is an eQTL for DSE/TSPYL1 mRNA expression in hippocampus tissue and for ROS1 expression in temporal cortex tissue. Both of these brain regions may be particularly relevant to beta EEG. It has been suggested that the beta rhythm may serve to establish transient physiological connections, reflected in coherence at the beta frequency among neurons in the hippocampus and related structures (Leung, 1992b; Vecchio et al., 2016b). Further, the high-frequency (i.e., fast beta) oscillations, often referred to as ‘rapid discharges’, have often been associated with seizure generation. Evidence from hippocampal slices shows that neuroelectric bursts in CA1 pyramidal cells are caused by highly synchronized β-band activity (Netoff and Schiff, 2002). Further, impairments observed in temporal lobe epilepsy (TLE) and Alzheimer’s disease have links to these brain structures and fast beta EEG. For example, lesions due to TLE typically involve mesial structures of the temporal lobe, particularly the amygdala and hippocampus (Kiernan, 2012). These structures play a central role in learning and memory (Leritz et al., 2006), while additionally involving sub-domains of working memory and executive functions (Stretton and Thompson, 2012; Zhao et al., 2014). Seizures caused by mesial TLE often involve fast frequency oscillations in the range of EEG fast-beta (Spencer et al., 1992; Bartolomei et al., 2004; Bartolomei et al., 2008). Further, researchers have hypothesized that cognitive impairments observed in Alzheimer’s disease may involve disrupted functional connectivity between frontotemporal and frontoparietal regions related to beta (and alpha) frequency EEG (Hsiao et al., 2013). These findings highlight that the brain regions implicated by the mRNA expression associated with DSE variant rs2252790 (hippocampus, temporal cortex) are related to beta band activity as well as higher cognitive functions, increasing the biological plausibility of this study’s findings. However, further studies are needed to understand the relationship of the DSE variant rs2252790, mRNA expression in hippocampus and temporal cortex, and beta EEG.
Variation within DSE has been associated with several cancers (Gouignard et al., 2016a; Thelin et al., 2013, 2012), Heschl’s Gyrus thickness (Cai et al., 2014), and is also a notable risk factor for Ehlers-Danlos syndrome, with a subtype specifically linked to dysfunction of DSE (Müller et al., 2013). Recent work by Gouignard et al. (Gouignard et al., 2016b) demonstrates a functional role for dse (the protein encoded by DSE) in cranial neural crest cell migration and in cell adhesion providing a potential biological mechanism linking DSE dysfunction to Ehlers-Danlos syndrome and other neural crest related disorders (i.e., neurocristopathies); the knockdown of dse impaired the correct activation of transcription factors involved in the epithelial-mesenchymal transition and reduced the extent of neural crest cell migration, subsequently leading to a decrease in neural crest-derived craniofacial skeleton, melanocytes and dorsal fin structures.
Given the association observed between AUDs and beta EEG in this and previous studies (Rangaswamy et al., 2004), we conducted a secondary analysis in which we repeated the initial GWAS of beta EEG, adjusting for DSM-V AUD severity. Results produced similar findings as the primary analyses; however, all p-values were slightly less significant. This suggests that the association of rs2252790 and beta EEG is not explained entirely by AUD; however, there may be an interaction among DSE variants, DSM-V AUD severity, and beta EEG. To comment on this more conclusively, future studies employing longitudinal data should assess the interaction of DSM-V AUD symptoms, DSE variants, and beta EEG.
Based on previous physical, co-expression, co-localization and pathway gene-gene interactions observed in the literature, GeneMANIA indicated that DSE is involved in a network of genes integral to membrane organization (Table 2). Given that scalp electrodes record potential differences that are caused by postsynaptic potentials in the cell membrane of cortical neurons, the relation between genes critical in cellular membrane organization and EEG seems broadly plausible. Several genes in this network have been previously linked to phenotypes of relevance to beta EEG and related traits (i.e., cognitive performance, bipolar disorder, AUD). For example, variants within DSEL (Dermatin Sulfate epimerase-like) have been associated with cognitive performance (Need et al., 2009), depression (Shi et al., 2011), and bipolar disorder (Goosens et al., 2003). Each of these phenotypes have been linked to variation in beta rhythms (cognitive performance (Klimesch, 1999); depression (Zotev et al., 2014); bipolar disorder (Andersson et al., 2008). In the present study, we find evidence of association among many variants within this DSE gene network and beta EEG, with the most robust associations (empirical p<0.05) observed for ZEB2, MCTP1, RND3, and CTBP2. Of these genes, ZEB2 and CTBP2 were also associated (empirical p-value>0.05) with DSM-V AUD (Table 3). Both ZEB2 and CTBP2 have been shown to influence gene expression in the brain, particularly during brain development, and have been previously correlated with traits of relevance to beta EEG and/or AUD.
