Abstract
The European Genome-phenome Archive (EGA) is a permanent archive that promotes distribution and sharing of genetic and phenotype data consented for specific approved uses, but not fully open public distribution. The EGA follows strict protocols for information management, data storage, security and dissemination. Authorized access to the data is managed in partnership with the data providing organizations. The EGA includes major reference data collections for human genetics research.
The technical ability to identify regions of the human genome that harbor variants influencing disease risk is one of the most important recent advances in genomics. Many studies use large disease cohorts including the Wellcome Trust Case Control Consortium1, and the UK10K project. At the same time, the International Cancer Genome Consortium (ICGC) is generating complete genomes of matching tumor and normal samples for a number of cancers in an effort to understand the genomics of the disease. Published genetic variants are collated into fully public resources such as the NHGRI Catalogue of Published Genome-Wide Association Studies2 or Ensembl3. In addition to the public variants, the individual-level genetic and phenotypic data or summary statistics from the research projects are often required for replication4, meta-analysis5 and many other secondary uses such as methods development6 or use as control samples7. However, these data must be processed, archived and transferred in a manner that respects the consent agreements signed by the study subjects8. This often means that data can only be provided to bona fide researchers and used for specific research aims9.
The existing public data archives that provide unrestricted access to data are incompatible with these requirements and so the European Genome-phenome Archive (EGA) was launched in 2008 by the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) to support the voluntary archiving and disseminating of data requiring secure storage and distribution only to authorized users. Recently the EGA has expanded from an exclusively EMBL-EBI project to a collaboration with the Centre for Genome Regulation (CRG) in Barcelona, Spain, in what may be a first step toward a larger distributed network of data archive and dissemination services. Both EMBL-EBI and the CRG are publicly funded organizations, and the former is an intergovernmental organization formed by a collection of mostly European member, and associate member, states.
Since its launch, researchers from around the world have deposited and accessed data from over 450 studies in the EGA of various types (Table 1; Figure 1). These studies vary from large-scale array-based genotyping experiments on thousands of samples in case-control1,10 or population based studies11,12 to sequencing-based studies designed to understand changes in the genome, transcriptome or epigenome in both normal tissue13 and in various diseases such as cancer14–16 . As a result, the EGA has grown from about 50 TB to 1000 TB during the last three years.
Table 1.
The EGA users are distributed throughout the world as of July 2014
| Geographic location | Number of Submitters | Archived Data | Number of authorized data users |
|---|---|---|---|
| North and South America | 41 | 96 TB | 1491 |
| Europe | 116 | 743 TB | 1988 |
| Asia | 22 | 55 TB | 346 |
| Australia | 4 | 65 TB | 124 |
| Total | 183 | 959 TB | 4829* |
The total number of authorized users includes 880 from commercial organizations, which have not been separated geographically.
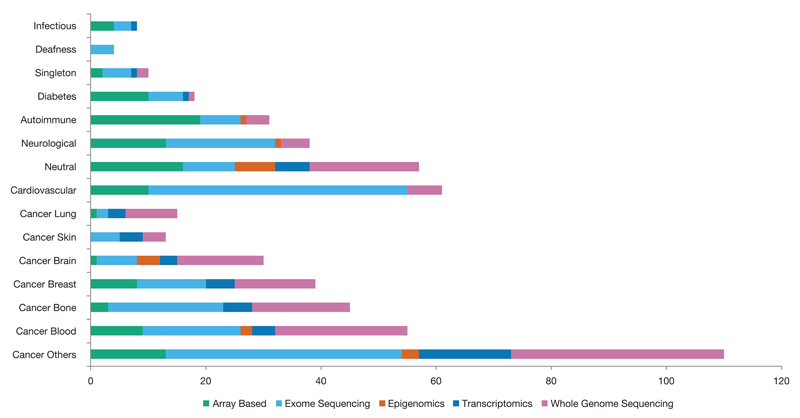
Figure 1.
