Abstract
The parent-child attachment relationship plays an important role in the development of the infant’s stress regulation system. However, genetic and epigenetic factors such as FK506 binding protein 51 (FKBP5) genotype and DNA methylation have also been associated with hypothalamic-pituitary-adrenal (HPA) axis functioning. In the current study, we examined how parent-child dyadic regulation works in concert with genetic and epigenetic aspects of stress regulation. We study the associations of attachment, extreme maternal insensitivity, FKBP5 SNP 1360780, and FKBP5 methylation, with cortisol reactivity to the Strange Situation Procedure in 298 14-month-old infants. Results indicate that FKBP5 methylation moderates the associations of FKBP5 genotype and resistant attachment with cortisol reactivity. We conclude that the inclusion of epigenetics in the field of developmental psychopathology may lead to a more precise picture of the interplay between genetic make-up and parenting in shaping stress reactivity.
Keywords: Attachment, FKBP5, DNA methylation, Cortisol, Stress
The attachment relationship between infant and parent is important in shaping the development of the child’s stress regulation system (Gunnar, Brodersen, Nachmias, Buss, & Rigatuso, 1996). In the first year of life, human infants are dependent on protective caregivers to regulate their temperature, food and fluid intake, and also to regulate stress in the face of threats and dangers (Bowlby, 1969). Sensitive parents, who promptly and adequately respond to their infants’ distress signals, help to create a safe haven from which the child can freely explore the environment (Cassidy, 2008). These infants are more likely to develop a secure attachment relationship and the associated expectation that, in times of need, their parent will be available to protect them (Ainsworth, Blehar, Waters, & Wall, 2015). Insensitive parents, however, may be less prompt and effective in buffering stressful events and settings for their infant. In turn, their infants will be less likely to develop trust and the expectancy of reassuring parental support in times of illness, threat, anxiety and other stressful situations. These infants are also more likely to develop an insecure attachment relationship and a more tenuous style of coping with stressors, potentially resulting in a more reactive hormonal stress system (Fox & Hane, 2008). Stress regulation takes place via the hypothalamic-pituitary-adrenal axis (HPA axis) and one of the crucial hormones involved is cortisol. Therefore, cortisol reactivity to stressors is usually considered to be a measure of the amount of stress experienced by children when confronted with challenges such as separation from the parent or entering an unknown environment or meeting with a stranger (Doom & Gunnar, 2013).
Extreme insensitivity of a parent, including displays of fright because of memories of traumatic experiences, or other threatening behaviors towards the infant, like physical abuse, may elicit even more disturbed attachment behaviors. In particular, extreme parental insensitivity or otherwise frightening behaviors may lead to disorganized/disoriented attachment, reflected in infant behavior, for example, in prolonged stilling, rapid approach–avoidance vacillation, sudden unexplained affect changes, severe distress followed by avoidance, or expressions of fear or disorientation upon return of a parent who has been away for a couple of minutes. Disorganized attachments are overrepresented in clinical samples and in samples with a high prevalence of child maltreatment and family violence (Carlson, Cicchetti, Barnett, & Braunwald, 1989; Cyr, Euser, Bakermans-Kranenburg, & Van IJzendoorn, 2010; Lyons-Ruth & Jacobvitz, 2008; Lyons-Ruth, Alpern, & Repacholi, 1993). Dysregulation of the hormonal stress system has been noted in infants with a disorganized attachment relationship to the parent (Hertsgaard, Gunnar, Erickson, & Nachmias, 1995; Spangler & Grossmann, 1993).
In the current study, we examined how attachment and extreme insensitivity interact with infants’ stress-related genetics to explain variability in their stress regulation. Specifically, we focus on the FK506 binding protein 51 (FKBP5) gene. FKBP5 has been shown to impede negative feedback of the HPA axis (Binder, 2009), and variants, amongst which the rs1360780 SNP in the FKBP5 gene has been related to recovery from psychosocial stress (Ising et al., 2008). Moreover, it was found that rs1360780 interacts with child abuse in the prediction of later development of posttraumatic stress disorder (Binder et al., 2008; Klengel et al., 2013) and of attempt of suicide after childhood trauma (Roy, Gorodetsky, Yuan, Goldman, & Enoch, 2010). In a Dutch subsample of the Generation R cohort, we previously found that rs1360780 interacts with variations in attachment quality in the prediction of stress reactivity. More specifically, infants with an insecure-resistant attachment to their mother—but not those with an insecure-disorganized attachment—had heightened cortisol reactivity to a mildly stressful situation (the Strange Situation Procedure, SSP, Ainsworth et al., 2015), especially if these children were carriers of the T allele in the rs1360780 SNP (Luijk, Velders, et al., 2010). Here we aim at extending our previous study in the Generation R subsample, by including extreme maternal insensitivity as an indicator of atypical parental caregiving behavior, as well as by taking DNA methylation into account.
Epigenetics is a relatively new venue in the field of developmental psychopathology. One of the most often studied epigenetic processes in cohort studies is DNA methylation, where a methyl group attaches to a cytosine nucleotide located next to a guanine in the DNA at a so-called CpG (cytosine-phosphate-guanine) site. Methylation can change the three-dimensional formation of the chromatin (Li & Reinberg, 2011), and subsequently affect gene transcription. DNA methylation is thought to be influenced by prenatal (Bouwland-Both et al., 2015; Cao-Lei et al., 2014; Mychasiuk, Ilnytskyy, Kovalchuk, Kolb, & Gibb, 2011; Rijlaarsdam et al., 2017) and postnatal life events (Hughes et al., 2009; Mehta et al., 2013; Murgatroyd et al., 2009), as well as by genetic background. It can therefore be seen as the dynamic interface between genes and the environment (Meaney, 2010; Van IJzendoorn, Bakermans-Kranenburg, & Ebstein, 2011). These genotype-by-methylation patterns may in turn affect associations between environmental factors and developmental outcomes (Van IJzendoorn, Caspers, Bakermans-Kranenburg, Beach, & Philibert, 2010). Hence, SNP associations with phenotypes such as stress reactivity may become more clearly apparent when DNA methylation is included in the analysis.
In rodents, maternal separation has been related to differential DNA methylation in a variety of HPA-axis related genes and altered stress-responsiveness (Kember et al., 2012; Murgatroyd et al., 2009; Wu, Patchev, Daniel, Almeida, & Spengler, 2014). In humans, similar results have been found. For example, in individuals who were adopted after stressful early life experiences, the short variant of the serotonin-transporter-linked polymorphic region predicted more unresolved loss or trauma, but only if methylation was low (Van IJzendoorn et al., 2010). Another study showed that prenatal exposure to maternal depressed mood was associated with nuclear receptor subfamily 3, group C, member 1 (NR3C1) gene methylation, which was in turn related to increased cortisol reactivity in 3-month-old infants (Oberlander et al., 2008). The NR3C1 gene codes for the glucocorticoid receptor (GR), and methylation is presumed to impede transcription of the NR3C1 gene into the GR protein, decreasing HPA-axis negative feedback through corticosteroid binding.
For the FKBP5 gene, which is associated with the binding of cortisol to the GR, Klengel et al. (2013) found that experienced early trauma was related to methylation of FKBP5, especially in carriers of the rs1360780 T-allele. The T-allele of rs1360780 facilitates gene transcription, which would lead to less sensitive GRs and ultimately to more or prolonged cortisol reactivity. Functionally, Klengel et al. (2013) showed that FKBP5 methylation affected cortisol reactivity as well and, in a separate sample, they found that GR sensitivity was especially affected in T-carriers of rs1360780 that had also experienced childhood abuse. Although these findings are elucidating, we do not know whether they generalize to the general population, where early traumatic experiences are relatively uncommon. Paquette et al. (2014) analyzed placental samples of the general population and infant neurodevelopment. They found an rs1360780 dependent effect of methylation of FKBP5 on mRNA expression in placental cells. Moreover, higher levels of placental FKBP5 methylation were found to be related to more arousal in 3-year-olds. However, it should be noted that arousal does not necessarily equate to cortisol regulation.
