Abstract
Nonalcoholic steatohepatitis (NASH) has been identified as a source of significant interindividual variation in drug metabolism. A previous ex vivo study demonstrated significant changes in hepatic Cytochrome P450 (CYP) activity in human NASH. This study evaluated the in vivo activities of multiple CYP isoforms simultaneously in prominent diabetic NASH mouse models. The pharmacokinetics of CYP selective substrates: caffeine, losartan, and omeprazole changed significantly in a diabetic NASH mouse model, indicating attenuation of the activity of Cyp1a2 and Cyp2c29, respectively. Decreased mRNA expression of Cyp1a2 and Cyp2c29, as well as an overall decrease in CYP protein expression, was found in the diabetic NASH mice. Overall, these data suggest that the diabetic NASH model only partially recapitulates the human ex vivo CYP alteration pattern. Therefore, in vivo determination of the effects of NASH on CYP activity should be conducted in human, and more appropriate models are required for future drug metabolism studies in NASH.
Keywords: Cytochrome P450, methionine and choline deficient, nonalcoholic steatohepatitis, variable drug responses
1 INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in the United States with a prevalence of 24% in adults[1] and 10%–20% in children.[2] NAFLD encompasses a spectrum of liver manifestations ranging from simple steatosis to nonalcoholic steatohepatitis (NASH),[3] which may progress to cirrhosis and hepatocellular carcinoma.[4,5] NAFLD is commonly referred to as the hepatic manifestation of the metabolic syndrome,[6,7] because of its association with risk factors such as central obesity, diabetes mellitus, and dyslipidemia.[8,9] It is estimated that the prevalence of NAFLD among type 2 diabetics is as high as 69%,[9] and the estimated prevalence of diabetes among NAFLD and NASH patients is 22.51% and 43.63%, respectively,[1] suggesting that NAFLD/NASH and diabetes are highly associated.
NASH has been identified as a potential contributing factor to variable drug response (VDR). It has been demonstrated that NASH can alter the metabolism and disposition of many drugs, including ezetimibe, pravastatin, methotrexate, morphine, acetaminophen, and metformin.[10–15] Remodeling of the hepatic absorption, distribution, metabolism, and excretion (ADME) pathways has been identified as the general mechanism for these changes, which can exacerbate the adverse drug reactions associated with these drugs.[12] The vast majority of altered ADME gene expression changes in NAFLD were observed after the progression to NASH, suggesting that NASH is where VDR will be most likely.[16] Previously, our group has shown that ex vivo activities of Cytochrome P450s (CYPs) are altered in human NASH liver.[17]
Various experimental models have been established to mimic the pathological conditions of human NASH, although to date there is no single animal model that can accurately encompass the full spectrum of human NASH progression.[18] The leptin-deficient ob/ob mice are widely used as a model of obesity, type II diabetes, and NAFLD, as they develop steatosis spontaneously.[19,20] In addition, they exhibit metabolic signatures including insulin resistance, hyperinsulinemia, hyperglycemia, and hyperlipidemia.[21] However, additional stimuli are required to drive the pathological development of NASH features, such as inflammation and fibrosis.[22] The methionine- and choline-deficient (MCD) diet, which disrupts the hepatic β-oxidation and the production of very low-density lipoprotein in animals, is widely employed to induce NASH in animal models.[18,23] The ob/ob MCD model develops mRNA and protein expression profiles of hepatic transporters that are consistent with human NASH[24] and was employed as a model for NASH in several mechanistic studies.[15,25,26] A 4-week MCD diet was fed to ob/ob mice to create our experimental diabetic NASH model.
In vivo analysis of CYP activity is the most accurate way to assess CYP function.[27] Administration of probe drugs in a cocktail approach is a preferable method to analyze in vivo activity of multiple CYP isoforms simultaneously.[28] Probe drugs used in this study include caffeine, losartan, omeprazole, midazolam. and dextromethorphan, which are the selective substrates of human CYP1A2, CYP2C9, CYP2C19, CYP3A4, and CYP2D6 isoforms respectively. In the present study, we characterized the reported mouse orthologs, or highly potential counterparts of human CYP isoforms listed above: Cyp1a2, Cyp2c29, Cyp2d22, and Cyp3a11, to reflect NASH-associated changes in CYP activity in mouse models.[29–32]
The purpose of the present study was to determine the in vivo alterations of the major CYP enzymatic activities in ob/ob and ob/ob MCD mouse models. By comparing these data to a previous ex vivo human study,[17] we determined whether this model can accurately replicate the CYP remodeling seen in human NASH.
2 MATERIAL AND METHODS
2.1 Chemicals and reagents
Caffeine, losartan, omeprazole, midazolam, 1′-hydroxymidazolam, phenacetin dextromethorphan, and dextrophan were purchased from Sigma-Aldrich (St Louis, MO). Paraxanthine, E3174 (losartan carboxylic acid), and 5-hydroxyomeprazole were purchased from Toronto Research Chemicals (Toronto, Ontario Canada). All other chemicals were obtained from commercial sources.
2.2 Animals
C57BL/6J wild-type (WT) and leptin-deficient (ob/ob) male mice were purchased from Jackson Laboratory (Bar Harbor, ME) at 8 weeks of age (n = 5 for each group) and housed on a 12-h light and 12-h dark cycle in the University of Arizona animal care facility. The animal studies were approved by the University of Arizona Animal Care and Use Committee. The mice in WT and ob/ob groups were fed a methionine and choline resupplemented diet (Dyets, Bethlehem, PA) for 4 weeks, whereas the mice in the ob/ob MCD group were fed a MCD diet for 4 weeks to induce NASH (Dyets, Bethlehem, PA).
