Abstract
Adaptation of neural circuits to changes in sensory input can modify several cellular processes within neurons, including neurotransmitter biosynthesis levels. For a subset of olfactory bulb interneurons, activity-dependent changes in GABA are reflected by corresponding changes in Glutamate decarboxylase 1 (Gad1) expression levels. Mechanisms regulating Gad1 promoter activity are poorly understood, but here we show that a conserved G:C-rich region in the mouse Gad1 proximal promoter region both recruits heterogeneous nuclear ribonucleoproteins (hnRNPs) that facilitate transcription and forms single-stranded DNA secondary structures associated with transcriptional repression. This promoter architecture and function is shared with Tyrosine hydroxylase (Th), which is also modulated by odorant-dependent activity in the olfactory bulb. This study shows that the balance between DNA secondary structure formation and hnRNP binding on the mouse Th and Gad1 promoters in the olfactory bulb is responsive to changes in odorant-dependent sensory input. These findings reveal that Th and Gad1 share a novel transcription regulatory mechanism that facilitates sensory input-dependent regulation of dopamine and GABA expression.
SIGNIFICANCE STATEMENT Adaptation of neural circuits to changes in sensory input can modify several cellular processes within neurons, including neurotransmitter biosynthesis levels. This study shows that transcription of genes encoding rate-limiting enzymes for GABA and dopamine biosynthesis (Gad1 and Th, respectively) in the mammalian olfactory bulb is regulated by G:C-rich regions that both recruit heterogeneous nuclear ribonucleoproteins (hnRNPs) to facilitate transcription and form single-stranded DNA secondary structures associated with repression. hnRNP binding and formation of DNA secondary structure on the Th and Gad1 promoters are mutually exclusive, and odorant sensory input levels regulate the balance between these regulatory features. These findings reveal that Th and Gad1 share a transcription regulatory mechanism that facilitates odorant-dependent regulation of dopamine and GABA expression levels.
Keywords: DNA secondary structure, GABAergic interneurons, heterogeneous ribonucleoproteins, olfactory bulb, transcription
Introduction
A fundamental question in neurobiology is how neurons adapt to changes in circuit activity. Identifying molecular mechanisms that underlie activity-dependent adaptations is important for understanding neural circuit plasticity. In the mammalian olfactory bulb (OB), changes in odorant sensory input can alter dopamine and GABA biosynthesis levels in interneurons. This sensory input-dependent regulation of dopamine and GABA is suggested to be integral for modifying the detection thresholds and discrimination of odorants (Parrish-Aungst et al., 2011). Activity-dependent changes of dopamine levels in the OB are reflected by corresponding changes in the expression of Tyrosine hydroxylase (Th), which encodes the rate-limiting enzyme for dopamine biosynthesis (Baker et al., 1983, 1993; Baker and Farbman, 1993). OB dopaminergic neurons also contain GABA and preferentially coexpress Glutamate decarboxylase 1 (Gad1) (Parrish-Aungst et al., 2007; Kiyokage et al., 2010), which encodes one of two rate-limiting enzyme isoforms for GABA biosynthesis. Activity-dependent changes of GABA levels in the OB are reflected by corresponding changes in the expression of Gad1 (Parrish-Aungst et al., 2011; Lau and Murthy, 2012; Banerjee et al., 2013).
The molecular mechanisms responsible for the sensory input-dependent expression of Gad1 and Th in OB dopaminergic neurons are not fully understood. For Th, odorant-dependent transcription requires AP-1 and CRE cis-regulatory elements in the proximal promoter (Kim et al., 1993; Nagamoto-Combs et al., 1997; Trocmé et al., 1997; Baker et al., 2001; Lewis-Tuffin et al., 2004; Wang et al., 2015). These elements straddle highly conserved G:C-rich regions that regulate transcription by adopting DNA secondary structures (G-quadruplexes and i-motifs) and recruiting heterogeneous ribonucleoprotein (hnRNP) transcription factors (Banerjee et al., 2014). For Gad1, however, the cis-regulatory regions and transcription factors required for activity-dependent transcription are not known. In this study, we show that the Gad1 proximal promoter, like Th, contains a conserved G:C-rich region that modulates transcription by adopting DNA secondary structures and recruiting hnRNPs. Using a unilateral naris occlusion mouse model of odor deprivation, we demonstrate for both the Gad1 and Th promoters that changes in odorant sensory input levels shift the balance between the protranscription recruitment of hnRNPs and repression mediated by DNA secondary structure formation. Together, these findings reveal a novel activity-dependent mechanism for regulating gene transcription.
Materials and Methods
Animals.
All mice were housed in humidity-controlled cages at 22°C under a 12:12 h light/dark cycle and provided with food and water ad libitum. All procedures were performed under protocols approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee and conformed to National Institutes of Health guidelines. Gad1-GFP mice were heterozygous for a GFP reporter knock-in allele in the Gad1 loci, and details for the generation of this line have been described previously (Tamamaki et al., 2003). Tissue from males and females was used for all analyses.
Unilateral naris closure mouse model of odor deprivation.
One nostril of wild-type (WT) C57BL/6J mice (aged 1–3 months) was permanently closed using a spark-gap cautery under isoflurane anesthesia. Naris occlusion was confirmed at 1 month after closure and tissue was harvested 1–3 months after confirmation of naris closure.
Organotypic slice cultures.
The preparation and culture of forebrain organotypic slices from postnatal day 4 mouse pups have been previously described (Akiba et al., 2009). Slices were cultured in depolarizing conditions containing 25 mm KCl. The slices cultured with 25 mm KCl were also treated either with 100 μm TMPyP4, 100 μm TMPyP2, or no drug. All slices were cultured for 24 h at 37°C and 5% CO2 and were imaged with a Nikon Eclipse 80i fluorescence microscope at 0 and 24 h.
Cell culture.
The Gad1-expressing human NT2 cell line (ATCC) was maintained at 37°C at 5% CO2 on primaria-coated culture dishes in DMEM/F12 cell culture medium supplemented with 10% FBS and 1% penicillin/streptomycin.
Transcription assays with NT2 cells overexpressing hnRNP LL were conducted using transient transfections with Lipofectamine LTX transfection reagent (Invitrogen). These experiments used the pGL3.0 basic plasmid (Promega) with a firefly luciferase reporter gene under the control of the WT rat 3.4 kb Gad1 upstream region and a pEFnFlag-hnRNP LL (a generous gift from Dr. Kristen Lynch, University of Pennsylvania). Four hours after transfection, cells transfected with pGL3.0-Gad1 3.4 were treated with either saline or 100 μm of either TMPyP2 or TMPyP4. Cells were transfected according to the manufacturer's protocol and maintained for 24 h before firefly luciferase activities were measured using the Luciferase Assay System (Promega). Luminescence was measured with LMaxII illuminometer (Molecular Devices). Luciferase activity was reported as the mean of three independent transfection experiments with error bars representing the SEM. Significant changes in activity were assessed by two-tailed Student's t tests.
For studies with membrane depolarization of human SH-SY5Y cultures, cells were plated in media containing either 50 mm KCl or NaCl. By contrast, NT2 cells were plated and were cultured for 24 h before the media was supplemented with either 50 mm KCl or NaCl. Both SH-SY5Y and NT2 cells were cultured for 42 h after treatment with either KCl or NaCl before harvested for either qRT-PCR or chromatin immunoprecipitation (ChIP) assays.
Immunohistology and antibodies.
All immunohistological analyses used adult C57BL/6 mice that were perfused transcardially with phosphate-buffered 4% formaldehyde. Brains were removed and postfixed for 4 h before being cryoprotected overnight in 30% sucrose and cut in 30 μm sagittal sections. Sections were blocked with 1% BSA in PBS for 1 h before overnight incubation with primary anti-hnRNP LL antibody (Cell Signaling Technology) at a 1:50 dilution. The primary antibody was visualized with an AlexaFluor-594 secondary antibody (Invitrogen) at a 1:400 dilution. Sections were imaged on either a Nikon 80i Eclipse fluorescence microscope or a Meta 500 scanning laser confocal microscope (Carl Zeiss).
Genomic sequence alignment.
All Gad1 proximal promoter genomic sequences were obtained from http://ensembl.org, and the Gad1 gene identifiers for each species used are provided below. For all species, 500 bp sequences upstream of the translation start site were aligned and visualized using Multi-LAGAN and mVISTA web-based services (http://genome.lbl.gov/vista/mvista/submit.shtml) (Brudno et al., 2003; Frazer et al., 2004). The Gad1 gene identifiers for each species used were as follows: armadillo, Dasypus novemcinctus ENSDNOG00000005509; cat, Felis catus ENSFCAG00000007099; chimpanzee, Pan troglodytes ENSPTRG00000012626; cow, Bos taurus ENSBTAG00000007258; dog, Canis familiaris ENSCAFG00000012560; dolphin, Tursiops truncatus ENSTTRG00000004553; elephant, Loxodonta africana ENSLAFG00000002694; ferret, Mustela putorius furo ENSMPUG00000000808; gibbon, Nomascus leucogenys ENSNLEG00000005195; gorilla, Gorilla gorilla ENSGGOG00000016598; guinea pig, Cavia porcellus ENSCPOG00000004156; horse, Equus caballus ENSG00000128683; human, Homo sapiens ENSG00000128683; macaque, Macaca mulatta ENSMMUG00000015198; mouse, Mus musculus ENSMUSG00000070880; pig, Sus scrofa ENSSSCG00000022233; rabbit, Oryctolagus cuniculus ENSOCUG00000012644; rat, Rattus norvegicus ENSRNOG00000000007; sheep, Ovis aries ENSOARG00000002329; and sloth, Choloepus hoffmanni ENSCHOG00000006753.
