Abstract
What pathways specify retinal ganglion cell (RGC) fate in the developing retina? Here we report on mechanisms by which a molecular pathway involving Sox4/Sox11 is required for RGC differentiation and for optic nerve formation in mice in vivo, and is sufficient to differentiate human induced pluripotent stem cells into electrophysiologically active RGCs. These data place Sox4 downstream of RE1 silencing transcription factor in regulating RGC fate, and further describe a newly identified, Sox4-regulated site for post-translational modification with small ubiquitin-related modifier (SUMOylation) in Sox11, which suppresses Sox11's nuclear localization and its ability to promote RGC differentiation, providing a mechanism for the SoxC familial compensation observed here and elsewhere in the nervous system. These data define novel regulatory mechanisms for this SoxC molecular network, and suggest pro-RGC molecular approaches for cell replacement-based therapies for glaucoma and other optic neuropathies.
SIGNIFICANCE STATEMENT Glaucoma is the most common cause of blindness worldwide and, along with other optic neuropathies, is characterized by loss of retinal ganglion cells (RGCs). Unfortunately, vision and RGC loss are irreversible, and lead to bilateral blindness in ∼14% of all diagnosed patients. Differentiated and transplanted RGC-like cells derived from stem cells have the potential to replace neurons that have already been lost and thereby to restore visual function. These data uncover new mechanisms of retinal progenitor cell (RPC)-to-RGC and human stem cell-to-RGC fate specification, and take a significant step toward understanding neuronal and retinal development and ultimately cell-transplant therapy.
Keywords: RGC, SoxC, stem cell
Introduction
What are the molecular signals that regulate neural cell fate? For example, retinal ganglion cells (RGCs) differentiate from multipotent retinal progenitor cells (RPCs) but little is known about the cell-autonomous mechanisms and environmental signals that specify RGC fate. The bHLH transcription factor Math5 is necessary but not sufficient for RGC fate, as Math5 expression is found in RPCs that differentiate into nearly all the cell types in the retina (Brown et al., 2001; Wang et al., 2001). Later, the POU-domain transcription factor Brn3b is downstream of Math5 in the regulatory hierarchy for RGC differentiation (Gan et al., 1996), and is a highly specific marker for RGCs in the retina. However, although Brn3b is required for RGC survival after differentiation, it is not required for RGC cell-fate specification, as the number of RGCs born in Brn3b-null retinas resembles that observed in wild-type retinas (Gan et al., 1999; Badea et al., 2009). A recent study has identified a hierarchical network of genes regulating RGC formation, where Math5 acts upstream for RGC competence, while Brn3b and Isl1 are downstream for RGC differentiation. Enforced expression of Brn3b and Isl1 can rescue RGC loss in Math5-knock-out retina, suggesting these two transcription factors are minimally sufficient to specify RGC fate (Wu et al., 2015).
Members of the SoxC family, consisting of three closely related transcription factors, Sox4, Sox11, and Sox12, are oncogenes (Penzo-Méndez, 2010) that also play a role in differentiation in the nervous system (Dy et al., 2008), where they have been identified as regulators of spinal motoneuron development (Thein et al., 2010), adult hippocampal neurogenesis (Mu et al., 2012; Miller et al., 2013), cortical lamination (Shim et al., 2012), corticospinal tract formation (Shim et al., 2012), and development of the inner ear (Gnedeva and Hudspeth, 2015). In each of these cases, knocking out expression of ≥2 SoxC family members was required to see significant loss of neuronal differentiation, but molecular mechanisms for cross-compensation have not been proposed. Sox4 and Sox11 were explored in retinal development in Xenopus Laevis (Cizelsky et al., 2013) and mouse (Jiang et al., 2013; Usui et al., 2013a,b; Kuwajima et al., 2017), although the Sox11 knock-out allele is embryonic lethal and demonstrates numerous other developmental defects, including microphthalmia and cardiovascular maldevelopment (Penzo-Méndez, 2010).
By using floxed alleles of Sox4 and Sox11 and a null allele for Sox12, and two cre-expressing mice with different retinal promoters, we now characterize these transcription factors as necessary and sufficient for RGC differentiation and optic nerve formation in vitro and in vivo. Furthermore, through a newly identified post-translational modification with small ubiquitin-related modifier (SUMOylation) that regulates Sox11 nuclear localization and activity in a Sox4-dependent manner, we identify a novel molecular mechanism for compensatory activity of Sox11 in the absence of Sox4. The conservation of this pro-RGC activity in human induced pluripotent stem cells (iPSCs) suggests a robust phenotype that may have implications for therapeutic approaches.
Materials and Methods
Animals.
All use of animals conformed to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Research, and was approved by the Institutional Animal Care and Use Committees and the Institutional Biosafety Committees of the University of Miami; University of California, San Diego; and Stanford University. Sprague Dawley rats and C57BL/6 mice of varying ages and of either sex were obtained from Harlan Laboratories or Charles River. Mice were bred from the following strains: C57BL/6-Tg(CAG-EGFP)10sb/J (stock #003291, Jackson Laboratory), Math5-Cre (generous gift from Lin Gan), floxed Sox4, floxed Sox11, Sox12−/− (generous gifts from Veronique Lefebvre), and Chx10-Cre (stock #005105, Jackson Laboratory).
Mice genotyping.
Genotyping was performed as follows using standard tail-derived genomic DNA preparations, followed by PCR: Sox4 (Penzo-Méndez et al., 2007): forward primer: 5-GAAGGAGGCGGAGAGTAGACGG; reverse primer: 5-CATAGCTCAACACAAATGCCAACGC; standard buffer supplemented with 2% DMSO; a denaturation step at 94°C for 1.5 min was followed by 35 cycles at 94°C for 30 s, 65°C for 75 s, and 72°C for 90 s, and an extension step for 10 min at 72°C. The Sox4+ PCR product is 450 bp. The Sox4 floxed PCR product is 520 bp; Sox11 (Bhattaram et al., 2010): forward primer: TTCGTGATTGCAACAAAGGCGGAG; reverse primer: GCTCCCTGCAGTTTAAGAAATCGG; standard buffer supplemented with 2 mm MgCl2; a denaturation step at 94°C for 3 min was followed by 35 cycles of 94°C for 30 s, 65°C for 75 s, and 72°C for 60 s, followed by a final extension step at 72°C for 7 min; the Sox11+ PCR product was 319 bp; the Sox11 floxed PCR product was 467 bp; Sox12 (Bhattaram et al., 2010): forward primer: CCTTCTTGCGCATGCTTGATGCTT; reverse primer: GGAAATCAAGTTTCCGGCGACCAA; standard buffer supplemented with 2.75 mm MgCl2; a denaturation step at 94°C for 3 min was followed by 35 cycles of 94°C for 30 s, 65°C for 75 s, and 72°C for 60 s, followed by a final extension step at 72°C for 7 min; the Sox12+ PCR product is 324 bp; Math5-Cre (Brown et al., 2001): For Math5 wild-type (WT) allele: forward primer: CGC CGC ATG CAG GGG CTC AAC ACG; reverse primer: GAT TGA GTT TTC TCC CCT AAG ACC C; 2% DMSO in 10× MasterAmp (Epicenter), with a denaturation step at 94°C for 5 min followed by 40 cycles at 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min, and an extension step for 7 min at 72°C; the Math5 PCR product is 243 bp; for Cre (Moore et al., 2011) and Cre genotyping from Jackson Labs https://www2.jax.org/protocolsdb/f?p=116:5:0::NO:5:P5_MASTER_PROTOCOL_ID,P5_JRS_CODE:288,006143 oIMR0042): CTA GGC CAC AGA ATT GAA AGA TCT; oIMR0043: GTA GGT GGA AAT TCT AGC ATC ATC C; oIMR1084: GCG GTC TGG CAG TAA AAA CTA TC; oIMR1085: GTG AAA CAG CAT TGC TGT CAC TT; a denaturation step at 94°C for 3 min was followed by 35 cycles at 94°C for 30 s, 51.7°C for 1 min, and 72°C for 1 min, and an extension step for 2 min at 72°C; the Cre transgene PCR product is ∼100 bp, the internal positive control is 324 bp; CHX10-Cre PCR as above, forward primer: GCG GTC TGG CAG TAA AAA CTA TC; reverse primer: GTG AAA CAG CAT TGC TGT CAC TT.
Retinal cell dissociation.
Timed pregnant or postnatal mice were euthanized and retinas were dissected and dissociated with papain (Worthington) in Dulbecco's PBS (Life Technologies) incubated at 37°C for 30 min. Retinas were then gently triturated into single-cell suspensions with ovomucoid inhibitors (Roche). The cell suspensions were counted by hemocytometer, spun down, and resuspended in either media for cell culture or protein lysis buffer for protein analysis (see below).
Lipofectamine-based overexpression.
Following dissociation, retinal cells were plated at 100 cells/μl on dishes coated with poly-d-lysine (PDL; 70 kDa, 10 μg/ml; Sigma-Aldrich) and laminin (2 μg/ml; Telios/Invitrogen) in a serum-free, defined medium as described containing BDNF (50 ng/ml; Peprotech), CNTF (10 ng/ml; Peprotech), insulin (5 μg/ml; Invitrogen), and forskolin (5 μm; Sigma-Aldrich; Barres et al., 1988; Meyer-Franke et al., 1995). Following overnight culture, cells were transfected with either GFP plasmid for control or double transfected with GFP and gene of interest with Lipofectamine LTX (Invitrogen). Cells were cultured for 4 d, fixed with PFA, counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) for nuclei and for the RGC marker Brn3 (pan-Brn3abc antibody; Santa Cruz Biotechnology, #sc-6026; see below for immunostaining protocol). Cells were imaged with fluorescence microscopy (Zeiss) and the Brn3+,GFP+ cells out of total GFP+ cells were quantified.
Lentiviral-based overexpression and shRNA knockdown.
For viral transduction-based overexpression, retinal cells were plated at 50 cells/μl on dishes coated with PDL (70 kDa, 10 μg/ml; Sigma-Aldrich) and laminin (2 μg/ml; Telios/Invitrogen) in a serum-free, defined medium as described containing BDNF (50 ng/ml; Peprotech), CNTF (10 ng/ml; Peprotech), insulin (5 μg/ml; Invitrogen), forskolin (5 μm; Sigma-Aldrich), and 5-ethynyl-2′-deoxyuridine (EdU; 5 μm, Invitrogen; Barres et al., 1988; Meyer-Franke et al., 1995). Following overnight culture, cells were exposed to GFP (control) or gene of interest viral particles (∼1 μl of virus with titers 107–108 into each well of each 24-well plate) for overexpression experiments, followed by a rinse into fresh media at 6 h. For knockdown experiments, scrambled shRNA (Santa Cruz Biotechnology) was used as a control and commercially available shRNA lentiviral particles against Sox11 mRNA were used at a threefold multiple of infection (see below for viral particle production and titer determination). Cells were cultured for 5 d, fixed with PFA, counterstained with DAPI (Invitrogen), and immunostained for Brn3 (Santa Cruz Biotechnology) and EdU (Invitrogen; see below for immunostaining and EdU Click-iT protocol). Cells were imaged with fluorescence microscopy (Zeiss). We quantified the cells that were Brn3+, EdU+, and GFP+ out of all Edu+ and GFP+ cells, and the cells that were Brn3+, Edu−, and GFP+ cells out of all GFP+ cells.
