Summary
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by breakdown of tolerance to nucleic acids and highly diverse clinical manifestations. To assess its molecular heterogeneity, we longitudinally profiled the blood transcriptome of 158 pediatric patients. Using mixed models accounting for repeated measurements, demographics, treatment, disease activity (DA) and nephritis class, we confirmed a prevalent IFN signature and identified a plasmablast signature as the most robust biomarker of DA. We detected gradual enrichment of neutrophil transcripts during progression to active nephritis, and distinct signatures in response to treatment in different nephritis subclasses. Importantly, personalized immunomonitoring uncovered individual correlates of disease activity that enabled patient stratification into seven groups, which were supported by patient genotypes. Our study uncovers the molecular heterogeneity of SLE and provides an explanation for the failure of clinical trials. This approach may improve trial design and implementation of tailored therapies in genetically and clinically complex autoimmune diseases.
Introduction
SLE is an incurable systemic autoimmune disease predominantly affecting young women. Patients characteristically produce autoantibodies against double-stranded DNA (dsDNA), ribonucleoproteins (RNPs), cardiolipin and phospholipids, among others. Autoantibodies form immune complexes (ICs) that deposit in many different organs such as the skin, joints and kidneys, leading to rashes, arthritis and lupus nephritis (LN). The disease course is unpredictable, with periods of remission and flares that lead to cumulative damage over time (Tsokos, 2011). This clinical heterogeneity calls for treatment personalization according to underlying molecular mechanisms.
Classification of SLE requires the presence of four out of eleven criteria. Disease activity (DA) is measured using any of six validated composite scores, including the SLE Disease Activity Index (SLEDAI), a weighted metric combining 24 components (Bombardier et al., 1992). These scoring systems, while useful, only quantify a non-exhaustive set of parameters. Because SLE is heterogeneous, not all manifestations are included in the SLEDAI, making reliable patient assessment challenging. Thus, there is a significant need for biomarkers of disease pathogenesis and DA.
SLE is primarily managed with hydroxychloroquine (HC), corticosteroids and immunosuppressive agents such as mycophenolate mofetil (MMF) and cyclophosphamide (Bernatsky et al., 2006). An anti-BAFF human monoclonal antibody (belimumab) was recently approved (Navarra et al., 2011), representing the only novel therapy with SLE indication in more than 50 years. Failure to achieve primary endpoints in clinical trials targeting CD20 with rituximab and IFNα with rontalizumab (Ugarte-Gil and Alarcon, 2014) suggests that SLE is molecularly heterogeneous. Therefore, stratification of patients according to immune networks that associate with DA may facilitate the development of customized therapies using rational clinical trial designs.
SLE patients display unique blood transcriptional signatures linked to type I interferon (IFN) and granulocytes (Baechler et al., 2003; Bennett et al., 2003; Chaussabel et al., 2008; Chiche et al., 2014). Preliminary work suggests that these signatures can be used to assess DA (Chaussabel et al., 2008). Most studies have focused on IFN-induced transcripts or proteins as biomarkers (Chiche et al., 2014; Kirou et al., 2005; Petri et al., 2009) and suffer from sample size limitations and disease-inherent clinical and therapeutic heterogeneity, making data interpretation difficult. This underscores the need to assess larger cohorts longitudinally, to use unbiased approaches that incorporate all elements of the signature, and to account for disease heterogeneity and clinical covariates during data interpretation.
We aimed to identify immune correlates of DA in children with SLE, who present with aggressive disease and lack co-morbidities that might confound data interpretation in older age groups. To this end, we transcriptionally profiled 924 longitudinal blood samples from 158 pediatric patients. To analyze these data, we developed models that incorporate DA, demographics, treatments, SLEDAI components and nephritis classes. Finally, we developed a personalized transcriptional immunomonitoring approach. This enabled patient stratification based on the immune networks best correlating with DA in each patient. The accuracy of this stratification was supported by single nucleotide polymorphism (SNP) analysis and provides a rationale for tailored therapeutic interventions.
Results
A longitudinal study of 158 SLE patients
We collected clinical and blood transcriptional profiles from 158 pediatric SLE patients followed longitudinally for up to 1412 days, representing 924 unique visits. The 24 components of the SLEDAI were collected at all time points to assess DA. Samples were categorized as DA1 (SLEDAI: 0-2), DA2 (SLEDAI: 3-7) or DA3 (SLEDAI > 7), based on SLEDAI distribution across the cohort. Treatment was recorded at blood draw and categorized as follows: no treatment (NT), HC only, oral steroids ± HC (OS), MMF ± HC/OS, and intravenous therapy consisting of cyclophosphamide and/or methylprednisolone ± HC/OS (CIV). Forty-eight healthy pediatric individuals were enrolled as controls. For validation, patients were separated into independent training and test sets in a two-to-one sample-size ratio. No significant difference was observed in the distribution of race, DA or treatment between the sets. Effects of circadian rhythm, season, treatment dose and frequency on the transcriptional variance were either negligible or accounted for. Cohort characteristics are summarized in Table S1.
The SLE whole blood fingerprint
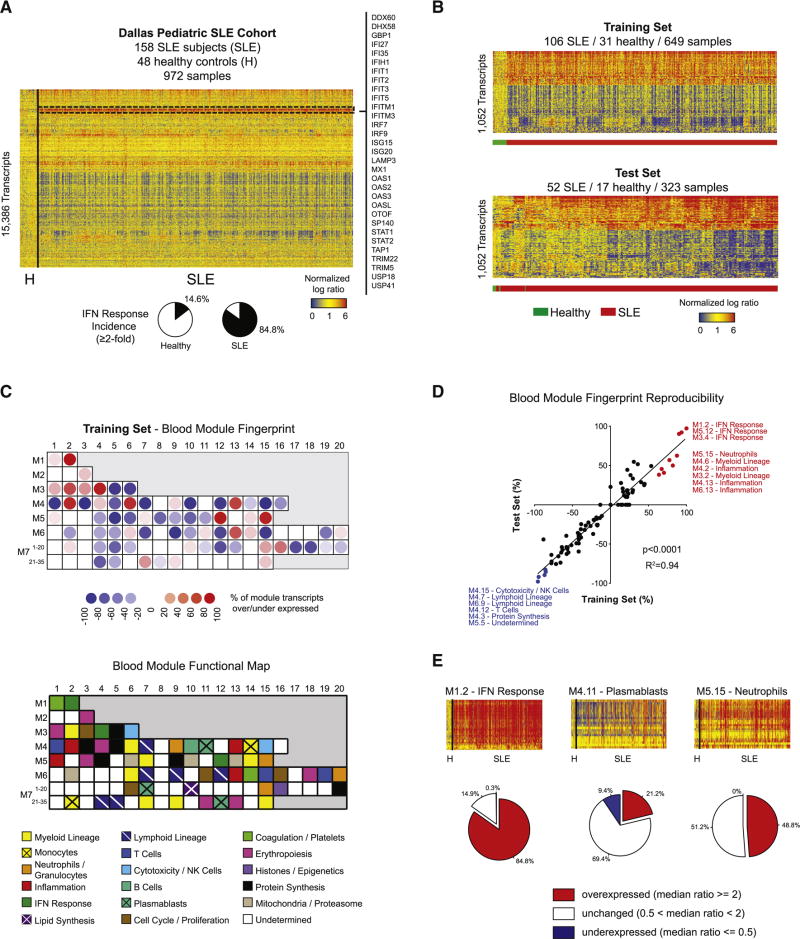
We first defined the global SLE signature by comparing all patient samples to healthy controls. Unsupervised hierarchical clustering of the 15,386 transcripts detected revealed a prevalent IFN signature in 784 out of 924 SLE samples (84.8%) (Figure 1A). To identify differentially expressed transcripts (DETs) between SLE patients and healthy controls, we developed a linear mixed model. These models include both fixed effects, such as disease, and random effects such as individual. They account for repeated measurements and enable the use of unbalanced data without excluding individuals with missing records. This model yielded 1,052 DETs in the training set, with high reproducibility in the test set (Figure 1B).