Circular dichroism (CD).
All CD experiments were performed with an Aviv 414 CD spectrophotomer (Aviv Biomedical) with a wavelength range from 200 to 330 nm using a 1 nm step width and 1 s dwell time. Spectra for each sample are shown as an average of three scans. All DNA oligonucleotide sequences were used at concentrations of 5 mm. Both WT and mutant G-rich oligonucleotides were in Tris buffer at pH 7.4 with either 0 or 100 mm KCl. Both WT and mutant C-rich oligonucleotides were in sodium citrate/phosphate buffer at either pH 7.6 or 4.6. Oligonucleotide sequences used for CD experiments as follows: Gad1 C-rich WT (the methylated oligonucleotide used the same sequence, except all cytosines within CpG motifs were methylated), 5′-CCTCGTGATTCCCCCCGCCGAGCGGGTCCCCGCCTCCCCACTCCGCCCCCGCCTCCCCCAAGCCCAG-3′; Gad1 G-rich WT, 5′-CTGGGCTTGGGGGAGGCGGGGGCGGAGTGGGGAGGCGGGGACCCGCTCGGCGGGGGGAATCACGAGG-3′; Gad1 C-rich Mut (mutated positions italicized), 5′-CCTCGTGATTATGTGAGCCGAGCGGGTCCCCGCCTATGTACTCCGATGTAGTATGTACCAAGCCCAG-3′; and Gad1 G-rich Mut (mutated positions italicized), 5′-CTGGGCTTGGTACATACTACATCGGAGTACATAGGCGGGGACCCGCTCGGCTCACATAATCACGAGG-3′.
PCR interference assays with TMPyP4 and TMPyP2.
All PCRs were performed on an ABI 7500 Fast Real-time PCR System (Applied Biosystems) using SYBR Green PCR master mix (Applied Biosystems) with 0, 5, 10, or 25 nm of either TMPyP4 (Sigma-Aldrich) or TMPyP2 (Frontier Scientific). Data are shown as the mean of three independent trials with error bars representing the SEM. The WT and mutant oligonucleotide template strands used were the same as those in the CD experiments. The oligonucleotide sequences for the template and primer strands are as follows: primer for the Gad1 WT and mutant forward strand: 5′-AGTCAGTCCTGGGCTT-3′; and primer for the Gad1 WT and mutant reverse strand: 5′-AGTCAGTCCCTCGTGA-3′.
hnRNP pull-down assays.
A total of 1 μg of single-stranded biotinylated DNA oligonucleotide was incubated with 5 mg of streptavidin-coated Dynabeads (Invitrogen) in 300 ml of streptavidin-biotin-binding buffer (10 mm Tris, pH 7.5, 1 mm EDTA, 1 m NaCl, 0.003% NP40) at room temperature for 20 min. To minimize nonspecific interactions, the oligo-bead complexes were incubated for 30 min with a blocking buffer (2.5 mg/ml BSA in 10 mm HEPES, pH 7.6, 100 mm potassium glutamate, 2.5 mm DTT, 10 mm magnesium acetate, 5 mm EGTA, 3.5% glycerol with 0.003% NP40, and 5 mg/ml polyvinylpyrrolidone). Gad1-expressing NT2 cells were grown in 10 mm dishes and collected at 80% confluence. A total of 3 × 107 cells were lysed, along with the freshly harvested mouse OB or cortex tissue, and the nuclei were extracted with NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific). Immobilized oligonucleotides were incubated with 100 mg of nuclear extract for 1 h at 4°C with constant rotation in 400 μl of protein-binding buffer (10 mm HEPES, pH 7.6, 100 mm potassium glutamate, 80 mm KCl, 2.5 mm DTT, 10 mm magnesium acetate, 5 mm EGTA, 3.5% glycerol with 0.001% NP40, and 1 mg of nonspecific carrier DNA), in the presence or absence of 100 μm TMPyP2 or TMPyP4 treatment. Protein–DNA complexes were then washed three times (10 mm HEPES, pH 7.6, 100 mm potassium glutamate, 2.5 mm DTT, 10 mm magnesium acetate, 5 mm EGTA, 3.5% glycerol, 0.5 mg/ml BSA, and 0.05% NP40). Proteins bound to the oligonucleotides were then eluted with 50 μl of denaturing Laemmli sample loading buffer at 37°C for 15 min. hnRNP K protein in the supernatant was visualized by Western blot with a rabbit anti-hnRNP K antibody (Abcam) at 1:2000 dilution or a rabbit anti-hnRNP LL antibody (AssayBioTech) at 1:200 dilution using an Odyssey imaging system (Li-Cor Biosciences). Band intensities were quantified using ImageJ software (National Institutes of Health) and are reported as the mean of duplicate pull-down assays with error bars representing the difference between the mean and the individual trials. The oligonucleotide sequences used in the experiments were (mutated positions are italicized): Gad1 WT strand, BIO-5′-CCTCGTGATTCCCCCCGCCGAGCGGGTCCCCGCCTCCCCACTCCGCCCCCGCCTCCCCCAAGCCCAG-3′; Gad1 hnRNP K binding site mutant, BIO-5′-CCTCGTGATTCCCCCCGCCGAGCGGGTCCCCGCCTAAAAACTCCGCCCCCGCCTAAAAAAAGCCCAG-3′; and Gad1 hnRNP LL binding site mutant, BIO-5′-CCTCGTGATTAATAATATTGAGCGGGTAATTAAATCCCCACTCCGCAAATTAATCCCCCAAGCCCAG-3′.
ChIP assays.
Adult C57BL/6 mice (3–6 months of age) were used for ChIP assays to examine promoter occupancy in OB and cortical tissue. ChIP assays in cultured cells were performed with NT2 cells, which were grown in 10 mm culture dishes to 70% confluence before being transfected with pEFnFlag-hnRNP LL (a gift from Dr. Kristen Lynch, University of Pennsylvania). For experiments in the presence of either TMPyP4 or TMPyP2, cells were treated with 100 mm TMPyP4 or TMPyP2 for 24 h before being harvested. For ChIP assays with depolarized SH-SY5Y and NT2 cells, cells were cultured with either 50 mm KCl or 50 mm NaCl for 42 h before being harvested.
All tissues and cells were washed with ice-cold PBS and then cross-linked with 1% formaldehyde in PBS on ice for 15 min. Fixation was terminated by addition of 125 mm glycine, and samples were washed with PBS before application of SDS lysis buffer (Millipore). Sample chromatin was sheared with a Bioruptor (Diagenode), and immunoprecipitation of cross-linked protein/DNA complexes used the Magna ChIP Protein-A/G kit (Millipore) following the manufacturer's instructions. Immunoprecipitation reactions used either 1 μg of rabbit anti-hnRNP LL (Cell Signaling Technology), 3 μg of mouse anti-hnRNP K (Abcam), or 4 μg of mouse anti-Flag M2 antibody (Sigma). An equivalent amount species-matched IgG (Santa Cruz Biotechnology) was used for control assays. Reverse cross-linking was done overnight in the presence of Proteinase K at 62°C with shaking. Immunoprecipitated Gad1 promoter genomic DNA fragments were purified with MinElute PCR purification columns (QIAGEN). The DNA fragments were amplified and quantified using a 7500 Fast Real-time PCR System (Applied Biosystems) and SYBR Green PCR master mix (Applied Biosystems). For all ChIP experiments, three independent experiments were conducted. The mean relative enrichment of the Gad1 proximal promoter is reported, with error bars representing the SEM. Statistical significance was assessed using the two-tailed Student's t test.
ChIP experiments to detect G-quadruplex secondary structures on the Th or Gad1 promoters used a His/FLAG-tagged BG4 single chain phage display antibody. The expression plasmid for this antibody was generously provided by Dr. Shankar Balasubramnian (Cambridge, UK). Control experiments used His-tagged thioredoxin expressed from a pET-32a expression plasmid (Novagen/EMD Millipore). Both His-tagged BG4 and Thioredoxin proteins were generated with BL21(DE3) Escherichia coli cells. An overnight growth was used to inoculate 300 ml of LB media that was incubated at 37°C with shaking to an OD600 of 0.1, at which point protein expression was induced with 1 mm IPTG (Calbiochem). Six hours after induction, the cells were spun down and the pellet was washed with PBS before being resuspended in 1.6 ml of equilibration buffer (PBS, with 10 mm imidazole, pH 7.4, and 1 mm PMSF). The resuspended cells were placed on ice, lysed by sonication, and cellular debris was removed by centrifugation. Lysate with equivalent loads of either BG4 or Thioredoxin was incubated on HisPur Ni-NTA resin (Thermo Scientific) at 4°C for 2 h. Sheared chromatin from either mouse OB or cortex tissue was added and incubated at 4°C overnight. The BG4/Thioredoxin-bound resin was then washed three times with 1× PBS containing 25 mm imidazole and eluted in PBS with 1 mm EDTA and 25 mm imidazole. Samples were reverse cross-linked overnight at 62°C on a shaker with 1 mg of Proteinase K. DNA was purified using MinElute PCR purification columns (QIAGEN). Th and Gad1 promoter genomic DNA fragments were amplified and quantified using a 7500 Fast Real-time PCR System (Applied Biosystems) and SYBR Green PCR master mix (Applied Biosystems). Data are presented as the mean from three independent ChIP experiments, with error bars representing the SEM. Statistical significance was assessed using two-tailed Student's t tests.