2.3 Histopathological analysis and plasma chemistries
Formalin-fixed and paraffin-embedded liver tissue were stained with hematoxylin and eosin (H&E) and pathology scored by a veterinary pathologist. Liver tissues were scored for steatosis, necrosis, inflammation, fibrosis, and biliary hyperplasia. Pathology scores were as follows: 0, no significant lesions (0%); 1, minimal (<10%); 2, mild (10~25%); 3, moderate (25~40%); 4, marked (40~50%); 5, severe (>50%). Whole blood was centrifuged for 15 min at 5000 rpm, 4°C to get plasma. Plasma collected prior to gavage was used to quantify peripheral blood insulin and glucose concentrations. Insulin was measured using an ELISA kit (Millipore, Billerica, MA), and glucose was measured using an absorbance-based assay kit (Abcam, Cambridge, MA), according to the manufactures’ protocol.
2.4 In vivo pharmacokinetics of probe drugs
Mice were dosed by oral gavage with five-drug cocktail comprising caffeine (5 mg/kg), losartan (40 mg/kg), omeprazole (15 mg/kg), dextromethorphan (20 mg/kg), and midazolam (5 mg/kg). In all cases, whole blood (30 μL) was taken from the tail vein at intervals of drug postadministration (30, 60, 120, 240, and 360 min) into a tube coated with heparin. Plasma samples were obtained by centrifugation.
2.5 Sample preparation and LC-MS/MS analysis
The plasma samples (5 μL) were spiked with an internal standard (10 μL of 400 ng/mL phenacetin) and mixed with 100 μL of acetonitrile for protein precipitation. Chromatographic separation was achieved using a gradient system of 0.1% (v/v) formic acid in water and 0.1%(v/v) formic acid in acetonitrile as follows: The HPLC column is a Luna® 3μm C18(2) 100Å LC column 100 × 4.6mm (Phenomenex, Torrance, CA) with a C-18 guard cartridge 3*4 mm. Analysis was performed on a TSQ Quantum Ultra triple quadrupole mass spectrometer with Surveyor MS pump plus and Surveyor Antosampler plus (Thermo Electron, San Jose, CA). The mass spectrometer was operated in electrospray positive ionization mode with a spray voltage of 4900 V, sheath gas (nitrogen) flow rate of 49 arbitrary units, and auxiliary gas flow rate of 5 arbitrary units. The capillary temperature is 320°C. Argon was used as the collision gas at a pressure of 0.8 mTorr. The mass transitions were as follows: m/z 195→138 for Caffeine, m/z 181→124 for paraxanthine, m/z 346→198 Omeprazole, m/z 362→214 for 5-hydroxyomeprazole, m/z 326→291 for midazolam, m/z 342→324 for 1-hydroxymidazolam, m/z 423→207 for losartan, m/z 437→207 for EXP-3174, m/z 272→215 for dextromethorphan, m/z 258→157 for dextrorphan, and m/z 180→110 for phenacetin.
2.6 RNA isolation and mRNA analysis
Total RNA was extracted from liver tissues using RNAzol B reagent from Tel-Test (Friendswood, TX) according to the manufacture’s protocol. Equal amounts of RNA (1 μg) were used for reverse transcription using the ReadyScriptTM cDNA synthesis mix (Sigma). The following forward and the reverse primers were obtained from Sigma Aldrich: mCyp1a2, forward (5′-CAATGACATCTTTGGAGCTG-3′) and reverse (5′-GATGAAGGCCTCTAGATATGG-3′); mCyp2c29, forward (5′-CAGATGTCACAGCTAAAGTC-3′) and reverse (5′-TTTAATG-TCACAGGTCACTG-3′); mCyp2d22, forward (5′-GTTGTACTAAAT-GGGCTGAC-3′) and reverse (5′-GCTAGGACTATACCTTGAGAG-3′); mCyp3a11, forward (5′-CTGACACCAGTATATGAGATG-3′) and reverse (5′-GGCTTTATGAGAGACTTTGTC-3′); mActin-beta, forward (5′-ACGGCCAACCGTGAAAAGAT-3′) and reverse (5′-GTGGTAC-GACCAGAGGCATAC-3′). qRT-PCR was performed on the LightCycler 480 system (Roche) as follows: one cycle of initial denaturation (4 min at 95°C), 45 cycles of amplification (10 s at 95°C and 30 s at 60°C), and a cooling period. The data presented are relative mRNA levels normalized to the level of Actin-beta, and the average value from the WT control group was set as 1.
2.7 Immunoblot protein preparation and analysis
Three hundred milligrams of liver tissue was homogenized in NP40 lysis buffer (20 mM Tris–HCl, 137 mM NaCl, 10% glycerol, 1% non-idet P-40, and 2 mM ethylenediaminetetraacetic acid EDTA with one protease inhibitor cocktail tablet (Roche, Indianapolis, IN) per 25 mL) on the ice. Homogenized tissue was then centrifuged at 10,000×g for 30 min, and then the supernatant was transferred to a new collection vial. Protein concentrations in the collected supernatant were quantified by the Pierce BCA Protein Quantitation Assay kit (Thermo Fisher Scientific, Waltham, MA) following the manufacture’s protocol.