Figure 1. The SLE blood transcriptional fingerprint.
A. Hierarchical clustering of the 15,386 transcripts detected across the 972 samples composing the Dallas pediatric SLE cohort. The IFN response is highlighted in the dashed rectangle, and representative transcripts are listed. B. Heatmap representing the 1,052 DETs between healthy and SLE in the training (upper panel) and test (lower panel) sets. C. Upper panel: Blood module fingerprint of SLE in the training set. Lower panel: Blood modules functional annotation key. D. Linear regression of blood module expression in the training (x-axis) and test (y-axis) sets. E. Frequency of over/under-expression of IFN, plasmablast and neutrophil signatures in SLE samples. See also Figure S1.
To functionally interpret these DETs, we conducted modular analysis (Chaussabel et al., 2008). Briefly, we used a framework of 260 modules of transcripts co-expressed in blood across various immunological conditions to reduce data dimensionality. These modules were annotated using knowledge-based and data-driven approaches. To assess their specific enrichment in sorted hematopoietic cell populations, we applied our modular analysis to two public datasets (Figures S1A and B, (Novershtern et al., 2011; Streicher et al., 2014). In addition, a correlation matrix of module expression across our cohort was built and clustered (Figure S1C). Functional annotation of several undetermined modules could be inferred based on their proximity to annotated modules from lymphoid, myeloid or erythropoietic lineages.
The SLE modular fingerprint from the training set revealed overexpression of IFN response, neutrophil, inflammation, cell cycle, erythropoiesis and histone modules. Conversely, modules linked to NK cell/cytotoxicity, lymphoid lineage, B cells, T cells and protein synthesis were underexpressed (Figure 1C). The SLE module fingerprint was highly reproducible in the test set as shown by linear regression (R2=0.94) (Figure 1D). Frequency analysis of IFN, plasmablast and neutrophil signatures, which were previously associated with SLE, revealed a more transient plasmablast signature (positive in 21.2% of samples) than IFN (84.8%) or neutrophil (48.8%), explaining the absence of plasmablast-related modules in the global SLE signature (Figure 1E). These results highlight the breadth and frequency of systemic immune signatures altered in SLE.
A plasmablast signature best correlates with disease activity
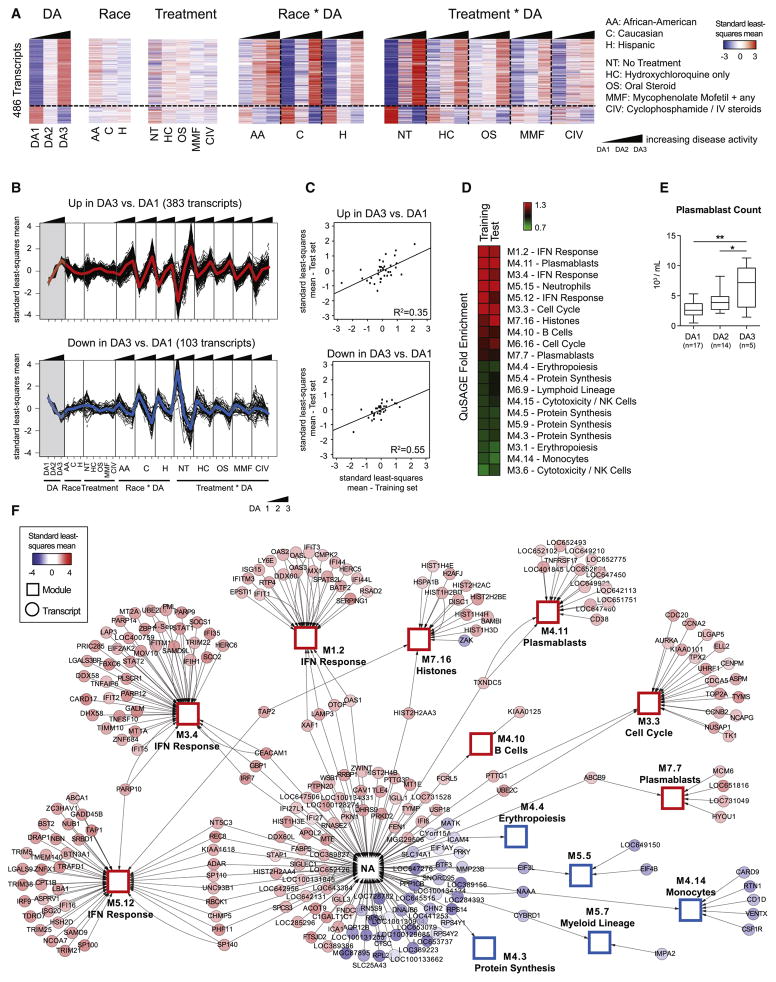
We next assessed how the SLE signature changes with DA. To do so, we developed a second model that incorporated both race and treatment, as these parameters impact the clinical course of SLE. It identified 3,501 DETs, which were clustered to highlight their individual behavior across DA, race and treatment groups, in training and test sets (Figure S2A and S2B).
Comparing DA3 vs. DA1 yielded 486 transcripts, of which 383 positively and 103 negatively correlated with DA (Figure 2A). These transcripts were differentially expressed between DA levels regardless of race or treatment, as shown by their sustained upward or downward expression pattern across Race*DA and Treatment*DA interaction groups (subgroups of samples organized by DA levels 1, 2 and 3 for each value of the Race or Treatment variables) (Figure 2B). Their expression pattern was consistent in the test set (p<0.0001, overexpressed: R2=0.35; underexpressed: R2=0.55) (Figures 2C and S2C).
Figure 2. Transcriptional correlates of disease activity.
A. Hierarchical clustering of the 486 DETs between DA3 and DA1, organized by DA, race, treatment and interaction terms. B. Line charts displaying the transcripts over- (upper panel) or under- (lower panel) expressed in DA3 vs. DA1. C. Linear regressions between training and test sets for over- and under-expressed transcripts. D. Heatmap representing the QuSAGE fold enrichment for these transcripts using blood modules as gene sets. E. Box plots representing the absolute count of circulating plasmablasts by FACS in a subset of patients, grouped by DA level. Plasmablast counts were found significantly different in DA3 vs. DA1 (p=0.0069) and DA3 vs. DA2 (p=0.045) by a mixed model that adjusted for age and treatment (*: p<0.05; **: p<0.01). F. Cytoscape network displaying the connectivity of DETs in DA3 vs. DA1 with blood modules, which are represented as hubs (squares). Genes not connected to a blood module are linked to the “NA” hub. Nodes are colored by the standard least-squares mean expression of the transcript in DA3. See also Figure S2.
To functionally interpret these gene lists, we applied quantitative set analysis for gene expression (QuSAGE) (Turner et al., 2015; Yaari et al., 2013), using the blood modules as gene sets (Figures 2D and S2D). Positive correlates of DA were enriched for IFN response, plasmablast, cell cycle, neutrophil, histone and B cell modules. Reproducibility of module enrichment was assessed by Pearson correlation analysis of a module’s eigengene profile in training and test sets (Figure S2E). The plasmablast signature was the most reproducible (M7.32: R2=0.86; M4.11: R2=0.65; M7.7: R2=0.61), while the IFN response showed lower reproducibility (M1.2: R2=0.07; M3.4: R2=0.25; M5.12: R2=0.22). Transcripts negatively correlated with DA were enriched for NK cell/cytotoxicity, protein synthesis and erythropoiesis modules. FACS analysis of circulating plasmablasts in a subset of patients (n=36) confirmed their increasing frequency with increasing DA (Figure 2E).