For ChIP assays, the primer sequences used for amplifying the G:C-rich regions of the Gad1 proximal promoter were as follows: 5′-TCGCCCTACAAAGCTCCAGAGGC-3′; and 5′-AGGGCTGCTTCCTTGCTTGCAC-3′. Primer sequences used to amplify the G:C-rich regions of the mouse Th proximal promoter were as follows: 5′-GTCGCCCTCGCTCTGTGCCCA-3′; and 5′-GGCTGACGTCAAAGCCCCTCTGG-3′.
qRT-PCR.
For either cultured cells or the OB tissue ipsilateral and contralateral to the closed nares, RNA was isolated using an RNA mini-prep kit (Epoch) following the manufacturer's protocol. First strand reactions were conducted using SuperScript III first strand synthesis kit (Invitrogen) following the manufacturer's protocol. Quantitative PCRs were performed on a 7500 Fast Real-time PCR System (Applied Biosystems). Quantitative PCRs used TaqMan Universal PCR Master Mix (Applied Biosystems) and TaqMan Gene Expression Assays primer sets (Applied Biosystems) Mm00447557_m1, Mm00725661_s1, Mm04409831_s1, and Mm00804896_m1 for Th, Gad1, hnRNP K, and hnRNP LL, respectively. All gene expression levels were normalized to β-actin (Mm01205627_g1) levels. Data are reported as the mean, with error bars representing the SD. Measures of significance in the expression levels between the ipsilateral and contralateral OBs were assessed using two-tailed Student's t tests.
Results
Regulation of Gad1 promoter activity by DNA secondary structure
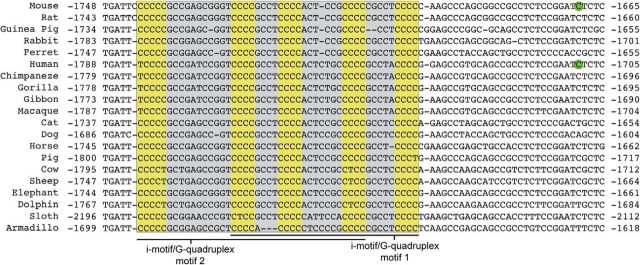
The human and rodent Gad1 proximal promoter contains a G:C-rich region upstream of the transcription start site (Erlander and Tobin, 1992; Szabó et al., 1996). Analysis of this G:C-rich region revealed two overlapping consensus sequences for i-motifs (C≥3N1–12C≥3N1–12C≥3N1–12C≥3) and G-quadruplexes (on the opposite strand; G ≥3N1–12G≥3N1–12G≥3N1–12G≥3) that are highly conserved in mammals (Fig. 1). By contrast, an analysis of the mammalian Gad2 proximal promoter (≤1 kb upstream from the transcription start site) did not find conservation of either G:C-rich regions or G-quadruplex/i-motif consensus sequences (data not shown).
Figure 1.
Conservation of a G:C-rich region in the mammalian Gad1 promoter. Nucleotide alignment of mammalian Gad1 proximal promoter DNA genomic sequences showed a highly conserved G:C-rich region (gray) containing two overlapping G-quadruplex/i-motif consensus sequences (highlighted in yellow). The human and mouse transcription start sites (Erlander and Tobin, 1992; Szabó et al., 1996) are circled in green, and nucleotide positions are numbered relative to the translation start site.
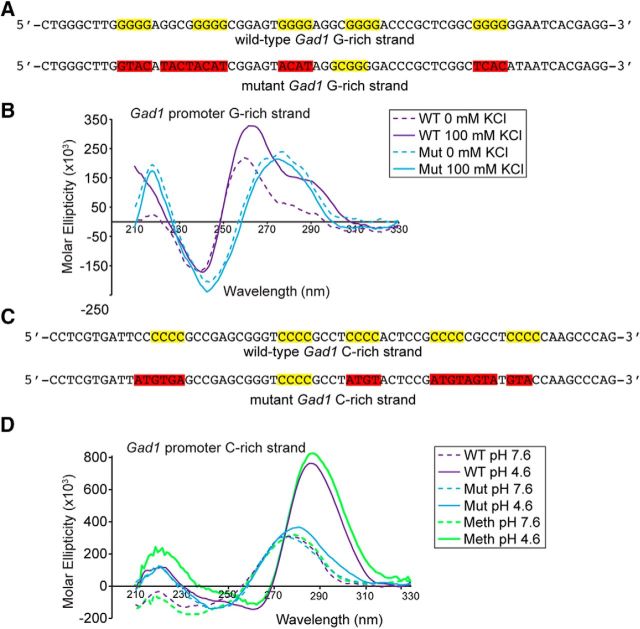
To determine whether the Gad1 G:C-rich region can adopt DNA secondary structure, single-stranded oligonucleotides from the mouse Gad1 promoter were examined by CD spectroscopy. G-quadruplexes are stabilized in vitro by high potassium concentrations (≥100 mm) (Burge et al., 2006), and CD spectra for the WT G-rich single strand displayed a potassium-dependent enhancement that was absent in a mutant strand lacking G-quadruplex consensus sequences (Fig. 2A,B). On C-rich single strand oligonucleotides, i-motif structures are stabilized by low pH (pH < 5) (Gehring et al., 1993). The CD spectra for the Gad1 C-rich strand showed a pH dependence that was absent when the i-motif consensus sequences were mutated (Fig. 2C,D). Together, the CD data show that the G- and C-rich strands of the Gad1 proximal promoter can form G-quadruplex and i-motif DNA secondary structures, respectively.
Figure 2.
CD spectra for G- and C-rich single oligonucleotide strands from the Gad1 promoter. A, C, WT and mutant oligonucleotides for analysis of the Gad1 promoter G-rich and C-rich strands, respectively. Yellow represents conserved G-quadruplex and i-motif consensus sequences. Red represents mutated positions. B, Spectra for the WT G-rich strand were potassium-dependent and consistent with G-quadruplex formation. By contrast, spectra for the mutant strand were not potassium-dependent, indicating that G-quadruplexes were not formed. D, Spectra for the WT C-rich strand were pH-dependent and consistent with i-motif formation. Spectra for the mutant strand did not show any pH dependence, indicating that i-motifs were not formed. By contrast, methylation of all cytosines in CpG motifs within the WT oligonucleotide enhanced the signal maxima of the spectra, indicating that methylation increased i-motif formation.
In schizophrenia, bipolar disorder, and autism spectrum disorder, reduced Gad1 expression is associated with increased methylation of its proximal promoter (Chen et al., 2011; Zhubi et al., 2014). To establish whether methylation of the Gad1 C-rich strand modified i-motif secondary structure formation, an oligonucleotide of the Gad1 C-rich strand with methylated CpG motifs was analyzed by CD spectroscopy. Under conditions that favor i-motif formation, the signal maximum at 290 nm for a methylated sequence was enhanced compared with the WT oligonucleotide (Fig. 2D). These results show that methylation does not disrupt i-motif formation, and suggest that it increases i-motif stability, which is consistent with recent thermodynamic studies (Bhavsar-Jog et al., 2014).
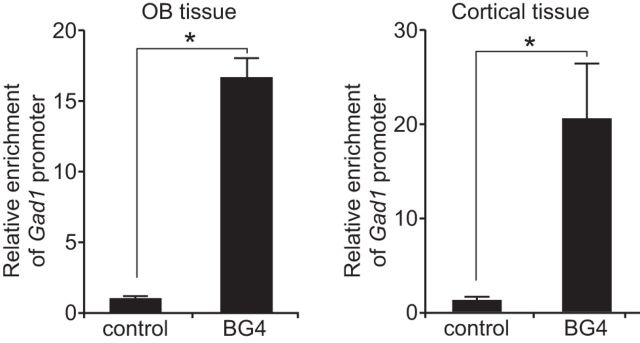
To test whether the Gad1 promoter contains DNA secondary structure in vivo, ChIP studies were conducted using an antibody (BG4) that specifically recognizes G-quadruplexes (Biffi et al., 2013). Assays with mouse OB and cortical tissue showed significant enrichment of the Gad1 proximal promoter (Fig. 3), revealing that DNA secondary structure is present on the Gad1 promoter in vivo in both OB and cortical GABAergic neurons.
Figure 3.
DNA secondary structures are present on the Gad1 promoter in vivo. ChIP assays with the BG4 antibody detected G-quadruplex secondary structure on the Gad1 promoter in both OB and cortical tissue. *p < 0.01.