Exogenous factor differentiation assay.
To test the effects of exogenous factors on RGC differentiation, cells were cultured at 50 cells/μl on dishes coated with PDL (70 kDa, 10 μg/ml; Sigma-Aldrich) and laminin (2 μg/ml; Telios/Invitrogen) in a serum-free, defined medium as described containing BDNF (50 ng/ml; Peprotech), CNTF (10 ng/ml; Peprotech), insulin (5 μg/ml; Invitrogen), forskolin (5 μm; Sigma-Aldrich), and EdU (5 μm, Invitrogen). Cells were cultured for 5 d, fixed with PFA, counterstained with DAPI (Invitrogen), Brn3 (Santa Cruz Biotechnology), and EdU (Invitrogen; see below for immunostaining and EdU Click-iT protocol). Cells were imaged with fluorescence microscopy (Zeiss) and cells that were both the Brn3+ and EdU+ out of all Edu+ cells were quantified.
Immunostaining.
Cells in culture were fixed with room temperature 4% PFA for 15 min and washed 3× with PBS. Cells were permeabilized with 0.1% Triton X-100 along with the primary antibodies and incubated overnight at 4°C. Secondary detection was performed with Alexa-488, Alexa-596, or Alexa-647 fluorescent antibodies (Life Technologies) at 1:500 dilution incubated overnight at 4°C. Nuclei were counterstained with DAPI (Life Technologies) in PBS for 5 min before a 3× rinse with PBS (Carl Zeiss Meditec) to remove unbound secondary antibodies. Cells immunostained with the same antibody were imaged with fluorescence microscopy (Zeiss) and exposed to the same fluorescence excitation exposure and emission time to maximize comparison between control and experimental groups.
Flatmount retinas were fixed in 4% paraformaldehyde for 30 min and either placed on glass slides or further processed for cryosectioning. The samples isolated for cryosectioning were incubated in 30% sucrose overnight and then frozen in optimal cutting temperature solution (OCT, TissueTek) using liquid nitrogen. Both flat-mount samples as well as retinal sections were simultaneously blocked and permeabilized with 20% donkey serum and 0.4% Triton X-100, respectively, for 30 min. Flatmount retinas and retinal sections were then incubated in primary antibodies, including anti-Brn-3 (1: 200; Santa Cruz Biotechnology; goat polyclonal), anti-βIII tubulin (1:200; Covance; mouse monoclonal), and rat anti-melanopsin (1:100; generous gift from K.-W. Yau) overnight at 4°C. Retinal samples were rinsed 3× with PBS and incubated with the matching secondary antibody overnight at 4°C. The retinal samples were then sealed with coverslips on slides with Vecta-Shield (Vector Labs) solution containing DAPI nuclear stain. EdU staining was conducted using the Click-iT EdU cell proliferation assay (Life Technologies). Other antibodies included Sox4 (1:100; Santa Cruz Biotechnology), Sox11 (1:100; Santa Cruz Biotechnology), Chx10 (1:100; Millipore), Pax6 (1:100; Developmental Studies Hybridoma Bank), glutamine synthase (1:10,000; Sigma-Aldrich), cone-specific arrestin (1:100; generous gift from Cheryl Craft), recoverin (1:500; Millipore), and cleaved caspase-3 (1:300; Cell Signaling Technology). Fixed retinas were imaged using confocal microscopy (Leica). Confocal microscopy was used because neurite outgrowth occurred in three dimensions, and tissue sections would only contain a limited view of the neurite outgrowth of each cell. Thus, whole-mount retinas were imaged to allow a broader, clearer image in the xy plane while retaining z-plane capabilities.
Quantification of cell numbers measured in retinal sections was normalized to linear micrometer of retinal cell layer, and quantification of cell numbers in flat-mounted retinas were normalized to square millimeter of retinal area.
In vivo EdU tagging.
Embryonic day (E) 16 timed pregnant animals were injected with 1 ml of 10 mm EdU solution. Animals were euthanized 1 h later. Embryos were fixed in 4% PFA, embedded in OCT, and cryosectioned. EdU staining was conducted using the Click-iT EdU cell proliferation assay (Invitrogen) in conjunction with Brn3 staining (see immunostaining protocol).
Retinal explant culture.
Retinal explant culture was performed as described previously (Johnson and Martin, 2008). Briefly following mouse euthanasia, adult mouse eyes were enucleated and transferred to cold 4% PFA for 1 h. Eyes were then rinsed in PBS 3× for 5 min each. The cornea and lens were removed and the neural retina was teased off the retinal pigment epithelium with special care taken not to disturb the RGC side of the retina. The retinas were than mounted onto slides with the ganglion cell layer (GCL) upward. The chamber was transferred to a six-well culture plate containing RGC medium as above.
RGC purification and culture.
RGCs were acutely purified from dissociated retina (see above) by immunopanning with the anti-CD90 (Thy1.2; AbD Serotec) antibody, yielding 99.5% pure RGCs (Meyer-Franke et al., 1995; Goldberg et al., 2002). Purified primary RGCs were cultured overnight in serum-free, defined medium as described containing BDNF (50 ng/ml; Peprotech), CNTF (10 ng/ml; Peprotech), insulin (5 μg/ml; Invitrogen), and forskolin (5 μm; Sigma-Aldrich; Barres et al., 1988; Meyer-Franke et al., 1995). Cells were fixed and immunostained following the protocol described above.
Quantitative RT-PCR.
RNA was isolated from mouse RPCs (E14) with RNeasy Kit (Qiagen). DNase treatment was performed for all the RNA samples before reverse transcription (RT). Then the equal amount of RNAs was used for RT reaction according to the manufacturer's instruction (iScript, cDNA Synthesis Kit, Bio-Rad). The quantitiative PCR (qPCR) was performed using primers listed below and the SYBR Green Master Mix Kit (iQ SYBR Green, Bio-Rad) in the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). Gene expression levels were normalized to reference gene cyclophilin A. Primer pairs used for qPCR are as follows: Sox4: forward: 5′-atgaacgcctttatggtgtggtcg-3′, reverse: 5′-acggaatcttgtcgctgtccttga-3′; Brn3b: forward: 5′-tctgcaaccagaggcagaaacaga-3′, reverse: 5′-tggtctgggttcacatttaccgga-3′; cyclophilin A: forward: 5′-agcatacaggtcctggcatc-3′, reverse: 5′-ttcaccttcccaaagaccac-3′.
SUMOylation prediction.
FastA (Fast Alignment) sequences for mouse Sox4 and Sox11 were acquired from EntrezGene and entered into SUMOsp2.0 SUMOylation prediction software (Cuckoo Workgroup). Scores were tabulated and compared with known SUMOylated sample proteins. The SUMOsp 2.0 software is freely available at http://sumosp.biocuckoo.org.
Coimmunoprecipitation.
Mouse RPCs (E14–E15) were resuspended and lysed in lysis buffer (125 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1% genapol C-100, 0.1% trehalose), supplemented with 20 mm N-ethylmaleimide (Sigma-Aldrich), 10 mm iodoacetamide (Sigma-Aldrich), and protease inhibitor (Roche Applied Science). After incubating on ice for 5 min, the protein lysates were spun at 4°C and 15,000 rpm for 5 min, and the supernatants were collected. For coimmunoprecipitation, the cell extracts were incubated with anti-SUMO-1 antibody (Santa Cruz Biotechnology) at 4°C overnight, then with protein-G Sepharose beads (EMD Millipore) for 2–4 h. The beads were washed 3× with lysis buffer and the proteins were eluted in 2× Laemmli buffer. For Western blots, the samples were boiled at 100°C for 5 min, and equal amount of proteins from cell lysates were loaded on the SDS PAGE. Then proteins were transferred to PVDF membrane and incubated with primary antibody anti-Sox11 (Santa Cruz Biotechnology) or anti-Sox4 (Santa Cruz Biotechnology) at 4°C overnight. The chemiluminescent detection of horseradish peroxidase-conjugated secondary antibodies was performed using the West Pico or West Femto Substrate Kit (Thermo-Pierce) according to the manufacturer's instructions.
Sox4, Sox11, and Math5 lentiviral plasmids construction and Sox11 mutagenesis.
The 1323 bp Sox4 (GenBank accession: NM_009238) or the 450 bp Math5 (GenBank accession: BC092234) coding region sequences were each subcloned into lentiviral expression vector pLenti-RRLsinPPT. The cloning sites were XbaI and AgeI. And the reporter gene m-cherry was fused in-frame into the 3′ end of Sox4 or Math5 at AgeI and SalI sites to replace EGFP gene. For Sox11, coding region sequence (GenBank accession: NM_009234) was subcloned into lentiviral expression vector pLenti-Jess2A (generous gift of the viral vector core facility at the University of Miami), 5′ to the GFP gene. The insertion sites were BamHI and SalI, which replaced the m-cherry. The Sox11 point mutant (K91R) was generated using the Quick II XL Site-Directed Mutagenesis Kit (Stratagene). The WT Sox11 plasmid was used as a template to construct the N-terminal mutation at encoding amino acid sequence 91 changing from Lys to Arg (nuclear acid coding sequence from AAG to AGG). Then Sox11K91R was cloned into pLenti-Jess2A vector with restriction sites at BamHI and SalI. The Sox4 coding region was cloned into the same pLenti-Jess2A plasmid at the same BamHI and SalI restriction sides too. All the constructs were verified by sequencing. All the oligo primers used for DNA constructs and generation of mutant are as follows: Sox4 (with pLenti-RRLsinPPT vector), forward: 5′-accgactctagagccatggtacaacagaccaacaac-3′, reverse: 5′-catggtaccggtgtaggtgaagaccaggttagagatgc-3′; Math5, forward: 5′-accgactctagaatgaagtcggcctgcaaaccc-3′, reverse: 5′-catggtaccggtgctggccatggggaagg-3′; mCherry, forward: 5′-gccagcaccggtaccatggtgagcaagggc-3′, reverse: 5′-ttgattgtcgacctacttgtacagctcgtccatgcc-3′; sequencing primer #1, 5′-gctaggctccaggtagaccctgag-3′; sequencing primer #2, 5′-ggcattaaagcagcgtatccacatag-3′; sequencing primer #3, 5′-tcgaactcgtggccgttcacggagc-3′; Sox4 (with pLenti-Jess-2A vector), forward: 5′-cgtacgggatccgccatggtacaacagaccaacaac-3′, reverse: 5′-cttccgactagtgtaggtgaagaccaggttagagatgc-3′; Sox11, forward: 5′-ggcgtacgggatccatggtgcagcaggccga-3′, reverse: 5′-ggtaccccggtcgacatacgtgaacaccaggtcggaga-3′; K91RSox11 (to make Sox11 mutant), forward: 5′-cgctggaagatgctgagggacagcgagaagatcccg-3′, reverse: 5′-cgggatcttctcgctgaccctcagcatcttccagcg-3′; sequencing primer #1, 5′-acacgctgaacttgtggccgtttacg-3′; sequencing primer #2, 5′-ggcattaaagcagcgtatccacatag-3′.
