Abstract
Prolactin (PRL) and its receptor (PRLR) are implicated in breast cancer invasiveness, although their exact roles remain controversial. The Na+/H+ exchanger (NHE1) plays essential roles in cancer cell motility and invasiveness, but the PRLR and NHE1 have not previously been linked. Here we show that in T47D human breast cancer cells, which express high levels of PRLR and NHE1, exposure to PRL led to the activation of Janus kinase-2 (JAK2)/signal transducer and activator of transcription-5 (STAT5), Akt, and ERK1/2 signaling and the rapid formation of peripheral membrane ruffles, known to be associated with cell motility. NHE1 was present in small ruffles prior to PRL treatment and was further recruited to the larger, more dynamic ruffles induced by PRL exposure. In PRL-induced ruffles, NHE1 colocalized with activated Akt, ERK1/2, and the ERK effector p90Ribosomal S kinase (p90RSK), known regulators of NHE1 activity. Stimulation of T47D cells with PRL augmented p90RSK activation, Ser703-phosphorylation of NHE1, NHE1-dependent intracellular pH recovery, pericellular acidification, and NHE1-dependent invasiveness. NHE1 activity and localization to ruffles were attenuated by the inhibition of Akt and/or ERK1/2. In contrast, noncancerous MCF10A breast epithelial cells expressed NHE1 and PRLR at lower levels than T47D cells, and their stimulation with PRL induced neither NHE1 activation nor NHE1-dependent invasiveness. In conclusion, we show for the first time that PRLR activation stimulates breast cancer cell invasiveness via the activation of NHE1. We propose that PRL-induced NHE1 activation and the resulting NHE1-dependent invasiveness may contribute to the metastatic behavior of human breast cancer cells.
Prolactin (PRL) is an α-helical hormone (1) expressed at high levels in the lactotroph cells of the anterior pituitary gland but is also produced locally elsewhere in the brain as well as in the peripheral tissues including the immune system, prostate, uterus, and mammary gland (2). PRL signals via the PRL receptor (PRLR), a class 1 cytokine receptor, which exists in several isoforms, of which the longest, an 85- to 95-kDa isoform, is by far the best described in terms of downstream signaling (3–5). Most cytokine receptors including the PRLR have been shown to be predimerized prior to hormone binding (6, 7), and hormone binding leads to subtle conformational changes (7–9) that initiate multiple intracellular signaling pathways. For PRLR activation by PRL these include the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway (primarily JAK2 and STAT5), the phosphatidyl-inositol-3 kinase (PI3K)-Akt pathway, and the MAPK pathways ERK1/2, and p38 MAPK (3, 4). In addition, PRLR signaling activates Src kinases including Fyn and Src (10, 11), Focal adhesion kinase (FAK) (11), and a Vav2-Nek3-Rac signaling axis (12). Further downstream, PRLR regulates the transcription of numerous genes, including receptor activator of nuclear factor-κB ligand, which is important for mammary gland development (13) and the suppressor of cytokine signaling-3, a major negative feedback regulator of cytokine signaling (14). Furthermore, PRLR signaling potentiates signaling via estrogen receptors (15, 16) and growth factor receptors of the human epidermal growth factor receptor (ErbB) family (17, 18).
In addition to its roles in regulation of lactation and mammary gland growth and differentiation, the biological functions of PRLR signaling include modulation of cell proliferation and survival (19–21). Although this remains controversial (eg, reference 22), a link between PRL signaling and cancer, especially breast cancer, has been proposed (23–25). Supporting this notion, constitutively active PRLR variants were identified in patients presenting with rare forms of benign breast tumors (26, 27); increased systemic or mammary PRL expression was found to induce mammary cancer in mouse models (23, 28); disruption of PRL or PRLR expression delayed oncogene-induced mammary tumorigenesis (29, 30); and plasma PRL levels correlate with breast cancer risk in postmenopausal women (25). The involvement of PRL signaling in the regulation of apoptosis, autophagy, clonogenic potential, proliferation, and cell motility has been reported in various cancer types and settings (4, 31–34). Most studies propose a stimulatory effect of PRLR signaling on breast cancer cell motility (12, 33, 35, 36), but inhibitory effects have also been reported (37). The mechanisms via which PRL regulates motility are incompletely described, but it has been shown that the PRLR forms a complex with integrins (38), and that PRLR signaling stimulates filamin A phosphorylation via p21-activated kinase 1 (32) and via Nek3-dependent Rac activation and paxillin phosphorylation (12). The link between PRLR signaling and breast cancer metastasis is substantiated by recent findings that the stiff extracellular matrix (ECM) typical of the breast cancer microenvironment potentiates PRLR signaling (39, 40). On the other hand, PRLR signaling is inhibited under acidotic extracellular conditions such as those occurring in solid tumors (41) due to the inhibition of PRL-PRLR interaction at acidic pH (42, 43).
