Abstract
Background:
The Wilms tumor 1 (WT1) gene is originally defined as a tumor suppressor gene and a transcription factor that overexpressed in leukemic cells. It is highly expressed in more than 80% of acute myeloid leukemia (AML) patients, both in bone marrow (BM) and in peripheral blood (PB), and it is used as a powerful and independent marker of minimal residual disease (MRD); we have determined the expression levels of the WT1 by real-time quantitative polymerase chain reaction (RQ-PCR) in PB and BM in 126 newly diagnosed AML patients.
Materials and Methods:
This study was done in molecular pathology and cancer research center from April 2014 to June 2015, RQ-PCR method was used to determine the WT1 gene expression in BM and/or PB samples from 126 patients of AML, we cloned both WT1 and ABL genes for creating a standard curve, and we calculate copy number of WT1 genes in patients.
Results:
A total of 126 AML patients consist of 70 males (55.6%) and 56 females (44.4%), with a median age of 26 years; 104 (81%) patients out of 126 show overexpression of WT1 gene. We also concomitant monitoring of fusion transcripts (PML RARa, AML1-ETO, MLL-MLL, CBFb-MYH11, or DEK-CAN) in our patients, the AML1-ETO group showing remarkably low levels of WT1 compared with other fusion transcript and the CBFB-MYH11 showing high levels of WT1.
Conclusion:
We conclude that WT1 expression by RQ-PCR in AML patients may be employed as an independent tool to detect MRD in the majority of normal karyotype AML patients.
Keywords: Acute myeloid leukemia, minimal residual disease, real-time quantitative polymerase chain reaction, Wilms tumor gene
INTRODUCTION
Most of the time, chemotherapy works in leukemia patients; however, despite high remission rate after chemotherapy, relapse always exists. Hence, it is highly necessary to find markers that enable the prediction of relapse. Monitoring minimal residue disease (MRD) is an important clinical factor in the assessment of response to chemotherapy, guiding therapeutic intervention and predicting relapse. There are pressing needs to choose a marker to detect MRD for a greater population of patients.[1] Wilms tumor 1 (WT1) was first reported as a tumor suppressor gene. Later findings demonstrated that WT1 had oncogenic functions in other malignancies. The WT1 gene is located on the short arm of chromosome 11 in locus 11p13, encoded as a zinc-finger transcriptional factor that has recognized as an important regulator of normal and malignant hematopoiesis, and also, this gene is known to control cellular apoptosis. The expression of WT1 gene in blast leukemia may cause resistance of blast to apoptosis and cause poor clinical outcomes.[2] The normal expression of WT1 is restricted in adults to a few numbers of tissues. In the normal BM, WT1 expression is limited to CD34+ cells and there are not in peripheral blood (PB) cells; therefore, expression of WT1 is limited to early progenitors of the blood system, several studies have demonstrated that WT1 is overexpressed in acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), myelodysplastic syndrome (MDS), and blast crisis of chronic myeloid leukemia (CML), and this gene is highly expressed in more than 80% of AML patients in bone marrow (BM) and PB in comparison with normal control. This overexpression of WT1 in acute leukemia could thus represent a universal molecular marker of malignant hematopoiesis and is suggested as the usefulness of quantitative assessment of WT1 expression as a marker for MRD. The biological functions of WT1 overexpression in patients with acute leukemia are not clearly understood, but it has been suggested that WT1 affects the pathogenesis of human leukemia during cellular differentiation and growth arrest. Low expression of WT1 and high expression of it are accompanied with clinical remission and relapse, respectively, which shows the fact that WT1 may be a potential prognostic factor and a therapeutic target in AML.[3] Here, we analyzed WT1 expression in a cohort of 126 patients with AML using real-time quantitative reverse transcription polymerase chain reaction (RQ-RT-PCR) and considered the usefulness of this marker for predicting relapse in our regions.