Overexpression in HEK cells.
293T cells (American Type Culture Collection) were cultured in 10% FBS/DMEM (Invitrogen) until 80% confluency was reached. Cells were than transfected with the gene of interest using the Lipofectamine LTX kit (Invitrogen). Cells were cultured for 2 d and then either fixed for immunostaining or lysed for protein.
Human iPSC culture and RGC differentiation.
Commercially available iPSCs (System Biosciences) derived from human foreskin fibroblasts were cultured (1) on irradiated mouse embryonic fibroblast feeder layers (GlobalStem) and (2) in feeder-free conditions on BD Matrigel in mTeSR1 medium (Stem Cell Technologies) supplemented with 5 μm Y-27632 ROCK inhibitor (Stemgent) and 1 μm Thiazovivin (Stemgent). iPS colonies were dissociated to single-cell suspension in ultralow attachment plates (Corning). Media was supplemented with Lefty A (R&D Systems) and DKK1 (R&D Systems). To confirm RPC induction, a subset of cells was fixed and immunostained for the RPC markers Pax6 (Developmental Studies Hybridoma Bank) and Rx (Santa Cruz Biotechnology). Embyroid bodies were dissociated into ≤100 μm aggregates and single cells by a 1:1 ratio treatment with Accutase and Accumax (StemGent) and plated onto 35 mm dishes coated with PDL and laminin and onto 24-well plates. Cells were then infected with 6 μl of lentiviral particles (3.5 × 107 pg/ml p24) containing either Lenti-eGFP (control virus), Lenti-Math5-RFP, Lenti-SOX4-2A-eGFP, or Lenti-Math5-RFP plus Lenti-SOX4-2A-eGFP at a 1:1 ratio. Cells were cultured in serum-free, defined medium as described containing BDNF (50 ng/ml; Peprotech), CNTF (10 ng/ml; Peprotech), insulin (5 μg/ml; Invitrogen), and forskolin (5 μm; Sigma-Aldrich; Barres et al., 1988; Meyer-Franke et al., 1995). Following 7 d in culture, cells were fixed and immunostained for the RGC marker Brn3 (see immunostaining section).
Patch-clamp electrophysiology.
Whole-cell patch-clamp recordings were performed on Math5-overexpressing and Sox4-overexpressing iPSC-derived neurons cultured on tissue culture plates coated with PDL and laminin. The pipettes were pulled from borosilicate glass on a Sutter P-97 puller (Sutter Instrument) to tip resistances of 4–6 MΩ. The external bath solution contained NaCl (140 mm), CaCl2 (2 mm), MgCl2 (1 mm), HEPES (5 mm), and dextrose (3 mm); the pipette solution contained K-gluconate (100 mm), CaCl2 (5 mm), EGTA (10 mm), and HEPES (10 mm). Current-clamp recordings were made using an Axopatch 200B amplifier (Molecular Devices), filtered at 10 kHz with a low-pass Bessel filter (Ithaco), and digitized with a Digitimer (Molecular Devices). Pipette resistance and offset were both compensated. After break-in, the cells were held at −60 mV and stimulated with 10 and 200 ms current pulses over a range of amplitudes. Traces were analyzed using Clampfit 9.2 (Molecular Devices).
Results
Sox4 and Sox11 are required for RGC development
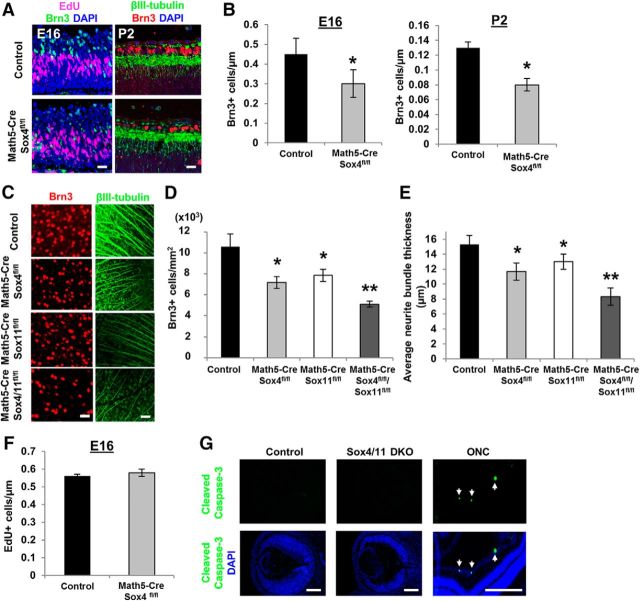
SoxC transcription factors, such as Sox4 and Sox11, were recently published to regulate RGC fate (Jiang et al., 2013). Here, we explored this regulation using two new Cre promoters, Chx10 and Math5. Chx10 expression comes up in progenitor cells very early in retinal development (Rowan and Cepko, 2004) and thus drives cre expression in progenitors that give rise to most retinal cells; Math5 expression in progenitor cells comes up slightly later (Yang et al., 2003) and in our hands was found to drive Cre expression in 40–50% of retinal cells when tested with a GFP reporter allele. We hypothesized that because Math5 is required for RGC specification and is expressed only transiently in retinal development (Brzezinski et al., 2012; Prasov and Glaser, 2012; Prasov et al., 2012), Math5 could be an appropriate promoter for Cre expression in RGC progenitors, although these progenitors also give rise to retinal cells other than RGCs (Brown et al., 2001; Wang et al., 2001). Immunostaining for RGC-specific markers (Brn3+, Pan-Brn3abc antibody) and their neurites (βIII-tubulin+) in E16 and postnatal day (P) 2 retinal sections (Fig. 1A) demonstrated a significant loss (∼25%) in the number of RGCs in both E16 and P2 Math5-Cre/Sox4fl/fl conditional knock-out (cKO) mice in vivo, normalized to GCLs measured in linear micrometers in retinal sections (Fig. 1B).
Figure 1.
Sox4 and Sox11 are required for normal RGC differentiation in vivo. A, Left, E16 timed pregnant mice were injected with 50 μm EdU in PBS and euthanized 1 h later. Embryos were fixed and retinal sections were immunostained for EdU (purple) and Brn3 (green, pan-Brn3abc antibody) and nuclei counterstained with DAPI (blue). Right, P2 mice were euthanized and enucleated, eyes were fixed, and retinal sections were immunostained for Brn3 (red), βIII-Tubulin (green), and DAPI (blue). B, There was a significant reduction in the number of RGCs (Brn3+) noted at E16 and P2 Math5-Cre/Sox4fl/fl mice compared with littermate controls without Cre expression (*p < 0.02, unpaired t test; mean ± SEM). C–E, Control, Math5-Cre/Sox4fl/fl, Math5-Cre/Sox11fl/fl, and Math5-Cre/Sox4fl/fl/Sox11fl/fl retinal explants were immunostained for the RGC marker Brn3+ (red) and the neurite marker βIII-tubulin+ (green). By adulthood, a similar decrease in RGCs was detected in Math5-Cre/Sox4fl/fl or Math5-Cre/Sox11fl/fl mice, and a significantly greater loss of RGCs in Math5-Cre/Sox4fl/fl/Sox11fl/fl mice. Similarly, the loss of Sox4, Sox11, or both resulted in thinner average intraretinal axon bundle thickness (*p < 0.02 from control; **p < 0.02 from control and from single cKO mice; N = 3, ANOVA with post hoc Dunnett's test; mean ± SEM shown). F, There was no change in the number of proliferating (EdU+) cells in Math5-Cre/Sox4fl/fl mice. G, There was no detectable cleaved caspase-3 staining in control or Sox4/11 dcKO E16 retina, although apoptotic cells were detected in adult optic nerve crush (ONC) retinas as a positive control. Scale bars: A, C, 30 μm; G, 200 μm.
Because the other closely related SoxC family members, Sox11 and Sox12, compensate for the loss of Sox4 in other systems (Hong and Saint-Jeannet, 2005; Bergsland et al., 2006; Hoser et al., 2007; Bhattaram et al., 2010; Penzo-Méndez, 2010), we hypothesized that Sox11 and Sox12 might be responsible for the remaining RGC differentiation observed in the Math5-Cre/Sox4fl/fl mice. We found that the Math5-driven conditional loss of Sox11 in the adult retina yielded a similar phenotype to the loss of Sox4, but the Math5-Cre/Sox4fl/fl/Sox11fl/fl double cKO (dcKO) showed a more severe absence of RGCs (∼60%; Fig. 1C,D). We also observed thinner intraretinal axon bundles in both Math5-Cre/Sox4fl/fl and Math5-Cre/Sox11fl/fl cKO animals compared with control, and thinner still in the Math5-Cre/Sox4fl/fl/Sox11fl/fl dcKO mice (Fig. 1C,E), consistent with the decrease in RGC number. The loss of Sox4 had no effect on the proliferation rate of embryonic RPCs in vivo, determined by injecting EdU into timed pregnant animals (Fig. 1F). Cleaved caspase-3 immunostaining also confirmed that the loss of Sox4 and Sox11 had no effect on apoptosis in the embryonic retina (Fig. 1G), in contrast to the retina after optic nerve crush immunostained as a positive control. We also studied a Sox12 KO allele, but there was no significant loss of RGCs in the Sox12 KO nor did Sox12 deficiency compound the loss of RGCs in the Sox4 or Sox11 cKOs or dcKOs. Together these data argue for an effect of Sox4 and Sox11 in regulating fate specification, as opposed to regulating survival of RGCs after differentiation.