Stimulation with PRL has been shown to elicit the formation of membrane ruffles (12, 35), Ruffles are sheet-like membrane protrusions that, in contrast to lamellipodia, do not attach to the substratum (44). They are highly dynamic, with a half-life of minutes, and at least two forms are distinguished, first in early studies by Abercrombie et al (45): peripheral ruffles, which form at the front of motile cells and move rearward, and dorsal ruffles, which as the name implies form dorsally, forming circular structures before they disappear (46). Although there are many similarities between them, ruffles are distinct from lamellipodia (44, 47), and different processes are involved in their formation (48). Dorsal and peripheral ruffles are also formed by distinct mechanisms (49) and are distinct structures, the former being assigned roles in pinocytosis and receptor endocytosis and the latter associated with cell motility (46, 47), and are proposed to be compartments of actin reorganization and to result from inefficient lamellipodial adhesion (47). We and others have shown that the Na+/H+ exchanger (NHE1; SLC9A1) plays important roles in stimulating cell motility in general and cancer cell invasiveness in particular (50–54). Thus, NHE1 is frequently up-regulated in cancer cells (55, 56) and accumulates in the leading edge lamellipodia of migrating cancer cells in which it contributes to the formation of intra- and extracellular pH gradients along the axis of migration (57–59). In fibroblasts, NHE1 is essential for regulation of directional cell motility after stimulation of platelet-derived growth factor receptor-α in the primary cilium (50) in a manner that involves Akt- and ERK1/2-dependent localization of NHE1 to the leading edge lamellipodia (51).
The roles of NHE1 in migration are thought to involve regulation of intra- as well as pericellular pH, in turn modulating integrin-dependent adhesion to the ECM (60, 61), and dynamics of F-actin, at least in part via the pH-dependence of actin dynamics-regulating proteins such as talin (62), Cdc42 (63), cofilin (64), and FAK (65). NHE1 also localizes to invadopodia, in which it regulates the invasion by facilitating ECM digestion (due to its acidic pH optimum) and increasing cofilin dynamics (53, 66). A recent study of proteins phosphorylated in response to stimulation of 3T3-F442A preadipocytes with GH revealed that human NHE1 was phosphorylated on Ser703 in response to GH (67). Importantly, Ser703 phosphorylation of NHE1, which is mediated by the ERK1/2 effector, p90 ribosomal S kinase (p90RSK), is associated with NHE1 activation (68). Because the GH receptor and downstream signaling events exhibit high similarity to PRLR and its downstream targets (69); this leads us to hypothesize that PRLR signaling might stimulate Ser703 phosphorylation of NHE1 in breast cancer cells, in turn eliciting increased invasiveness.
The aim of this study was therefore to determine the impact of PRL-mediated PRLR signaling on NHE1 and NHE1-dependent invasiveness in breast cancer cells. We provide here the first report demonstrating that exposure to PRL elicits the phosphorylation, activation, and peripheral ruffle-targeted localization of NHE1 in breast cancer cells and stimulates their invasion through matrigel in an NHE1-dependent manner. This suggests that the PRL-PRLR-NHE1 signaling axis is of potential therapeutic interest in breast cancer.