Fifty micrograms of the whole cell lysate in each well was separated on 7.5% SDS–PAGE gels and transferred to polyvinylidene difluoride membranes. The following antibodies were used for the detection of proteins: Erk2 (Santa Cruz Biotechnology, Dallas, TX, sc-154), Cyp1a2 (sc-9835), Cyp3a (sc-25845), Cyp2d22 (Abcam, Cambridge, MA, Ab62204), and Cyp2c29 (Ab137015). Densitometry analysis of the Western blot was performed using ImageJ software (National Institutes of Health, Bethesda, MD), and relative protein expression was calculated by normalizing to the housekeeping protein Erk2 (sc-154).
2.8 Concordance analysis across human and ob/ob MCD mice
Concordance analysis was performed to compare diabetic NASH mice to human NASH on the enzymatic activity, mRNA expression, and protein expression of the CYP isoforms: CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4. The analysis was conducted by measuring the effect size of NASH versus control, in the mice is the ob/ob MCD versus WT whereas in human is NASH with/without fatty versus normal, of either enzymatic activity, mRNA expression or protein expression. The effect size (estimated by Glass’s D) is defined as the standardized mean difference between two populations and can be calculated using the following equation:
where μ1 and μ2 are the sample means for two groups, and Sp is the pooled standard deviation for both. Data of this study were compared to the published data in human.[17] The analysis was performed using R version 3.2.3 with packages of gdata_2.17.0, xtable_1.8-0, ggplot2_1.0.1, and knitr_1.11 (http://www.r-project.org/).
2.9 Statistical analysis
All results are represented as mean ± standard error of the mean (SEM). For all comparisons within this study, one-way analysis of variance (ANOVA) statistical analysis was employed with a Tukey’s multiple comparisons posttest to compare WT versus ob/ob mice, WT versus ob/ob MCD mice, and ob/ob versus ob/ob MCD mice. All analysis was carried out using GraphPad Prism software (GraphPad Software, La Jolla, CA).
3 RESULTS
3.1 NASH phenotype in mouse models
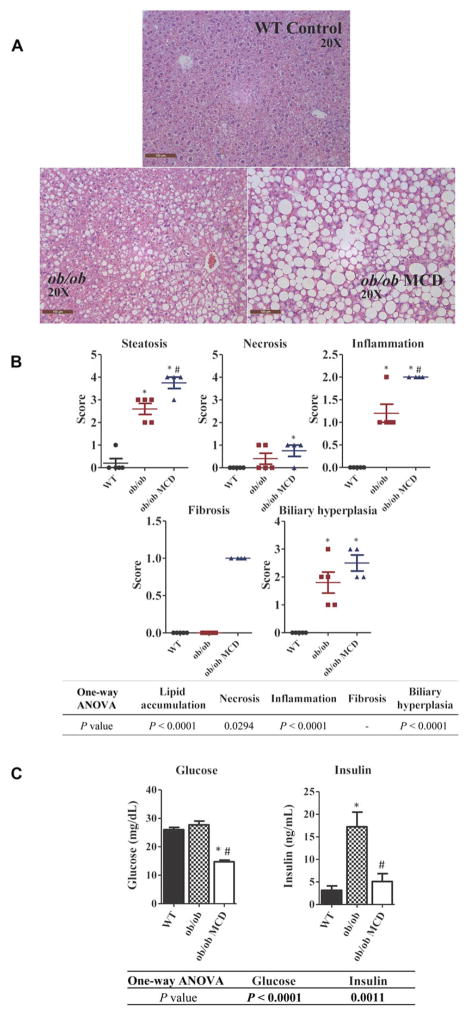
H&E stained liver tissue sections from representative WT, ob/ob and ob/ob MCD mice are shown in Figure 1A. Livers were evaluated, and incidence/severity was scored by a board-certified veterinary pathologist (Figure 1B). Macrovesicular steatosis, a distinguishing characteristic of NASH, was only observed in the ob/ob MCD mice. The ob/ob MCD exhibited greater incidence/severity scores on each of the typical NASH histological diagnoses, including hepatic steatosis, inflammation, fibrosis, biliary hyperplasia, and hepatocyte necrosis compared to the WT (Figure 1B). To determine the diabetic phenotype, plasma glucose and insulin levels were examined in each group of mice. ob/ob mice displayed a diabetic phenotype with higher levels insulin. The glucose levels significantly decreased in ob/ob MCD mice, but a significant change of insulin level was not observed in ob/ob MCD mice (Figure 1C).
FIGURE 1.
Histological and pathological features of NASH in WT, ob/ob, and ob/ob MCD mice. Pictures of representative H&E-stained liver sections from WT, ob/ob, and ob/ob MCD mice were taken in original magnification of 20× (A). Histopathological analysis of liver from WT, ob/ob, and ob/ob MCD mice on steatosis, necrosis, inflammation, fibrosis, and biliary hyperplasia (B). Plasma collected from WT, ob/ob, and ob/ob MCD mice was analyzed for peripheral blood glucose and insulin concentrations (C). Data represent mean ± SEM. WT n = 5, ob/ob n = 5, ob/ob MCD n = 5. One-way ANOVA with Tukey’s multiple comparisons posttest were performed to determine the statistical significance. * Indicates a significant difference between ob/ob and/or ob/ob MCD with WT mice, whereas # indicates a significant difference between ob/ob and ob/ob MCD mice.