The goal for this report was to further explore the relationship between extreme maternal insensitivity, attachment, and cortisol reactivity, for the first time including both genetic and epigenetic factors. In the findings of Luijk, Velders, et al. (2010), it remained puzzling why insecure-resistant attached infants seemed most affected by the SSP in terms of their cortisol reactivity, more so than disorganized infants. Resistant attachment behavior is usually accompanied with explicit signs of distress such as crying and the display of anger to the parent on return after a brief separation. As a result, children with insecure-resistant attachments might show higher cortisol stress reactivity to this challenge than securely attached infants. But infants with insecure-disorganized attachments might have even more difficulties with coping, and may be more dysregulated than insecure-resistant children because their previous experiences with extremely insensitive and frightening parental behaviors may have made them hypersensitive to stress and to lack of parental support when badly needed (Hesse & Main, 2006; Main & Solomon, 1990). Including genetic as well as epigenetic factors influencing the expression of the FKBP5 gene might be necessary to uncover the associations between parenting, attachment, and allelic differences. G×E interactions might emerge more clearly when epigenetic variance is taken into account.
In sum, in this study, we aim to clarify if DNA methylation interacts with genetic effects and parenting on cortisol reactivity. We expand the study by Luijk, Velders, et al. (2010) by investigating if and how FKBP5 methylation affects the rs1360780 SNP-by-resistant attachment interaction reported in that study. Moreover, by including extreme maternal insensitivity, we take a broader perspective on the caregiver-child interaction. We hypothesize that the group with the highest risk for increased stress reactivity includes infants who show resistant or disorganized attachment behaviors, whose mothers display signs of extreme insensitive parenting, who are rs1360780 T-carriers, and who have the highest levels of FKBP5 methylation.
Methods
Setting
The current study is embedded in Generation R, a prospective population-based cohort from fetal life onwards. Pregnant women living in the study area of Rotterdam, the Netherlands, with an expected delivery date between April 2002 and January 2006 were invited to participate. A more detailed description of the Generation R Study can be found elsewhere (Jaddoe et al., 2012; Kruithof et al., 2014). In a randomly assigned subgroup of Dutch pregnant women and their infants, detailed assessments were performed, including the Strange Situation Procedure (SSP). This subgroup is ethnically homogenous (all with European ancestry) to exclude confounding or ethnic stratification effects. The Generation R Study is conducted in accordance with the World Medical Association Declaration of Helsinki and has been approved by the Medical Ethics Committee of the Erasmus Medical Center, Rotterdam. Written informed consent was obtained from the parents of all participating infants.
Study population
DNA was collected from cord blood samples at birth. Information on rs1360780 genotype and FKBP5 methylation levels was available for 956 infants. At the age of 14 months (mean = 14.58, SD = 0.87), 568 of them participated in a lab visit, during which the SSP, extreme maternal insensitivity, and salivary cortisol samples were obtained. We were able to retrieve salivary cortisol samples from a total of 298 of these infants. This sample is nearly identical to the sample used by Luijk, Velders, et al. (2010) (N = 310), with the discrepancy primarily caused by missing FKBP5 methylation data. Unsuccessful cortisol sampling was mainly due to the infants’ unwillingness to chew on the cotton swabs, and was especially seen in infants who were unfamiliar with pacifiers or who ceased using them. Sample characteristics are presented in Table 1. Excluded infants (i.e., infants without data on salivary cortisol, N = 270) did not differ from included children (N = 298) on resistant behavior during the SSP (t(566) = 0.16, p = .87, d = 0.01), disorganized attachment behavior (t(566) = 1.06, p = .29, d = 0.09), extreme maternal insensitivity (t(513)= -0.87, p = .39, d = 0.08) or maternal smoking during pregnancy (χ2(1) = 0.06, p = .80, d = 0.02). However, excluded infants differed from included infants in terms of age at the time of the SSP (t(566) = 2.37, p = .02, d = 0.20), gender (χ2(1) = 7.98, p < .01, d = 0.24) and maternal education (χ2(1) = 4.64, p = .03, d = 0.19). Specifically, infants with successful cortisol sampling were younger (mean age was 14.6 months in the included group, versus 14.8 months in the excluded group), were more often boys (57.0% in the included group, versus 45.2% in the excluded group) and their mothers were more often lower educated (39.5% of the mothers in included group had no formal higher education, versus 30.9% of the excluded group).
Table 1.
Sample Characteristics (N = 298)
| Variable | Mean (SD) |
|---|---|
| Infant characteristics | |
| Age at assessment of SSP, months | 14.6 (0.9) |
| Gender, % girls | 43.0 |
| FKBP5 rs1360780 variant, % | |
| CC | 47.0 |
| CT | 45.0 |
| TT | 8.1 |
| FKBP5 methylation factor 1, score | 0.15 (0.02) |
| FKBP5 methylation factor 2, score | 0.31 (0.04) |
| Resistant behavior, continuous score | 2.2 (1.3) |
| Resistant attachment, % resistant | 24.5 |
| Disorganized attachment behavior, score | 3.4 (1.8) |
| Cortisol reactivity in Δ nmol/L | 0.7 (6.2) |
| Mother characteristics | |
| Educational level, % lower | 39.9 |
| Smoking during pregnancy, % yes | 11.4 |
| Extreme insensitivity, continuous score | 1.4 (1.0) |
| Extreme insensitive behaviors, % one or more | 16.1 |
Note. SSP = Strange Situation Procedure
In 12 of the 298 infants for whom cortisol samples were available, observations of extreme maternal insensitivity were missing, due to procedural problems. To avoid reducing the group size of infants with the rs1360780 TT genotype (the hypothesized risk group), extreme maternal insensitivity scores were imputed using the expectation-maximization (EM) algorithm, using all other variables as well as prenatal maternal lifetime depression and breastfeeding at 6 months. Imputation with the EM algorithm was also performed to impute two missing values on the amount of crying during the SSP. Results remained essentially unchanged when rerunning the analyses using listwise deletion.
Measures
Genotyping
Cord blood DNA was genotyped for the rs1360780 single nucleotide polymorphism (SNP) of the FK506 Binding Protein 5 (FKBP5) with the TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA) and Abgene QPCR mix (Abgene, Hamburg, Germany). Polymerase chain reaction (PCR) took place with the GeneAmp® PCR system 9600, at 95 °C for 15 min, followed by 40 cycles of 94 °C for15s and 60 °C for 1 min. The 7900HT Fast Real-Time PCR System (Applied Biosystems) was used for fluorescence detection and genotypes were determined with SDS software (version 2.3, Applied Biosystems).
Contamination with the mother’s blood was checked for the boys, by examination of the sex chromosomes. Samples in which contamination had occurred were excluded (< 1%). Furthermore, genotyping of the FKBP5 SNP was successful in 97-99% of the cases and reanalysis of 276 randomly selected samples showed an error rate of < 1%. Genotype frequencies were in Hardy-Weinberg equilibrium (χ2 = 1.07, p = .30).