MATERIALS AND METHODS
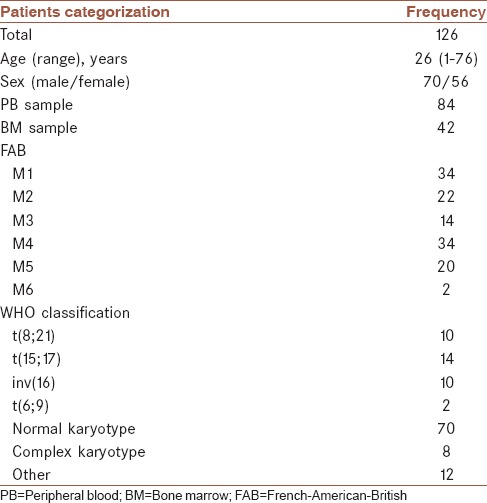
BM and PB samples were referred to our laboratory for central leukemia diagnosis. A total of 126 AML patients consisting of 70 males (55.6%) and 56 females (44.4%) with a median age of 26 years (age range of 1–76 years) were recruited for the WT1 overexpression study. All patients were diagnosed with AML at the molecular pathology and cancer research center from April 2014 to June 2015, a total of 14 months. This clinical study was approved by the Ethics Committee of participating institutions and each patient provided informed consent before the initiation of the studies, 2 ml of BM aspiration or PB sample was collected in the ethylenediaminetetraacetic acid and heparin vacutainer from all 126 patients for the molecular and cytogenetic study, respectively, before treatment. The WT1 overexpression was defined in this study as ≥50 copies WT1/104 ABL in PB sample and ≥250 copies WT1/104 ABL in BM sample. The patient characteristics of the total cohort are shown in Table 1. AML was defined based on criteria which are written below and definitive diagnosis was done by an expert hematologist.
Table 1.
Patient characteristics
Presence of ≤20% myeloid blasts in BM or PB specimen
Positive myeloperoxidase, specific and nonspecific esterase, Sudan black, and periodic acid–Schiff staining of patient's slide
Positive markers such as CD13, CD117, CD64, CD14, and CD117 in peripheral samples
Discovering AML-related genetic disorders such as t(8;21), t(15;17), and t(6;9).
Samples without above-mentioned criteria were excluded from this study.
Preparation of RNA and cDNA synthesis
Mononuclear cells, including leukemic blasts, were separated from BM or PB samples on a Ficoll–Histopaque 1077 (Sigma-Aldrich Company, Saint Louis, MO, USA) density gradient. Total RNA was extracted using RNeasy mini kit following the manufacturer's protocol (Qiagen, Hilden, Germany). The RT step was adapted from the BIOMED 1 protocol.[4] Samples were incubated for 10 min at 20°C, 45 min at 42°C, and 3 min at 99°C, followed by 10 min at 4°C.
Generation of plasmid standard curves
To construct a WT1 plasmid and ABL plasmid for the standard curve in the RQ-PCR assay, we added the following primers and reagents with a final volume of 50 μL to amplify a WT1 and ABL RT-PCR product:
WT1-forward primer: Exon 7-5_-GGCATCTGAGA CCAGTGAGAA-3_
WT1-reverse primer: exon 10 5_-GGACTAATTCA TCGACCGGG-3
ABL-F: TGG AGA TAA CAC TCT AAG CAT AAC TAA AGGT
ABL-R: GAT GTAGTT GCT TGG GAC.
PCR buffer 1X: 10 mM Tris-HCl, 50 mM KCl (pH 8.3), MgCl2:2.5 mM (final concentration), dNTP: 200 μM (final concentration), 400 nM of each primer (final concentration), Taq enzyme: 1 U and 3 μL of cDNA product.
Use the following thermal cycler temperatures and time conditions:
Initial melting at 95°C for 30 s, 35 PCR cycles at the following conditions: 94°C for 30 s, 65°C for 1 min, 72°C for 1 min.