3.2 In vivo pharmacokinetics of a drug cocktail in NASH models
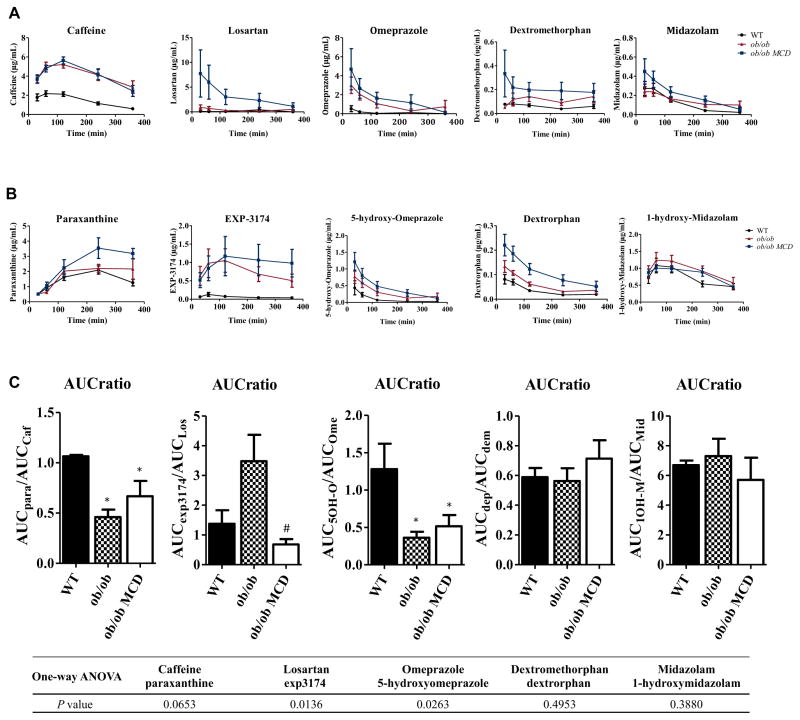
A single dose oral pharmacokinetics study was performed using a cocktail of CYP probe drugs to determine the activity of the respective CYP isoforms. The plasma concentration of the parent drugs (caffeine, losartan, omeprazole, midazolam, and dextromethophan) and their metabolites (paraxanthine, EXP3174, 5-hydroxyomeprazole, 1-hydroxymidazolam, and dextrorphan) was measured by LC-MS/MS. The pharmacokinetic curves of each probe drug and metabolite are shown in Figures 2A and 2B, respectively, and the pharmacokinetic parameters are summarized in Table 1. The maximum concentration (Cmax) and the area under curve (AUC) of caffeine in diabetic and diabetic NASH mice were significantly increased, although the drug half-life (t1/2) was not significantly changed in diabetic or diabetic NASH mice (Table 1). The metabolism AUC ratio, defined as the ratio of metabolite AUC to parent drug AUC, of each probe drug was calculated to represent the respective enzymatic activity. The metabolism AUC ratio of caffeine and omeprazole was significantly decreased in diabetic and diabetic NASH mice compared to WT mice. The metabolism AUC ratio of losartan was significantly decreased in the diabetic NASH mice compared to diabetic mice (Figure 2C).
FIGURE 2.
Pharmacokinetics of the five probe drugs and their metabolites. Pharmacokinetics curves of the five probe drugs: caffeine, omeprazole, midazolam, losartan, and dextromethorphan (A) and their metabolites: paraxanthine, 5-hydroxy-omeprazole, 1-hydroxy-midazolam, EXP-3174, and dextrotphan (B) were graphed based on the time-course plasma concentrations. The AUC ratio of each pair of probe drug and the metabolite, calculated based on the AUC of each pharmacokinetic curve of each compound, represents the relative enzymatic activity of the corresponding CYP isoform (C). Data represent mean ± SEM. WT n = 5, ob/ob n = 5, ob/ob MCD n = 5. One-way ANOVA with Tukey’s multiple comparisons posttest were performed to determine the statistical significance. * Indicates a significant difference between ob/ob and/or ob/ob MCD with WT mice, whereas # indicates a significant difference between ob/ob and ob/ob MCD mice.
TABLE 1.
Comparison of Pharmacokinetics Parameters of the Probe Drugs
| Group | Cmax (μg/mL) | AUC (min·μg/mL) | AUCmetabolite/AUCdrug | t1/2 (min) | |
|---|---|---|---|---|---|
| Caffeine | WT | 2.0 ± 1.1 | 491.1 ± 91.3 | 1.06 ± 0.03 | 158 ± 71 |
| ob/ob | 4.9 ± 0.9 * | 1418.0 ± 341.8 * | 0.46 ± 0.17 * | 342 ± 236 | |
| ob/ob MCD | 5.1 ± 1.2 * | 1431.0 ± 325.7 * | 0.67 ± 0.34 * | 221 ± 131 | |
| Omeprazole | WT | 0.6 ± 0.7 | 36.2 ± 31.7 | 1.28 ± 0.76 | 70 ± 45 |
| ob/ob | 3.1 ± 1.9 | 314.5 ± 195.4 | 0.36 ± 0.18 * | 56 ± 13 | |
| ob/ob MCD | 4.7 ± 4.8 | 486.8 ± 450.7 | 0.52 ± 0.34 * | 77 ± 31 | |
| Midazolam | WT | 0.30 ± 0.15 | 36.7 ± 11.8 | 6.70 ± 0.69 | 88 ± 38 |
| ob/ob | 0.26 ± 0.10 | 47.9 ± 18.5 | 7.30 ± 2.60 | 140 ± 53 | |
| ob/ob MCD | 0.47 ± 0.27 | 66.1 ± 37.2 | 5.71 ± 3.33 | 113 ± 32 | |
| Losartan | WT | 0.15 ± 0.13 | 64.6 ± 115.1 | 1.38 ± 1.00 | 114 ± 51 |
| ob/ob | 0.94 ± 1.12 | 123.9 ± 109.0 | 3.48 ± 1.97 | 90 ± 32 | |
| ob/ob MCD | 7.8 ± 10.50 | 1006.0 ± 1135.0 | 0.68 ± 0.41 # | 132 ± 69 | |
| Dextromethorphan | WT | 0.08 ± 0.03 | 19.1 ± 7.8 | 0.59 ± 0.14 | 123 ± 30 |
| ob/ob | 0.17 ± 0.10 | 37.0 ± 23.3 | 0.56 ± 0.19 | 120 ± 35 | |
| ob/ob MCD | 0.36 ± 0.43 | 65.9 ± 55.3 | 0.71 ± 0.28 | 172 ± 131 |
Noncompartmental analysis was applied to calculate AUC, terminal half-life (t1/2), maximum plasma concentration (Cmax), and the AUC ratio was calculated based on the AUC value of the metabolite and the parent drug.