DNA methylation
Per sample, 500 nanogram of leukocyte DNA was extracted from cord blood and underwent bisulfite conversion with the EZ-96 DNA Methylation kit (Shallow) (Zymo Research Corporation, Irvine, USA). Methylation was analyzed with the Illumina Infinium Human Methylation 450K BeadChip (Illumina Inc., San Diego, USA). Quality control of samples was performed using standardized criteria. Samples were checked for <99% call rate (six samples were excluded), color balance >3, staining efficiency, extension efficiency, hybridization performance, stripping efficiency after extension (no samples excluded in each case), and bisulfite conversion (one sample excluded). Also, two samples were removed due to a gender mismatch, leaving a total of 969 samples that passed quality control. Dasen normalization was ran using a pipeline adapted from Touleimat and Tost (2012), as described by Pidsley et al. (2013), and samples were dye bias corrected.
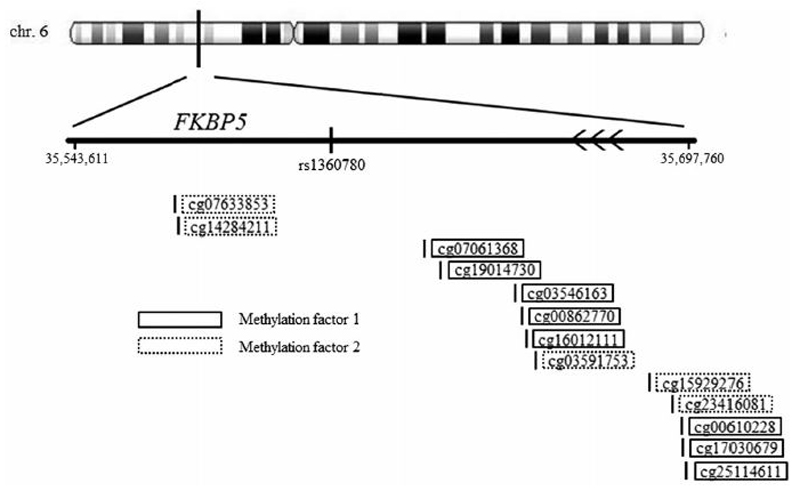
We extracted the beta values of 32 CpGs that mapped to the FKBP5 gene or overlapping regions adjacent to FKBP5 (i.e. position 35543611 to 35697760; see Figure 1). Beta values represent the ratio of methylated signal relative to the sum of the methylated and unmethylated signals, per CpG. To avoid multiple testing issues due to the large number of CpG beta values, we decided to examine the dimensional structure of the data (mean r = .01, range r [-.63, .77]) by using factor analysis in MPlus Version 7.31 (Muthén & Muthén, 2012). The factor analysis took place in the full DNA methylation sample, using the 29 CpG beta values with sufficient variation (SD > 0.01). Factor analysis proceeded in two steps. First, exploratory factor analysis (EFA) was performed. The optimal number of underlying factors was assessed by inspecting the Scree-plot and by comparing fit statistics between models estimating one to five factors. CpGs with a Geomin (oblique) rotated absolute loading of > .40 to one of the factors were included. Model fit was established using the chi-square statistic. In the event of significant chi-square values, we further examined relative fit indices, including the mean square error of approximation (RMSEA; acceptable fit ≤ 0.08), as well as the comparative fit index and the Tucker-Lewis index (CFI and TLI respectively; acceptable fit ≥ 0.90). A two-factor model was identified (χ2[53] = 367.56, p < .001; RMSEA = 0.078; TLI = 0.949; CFI = 0.926). The first factor had an eigenvalue of 5.2 and contained eight CpGs, of which five had positive and three had negative factor loadings. The second factor had an eigenvalue of 2.4 and contained five CpGs, all of which had positive factor loadings (see Figure 1). In the second step, we used confirmatory factor analysis (CFA) to validate the two-factor model (χ2(64) = 438.60; RMSEA = .077; CFI = .913; TLI = .894). For each FKBP5 methylation factor, we computed average methylation scores based on the relevant CpGs, using reversed scores for those with negative loadings on factor 1. These average methylation factor scores were used throughout. In an exploratory analysis, regression analyses were repeated for each CpG individually, to gauge if our main finding was caused by only one or a few CpGs, or rather by the combined effect of all CpGs.
Figure 1.
Beta values of 32 CpGs that mapped to the FKBP5 gene or overlapping regions adjacent to FKBP5.
Attachment
Mother-infant dyads were observed in the Strange Situation Procedure (SSP). During the SSP, mild stress evokes attachment behavior in the infant by the unfamiliar lab environment, a stranger entering the room and engaging with the infant, and the parent briefly leaving the room twice. The total procedure consists of seven three-minute episodes, with the preseparation and separation in our study shortened by 1 min each, keeping the critical reunion episodes intact (Kok et al., 2013; Luijk, Saridjan, et al., 2010).
Two reliable coders, trained at the University of Minnesota, coded the SSP recordings, according to the Ainsworth et al. (2015) and Main and Solomon (1990) coding systems. For each of two reunions with the mother, the infant received a resistant behavior score ranging from 1 to 7. These scores were averaged to create a resistant behavior score. Examples of resistant behavior include (i) a struggle against being held or (ii) throwing away toys that are handed to the infant. Intercoder reliability (intraclass correlation or ICC, single measure, absolute agreement) for resistant behavior was .86 (n = 70). For a sensitivity analysis (see below), a resistant attachment classification was derived from a pattern of attachment behaviors during the reunion periods. A typically resistant infant actively seeks proximity to the mother and tries to maintain contact with her, while at the same time showing obvious signs of resistance to her attempts of consolidation. Intercoder agreement for resistant attachment was 77% (κ = .63, n = 70). Resistant behavior in the reunion episodes and resistant attachment classification were strongly correlated (r = .78, p < .01). Disorganization of attachment behavior was rated using the 9-point Main and Solomon (1990) coding system. Examples of disorganized/disoriented behaviors are prolonged stilling, rapid approach–avoidance vacillation, sudden unexplained affect changes, severe distress followed by avoidance, and expressions of fear or disorientation upon return of mother. The ICC for the disorganization rating scale was .88 (n = 70) (Luijk et al., 2011).
Extreme maternal insensitivity
Extreme maternal insensitivity was observed during the psychophysiological assessment and during the break of the 14-month lab visit and was rated by coders unaware of the attachment coding. During the psychophysiological assessment, the child had ECG-measurement equipment attached while sitting on the mothers lap and watching an episode of the Teletubbies© (BBC/Ragdoll Limited). The break was unstructured, and mother and child interacted freely. The extreme maternal insensitivity scale includes: (1) withdrawal and neglect; and (2) intrusive, negative, aggressive or otherwise harsh parental behaviors (Out, Bakermans-Kranenburg, & Van IJzendoorn, 2009). Extremely insensitive behaviors were coded on a 9-point scale, with higher scores indicating more extreme insensitivity. The ICC was .63 (n = 36).
Cortisol reactivity
Saliva samples were taken during the 14-month lab visit with Salivette sampling devices (Sarstedt, Rommelsdorf, Germany). Samples were centrifuged and frozen at -80 °C and analyzed by the Kirschbaum laboratory (Technical University of Dresden, Biological Psychology, Germany). Salivary cortisol concentrations were assessed with a chemiluminescence imunnoassay (CLIA; IBL Hamburg, Germany). Intra- and interassay coefficients of variation were below 7% and below 9%, respectively. Cortisol concentrations above the 99th percentile (>200 nmol/L) were excluded (n = 12) from the analyses. Cortisol reactivity was determined by calculation of the difference between cortisol concentration 15 minutes after the SSP (post-SSP cortisol) and cortisol concentrations prior to the SSP (pre-SSP cortisol). Mean sampling time of pre-SSP cortisol was 11:26 a.m. (SD = 2:01 h), mean sampling time of post-SSP cortisol was 12:22 p.m. (SD = 2:00 h). We had information on corticosteroid medication for 248 infants. None of these infants used systemic corticosteroid medication, but five infants used other corticosteroid-containing medication. Since these infants did not differ significantly in cortisol reactivity from infants without corticosteroid-containing medication (t(246) < .01, p > .99, d < 0.01), they were included in all further analyses.