Then, we cloned the PCR products into the InsTAclone PCR Cloning Kit (Thermo Scientific) according to the manufacturer's instructions.
The selected clones were screened for the presence of the insert by PCR and then sequenced for confirmation. The plasmid was extracted using the GeneJET Plasmid Miniprep Kit (Thermo Scientific) and quantified spectrophotometrically. The copy number for 1 μg is estimated according to the molecular weight of the vector plus the insert.
Six successful serial dilutions (10,0000, 10,000, 1000, 100, and 10 copies) of each genes were prepared.
Quantitative analysis of WT1 gene expression was done by standard curve analysis. The expression of WT1 was normalized against the control gene ABL. A method for ABL amplification was according to BIOMED concerted action.[4] The expression ratio was given in 100 × WT1/ABL, and WT1 overexpression was defined in this study as ≥50 copies WT1/104 ABL in PB sample and ≥250 copies WT1/104 ABL in BM sample as previously recommended.[5] A dilution series of a known concentration of ABL and WT1 plasmid was used for the creation of a standard curve. PCR efficiencies of WT1 and ABL were measured at 97% and 98%, respectively. The PCR conditions, primer, and probe for the detection and amplification of PML-RARA, AML1-ETO, CBFB-MYH11, and NUP-DEK samples have been described elsewhere.[4]
Real-time quantitative reverse transcription polymerase chain reaction
All the RT-RT-PCR reactions were performed on a 7700 ABI platform. In each reaction, 5 μL of cDNA was amplified. Each reaction contained 12.5 μL of Mastermix (TaqMan Universal PCR Master Mix), 200 nM hybridization probe (WT1-probe: CTTACAGATGCACAGCAGGAAGCACACTG), 300 nM of each primer ([WT1-F: CAGGCTGCAA TAAGAGATATTTTAAGCT] and [WT1-R: GAAGTCACACTGGTATGGTTTCTCA]). The RQ-PCR primers and probe for ABL are as follows: forward primer 5_-TGGAGATAACACTC TAAGCATAACTAAAGGT-3, reverse primer 5_-GATGTAGTTGCTTGGGACCCA-3, TaqMan probe 5_-CCATTTTTGGTTTGGGCTTCACACCATT-3. Sterilized water to reach a final volume of 25 μL 50 cycles of denaturation at 95°C for 15 s followed by annealing/extension at 60°C for 60 s was added.
Cytogenetic study
Chromosome analysis was performed on synchronized 24 h and 48 h using fdur/uridine block with cell density of 106 cells/mL. Aliquots of BM aspirate were suspended in RPMI 1640 supplemented with 20% fetal calf serum, l-glutamine, and antibiotics. After 24 h incubation at 37°C, colcemid was added to a final concentration of 0.05 μg/mL for 30 min. The cells were exposed to a hypotonic solution (0.075 mol/L KCl) for 30 min at 37°C and then fixed three times in methanol: acetic acid (3:1, v/v) for 30 min each. Slides were prepared and then air dried. Preparations were banded with the G-banding technique. Clonal karyotypic abnormalities were identified and described according to the 2013 International System of Human Cytogenetic Nomenclature.
Statistical analyses
Statistical analysis was done using SPSS V 13 (SPSS Inc., Chicago, IL, USA). Parametric and nonparametric tests included, Mann–Whitney, Student's t-test, and ANOVA, were applied according to data distribution and the results interpretations are described in the next section. Data were reported in the form of mean ± standard deviation in the present study and. P < 0.05 was considered statistically significant.
RESULTS
Sensitivity of Wilms tumor 1 monitoring
To generate a standard curve and define the sensitivity of WT1 monitoring, a dilution series of RNA from the cell line K562 expressing WT1 was performed. K562 RNA could be successfully detected in normal PB RNA in a dilution of 1:20,000. The mean slope of 3.3 ± 0.3 (correlation coefficient range: 0.984–0.993) was close to optimal PCR reaction. DRn threshold of 0.1, the intercept of the standard curve was 40.3 ± 0.3, which was compatible with our defined detection limit of Ct = 39. Thereby, we found our designed primers and probe suitable for WT1 quantification.