CTBP2 is from a family of COOH-terminal binding proteins (CtBPs), which are widely expressed during several developmental processes, and have been linked to various complex traits, including cancers and Alzheimer’s disease (Liu et al., 2014; Zhang et al., 2014; Zheng et al., 2015). Importantly, two previous studies have found associations among CTBP2 and alcohol related phenotypes. A linkage analysis of alcohol and cigarette consumption (maximum cigarettes/grams of alcohol consumed per day) conducted in 1,390 individuals from 41 extended Mexican American families indicated a linkage peak on Chromosome 10. Subsequently, an expression profile analyses of 342 RNA transcripts under the linkage peak pointed to two genes, one of which was CTBP2. Further, CTBP2 was shown to influence RNA levels, which was negatively correlated with smoking and/or drinking (Viel et al., 2008). In addition, an early GWAS of alcohol and nicotine dependence reported an association of CTBP2 and DSM-IV Alcohol Dependence in an Australian cohort of 1,224 cases and 1,162 controls, although the association did not meet genome-wide significant criteria (p-value: 3.91×10−7;(Lind et al., 2010)). Further support for the role of CTBP2 in alcohol related behavior comes from model organism work (Grotewiel and Bettinger, 2015); a genetic screen in nematode Caenorhabditis elegans identifies ctbp-1 (the ortholog of CTBP2) as a key regulator required for the development of acute functional tolerance to ethanol (Bettinger et al., 2012; Reid et al., 2015). While this previous literature lends corroborating support for the association of CTBP2 and AUD reported here, this finding should clearly be replicated in independent, larger samples.
ZEB2 (zinc finger E-box binding homeobox 2) encodes the Smad Interacting Protein 1, which is involved in the TGF-β/BMP/Smad signaling cascade (Babkina et al., 2016). ZEB2 mRNA is expressed during early embryogenesis in brain tissue, and is thought to play an important role in neural crest cell migration (Van de Putte et al., 2003) and in the regulation of corticogenesis (Seuntjens et al. 2009). Mutations in ZEB2 have been linked with epilepsy (EPICURE Consortium et al., 2012) and related disorders, such as Hirschsprung disease/Mowat-Wilson syndrome (MWS; (Cordelli et al., 2013)). MWS is caused by heterozygous mutations or deletions of ZEB2 and is characterized by epilepsy, moderate to severe intellectual disability, corpus callosum abnormalities and other congenital malformations. Recent studies (Cordelli et al., 2013) suggest that a distinct “electroclinical” phenotype, characterized by age-dependent EEG changes, can be recognized in most patients with MWS. While the mechanism underlying epilepsy in individuals with ZEB2 mutations is not well understood, studies by McKinsey and Van den Berghe (McKinsey et al., 2013; van den Berghe et al., 2013) show the influence of ZEB2 on the neurogenesis of cortical γ-aminobutyric acid (GABA)ergic interneurons. Further, lack of ZEB2 prevents the repression of NKX2-1 homeobox transcription factor, the expression of which induces the differentiation of progenitor cells into striatal interneurons rather than cortical neurons (McKinsey et al., 2013; van den Berghe et al., 2013). Subsequently, deficit of GABAergic inhibition is thought to result in seizures (Yalçın, 2012). A recent exome sequencing study identified a de novo missense variant in ZEB2 and early infant epileptic encephalopathy characterized by burst-suppression EEG (Babkina et al., 2016). In addition, independent genetic studies have also found an association of ZEB2 variants, obesity related traits (Comuzzie et al., 2012), depression, bipolar disorder, and schizophrenia (Ripke et al., 2014). Taken together, this may suggest that variation in ZEB2 may have broader neurobiological implications, beyond epilepsy. While no previous genetic association studies have provided a clear association of either ZEB2 or CTBP2 and beta EEG, mRNA expression in key brain regions for beta EEG, along with associations with potentially related traits (i.e., AUD, alcohol and cigarette use frequency, epilepsy) could provide a potential link for the association of ZEB2, CTBP2 and beta EEG. These conclusions however, are beyond the scope of the present study and must be explored in future, independent studies.
Since the three genome-wide associated variants were located within an intron of DSE, this discussion has primarily focused on DSE and related gene networks. However, it should be noted that the pattern of association observed for beta EEG could implicate several nearby genes (see Figure 2); several variants in high linkage disequilbirum with rs2252790 are located within TSPYL4. For example, rs910391, which is in perfect linkage disequilbirum with rs2252790, is located in the promoter region of TSPYL4. Further, rs2252790 is nominally associated with the mRNA expression of several neighboring genes including TSPYL1, TSPYL4, ROS1, NT5DC1, and FRK, although the most robust effects were observed for DSE expression. Therefore, it is possible that associations observed in this study are due to the influence of these (or other) genes on beta EEG.
This study also confirmed that several variants within GABRA2 were modestly (p<0.05) associated with fast beta EEG in the fronto-central region (Table 4). The association of GABRA2 and beta EEG was initially reported with linkage and association analysis in COGA AD families using the first spatial/spectral component of 11 bipolar electrode pairs and 3 beta frequency bands ranging from 12.5–28 Hz (Porjesz et al., 2002; Edenberg et al., 2004). Subsequently, an association between GABRA2 variants and bipolar beta (13.5–27 Hz) EEG, assessed at fronto-central electrodes, was also reported in a case-control study of AD (Lydall et al., 2011). More recently, Malone et al., (2014) reported an association of GABRA2 and monopolar beta EEG, assessed at the central electrode (i.e., CZ) using GWAS data on a community sample of adolescent twins and their parents (Malone et al., 2014). In the present study, we build upon this prior literature in providing additional support for the association of GABRA2 variants and bipolar fast beta (20–28 Hz) EEG, assessed at fronto-central electrodes, in a family sample enriched for AD.