Materials and Methods
Reagents and antibodies
Human prolactin was recombinantly expressed and purified as described (1) and, unless otherwise mentioned, used at 10 nM (∼230 ng/mL). 5-(N-ethyl-isopropyl)amiloride (EIPA) was from Life Technologies, and cariporide was a kind gift from Sanofi-Aventis. U0126 and Akti-1/2 were from Merck. Antibodies against Akt, phospho-Akt (pSer473), ERK1/2, phospho-ERK1/2 (pThr202/Tyr204), phospho-p90RSK (pSer380), phospho-FAK (pTyr576/577), and phospho-STAT5 (pTyr694) were from Cell Signaling Technology; antibodies against JAK2 and phospho-JAK2 (pTyr1007/1008) were from Millipore, antibodies against β-actin and vinculin were from Sigma-Aldrich, antibodies against the NaHE1, STAT5, p90RSK1, and the intracellular domain of PRLR (α-PRLR-ICD (H300)) were from Santa Cruz Biotechnology, and an antibody against p150Glued was from BD Biosciences. Antibody against phospho-S703-NHE1 was from DSTT, University of Dundee, Dundee, Ireland. Rhodamine-phalloidin was from Invitrogen.
Cell lines and media
MCF10A cells were grown in a 1:1 mix of DMEM and Ham's F12 nutrient mixture medium, 5% horse bovine serum (Invitrogen), 1% penicillin/streptomycin (Invitrogen); 20 ng/mL recombinant human epidermal growth factor (Sigma-Aldrich), 0.25 ng/mL hydrocortisone (Sigma-Aldrich), and 10 μg/mL bovine insulin-transferrin-selenium (Gibco). T47D cells were grown in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 5 μg/mL bovine insulin-transferrin-selenium. Cells were maintained at 37°C, 5% CO2, passaged every 3–4 days (maximum 22 passages), and serum starved for 2 hours prior to all experiments.
MDCK GII cells were transfected with pcDNA-DEST47 (Invitrogen) encoding mouse NHE1-GFP, prepared as follows: mouse NHE1 (NM-016981) was cloned into the Gateway pDONR vector (Invitrogen), using the following primers: forward primer with AttB recombination-sequence, kozak sequence and start codon in-frame with the attB sequence, 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TTC ACC ATG ATG CTT CGG TGG TCC GGC GT; reverse primer, 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC CTG TCC TTT GGG GAT GAA AGG CTC T. This was followed by recombination into pcDNA-DEST47 for the addition of a C-terminal green fluorescent protein (GFP) tag, following the manufacturer's instructions. Selection for stable expression was performed with G418. Expression was verified by Western blotting and fluorescence microscopy. NHE1-GFP cells were used for time-lapse imaging of NHE1 and plasma membrane dynamics during PRL-induced ruffling. Cells were grown in DMEM with 1 g/L glucose (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 0.5 U/mL penicillin (Sigma), 0.5 g/mL streptomycin (Gibco), 1 mg/mL kanamycin (Gibco). MDCK cells were maintained at 37°C and 5% CO2, split every 2–4 days for up to 1 month, and serum starved for 2 hours prior to experiments.
Immunoblotting
MCF10A and T47D cells were grown to approximately 80% confluence in 10-cm Petri dishes, washed in ice-cold PBS (NaCl 140 mM, KCl 2 mM, Na2HPO4 8 mM,KH2PO4 1 mM), lysed in boiling lysis buffer (1% sodium dodecyl sulfate; 10 mM Tris HCl, pH 7.5), sonicated, and centrifuged to clear debris. 200 000 NHE1-GFP-expressing MDCK-II cells were seeded in 35-mm dishes and grown overnight. They were serum starved for 2 hours followed by stimulation for 5 or 15 minutes with 10 nM PRL in PBS. Cells were washed in PBS and lysed in boiling lysis buffer as described for MCF10A and T47D cells. Identical amounts of protein (20 or 40 μg) diluted in sample buffer (141 mM Tris base, 106 mM Tris HCl, 69 mM LDS, 0.51 mM EDTA, 10% glycerol, 0.007% Phenol Red) were boiled for 5 minutes, separated on Biorad 10% BIS-TRIS gels (Bio-Rad Laboratories), and transferred onto nitrocellulose membranes. Membranes were stained with Ponceau S, blocked for 1 hour at 37°C in blocking buffer (120 mM NaCl, 10 mM Tris HCl, 1% Tween 20, and 5% nonfat dry milk), and incubated with primary antibodies in blocking buffer overnight at 4°C. After washing in TBS (0.01 M Tris/HCl, 0.15 M NaCl) + 0.1% Tween 20 (TBST), membranes were incubated with alkaline-phosphatase-conjugated secondary antibodies (1:2000), washed in TBST, and visualized using 5-bromo-4-chloro-3-indoyl-phosphate, 4-toluidine salt/4-nitro blue tetrazolium chloride (BCIP/NBT) (KPL). Bands were quantified by densitometric analysis (UN-SCAN-IT; Silk Scientific).