Covariates
Information on family background characteristics was obtained by questionnaire during pregnancy. We included as covariates infant’s age at the SSP, infant gender, mothers’ highest attained educational level (no formal higher education versus higher vocational training or higher academic education), maternal smoking during pregnancy (never smoked or quit when pregnancy was known versus continued smoking during pregnancy), technical covariates (sample array number and position on the array, and leukocyte cell type proportions [CD4+ T-lymphocytes, CD8+ T-lymphocytes, natural killer cells, B-lymphocytes, monocytes, and granulocytes] (Houseman et al., 2012). To account for the negative association (which might be interpreted as a ceiling effect) between the initial cortisol value and the slope of the cortisol reactivity, the cortisol concentration prior to the SSP (pre-SSP cortisol) was also included as a covariate. Finally, to exclude the possibility that resistant behavior and cortisol reactivity are related through the physiologically arousing nature of crying that often accompanies resistant behavior, we performed the regression analysis with and without the inclusion of percentage of crying time during the SSP as a covariate.
Statistical analyses
Hierarchical linear regressions were performed using SPSS version 23 (IBM Corporation, Chicago, USA) to examine the associations of FKBP5 rs1360780, FKBP5 methylation and attachment (resistant or disorganized) with infant cortisol reactivity during the SSP. These regression analyses were performed separately for the two FKBP5 methylation factors and for each of the two attachment variables.
In the first step of the regression equation, FKBP5 rs1360780, FKBP5 methylation, attachment, extreme maternal insensitivity, and the covariates were entered. In the second step, all two-way interactions between FKBP5 rs1360780, FKBP5 methylation, attachment, and extreme maternal insensitivity were entered. In the third step, all three-way interactions were entered. In the interest of statistical power, the four-way interaction with all possible predictors was not included. When one of the main predictors was not found to have a significant main or interaction effect on cortisol reactivity, the steps were repeated excluding this variable.
To reduce the influence of extreme scores on the results, two outliers (z-score >3.29) for FKBP5 methylation factor 1, four for FKBP5 methylation factor 2, six for cortisol reactivity, and 10 for extreme insensitivity were winsorized (i.e. transformed to match the next highest value). FKBP5 rs1360780, the FKBP5 average methylation factors, resistant and disorganized behavior, and extreme maternal insensitivity, were mean-centered in order to reduce collinearity due to the scaling of variables.
Sensitivity analyses
Two sensitivity analyses were performed. First, in order to examine whether associations were dependent on the continuous resistance scale, we also used the resistant versus non-resistant attachment classification (Luijk, Velders, et al., 2010) as a predictor instead of the continuous resistant behavior score. Second, since most mothers had the lowest possible score on extreme insensitivity, which resulted in a skewed distribution of scale scores, we performed a sensitivity analysis with a dichotomized extreme insensitivity variable. Mothers not showing any extremely insensitive behaviors were contrasted with mothers presenting one or more extremely insensitive behaviors.
Results
Extreme maternal insensitivity
As can be seen in Table 2, none of the main predictors were correlated, with the exception of FKBP5 methylation factors 1 and 2 (r = .34, p < .01), and disorganized and resistant behavior (r = .19, p < .01). The regression analyses did not show an association of cortisol reactivity with extreme maternal insensitivity (β = -.04, p = .43) in the first step, nor a with a two-way interaction of extreme maternal insensitivity and FKBP5 rs1360780, FKBP5 methylation factor 1, or resistant behavior (strongest interaction with FKBP5 rs1360780: β = -.02 , p = .69) in the second step, nor with a three-way interaction with extreme maternal insensitivity and any combination of these predictors (strongest interaction with FKBP5 methylation factor 1 and resistant behavior: β = -.04, p = .51) in the final step. This was also the case for the analyses with FKBP5 methylation factor 2.
Table 2.
Pearson correlations (N =298)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| 1. FKBP5 rs1360780 | ||||||||
| 2. FKBP5 methylation factor 1 | < -.01 | |||||||
| 3. FKBP5 methylation factor 2 | -.03 | .34*** | ||||||
| 4. Resistant behavior | -.02 | -.03 | -.05 | |||||
| 5. Disorganized behavior | .01 | -.02 | < .01 | .19** | ||||
| 6. Extreme insensitivity | -.01 | .05 | .03 | .01 | .02 | |||
| 7. Crying | .02 | .01 | -.03 | .55*** | .01 | .02 | ||
| 8. Cortisol reactivity | .14* | -.04 | < .01 | .26*** | -.07 | < .01 | .36*** |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2.
* p < .05
** p < .01
*** p < .001
Resistant attachment, FKBP5 rs1360780, and FKBP5 methylation
Table 3 shows that both FKBP5 rs1360780 (β = .13, p < .01) and resistant behavior (β = .30, p < .01), but not FKBP5 factor 1 methylation (β = -.06, p =.40), were positively associated with infant cortisol reactivity. The two-way interaction of rs1360780 and FKBP5 methylation factor 1 was significant (β = .11, p = .03), as was the three-way interaction FKBP5 rs1360780 × FKBP5 methylation factor 1 × resistant behavior (β = .14, p < .01). Tallele carriers of FKBP5 rs1360780 with high FKBP5 methylation factor 1 scores and high levels of resistant behavior had the highest cortisol reactivity.
Table 3.
Associations between FKBP5 rs1360780, FKBP5 methylation factor 1, and resistant behavior on cortisol reactivity during the Strange Situation Procedure (N = 298)
| Model | B (95% CI) | β |
|---|---|---|
| FKBP5 rs1360780 | 1.13 (0.30; 1.97) | .13 ** |
| FKBP5 methylation factor 1 | -12.99 (-43.50; 17.53) | -.06 |
| Resistant behavior | 1.82 (1.20; 2.45) | .30 *** |
| FKBP5 rs1360780 × FKBP5 methylation factor 1 | 39.11 (3.38; 74.85) | .11 * |
| FKBP5 rs1360780 × resistant behavior | 0.89 (-0.14; 1.93) | .09 |
| FKBP5 methylation factor 1 × resistant behavior | 10.43 (-19.47; 40.32) | .04 |
| FKBP5 rs1360780 × FKBP5 methylation factor 1 × resistant behavior | 64.22 (17.34; 111.10) | .14 ** |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2. Analyses are adjusted for technical methylation covariates, cell type proportions of DNA methylation sample, infant age at assessment of SSP, infant gender, educational level of the mother and maternal smoking during pregnancy. The statistics are derived from the final block of the regression model.
p < .05
p < .01
p < .001.
Similarly, FKBP5 methylation factor 2 was unrelated to cortisol reactivity (β = .04, p = .77; Table 4). The interaction between FKBP5 rs1360780 and FKBP5 methylation factor 2 did not reach significance, but there was again a positive association between cortisol reactivity and resistant behavior (β = .28, p < .01) and a significant three-way interaction of FKBP5 rs1360780 × FKBP5 methylation factor 2 × resistant behavior (β = .13, p = .01), again suggesting that T-allele carriers of rs1360780, with high FKBP5 methylation factor 2 levels and high resistant behavior had the highest cortisol reactivity to the SSP.
Table 4.