Breakdown of EGA studies by disease topic. The majority (57%) of EGA studies investigate cancers of various type. EGA experimental data describes the methodology employed for each study and is shown by the different colours in each column. Exome sequencing (38%) is the most common followed by whole genome sequencing (29%), array based technologies (20%), transcriptomics (9%) and epigenomics (3%). As one may expect, array based experiments are typically from older studies whereas both transcriptomic and epigenetic investigations are more recent. A complete list of the meta-data collated to create this information is provided at our website.
Since 2011, the bulk of EGA submissions have transitioned from array-based genotyping to next generation sequencing studies. Summary level genetic information is also accepted if such data cannot by publicly released. Phenotype data are currently most often provided at the dataset rather than the individual level, for example a group of samples may be reported to have the same disease phenotype. However, submission of individual level detailed phenotypes is increasing and encouraged.
In this report we describe the roles and policies of the EGA, provide information on how access decisions are made, outline the methods for data submission and dissemination, and describe the EGA system infrastructure. The EGA has similarities and differences to the dbGaP database provided by the NCBI17 and, where appropriate, we describe the features and procedures that distinguish these two databases.
Roles and policies of the EGA
The role of the EGA is to promote the distribution and sharing of biomolecular and phenotypic data collected from human subjects that have consented for them to be shared for research uses—but not for full, open public release—by providing a system for secure archiving and dissemination of such data. We require submitters to certify that the data they have deposited in the EGA has been produced and made available in a manner that is consistent with the original consent agreements, national laws and applicable regulations. We further require that data sets submitted to the EGA are made accessible in a timely fashion to all bona fide researchers whose use of the data is consistent with the original consent agreements. As described below, the EGA brokers data access on behalf of the submitting organization and provides data management and distribution services for users of the database. Any security breach or data misuse by users is reported to the relevant Data Access Committee (DAC) immediately upon becoming known to the EGA following a standard operating procedure.
The EGA supports prepublication data release in accordance with the Toronto agreement for community resource projects18 and for other research organizations and funding agencies that require or encourage data release. For example, the data from the UK10K project are made available to authorized users as regular data updates during the project. In addition, the ICGC uses EGA for providing access to the raw sequence and other appropriate data generated by many of the international partner projects19. The EGA also archives and distributes a wide range of datasets in support of scientific publications providing published datasets as a permanent part of the scientific record.
Biomolecular databases and archives are distributed worldwide with significant concentrations at dedicated institutes including the NCBI and EMBL-EBI. Within this landscape, the EGA serves as a secure authorized access mechanism for data types that, if consented for fully open release, could otherwise have been deposited into the EMBL-EBI resources tailored to store DNA, RNA, protein or sample data20–22. In some cases, the EGA stores sample-level raw data files and detailed phenotype information while aggregated results such as disease associated variants or other non-sensitive data are stored in the public archives with dataset linking to enable discovery.
The EGA security policy includes development of a safe computing facility and a comprehensive suite of protocols for information management.
Access to Data
The EGA has a distributed access-granting policy in which data access decisions are made by the nominated DAC for a given submission and not by the EGA. The DAC may consist of a dedicated committee formed by the funding or governmental organization that approved and monitored the initial study, an institutional committee, or an individual primary investigator. Regardless of the scope or composition of the DAC, the EGA only provides services for studies when access decisions are made exclusively on basis of scientific and ethical criteria in compliance with the original informed consent agreements. The EGA will withdraw service if data access is being denied selectively because of scientific competitiveness or other reasons not based in the original informed consent.