Limitations
Most notable is the relatively small sample size and related lack of statistical power to detect subtle genotypic effects. A recent article described the large projected sample sizes needed for a well powered genetic study of EEG, and highlighted the concerns that statistically underpowered genetic studies raise (Iacono et al., 2016). However, GWAS results seem reliable based on corroborating information (i.e., multiple genome-wide significant SNPs in high LD, biological plausibility). Nevertheless, genetic associations reported in this study must be replicated in a large, independent sample. Furthermore, given the nominal associations observed in eQTL analyses, these findings must also be replicated in larger samples. In addition, the current study includes participants with a wide age range (ages 7–74), which introduces potential for unmeasured confounding effects due to age-related changes in beta EEG; GWAS analyses were adjusted for age and age2 in an effort to minimize age related differences in beta EEG genetic association findings. However, future studies should examine the effects of genetic variants on trajectories of beta EEG during development in order to delineate age-specific effects, and the links between these effects and/or the onset of psychopathology, such as AUD. Finally, the analytic software employed for this genome-wide analysis of family based samples (GWAF) does not currently allow for the analysis of sex chromosomes.
Conclusions
To date, there have been relatively few genetic studies examining beta EEG, and only one finding that has been replicated. This study reports association between intronic SNPs located within DSE on 6q22 and fronto-central fast beta EEG in a sample of related individuals of EA. The most significant SNP is an eQTL for DSE, a gene encoding a protein important in cranial neural crest development, previously implicated in several complex traits (e.g., Ehlers Danlos syndrome, bipolar disorder, brain morphology) and expressed in hippocampus and temporal cortex, brain regions of importance to beta EEG. Further, GeneMANIA has indicated that DSE interacts with a network of genes integral to membrane organization. In the present study, gene-based tests of association suggest that several variants within this network (i.e., variants within DSE, ZEB2, MCTP1, RND3, and CTBP2) were associated with beta EEG, and ZEB2 and CTBP2 were also associated with DSM-V AUD. Converging data from GWAS, gene expression, and gene-networks presented in this study provide support for the role of genetic variants within DSE and related genes in neural hyperexcitability, and has highlighted two genes potentially related to AUD. While results presented in this study are intriguing, findings clearly need additional support including replication in larger, independent studies.
Supplementary Material
Highlights.
GWAS of resting state fast beta (20–28Hz) EEG (F3-C3) in COGA families of European Ancestry.
Fast beta EEG is genome-wide associated with three variants within DSE (6q22).
rs2252790 influences mRNA expression in brain tissue.
Related genes ZEB2, RND3, MCTP1, and CTBP2 are associated with beta EEG
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); University of Texas Health Science Center at San Antonio (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, J. McClintick, S. O’Connor, L. Wetherill, X. Xuei, Y. Liu (Indiana University); G. Chan (University of Iowa; University of Connecticut); D. Chorlian, N. Manz, C. Kamarajan, A. Pandey (SUNY Downstate); J.-C. Wang, M. Kapoor (Icahn School of Medicine at Mount Sinai) and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Andersson S, Barder HE, Hellvin T, Løvdahl H, Malt UF. Neuropsychological and electrophysiological indices of neurocognitive dysfunction in bipolar II disorder. Bipolar Disord. 2008;10:888–99. doi: 10.1111/j.1399-5618.2008.00638.x. [DOI] [PubMed] [Google Scholar]