Associations between FKBP5 rs1360780, FKBP5 methylation factor 2, and resistant behavior on cortisol reactivity during the Strange Situation Procedure (N = 298)
| Model | B (95% CI) | β |
|---|---|---|
| FKBP5 rs1360780 | 1.14 (0.30; 1.98) | .13 ** |
| FKBP5 methylation factor 2 | 5.19 (-29.48; 39.86) | .04 |
| Resistant behavior | 1.75 (1.14; 2.36) | .28 *** |
| FKBP5 rs1360780 × FKBP5 methylation factor 2 | 9.69 (-14.00; 33.37) | .04 |
| FKBP5 rs1360780 × resistant behavior | 0.41 (-0.58; 1.39) | .04 |
| FKBP5 methylation factor 2 × resistant behavior | -9.44 (-23.80; 4.91) | -.07 |
| FKBP5 rs1360780 × FKBP5 methylation factor 2 × resistant behavior | 31.06 (6.60; 55.51) | .13 * |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2. Analyses are adjusted for technical methylation covariates, cell type proportions of DNA methylation sample, infant age at assessment of SSP, infant gender, educational level of the mother and maternal smoking during pregnancy. The statistics are derived from the final block of the regression model.
p < .05
p < .01
p < .001.
Although resistant behavior was positively correlated with crying (r = .48, p < .001), adding crying as a covariate to the model did not meaningfully change the results. The three-way interactions of (i) FKBP5 rs1360780 × FKBP5 methylation factor 1 × resistant behavior (β = .14, p < .01) and (ii) FKBP5 rs1360780 × FKBP5 methylation factor 2 × resistant behavior (β = .11, p = .03) remained significant.
Finally, to explore whether the results for the methylation factor scores were localized in just one or a few CpGs, or were based on the combined effect of all CpGs, the analyses were repeated for each CpG separately. For methylation factor 1, three out of eight CpGs were associated with cortisol reactivity in the FKBP5 rs1360780 × FKBP5 CpG × resistant behavior interaction at the p < .05 level (Table 5). However, two of the other five CpGs may also have contributed to the FKBP5 methylation factor 1 involvement in the three-way interaction, as the interaction terms for two CpGs were associated with cortisol reactivity at p < .10. For methylation factor 2, four out of five CpGs were associated on the p < .05 with cortisol reactivity in interaction with FKBP5 rs1360780 and resistant behavior (Table 6). For CpGs of both FKBP5 methylation factors, no clear localization pattern of p < .05 results could be distinguished, as they were relatively scattered over the FKBP5 gene.
Table 5.
Characteristics of the individual FKBP5 methylation factor 1 CpGs and β and p-value of the FKBP5 rs1360780 x FKBP5 CpG beta value x resistant behavior in a regression analysis of the associations between FKBP5 rs1360780, FKBP5 CpG, and resistant behavior on cortisol reactivity during the Strange Situation Procedure
| Three-way interaction values with cortisol reactivity |
||||
|---|---|---|---|---|
| CpG | mean beta value (SD) | Factor 1 loadings | β | p-value |
| cg07061368 | .89 (.03) | -0.58 | -.01 | .811 |
| cg19014730 | .80 (.05) | -0.76 | -.14 | .007 |
| cg03546163 | .79 (.06) | -0.71 | -.12 | .017 |
| cg00862770 | .07 (.01) | 0.72 | .09 | .071 |
| cg16012111 | .10 (.01) | 0.63 | .04 | .453 |
| cg00610228 | .14 (.04) | 0.93 | .09 | .092 |
| cg17030679 | .07 (.02) | 0.55 | .07 | .210 |
| cg25114611 | .35 (.04) | 0.73 | .12 | .014 |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2. Analyses are adjusted for technical methylation covariates, cell type proportions of DNA methylation sample, infant age at assessment of SSP, infant gender, educational level of the mother and maternal smoking during pregnancy. The statistics are derived from the final block of the regression model.
Table 6.
Characteristics of the individual FKBP5 methylation factor 2 CpGs and β and p-value of the FKBP5 rs1360780 x FKBP5 CpG beta value x resistant behavior in a regression analysis of the associations between FKBP5 rs1360780, FKBP5 CpG, and resistant behavior on cortisol reactivity during the Strange Situation Procedure
| Three-way interaction values with cortisol reactivity |
||||
|---|---|---|---|---|
| CpG | mean beta value (SD) | Factor 2 loadings | β | p-value |
| cg07633853 | .33 (.07) | 0.63 | .05 | .313 |
| cg14284211 | .26 (.06) | 0.90 | .14 | .006 |
| cg03591753 | .55 (.03) | 0.66 | .11 | .036 |
| cg15929276 | .13 (.04) | 0.47 | .13 | .008 |
| cg23416081 | .27 (.05) | 0.86 | .11 | .033 |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2. Analyses are adjusted for technical methylation covariates, cell type proportions of DNA methylation sample, infant age at assessment of SSP, infant gender, educational level of the mother and maternal smoking during pregnancy. The statistics are derived from the final block of the regression model.
Disorganized attachment, FKBP5 rs1360780, and FKBP5 methylation
When resistant behavior was replaced by disorganized attachment behavior in the regression analyses including FKBP5 methylation factor 1 and maternal extreme insensitivity, neither an association between cortisol reactivity and attachment disorganization (β = -.03, p = .56), nor any two- or three-way interactions (strongest interaction with FKBP5 methylation factor 1: β = -.06, p = .31) with disorganized behavior was found. Results were found to be similarly non-significant for the analyses with FKBP5 methylation factor 2 (see also Supplemental Table 1 and Supplemental Table 2, respectively). Moreover, a z-test indicated that the main effect for disorganized attachment behavior and the FKBP5 rs1360780 × FKBP5 methylation factor 1 × disorganized attachment behavior interaction differed significantly from the main effect for resistant behavior (z = 5.11, p < .01) and the FKBP5 rs1360780 × FKBP5 methylation factor 1 × resistant behavior interaction (z = 2.78, p < .01).
Sensitivity Analyses
When repeating the analyses using the resistant attachment classification variable(categorical, resistant versus nonresistant) instead of the continuous resistant behavior score, a similar pattern of findings for FKBP5 methylation factor 1 was observed. That is, we observed a significant three-way interactions of FKBP5 rs1360780 × FKBP5 methylation factor 1 × resistant attachment (β = .19, p < .01) in the prediction of infant cortisol reactivity (Table 7). The three-way interaction of FKBP5 rs1360780 × FKBP5 methylation factor 2 × resistant attachment was also significant (Table 8; β = .13, p = .04).
Table 7.
Associations between FKBP5 rs1360780, FKBP5 methylation factor 1, and resistant attachment classification on cortisol reactivity during the Strange Situation Procedure (N = 298)
| Model | B (95% CI) | β |
|---|---|---|
| FKBP5 rs1360780 | 1.72 (0.79; 2.65) | .20*** |
| FKBP5 methylation factor 1 | -4.14 (-39.10; 30.81) | -.02 |
| Resistant attachment | 1.86 (1.26; 2.47) | .30*** |
| FKBP5 rs1360780 × FKBP5 methylation factor 1 | 72.95 (32.75; 113.15) | .20*** |
| FKBP5 rs1360780 × resistant attachment | 1.12 (0.20; 2.05) | .13* |
| FKBP5 methylation factor 1 × resistant attachment | 18.15 (-10.02; 46.32) | .08 |
| FKBP5 rs1360780 × FKBP5 methylation factor 1 × resistant attachment | 69.22 (29.10; 109.34) | .19** |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2. Analyses are adjusted for technical methylation covariates, cell type proportions of DNA methylation sample, infant age at assessment of SSP, infant gender, educational level of the mother and maternal smoking during pregnancy. The statistics are derived from the final block of the regression model.
p < .05
p < .01
p < .001.