G-quadruplex and i-motif secondary structures are stabilized by interactions with the porphyrin compound, TMPyP4 (Fedoroff et al., 2000; Phan et al., 2005). By contrast, the isomer, TMPyP2, is ineffective at stabilizing these structures (F. X. Han et al., 1999; H. Han et al., 2001; Qin and Hurley, 2008). We previously showed that TMPyP4-mediated stabilization of DNA secondary structures can impede PCR amplification of template oligonucleotide strands (Banerjee et al., 2014). PCRs using the oligonucleotide sequences from the CD studies as template strands confirmed that TMPyP4, but not TMPyP2, can stabilize secondary structures formed by the Gad1 proximal promoter (Fig. 4A).
Figure 4.
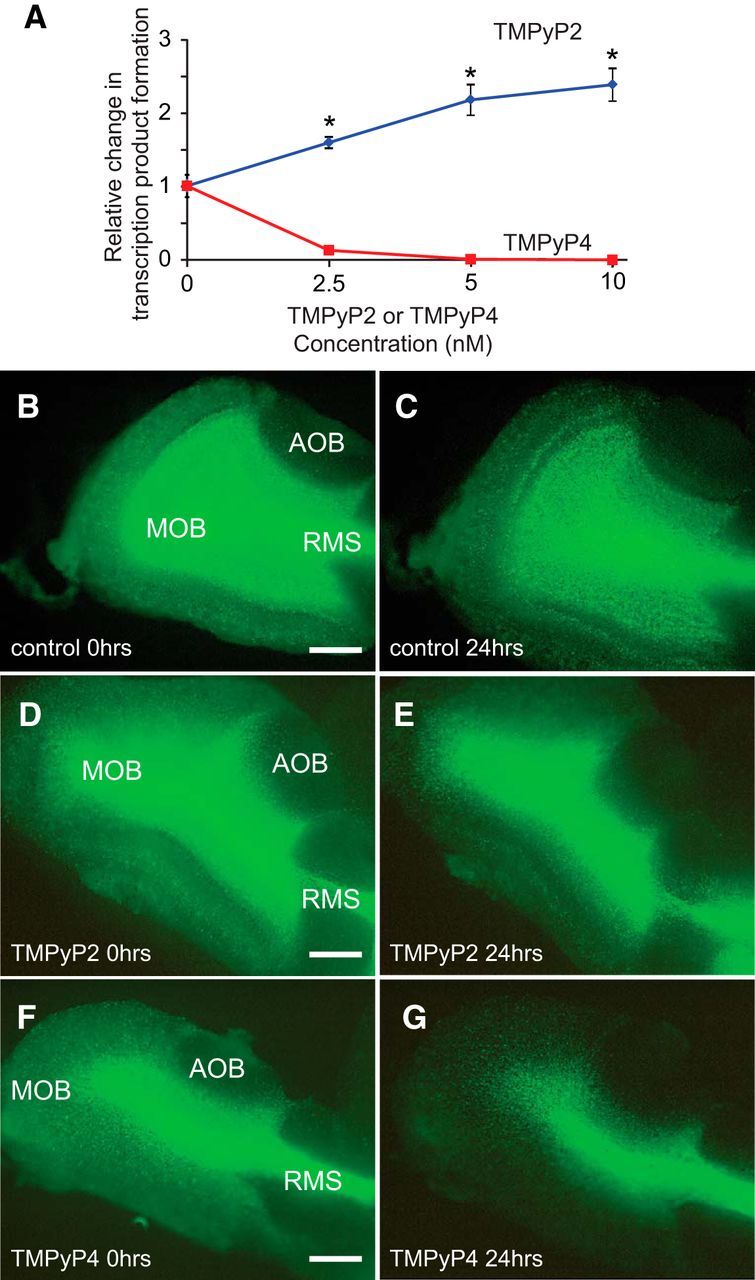
Regulation of Gad1 promoter activity by small molecules that stabilize nucleic acid secondary structure. A, PCR interference assays showed that increasing concentrations of TMPyP4, but not TMPyP2, blocked transcription and PCR amplification of WT template oligonucleotides. *p < 0.01. B–G, Gad1-GFP expression was strongly repressed in the main OB (MOB) of depolarized forebrain slice cultures after 24 h exposure to 100 μm of TMPyP4, but not TMPyP2. Gad1-GFP is not expressed in the accessory OB (AOB), and Gad1-GFP expression in the rostral migratory stream (RMS) does not show the same sensitivity to TMPyP4. Scale bar, 100 μm.
Because of toxicity (Fujiwara et al., 2015), we were unable to test whether modifying the stability of Gad1 promoter DNA secondary structures with TMPyP4 could alter Gad1 expression levels in vivo. As an alternative, Gad1 promoter activity was monitored in forebrain organotypic slice cultures from Gad1-GFP mice. Compared with either control or TMPyP2-treated cultures, GFP expression in the OB was strongly attenuated in cultures treated with TMPyP4 (Fig. 4B–G). These findings show that small molecules that alter nucleic acid secondary structure stability can modulate Gad1 expression levels in brain tissue. Interestingly, TMPyP4 did not reduce GFP expression in the rostral migratory stream, which contains neuroblasts migrating to the OB, suggesting that there is differential regulation of Gad1 transcription between migrating neuroblasts and mature interneurons in the OB (Fig. 4B–G).
Regulation of Gad1 promoter activity by hnRNP proteins
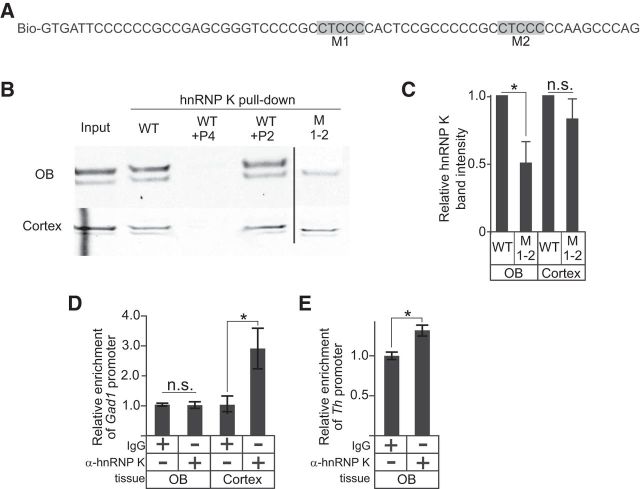
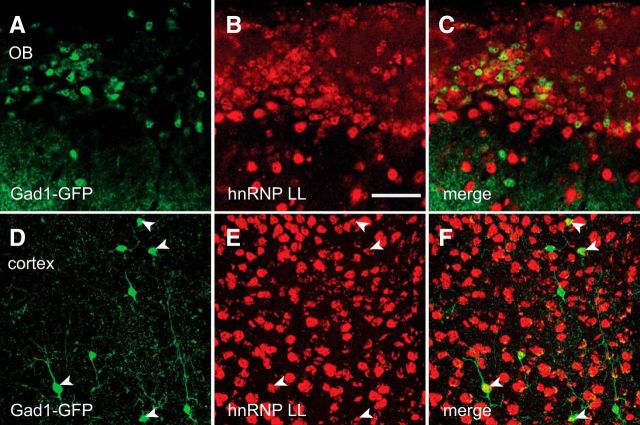
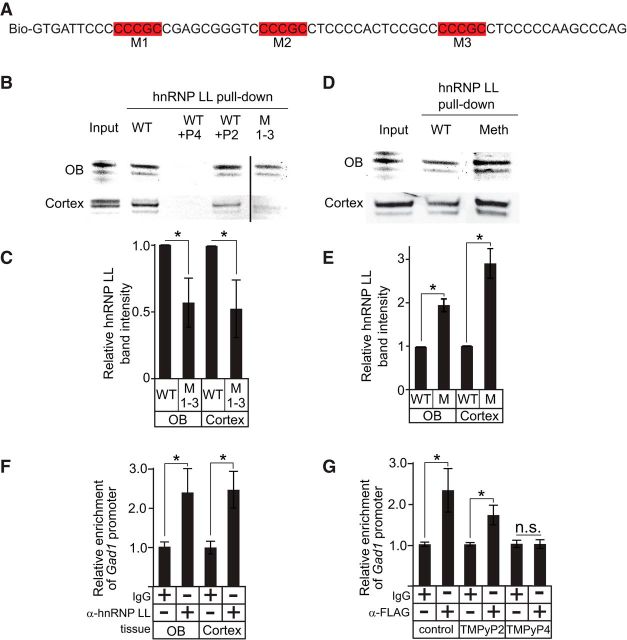
We previously showed that hnRNP K binds to C-rich regions in the Th proximal promoter that form secondary structures (Banerjee et al., 2014), but we found that hnRNP K was not recruited to the Gad1 promoter in the OB in vivo, and only indirectly associated with Gad1 promoter in the cortex (Fig. 5). hnRNP LL binds to CG elements (5′-CCCGC-3′) (Kang et al., 2014), and the presence of several CG elements in the Gad1 C-rich strand suggested that hnRNP LL targets the Gad1 C-rich strand. Immunofluourescence studies with Gad1-GFP mice showed that hnRNP LL was expressed in nearly all Gad1-containing neurons in the OB glomerular layer (Fig. 6A–C). Coexpression was also observed in a subset of Gad1-expressing neurons in the cortex (Fig. 6D–F). Transcription assays in NT2 cells showed that hnRNP LL overexpression was also sufficient to increase Gad1 reporter gene expression levels 48 ± 10% (p = 0.01) relative to controls containing an empty expression plasmid. Western blots of protein pull-down experiments showed that a biotinylated single-stranded Gad1 C-rich oligonucleotide interacted with hnRNP LL protein from either OB or cortical nuclear lysate (Fig. 7A,B). Mutation of hnRNP LL recognition motifs substantially diminished the pull-down of hnRNP LL from OB and cortical tissue (Fig. 7B,C), suggesting that hnRNP LL directly associates with the Gad1 promoter through these recognition sites in both brain regions. To address whether methylation of the Gad1 promoter altered recruitment of hnRNP LL, protein pull-down assays were performed with the biotinylated single-stranded Gad1 C-rich oligonucleotide that had all CpG motifs methylated. Nuclear lysate from either the OB or the cortex showed that the methylated strand had an increased association with hnRNP LL compared with the unmethylated strand (Fig. 7D,E), indicating that methylation of the CpG sites on the Gad1 promoter enhances recruitment of hnRNP LL.