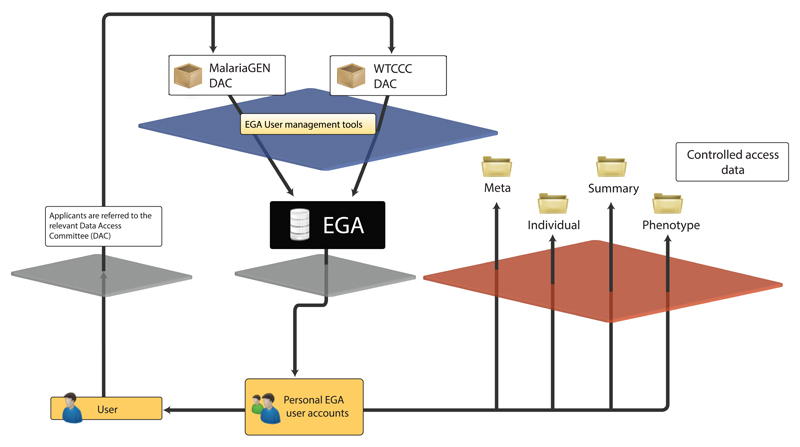
In a typical case, users wishing to access a specific dataset apply directly to the corresponding DAC (Figure 2) following contact instructions on the EGA website. Assuming approval, the Data Access Agreement (DAA) is made directly between the prospective user and the DAC and it dictates data management policies, security arrangements and other potential limitations on data use. For example, some data may not be used for commercial purposes and users may be subject to a temporary publication embargo for projects participating in pre-publication data release. In accordance with accepted practice, the EGA provides data access at the level of granularity that is appropriate for the submitted study. As an example, in a case-control study the user may separately request to access individual level data only for the control dataset.
Figure 2.
The EGA distributed data access model. The EGA refers applicants to appropriate DACs at the website. Each DAC grants access to their data independently. The EGA will create a personal account for each approved applicant listed in the application. The account holds all approved data access permissions and allows account holders to request services from the EGA team, such as downloading of encrypted files from the archive or support for any technical or data content related questions. The data downloaded from the EGA website is provided under a DAA, which is a legal agreement between each approved user and the data governing DAC.
Once approved for access, a user will be issued with an EGA account, which is subject to a number of conditions including that the account information is not shared. EGA accounts can be updated with additional access rights upon each successful application. To ensure that the DAA remains valid, the EGA requires DAC authorization for changes to user details, such as institutional affiliation. The EGA offers support and online tools for the DACs to manage the access rights for their datasets directly within our system.
The policy of distributed access-granting is the most important distinguishing feature of EGA compared with dbGaP. Authorization decisions for dbGaP’s datasets are made by the NIH institute that sponsored the study in question. In the United States, the NIH serves as both a funding and policy making agency and, through the NCBI, a mechanism for data distribution. This allows the NIH to specify dbGaP as a required (although non-exclusive) data distribution channel for specific studies. In contrast, the rest of the world has a diversity of funding agencies and national regulations and these are very often compatible with the distributed data access policy of the EGA for data archiving and distribution. Indeed, this distributed model is an especially good fit for the European research structures that provide support to the EGA.
The EGA and dbGaP share meta-data to improve the discoverability of data deposited in either repository. This sharing of publicly available information such as study name and publication information enables researchers to search for data sets and find the relevant starting point for the access approval process regardless of whether the data is in the EGA or dbGaP. Data are only disseminated from the archive that accepted the original submission since the actual data files are not exchanged between the EGA and dbGaP.
The EGA Websites
Users can access the full EGA service from its instance at either EMBL-EBI or the CRG. Both current EGA websites are arranged around the study concept. A study is typically an experimental investigation of a particular phenomenon, for example a genome wide association study or a matched tumor-normal cancer genome project. The EGA study page describes how the study was conducted and all the associated datasets. The page also includes links to other relevant data resources at the EMBL-EBI or NCBI, the primary publication when available and the data provider. Studies, datasets, DACs and data providers are assigned stable identifiers that should be referred to in the publication and are used to link information together within the EGA. These identifiers provide direct access to the relevant EGA web page through the central EMBL-EBI search engine and serve as stable URLs.