Finally, transcripts linked to DA changes were represented as a Cytoscape network (Shannon et al., 2003) (Figure 2F). Along with blood module transcripts, this network displays DETs that do not belong to pre-established modules (connected to “NA” hub). Within this group, numerous IFN-regulated transcripts (IFI6, IFI27, IFI27L1, DDX60L, SIGLEC1), histone (HIST1H3E, HIST2H2AA4, HIST2H4B) and B cell-related (STAP1, LOC652126) genes were found, thereby expanding the results from modular enrichment analysis.
These observations highlight the plasmablast signature as the most robust biomarker of SLE DA across independent datasets.
Increased plasmablast responses in African-American patients
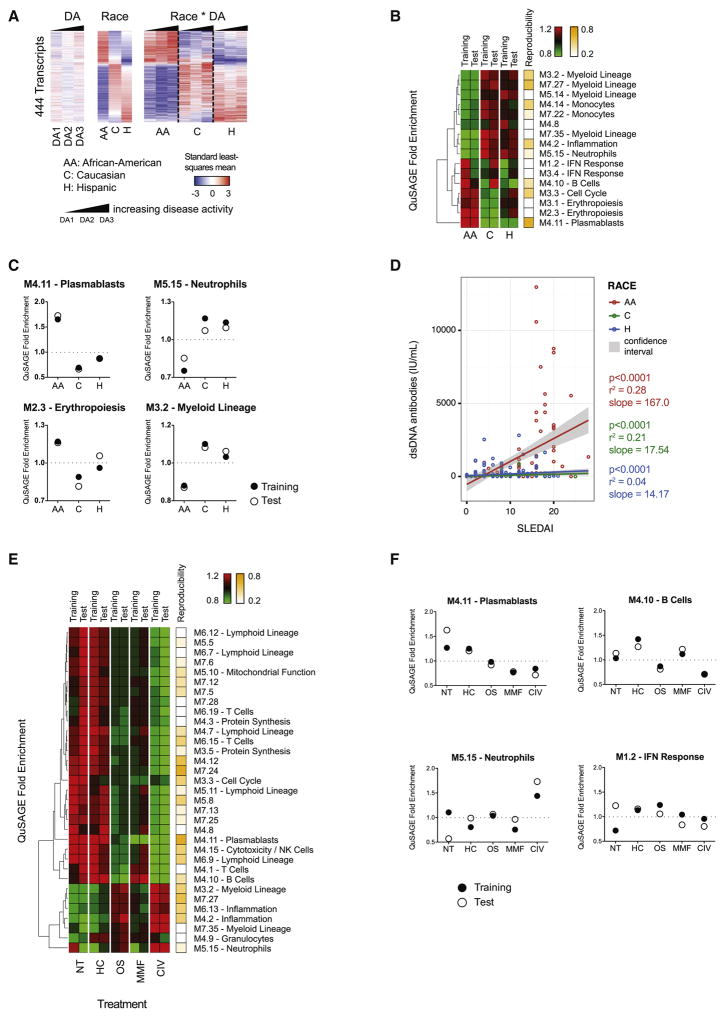
The influence of race on SLE clinical severity is well documented, with African-Americans often presenting with more severe disease than Caucasians (Korbet et al., 2007). To assess whether transcriptional differences among individuals of various races exist, we compared the transcriptomes from African-American, Hispanic, and Caucasian patients in paired analyses, i.e., i) African-American, ii) Caucasian and iii) Hispanic vs. Others. We uncovered 444 DETs among the three groups (Figure 3A). Functional interpretation by QuSAGE identified an enrichment of plasmablast, cell cycle and erythropoiesis modules in African-Americans (Figures 3B and 3C). Conversely, Hispanics and Caucasians showed enrichment of neutrophil, myeloid lineage and inflammation-related modules.
Figure 3. Influence of race and treatment on the SLE blood transcriptional fingerprint.
A. Hierarchical clustering of the 444 DETs between the three race groups, organized by DA, race, treatment and interaction terms. B. Heatmap representing the QuSAGE fold enrichment of each group using blood modules as gene sets. The reproducibility of each module between training and test sets is displayed on the right. C. Dot plots representing the QuSAGE fold enrichment for four modules from B, with training and test set results combined. D. X-Y plot representing the linear regressions of SLEDAI vs. anti-dsDNA antibody titers by race group. E. Heatmap representing the QuSAGE fold enrichment for each treatment group versus others combined. Reproducibility between training and test sets is also displayed. F. Dot plots representing the QuSAGE fold enrichment for neutrophil, B cell, plasmablast and IFN modules. See also Figure S3.
To complement these observations, we analyzed the distribution of SLEDAI, anti-dsDNA antibody titers, C3 and ESR among these races by DA level. African-Americans displayed a significantly higher SLEDAI in the DA3 group and higher systemic inflammation as measured by ESR and C3 levels, consistent with increased clinical burden (Figure S3A). Accordingly, their anti-dsDNA antibody titers were increased compared to Caucasians and Hispanics in the DA3 group, as substantiated by the slope of the linear regression of SLEDAI vs. anti-dsDNA antibody titers (p<0.0001) (Figure 3D). Therefore, increase in DA is connected to enrichment in plasmablast signatures and circulating anti-dsDNA antibodies, particularly in African-Americans.
Effect of therapy on SLE blood signatures
We next assessed how treatment influences SLE blood transcriptional signatures by categorizing each sample into one of five treatment categories (NT, HC, OS, MMF, CIV). We identified 622 DETs in any treatment vs. others. Patients receiving HC only, given as primary therapy for milder clinical manifestations, displayed a module enrichment profile similar to that of untreated patients, who for the most part were in remission. Treatment groups including corticosteroids (OS and CIV) displayed an increased neutrophil signature, consistent with increased numbers of circulating neutrophils following these treatments. The plasmablast signature was decreased by all treatments compared to NT, but most strongly by MMF and CIV, two cytostatic drugs that suppress activated lymphocytes. These two therapies were also the most effective in decreasing the IFN response (Figures 3E and 3F). These results highlight changes in circulating leukocyte ratios as well as the association of more potent therapies with significant decrease in plasmablast and IFN response.