Immunoprecipitation
T47D cells were grown to 90% confluence, washed in PBS, and lysed with Nonidet-P40 (NP40) lysis buffer (1% NP40, 1 mM NaF, 1 mM Na3VO4, protease and phosphatase inhibitors). Cells were incubated for 1 hour on ice prior to 3- × 15-second sonication cycles and centrifuged at 12 000 × g, 10 minutes, 4°C to clear debris. Protein concentration in the supernatant was quantified by bicinchoninic assay and identical amounts of protein (2 mg/condition) were used in all samples. To minimize unspecific binding of proteins to the immunoprecipitating beads, the lysates were precleared with a slurry of recombinant protein A/G-sepharose (1 μL per 100 μL; GE Healthcare) and incubated under gentle agitation for 1 hour at 4°C. The beads were centrifuged at 2000 × g for 2 minutes at 4°C. The lysates were then incubated with 2 μg of the primary antibody of interest overnight at 4°C under gentle agitation or rabbit IgG as a negative control testing. The next day, lysates were incubated with protein A/G-sepharose slurry beads for 3 hours, 4°C, in rotation, spun down (2000 × g, 2 min, 4°C). Subsequently the beads were washed three times with NP40 lysis buffer, sample buffer was added to elute the proteins attached to the beads, and the beads were boiled for 5 minutes, vortexed, and spun down (15 000 × g, 4 min, 4°C) to obtain the immunoprecipitated fraction. Results were analyzed by immunoblotting using antibodies against NHE1 and PRLR.
Immunocytochemistry
Cells were grown to 60% confluence on glass coverslips and treated ± PRL for 5 minutes as indicated. Although NHE1 is maximally phosphorylated at 5 minutes after PRL addition (see Figure 5A), the 5-minutes time point was chosen for immunocytochemistry because we observed that the ruffling is a very rapid process, as is NHE1 activation. Preparations were washed in ice-cold PBS, fixed in 2% paraformaldehyde for 15 minutes at room temperature and 30 minutes on ice, washed in TBST (2 × 5 min), permeabilized for 5 minutes in 0.1% Triton X-100 in TBST, blocked for 30 minutes in 5% BSA in TBST, and incubated overnight at 4°C in a humid chamber with primary antibodies diluted in TBST + 1% BSA. The next day, preparations were washed in TBST + 1% BSA (3 × 5 min), incubated for 1 hour at room temperature protected from light with fluorophore-conjugated secondary antibodies diluted in TBST + 1% BSA, and washed in TBST + 1% BSA for 5 × 5 minutes. The second wash contained 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Coverslips were gently dried and mounted with a drop of N-propyl-gallate antifade mounting medium on glass slides and sealed. The slides were stored at 4°C in lightproof boxes. Cells were visualized using the 40× 1.0 NA, 60× 1.2 NA, or 100× 1.4 NA objective of an Olympus Bx63 epifluorescence microscope. No or negligible labeling was seen in the absence of primary antibody at the settings used. A z-stack analysis (0.5–0.6 μm step size) and isosurface projections were carried out using Olympus software. Overlays and brightness/contrast adjustment was carried out using Adobe Photoshop software. No other image adjustment was performed.