Table 8.
Associations between FKBP5 rs1360780, FKBP5 methylation factor 2, and resistant attachment classification on cortisol reactivity during the Strange Situation Procedure (N = 298)
| Model | B (95% CI) | β |
|---|---|---|
| FKBP5 rs1360780 | 1.61 (0.66; 2.56) | .19 ** |
| FKBP5 methylation factor 2 | 1.64 (-34.35; 37.63) | .01 |
| Resistant attachment | 1.80 (1.19; 2.41) | .29 *** |
| FKBP5 rs1360780 × FKBP5 methylation factor 2 | 25.82 (-3.06; 54.70) | .11 |
| FKBP5 rs1360780 × resistant attachment | 0.99 (0.04; 1.94) | .12 * |
| FKBP5 methylation factor 2 × resistant attachment | -8.64 (-25.60; 8.33) | -.06 |
| FKBP5 rs1360780 × FKBP5 methylation factor 2 × resistant attachment | 30.07 (1.63; 58.52) | .13 * |
Note. FKBP5 rs1360780: CC = 0, CT = 1, TT = 2. Analyses are adjusted for technical methylation covariates, cell type proportions of DNA methylation sample, infant age at assessment of SSP, infant gender, educational level of the mother and maternal smoking during pregnancy. The statistics are derived from the final block of the regression model.
p < .05
p < .01
p < .001.
Last, we inserted the dichotomous extreme insensitivity variable in the model instead of the continuous variant. This yielded no significant additive or interactive associations of variables with the dichotomous extreme insensitivity variable involved with cortisol reactivity. This was the case for the analysis with FKBP5 methylation factor 1 as well as for the analysis with FKBP5 methylation factor 2.
Discussion
In this population-based cohort study, we found that resistant attachment behavior and FKBP5 rs1360780 genotype were associated with cortisol reactivity both in an additive and in an interactive manner. Methylation of the FKBP5 gene moderated the relationship between FKBP5 rs1360780 genotype and cortisol reactivity, in that rs1360780 T-carriers had an even higher chance of increased cortisol reactivity, when they also had a high FKBP5 methylation factor 1 score. This might suggest that DNA methylation patterns affect transcription of the FKBP5 gene to influence subsequent stress responses. The modification of the rs1360780 association with cortisol reactivity by FKBP5 methylation factor 1 score, seemed especially pronounced in infants who displayed resistant attachment behavior towards their mother. Results for the analysis with methylation factor 2 and with the resistant attachment classification corroborated these findings. It is noteworthy that the interaction between rs1360780 and methylation was specifically modified by resistant attachment behavior, and not by disorganized attachment.
Although our study shows promising findings and does support the potentially important role of DNA methylation in infant cortisol reactivity, it should be emphasized that the study must be firmly placed in the context of discovery (Popper, 1959). For several reasons it is too early for this and related human development studies on DNA methylation to provide more definite confirmation or falsification of hypotheses or theories in the context of justification. First, little is known about the metric qualities of DNA methylation indices. For example, stability of DNA methylation across time has not yet been examined thoroughly for most genes and developmental periods (see Wong et al., 2010, for an exception). Second, it is still not fully clear whether and how strongly DNA methylation patterns in blood and brain regions are associated (Van IJzendoorn et al., 2011). Some recent studies show significant convergence between FKBP5 methylation derived from peripheral blood and brain tissue (Ewald et al., 2014; Hannon, Lunnon, Schalkwyk, & Mill, 2015), but more research is certainly needed. Third, most studies on DNA methylation in the domain of developmental psychopathology are severely underpowered with potentially quite a few false positive findings that may turn out to be impossible to replicate. In an epigenome-wide study of a large Generation R sample of 912 families, we were unable to replicate our suggestive findings on the association between maternal prenatal stress and neonatal DNA methylation in another large sample of 828 families, the Avon Longitudinal Study of Parents and Children, ALSPAC (Rijlaarsdam et al., 2016). Our current study on almost 300 children is one of the largest candidate-(epi)gene studies on DNA methylation, but still underpowered in view of the Gene × Methylation × Environment (G × M × E, Van IJzendoorn et al, 2010; Bakermans-Kranenburg & Van IJzendoorn, 2015) three-way interactions. Independent replication is therefore badly needed (Rijlaarsdam et al., 2016).
It is somewhat assuring that our findings are in line with Klengel et al. (2013), who found an association between FKBP5 methylation and HPA axis regulation, particularly in FKBP5 rs1360780 T allele carriers. However, their sample size for these analyses was only 76 with 30 highly traumatized cases and 46 controls in one of their central epigenetic analyses. Paquette et al. (2014) also found an association between placental FKBP5 methylation and postnatal infant arousal, but this was specific for infants with the FKBP5 rs1360780 CC genotype. One explanation for this diverging result might be that arousal is regulated by the autonomic nerve system, which is related to the HPA axis, but does not completely overlap in its function and activity. Also, whereas Klengel et al. (2013) and Paquette et al. (2014) specifically found effects of methylation of CpG sites in intron 7 of the FKBP5 gene, we considered methylation of all FKBP5 CpG sites that contributed meaningfully to one of two factors (as in Philibert et al., 2010). Since these factors, which included CpG sites with positive as well as negative factor loadings, were found to be associated with cortisol reactivity, it might be that the effects of DNA methylation are less unidirectional than assumed previously. Exploratory analyses also showed that the individual CpGs contributing to the FKBP5 methylation factor scores were quite scattered along the FKBP5 gene, rather than being localized in a specific part. Unfortunately, the different methodologies for DNA methylation detection employed do not allow for direct comparison of our approach with those of Klengel et al. (2013) and Paquette et al. (2014).
The interaction between FKBP5 rs1360780 and FKBP5 methylation was only found in children with resistant but not with disorganized attachment behavior. This is somewhat unexpected, since disorganized attachment has been related to dysregulation of the HPA axis functioning in a number of studies (Bernard & Dozier, 2010; Hertsgaard et al., 1995; Spangler & Grossmann, 1993). Another remarkable result is the negligible role of maternal extreme insensitivity in the prediction of cortisol reactivity, since we had expected that infants of mothers displaying extreme insensitive parenting behaviors would show increased cortisol reactivity. Perhaps our relatively brief observation in a lab setting was not optimal to register maternal extreme insensitivity. Moreover, the non-clinical nature of the sample may also have contributed to the skewedness of the distribution, thereby hindering detection of associations with maternal extreme insensitivity. This might also explain the lack of association between maternal extreme insensitivity and disorganized attachment. Future research on maternal extreme insensitivity therefore might include more high-risk populations than the one examined here, which could possibly also help in further exploring the (epi-)genetic differences in stress regulation between children with disorganized and resistant attachments.
Some limitations of the current study should be mentioned. First, DNA methylation levels were measured in cord blood at birth, whereas cortisol reactivity was measured at 14 months. Neonatal DNA methylation might be influenced by prenatal environmental factors such as maternal smoking (Bouwland-Both et al., 2015; Richmond et al., 2014) or prenatal stress (Mulligan, D'Errico, Stees, & Hughes, 2012; Rijlaarsdam et al., 2016). An important question that remains unanswered, is whether DNA methylation levels are stable between birth and our behavioral observations at 14 months. However, based on Klengel et al.’s (2009) finding that trauma during childhood affects FKBP5 methylation in a way that dysregulates HPA axis functioning, one might speculate that insecure-resistant mother-child attachment—although not traumatizing in and of itself of course—could affect FKBP5 methylation over the first year of life, so that its associations with cortisol reactivity would have been even stronger with FKBP5 methylation measured at 14 months than at birth. In order to attain a more complete picture of the role of epigenetics in shaping the relations between parenting, attachment and stress regulation, longitudinal and experimental research is needed to test whether the quality of parenting (i.e., sensitivity) and the attachment relationship in itself can affect DNA methylation. Longitudinal data on DNA methylation of stress-related genes at multiple time-points may be informative, as well as pre- and posttest assessments of DNA methylation patterns in randomized controlled trials aiming at enhancing the quality of parent-child interactions and relationship (Bakermans-Kranenburg, Van IJzendoorn, Mesman, Alink, & Juffer, 2008).