The primary point-of-entry for accessing the controlled-access data stored in the EGA is provided through the dataset page. Each dataset includes publicly available information about the technology used for assaying the samples and guidelines describing how to apply for data access. Once the access has been granted and users have logged into the secure EGA website, the same page will show all the associated manufacturer raw data files, processed information such as the variants or genotypes or any associated study summary data. Logging in to the EGA account facilitates data requests from the archive and allows users to track their current requests within the system.
All data are encrypted for dissemination and made available to each authorized user through FTP as well as fast Internet transfer protocols such as Aspera and UDT. Data transmission methods for submission and dissemination have evolved as data volumes have increased, and now include a custom java client making use of the UDT protocol and performing automatic MD5-checksum validation and encryption. This automation has increased user-friendliness and reduced error.
Data Submission
Complete up-to-date information about submitting data to the EGA is available from its websites. Briefly, submitters first request a private submission account from the EGA to access the range of tools available for file and meta-data upload. We recommend that all primary data files be uploaded using the secure EGA application that automatically provides data encryption and transfer integrity checks. Meta-level information about a study should be submitted using either the Webin online tool22 for experiments using next-generation sequencing technology or an EGA-provided spreadsheet-based meta-data submission template for other assay types. It is also possible for submitters to connect local information management systems directly to the EGA for automatic submission support. The EGA submission guidelines provide detailed information about each stage (Table 2). To ensure that all possible submitters can be served by the EGA, we will also accept encrypted data on hard drives if data size or submitter bandwidth necessitates.
Table 2.
More information about the EGA is available at our website.
| Guidelines | Description | Web address |
|---|---|---|
| Introduction to EGA | Documents related to EGA processes, stable identifiers and Frequently Asked Questions. | www.ebi.ac.uk/ega/about/introduction/ |
| Tutorial videos | Video library for EGA account holders, data submitters or DAC members. | www.ebi.ac.uk/ega/about/videos/ |
| Submissions to EGA | Submission manual for all experiment types and Frequently Asked Questions. | www.ebi.ac.uk/ega/submission/ |
| Data Access Committee (DAC) | Documentation explaining how to establish and manage a DAC effectively | www.ebi.ac.uk/ega/submission/data_access_committee/ |
| EGA applications | Collection of EGA tools for data download or submission process | www.ebi.ac.uk/ega/submission/applications/ |
| EGA security | EGA security policies | https://www.ebi.ac.uk/ega/about/security/ |
Once the submission has been completed, the EGA will confirm the integrity of each submitted file, transfer the data into a secure computing area, decrypt and upload it into archival databases. The EGA staff work directly with the submitter to make sure that the data are correctly uploaded into our system, pass quality checks and are accurately represented at our website. While the data are being collected and analyzed, all uploaded files and the website may be made visible to research collaborators, referees for manuscripts under review provided they are willing to reveal their identity and make an access application to the appropriate DAC, or any other approved users. The EGA supports a “hold until publication” (HUP) status of 6-12 months to enable a study to be submitted and verified but kept private until it is released simultaneously with a journal publication. There is no defined maximum time that a data set can remain HUP, but extensions beyond one year require justification. Although all published data are made available as soon as possible, actual initiation of data dissemination from the EGA requires authorization from the submitting organization.
Future Directions
The recent expansion of the EGA to an EMBL-EBI / CRG collaboration will help support major new EGA datasets including genomic data from the Genome of the Netherlands23 and the Deciphering Developmental Disorders projects24; epigenetic and functional data from the Blueprint25 and HipSci consortia and data relevant to the genetic basis of rare disease from UK BRIDGE project.
The EGA is also working on several new added-value services that will increase the usability of the submitted data. For example, submitted sample phenotypes will be described using ontology-based terms to facilitate better search functionality and assist users looking to merge data across studies. Links are being established with literature databases such as Europe PubMed Central to more closely track secondary publications based on data from the EGA. The EGA will also provide a variant calling and imputation service for limited sets of data submitted into our database. Finally, with the support of Barcelona Supercomputing Center (BSC) and user-facing EMBL-EBI computational resources, the EGA is exploring cloud-based data analysis options.