Data represent mean ± SEM. WT n = 5, ob/ob n = 5, ob/ob MCD n = 5. One-way ANOVA with Tukey’s multiple comparisons posttest were performed to determine the statistical significance
Indicates a significant difference between ob/ob and/or ob/ob MCD with WT mice, whereas # indicates a significant difference between ob/ob and ob/ob MCD mice.
3.3 Hepatic CYP mRNA expression alteration in diabetic NASH models
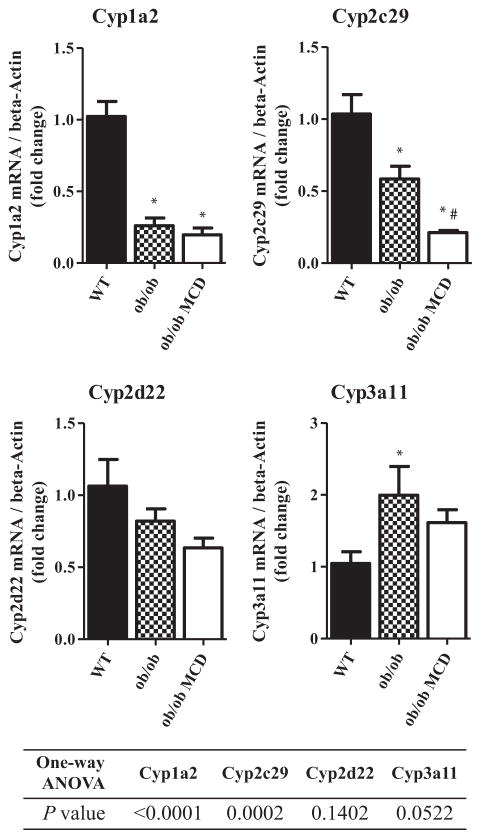
The hepatic mRNA expression of the major CYP isoforms, Cyp1a2, Cyp2c29, Cyp2d22, and Cyp3a11 was normalized to mRNA expression of beta-actin, and the fold changes are shown in Figure 3. The Cyp1a2 mRNA expression level was significantly decreased in diabetic and diabetic NASH mice. The diabetic mice had a decreased mRNA level of Cyp2c29 compared to the WT mice. Additionally, the mRNA level of Cyp2c29 was significantly decreased in the diabetic NASH mice compared to both the WT and the diabetic mice. The mRNA level of Cyp3a11 was significantly increased in the diabetic mice compared to wild type.
FIGURE 3.
Hepatic mRNA expression of murine CYP isoforms in WT, ob/ob, and ob/ob MCD mice. Hepatic mRNA expression of Cyp1a2, Cyp2c29, Cyp2d22, and Cyp3a11 was quantified and normalized to beta-Actin in WT, ob/ob, and ob/ob MCD mice. Data represent mean ± SEM. WT n = 5, ob/ob n = 5, ob/ob MCD n = 5. One-way ANOVA with Tukey’s multiple comparisons posttest were performed to determine the statistical significance. * Indicates a significant difference between ob/ob and/or ob/ob MCD with WT mice, whereas # indicates a significant difference between ob/ob and ob/ob MCD mice.