A neutrophil signature associates with lupus nephritis
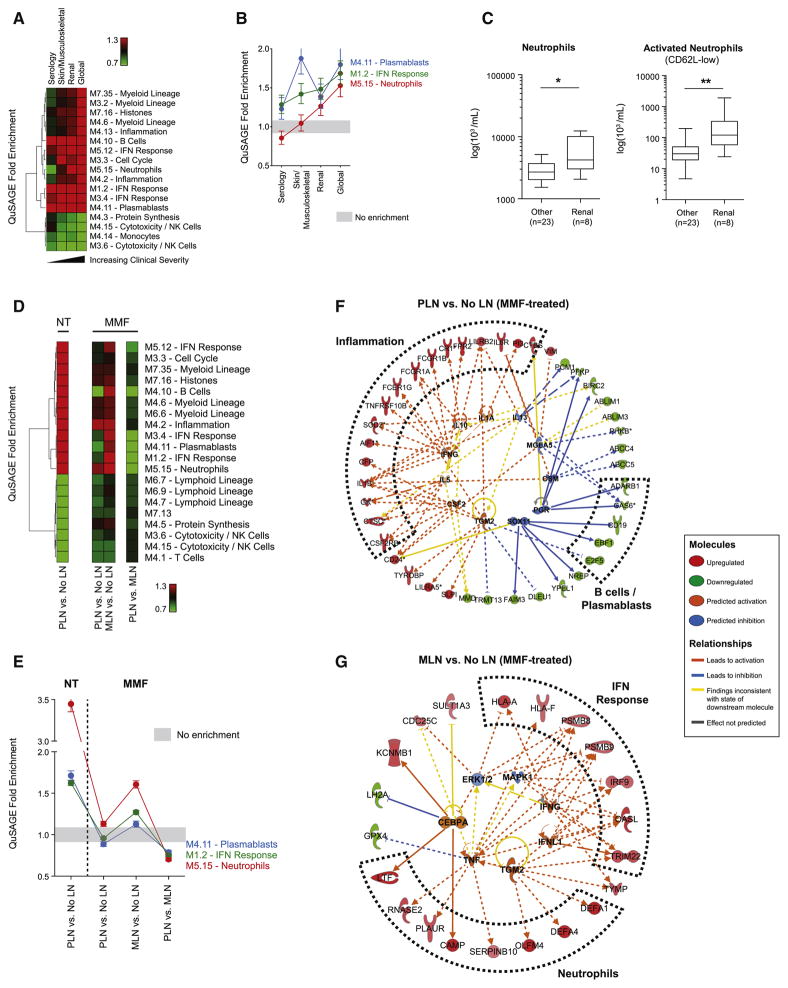
Because the SLEDAI is a composite score evaluating 24 parameters, we assessed whether transcriptional differences could be detected as a function of discrete SLEDAI categories. We classified samples according to five groups of SLEDAI components: no SLEDAI parameters (“none”), alterations in serum parameters only (“serology”), connective tissue +/− serology (“skin/musculoskeletal”), kidney +/− serology (“renal”), or all combined (“global”) reflecting the highest disease severity (Figure S3B). SLEDAI distribution across these groups confirmed a pattern of increasing DA from “none” to “global” (Figure S3C). To compare these groups, we developed a third model accounting for treatment, and obtained estimates for the comparisons between each active component group and “none”. QuSAGE detected enrichment in IFN response, plasmablast and B cell modules in all comparisons, suggesting early involvement of these pathways in disease progression. Conversely, the neutrophil, myeloid lineage and inflammation modules were only enriched in the renal and global component groups, suggesting an association with kidney involvement (Figures 4A and 4B). FACS analysis in a subset of patients (n=31) revealed increased total and activated CD62L-low circulating neutrophil counts in patients with active nephritis (Figure 4C). These data support a model of gradual disease progression, whereby an initial increase in IFN response and differentiation of B cells into short-lived plasmablasts, which may occur before clinical onset (Arbuckle et al., 2003), is followed by kidney disease and full-blown systemic inflammation fueled by myeloid cells, including neutrophils.
Figure 4. A neutrophil signature associates with lupus nephritis.
A. Heatmap representing the QuSAGE fold enrichment for each SLEDAI component group as compared to None. B. Line chart of the QuSAGE fold enrichment for IFN response, plasmablast and neutrophil modules. Whiskers represent the 95% confidence intervals. C. Box plots representing the absolute counts of bulk (left panel) and activated CD62L-low (right panel) neutrophils by FACS in a subset of patients with (n=8) or without (n=23) active LN. (*: p<0.05; **: p<0.01). Data were adjusted for age through a mixed model. Whiskers represent the minimum and maximum values. D. Heatmap representing the QuSAGE fold enrichment for PLN and MLN either compared to no nephritis (No LN) or directly to each other, with (MMF) or without (NT) treatment. E. Line chart representing the QuSAGE fold enrichment for IFN response, plasmablast and neutrophil modules across the four comparisons. Whiskers represent the 95% confidence intervals. F–G. Ingenuity Pathway Analysis (IPA) networks of DETs from the estimates of PLN vs. no LN and MLN vs. no LN treated with MMF. DETs are represented on the outer circle and colored by fold change (red: overexpressed; green: underexpressed). Predicted upstream and downstream regulators (absolute z-score > 1) are represented on the inner circle.
Different transcriptional responses to treatment according to nephritis class
The term lupus nephritis (LN) includes a heterogeneous group of kidney pathologies histologically classified into six major types (I-VI) (Weening et al., 2004). Severe cases comprise proliferative (PLN, III and IV) and membranous (MLN, V) nephritis, or a combination of the two (VI). MMF is used to treat both PLN and MLN, though the pathogenesis of these two nephrites is different. To determine whether blood transcriptomics could inform on these differences, we developed a fourth model accounting for interactions between treatment and nephritis class, while adjusting for SLEDAI differences between groups. Samples were categorized into no LN (I), mesangial nephritis (II), PLN (III and IV) and MLN (V) (type VI was excluded from the analysis). We focused on three comparisons: i) untreated patients without LN vs. untreated PLN (untreated MLN samples were underrepresented in our cohort), ii) MMF-treated patients without LN vs. PLN or MLN and iii) direct comparison of PLN vs. MLN on MMF (Figure S3D).
MMF-treated PLN and MLN groups showed no difference in SLEDAI or neutrophil percentage (Figure S3E). Untreated PLN displayed enrichment (FDR<0.001) for modules correlating with DA, including neutrophils (3.49), plasmablasts (1.71), the IFN response (1.63) and B cells (1.33). Upon MMF treatment, enrichment of these modules decreased significantly more in PLN than in MLN (1.42, 1.27, 1.27 and 1.41-fold respectively, FDR<0.001) (Figures 4D and 4E).
In silico Ingenuity pathway analysis (IPA) further revealed that MMF-treated PLN overexpressed transcripts linked to myeloid lineage and inflammation (FcRs, CR1, IL1B, IL6R, CTSC) and underexpressed B cell and plasmablast transcripts (CD19, EBF1, E2F5, GAS6, ADARB1) (Figure 4F). Conversely, MMF-treated MLN overexpressed transcripts linked to activated neutrophils (DEFA1, DEFA4, CAMP, RNASE2, LTF) and the IFN response (MHC class I, IRF9, PSMBs, OASL, TRIM22) (Figure 4G). These data highlight a differential enrichment of molecular signatures in MMF-treated PLN and MLN, perhaps reflecting different pathogenic mechanisms leading to disease.
Personalized SLE transcriptional immunomonitoring
SLE is a heterogeneous disease, for which no single treatment is curative. Stratification of patients will be important to formulate customized therapies and improve clinical trial design. To address this, we leveraged weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008; Zhang and Horvath, 2005) to identify modules of longitudinally co-expressed transcripts for individual patients and uncover associated clinical traits. Briefly, the entire dataset was divided into subsets of samples corresponding to each subject. Patient-specific modules of co-expressed transcripts over time were identified by WGCNA and continuous clinical traits were correlated to module eigengenes (Figure S4A). A module/trait correlation matrix was built for each patient and the module that best correlated with the SLEDAI was selected (hereafter SLEDAI WGCNA module) (Figure S4B).