In addition, using a Chx10-Cre line, we asked whether the Sox4/11 dcKO effect on reducing RGC differentiation would be exacerbated by earlier and broader retinal Cre expression. We found that in the Chx10-Cre/Sox4fl/fl/Sox11fl/fl mice, there was a nearly complete absence of RGCs, substantiated by immunostaining against two RGC markers, Brn3 and RBPMS (RNA-binding protein with multiple splicing; Fig. 2A–C). An intermediate phenotype was again observed in each of the single Sox4 and Sox11 cKO mice. The few RGCs occasionally counted in Chx10-Cre/Sox4/Sox11 dcKO animals could reflect incomplete dependence on Sox4 and Sox11, or incomplete expression of Cre in progenitor cells. To address this question, we examined Cre expression at E14 and E18, and found it was expressed in most retinal cells at those ages (Fig. 2D), coinciding with the expression of Chx10. The few Brn3-positive cells observed in E14 and E18 Chx10-Cre/Sox4fl/fl/Sox11fl/fl dcKO retinas were Cre-negative (Fig. 2D), suggesting that they may have derived from Cre-negative progenitor cells. Interestingly, we also checked the compound heterozygous deletion of Sox4 and Sox11 and found no effect of single allele deficiency at both loci on RGC fate specification, suggesting that this is not merely a gene dosage-mediated effect. Together, these data showing that a short duration of early Sox4 or Sox11 expression (i.e., between Chx10-driven and Math5-driven cre expression) lets as many as half of the RGCs differentiate, suggest SoxC transcription factors are critical at the earliest time points of RGC fate specification.
Figure 2.
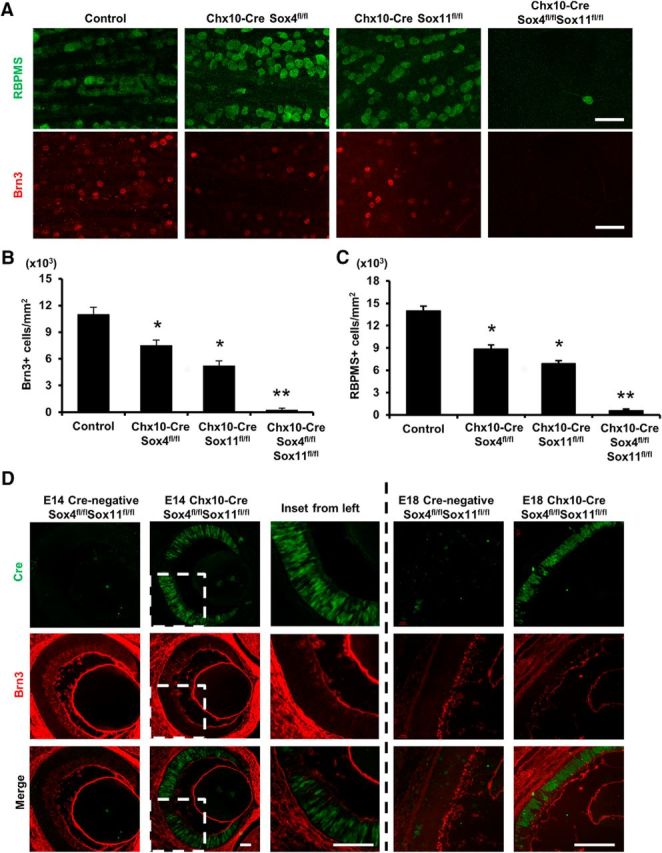
Sox4 and Sox11 deletion during earliest retinal development demonstrates requirement for RGC differentiation in vivo. A–C, Control, Chx10-Cre/Sox4fl/fl, Chx10-Cre/Sox11fl/fl, and Chx10-Cre/Sox4fl/fl/Sox11fl/fl retinal explants were immunostained for the RGC markers RBPMS (green) and Brn3+ (red). There was a similar decrease in RGCs detected in Chx10-Cre/Sox4fl/fl or Chx10-Cre/Sox11fl/fl mice, and a near complete loss of RGCs in the Chx10-Cre/Sox4fl/fl/Sox11fl/fl mice. D, Eye sections of Cre-negative or Cre-expressing mice from E14 and E18 were immunostained for Cre recombinase (green) and Brn3 (red). Magnified images from E14 retina are shown as insets. Scale bars: A, 30 μm; D, 100 μm (*p < 0.05; **p < 0.01 by Student's t test).
Effect of Sox4 and Sox11 on other retinal cells during retinal development
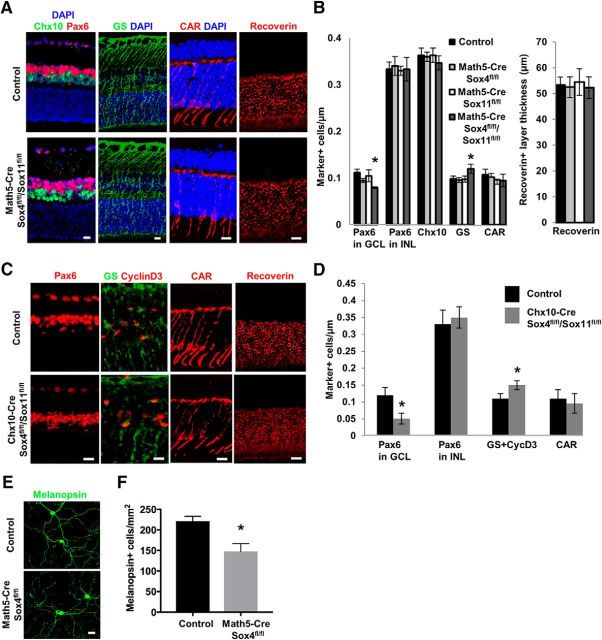
Next we asked whether other retinal cell types are affected by the loss of either Sox4, Sox11, or both, using markers in the inner nuclear layer (INL) for amacrine cells (Pax6+), bipolar cells (Chx+), and Muller glia; and in the outer nuclear layer for cone photoreceptors and all photoreceptors (Fig. 3A). SoxC transcription factor deletion did not change the numbers of any of the other retinal neurons, but there was a statistically significant increase in the number of Muller glial cells (Fig. 3B), consistent with data showing that RPCs that do not differentiate into neurons throughout retinal development default to Muller glial cell fate (Jadhav et al., 2009). Similar data were derived when studying Chx10-Cre-driven knock-out mice, that the loss of RGCs was accompanied by a slight but significant increase in Muller glia cells (Fig. 3C,D). We did see decreased Pax6+ cells in the GCL (which has both RGCs and amacrine cells), but we did not see a difference in Pax6+ cells in the INL, which comprises amacrine cells (and others), suggesting that amacrine cells as a whole were not affected, although a loss of a subtype of amacrine cells found primarily in the GCL remains possible. No microphthalmia was observed in these animals, unlike that seen in the Sox11-null mouse (Wurm et al., 2008). We did find a preferential loss of melanopsin-expressing intrinsically photosensitive RGCs (Provencio et al., 2000, 2002; Berson et al., 2002; Hattar et al., 2002) in the Math5-cre/Sox4fl/fl retina (Fig. 3E,F). Thus, RGC differentiation dependence on SoxC transcription factors is not due to failure in progenitor cell proliferation, and was preferentially exhibited in at least this one important RGC subpopulation.
Figure 3.
Sox4/11 deletion does not affect other neuronal cell numbers in the developing retina. A, B, Using markers for amacrine cells (Pax6+) in the inner nuclear layer (INL), bipolar cells (Chx10+), Muller glia (GS+), cone photoreceptors (CAR+), and photoreceptors (Recoverin+) in control, single-KO, or double-KO mice. In addition to significant decrease of Pax6+ cells in GCL, there was no change in layer thickness or cell number of any of the other retinal neurons, but there was a statistically significant increase in the number of Muller glial cells in the Math5-Cre/Sox4fl/fl/Sox11fl/fl group. C, D, Immunofluorescence staining for other retinal cell types, as marked, confirms decrease in GCL Pax6+ cells and a small increase in Muller glia, without any effect on amacrine cells or photoreceptors. E, F, Control and Math5-Cre/Sox4fl/fl retinal explants were immunostained for melanopsin to label intrinsically photosensitive RGCs (ipRGCs); there was preferential loss of melanopsin-expressing ipRGCs in the Math5-Cre/Sox4fl/fl mutant retina. Scale bars: A, C, E, 30 μm (*p < 0.05 by Student's t test; data in B, D normalized to GCLs measured in linear micrometers in retinal sections; data in F normalized to retina areas measured in square millimeters in flat-mounted retinas.
Loss of Sox4 and Sox11 impairs optic nerve development
In many cases in the nearly complete absence of RGCs in the Chx10-Cre dcKO mice, the optic nerve completely failed to form (Fig. 4A). With the few hypoplastic nerves that were observed in limited cases, we used electronic microscopy to investigate the morphology and structure of optic nerve in control, Chx10-Cre/Sox4fl/fl cKO, and Chx10-Cre/Sox4fl/fl/Sox11fl/fl dcKO mice. We found that the size of cross section of optic nerve in Chx10-Cre/Sox4fl/fl cKO group was slightly decreased compared with controls, but the dcKO optic nerves were severely hypoplastic (Fig. 4B,C). Cross sections of control and Chx10-Cre/Sox4fl/fl cKO optic nerves demonstrated well organized myelination of axons (Fig. 4D,E). However, in cases in which Chx10-Cre/Sox4fl/fl/Sox11fl/fl dcKO optic nerves could be recovered, they demonstrated little myelination around RGC axons (Fig. 4D,E). Although the single cKO nerves appeared grossly normal, using a frequency plot for individual axon size, Chx10-Cre/Sox4fl/fl cKO optic nerves showed slightly smaller axon diameters compared with control nerves (Fig. 4F). In these optic nerves, we quantified a 20% reduction in axon number and 30% reduction in total axon area in Chx10-Cre/Sox4fl/fl cKO nerves (Fig. 4G); no axon number or area could be determined in Chx10-Cre/Sox4fl/fl/Sox11fl/fl dcKO optic nerves. Thus, concomitant with RGC developmental failure, there was a significant decrease in optic nerve axons, although the axons that did grow down the optic nerve were myelinated, suggesting that RGC–oligodendrocyte interactions were preserved in remaining axons.
Figure 4.