In this study, we evaluated the WT1 gene expression in AML patients. In a total of 126 patients, 55.6% and 44.4% were men and women, respectively, where the patients with ages under 15 years were considered as children who were 31.7% and patients with ages above 15 years were considered as adults who were 68.3%. In this study, there were 66.7% PB samples and 33.3% BM samples. The age of patients varied from 1 to 76 years, with the median of 26 years. The WT1 gene overexpression was detected in 81% (104 of 126) of all patients, with frequency of 17.5% copies of WT1/104 ABL in BM and 63.5% copies of WT1/104 ABL in PB samples (P = 0.0001, P = 0.0001, respectively).
Each patient provided informed consent. The diagnosis of AML was made according to the French–American–British (FAB) and WHO classification. The standard Giemsa banding techniques were applied in the karyotyping of leukemia. In 126 patients, WT1 levels were quantified at primary diagnosis. Ten patients were positive for AML1-ETO, ten patients for CBFB-MYH11, 14 for PML-RARA, and two for NUP-DEK. A total of seventy patients had not any of recurrent translocation, 12 patients had other cytogenetic abnormalities, and eight patients had complex aberrant karyotypes. In addition, WT1 expression levels were analyzed in 12 blood and two BM samples of healthy donors.
The mean expression level of WT1 in normal BMs samples was 19 (range 17–21). In all ten analyzed PB samples, WT1 expression was very low and undetectable [Table 2].
Table 2.
Wilms tumor 1 expression levels in the peripheral blood and bone marrow of healthy controls as compared to acute myeloblastic leukemia patients at diagnosis

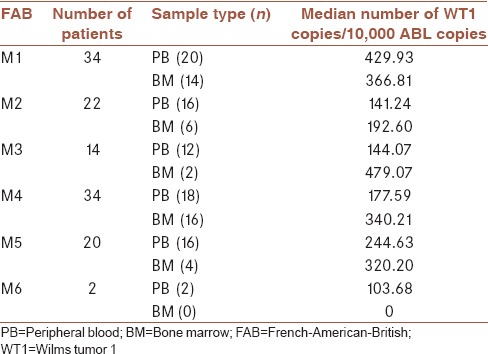
We also detect the median WT1 gene expression in each WHO classification and FAB subtype [Tables 3 and 4], highest levels of WT1 expression were seen first in patients with normal karyotype at diagnosis and second in CBFB-MYH1, and lowest levels were detected in patients with AML1-ETO positive. In FAB subtype, the highest levels of WT1 at diagnosis were assessed in the M1 subtype and the lowest levels were observed in the M6 subtype.
Table 3.
Wilms tumor 1 expression at diagnosis, according to different cytogenetic or molecular subgroups
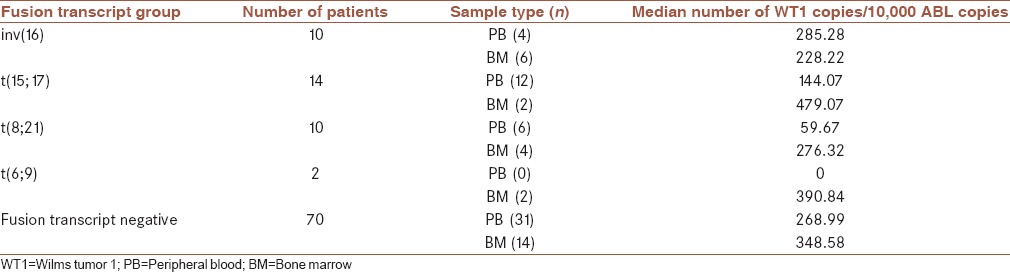
Table 4.