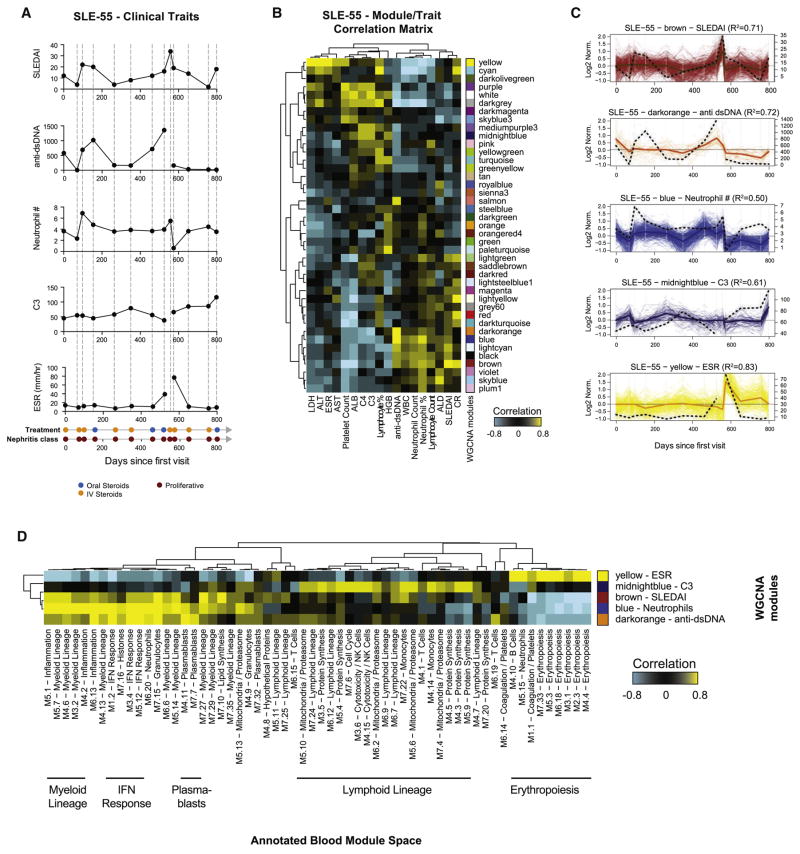
An example is provided for patient SLE-55, an African-American female with PLN immunomonitored 13 times over 798 days. The SLEDAI indicated two flares at visits 3 and 9. Both were accompanied by increases in anti-dsDNA antibody titers and neutrophil counts, but only the second flare was accompanied by an increase in ESR (Figure 5A). WGCNA identified 41 modules specific to her immunomonitoring time course, which were correlated with clinical traits (Figure 5B). The modules that best correlated with SLEDAI (brown), anti-dsDNA antibody titers (darkorange), neutrophil count (blue), C3 (midnightblue) and ESR (yellow) were selected (Figure 5C). To biologically interpret these modules, we projected their expression profile in the annotated blood module space, by correlating their eigengenes to those of blood modules. This approach allows us to automate the annotation of hundreds of WGCNA modules by comparing their expression pattern and transcript content to those of our reference blood modules. In this patient, the profile of the brown (SLEDAI) and blue (neutrophil %) modules positively correlated with myeloid lineage, inflammation and IFN response blood modules. The darkorange module that best fitted anti-dsDNA antibody titers correlated with plasmablast, cell cycle and IFN modules. Finally, the yellow module (ESR) highly correlated with erythropoiesis (Figure 5D).
Figure 5. Individual immunomonitoring by WGNCA.
A. Clinical summary for patient SLE-55. SLEDAI, anti-dsDNA antibody titers, neutrophil count, C3 and ESR are displayed as line charts as a function of days since the first visit (x-axis). Treatment and nephritis class are also displayed. B. Hierarchical clustering of the module/trait correlation matrix for SLE-55. C. Line charts representing the modules that best correlate with SLEDAI, anti-dsDNA antibody titers, neutrophil count, C3 and ESR. The profile of the clinical trait is overlaid on the plot and represented as an interrupted black line. D. Hierarchical clustering of the correlation matrix between WGCNA and blood module eigengenes for SLE-155. See also Figure S4.
To facilitate access to these data, we developed a web interface available at http://websle.com. For each patient, users can i) follow clinical trait changes over time; ii) analyze the profile and content of each WGCNA module; iii) identify modules and transcripts best correlating with clinical traits; iv) identify major module hubs through module membership quantification. The interface’s features are detailed in Figure S5.
To assess the heterogeneity of immune signatures associating with DA across patients, SLEDAI WGCNA modules were then recombined into an interindividual correlation matrix between WGCNA (y-axis) and whole blood (x-axis) modules, and hierarchically clustered. Subgroups of patients were identified based on the combination of blood immune signatures that best correlated with the SLEDAI. Finally, to assess whether there is a genetic basis for these clusters, we conducted SNP analysis (Figure S4C). We reasoned that the identification of individual immune correlates of DA could inform personalized therapeutic interventions (Figure S4D).
Patients stratify into seven groups based on transcriptional correlates of SLEDAI
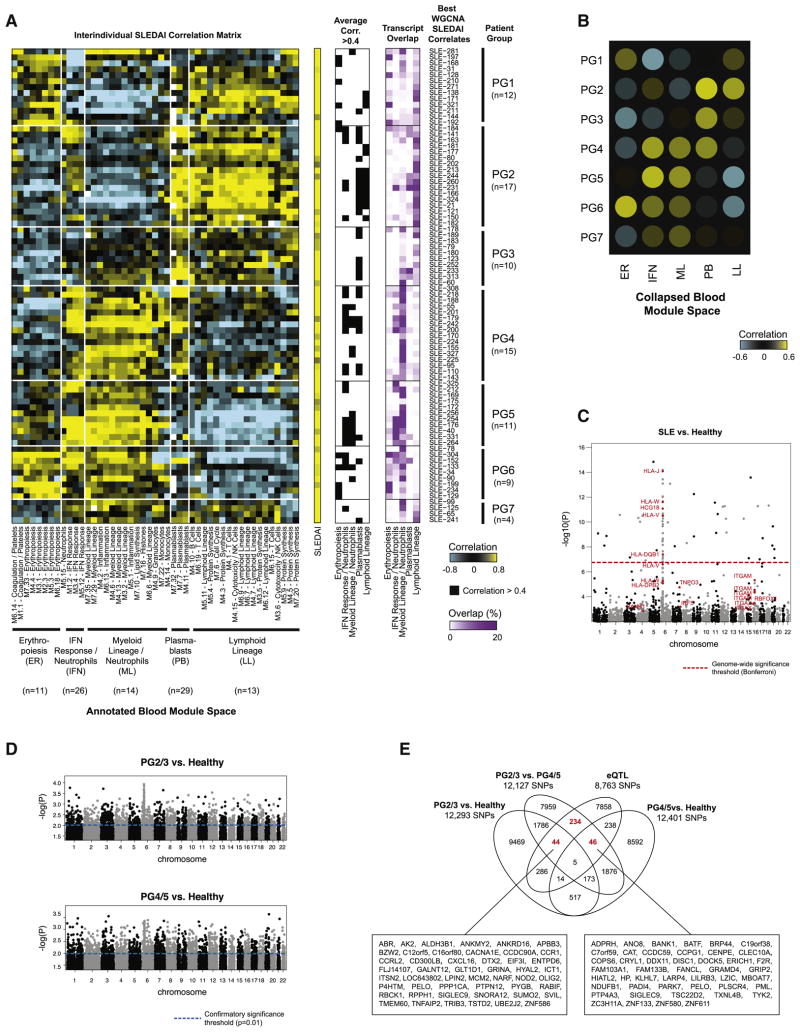
We automated this analytical approach to stratify 80 patients with five or more visits. Clustering of the interindividual SLEDAI correlation matrix identified seven patient groups (PG1-7), each displaying a specific combination of five immune signatures correlating with the SLEDAI, including erythropoiesis, IFN response, myeloid lineage/neutrophils, plasmablasts and lymphoid lineage (Figure 6A). Group-specific signature enrichment patterns are summarized in Figure 6B. Only plasmablast modules correlated with the SLEDAI in PG3, while only IFN response and myeloid lineage modules did in PG5. As a positive control, we identified WGCNA modules that best correlated with neutrophil percent in each patient, revealing a homogenous correlation pattern with myeloid lineage/neutrophil modules across patients (Figure S6A).
Figure 6. Stratification of SLE patients based on transcriptional correlates of SLEDAI.