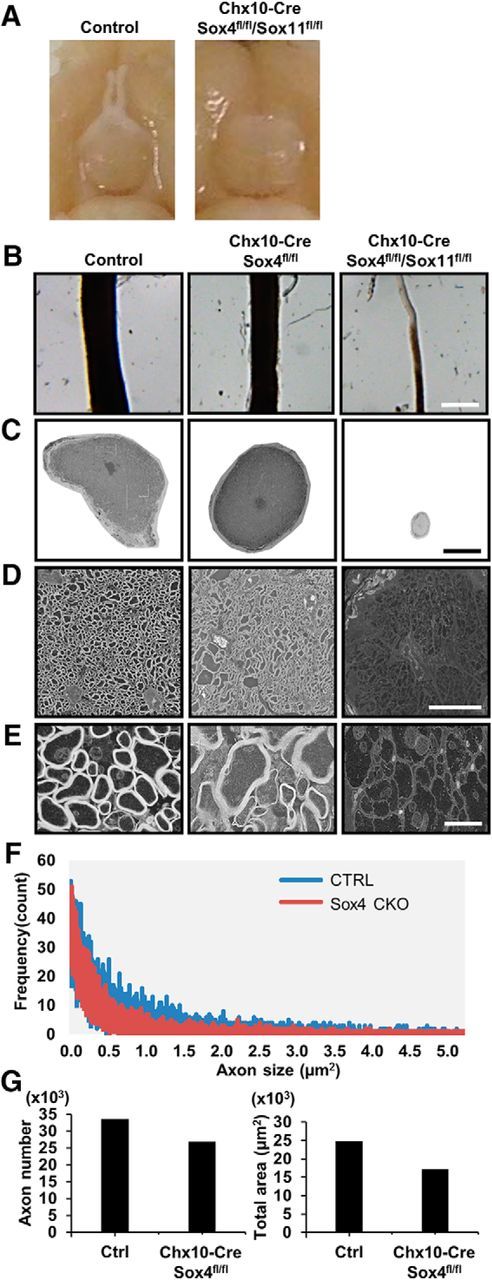
Sox4 and Sox11 are required for normal optic nerve development. A, Examination of perfusion-fixed control and Chx10-Cre/Sox4fl/fl/Sox11fl/fl mice revealed a complete loss of optic nerve in the dcKO mice. B, Optic nerves of control, Chx10-Cre/Sox4fl/fl, and Chx10-Cre/Sox4fl/fl/Sox11fl/fl mice mounted on agarose gel plates for electron microscopy demonstrated optic nerve atrophy in the Sox4 knock-out and severe atrophy in the subset of Sox4/11 dcKO mice with any optic nerves (scale bar, 500 μm). C–E, Cross sections of optic nerves from the samples shown in B were imaged with electron microscopy. Scale bars: C, 150 μm; D, 15 μm: E, 2 μm. F, Frequency plot for individual axon size. G, Analysis of individual axon number and axon area of control and Chx10-Cre/Sox4fl/fl optic nerves demonstrates significant reductions in these two measures.
Sox4 and Sox11 are expressed and developmentally regulated in RGCs
We next examined when and where Sox4 and Sox11 mRNA and protein are present in the developing retina. In situ hybridization for Sox4 (Fig. 5A) and Sox11 (Fig. 5B) mRNA demonstrated that both genes are expressed in embryonic retina and concentrated in the innermost part of the developing retina where the GCL emerges (images courtesy of www.genepaint.org). The developmental time series of Sox4 and Sox11 expression in the retina were previously published (Jiang et al., 2013). Developmental expression profiling of acutely purified RGCs revealed high embryonic Sox4 (Fig. 5C) and Sox11 (Fig. 5D) mRNA expression (peaking at E18) that went down throughout development but remained expressed in postnatal and adult RGCs (Wang et al., 2007). We next examined the spatiotemporal expression of Sox4 and Sox11 protein through retinal development. To show subcellular localization at a time point when all RGCs are terminally differentiated, Sox4 and Sox11 were immunodetected in cells positive for the RGC marker Brn3 (Fig. 5E,F) at P10; antibody specificity was confirmed using Sox4 knock-out tissues as negative controls (Fig. 5G). Interestingly, we observed Sox4 mostly present in RGC nuclei (Fig. 5E, right) while Sox11 was observed in RGC cytoplasm (Fig. 5F, right). We also observed occasional Sox4 and Sox11 in displaced amacrine cells in the GCL and in amacrine cells of the INL (Fig. 5G), although as noted above the differentiation of these cells did not depend on Sox4/Sox11 expression (Fig. 3). In cultured E14 retinal cell suspension, Sox4 was coexpressed only in βIII-tubulin-positive cells (∼30% of the total cells; Fig. 5H) and colocalized with Brn3 (Fig. 5I) in the nuclei of RGCs (which are ∼15% of the retinal cells at this stage in development).
Figure 5.
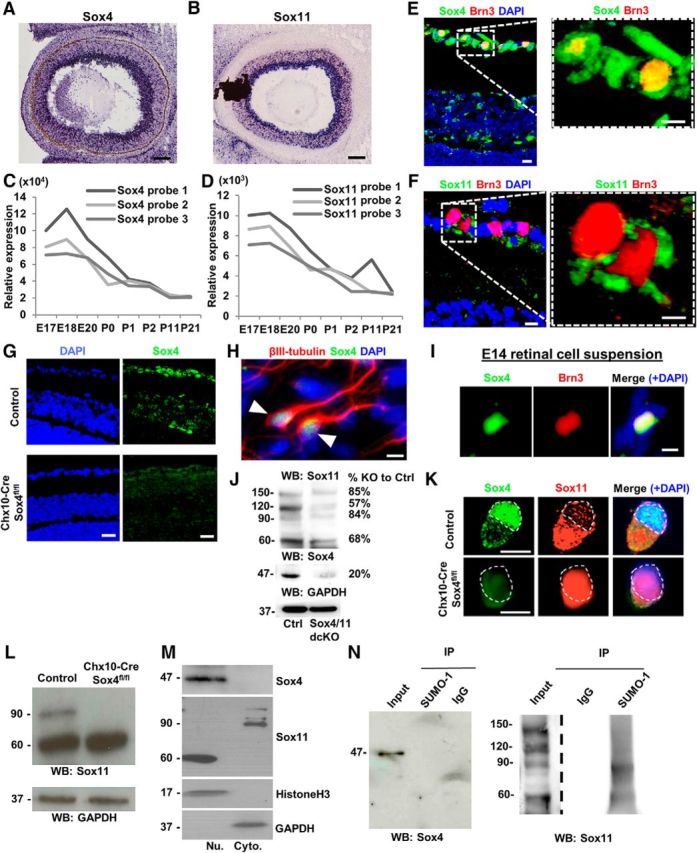
Developmental expression and regulation of compensation by SUMOylation in SoxC control of RGC differentiation. A–F, Sox4 and Sox11 are detected in RGCs by in situ hybridization of E14 embryos (A, B; www.genepaint.com), by microarray of E18 embryos (C, D; Wang et al., 2007), and by immunofluorescence in P10 retinal sections (E, F). G, Sox4 antibody specificity was confirmed by immunostaining on control and Chx10-Cre/Sox4fl/fl tissue. H, I, Dissociated E14 retinal cells showed colocalization of Sox4 with the RGC-specific markers βIII-tubulin and Brn3, as marked. J, Western blot of adult retinal protein lysates detected Sox4 and a series of Sox11-reactive bands laddering on the blot. K, Immunostaining for Sox4 and Sox11 in control and Sox4 cKO embryonic retinal cells. Nuclei outlined with dashed line. L, Western blot for Sox11 in control and Sox4 cKO whole-retinal protein lysate, as marked. M, Nuclear fractionation of RGCs followed by Western blot against Sox4 and Sox11 showed Sox4's predominantly nuclear and Sox11's predominantly cytoplasmic localizations. GADPH and histone H3 were used as internal controls for cytoplasm and nucleus, respectively. N, Embryonic retinal lysate was immunoprecipitated with antibodies to SUMO-1 and probed with Sox4 (left) and Sox11 (right) using IgG as a negative control. Because input concentrations were much lower than post-immunoprecipitation concentrations, the exposure times were different (dotted line to indicate different exposure time). Scale bars: E, F, H, I, K, 30 μm; A, G, 50 μm.
We hypothesized that the differential protein localization of Sox4 and Sox11 might be explained by differences in post-translational modifications. Western blot of embryonic retinal lysate for Sox4 (Fig. 5J, bottom) showed a single 47 kDa band; Western blot for Sox11 (Fig. 5J, top) demonstrated multiple bands including 60, 90, 120, and 150 kDa bands. In purified mouse E18 RGCs, Sox11 was present mostly in RGC cytoplasm (Fig. 5K). Furthermore, nuclear fractionation in P7 retinas demonstrated that the higher molecular weight isoforms of Sox11 were only found in the cytoplasmic fraction, and that most Sox11 was found in the cytoplasm (Fig. 5J,M). In contrast, Sox4 was detected only in the nuclear fraction (Fig. 5K,M). Interestingly, we found that Sox11 protein shifted to the nucleus in Chx10-Cre/Sox4fl/fl cKO RGCs (Fig. 5K,L), suggesting the hypothesis that Sox11 translocation compensates for the loss of Sox4.
Since the Sox11 banding suggested laddering with ∼30 kDa additions, we explored for consensus sequences bioinformatically. SUMOsp 2.0t SUMOylation prediction software suggested a putative SUMOylation site on Sox11 at lysine 91 (K91) not found on Sox4. To determine whether Sox4 or Sox11 were SUMOylated, we performed anti-SUMO immunoprecipitations. A higher molecular weight isoform of Sox11 but not Sox4 immunoprecipitated from acutely dissociated embryonic retina, demonstrating the existence of SUMOylated Sox11 in the developing retina (Fig. 5N). Thus, SUMOylated Sox11 is predominantly cytoplasmic, whereas Sox4, as well as a lesser amount of non-SUMOylated Sox11 are localized to the nucleus. The dissimilar subcellular compartmentalization of Sox4 (nuclei) and Sox11 (cytoplasm) observed both in vivo and in vitro as well as the disparate number of molecular weight forms raised interesting questions about whether each performs specific, overlapping, or redundant roles during development.