3.4 Hepatic CYP protein content alteration in diabetic NASH models
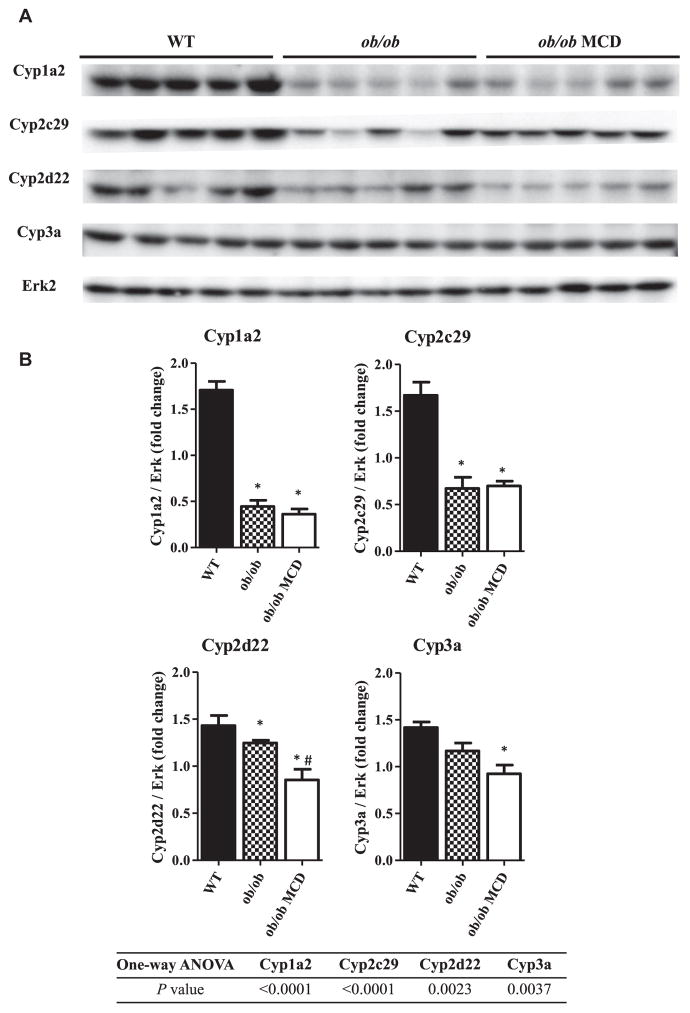
Western blot analysis was performed to determine the relative protein content of the major CYPs in mice of each group (Figure 4A). The fold change of protein content of each CYP isoform, which is normalized to Erk protein content, is shown in Figure 4B. In the diabetic mice, the protein expression of Cyp1a2, Cyp2c29, and Cyp2d22 was significantly decreased. Furthermore, a significant decrease in protein content of Cyp1a2, Cyp2c29, Cyp2d22, and Cyp3a was observed in the diabetic NASH mice compared to the WT mice. However, the only significant change in the diabetic NASH mice compared to the diabetic mice was the decreased Cyp2d22 protein content.
FIGURE 4.
Hepatic protein content of murine CYP isoforms in WT, ob/ob and ob/ob MCD mice. Hepatic protein expression of Cyp1a2, Cyp2c29, Cyp2d22, and Cyp3a11 were measured in WT, ob/ob, and ob/ob MCD mice by Western blot (A). Densitometry analysis of Western blot was performed, and relative protein expression of each CYP isoform to Erk protein was shown (B). Data represent mean ± SEM. WT n = 5, ob/ob n = 5, ob/ob MCD n = 5. One-way ANOVA with Tukey’s multiple comparisons posttest were performed to determine the statistical significance. * Indicates a significant difference between ob/ob and/or ob/ob MCD with WT mice, whereas # indicates a significant difference between ob/ob and ob/ob MCD mice.
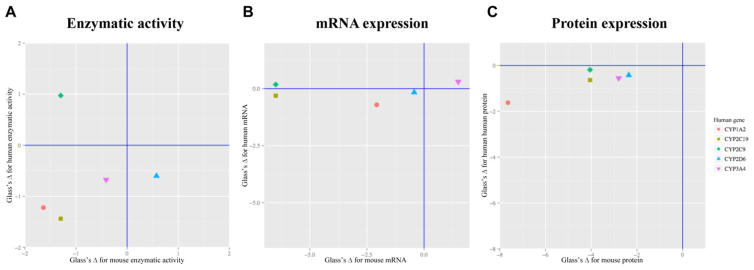
3.5 Concordance analysis across human and mouse CYP activities, mRNA, and protein expression in NASH
To determine the extent to which the diabetic NASH model recapitulates the alterations in CYP mRNA, protein expression and enzymatic activities in human NASH, a concordance analysis was performed using previously published human NASH data.[17] Since Cyp2c29 was reported as the mouse ortholog of both CYP2C9 and CYP2C19 in human,[29,30] Cyp2c29 was compared to both CYP2C9 and CYP2C19. Effect sizes in the same direction (the lower left or upper right quadrant) represent similar trends of change, whereas the magnitude of change corresponds to the statistical power in the detection of expression difference in NASH versus control. Figure 5A represents the comparison of enzymatic activities. Three pairs of CYP isoforms changed in the same direction, which are CYP1A2, CYP2C19, and CYP3A4 in human. The mRNA expression of Cyp1a2, Cyp2c29, and Cyp2d22 changed in the same direction as their corresponding human genes CYP1A2, CYP2C19, and CYP2D6, though the magnitude of that change varied widely (Figure 5B). While all the five comparisons pairs indicated the same direction of decreased protein expression (Figure 5C), there was large variability in the magnitude of that change.
FIGURE 5.
Effective size analysis of ob/ob MCD mouse and human enzymatic activity, mRNA expression, and protein expression of five CYP isoforms. Comparison of effect size of human NASH on CYP isoform enzymatic activity (A), mRNA expression (B), and protein expression (C) changes versus the diabetic NASH mouse model changes. Positive effect reflects increase, whereas negative effect changes reflect decrease. The points in the lower left and upper right quadrant represent the same direction of changes, whereas the dots in the upper left or lower right quadrant represent the opposite direction of changes. Values were acquired by the method described in the Materials and Method section.
4 DISCUSSION
An ideal animal model that can accurately reflect the diverse population of patients with NASH is lacking, which limits preclinical risk assessment or predictability of VDRs in NASH patients. In this present study, we utilized the ob/ob MCD mouse model as our investigative tool, because it may predict both histomorphological and some of the metabolic or biochemical features of human NASH. The typical lesion of macrovesicular steatosis as well as a certain degree of inflammation and fibrosis was repeated in the ob/ob MCD mice in this study as previously demonstrated[15,33] (Figures 1A and 1B). Additionally, insulin and glucose levels were also consistent with the development of diabetes as previously shown[15] (Figure 1C).