A. Left panel: hierarchical clustering of the interindividual correlation matrix between the SLEDAI WGCNA modules for 80 patients (rows) and blood modules (columns). Center panel: correlation of blood modules averaged by immune group for each SLEDAI WGCNA module. A black square indicates a correlation ≥0.4. Right panel: transcript overlap between each WGCNA module and the combined list of blood module transcripts from the five groups (PG: patient group). B. Summary heatmap of patient stratification based on SLEDAI correlates. (ER: erythropoiesis; IFN: IFN response/neutrophils; ML: myeloid lineage/neutrophils; PB: plasmablasts; LL: lymphoid lineage). C–D. Manhattan plots representing the results from the comparative SNP analysis between SLE (n=80), PG2/3 (n=27) or PG4/5 (n=26) and healthy controls. Loci related to genes previously associated with SLE are highlighted in red. E. Venn diagram of overlapping SNPs between PG2/3 vs. healthy, PG4/5 vs. healthy, PG2/3 vs. PG4/5 and eQTLs (p<0.05). Genes in cis with these SNPs are displayed in boxes for relevant lists. See also Figure S6 and Tables S2, S3 and S4.
We considered a different stratification approach that used samples with high DA only (Supplemental Experimental Procedures). Clustering of transcripts modulated in this dataset revealed partial conservation of the original PG stratification, in particular for PG4 and PG5 (Figure S6B). High DA and longitudinal correlation analyses were then conducted in parallel (Figure S6C). Similarities between methods were observed for the plasmablast signature (R2=0.47, Figure S6D). More pronounced differences were observed for IFN, erythropoiesis and myeloid lineage signatures, suggesting that the longitudinal approach contributes additional stratification information by incorporating molecular status during quiescent disease.
We analyzed the distribution of renal SLEDAI components, patient demographics, laboratory values and treatment between groups (Table S2). No difference was found between PGs for age, gender, race, sampling season or visit number. PG4 patients, for whom SLEDAI best correlated with IFN response, myeloid lineage and plasmablasts, all had LN, displayed the highest proportion of PLN, were on aggressive therapy and had the highest percentage of neutrophils, suggesting higher disease burden. Conversely, PG1 and PG6 patients, for whom SLEDAI best correlated with erythropoiesis, displayed the lowest proportion of active nephritis.
To assess the contribution of genetics to this distribution, we genotyped 135 patients by SNP array and collected race-matched healthy controls from both our cohort and the 1000 Genomes Project (G1K) (Figure S6E, Supplemental Experimental Procedures). We detected SNPs previously associated with SLE among numerous members of the HLA cluster on chromosome 6, as well as ITGAM, ITGAX, IRF5, TNPO3 and BANK1 with confirmatory significance (p<0.001) (Figure 6C) (Bentham et al., 2015). These results were validated by random permutation analysis (Supplemental Experimental Procedures).
To assess potential SNP differences between PGs while maximizing sample size, we grouped patients from PG2 and PG3 (PG2/3), where DA correlated with plasmablasts and/or lymphoid lineage, and patients from PG4 and PG5 (PG4/5), where DA correlated with IFN response and myeloid lineage. We compared these groups to race-matched healthy controls (Figure 6D) and to each other. In parallel, we conducted expression quantitative trait loci (eQTL) analysis to identify SNPs acting in cis on genes that change with DA (Supplemental Experimental Procedures, Table S3). We then overlapped the results from these analyses, focusing on 362 group-specific SNPs that overlapped with eQTLs (Figure 6E, Table S4). Numerous genes in cis with SNPs and distinct between PG2/3 and PG4/5 were related to lymphoid lineage/B cells (BANK1, CD3E, CD3G), myeloid lineage/neutrophils (BPI, CD300A, CXCL16, CCR1, RXRA, TKT, LILRB3, NOD2, SIGLEC9, PGLYRP1, MYD88, IL8, TNFAIP2, TNFRSF1A) or both (CD40). Overall, 126 out of 362 (35%) SNPs were acting in cis with an IFN-inducible gene (Rusinova et al., 2013). While studies in larger cohorts are warranted, these data suggest an association between genotype and transcriptional correlates of DA in discrete groups of SLE patients, which in combination might accelerate the identification of targets for personalized treatment.
A transcript panel for the molecular stratification of SLE patients
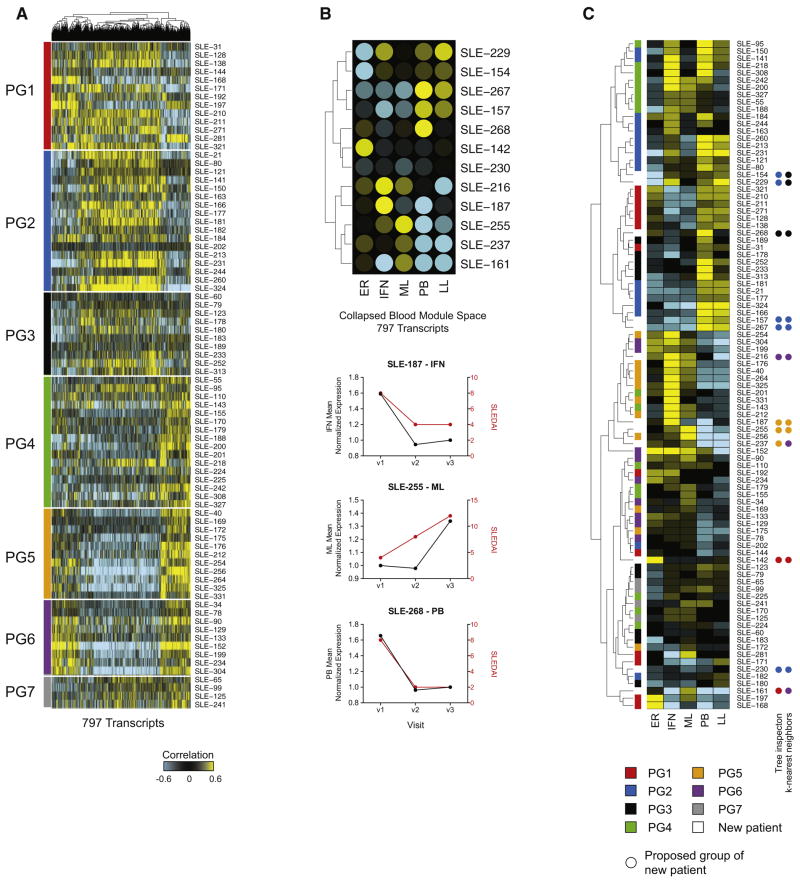
Finally, we sought a gene set that could stratify SLE patients with shorter follow-up. To do so, we conducted ANOVA (p<0.001) on the 11,457 transcripts that form the union of the 80 WGCNA SLEDAI modules. 3,011 transcripts were differentially correlated between the seven patient groups. Of these, 797 intersected with the blood module space (erythropoiesis: 149 transcripts; IFN response: 40; myeloid lineage/neutrophils: 163; plasmablasts: 9; lymphoid lineage: 436) (Figure 7A, Table S5). Classification accuracy of these transcripts in cross-validation analysis ranged between 0.81 and 0.91 depending on PG (Supplemental Experimental Procedures). We next correlated the SLEDAI of 12 additional patients followed during three or four clinic visits with the expression of these 797 transcripts. Specific immune correlates of the SLEDAI were identified in these individuals (e.g. IFN in SLE-187, myeloid lineage/neutrophils in SLE-255 and plasmablasts in SLE-268) (Figure 7B). Finally, we assigned PG groups to these 12 donors using both hierarchical cluster tree inspection and k-nearest neighbors (k=5) approaches (Figure 7C). The two methods agreed for 8 out of 12 patients. SLE-157, 230 and 267 reproducibly associated with PG2, while SLE-187 and 255 associated with PG5. This suggests that these transcripts can stratify SLE patients and could be used in a targeted cost-efficient bioassay. Overall, our approach highlights immune signatures correlating with SLEDAI across different genotypes and phenotypes and supports the development of customized treatment strategies.
Figure 7. A targeted panel for SLE patient stratification.