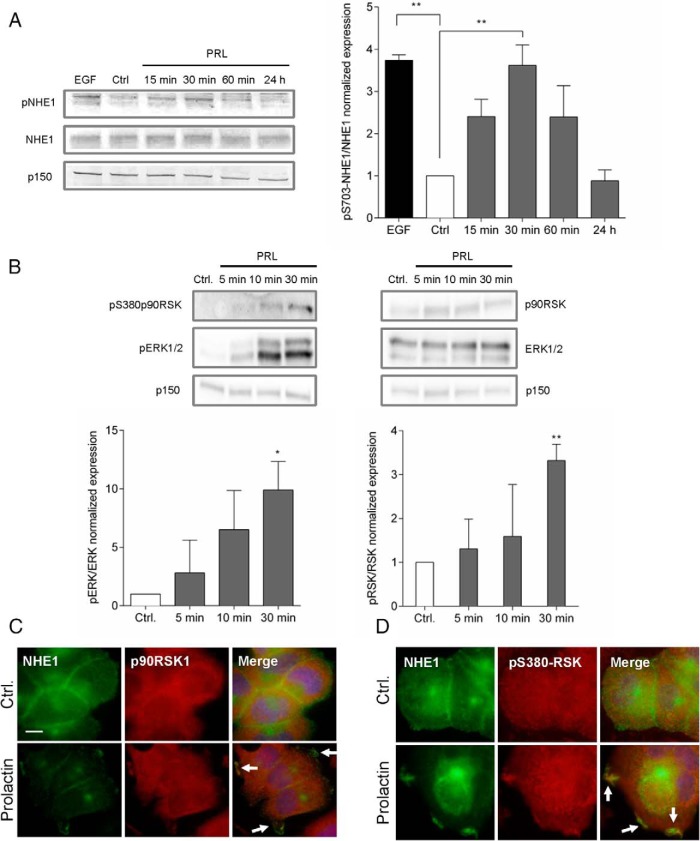
Figure 5. PRL elicits NHE1 phosphorylation on S703, and active p90RSK colocalizes with NHE1 in PRL-induced ruffles.
A, Left panel, Representative immunoblots of Ser703-phosphorylated and total NHE1, with p150 as a loading control. Right panel, Quantification of pSer703-NHE1 relative to total NHE1. Data are based on three independent experiments. **, P ≤ .01, one-way ANOVA with Tukey posttest. B, Top panel, Representative immunoblots of p380-p90RSK, p90RSK1, pThr202/Tyr204-ERK, ERK, and p150 (loading control). Bottom panel, Quantification of pERK relative to total ERK and p380p90RSK relative to total p90RSK. Data are based on three independent experiments. *, P ≤ .05, **, P ≤ .01, one-way ANOVA with Tukey posttest. C and D, IF analysis of T47D cells treated or not with PRL (10 nM, 5 min), as indicated. Arrows mark regions of prominent colocalization. C, NHE1 (green) and p90RSK1 (red). D, NHE1 (green) and pSer380-RSK (red). Images are merged and DNA (blue) is stained by DAPI. Scale bar, 10 μm. data are representative of three independent experiments per condition. Ctrl, control.
Measurements of intracellular pH
Cells were seeded in collagen-I-coated WillCo glass-bottom dishes (WillCo Wells). Cells were serum starved for 1.5 hours and washed once in 37°C isotonic ringer (IR: 143 mM NaCl, 5 mM KCl, 1 mM MgSO4-7H2O, 1 mM Na2HPO4-2H2O, 1 mM CaCl2-2H2O, 3.3 mM 3-[N-morpholino]propanesulfonic acid, 3.3 mM 2-[Tris[hydroxymethyl]-methylamino]-ethanesulfonic acid, 5 mM HEPES, adjusted with NaOH to pH 7.4 at 37°C), followed by incubation with 1.6 μM 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (BCECF) acetoxymethyl ester in a CO2-free incubator for 30 minutes at 37°C and protected from light. After incubation, the dishes were washed twice in IR and placed in a temperature-controlled chamber at 37°C at the stage of a Nikon Eclipse Ti microscope. A group of cells were focused and monitored using a 40× oil 1.4 NA objective, and the imaging software EasyRatioPro (PTI). Emission was measured at 520 nm after excitation at 440 and 485 nm for 8 minutes in IR, 10 minutes in 20 mM NH4Cl in IR, 1 minute in Na+-free Ringer (143 mM NaCl replaced by N-methyl-d-glucamine chloride), and return to IR. Calibration using the high K+/nigericin method was obtained for each cell line. Briefly, cells were perfused with KCl Ringer (156 mM KCl) at pH 6.7, 7.0, 7.2, and 7.4, and nigericin (10 μM; Sigma-Aldrich) was added. For intracellular pH (pHi) measurements using Akti-1/2 or U0126 to inhibit Akt or ERK1/2 activity, respectively, cells were preincubated for 1 hour with 10 μM of Akti-1/2 or U0126 or both compounds, respectively (30 min prior to BCECF loading, and 30 min together with the BCECF), and cells were exposed to NH4Cl for 10 minutes in the presence of 10 nM PRL. Because the most acidic pHi obtained after NH4Cl removal, was not significantly different between experiments, pHi recovery rates could be directly compared and were obtained by fitting a linear line to the initial phase of the pHi recovery curves.