- Babkina N, Deignan JL, Lee H, Vilain E, Sankar R, Giurgea I, Mowat D, Graham JM. Early Infantile Epileptic Encephalopathy with a de novo variant in ZEB2 identified by exome sequencing. Eur J Med Genet. 2016;59:70–74. doi: 10.1016/j.ejmg.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Bauer LO. Predicting relapse to alcohol and drug abuse via quantitative electroencephalography. Neuropsychopharmacology. 2001;25:332–40. doi: 10.1016/S0893-133X(01)00236-6. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B. What is inherited in the predisposition toward alcoholism? A proposed model. Alcohol Clin Exp Res. 1999;23:1125–35. doi: 10.1111/j.1530-0277.1999.tb04269.x. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, Leung K, Bolling MH, Goldsmith AD, Davies AG. Lipid environment modulates the development of acute tolerance to ethanol in Caenorhabditis elegans. PLoS One. 2012;7:e35192. doi: 10.1371/journal.pone.0035192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D-C, Fonteijn H, Guadalupe T, Zwiers M, Wittfeld K, Teumer A, Hoogman M, Arias-Vásquez A, Yang Y, Buitelaar J, Fernández G, Brunner HG, van Bokhoven H, Franke B, Hegenscheid K, Homuth G, Fisher SE, Grabe HJ, Francks C, Hagoort P. A genome-wide search for quantitative trait loci affecting the cortical surface area and thickness of Heschl’s gyrus. Genes Brain Behav. 2014;13:675–85. doi: 10.1111/gbb.12157. [DOI] [PubMed] [Google Scholar]
- Choi J-S, Park SM, Lee J, Hwang JY, Jung HY, Choi S-W, Kim DJ, Oh S, Lee J-Y. Resting-state beta and gamma activity in Internet addiction. Int J Psychophysiol. 2013;89:328–33. doi: 10.1016/j.ijpsycho.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel Genetic Loci Identified for the Pathophysiology of Childhood Obesity in the Hispanic Population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordelli DM, Garavelli L, Savasta S, Guerra A, Pellicciari A, Giordano L, Bonetti S, Cecconi I, Wischmeijer A, Seri M, Rosato S, Gelmini C, Della Giustina E, Ferrari AR, Zanotta N, Epifanio R, Grioni D, Malbora B, Mammi I, Mari F, Buoni S, Mostardini R, Grosso S, Pantaleoni C, Doz M, Poch-Olivé ML, Rivieri F, Sorge G, Simonte G, Licata F, Tarani L, Terazzi E, Mazzanti L, Cerruti Mainardi P, Boni A, Faravelli F, Grasso M, Bianchi P, Zollino M, Franzoni E. Epilepsy in Mowat-Wilson syndrome: delineation of the electroclinical phenotype. Am J Med Genet A. 2013;161A:273–84. doi: 10.1002/ajmg.a.35717. [DOI] [PubMed] [Google Scholar]
- Deckel AW, Hesselbrock V, Bauer L. Antisocial personality disorder, childhood delinquency, and frontal brain functioning: EEG and neuropsychological findings. J Clin Psychol. 1996;52:639–50. doi: 10.1002/(SICI)1097-4679(199611)52:6<639::AID-JCLP6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane H, Lempicki RA, Quackenbush J, Wheeler D, Chappey C, Lash A, Leipe D, Madden T, Schuler G, Tatusova T, Rapp B, Wu C, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z-Z, Ledley R, Lewis K, Mewes H-W, Orcutt B, Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D, Costanzo M, Crawford M, Hirschman J, Kranz J, Olsen P, Robertson L, Skrzypek M, Braun B, Hopkins K, Kondu P, Kanehisa M, Goto S, Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Gasteiger E, Jung E, Bairoch A, Cicala C, Arthos J, Selig S, Dennis G, Hosack D, Ryk D, Van Spangler M, Steenbeke T, Khazanie P, Gupta N, Sonnhammer E, Eddy S, Durbin R, Hosack D, Dennis G, Sherman B, Lane H, Lempicki R. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Latendresse S, Porjesz B, Schuckit M, Rangaswamy M, Hesselbrock V, Edenberg H, Nurnberger J, Agrawal A, Bierut L, Wang J, Bucholz K, Kuperman S, Kramer J. How phenotype and developmental stage affect the genes we find: GABRA2 and impulsivity. Twin Res Hum Genet. 2013;16:661–9. doi: 10.1017/thg.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li T-K, Nurnberger JI, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR. Evidence for a genetic component for substance dependence in Native Americans. Am J Psychiatry. 2013;170:154–64. doi: 10.1176/appi.ajp.2012.12010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Shen PH, Goldman D, Roy A. The influence of GABRA2, childhood trauma, and their interaction on alcohol, heroin, and cocaine dependence. Biol Psychiatry. 2010;67:20–7. doi: 10.1016/j.biopsych.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPICURE Consortium S, EMI Net Consortium L. Steffens M, Leu C, Ruppert A-K, Zara F, Striano P, Robbiano A, Capovilla G, Tinuper P, Gambardella A, Bianchi A, La Neve A, Crichiutti G, de Kovel CGF, Kasteleijn-Nolst Trenité D, de Haan G-J, Lindhout D, Gaus V, Schmitz B, Janz D, Weber YG, Becker F, Lerche H, Steinhoff BJ, Kleefuß-Lie AA, Kunz WS, Surges R, Elger CE, Muhle H, von Spiczak S, Ostertag P, Helbig I, Stephani U, Møller RS, Hjalgrim H, Dibbens LM, Bellows S, Oliver K, Mullen S, Scheffer IE, Berkovic SF, Everett KV, Gardiner MR, Marini C, Guerrini R, Lehesjoki A-E, Siren A, Guipponi M, Malafosse A, Thomas P, Nabbout R, Baulac S, Leguern E, Guerrero R, Serratosa JM, Reif PS, Rosenow F, Mörzinger M, Feucht M, Zimprich F, Kapser C, Schankin CJ, Suls A, Smets K, De Jonghe P, Jordanova A, Caglayan H, Yapici Z, Yalcin DA, Baykan B, Bebek N, Ozbek U, Gieger C, Wichmann H-E, Balschun T, Ellinghaus D, Franke A, Meesters C, Becker T, Wienker TF, Hempelmann A, Schulz H, Rüschendorf F, Leber M, Pauck SM, Trucks H, Toliat MR, Nürnberg P, Avanzini G, Koeleman BPC, Sander T. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum Mol Genet. 2012;21:5359–72. doi: 10.1093/hmg/dds373. [DOI] [PubMed] [Google Scholar]