Wilms tumor 1 expression according to cytomorphological subtypes
DISCUSSION
WT1 gene is a tumor suppressor gene that is associated with WT and other related syndromes such as WAGR and Denys–Drash.[6] Unlike other tumor suppressors such as P53 or RB1, WT1 expression in normal adult tissues is limited to a number of mainly genitourinary system. In normal BM, progenitor cells expression of WT1 gene is very low and limited only in the beginning; however, numerous studies suggest that increased expression of WT1 in AML, ALL, MDS, and blastic phase of CML can be seen.[7] The WT1 gene expression can be used as a molecular marker for hematopoietic malignancies. Many studies have mentioned that using quantitative gene expression can be used to assess the exact extent of residual disease (MRD),[8,9] with respect to the biological role of WT1 gene expression in leukemia patients; this gene involved in pathogenesis of leukemia as stopping cell growth and differentiation by suppressing duplication numerous studies have indicated that the prognosis in leukemia patients is inversely associated with the expression of WT1.[10,11] Since the majority of AML patients express very high values of WT1 at diagnosis, the assessment of WT1 expression provides a strong method for monitoring the persistence of the disease after chemotherapy or BM transplantation or during the treatment.
WT1 gene is expressed in more than 80% of patients with AML in both PB and BM samples. WT1 overexpression is used as a panleukemic marker to determine the amount of residual disease (MRD) and is a good marker for patients with no cytogenetic or molecular abnormalities (about 50%).[12]
The overexpression of WT1 gene in this study was 81%, which our results were similar to other studies. For example, in one study, the expression of this gene were shown in more than 90% of AML patients and they mentioned WT1 can be used as a marker for MRD; in this study, the expression of WT1 gene was performed before and after transplantation, and the results of chimerism was compared with flow cytometry and concluded that the WT1 expression in AML patients is associated with the amount of residual disease.[13] Another study conducted in 2014 indicated that WT1 expression was limited to CD34+/CD38− cells and this feature makes WT1 useful as a unique marker for determining the amount of residual disease in AML patients.[14]
In another study, Baba et al. conducted the level of WT1 which was significantly related to the percentage of blast in BM among MDS cases, and the refractory anemia (RA) cases showed significantly lower WT1 level than patients with RA with excess blast and concluded that WT1 might be a good marker to differentiate low blast percentage MDS and cytopenia.[15] In our study, the expression of WT1 in AML was studied, but in other studies, the overexpression of WT1 gene was seen in 75% of acute leukemia and blastic phase of chronic myeloid.[16] The WT1 gene had been highly expressed in most patients of AML and ALL and in blastic phase of CML and MDS. The degree of mRNA expression of WT1 gene are increased with the progression of the disease; furthermore, WT1 gene is known as a tumor suppressor and there is evidence indicating that WT1 has oncogenic role in leukemogenesis and tumorigenesis.[17]
AML is a hematopoietic stem cells malignancy known by increasing clonal myeloid blast, which categorizes mainly into three different groups, good, moderate, and bad based on chromosomal abnormalities. About half (50%–40%) of AML patients with normal karyotype or normal cytogenetic classified in the medium-risk group; however, there is a paradox in this group on how to respond to chemotherapy and prognosis for the treatment that makes it difficult to make decisions about continuing the treatment.[18] A number of other molecular markers in response to the therapy and disease prognosis in AML with normal cytogenetics (CN-AML) is important such as nucleophosmin gene (NPM1) and FLT3 gene. Identification of mutations in these genes to assess how to treat and prognosis in patients with CN-AML are important. The WT1 is an important regulatory factor of normal and malignant blood cells. On the other hand, the resistance of leukemic blasts against apoptosis leads to poor clinical status of the patient, so regulating the apoptotic and antiapoptotic pathways is associated with improved treatment and survival in AML patients.[2,8]