Figure 5.
Indirect regulation of Gad1 promoter activity by hnRNP K. A, Biotinylated oligonucleotide sequence of the WT Gad1 promoter C-rich region with CT elements that were mutated highlighted in gray (M1 and M2). B, Western blots for hnRNP K from protein pull-down assays using WT and mutant (M1–2) oligonucleotides with either mouse OB or cortical nuclear lysate. Vertical line indicates the removal of intervening lanes. C, Relative Western blot band intensities for hnRNP K pulled down with either the WT or M1–2 oligonucleotides in B. D, ChIP assays for hnRNP K show occupancy on the Gad1 promoter in cortical, but not OB, tissue. E, ChIP assays for hnRNP K show occupancy on the Th promoter in OB tissue. *p < 0.01. Together, these findings indicate that hnRNP K is not recruited to the Gad1 promoter in the OB in vivo, and only indirectly associated with Gad1 promoter in the cortex.
Figure 6.
Coexpression of hnRNP LL and Gad1. A–C, Immunofluorescence analysis showed that most Gad1-GFP cells in the mouse OB coexpress hnRNP LL. D–F, Immunofluorescence analysis of the mouse cortex showed that a subset of cells with Gad1-GFP coexpress hnRNP LL (indicated with arrowheads). Scale bar, 50 μm.
Figure 7.
hnRNP LL interacts with the Gad1 promoter C-rich strand. A, WT sequences of the biotinylated oligonucleotide used for pulldown of hnRNP LL. Red boxes indicate CG elements that were mutated. B, Western blots for hnRNP LL from protein pull-down assays using WT and mutant (M1–3) oligonucleotides with either mouse OB or cortical nuclear lysate. Vertical line indicates the removal of intervening lanes. C, Relative Western blot band intensities for hnRNP LL pulled down with either the WT or mutant (M1–3) oligonucleotides. D, Western blots for hnRNP LL from protein pull-down assays using WT and methylated oligonucleotides with either mouse OB or cortical nuclear lysate. The methylated oligonucleotide sequence was identical to the WT, except that all cytosine nucleotides in CpG motifs were methylated. E, Relative Western blot band intensities for hnRNP LL pulled down with either the WT or methylated oligonucleotides. F, ChIP assays for hnRNP LL show occupancy on the Gad1 promoter in both cortical and OB tissue. G, ChIP assays show that hnRNP LL occupancy on the Gad1 promoter in NT2 cells is blocked when TMPyP4, but not TMPyP2, is present. *p < 0.01.
ChIP assays with OB and cortical tissue showed that hnRNP LL associated with the Gad1 promoter in vivo (Fig. 7F). The pull-down of hnRNP LL from either OB or cortical lysate was blocked by the presence of TMPyP4, but not TMPyP2 (Fig. 7B), indicating the secondary structure formation and hnRNP LL protein binding were mutually exclusive. Consistent with these findings, ChIP assays using NT2 cells expressing FLAG-tagged hnRNP LL showed that hnRNP LL occupancy on the Gad1 promoter was blocked when the cells were treated with TMPyP4, but not with TMPyP2 (Fig. 7G). Together, the studies indicate that hnRNP LL directly interacts with the Gad1 C-rich strand in both the OB and cortex to facilitate Gad1 expression.
Odorant sensory input-dependent DNA secondary structure formation and hnRNP occupancy on the Gad1 and Th proximal promoters
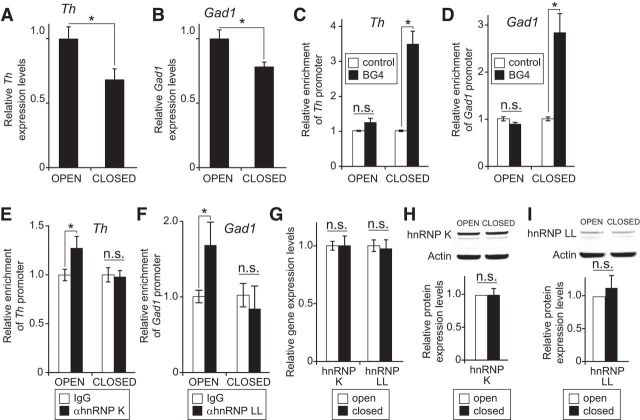
Circuit activity levels in the OB are dependent on odorant sensory input from the olfactory epithelium. A subset of glomerular layer interneurons, which includes interneurons coexpressing dopamine and GABA, receives direct afferent input through synaptic connections with the olfactory sensory neuron axons (Kosaka and Kosaka, 2005, 2007). In addition, both Gad1 and Th expression levels in the OB are dependent on odorant sensory input (Baker et al., 1983; Wilson and Wood, 1992; Baker et al., 1993, 1999; Cigola et al., 1998; Puche and Shipley, 1999; Akiba et al., 2009; Parrish-Aungst et al., 2011; Banerjee et al., 2013). In mice with unilateral naris occlusion, the permanent closure of one nostril strongly reduces odorant-dependent circuit activity in the ipsilateral (closed) OB compared with the contralateral (open) OB (Coppola, 2012). Both Th and Gad1 expression levels are significantly reduced in the closed OB of adult mice 1 month after naris occlusion (Fig. 8A,B). To address whether this reduction in gene expression was associated with an increase in secondary structure on the Th and Gad1 proximal promoters, ChIP assays were performed with the G-quadruplex-specific BG4 antibody. These assays showed that both the Gad1 and Th promoters in the closed OB contained much higher levels of secondary structure (Fig. 8C,D). Because the pull-down and ChIP assays suggested that the interaction between hnRNP LL and the Gad1 promoter was mutually exclusive to secondary structure formation (Fig. 7B,G), ChIP assays were performed to test whether hnRNP occupancy was diminished in the closed OBs. These assays showed a significant reduction in hnRNP K and LL occupancy on the Th and Gad1 proximal promoters, respectively, in the closed OB (Fig. 8E,F). Western blot and qRT-PCR studies confirmed that this reduced occupancy of hnRNP K and LL was not due to a decrease in either gene or protein expression levels (Fig. 8G–I). Together, these studies show that odorant sensory input-dependent changes in Th and Gad1 transcription are associated with a shift in the balance between secondary structure formation and hnRNP protein occupancy on each respective promoter.
Figure 8.
DNA secondary structure formation and hnRNP recruitment on Gad1 and Th promoters in the OB are regulated by odorant-dependent sensory input. A, B, Relative Th and Gad1 expression levels, respectively, measured by qRT-PCR in mouse OB tissue either ipsilateral (closed) or contralateral (open) to unilateral naris occlusion. C, D, ChIP assays with the BG4 antibody detected significantly more G-quadruplex secondary structure on the Th and Gad1 promoters, respectively, in the closed versus open OB. E, ChIP assays showed a significant increase in hnRNP K occupancy on the Th promoter in open versus closed OB. F, ChIP assays showed a significant increase in hnRNP LL occupancy on the Gad1 promoter in the open versus closed OB. G, qPCR analysis indicated that there was no significant difference in either hnRNP K or LL expression levels in the open versus closed OB. H, I, Western blots showed no significant difference in hnRNP K or LL protein expression levels, respectively, in the open versus closed OB. *p < 0.01.