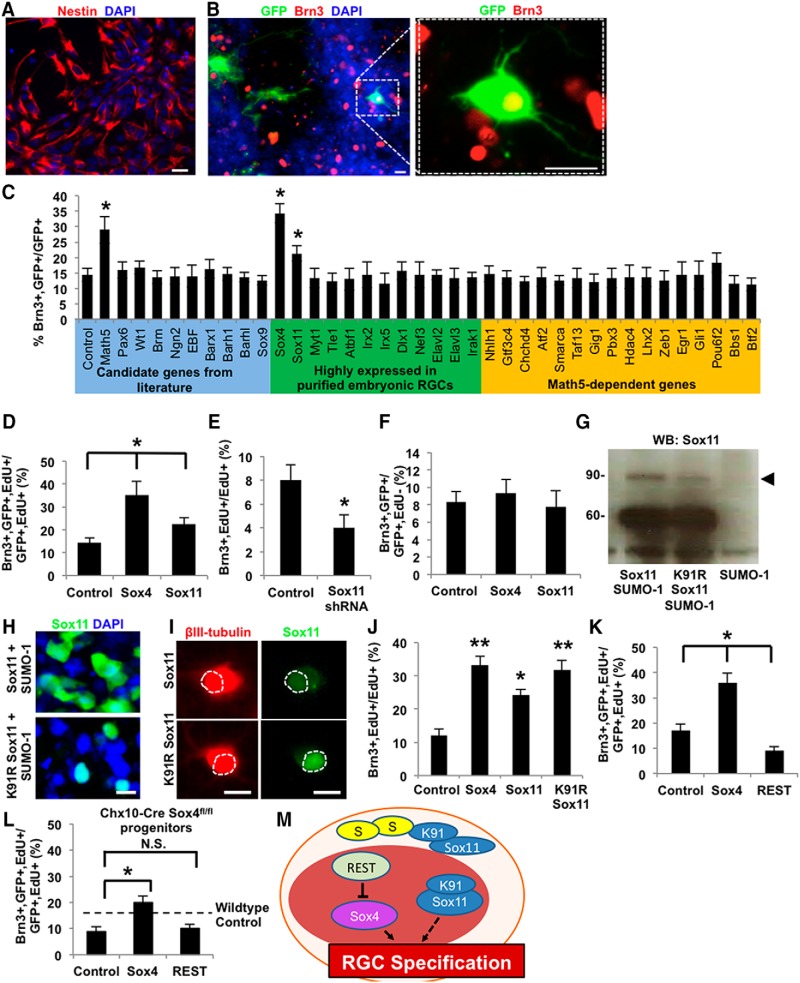
Overexpression of Sox4 and Sox11 in retinal progenitors increase RGC specification in vitro
Thus Sox4 and Sox11 are necessary for RGC specification, but it is not known whether they act directly on RPCs to promote RGC fate specification. To test whether these or other transcription factors either demonstrated or suggested to be necessary for RGC specification, or transcription factors strongly expressed in embryonic RGCs (Mu et al., 2005; Wang et al., 2007), were sufficient to promote RGC differentiation, we performed an in vitro RPC overexpression screen (Fig. 6A,B). Although we cannot rule out whether factors negative in the screen might still play a role in RGC differentiation, the screen demonstrated Sox4 and Sox11 transcription factors increased RGC differentiation (Fig. 6C). To confirm that the effect of Sox4 and Sox11 on RGC differentiation occurs before RPCs exit their final cell cycle, E14 retinal cells were cultured with the thymidine analog EdU. Exogenous Sox4 or Sox11 expression in EdU-tagged embryonic RPCs increased RGC differentiation compared with control GFP expression (Fig. 6D). Conversely, shRNA targeting Sox11 reduced RGC fate specification (Fig. 6E). In both assays, Sox4 had a significantly stronger positive effect on RGC differentiation compared with Sox11. Neither Sox4 nor Sox11 overexpression in EdU−/Brn3+ RGCs (that is, E14 primary RGCs differentiated in vivo before plating into culture) affected RGC numbers (Fig. 6F), suggesting no effect on survival of differentiated RGCs. Together, the increase in differentiated RGCs following Sox4 and Sox11 overexpression suggest that the SoxC subfamily of transcription factors is sufficient for directing embryonic RPCs toward RGC fate.
Figure 6.
Overexpression of Sox4 and Sox11 is sufficient to promote RGC differentiation from RPCs. A, B, Nestin+ (A, red) E14 RPCs were transfected with a GFP reporter plasmid (B, green, marking transfected cells) and candidate differentiation genes and counterstained with DAPI for nuclei (blue) and for the RGC-specific marker Brn3 (red), as marked. C, Following transfection with candidate genes as marked, RPCs were cultured in pro-RGC differentiation and survival conditions for 4 d, fixed, and immunostained. Brn3+ and GFP+ cells out of all GFP+ cells were quantified. Overexpression of Math5, Sox4, and Sox11 increased the total number of GFP+ and Brn3+ cells (mean ± SEM, *p < 0.01, N = 3, ANOVA with post hoc Dunnett's test, mean ± SEM). D, RPCs were differentiated in the presence of EdU (5 μm) for 5 d and immunostained for the RGC marker Brn3 and EdU. Sox4 and Sox11 overexpression with lentivirus increased RGC differentiation (Brn3+, GFP+, and EdU+ cells) of proliferative, transfected RPCs (GFP+ and EdU+ cells) compared with control GFP overexpression (*p < 0.02, paired t test; mean ± SEM; all samples differ significantly from each other). E, Embryonic retinal cells from Math5-Cre/Sox4fl/fl mice were differentiated in the presence of control or Sox11 shRNA; Sox11 knockdown further decreased RGC differentiation of Math5-Cre/Sox4fl/fl progenitors compared with control mice. F, Sox4 and Sox11 overexpression with lentivirus had no effect on the survival or total number of RGCs that were postmitotic in vivo and then placed in culture (Brn3+, GFP+, and EdU− cells; *p < 0.02, paired t test; mean ± SEM). G, Overexpression of Sox11-SUMO-1, Sox11K91R-SUMO-1, and SUMO-1 in HEK cells and probed with Sox11 for Western blot. H–J, RPCs were further infected with Sox11 mutant gene (Sox11K91R) for immunofluorescence (H), or probed with βIII-tubulin (I), or probed with Brn3 and EdU (J). K, Overexpression of REST in WT E14 RPCs decreased RGC differentiation compared with controls as measured by immunostaining of differentiated RGCs (Brn3+, GFP+, EdU+/GFP+, and EdU+; *p < 0.05, **p < 0.01 paired t test; mean ± SEM; all samples differ significantly from each other). L, REST overexpression in E14 RPCs derived from retinal-specific Sox4 cKO mice elicited no changes in RGC fate specification. M, Integrated model for regulatory mechanisms of RGC fate specification. Scale bars, 30 μm.
We also hypothesized that the Sox11 SUMOylation discovered above (Fig. 5) might affect its subcellular localization and function in promoting RGC differentiation from RPCs. To test the physiologic role of SUMOylation at the Lys91 site, we generated mutant Sox11 cDNA clones with an arginine (K91R) substitution, to render the site un-SUMOylatable. We expressed Sox11 (native or K91R) and observed an expected band at 60 kDa (Fig. 6G, lanes 1, 2), but the expression of the Sox11K91R non-SUMOylatable form led to the decreased detection of the 90 kDa isoform (Fig. 6G, compare lanes 1, 2). This confirmed that the K91R mutant indeed demonstrates lower SUMOylation as hypothesized. Immunofluorescence of the non-SUMOylatable Sox11K91R showed a concomitant shift from cytoplasmic to nuclear localization (Fig. 6H). A similar shift was apparent after expressing Sox11 versus Sox11K91R in embryonic RPCs differentiated to β-tubulin-positive RGCs (Fig. 6I). Finally, in an RGC differentiation assay, Sox11K91R increased RGC differentiation compared with WT Sox11, to a level similar to Sox4 (Fig. 6J). Together these data suggest that native, K91-SUMOylated Sox11 is localized in the cytoplasm where it plays less of a pro-RGC differentiation role (Fig. 6M).
RE1 silencing transcription factor (REST) is a zinc finger repressor protein that has been known to repress many genes encoding neuronal proteins (Chong et al., 1995; Schoenherr and Anderson, 1995). REST knock-out promotes RGC differentiation, even in the absence of Math5 (Mao et al., 2011), raising the hypothesis that REST may act through a Math5-independent pathway. We asked whether REST suppression of RGC differentiation depends on Sox4 expression. We observed that overexpression of REST gene reduces Brn3+/EdU+ cell production in E14 RPCs (Fig. 6K). However, no reduction was observed with REST expression in Sox4 cKO RPCs (Fig. 6L). These data support a model in which REST suppresses RGC fate specification through a Sox4-dependent mechanism (Fig. 6M).
Sox4 promotes RGC differentiation from human iPSCs
Finally we asked, does overexpression of Sox4 alone or in combination with Math5 potentiate the differentiation of functional RGCs from other stem cell populations? For example, human iPSCs can generate RGC-like cells in vitro (Chen et al., 2010; Parameswaran et al., 2010). We cultured human iPSCs and confirmed their identity by morphology (Fig. 7A), by expression of typical stem cells markers (Fig. 7B,C), and by lack of expression of differentiation markers or neurites (Fig. 7D). After adjusting culture conditions on substrates coated with PDL and laminin for 24 h to promote retinal progenitor-like differentiation (Joo et al., 2015), PAX6 and Rx RPC markers were detected in most cells (Fig. 7E). Human iPSC-derived RPCs were then exposed to lentiviral vectors for Math5-RFP and Sox4-GFP genes (Fig. 7F). Differentiating cultures were assayed for neurite morphology (Fig. 7G) and Brn3 expression (Fig. 7H). We found that Math5 and Sox4 greatly potentiated the differentiation of RGCs from human iPSCs (Fig. 7I). The combination of both genes in particular showed a synergistic effect on potentiating RGC differentiation, with as many as 25% of progeny demonstrating RGC-marker expression and morphology. We used patch-clamp electrophysiology to test whether these RGC-like neurons were electrically excitable. Before differentiation, human iPSC-derived RPCs failed to generate action potentials spontaneously or in response to current injection (data not shown). Following differentiation, RGC-like progeny generated single and multiple burst action potentials (Fig. 7J,K) with properties similar to those of purified primary rodent RGCs (Meyer-Franke et al., 1995). Together, these data suggest that Sox4 and Math5 strongly promote human stem cell differentiation into functional RGC-like neurons with an efficiency and electrophysiologic function not previously described.
Figure 7.
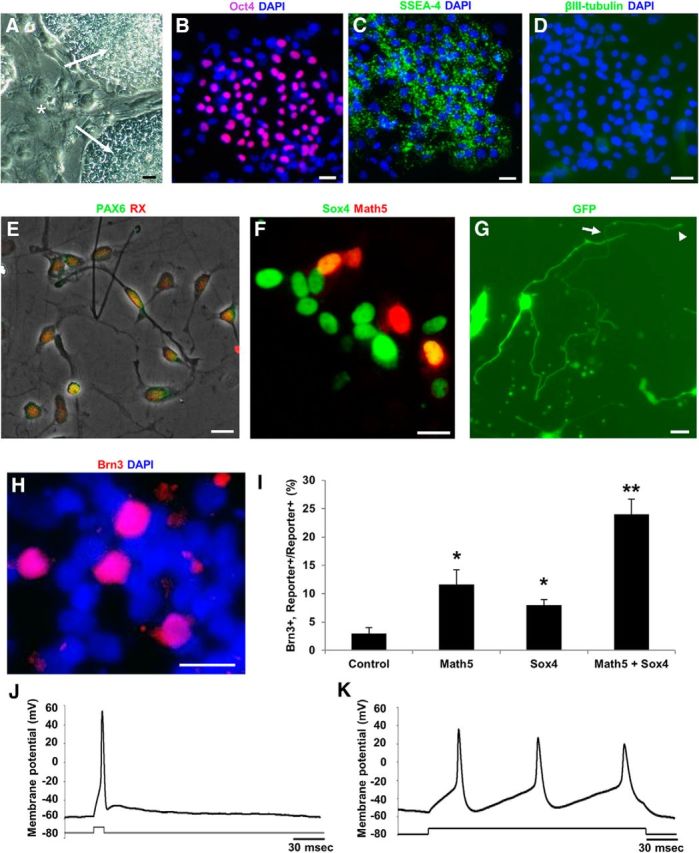
Sox4 promotes human stem cell differentiation into functional RGCs. A–D, Human iPSCs (A, arrows) cultured on eradicated mouse embryonic fibroblast (MEF) feeder layers (A, asterisks) expressed embryonic stem cell markers Oct4 (B) and SSEA-4 (C), but not the neuronal marker βIII-tubulin (D). E, Embryoid bodies dissociated into ≤100 μm aggregates and single cells and plated on PDL and laminin for 24 h showed RPC marker coexpression of Pax6 and Rx. F, During 5 d of differentiation, cells were infected with eGFP (control), Math5-RFP, SOX4-2A-eGFP, or both (example shown in F). G–I, After 5 d, overexpression of SOX4-GFP (G) potentiated RGC morphologic differentiation including long neurites (arrow) with growth cones (arrowhead), and (H and I) increased expression of the RGC marker Brn3. Math5 and Sox4 co-overexpression in human iPSCs together synergized in potentiating RGC-like differentiation (*p < 0.05 from control; **p < 0.01 from control, Math5-alone, and Sox4-alone conditions; N = 3, ANOVA with post hoc Dunnett's test, mean ± SEM shown). J, K, RGC-like progeny generated (J) single and (K) multiple burst action potentials with properties similar to those of purified primary rodent RGCs. Scale bars, 30 μm.