In our study, we first examined the level of WT1 in 12 PB samples and two BM samples from healthy donors and determined that the WT1 expression in normal PB and BM samples were very low, these data provide a stable basis for the use of WT1 as MRD target, all AML patients were classified according to WHO criteria and the frequency of the most common translocations involving: t(15; 17), inv (16), t(8; 21), and t(6; 9) are calculated, patients who had none of these recurrent translocations were 71.4% of the patients. In our cases, patients who had t(15; 17) had the highest frequency (11.1%), and an equal number of AML patients with t(8; 21) and inv (16) had a frequency of 7.9% and two patients had t(6; 9). While Østergaard et al. compared the WT1 expression level between patients who were positive for the selected fusion gene, the average WT1 gene copies in each of the 104 copies of the ABL gene is calculated, most WT1 gene copy number was observed in the group of patients who had normal cytogenetic, and then, in patients with inv (16) and the lowest WT1, gene copy was observed in patients with t(8; 21). The average of WT1 gene copy in patients is shown in Table 3. Our findings are somewhat different with Candoni et al., most of the copy of the WT1 gene was observed in patients who had inv (16) with an average of 23,100 copies and then in patients who had normal cytogenetic with NPM1 mutations, with an average of 14,400 copies.[7] In another study by Østergaard et al. had somewhat different results and the maximum amount of copies of the WT1 gene were in patients with t(15; 17) and the lowest amount of copies were in patients with t(8; 21).[19] In this study, we also examined WT1 gene copies in each of the morphological groups based on the FAB classifications, maximum copy of the WT1 gene were in the M1 with an average of 403.94 and the lowest were in the M6; increased expression of WT1 in M1 can be explained by an increase in the number of blasts in this group.
The data of patients presented in this study show that an accurate quantitative amount of the WT1 transcript provided by the RQ-PCR methods allows us to distinguish between normal and abnormal expression levels of WT1 gene, we significantly observed a great variation in WT1 level in de novo AML patients, which in this study did not show clear correlations to cytogenetically prognostic subgroups.
CONCLUSION
With the concordance between WT1 gene MRD and hematological findings in both groups of fusion transcript-negative and transcript-positive patients, our study extremely supports the use of WT1 as a strong MRD marker.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
This study was the result of a master student thesis proposal supported financially by the research vice chancellor of Mashhad University of Medical Sciences and cancer molecular pathology research center.
REFERENCES
- 1.Malagola M, Skert C, Ruggeri G, Turra A, Ribolla R, Cancelli V, et al. Peripheral blood WT1 expression predicts relapse in AML patients undergoing allogeneic stem cell transplantation. Biomed Res Int 2014. 2014 doi: 10.1155/2014/123079. 123079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyu X, Xin Y, Mi R, Ding J, Wang X, Hu J, et al. Overexpression of Wilms tumor 1 gene as a negative prognostic indicator in acute myeloid leukemia. PLoS One. 2014;9:e92470. doi: 10.1371/journal.pone.0092470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X. Wilms’ Tumor Gene 1 in Diff erent Types of Cancer. Umeå University medical dissertations. 2015;1717:59. [Google Scholar]
- 4.Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – A Europe Against Cancer program. Leukemia. 2003;17:2318–57. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 5.Cilloni D, Renneville A, Hermitte F, Hills RK, Daly S, Jovanovic JV, et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: A European LeukemiaNet study. J Clin Oncol. 2009;27:5195–201. doi: 10.1200/JCO.2009.22.4865. [DOI] [PubMed] [Google Scholar]