Significant changes in probe drug pharmacokinetics were found in the diabetic NASH mice, suggesting alterations in the in vivo activities of their corresponding CYP isoforms. The pharmacokinetics of caffeine, the probe drug of Cyp1a2, as well as omeprazole and losartan, the two probe drugs of the Cyp2c subfamily, were significantly changed, directly indicating a decrease in enzymatic activity (Figure 2C). The mRNA expression of Cyp1a2 and Cyp2c29 was significantly decreased in both the obese diabetic mice and the diabetic NASH mice (Figure 3), whereas an overall decrease of protein expression of Cyp1a2, Cyp2c29, Cyp2d22, and Cyp3a11 was found in the diabetic NASH mice (Figure 4). Accordingly, data of enzymatic activity, mRNA expression, and protein expression of the murine CYP isoforms are not in complete concordance with each other, which may be indicative of the heterogeneity in the regulation mechanisms known for each isoform. Others have reported altered CYP activities and expression levels in multiple rodent models of diabetes and NASH. A study by Watson et al. reported no significant change of Cyp1a activity in ob/ob mice.[34] A separate study in MCD mice reported a decrease in mRNA expression of Cyp1a2 and an increase in Cyp3a11.[35] Lam et al reported a significant increase in Cyp2c29 mRNA expression in 10-week old db/db mice, which are the leptin receptor deficient mice exhibiting similar diabetic phenotype as ob/ob mice, and a significant decrease in expression in 25-week old db/db mice compared to their 25-week old control.[36] Yoshinari and co-workers observed an increase in both mRNA and protein expression of Cyp2c29 in 10-week old db/db mice.[37] Finally, a microarray study, intended to identify the general gene expression alterations in the ob/ob, db/db, and HFD-fed C57BL/6J mice, revealed that the dramatically changed genes were highly interconnected and significantly enriched for processes involving metabolism by CYPs.[38] Collectively, these data indicate that, while there are similarities between mouse models of diabetes and NASH, they are not all the same and care should be taken to understand the changes in CYP-mediated drug metabolism.
The concordance analysis between mouse in vivo and previously published human ex vivo data for CYP activity shows that even though the diabetic NASH mouse model we used in this study has been demonstrated to accurately reflect the drug transporter alteration profiles in humans, it partially recapitulates the alteration pattern of CYP1A2, CYP2C19, and CYP3A4, but fails to resemble the alterations of CYP2C9 and CYP2D6 observed in human NASH.
In conclusion, while the ob/ob MCD model accurately recapitulates certain aspects of human NASH including insulin insensitivity, pathological hallmarks, and transporter expression profiles, it cannot precisely reflect the alteration in the major CYP activities for drug metabolism. Studies modeling the effect of NASH on the disposition of drugs through the action of drug transporters may be highly relevant to human NASH patients, but caution should be used when extrapolating any CYP-mediated drug metabolism. More comprehensive human studies should be performed to evaluate the in vivo CYP activity in NASH patients. Additional effort should be devoted to establish a more accurate model of CYP regulation in the progression of NASH to ensure the safety of this at risk patient population.
Acknowledgments
Contract Grant Sponsor: National Institutes of Health.
Contract Grant Numbers: ES006694 and HD062489.
Contract Grant Sponsor: National Institute of Environmental Health Science. Toxicology Training Grant.
Contract Grant Number: ES007091.
The authors thank Wade Chew for helping with drug quantification using LC-MS/MS.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Hepatology. 2015 doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Schwimmer JB, Deutsch R, Kahen T, Lavine JE, Stanley C, Behling C. Pediatrics. 2006;118:1388–1393. doi: 10.1542/peds.2006-1212. [DOI] [PubMed] [Google Scholar]
- 3.Tiniakos DG, Vos MB, Brunt EM. Annu Rev Pathol. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 4.Wree A, Broderick L, Canbay A, Hoffman HM, Feldstein AE. Nat Rev Gastroenterol Hepatol. 2013;10:627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 5.Michelotti GA, Machado MV, Diehl AM. Nat Rev Gastroenterol Hepatol. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 6.Cortez-Pinto H, Camilo ME, Baptista A, De Oliveira AG, De Moura MC. Clin Nutr. 1999;18:353–358. doi: 10.1016/s0261-5614(99)80015-6. [DOI] [PubMed] [Google Scholar]
- 7.Friis-Liby I, Aldenborg F, Jerlstad P, Rundström K, Björnsson E. Scand J Gastroenterol. 2004;39:864–869. doi: 10.1080/00365520410006431. [DOI] [PubMed] [Google Scholar]
- 8.Karim MF, Al-Mahtab M, Rahman S, Debnath CR. Mymensingh Med J. 2015;24:873–880. [PubMed] [Google Scholar]