Discussion
SoxC transcription factors are necessary for RGC and optic nerve development
Together, these data have important implications. First, they describe a novel signaling pathway for RGC fate specification in vitro and in vivo through Sox4 and Sox11, consistent with a report examining a Six3-cre-driven double Sox4/Sox11 knock-out (Jiang et al., 2013). A recent paper further indicated that SoxC transcription factors promote contralateral but not ipsilateral RGC differentiation by binding to Hes5 and thus repressing Notch signaling (Kuwajima et al., 2017); as >95% of RGCs are contralateral-projecting in rodents, these data are consistent with our findings here. Previous lineage-tracing experiments demonstrated the necessity for but not sufficiency of Math5 in RGC fate specification: Math5 is required for RGC development, but precursors for most early-born and some late-born retinal neurons express Math5, which suggests that Math5 contributes to the competence of early RPCs toward RGC fate. In this initial screening, we only used single-factor screening. Combinations of transcription factors might be better and could be tested in the future. We further found that Sox4 and Math5 can synergistically promote RGC differentiation from iPSC-derived RPCs, again suggesting that combinations of transcription factors might work better. Other protein subfamilies, such as the Brn3 family of proteins and WT1, are critical for the later survival but not the initial cell fate specification of RGCs. In contrast, these data describe a regulatory pathway in which SoxC transcription factors are necessary and sufficient to generate RGCs from RPCs, and in their absence there is a complete loss of the optic nerve. Because Brn3 expression was decreased in E14 retina in the Sox4/Sox11 dcKO, and conversely Sox4 or Sox11 led to increased Brn3+/EdU+ expression in E14 RPC culture or in human stem cell culture, we place Sox4/Sox11 upstream of Brn3, although perhaps not directly.
Whether Sox4 or Sox11 can rescue RGC fate specification downstream of or parallel to Math5 remains to be addressed (Jiang et al., 2013). Interestingly, a recent report found that Brn3b and Isl1 constitute a minimal transcription factor pair for RGC fate specification in the absence of Math5, suggesting a master core of transcription factors that regulate RGC differentiation (Wu et al., 2015). Whether Sox4 and Sox11 regulate the same gene expression program for RGC specification could be studied next using transcriptome profiling in the RPC-to-RGC transition.
Why is there a complete loss of RGCs and optic nerve formation in the Chx10-Cre (this paper) or Six3-Cre (Jiang et al., 2013) Sox4fl/fl/Sox11fl/fl line while the Math5- Cre/Sox4fl/fl/Sox11fl/fl mice still retain ∼40% of their RGCs? We hypothesize that the stronger phenotype observed is due to the timing of Math5 expression. Math5 is expressed in progenitors in transition to RGCs, which may be slightly late to elicit the full SoxC KO phenotype, especially if there is already sufficient SoxC protein expression at the time of gene excision. Thus, even brief SoxC transcription factor expression in the Math5-Cre dcKO can promote differentiation of approximately half of the normal complement of RGCs.
The importance of early Sox4 or Sox11 knock-out is also captured in the previous failure to identify Sox11 as a regulator of RGC differentiation in experiments in which Sox11 was overexpressed at E17, which may be too late to revert later RPCs to RGC fate (Usui et al., 2013b). The loss of Muller glia in those experiments is consistent with the increase in Muller glia seen in our dcKO data. Similarly, the Sox11-null allele was embryonic lethal with major cardiovascular system defects, microphthalmia, and gross defects in lens development, and thus failed to capture the importance of Sox11 in RGC fate specification. This failure may be due to the presence of Sox4 or to additional indirect effects of microphthalmia and lens maldevelopment. Also, if Sox11 is responsible for a later-born subset of RGCs, any effect may not have been observable in these E18 embryonic lethal lines. We observed a loss of RGCs at P1 that persists into adulthood. Here, we did not look at different transcription factors for production of different RGC subtypes. For rodent progenitor cells and for human iPSCs, there are no validated marker antibodies for RGC subtypes other than for melanopsin-positive intrinsically photosensitive RGCs studied in rodent cells, so further exploration of subtype differentiation, though important, must be left to future experiments. Together, these data suggest a model in which RGC fate specification may occur in the earliest stages of retinal development but may not be evident until the full wave of RGC generation is completed, around the time of birth.
Both Sox4 and Sox11 on their own contribute to RGC differentiation in vivo (as evidenced by RGC loss in the single-cKO mice). Whether they contribute in a dose dependence or by regulating different genes is not clear from these data, although the compound heterozygous Sox4+/−/Sox11+/− mice did not show a phenotype as strong as when either gene was deficient at both alleles. In occasional animals with single alleles of either knocked out, we did see a small decrease in RGC numbers (Figs. 1, 2). We did not assay co-overexpression of both Sox4 and Sox11 at the same time due to the lower efficiency of double transfection, but we speculate that co-overexpression of both genes could further promote RGC fate in RPC culture.
SUMOylation as a novel mechanism for regulating of transcription factor compensation
How do SoxC transcription factors regulate their cross-compensatory activities? Here we delineate a new biological interaction whereby Sox4's normal expression is associated with suppression of Sox11 nuclear localization and function through SUMOylation, a post-translational modification that may regulate function of other Sox transcription factors (Gill, 2005). Ubc9, found in SUMOylation complexes, has been shown to interact with Sox4 through SUMOylation-independent interactions (Bergsland et al., 2006). This suggests a potential feedback loop in which one transcription factor family member, Sox4, controls the SUMOylation-dependent subcellular localization of a closely related and highly redundant transcription factor family member, Sox11. This explanation for compensatory transcription factor function may similarly apply to the regulation of other transcription factors, including myc (Kessler et al., 2012) and neural retina leucine zipper factor (Roger et al., 2010). Such compensation raises the hypothesis that Sox4 is less likely to be found mutated in human optic nerve hypoplasias, if Sox11 activity is increased in its absence. These data may also explain why, in certain cancers, such as mantle cell lymphoma, Sox11 protein location changes from cytoplasmic to nuclear (Ek et al., 2008). Whether such protein regulation of one transcription factor by a related family member is also used by other transcription factor families to modulate the level of transcriptional activation or repression may have implications for gene duplication into multigene families during evolution. Indeed, Acropora millepora SoxC, one of the evolutionarily oldest SoxC family members found in coral, is not SUMOylated (Shinzato et al., 2008), nor is Drosophila SoxC (Savare and Girard, 2005). Looking for direct interactions at the SoxC promoters and identifying gene targets will be an interesting topic for future work.
Sox4 and RGC differentiation for stem cell therapy
Finally, identifying regulatory mechanisms not only necessary, but also sufficient to generate RGCs may be a useful strategy to generate RGCs from human stem cell populations for enhancing cell-replacement therapy for degenerative disease. For example, stem cell therapies hold promise for replacing degenerating RGCs in glaucoma and other optic neuropathies (Hertz and Goldberg, 2012). In the eye, subretinal stem cell and retinal progenitor-derived grafts have been used to achieve functional photoreceptor replacement in mouse models of retinal degeneration (MacLaren et al., 2006; West et al., 2009) and embryonic stem cell-derived retinal pigment epithelium has entered human clinical trials (Schwartz et al., 2012). However, RGC replacement is significantly more challenging as transplanted RGCs would need to integrate into the more complex circuitry of the inner retina and project lengthy axons capable of synapsing at precise brain targets.
Recent advances in enhancing RGC migration into the retina following intravitreal cell delivery suggests that RGCs may survive, migrate into the GCL, send local dendrites into the inner plexiform layer, and elongate axons (Hertz et al., 2014; Venugopalan et al., 2016). Furthermore, RGC axons injured in the optic nerve can be coaxed to regenerate long distances in the optic nerve and to the brain (de Lima et al., 2012), suggesting that a transplantation therapy may yet be possible. However, a limitation for RGC cell therapies is the low overall numbers of RGCs that normally differentiate from progenitor or stem cells in normal development or in vitro, often in the low, single-digit percentages (Jagatha et al., 2009; Chen et al., 2010; Parameswaran et al., 2010). Manipulation of the SoxC transcriptional network by overexpression of SoxC genes or by manipulating SUMOylation may be used to more efficiently generate RGC donor cells for cell-based therapies in these debilitating diseases.
Footnotes
This work was supported by the Hope for Vision Foundation, the National Eye Institute (P30 EY014081, University of Miami), an unrestricted grant from Research to Prevent Blindness, and the Walter G. Ross Distinguished Chair in Ophthalmic Research. J.H. was supported in part by National Institutes of Health Grant T32-NS07492, and by a Lois Pope Life Fellowship. We thank Cheryl Craft for generously donating the cone-specific arrestin antibody; Veronique Lefebvre for floxed Sox4, floxed Sox11, and Sox12-null alleles; Eleut Hernandez for animal husbandry; Gabriel Gaidosh for assistance with microscopy; Waitt Advanced Biophotonics Center at the Salk Institute for assistance with electron microscopy and data analysis; and Alexander Kreymerman, Suqian Wu, and Xin Xia for their technical help.
The authors declare no competing financial interests.