Acknowledgements
We thank A. Ducanson, R. Banerjee, N. Walker, E. Birney and S. Potter for helpful discussions and comments. The EGA has received support from the European Molecular Biology Laboratory, the EU ELIXIR Technical Feasibility Study, the Wellcome Trust [grant number WT 085475/C/08/Z], the UK Medical Research Council [grant number G0800681], the Spanish Instituto de Salud Carlos III Instituto Nacional de Bioinformatica [grant number PT13/0001/0026]; the Spanish Ministerio de Economía y Competitividad (MINECO) and Centro de Excelencia Severo Ochoa [grant number SEV-2012-0208], the Fundació La Caixa and the Barcelona Supercomputing Centre. The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013 under the grant agreement numbers 211601—ELIXIR; 200754—GEN2PHEN; 262055—ESGI; 242006—BASIS; 261376—IHMS; 305444—RD-CONNECT).
Footnotes
URLs
EGA Website: http://www.ebi.ac.uk/ega/
EGA Website: http://ega.crg.eu
UK10K Project: http://www.uk10k.org
NIH Data Sharing Policies: http://www.nlm.nih.gov/NIHbmic/nih_data_sharing_policies.html
HipSci Project: http://www.hipsci.org
UK BRIDGE Project: https://bridgestudy.medschl.cam.ac.uk/
Europe PubMed Central: http://europepmc.org
Author contributions
IK, AN, JRdA, JP and PF provided project leadership and management. VK, AS, JDS, SuR, MC, RL, PM-G, IM, OML and SdlT developed software. JA-K, MSP and GS provided user support. JK, BV, TL, FR and MA developed the EGA web site. JB, ACT and PJ provided systems support. PF and IK wrote the paper with contributions from all other authors.
There are no competing interests.
References
- 1.Wellcome Trust Case Control Consortium. Nature. 2007;447:661–78. [Google Scholar]
- 2.Welter D, et al. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, et al. Nucleic Acids Res. 2014;42:D749–55. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ban M, et al. Eur J Hum Genet. 2009;17:1309–13. doi: 10.1038/ejhg.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berndt SI, et al. Nat Genet. 2013;45:501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, et al. Hum Mol Genet. 2014 [Google Scholar]
- 8.Muddyman D, Smee C, Griffin H, Kaye J, the UK10K Project Genome Med. 2013;5:100. doi: 10.1186/gm504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaye J. Annu Rev Genomics Hum Genet. 2012;13:415–31. doi: 10.1146/annurev-genom-082410-101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trynka G, et al. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEvoy BP, et al. Genome Res. 2009;19:804–14. doi: 10.1101/gr.083394.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surakka I, et al. Genome Res. 2010;20:1344–51. doi: 10.1101/gr.106534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zilbauer M, et al. Blood. 2013;122:e52–60. doi: 10.1182/blood-2013-05-503201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegand KC, et al. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulis M, et al. Nat Genet. 2012;44:1236–42. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y, et al. Nat Genet. 2013;45:860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 17.Mailman MD, et al. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toronto International Data Release Workshop Authors et al. Nature. 2009;461:168–70. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Cancer Genome Consortium. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gostev M, et al. Nucleic Acids Res. 2012;40:D64–70. doi: 10.1093/nar/gkr937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizcaíno JA, et al. Nucleic Acids Res. 2013;41:D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pakseresht N, et al. Nucleic Acids Res. 2014;42:D38–43. doi: 10.1093/nar/gkt1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Genome of the Netherlands Consortium. Nat Genet. 2014 [Google Scholar]
- 24.Firth HV, Wright CF, for the DDD Study Dev Med Child Neurol. 2011 doi: 10.1111/j.1469-8749.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 25.Adams D, et al. Nat Biotechnol. 2012;30:224–6. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]