Second, another limitation may be the candidate-(epi-)gene approach that limits the analysis to one specific gene, i.e. the FKBP5 gene, in combination with a single SNP, i.e. rs1360780, which was a logical follow-up on the study performed by Luijk, Velders, et al. (2010). It would be interesting to try and obtain a more complete picture of DNA methylation in stress regulation by including more SNPs of the FKBP5 gene as well as other genes related to cortisol reactivity (e.g., NR3C1; Mulligan, D'Errico, Stees, & Hughes, 2012; or KITLG; Houtepen et al., 2016). Combinations of such HPA-axis related genes into a genetic pathway might provide a better basis for a wider epigenetic search into the influence of DNA methylation patterns on stress regulation. It should be noted, however, that focusing on methylation patterns of a single gene with documented functionality for the phenotype of interest has the advantage of better localization of the effect and of optimizing the statistical power that is often lacking in hypothesis-free approaches. Nevertheless, our results are based on a complicated three-way interaction (G ×M × E) and should be replicated in independent samples. In such studies, the factor-analytic method to examine the dimensionality of an interrelated set of CpG beta values may reduce the number of tests that otherwise would lower statistical power. More specifically, this G × M × E study shows that stress regulation in an infant with a resistant attachment to their mother is more likely to be problematic when the infant is a FKBP5 rs1360780 T-carrier and even more so when it also has a higher methylation factor score.
In sum, the current findings are a valuable extension of our earlier results on attachment, FKBP5 rs1360780 and cortisol reactivity in that genetic effects on child outcomes may be better specified when DNA methylation is taken into account. Moreover, whereas most research on FKBP5 methylation focuses on extreme circumstances in early life, this study reveals that DNA methylation plays a role in coping with everyday stressors in a non-clinical population. Although we emphasize that epigenetic studies on (child developmental) psychopathology are still in an exploratory stage, neglect of DNA methylation and other regulatory mechanisms in molecular genetic studies increases the risk that an incomplete picture of associations between genes, environment and development is created (Meaney, 2010). The study of epigenetics is therefore an important asset to the field of developmental psychopathology, and a crucial move to the biological level of gene-by-environment interplay.
Supplementary Material
Acknowledgements
The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the Erasmus University Rotterdam, School of Law and Faculty of Social Sciences, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam, and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR), Rotterdam. We gratefully acknowledge the contribution of general practitioners, hospitals, midwives and pharmacies in Rotterdam. The Generation R Study is made possible by financial support from: Erasmus Medical Center, Rotterdam, Erasmus University Rotterdam and the Netherlands Organization for Health Research and Development (ZonMw). The generation and management of the Illumina 450K methylation array data (EWAS data) for the Generation R Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. The EWAS data was funded by a grant from the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA; project nr. 050-060-810), by funds from the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, by a grant from the Netherlands Organization for Health Research and Development (VIDI 016.136.361) and a Consolidator Grant from the European Research Council (ERC-2014-CoG-64916). We thank Mr. Michael Verbiest, Ms. Mila Jhamai, Ms. Sarah Higgins, Mr. Marijn Verkerk and Dr. Lisette Stolk for their help in creating the EWAS database. Janine Felix has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 633595 (DynaHEALTH). Marinus van IJzendoorn and Marian Bakermans-Kranenburg were supported by the Dutch ministry of Education, Culture, And Science and the Netherlands Organization for Scientific Research (Gravitation program, SPINOZA, VICI).
References
- Ainsworth MDS, Blehar MC, Waters E, Wall SN. Patterns of attachment: A psychological study of the strange situation. Psychology Press; 2015. [Google Scholar]
- Bakermans-Kranenburg MJ, Van IJzendoorn MH. The hidden efficacy of interventions: Gene × environment experiments from a differential susceptibility perspective. Annual Review of Psychology. 2015;66:381–409. doi: 10.1146/annurev-psych-010814-015407. [DOI] [PubMed] [Google Scholar]
- Bakermans-Kranenburg MJ, Van IJzendoorn MH, Mesman J, Alink LRA, Juffer F. Effects of an attachment-based intervention on daily cortisol moderated by dopamine receptor D4: A randomized control trial on 1-to 3-year-olds screened for externalizing behavior. Development and psychopathology. 2008;20:805–820. doi: 10.1017/S0954579408000382. [DOI] [PubMed] [Google Scholar]
- Bernard K, Dozier M. Examining infants' cortisol responses to laboratory tasks among children varying in attachment disorganization: stress reactivity or return to baseline? Developmental psychology. 2010;46:1771. doi: 10.1037/a0020660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34:S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, et al. Nemeroff CB. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwland-Both MI, Van Mil NH, Tolhoek CP, Stolk L, Eilers PH, Verbiest MM, et al. Van IJzendoorn MH. Prenatal parental tobacco smoking, gene specific DNA methylation, and newborns size: the Generation R study. Clinical epigenetics. 2015;7:1. doi: 10.1186/s13148-015-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlby J. Attachment, Vol. 1 of Attachment and loss. New York: Basic Books; 1969. [Google Scholar]
- Cao-Lei L, Massart R, Suderman MJ, Machnes Z, Elgbeili G, Laplante DP, et al. King S. DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: project ice storm. PLoS ONE. 2014;9:e107653. doi: 10.1371/journal.pone.0107653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson V, Cicchetti D, Barnett D, Braunwald K. Disorganized/disoriented attachment relationships in maltreated infants. Developmental psychology. 1989;25:525. [Google Scholar]
- Cassidy J. The nature of the child's ties. 2008 [Google Scholar]
- Cyr C, Euser EM, Bakermans-Kranenburg MJ, Van IJzendoorn MH. Attachment security and disorganization in maltreating and high-risk families: A series of meta-analyses. Development and psychopathology. 2010;22:87–108. doi: 10.1017/S0954579409990289. [DOI] [PubMed] [Google Scholar]
- Doom JR, Gunnar MR. Stress physiology and developmental psychopathology: past, present, and future. Development and psychopathology. 2013;25:1359–1373. doi: 10.1017/S0954579413000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald ER, Wand GS, Seifuddin F, Yang X, Tamashiro KL, Potash JB, et al. Lee RS. Alterations in DNA methylation of Fkbp5 as a determinant of blood–brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NA, Hane AA. Studying the biology of human attachment. Handbook of attachment: Theory, research, and clinical applications. 2008;2:217–240. [Google Scholar]
- Gunnar MR, Brodersen L, Nachmias M, Buss K, Rigatuso J. Stress reactivity and attachment security. Developmental Psychobiology. 1996;29:191–204. doi: 10.1002/(SICI)1098-2302(199604)29:3<191::AID-DEV1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–1032. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertsgaard L, Gunnar M, Erickson MF, Nachmias M. Adrenocortical responses to the strange situation in infants with disorganized/disoriented attachment relationships. Child development. 1995;66:1100–1106. [PubMed] [Google Scholar]