References
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J (2009) Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61:852–864. 10.1016/j.neuron.2009.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA, Silverstein BE, Corey DP, Chun LL (1988) Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron 1:791–803. 10.1016/0896-6273(88)90127-4 [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J (2006) The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev 20:3475–3486. 10.1101/gad.403406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M (2002) Phototransduction by retinal ganglion cells that set the circadian clock. Science 295:1070–1073. 10.1126/science.1067262 [DOI] [PubMed] [Google Scholar]
- Bhattaram P, Penzo-Méndez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, Depamphilis ML, Wegner M, Lefebvre V (2010) Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T (2001) Math5 is required for retinal ganglion cell and optic nerve formation. Development 128:2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA 4th, Prasov L, Glaser T (2012) Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol 365:395–413. 10.1016/j.ydbio.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chen Q, Sun X, Shen W, Liu B, Zhong X, Leng Y, Li C, Zhang W, Chai F, Huang B, Gao Q, Xiang AP, Zhuo Y, Ge J (2010) Generation of retinal ganglion-like cells from reprogrammed mouse fibroblasts. Invest Ophthalmol Vis Sci 51:5970–5978. 10.1167/iovs.09-4504 [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80:949–957. 10.1016/0092-8674(95)90298-8 [DOI] [PubMed] [Google Scholar]
- Cizelsky W, Hempel A, Metzig M, Tao S, Hollemann T, Kühl M, Kühl SJ (2013) sox4 and sox11 function during Xenopus laevis eye development. PLoS One 8:e69372. 10.1371/journal.pone.0069372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima S, Koriyama Y, Kurimoto T, Oliveira JT, Yin Y, Li Y, Gilbert HY, Fagiolini M, Martinez AM, Benowitz L (2012) Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc Natl Acad Sci U S A 109:9149–9154. 10.1073/pnas.1119449109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy P, Penzo-Méndez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V (2008) The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res 36:3101–3117. 10.1093/nar/gkn162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek S, Dictor M, Jerkeman M, Jirström K, Borrebaeck CA (2008) Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood 111:800–805. 10.1182/blood-2007-06-093401 [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J (1996) POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A 93:3920–3925. 10.1073/pnas.93.9.3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH (1999) POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol 210:469–480. 10.1006/dbio.1999.9280 [DOI] [PubMed] [Google Scholar]
- Gill G. (2005) SUMO changes Sox for developmental diversity. Mol Cell 20:495–496. 10.1016/j.molcel.2005.11.004 [DOI] [PubMed] [Google Scholar]
- Gnedeva K, Hudspeth AJ (2015) SoxC transcription factors are essential for the development of the inner ear. Proc Natl Acad Sci U S A 112:14066–14071. 10.1073/pnas.1517371112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA (2002) Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron 33:689–702. 10.1016/S0896-6273(02)00602-5 [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW (2002) Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295:1065–1070. 10.1126/science.1069609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz J, Goldberg JL (2012) Stem cells in glaucoma. In: Stem cell biology and regenerative medicine (Tsang S, ed). New York: Humana; ). [Google Scholar]
- Hertz J, Qu B, Hu Y, Patel RD, Valenzuela DA, Goldberg JL (2014) Survival and integration of developing and progenitor-derived retinal ganglion cells following transplantation. Cell Transplant 23:855–872. 10.3727/096368913X667024 [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP (2005) Sox proteins and neural crest development. Semin Cell Dev Biol 16:694–703. 10.1016/j.semcdb.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Hoser M, Baader SL, Bosl MR, Ihmer A, Wegner M, Sock E (2007) Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci 27:5495–5505. 10.1523/JNEUROSCI.1384-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Roesch K, Cepko CL (2009) Development and neurogenic potential of Muller glial cells in the vertebrate retina. Prog Retin Eye Res 28:249–262. 10.1016/j.preteyeres.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagatha B, Divya MS, Sanalkumar R, Indulekha CL, Vidyanand S, Divya TS, Das AV, James J (2009) In vitro differentiation of retinal ganglion-like cells from embryonic stem cell derived neural progenitors. Biochem Biophys Res Commun 380:230–235. 10.1016/j.bbrc.2009.01.038 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Ding Q, Xie X, Libby RT, Lefebvre V, Gan L (2013) Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J Biol Chem 288:18429–18438. 10.1074/jbc.M113.478503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TV, Martin KR (2008) Development and characterization of an adult retinal explant organotypic tissue culture system as an in vitro intraocular stem cell transplantation model. Invest Ophthalmol Vis Sci 49:3503–3512. 10.1167/iovs.07-1601 [DOI] [PubMed] [Google Scholar]
- Joo S, Kim JY, Lee E, Hong N, Sun W, Nam Y (2015) Effects of ECM protein micropatterns on the migration and differentiation of adult neural stem cells. Sci Rep 5:13043. 10.1038/srep13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, Rao M, Yu P, Dominguez-Vidana R, Liang AC, Solimini NL, Bernardi RJ, Yu B, Hsu T, Golding I, Luo J, et al. (2012) A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335:348–353. 10.1126/science.1212728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T, Soares CA, Sitko AA, Lefebvre V, Mason C (2017) SoxC transcription factors promote contralateral retinal ganglion cell differentiation and axon guidance in the mouse visual system. Neuron 93:1110–1125.e5. 10.1016/j.neuron.2017.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren RE, Pearson RA, MacNeil A, Douglas RH, Salt TE, Akimoto M, Swaroop A, Sowden JC, Ali RR (2006) Retinal repair by transplantation of photoreceptor precursors. Nature 444:203–207. 10.1038/nature05161 [DOI] [PubMed] [Google Scholar]
- Mao CA, Tsai WW, Cho JH, Pan P, Barton MC, Klein WH (2011) Neuronal transcriptional repressor REST suppresses an Atoh7-independent program for initiating retinal ganglion cell development. Dev Biol 349:90–99. 10.1016/j.ydbio.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA (1995) Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron 15:805–819. 10.1016/0896-6273(95)90172-8 [DOI] [PubMed] [Google Scholar]
- Miller JA, Nathanson J, Franjic D, Shim S, Dalley RA, Shapouri S, Smith KA, Sunkin SM, Bernard A, Bennett JL, Lee CK, Hawrylycz MJ, Jones AR, Amaral DG, Šestan N, Gage FH, Lein ES (2013) Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development 140:4633–4644. 10.1242/dev.097212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Apara A, Goldberg JL (2011) Kruppel-like transcription factors in the nervous system: novel players in neurite outgrowth and axon regeneration. Mol Cell Neurosci 47:233–243. 10.1016/j.mcn.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L, Berti L, Masserdotti G, Covic M, Michaelidis TM, Doberauer K, Merz K, Rehfeld F, Haslinger A, Wegner M, Sock E, Lefebvre V, Couillard-Despres S, Aigner L, Berninger B, Lie DC (2012) SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J Neurosci 32:3067–3080. 10.1523/JNEUROSCI.4679-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH (2005) A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol 280:467–481. 10.1016/j.ydbio.2005.01.028 [DOI] [PubMed] [Google Scholar]
- Parameswaran S, Balasubramanian S, Babai N, Qiu F, Eudy JD, Thoreson WB, Ahmad I (2010) Induced pluripotent stem cells generate both retinal ganglion cells and photoreceptors: therapeutic implications in degenerative changes in glaucoma and age-related macular degeneration. Stem Cells 28:695–703. 10.1002/stem.320 [DOI] [PubMed] [Google Scholar]
- Penzo-Méndez AI. (2010) Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol 42:425–428. 10.1016/j.biocel.2009.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo-Méndez A, Dy P, Pallavi B, Lefebvre V (2007) Generation of mice harboring a Sox4 conditional null allele. Genesis 45:776–780. 10.1002/dvg.20358 [DOI] [PubMed] [Google Scholar]
- Prasov L, Glaser T (2012) Pushing the envelope of retinal ganglion cell genesis: context dependent function of Math5 (Atoh7). Dev Biol 368:214–230. 10.1016/j.ydbio.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L, Nagy M, Rudolph DD, Glaser T (2012) Math5 (Atoh7) gene dosage limits retinal ganglion cell genesis. Neuroreport 23:631–634. 10.1097/WNR.0b013e328355f260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD (2000) A novel human opsin in the inner retina. J Neurosci 20:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I, Rollag MD, Castrucci AM (2002) Photoreceptive net in the mammalian retina. This mesh of cells may explain how some blind mice can still tell day from night. Nature 415:493. [DOI] [PubMed] [Google Scholar]
- Roger JE, Nellissery J, Kim DS, Swaroop A (2010) Sumoylation of bZIP transcription factor NRL modulates target gene expression during photoreceptor differentiation. J Biol Chem 285:25637–25644. 10.1074/jbc.M110.142810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL (2004) Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol 271:388–402. 10.1016/j.ydbio.2004.03.039 [DOI] [PubMed] [Google Scholar]
- Savare J, Girard F (2005) SUMO modification represses transcriptional activity of Sox proteins (in French). Med Sci (Paris) 21:917–919. 10.1051/medsci/20052111917 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267:1360–1363. 10.1126/science.7871435 [DOI] [PubMed] [Google Scholar]
- Schwartz SD, Hubschman JP, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R (2012) Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 379:713–720. 10.1016/S0140-6736(12)60028-2 [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N (2012) Cis-regulatory control of corticospinal system development and evolution. Nature 486:74–79. 10.1038/nature11094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Iguchi A, Hayward DC, Technau U, Ball EE, Miller DJ (2008) Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol Biol 8:311. 10.1186/1471-2148-8-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein DC, Thalhammer JM, Hartwig AC, Crenshaw EB 3rd, Lefebvre V, Wegner M, Sock E (2010) The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J Neurochem 115:131–141. 10.1111/j.1471-4159.2010.06910.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui A, Iwagawa T, Mochizuki Y, Iida A, Wegner M, Murakami A, Watanabe S (2013a) Expression of Sox4 and Sox11 is regulated by multiple mechanisms during retinal development. FEBS Lett 587:358–363. 10.1016/j.febslet.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Usui A, Mochizuki Y, Iida A, Miyauchi E, Satoh S, Sock E, Nakauchi H, Aburatani H, Murakami A, Wegner M, Watanabe S (2013b) The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development 140:740–750. 10.1242/dev.090274 [DOI] [PubMed] [Google Scholar]
- Venugopalan P, Wang Y, Nguyen T, Huang A, Muller KJ, Goldberg JL (2016) Transplanted neurons integrate into adult retinas and respond to light. Nat Commun 7:10472. 10.1038/ncomms10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL (2007) Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci 27:8593–8603. 10.1523/JNEUROSCI.4488-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L (2001) Requirement for math5 in the development of retinal ganglion cells. Genes Dev 15:24–29. 10.1101/gad.855301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West EL, Pearson RA, MacLaren RE, Sowden JC, Ali RR (2009) Cell transplantation strategies for retinal repair. Prog Brain Res 175:3–21. 10.1016/S0079-6123(09)17501-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Kaczynski TJ, Sethuramanujam S, Li R, Jain V, Slaughter M, Mu X (2015) Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc Natl Acad Sci U S A 112:E1559–E1568. 10.1073/pnas.1421535112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm A, Sock E, Fuchshofer R, Wegner M, Tamm ER (2008) Anterior segment dysgenesis in the eyes of mice deficient for the high-mobility-group transcription factor Sox11. Exp Eye Res 86:895–907. 10.1016/j.exer.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L (2003) Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol 264:240–254. 10.1016/j.ydbio.2003.08.005 [DOI] [PubMed] [Google Scholar]