A. Hierarchical clustering of the 797 transcripts differentially correlating with SLEDAI between the seven patient groups. B. Upper panel: summary heatmap of immune signatures correlating with the SLEDAI of 12 independent patients with three to four visits. Lower panel: Linecharts representing the mean normalized expression of immune signatures (left Y-axis) and SLEDAI (right Y-axis) for three representative patients. C. Hierarchical cluster of the 92 SLE patients considered, according to individual SLEDAI correlation profile in the collapsed blood module space. Patients are colored by PG classification. PG assignment of the 12 new patients is highlighted by colored dots. See also Table S5.
Discussion
SLE is a heterogeneous disease characterized by a wide spectrum of clinical manifestations and degrees of severity. To gain insight into the molecular heterogeneity of SLE, we profiled the blood transcriptome of a longitudinal cohort of pediatric patients. We identified a plasmablast signature as the most robust biomarker of DA. We also detected a gradual enrichment of immune signatures during disease progression, linking neutrophils to active nephritis. Finally, personalized immunomonitoring enabled patient stratification based on correlates of DA. This resulted in a comprehensive view into the heterogeneity of molecular signatures associated with clinical progression that could not be predicted with cohort-level analyses.
Previous studies designed to identify blood transcriptional correlates of DA in SLE mainly focused on limited numbers of IFN-inducible transcripts that might be induced indistinctly by type I-III IFN family members. Through an unbiased approach, we confirmed that the majority of samples in our cohort over-expressed IFN-inducible transcripts. This signature, together with plasmablast, B cell, neutrophil and histone transcripts, correlated with DA. Of these, the plasmablast signature was the most robust DA biomarker. These observations complement known associations between DA and numbers of circulating plasmablasts (Odendahl et al., 2000), which are the source of anti-dsDNA antibodies that fluctuate with DA (Sanz and Lee, 2010). The plasmablast signature was reduced by all conventional SLE therapies, but most significantly by MMF and CIV. In addition to the anti-proliferative effect of both drugs, MMF is known to inhibit inosine 5′-monophosphate dehydrogenase, which is required for differentiation of mature B cells into plasma cells (Jonsson and Carlsten, 2003).
When accounting for ethnicity, the plasmablast signature showed stronger signal in African-Americans, who also displayed the highest DA levels. Accordingly, we found the best correlation between anti-dsDNA antibody titers and SLEDAI in this ethnic group. African-American patients respond better to B cell depletion therapies than Caucasian patients (Merrill et al., 2010). Conversely, they displayed lower responses to anti-BAFF treatment in a Phase III clinical trial (Navarra et al., 2011). This might be explained by their higher serum levels of BAFF (Ritterhouse et al., 2011), which may require higher drug doses for neutralization.
Neutrophil transcripts were enriched in patients with active renal disease. In lupus, these cells are primed by IFN and release both pro-inflammatory mediators and interferogenic DNA when exposed to SLE ICs (Garcia-Romo et al., 2011; Hakkim et al., 2010; Villanueva et al., 2011). Their specific contribution to different LN types has yet to be elucidated. Similarly to the plasmablast and IFN signatures, the neutrophil signature was extinguished after MMF treatment in PLN but not in MLN. Mechanistically, these observations are difficult to interpret. MMF may target an upstream event leading to induction of these signatures only in PLN. For example, a short-lived plasmablast-derived autoantibody responsible for pDC and neutrophil activation could drive PLN, as opposed to autoantibodies from long-lived non-proliferative plasma cells that would be resistant to this mode of therapy in MLN. Conversely, a pro-inflammatory signature including IL1A, IL1B and IL6R remained patent in PLN under MMF, suggesting that combination therapies including drugs targeting these pathways might be superior to current regimens against PLN.
The FDA approved only one drug for SLE treatment in the past 50 years. While several treatments improved disease in pre-clinical models and/or phase I-II clinical trials, larger phase III trials proved unsuccessful. A major hurdle in lupus therapy development is the lack of knowledge about molecular mechanisms driving DA in individual patients. Genomic approaches have identified various genetic pathways contributing to familial SLE, including immature B cell survival, early complement cascade components, regulation of IFN production and defects in cytoplasmic or extracellular DNA degradation (Cheng and Anderson, 2012). Allelic variants of genes involved in some of these pathways increase the risk of developing SLE in more common, non-familial cases. However, these alleles have so far not enabled the stratification of patients for customized treatment. Our longitudinal personalized immunoprofiling approach stratified patients into seven molecular groups according to DA correlates. This transcriptional stratification was complemented by SNP analysis that pointed at specific associations in distinct groups, despite limited sample size. Simultaneous eQTL analysis enabled the identification of group-specific SNPs that may directly affect the expression of neighboring genes. Further studies in larger cohorts are warranted and should include leukocyte profiling by flow cytometry to correct for blood population distribution in eQTL analyses.
By highlighting the spectrum of individual correlates of DA over time, our approach provides a rationale for the failure of clinical trials in SLE, and an opportunity for the development of personalized therapies. Indeed, prevalent signatures that correlated with DA at the cohort level, such as IFN or plasmablast, failed to do so at the individual level in two thirds of patients. Future studies should assess the response to treatments targeting B cells/plasmablasts, IFN and pro-inflammatory cytokines in these subgroups of patients.
While our longitudinal data could have been used to stratify patients solely based on molecular profiles, one of our main goals was to identify immune pathways associated with changes in clinical features. A potential caveat is the use of the SLEDAI to assess DA. This measure, although validated in all age and ethnic groups, still falls short of accurately capturing some relevant clinical aspects of the disease. In our pediatric cohort, however, the major contributor to SLEDAI is LN, which is more objectively quantified than other components of the index.
Biomarkers that can predict occurrence and frequency of flares will be of great value in the clinic (McKinney et al., 2015). The observational nature of this study, however, prevented the identification of flare predictors, as this would require more standardized and frequent sampling of individuals. Nevertheless, the molecular heterogeneity we unravel may inform the design of future studies to identify classifiers and predictors of DA.
While our study focused on SLE, our approach should be applicable to other clinically and genetically complex autoimmune diseases such as rheumatoid arthritis, inflammatory bowel disease, or inflammatory myopathies, where even approved therapies fail to induce remission in a significant number of patients. Future challenges include establishing the value of this approach to predict responses to targeted therapies and reducing the time to stratification. This might require integration of additional information such as epigenetic and proteomic data. Altogether, uncovering the molecular heterogeneity of complex diseases should enable a more rational use of available treatments, improve patient selection for clinical trials and guide the development of novel targeted drugs for precision medicine.
Experimental Procedures
Study design and patient characteristics
Children and adolescents with SLE were enrolled from the Rheumatology clinics at Texas Scottish Rite Hospital for Children and Children’s Medical Center Dallas. Study procedures were followed in accordance with protocols approved by the Institutional Review Boards at the University of Texas Southwestern Medical Center (092010-067) and Baylor University Medical Center (011-200). Informed consent was obtained from adults and the parents or guardians of those younger than 18 years of age. Assent was obtained from patients between 10 and 17 years of age. Patients were evaluated by a standardized protocol during routine morning clinic visits every three months and more frequently if clinical symptoms warranted evaluation. Blood was collected in Tempus blood RNA tubes (Life Technologies) for microarray and ACD tubes (BD) for flow cytometry studies, and laboratory measurements were recorded. A patient was considered under intravenous cyclophosphamide or methylprednisolone treatment if they were administered within four weeks prior to blood draw. Renal disease was defined by histopathological pattern on renal biopsy according to the International Society of Nephrology (ISN)/Renal Pathology Society (RPS) classification system. Active LN was defined by the presence of at least one component of the renal SLEDAI in patients with documented renal biopsy. Samples with hematuria attributable to menstruation were excluded. Because of low frequency, patients of Asian ethnicity were excluded.
RNA sampling, extraction and processing
Total RNA was extracted and amplified as previously described (Berry et al., 2010).