- Hesse E, Main M. Frightened, threatening, and dissociative parental behavior in low-risk samples: Description, discussion, and interpretations. Development and psychopathology. 2006;18:309–343. doi: 10.1017/S0954579406060172. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:1. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtepen LC, Vinkers CH, Carrillo-Roa T, Hiemstra M, Van Lier PA, Meeus W, et al. Mill J. Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nature Communications. 2016;7 doi: 10.1038/ncomms10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes LA, Van den Brandt PA, De Bruïne AP, Wouters KA, Hulsmans S, Spiertz A, et al. Weijenberg MP. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PloS ONE. 2009;4:e7951. doi: 10.1371/journal.pone.0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ising M, Depping AM, Siebertz A, Lucae S, Unschuld PG, Kloiber S, et al. Holsboer F. Polymorphisms in the FKBP5 gene region modulate recovery from psychosocial stress in healthy controls. European Journal of Neuroscience. 2008;28:389–398. doi: 10.1111/j.1460-9568.2008.06332.x. [DOI] [PubMed] [Google Scholar]
- Jaddoe VW, Van Duijn CM, Franco OH, Van der Heijden AJ, Van IJzendoorn MH, De Jongste JC, et al. Raat H. The Generation R Study: design and cohort update 2012. European Journal of Epidemiology. 2012;27:739–756. doi: 10.1007/s10654-012-9735-1. [DOI] [PubMed] [Google Scholar]
- Kember R, Dempster E, Lee T, Schalkwyk LC, Mill J, Fernandes C. Maternal separation is associated with strain-specific responses to stress and epigenetic alterations to Nr3c1, Avp, and Nr4a1 in mouse. Brain and Behavior. 2012;2:455–467. doi: 10.1002/brb3.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. Bradley B. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nature Neuroscience. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok R, Van IJzendoorn MH, Linting M, Bakermans-Kranenburg MJ, Tharner A, Luijk MP, et al. Verhulst F. Attachment insecurity predicts child active resistance to parental requests in a compliance task. Child: Care, Health and Development. 2013;39:277–287. doi: 10.1111/j.1365-2214.2012.01374.x. [DOI] [PubMed] [Google Scholar]
- Kruithof CJ, Kooijman MN, Van Duijn CM, Franco OH, De Jongste JC, Klaver CC, et al. Rings EH. The generation R study: Biobank update 2015. European Journal of Epidemiology. 2014;29:911–927. doi: 10.1007/s10654-014-9980-6. [DOI] [PubMed] [Google Scholar]
- Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Current Opinion in Genetics & Development. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijk MP, Roisman GI, Haltigan JD, Tiemeier H, Booth-LaForce C, Van IJzendoorn MH, et al. Hofman A. Dopaminergic, serotonergic, and oxytonergic candidate genes associated with infant attachment security and disorganization? In search of main and interaction effects. Journal of Child Psychology and Psychiatry. 2011;52:1295–1307. doi: 10.1111/j.1469-7610.2011.02440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijk MP, Saridjan N, Tharner A, Van IJzendoorn MH, Bakermans-Kranenburg MJ, Jaddoe VW, et al. Tiemeier H. Attachment, depression, and cortisol: Deviant patterns in insecure-resistant and disorganized infants. Developmental Psychobiology. 2010;52:441–452. doi: 10.1002/dev.20446. [DOI] [PubMed] [Google Scholar]
- Luijk MP, Velders FP, Tharner A, VanIJzendoorn MH, Bakermans-Kranenburg MJ, Jaddoe VW, et al. Tiemeier H. FKBP5 and resistant attachment predict cortisol reactivity in infants: gene–environment interaction. Psychoneuroendocrinology. 2010;35:1454–1461. doi: 10.1016/j.psyneuen.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Lyons-Ruth K, Jacobvitz D. Attachment disorganization: Genetic factors, parenting contexts, and developmental transformation from infancy to adulthood. 2008 [Google Scholar]
- Lyons-Ruth K, Alpen L, Repacholi B. Disorganized infant attachment classification and maternal psychosocial problems as predictors of hostile-aggressive behavior in the preschool classroom. Child development. 1993;64:572–585. doi: 10.1111/j.1467-8624.1993.tb02929.x. [DOI] [PubMed] [Google Scholar]
- Main M, Solomon J. Procedures for identifying infants as disorganized/disoriented during the Ainsworth Strange Situation. Attachment in the preschool years: Theory, research, and intervention. 1990;1:121–160. [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of gene× environment interactions. Child development. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, et al. Mercer KB. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proceedings of the National Academy of Sciences. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan C, D'Errico N, Stees J, Hughes D. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7:853–857. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, et al. Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Muthén LK, Muthén BO. Mplus User’s Guide. (7th edn) Muthén and Muthén; Los Angeles, CA: 2012. [Google Scholar]
- Mychasiuk R, Ilnytskyy S, Kovalchuk O, Kolb B, Gibb R. Intensity matters: brain, behaviour and the epigenome of prenatally stressed rats. Neuroscience. 2011;180:105–110. doi: 10.1016/j.neuroscience.2011.02.026. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Out D, Bakermans-Kranenburg MJ, Van IJzendoorn MH. The role of disconnected and extremely insensitive parenting in the development of disorganized attachment: Validation of a new measure. Attachment & Human Development. 2009;11:419–443. doi: 10.1080/14616730903132289. [DOI] [PubMed] [Google Scholar]
- Paquette AG, Lester BM, Koestler DC, Lesseur C, Armstrong DA, Marsit CJ. Placental FKBP5 genetic and epigenetic variation is associated with infant neurobehavioral outcomes in the RICHS Cohort. PloS ONE. 2014;9:e104913. doi: 10.1371/journal.pone.0104913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SR, Gunter TD, Brody GH, Madan A, Gerrard M. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153:619–628. doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper KR. The logic of scientific discovery. London: Hutchinson; 1959. [Google Scholar]
- Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, et al. Tilling K. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Human molecular genetics. 2014;739 doi: 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam J, Cecil AM, Walton E, Mesirow MSC, Relton CT, Gaunt TR, et al. Barker ED. Prenatal unhealthy diet, insulin-like growth factor 2 gene (IGF2) methylation and attention deficit hyperactivity disorder (ADHD) symptoms for early-onset conduct problem youth. The Journal of Child Psychology and Psychiatry. 2017;58:19–27. doi: 10.1111/jcpp.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam J, Pappa I, Walton E, Bakermans-Kranenburg MJ, Mileva-Seitz VR, Rippe RCA, et al. Felix JF. An epigenome-wide association meta-analysis of prenatal maternal stress in neonates: A model approach for replication. Epigenetics. 2016;11:140–149. doi: 10.1080/15592294.2016.1145329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Gorodetsky E, Yuan Q, Goldman D, Enoch M-A. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology. 2010;35:1674–1683. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler G, Grossmann KE. Biobehavioral organization in securely and insecurely attached infants. Child development. 1993;64:1439–1450. doi: 10.1111/j.1467-8624.1993.tb02962.x. [DOI] [PubMed] [Google Scholar]
- Touleimat N, Tost J. Complete pipeline for Infinium® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- Van IJzendoorn MH, Bakermans-Kranenburg MJ, Ebstein RP. Methylation matters in child development: Toward developmental behavioral epigenetics. Child Development Perspectives. 2011;5:305–310. [Google Scholar]
- Van IJzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R. Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biological Psychiatry. 2010;68:405–407. doi: 10.1016/j.biopsych.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCY, Caspi A, Williams B, Craig IW, Houts R, Ambler A, et al. Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Patchev AV, Daniel G, Almeida OF, Spengler D. Early-life stress reduces DNA methylation of the Pomc gene in male mice. Endocrinology. 2014;155:1751–1762. doi: 10.1210/en.2013-1868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.