- Gasser SM, Laemmli UK. Improved methods for the isolation of individual and clustered mitotic chromosomes. Exp Cell Res. 1987;173:85–98. doi: 10.1016/0014-4827(87)90334-x. [DOI] [PubMed] [Google Scholar]
- Gasser T, Köhler W, Jennen-Steinmetz C, Sroka L. The analysis of noisy signals by nonparametric smoothing and differentiation. IEEE Trans Biomed Eng. 1986;33:1129–33. doi: 10.1109/TBME.1986.325690. [DOI] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Iacono WG. Brain electrophysiological endophenotypes for externalizing psychopathology: a multivariate approach. Behav Genet. 2010a;40:186–200. doi: 10.1007/s10519-010-9343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens D, Van Gestel S, Claes S, De Rijk P, Souery D, Massat I, Van den Bossche D, Backhovens H, Mendlewicz J, Van Broeckhoven C, Del-Favero J. A novel CpG-associated brain-expressed candidate gene for chromosome 18q-linked bipolar disorder. Mol Psychiatry. 2003;8:83–89. doi: 10.1038/sj.mp.4001190. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gouignard N, Maccarana M, Strate I, von Stedingk K, Malmström A, Pera EM. Musculocontractural Ehlers-Danlos syndrome and neurocristopathies: dermatan sulfate is required for Xenopus neural crest cells to migrate and adhere to fibronectin. Dis Model Mech. 2016a;9:607–20. doi: 10.1242/dmm.024661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouignard N, Maccarana M, Strate I, von Stedingk K, Malmström A, Pera EM. Musculocontractural Ehlers-Danlos syndrome and neurocristopathies: dermatan sulfate is required for Xenopus neural crest cells to migrate and adhere to fibronectin. Dis Model Mech. 2016b;9:607–20. doi: 10.1242/dmm.024661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewiel M, Bettinger JC. Drosophila and Caenorhabditis elegans as Discovery Platforms for Genes Involved in Human Alcohol Use Disorder. Alcohol Clin Exp Res. 2015;39:1292–311. doi: 10.1111/acer.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Enoch M-A, Srivastava V, Cummins-Oman JS, Ferrier C, Iarikova P, Sankararaman S, Yamini G, Yuan Q, Zhou Z, Albaugh B, White KV, Shen P-H, Goldman D. Genome-wide association identifies candidate genes that influence the human electroencephalogram. Proc Natl Acad Sci U S A. 2010;107:8695–700. doi: 10.1073/pnas.0908134107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vrieze SI. Endophenotype best practices. Int J Psychophysiol. 2016 doi: 10.1016/j.ijpsycho.2016.07.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber L, Nunez P. Statistical mechanics of neocortical interactions: High-resolution path-integral calculation of short-term memory. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1995;51:5074–5083. doi: 10.1103/physreve.51.5074. [DOI] [PubMed] [Google Scholar]
- Ittiwut C, Yang B-Z, Kranzler HR, Anton RF, Hirunsatit R, Weiss RD, Covault J, Farrer LA, Gelernter J. GABRG1 and GABRA2 variation associated with alcohol dependence in African Americans. Alcohol Clin Exp Res. 2012;36:588–93. doi: 10.1111/j.1530-0277.2011.01637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimesch W. EEG alpha and theta oscillations reflect cognitive and memory performance: a review and analysis. Brain Res Brain Res Rev. 1999;29:169–95. doi: 10.1016/s0165-0173(98)00056-3. [DOI] [PubMed] [Google Scholar]
- Lee J, Hwang JY, Park SM, Jung HY, Choi SW, Kim DJ, Lee JY, Choi JS. Differential resting-state EEG patterns associated with comorbid depression in Internet addiction. Prog Neuropsychopharmacol Biol Psychiatry. 2014;50:21–6. doi: 10.1016/j.pnpbp.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Leung LS. Fast (beta) rhythms in the hippocampus: a review. Hippocampus. 1992a;2:93–8. doi: 10.1002/hipo.450020202. [DOI] [PubMed] [Google Scholar]
- Leung LS. Fast (beta) rhythms in the hippocampus: a review. Hippocampus. 1992b;2:93–8. doi: 10.1002/hipo.450020202. [DOI] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MHM, Smit AB, Hottenga J-J, Richter MM, Heath AC, Martin NG, Willemsen G, de Geus EJC, Vogelzangs N, Penninx BW, Whitfield JB, Montgomery GW, Boomsma DI, Madden PAF. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yao N, Qian J, Huang H. High expression and prognostic role of CAP1 and CtBP2 in breast carcinoma: associated with E-cadherin and cell proliferation. Med Oncol. 2014;31:878. doi: 10.1007/s12032-014-0878-7. [DOI] [PubMed] [Google Scholar]