Batch correction
To correct for batch effect identified by principal variance component analysis (PVCA) (Figure S7A) and principal component analysis (PCA) (Figure S7B), 24 healthy control samples were run in both batches. The median raw expression ratio between batch 1 and batch 2 was calculated for each transcript for these 48 samples. The raw expression values from batch 1 samples were then multiplied by that ratio for each probe. This method was more effective than grouped-batch-profile (GBP) normalization and ComBat.
Modular transcriptional analysis
We used a pre-existing framework of 260 transcriptional modules to analyze this dataset (Chaussabel and Baldwin, 2014). For clarity, we presented the 97 modules forming the first seven rounds of selection of the module set. For the global SLE module fingerprint, each probe was statistically tested for difference in SLE vs. healthy through the mixed model (FDR<0.05). For each module, the percentage of probes significantly up- or down-regulated in the SLE group was calculated. The module score was defined as the difference %up - %down. For module fingerprints of individual samples, transcripts that display a normalized fold change ≥1.5 fold and a raw data difference ≥100 compared to the median of healthy controls were considered differentially expressed. For datasets generated with the Affymetrix platform, the transcripts were mapped to Illumina IDs through Refseq ID before the module activity scores were calculated.
Linear mixed models
Detailed explanations, code and result summary are presented in Supplemental Experimental Procedures and at http://websle.com.
Patient genotyping and SNP analysis
Detailed explanations are presented in Supplemental Experimental Procedures.
Online data access
The dataset described in this manuscript is deposited in the NCBI Gene Expression Omnibus under GEO Series accession number GSE65391. Both background-substracted and batch-corrected expression datasets are presented.
Supplementary Material
Acknowledgments
This work was supported by grants P50 AR054083-01, U19 AIO82715, the Alliance for Lupus Research and the Baylor-Scott & White Health Care Research Foundation, and is dedicated to the memory of Dr. J. Donald Capra. We thank members of the sample, genomics and flow cytometry cores at BIIR for research support. We thank Dr. Anna Lisa Lucido for critical review of the manuscript and Drs. Katie Stewart, Julie Fuller and Ashley Cooper for their clinical expertise. Jacques Banchereau is a member of the Board of Directors and the chairman of the Scientific Advisory Board of Neovacs, a French biotechnology company focused on the development of Kinoids, therapeutic vaccines for the treatment of autoimmune and inflammatory diseases (in particular SLE) and cancer. Yong-Jun Liu is an employee of Medimmune, which is involved in the development of drugs to treat SLE.
Footnotes
Author contributions
RB and SH analyzed data and wrote the manuscript. NB and JRU analyzed data. BC, and SK conducted SNP analysis. ME procured and archived clinical data. JB managed regulatory aspects, sample and clinical data collection. AMC, EW, YJL and JB critically reviewed the manuscript. RB and PA developed the web interface. EA and PV processed samples. SH, JT and DB conducted statistical analysis and developed models. GO supervised FACS analysis. TW, AG and MP followed patients and collected clinical data. VP supervised the study, interpreted data and wrote the manuscript.
References
- Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. The New England journal of medicine. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. The Journal of experimental medicine. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW, Martin J, Fairfax BP, Knight JC, Chen L, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nature genetics. 2015 doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatsky S, Boivin JF, Joseph L, Manzi S, Ginzler E, Gladman DD, Urowitz M, Fortin PR, Petri M, Barr S, et al. Mortality in systemic lupus erythematosus. Arthritis and rheumatism. 2006;54:2550–2557. doi: 10.1002/art.21955. [DOI] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis and rheumatism. 1992;35:630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nature reviews Immunology. 2014;14:271–280. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MH, Anderson MS. Monogenic autoimmunity. Annual review of immunology. 2012;30:393–427. doi: 10.1146/annurev-immunol-020711-074953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche L, Jourde-Chiche N, Whalen E, Presnell S, Gersuk V, Dang K, Anguiano E, Quinn C, Burtey S, Berland Y, et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis & rheumatology. 2014;66:1583–1595. doi: 10.1002/art.38628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, Punaro M, Baisch J, Guiducci C, Coffman RL, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Science translational medicine. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkim A, Furnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V, Herrmann M, Voll RE, Zychlinsky A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson CA, Carlsten H. Mycophenolic acid inhibits inosine 5′-monophosphate dehydrogenase and suppresses immunoglobulin and cytokine production of B cells. International immunopharmacology. 2003;3:31–37. doi: 10.1016/s1567-5769(02)00210-2. [DOI] [PubMed] [Google Scholar]
- Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis and rheumatism. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- Korbet SM, Schwartz MM, Evans J, Lewis EJ Collaborative Study G. Severe lupus nephritis: racial differences in presentation and outcome. Journal of the American Society of Nephrology : JASN. 2007;18:244–254. doi: 10.1681/ASN.2006090992. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill JT, Neuwelt CM, Wallace DJ, Shanahan JC, Latinis KM, Oates JC, Utset TO, Gordon C, Isenberg DA, Hsieh HJ, et al. Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: the randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis and rheumatism. 2010;62:222–233. doi: 10.1002/art.27233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarra SV, Guzman RM, Gallacher AE, Hall S, Levy RA, Jimenez RE, Li EK, Thomas M, Kim HY, Leon MG, et al. Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377:721–731. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendahl M, Jacobi A, Hansen A, Feist E, Hiepe F, Burmester GR, Lipsky PE, Radbruch A, Dorner T. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. Journal of immunology. 2000;165:5970–5979. doi: 10.4049/jimmunol.165.10.5970. [DOI] [PubMed] [Google Scholar]
- Petri M, Singh S, Tesfasyone H, Dedrick R, Fry K, Lal P, Williams G, Bauer J, Gregersen P, Behrens T, et al. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009;18:980–989. doi: 10.1177/0961203309105529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritterhouse LL, Crowe SR, Niewold TB, Merrill JT, Roberts VC, Dedeke AB, Neas BR, Thompson LF, Guthridge JM, James JA. B lymphocyte stimulator levels in systemic lupus erythematosus: higher circulating levels in African American patients and increased production after influenza vaccination in patients with low baseline levels. Arthritis and rheumatism. 2011;63:3931–3941. doi: 10.1002/art.30598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic acids research. 2013;41:D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz I, Lee FE. B cells as therapeutic targets in SLE. Nature reviews Rheumatology. 2010;6:326–337. doi: 10.1038/nrrheum.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher K, Morehouse CA, Groves CJ, Rajan B, Pilataxi F, Lehmann KP, Brohawn PZ, Higgs BW, McKeever K, Greenberg SA, et al. The plasma cell signature in autoimmune disease. Arthritis & rheumatology. 2014;66:173–184. doi: 10.1002/art.38194. [DOI] [PubMed] [Google Scholar]
- Tsokos GC. Systemic lupus erythematosus. The New England journal of medicine. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- Turner JA, Bolen CR, Blankenship DM. Quantitative gene set analysis generalized for repeated measures, confounder adjustment, and continuous covariates. BMC bioinformatics. 2015;16:272. doi: 10.1186/s12859-015-0707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte-Gil MF, Alarcon GS. Systemic lupus erythematosus: a therapeutic challenge for the XXI century. Clinical rheumatology. 2014;33:441–450. doi: 10.1007/s10067-014-2531-4. [DOI] [PubMed] [Google Scholar]
- Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, Lin AM, Rubin CJ, Zhao W, Olsen SH, Klinker M, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. Journal of immunology. 2011;187:538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening JJ, D’Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, Balow JE, Bruijn JA, Cook T, Ferrario F, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. Journal of the American Society of Nephrology : JASN. 2004;15:241–250. doi: 10.1097/01.asn.0000108969.21691.5d. [DOI] [PubMed] [Google Scholar]
- Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic acids research. 2013;41:e170. doi: 10.1093/nar/gkt660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.