Differential occupancy of DNA secondary structure and hnRNPs on the Gad1 and Th proximal promoter in cultured cells with membrane depolarization
To test whether DNA secondary structure and hnRNPs differentially occupy the Gad1 and Th proximal promoters in an alternative model system, BG4 and hnRNP ChIP assays were performed in KCl depolarized human SH-SY5Y and NT2 cultured cell lines. Unlike primary cell culture alternatives, this simple model system provides a large and homogenous set of Th- or Gad1-expressing cells suitable for ChIP analyses. Th and Gad1 expression levels are increased in human SH-SY5Y and NT2 cells, respectively, depolarized with 50 mm KCl compared with cultures with equimolar treatment of NaCl (Fig. 9A,B). In the SH-SY5Y and NT2 cell lines, ChIP assays with the BG4 antibody showed that KCl-mediated depolarization reduced the presence of G-quadruplexes in the Th and Gad1 promoter regions, respectively (Fig. 9C,D). By contrast, ChIP assays for hnRNP K and LL showed increased occupancy on the Th and Gad1 promoter, respectively, in response to membrane depolarization (Fig. 9E,F). Together, these findings show that membrane depolarization induces changes in secondary structure formation and hnRNP protein recruitment on the Th and Gad1 promoters that are associated with increased transcription levels. Because membrane depolarization is an essential component of synaptic activity, the findings with the cultured cell lines complement and support the model generated with in vivo and in vitro studies using OB and cortical tissue.
Figure 9.
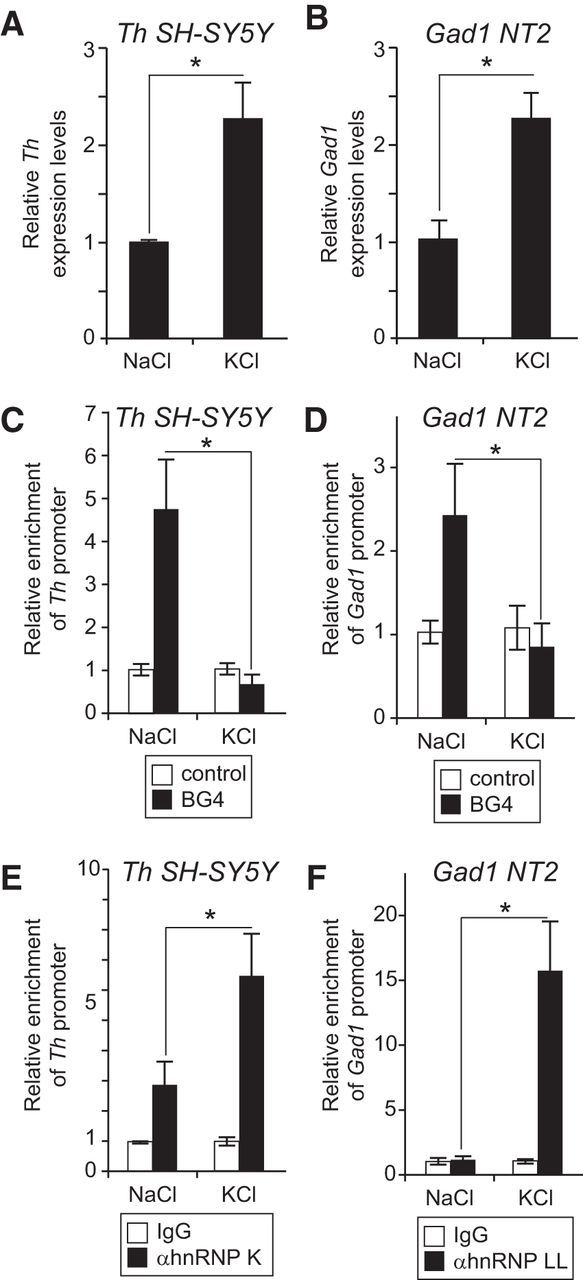
Differential occupancy of G-quadruplexes and hnRNPs on the Th and Gad1 promoters in response to membrane depolarization of cultured cells. A, B, Relative Th and Gad1 expression levels, respectively, as measured by qRT-PCR, were increased in SH-SY5Y (A) and NT2 (B) cells depolarized with 50 mm KCl compared with control cultures treated with 50 mm NaCl. C, D, ChIP assays with the BG4 antibody detected significantly more G-quadruplex secondary structure on the Th and Gad1 promoters in the SH-SY5Y and NT2 cells, respectively, depolarized with KCl compared with control cultures treated with NaCl. E, F, ChIP assays showed a significant increase in hnRNP K occupancy on the Th promoter in depolarized SH-SY5Y cells compared with NaCl-treated control cells. F, ChIP assays showed a significant increase in hnRNP LL occupancy on the Gad1 promoter in depolarized NT2 cells compared with NaCl-treated control cells. *p < 0.01.
Discussion
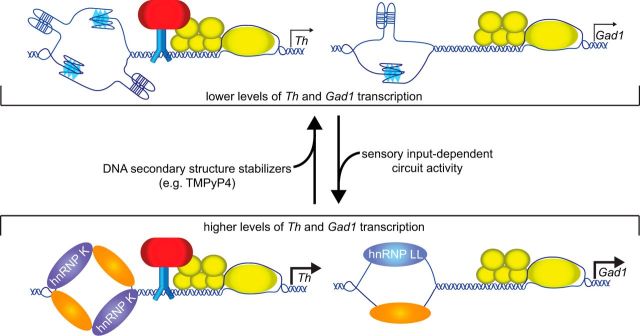
These studies have identified a novel activity-dependent molecular mechanism regulating Gad1 and Th transcription that is mediated, in part, by DNA secondary structure (summarized in Fig. 10). Single DNA strands in G:C-rich regions can either adopt secondary structures, such as G-quadruplexes and i-motifs, or recruit single-strand DNA-binding proteins, such as hnRNP K and LL. DNA secondary structure formation is associated with reduced gene transcription levels. The mechanisms underlying this secondary structure-dependent repression are not established but may include either disruption of transcription elongation, blocking the binding of transcription activator proteins, or recruitment of transcription repressor proteins (Brooks et al., 2010). By contrast, the recruitment of single-strand DNA-binding proteins, such as hnRNP K and LL, promote gene transcription by preventing secondary structure formation. hnRNP K can also facilitate transcription by contacting general transcription factors, such as TBP (Michelotti et al., 1996; Shnyreva et al., 2000). Both hnRNP K and LL preferentially bind C-rich strands, and the complementary G-rich strands are likely bound by related proteins, such as hnRNP A1 (Banerjee et al., 2014). The absence of hnRNP LL knock-out mice and the lethality of hnRNP K knock-out mice (Gallardo et al., 2015) limited the current study from further defining the role of these proteins, but future studies incorporating either tissue-selective knockdown or gene editing will better define their role in neural circuit activity-dependent regulation of gene expression.
Figure 10.
Proposed synaptic activity-dependent mechanism for hnRNPs and DNA secondary structure on the Th and Gad1 proximal promoters. Transcription of Th and Gad1 by the RNA polymerase transcription protein complex (yellow) drives the separation of the upstream G:C-rich regions. The single strands either are bound by proteins, such as hnRNPs, or adopt secondary structures, such as G-quadruplexes or i-motifs. The Th and Gad1 C-rich strands recruit hnRNP K and LL (purple), respectively, and the G-rich strand is bound to additional hnRNPs (orange). On the Th promoter, hnRNP K can interact with other transcription factors bound to the promoter (blue, red) as well as the RNA polymerase complex. Increased sensory input-dependent circuit activity levels favor hnRNP binding and promote higher expression levels. By contrast, secondary structure formation reduces gene expression levels, and small molecules, such as TMPyP4, favor this state.
The presence of either secondary structure or hnRNPs in the G:C-rich regions of the Gad1 and Th proximal promoters is mutually exclusive in the proposed mechanism. This is consistent with structural studies showing that hnRNP binding requires hydrogen bonding and stacking interactions with the nucleotide bases that would preclude secondary structure formation (Shamoo et al., 1997; Xu et al., 1997; Backe et al., 2005). Our studies indicate that sensory input-dependent activity levels can shift the balance between DNA secondary structure formation and hnRNP binding. This role for neural circuit activity is a previously unreported feature in DNA secondary structure-mediated regulation of gene transcription. Moreover, the strong conservation of the G-quadruplex/i-motif consensus sequences and recognition sites for hnRNPs in both Th and Gad1 throughout mammals suggests that this shared mechanism is functional in both rodents and humans.
The proposed sensory input-dependent mechanism is expected to be independent of regional patterning transcription factors required for expression of these genes in the OB and other brain regions. Regional transcription factors are likely necessary to maintain Th and Gad1 in a transcriptionally active state, whereas DNA secondary structure and hnRNPs modulate transcription levels in response to changes in odorant sensory input levels without disrupting the regional transcription factors. RNA polymerase generates negative supercoiling in upstream genomic DNA, and this negative supercoiling drives the formation of secondary structures, such as G-quadruplexes (Sun and Hurley, 2009; Zhang et al., 2013). Our finding that G-quadruplexes are present on Th and Gad1 promoters when sensory input-dependent activity levels are low indicates that regional transcription factors bound to the promoter and/or distal regulatory regions of these genes are able to recruit RNA polymerase, but these protein complexes are ineffective at resolving DNA secondary structure generated by RNA polymerase. High sensory input-dependent activity levels can resolve these structures and increase transcription levels, but the molecular details driving the shift from DNA secondary structure formation to hnRNP recruitment remain to be established. Additional studies are also required to determine whether this driving force is downstream of synaptically activated pathways within the neuron or, alternatively, by the activity-related release of growth factors or other signaling molecules from neighboring cells that can lead to modifications of gene promoter chromatin environments. Nevertheless, the integration of regulation from regional transcription factors with the sensory input-dependent shown in this study would provide the dynamic dopamine and GABA production in OB interneurons and short axon cells needed to adjust detection thresholds in response to changing odorant levels. Similarly, this regulation of Gad1 in cortical interneurons would allow for dynamic GABA expression required to regulate principal cells in cortical circuits.