- Lydall GJ, Saini J, Ruparelia K, Montagnese S, McQuillin A, Guerrini I, Rao H, Reynolds G, Ball D, Smith I, Thomson AD, Morgan MY, Gurling HMD. Genetic association study of GABRA2 single nucleotide polymorphisms and electroencephalography in alcohol dependence. Neurosci Lett. 2011;500:162–6. doi: 10.1016/j.neulet.2011.05.240. [DOI] [PubMed] [Google Scholar]
- Malone SM, Burwell SJ, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Heritability and molecular-genetic basis of resting EEG activity: a genome-wide association study. Psychophysiology. 2014;51:1225–45. doi: 10.1111/psyp.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, Huylebroeck D, Higashi Y, Rubenstein JLR. Dlx1&2-Dependent Expression of Zfhx1b (Sip1, Zeb2) Regulates the Fate Switch between Cortical and Striatal Interneurons. Neuron. 2013;77:83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, Mizumoto S, Suresh I, Komatsu Y, Vodopiutz J, Dundar M, Straub V, Lingenhel A, Melmer A, Lechner S, Zschocke J, Sugahara K, Janecke AR. Loss of dermatan sulfate epimerase (DSE) function results in musculocontractural Ehlers-Danlos syndrome. Hum Mol Genet. 2013;22:3761–72. doi: 10.1093/hmg/ddt227. [DOI] [PubMed] [Google Scholar]
- Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, Ge D, Heinzen EL, Maia JM, Shianna KV, Weale ME, Cherkas LF, Clement G, Spector TD, Gibson G, Goldstein DB. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–61. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermeyer E. A concept of consciousness. Ital J Neurol Sci. 1999;20:7–15. doi: 10.1007/s100720050004. [DOI] [PubMed] [Google Scholar]
- Nunez PL, Srinivasan R, Westdorp AF, Wijesinghe RS, Tucker DM, Silberstein RB, Cadusch PJ. EEG coherency: I: statistics, reference electrode, volume conduction, Laplacians, cortical imaging, and interpretation at multiple scales. Electroencephalogr Clin Neurophysiol. 1997;103:499–515. doi: 10.1016/S0013-4694(97)00066-7. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, Goate A, Rice JP, O’connor SJ, Rohrbaugh J, Kuperman S, Bauer LO, Crowe RR, Schuckit MA, Hesselbrock V, Conneally PM, Tischfield JA, Li T-K, Reich T, Begleiter H. Linkage disequilibrium between the beta frequency of the human EEG and a GABA A receptor gene locus. n.d doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Wang K, Almasy L, Chorlian DB, Stimus AT, Kuperman S, O’Connor SJ, Rohrbaugh J, Bauer LO, Edenberg HJ, Goate A, Rice JP, Reich T. Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol. 2002;61:229–48. doi: 10.1016/s0301-0511(02)00060-1. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Rangaswamy M, Kamarajan C, Jones KA, Padmanabhapillai A, Begleiter H. The utility of neurophysiological markers in the study of alcoholism. Clin Neurophysiol. 2005;116:993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Propping P, Krüger J, Mark N. Genetic disposition to alcoholism. An EEG study in alcoholics and their relatives. Hum Genet. 1981;59:51–9. doi: 10.1007/BF00278854. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Bauer LO, Rohrbaugh J, O’Connor SJ, Kuperman S, Reich T, Begleiter H. Beta power in the EEG of alcoholics. Biol Psychiatry. 2002;52:831–42. doi: 10.1016/s0006-3223(02)01362-8. [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Kuperman S, Rohrbaugh J, O’Connor SJ, Bauer LO, Reich T, Begleiter H. Resting EEG in offspring of male alcoholics: beta frequencies. Int J Psychophysiol. 2004;51:239–51. doi: 10.1016/j.ijpsycho.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Reid A, Sherry TJ, Yücel D, Llamosas E, Nicholas HR. The C-terminal binding protein (CTBP-1) regulates dorsal SMD axonal morphology in Caenorhabditis elegans. Neuroscience. 2015;311:216–30. doi: 10.1016/j.neuroscience.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, Pers TH, Agartz I, Agerbo E, Albus M, Alexander M, Amin F, Bacanu SA, Begemann M, Belliveau RA, Jr, Bene J, Bergen SE, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Byerley W, Cahn W, Cai G, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Chan RCK, Chen RYL, Chen EYH, Cheng W, Cheung EFC, Ann Chong S, Robert Cloninger C, Cohen D, Cohen N, Cormican P, Craddock N, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, Demontis D, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Durmishi N, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedl M, Friedman JI, Fromer M, Genovese G, Georgieva L, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, Golimbet V, Gopal S, Gratten J, de Haan L, Hammer C, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn JN, Hoffmann P, Hofman A, Hollegaard MV, Hougaard DM, Ikeda M, Joa I, Julià A, Kahn RS, Kalaydjieva L, Karachanak-Yankova S, Karjalainen J, Kavanagh D, Keller MC, Kennedy JL, Khrunin A, Kim Y, Klovins J, Knowles JA, Konte B, Kucinskas V, Ausrele Kucinskiene Z, Kuzelova-Ptackova H, Kähler AK, Laurent C, Lee Chee Keong J, Hong Lee S, Legge SE, Lerer B, Li M, Li T, Liang KY, Lieberman J, Limborska S, Loughland CM, Lubinski J, Lönnqvist J, Macek M, Jr, Magnusson PKE, Maher BS, Maier W, Mallet J, Marsal S, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Mors O, Murphy KC, Murray RM, Myin-Germeys I, Müller-Myhsok B, Nelis M, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nikitina-Zake L, Nisenbaum L, Nordin A, O’Callaghan E, O’Dushlaine C, O’Neill FA, Oh SY, Olincy A, Olsen L, Van Os J, Pantelis C, Papadimitriou GN, Papiol S, Parkhomenko E, Pato MT, Paunio T, Pejovic-Milovancevic M, Perkins DO, Pietiläinen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Schall U, Schubert CR, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Sigurdsson E, Silagadze T, Silverman JM, Sim K, Slominsky P, Smoller JW, So HC, Spencer CA, Stahl EA, Stefansson H, Steinberg S, Stogmann E, Straub RE, Strengman E, Strohmaier J, Scott Stroup T, Subramaniam M, Suvisaari J, Svrakic DM, Szatkiewicz JP, Söderman E, Thirumalai S, Toncheva D, Tosato S, Veijola J, Waddington J, Walsh D, Wang D, Wang Q, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wong EHM, Wormley BK, Simon Xi H, Zai CC, Zheng X, Zimprich F, Wray NR, Stefansson K, Visscher PM, Adolfsson R, Andreassen OA, Blackwood DHR, Bramon E, Buxbaum JD, Børglum AD, Cichon S, Darvasi A, Domenici E, Ehrenreich H, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jönsson EG, Kendler KS, Kirov G, Knight J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, McCarroll SA, McQuillin A, Moran JL, Mortensen PB, Mowry BJ, Nöthen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sham PC, Sklar P, St Clair D, Weinberger DR, Wendland JR, Werge T, Daly MJ, Sullivan PF, O’Donovan MC Trust Case-Control Consortium W, Endophenotypes International Consortium P. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh S, Matsushita S, Hara S, Maesato H, Matsui T, Suzuki G, Miyakawa T, Ramchandani VA, Li T-K, Higuchi S. Role of GABRA2 in moderating subjective responses to alcohol. Alcohol Clin Exp Res. 2011;35:400–7. doi: 10.1111/j.1530-0277.2010.01357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletu-Zyhlarz GM, Arnold O, Anderer P, Oberndorfer S, Walter H, Lesch OM, Böning J, Saletu B. Differences in brain function between relapsing and abstaining alcohol-dependent patients, evaluated by EEG mapping. Alcohol Alcohol. 39:233–40. doi: 10.1093/alcalc/agh041. [DOI] [PubMed] [Google Scholar]
- Salvatore JE, Meyers JL, Yan J, Aliev F, Lansford JE, Pettit GS, Bates JE, Dodge KA, Rose RJ, Pulkkinen L, Kaprio J, Dick DM. Intergenerational continuity in parents’ and adolescents’ externalizing problems: The role of life events and their interaction with GABRA2. J Abnorm Psychol. 2015;124:709–729. doi: 10.1037/abn0000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Umans L, Zwijsen A, Sampaolesi M, Verfaillie CM, Huylebroeck D. Transforming Growth Factor type beta and Smad family signaling in stem cell function. Cytokine Growth Factor Rev. n.d;20:449–58. doi: 10.1016/j.cytogfr.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, Lawson WB, DePaulo JR, Gejman PV, Sanders AR, Johnson JK, Adams P, Chaudhury S, Jancic D, Evgrafov O, Zvinyatskovskiy A, Ertman N, Gladis M, Neimanas K, Goodell M, Hale N, Ney N, Verma R, Mirel D, Holmans P, Levinson DF. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit DJA, Posthuma D, Boomsma DI, Geus EJC. Heritability of background EEG across the power spectrum. Psychophysiology. 2005;42:691–7. doi: 10.1111/j.1469-8986.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Chorlian DB, Rangaswamy M, O’Connor S, Taylor R, Rohrbaugh J, Porjesz B, Begleiter H. Heritability of bipolar EEG spectra in a large sib-pair population. Behav Genet. 2007a;37:302–13. doi: 10.1007/s10519-006-9133-0. [DOI] [PubMed] [Google Scholar]