- 9.Leite NC, Salles GF, Araujo ALE, Villela-Nogueira CA, Cardoso CRL. Liver Int. 2009;29:113–119. doi: 10.1111/j.1478-3231.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardwick RN, Fisher CD, Street SM, Canet MJ, Cherrington NJ. Drug Metab Dispos. 2012;40:450–460. doi: 10.1124/dmd.111.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke JD, Hardwick RN, Lake AD, Lickteig AJ, Goedken MJ, Klaassen CD, Cherrington NJ. J Hepatol. 2014;61:139–147. doi: 10.1016/j.jhep.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwick RN, Clarke JD, Lake AD, Canet MJ, Anumol T, Street SM, Merrell MD, Goedken MJ, Snyder SA, Cherrington NJ. Toxicol Sci. 2014;142:45–55. doi: 10.1093/toxsci/kfu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dzierlenga AL, Clarke JD, Hargraves TL, Ainslie GR, Vanderah TW, Paine MF, Cherrington NJ. Pharmacol Exp Ther. 2015;352:462–470. doi: 10.1124/jpet.114.220764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canet MJ, Merrell MD, Hardwick RN, Bataille AM, Campion SN, Ferreira DW, Xanthakos SA, Manautou JE, A-Kader HH, Erickson RP, Cherrington NJ. Drug Metab Dispos. 2015;43:829–835. doi: 10.1124/dmd.114.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke JD, Dzierlenga AL, Nelson NR, Li H, Werts S, Goedken MJ, Cherrington NJ. Diabetes. 2015;64:3305–3313. doi: 10.2337/db14-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, Cherrington NJ. Drug Metab Dispos. 2011;39:1954–1960. doi: 10.1124/dmd.111.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, Cherrington NJ. Drug Metab Dispos. 2009;37:2087–2094. doi: 10.1124/dmd.109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi Y, Soejima Y, Fukusato T. World J Gastroenterol. 2012;18:2300–2308. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindström P. ScientificWorldJournal. 2007;7:666–685. doi: 10.1100/tsw.2007.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austin BP, Garthwaite TL, Hagen TC, Stevens JO, Menahan LA. Exp Gerontol. 1984;19:121–132. doi: 10.1016/0531-5565(84)90015-9. [DOI] [PubMed] [Google Scholar]
- 21.Garthwaite TL, Martinson DR, Tseng LF, Hagen TC, Menahan LA. Endocrinology. 1980;107:671–676. doi: 10.1210/endo-107-3-671. [DOI] [PubMed] [Google Scholar]
- 22.Ibrahim SH, Hirsova P, Malhi H, Gores GJ. Dig Dis Sci. 2015;61:1325–1336. doi: 10.1007/s10620-015-3977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serviddio G, Giudetti AM, Bellanti F, Priore P, Rollo T, Tamborra R, Siculella L, Vendemiale G, Altomare E, Gnoni GV. PLoS One. 2011;6:e24084. doi: 10.1371/journal.pone.0024084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canet MJ, Hardwick RN, Lake AD, Dzierlenga AL, Clarke JD, Cherrington NJ. Drug Metab Dispos. 2014;42:586–595. doi: 10.1124/dmd.113.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lima VMR, de Oliveira CPMS, Sawada LY, Barbeiro HV, de Mello ES, Soriano FG, Alves VAF, Caldwell SH, Carrilho FJ. Liver Int. 2007;27:227–234. doi: 10.1111/j.1478-3231.2006.01405.x. [DOI] [PubMed] [Google Scholar]
- 26.Jha P, Knopf A, Koefeler H, Mueller M, Lackner C, Hoefler G, Claudel T, Trauner M. Biochim Biophys Acta. 2014;1842:959–970. doi: 10.1016/j.bbadis.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hisaka A, Ohno Y, Yamamoto T, Suzuki H. Pharmacol Ther. 2010;125:230–248. doi: 10.1016/j.pharmthera.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Spaggiari D, Geiser L, Daali Y, Rudaz S. Anal Bioanal Chem. 2014;406:4857–4887. doi: 10.1007/s00216-014-7915-4. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin LA, Dickmann LJ, Wolf CR, Henderson CJ. Drug Metab Dispos. 2008;36:1322–1331. doi: 10.1124/dmd.108.021261. [DOI] [PubMed] [Google Scholar]
- 30.Hart SN, Cui Y, Klaassen CD, Zhong X. Drug Metab Dispos. 2009;37:116–121. doi: 10.1124/dmd.108.023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann C, van Waterschoot RAB, Harmsen S, Maier A, Gutmann H, Schinkel AH. Eur J Pharm Sci. 2009;36:565–571. doi: 10.1016/j.ejps.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Singh S, Singh K, Patel DK, Singh C, Nath C, Singh VK, Singh RK, Singh MP. Rejuvenation Res. 2009;12:185–197. doi: 10.1089/rej.2009.0850. [DOI] [PubMed] [Google Scholar]
- 33.Canet MJ, Hardwick RN, Lake AD, Dzierlenga AL, Clarke JD, Cherrington NJ. Drug Metab Dispos. 2014;42:586–595. doi: 10.1124/dmd.113.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson AM, Poloyac SM, Howard G, Blouin RA. Drug Metab Dispos. 1999;27:695–700. [PubMed] [Google Scholar]
- 35.Yamazaki Y, Kakizaki S, Horiguchi N, Sohara N, Sato K, Takagi H, Mori M, Negishi M. Gut. 2007;56:565–574. doi: 10.1136/gut.2006.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam JL, Jiang Y, Zhang T, Zhang EY, Smith BJ. Drug Metab Dispos. 2010;38:2252–2258. doi: 10.1124/dmd.110.034223. [DOI] [PubMed] [Google Scholar]
- 37.Yoshinari K, Takagi S, Sugatani J, Miwa M. Biol Pharm Bull. 2006;29:1634–1638. doi: 10.1248/bpb.29.1634. [DOI] [PubMed] [Google Scholar]
- 38.Hennig EE, Mikula M, Goryca K, Paziewska A, Ledwon J, Nesteruk M, Woszczynski M, Walewska-Zielecka B, Pysniak K, Ostrowski J. J Cell Mol Med. 2014;18:1762–1772. doi: 10.1111/jcmm.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]