- 6.Barragán E, Cervera J, Bolufer P, Ballester S, Martín G, Fernández P, et al. Prognostic implications of Wilms’ tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica. 2004;89:926–33. [PubMed] [Google Scholar]
- 7.Candoni A, Tiribelli M, Toffoletti E, Cilloni D, Chiarvesio A, Michelutti A, et al. Quantitative assessment of WT1 gene expression after allogeneic stem cell transplantation is a useful tool for monitoring minimal residual disease in acute myeloid leukemia. Eur J Haematol. 2009;82:61–8. doi: 10.1111/j.1600-0609.2008.01158.x. [DOI] [PubMed] [Google Scholar]
- 8.Brieger J, Weidmann E, Maurer U, Hoelzer D, Mitrou PS, Bergmann L. The Wilms’ tumor gene is frequently expressed in acute myeloblastic leukemias and may provide a marker for residual blast cells detectable by PCR. Ann Oncol. 1995;6:811–6. doi: 10.1093/oxfordjournals.annonc.a059321. [DOI] [PubMed] [Google Scholar]
- 9.Cilloni D, Saglio G. Usefulness of quantitative assessment of Wilms tumor suppressor gene expression in chronic myeloid leukemia patients undergoing imatinib therapy. Semin Hematol. 2003;40(2 Suppl 2):37–41. doi: 10.1053/shem.2003.50040. [DOI] [PubMed] [Google Scholar]
- 10.Hirose M. The role of Wilms’ tumor genes. J Med Invest. 1999;46:130–40. [PubMed] [Google Scholar]
- 11.Bergmann L, Miething C, Maurer U, Brieger J, Karakas T, Weidmann E, et al. High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997;90:1217–25. [PubMed] [Google Scholar]
- 12.Polák J, Hájková H, Maalaufová-Soukupová J, Marková J, Sálek C, Schwarz J, et al. Estimation of molecular upper remission limit for monitoring minimal residual disease in peripheral blood of acute myeloid leukemia patients by WT1 expression. Exp Ther Med. 2012;3:129–33. doi: 10.3892/etm.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon M, Martínez-Laperche C, Infante M, Carretero F, Balsalobre P, Serrano D, et al. Evaluation of minimal residual disease by real-time quantitative PCR of Wilms’ tumor 1 expression in patients with acute myelogenous leukemia after allogeneic stem cell transplantation: Correlation with flow cytometry and chimerism. Biol Blood Marrow Transplant. 2012;18:1235–42. doi: 10.1016/j.bbmt.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Guillaumet-Adkins A, Richter J, Odero MD, Sandoval J, Agirre X, Catala A, et al. Hypermethylation of the alternative AWT1 promoter in hematological malignancies is a highly specific marker for acute myeloid leukemias despite high expression levels. J Hematol Oncol. 2014;7:4. doi: 10.1186/1756-8722-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba M, Hata T, Tsushima H, Mori S, Sasaki D, Turuta K, et al. The level of bone marrow WT1 message is a useful marker to differentiate myelodysplastic syndromes with low blast percentage from cytopenia due to other reasons. Intern Med. 2015;54:445–51. doi: 10.2169/internalmedicine.54.3123. [DOI] [PubMed] [Google Scholar]
- 16.Yamauchi T, Matsuda Y, Takai M, Tasaki T, Hosono N, Negoro E, et al. Wilms’ tumor-1 transcript in peripheral blood helps diagnose acute myeloid leukemia and myelodysplastic syndrome in patients with pancytopenia. Anticancer Res. 2012;32:4479–83. [PubMed] [Google Scholar]
- 17.Di Stasi A, Jimenez AM, Minagawa K, Al-Obaidi M, Rezvani K. Review of the results of WT1 peptide vaccination strategies for myelodysplastic syndromes and acute myeloid leukemia from nine different studies. Front Immunol. 2015;6:36. doi: 10.3389/fimmu.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, Sun HQ, Li G, Bai FY, Yang Y, Jing Q, et al. Detection of WT1 gene in acute myeloid leukemia children by real-time fluorescent quantitative RT-PCR and its clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2011;19:959–63. [PubMed] [Google Scholar]
- 19.Østergaard M, Olesen LH, Hasle H, Kjeldsen E, Hokland P. WT1 gene expression: An excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients – Results from a single-centre study. Br J Haematol. 2004;125:590–600. doi: 10.1111/j.1365-2141.2004.04952.x. [DOI] [PubMed] [Google Scholar]