- Tang Y, Chorlian DB, Rangaswamy M, Porjesz B, Bauer L, Kuperman S, O’Connor S, Rohrbaugh J, Schuckit M, Stimus A, Begleiter H. Genetic influences on bipolar EEG power spectra. Int J Psychophysiol. 2007b;65:2–9. doi: 10.1016/j.ijpsycho.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Thelin MA, Bartolini B, Axelsson J, Gustafsson R, Tykesson E, Pera E, Oldberg Å, Maccarana M, Malmstrom A. Biological functions of iduronic acid in chondroitin/dermatan sulfate. FEBS J. 2013;280:2431–46. doi: 10.1111/febs.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin MA, Svensson KJ, Shi X, Bagher M, Axelsson J, Isinger-Ekstrand A, van Kuppevelt TH, Johansson J, Nilbert M, Zaia J, Belting M, Maccarana M, Malmström A. Dermatan sulfate is involved in the tumorigenic properties of esophagus squamous cell carcinoma. Cancer Res. 2012;72:1943–52. doi: 10.1158/0008-5472.CAN-11-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–82. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco EM, Hicks BM, Villafuerte S, Nigg JT, Burmeister M, Zucker RA. Temperament and externalizing behavior as mediators of genetic risk on adolescent substance use. J Abnorm Psychol. 2016;125:565–75. doi: 10.1037/abn0000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Heritability of human brain functioning as assessed by electroencephalography. Am J Hum Genet. 1996;58:562–73. [PMC free article] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice Lacking Zfhx1b, the Gene That Codes for Smad-Interacting Protein-1, Reveal a Role for Multiple Neural Crest Cell Defects in the Etiology of Hirschsprung Disease–Mental Retardation Syndrome. The American Journal of Human Genetics. 2003 doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berghe V, Stappers E, Vandesande B, Dimidschstein J, Kroes R, Francis A, Conidi A, Lesage F, Dries R, Cazzola S, Berx G, Kessaris N, Vanderhaeghen P, van IJcken W, Grosveld FG, Goossens S, Haigh JJ, Fishell G, Goffinet A, Aerts S, Huylebroeck D, Seuntjens E. Directed Migration of Cortical Interneurons Depends on the Cell-Autonomous Action of Sip1. Neuron. 2013;77:70–82. doi: 10.1016/j.neuron.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Vecchio F, Miraglia F, Piludu F, Granata G, Romanello R, Caulo M, Onofrj V, Bramanti P, Colosimo C, Rossini PM. "Small World" architecture in brain connectivity and hippocampal volume in Alzheimer’s disease: a study via graph theory from EEG data. Brain Imaging Behav. 2016a doi: 10.1007/s11682-016-9528-3. [DOI] [PubMed] [Google Scholar]
- Vecchio F, Miraglia F, Piludu F, Granata G, Romanello R, Caulo M, Onofrj V, Bramanti P, Colosimo C, Rossini PM. "Small World" architecture in brain connectivity and hippocampal volume in Alzheimer’s disease: a study via graph theory from EEG data. Brain Imaging Behav. 2016b doi: 10.1007/s11682-016-9528-3. [DOI] [PubMed] [Google Scholar]
- Viel K, Charlesworth J, Tejero E, Dyer T, Cole S, Haack K, MacCluer J, Blangero J, Almasy L. A linkage analysis of cigarette and alcohol consumption in an unselected Mexican American population. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:983–6. doi: 10.1002/ajmg.b.30661. [DOI] [PubMed] [Google Scholar]
- Wang FL, Chassin L, Geiser C, Lemery-Chalfant K. Mechanisms in the relation between GABRA2 and adolescent externalizing problems. Eur Child Adolesc Psychiatry. 2016;25:67–80. doi: 10.1007/s00787-015-0703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill L, Agrawal A, Kapoor M, Bertelsen S, Bierut LJ, Brooks A, Dick D, Hesselbrock M, Hesselbrock V, Koller DL, Le N, Nurnberger JI, Salvatore JE, Schuckit M, Tischfield JA, Wang J-C, Xuei X, Edenberg HJ, Porjesz B, Bucholz K, Goate AM, Foroud T. Association of substance dependence phenotypes in the COGA sample. Addict Biol. 2015;20:617–27. doi: 10.1111/adb.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterer G, Klöppel B, Heinz A, Ziller M, Dufeu P, Schmidt LG, Herrmann WM. Quantitative EEG (QEEG) predicts relapse in patients with chronic alcoholism and points to a frontally pronounced cerebral disturbance. Psychiatry Res. 1998;78:101–13. doi: 10.1016/s0165-1781(97)00148-0. [DOI] [PubMed] [Google Scholar]
- Yalçın Ö. Genes and molecular mechanisms involved in the epileptogenesis of idiopathic absence epilepsies. Seizure. 2012;21:79–86. doi: 10.1016/j.seizure.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Zhang C, Gao C, Xu Y, Zhang Z. CtBP2 could promote prostate cancer cell proliferation through c-Myc signaling. Gene. 2014;546:73–9. doi: 10.1016/j.gene.2014.05.032. [DOI] [PubMed] [Google Scholar]
- Zheng X, Song T, Dou C, Jia Y, Liu Q. CtBP2 is an independent prognostic marker that promotes GLI1 induced epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget. 2015;6:3752–69. doi: 10.18632/oncotarget.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotev V, Phillips R, Yuan H, Misaki M, Bodurka J. Self-regulation of human brain activity using simultaneous real-time fMRI and EEG neurofeedback. Neuroimage. 2014;85(Pt 3):985–95. doi: 10.1016/j.neuroimage.2013.04.126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.