This study shows that promoter DNA secondary structure is a component of activity-dependent regulation of gene transcription, but the presence of G-quadruplexes or i-motifs in the proximal promoter does not necessarily indicate that transcription of a gene is dependent on synaptic activity. In both humans and mice, several genes with activity-dependent transcription (including Arc, Egr1, Nr4a1, and Rheb) have G-quadruplex motifs in their proximal promoters. Genomic analyses of G-quadruplex motif distribution in humans and rodents, however, have shown that these motifs have a preference for the proximal promoter of genes (Verma et al., 2008; Maizels and Gray, 2013; Chambers et al., 2015; Hänsel-Hertsch et al., 2016). Furthermore, ∼40% of genes in the human genome have G-quadruplex consensus motifs in their proximal promoter regions (Huppert and Balasubramanian, 2007), and most of these genes are not dependent on synaptic activity or membrane depolarization for transcription. Thus, although these findings show that DNA secondary structure can be a component of activity-dependent regulation of gene transcription, the presence of secondary structure in a gene promoter is not sufficient to make gene transcription activity-dependent.
The proposed mechanism provides a molecular basis for the shared odorant sensory input-dependent regulation of Th and Gad1 in the OB. The presence of DNA secondary structure and the hnRNP binding on the Gad1 promoter in the cortex, however, suggests that this mechanism is not just limited to OB GABAergic interneurons. Several studies have reported that Gad1 expression levels in the cortex and hippocampus are activity-dependent (Liang et al., 1996; Esclapez and Houser, 1999; Ramírez and Gutiérrez, 2001; Patz et al., 2003; Lau and Murthy, 2012). Although speculative, the mechanism established in this study is likely functional in the GABAergic neurons that coexpress either hnRNP LL in these regions as well. Additional studies to identify specific channels and receptors required for regulating Gad1 expression by DNA secondary structure and hnRNPs may provide insight into neurological disorders associated with impaired Gad1 activity and synaptic inhibition, such as epilepsy.
This study also significantly advances our molecular understanding of how Gad1 transcription is activated by demonstrating that hnRNP LL interacts with the Gad1 promoter in both OB and cortex and facilitating transcription in cultured cells. In the midbrain, hnRNP LL may interact with Pitx2 on the Gad1 promoter because Pitx2 both specifies midbrain GABAergic neurons and activates the Gad1 expression by binding its promoter (Westmoreland et al., 2001; Waite et al., 2011). In the forebrain, however, other factors directly bound to the Gad1 promoter that can interact with hnRNP LL are not presently known.
Increased methylation of the Gad1 promoter is associated with reduced gene expression levels in schizophrenia, bipolar disorder, and autism (Chen et al., 2011; Zhubi et al., 2014), but this study showed that methylated CpG motifs in the Gad1 promoter C-rich strand do not disrupt either secondary structure formation or hnRNP recruitment. These findings would suggest that promoter methylation does not block the activity-dependent regulation of gene expression by DNA secondary structure and hnRNPs, but further studies are needed to establish whether methylation impedes the ability of proteins, such as helicases, to resolve DNA secondary structures or if methylated C:G-rich regions recruit repressive factors that either block hnRNP binding or further stabilize the DNA secondary structures.
The Gad1 transcription start site used in this study was previously identified as the dominant start site and is conserved between mice and humans (Szabó et al., 1996; Westmoreland et al., 2001). There are, however, alternative Gad1 transcription start sites that are upstream (Szabó et al., 1996; Yanagawa et al., 1997; Westmoreland et al., 2001; Chen et al., 2011). Both hnRNP recruitment and DNA secondary structure formation by the G:C-rich region identified in this study would also modulate Gad1 expression from the alternative start sites. G-quadruplexes on the template strand, as is the case in Gad1, can impede or block RNA polymerase as it transcribes genes (Yu et al., 2009; Broxson et al., 2011; Agarwal et al., 2014). Proteins, such as hnRNP LL, that bind the single-stranded template can prevent secondary structure formation. Thus, regulating the balance between hnRNP recruitment and DNA secondary structure formation is likely to be important in the expression of all Gad1 transcripts.
Footnotes
This work was supported by National Institutes of Health Grant DC008955 and the Burke Medical Research Institute. We thank Dr. Shankar Balasubramanian (Cambridge University) for the BG4 antibody; and Dr. Kristen Lynch (University of Pennsylvania School of Medicine) for the hnRNP LL expression plasmid.
The authors declare no competing financial interests.
References
- Agarwal T, Roy S, Kumar S, Chakraborty TK, Maiti S (2014) In the sense of transcription regulation by G-quadruplexes: asymmetric effects in sense and antisense strands. Biochemistry 53:3711–3718. 10.1021/bi401451q [DOI] [PubMed] [Google Scholar]
- Akiba Y, Sasaki H, Huerta PT, Estevez AG, Baker H, Cave JW (2009) γ-Aminobutyric acid-mediated regulation of the activity-dependent olfactory bulb dopaminergic phenotype. J Neurosci Res 87:2211–2221. 10.1002/jnr.22055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backe PH, Messias AC, Ravelli RB, Sattler M, Cusack S (2005) X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure 13:1055–1067. 10.1016/j.str.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Baker H, Farbman AI (1993) Olfactory afferent regulation of the dopamine phenotype in the fetal rat olfactory system. Neuroscience 52:115–134. 10.1016/0306-4522(93)90187-K [DOI] [PubMed] [Google Scholar]
- Baker H, Kawano T, Margolis FL, Joh TH (1983) Transneuronal regulation of tyrosine hydroxylase expression in olfactory bulb of mouse and rat. J Neurosci 3:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker H, Morel K, Stone DM, Maruniak JA (1993) Adult naris closure profoundly reduces tyrosine hydroxylase expression in mouse olfactory bulb. Brain Res 614:109–116. 10.1016/0006-8993(93)91023-L [DOI] [PubMed] [Google Scholar]
- Baker H, Cummings DM, Munger SD, Margolis JW, Franzen L, Reed RR, Margolis FL (1999) Targeted deletion of a cyclic nucleotide-gated channel subunit (OCNC1): biochemical and morphological consequences in adult mice. J Neurosci 19:9313–9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker H, Liu N, Chun HS, Saino S, Berlin R, Volpe B, Son JH (2001) Phenotypic differentiation during migration of dopaminergic progenitor cells to the olfactory bulb. J Neurosci 21:8505–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Akiba Y, Baker H, Cave JW (2013) Epigenetic control of neurotransmitter expression in olfactory bulb interneurons. Int J Dev Neurosci 31:415–423. 10.1016/j.ijdevneu.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Wang M, Cai E, Fujiwara N, Baker H, Cave JW (2014) Regulation of tyrosine hydroxylase transcription by hnRNP K and DNA secondary structure. Nat Commun 5:5769. 10.1038/ncomms6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar-Jog YP, Van Dornshuld E, Brooks TA, Tschumper GS, Wadkins RM (2014) Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry 53:1586–1594. 10.1021/bi401523b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5:182–186. 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks TA, Kendrick S, Hurley L (2010) Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J 277:3459–3469. 10.1111/j.1742-4658.2010.07759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxson C, Beckett J, Tornaletti S (2011) Transcription arrest by a G quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry 50:4162–4172. 10.1021/bi2002136 [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13:721–731. 10.1101/gr.926603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34:5402–5415. 10.1093/nar/gkl655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 33:877–881. 10.1038/nbt.3295 [DOI] [PubMed] [Google Scholar]
- Chen Y, Dong E, Grayson DR (2011) Analysis of the GAD1 promoter: trans-acting factors and DNA methylation converge on the 5′ untranslated region. Neuropharmacology 60:1075–1087. 10.1016/j.neuropharm.2010.09.017 [DOI] [PubMed] [Google Scholar]
- Cigola E, Volpe BT, Lee JW, Franzen L, Baker H (1998) Tyrosine hydroxylase expression in primary cultures of olfactory bulb: role of L-type calcium channels. J Neurosci 18:7638–7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola DM. (2012) Studies of olfactory system neural plasticity: the contribution of the unilateral naris occlusion technique. Neural Plast 2012:351752. 10.1155/2012/351752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlander MG, Tobin AJ (1992) A transcriptional regulatory element of the gene encoding the 67,000-M(r) form of human glutamate decarboxylase is similar to a Drosophila regulatory element. J Neurochem 58:2182–2190. 10.1111/j.1471-4159.1992.tb10962.x [DOI] [PubMed] [Google Scholar]
- Esclapez M, Houser CR (1999) Up-regulation of GAD65 and GAD67 in remaining hippocampal GABA neurons in a model of temporal lobe epilepsy. J Comp Neurol 412:488–505. 10.1002/(SICI)1096-9861(19990927)412:3%3C488::AID-CNE8%3E3.0.CO%3B2-6 [DOI] [PubMed] [Google Scholar]
- Fedoroff OY, Rangan A, Chemeris VV, Hurley LH (2000) Cationic porphyrins promote the formation of i-motif DNA and bind peripherally by a nonintercalative mechanism. Biochemistry 39:15083–15090. 10.1021/bi001528j [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara N, Mazzola M, Cai E, Wang M, Cave JW (2015) TMPyP4, a stabilizer of nucleic acid secondary structure, is a novel acetylcholinesterase inhibitor. PLoS One 10:e0139167. 10.1371/journal.pone.0139167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M, Lee HJ, Zhang X, Bueso-Ramos C, Pageon LR, McArthur M, Multani A, Nazha A, Manshouri T, Parker-Thornburg J, Rapado I, Quintas-Cardama A, Kornblau SM, Martinez-Lopez J, Post SM (2015) hnRNP K is a haploinsufficient tumor suppressor that regulates proliferation and differentiation programs in hematologic malignancies. Cancer Cell 28:486–499. 10.1016/j.ccell.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring K, Leroy JL, Guéron M (1993) A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature 363:561–565. 10.1038/363561a0 [DOI] [PubMed] [Google Scholar]
- Han FX, Wheelhouse RT, Hurley LH (1999) Interactions of TMPyP4 and TMPyP2 with quadruplex DNA: structural basis for the differential effects on telomerase inhibition. J Am Chem Soc 121:3561–3570. 10.1021/ja984153m [DOI] [Google Scholar]
- Han H, Langley DR, Rangan A, Hurley LH (2001) Selective interactions of cationic porphyrins with G-quadruplex structures. J Am Chem Soc 123:8902–8913. 10.1021/ja002179j [DOI] [PubMed] [Google Scholar]
- Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S (2016) G-quadruplex structures mark human regulatory chromatin. Nat Genet 48:1267–1272. 10.1038/ng.3662 [DOI] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res 35:406–413. 10.1093/nar/gkl1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kendrick S, Hecht SM, Hurley LH (2014) The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J Am Chem Soc 136:4172–4185. 10.1021/ja4109352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Lee MK, Carroll J, Joh TH (1993) Both the basal and inducible transcription of the tyrosine hydroxylase gene are dependent upon a cAMP response element. J Biol Chem 268:15689–15695. [PubMed] [Google Scholar]
- Kiyokage E, Pan YZ, Shao Z, Kobayashi K, Szabó G, Yanagawa Y, Obata K, Okano H, Toida K, Puche AC, Shipley MT (2010) Molecular identity of periglomerular and short axon cells. J Neurosci 30:1185–1196. 10.1523/JNEUROSCI.3497-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka K, Kosaka T (2005) Synaptic organization of the glomerulus in the main olfactory bulb: compartments of the glomerulus and heterogeneity of the periglomerular cells. Anat Sci Int 80:80–90. 10.1111/j.1447-073x.2005.00092.x [DOI] [PubMed] [Google Scholar]
- Kosaka K, Kosaka T (2007) Chemical properties of type 1 and type 2 periglomerular cells in the mouse olfactory bulb are different from those in the rat olfactory bulb. Brain Res 1167:42–55. 10.1016/j.brainres.2007.04.087 [DOI] [PubMed] [Google Scholar]
- Lau CG, Murthy VN (2012) Activity-dependent regulation of inhibition via GAD67. J Neurosci 32:8521–8531. 10.1523/JNEUROSCI.1245-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Quinn PG, Chikaraishi DM (2004) Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol Cell Neurosci 25:536–547. 10.1016/j.mcn.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Liang F, Isackson PJ, Jones EG (1996) Stimulus-dependent, reciprocal up- and downregulation of glutamic acid decarboxylase and Ca2+/calmodulin-dependent protein kinase II gene expression in rat cerebral cortex. Exp Brain Res 110:163–174. [DOI] [PubMed] [Google Scholar]
- Maizels N, Gray LT (2013) The G4 genome. PLoS Genet 9:e1003468. 10.1371/journal.pgen.1003468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti EF, Michelotti GA, Aronsohn AI, Levens D (1996) Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol 16:2350–2360. 10.1128/MCB.16.5.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamoto-Combs K, Piech KM, Best JA, Sun B, Tank AW (1997) Tyrosine hydroxylase gene promoter activity is regulated by both cyclic AMP-responsive element and AP1 sites following calcium influx: evidence for cyclic AMP-responsive element binding protein-independent regulation. J Biol Chem 272:6051–6058. 10.1074/jbc.272.9.6051 [DOI] [PubMed] [Google Scholar]
- Parrish-Aungst S, Shipley MT, Erdelyi F, Szabó G, Puche AC (2007) Quantitative analysis of neuronal diversity in the mouse olfactory bulb. J Comp Neurol 501:825–836. 10.1002/cne.21205 [DOI] [PubMed] [Google Scholar]
- Parrish-Aungst S, Kiyokage E, Szabó G, Yanagawa Y, Shipley MT, Puche AC (2011) Sensory experience selectively regulates transmitter synthesis enzymes in interglomerular circuits. Brain Res 1382:70–76. 10.1016/j.brainres.2011.01.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz S, Wirth MJ, Gorba T, Klostermann O, Wahle P (2003) Neuronal activity and neurotrophic factors regulate GAD-65/67 mRNA and protein expression in organotypic cultures of rat visual cortex. Eur J Neurosci 18:1–12. 10.1046/j.1460-9568.2003.02702.x [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Gaw HY, Patel DJ (2005) Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol 1:167–173. 10.1038/nchembio723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puche AC, Shipley MT (1999) Odor-induced, activity-dependent transneuronal gene induction in vitro: mediation by NMDA receptors. J Neurosci 19:1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Hurley LH (2008) Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 90:1149–1171. 10.1016/j.biochi.2008.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez M, Gutiérrez R (2001) Activity-dependent expression of GAD67 in the granule cells of the rat hippocampus. Brain Res 917:139–146. 10.1016/S0006-8993(01)02794-9 [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA (1997) Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat Struct Biol 4:215–222. 10.1038/nsb0397-215 [DOI] [PubMed] [Google Scholar]
- Shnyreva M, Schullery DS, Suzuki H, Higaki Y, Bomsztyk K (2000) Interaction of two multifunctional proteins: heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J Biol Chem 275:15498–15503. 10.1074/jbc.275.20.15498 [DOI] [PubMed] [Google Scholar]
- Sun D, Hurley LH (2009) The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Med Chem 52:2863–2874. 10.1021/jm900055s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó G, Katarova Z, Körtvély E, Greenspan RJ, Urbán Z (1996) Structure and the promoter region of the mouse gene encoding the 67-kD form of glutamic acid decarboxylase. DNA Cell Biol 15:1081–1091. 10.1089/dna.1996.15.1081 [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T (2003) Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol 467:60–79. 10.1002/cne.10905 [DOI] [PubMed] [Google Scholar]
- Trocmé C, Mallet J, Biguet NF (1997) AP-1 mediates trans-synaptic induction of tyrosine hydroxylase gene expression in adrenal medulla but not in superior cervical ganglia. J Neurosci Res 48:489–498. 10.1002/(SICI)1097-4547(19970615)48:6%3C489::AID-JNR1%3E3.0.CO%3B2-6 [DOI] [PubMed] [Google Scholar]
- Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S (2008) Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem 51:5641–5649. 10.1021/jm800448a [DOI] [PubMed] [Google Scholar]
- Waite MR, Skidmore JM, Billi AC, Martin JF, Martin DM (2011) GABAergic and glutamatergic identities of developing midbrain Pitx2 neurons. Dev Dyn 240:333–346. 10.1002/dvdy.22532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Banerjee K, Baker H, Cave J (2015) Nucleotide sequence conservation of novel and established cis-regulatory sites within the tyrosine hydroxylase gene promoter. Front Biol (Beijing) 10:74–90. 10.1007/s11515-014-1341-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland JJ, McEwen J, Moore BA, Jin Y, Condie BG (2001) Conserved function of Caenorhabditis elegans UNC-30 and mouse Pitx2 in controlling GABAergic neuron differentiation. J Neurosci 21:6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DA, Wood JG (1992) Functional consequences of unilateral olfactory deprivation: time-course and age sensitivity. Neuroscience 49:183–192. 10.1016/0306-4522(92)90086-H [DOI] [PubMed] [Google Scholar]
- Xu RM, Jokhan L, Cheng X, Mayeda A, Krainer AR (1997) Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure 5:559–570. 10.1016/S0969-2126(97)00211-6 [DOI] [PubMed] [Google Scholar]
- Yanagawa Y, Kobayashi T, Kamei T, Ishii K, Nishijima M, Takaku A, Tamura S (1997) Structure and alternative promoters of the mouse glutamic acid decarboxylase 67 gene. Biochem J 326:573–578. 10.1042/bj3260573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Schonhoft JD, Dhakal S, Bajracharya R, Hegde R, Basu S, Mao H (2009) ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J Am Chem Soc 131:1876–1882. 10.1021/ja806782s [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu HH, Zheng KW, Hao YH, Tan Z (2013) DNA G-quadruplex formation in response to remote downstream transcription activity: long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res 41:7144–7152. 10.1093/nar/gkt443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR (2014) Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry 4:e349. 10.1038/tp.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]