Measurements of pericellular pH
Pericellular pH (pHe) was measured essentially as in (59), with a few modifications. Briefly, cells were grown in collagen-I coated WillCo dishes (WillCo Wells). Cells were serum starved for 1.5 hours and incubated with 1 μg/mL of N-(fluorescein-5-thiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Fluorescein-DHPE; ThermoFisher Scientific) for 40–45 minutes in serum-free medium. Prior to pHe measurements, cells were washed once in 37°C IR and transferred to the temperature-controlled chamber of a Nikon Eclipse Ti microscope. Measurements were carried out as described for pHi, except without continuous perfusion, and calibration was performed in the absence of nigericin. Where indicated, PRL was added to a final concentration of 10 nM. Data were calculated as the background-corrected 485:440 nm ratio and converted to pHe values using the calibration curves.
Cell invasion
The 80% confluent cells were starved for 2 hours in the presence of 0.1% BSA, whereas control cell culture inserts (BD Biosciences) and growth factor-reduced matrigel invasion chambers (BD Biosciences) were rehydrated with starvation medium for 2 hours at 37°C, 5% CO2 in 24-well companion plates (BD Biosciences). Cells were then trypsinized, resuspended in growth medium, counted, and centrifuged at 1000 × g, 5 minutes, 4°C. The pellet was resuspended in starvation medium to a concentration of 5 × 104 cells/mL (according to the manufacturer's protocol; BD Biosciences). Medium from the rehydrated chambers was aspirated, and experimental medium was added to the lower chambers of invasion and control inserts. Cell suspension was added to the upper chamber. The chambers were incubated for 22 hours at 37°C, 5% CO2. After the end of the incubation, cells remaining on the upper chamber side were gently removed, and filters with the migrated/invaded cells were fixed for 1 hour at 4°C in methanol and stained by 10% Giemsa (Sigma-Aldrich) in Gurr buffer (Gibco) for 30 minutes at room temperature. The membranes were washed three times in Gurr buffer, removed and placed cell side down on a microscope slide, covered, sealed, and analyzed. Migrated cells were counted at ×40 magnification using an Olympus BX63 microscope.
Time-lapse imaging of NHE1 and plasma membrane dynamics
MDCK cells stably expressing NHE1-GFP (NHE1-GFP-MDCK) were split onto glass coverslips and incubated for 5–8 hours to allow cells to attach. Medium was then changed to serum starvation medium, and the cells were incubated another 2 hours. The coverslip was mounted in an imaging chamber with PBS. Each cell was imaged every 2 seconds with wide-field fluorescence for 5 minutes, and then a final concentration of 10 nM PRL was added. After 5 minutes of incubation, the same cell was imaged again using the same settings for another 5 minutes. Time-lapse imaging was performed on a Nikon Ti Eclipse inverted microscope equipped with a Perfect Focus 3 system, a CF160 Apo TIRF ×100 objective, an Andor Zyla cMOS camera, and an OkoLAB heating chamber maintaining 37°C and 5% CO2.
Statistical analysis
Unless otherwise indicated, results are presented as mean with SEM error bars and based on at least three experiments. Statistical significance was analyzed using either a two-sided Student's t test (Holm-Sidak method) or a one- or two-way ANOVA followed by a Tukey or Dunnett multiple comparisons post test as relevant, all performed in GraphPad Prism (*, **, ***, and ****, P ≤ .05, P ≤ .01, P ≤ .001, and P ≤ .0001, respectively; ##, ###, P ≤ .01 and P ≤ .001 when comparing across cell lines, respectively).
Results
Stimulation with PRL induces peripheral membrane ruffling, cytoskeletal reorganization, and NHE1-PRLR colocalization to ruffles
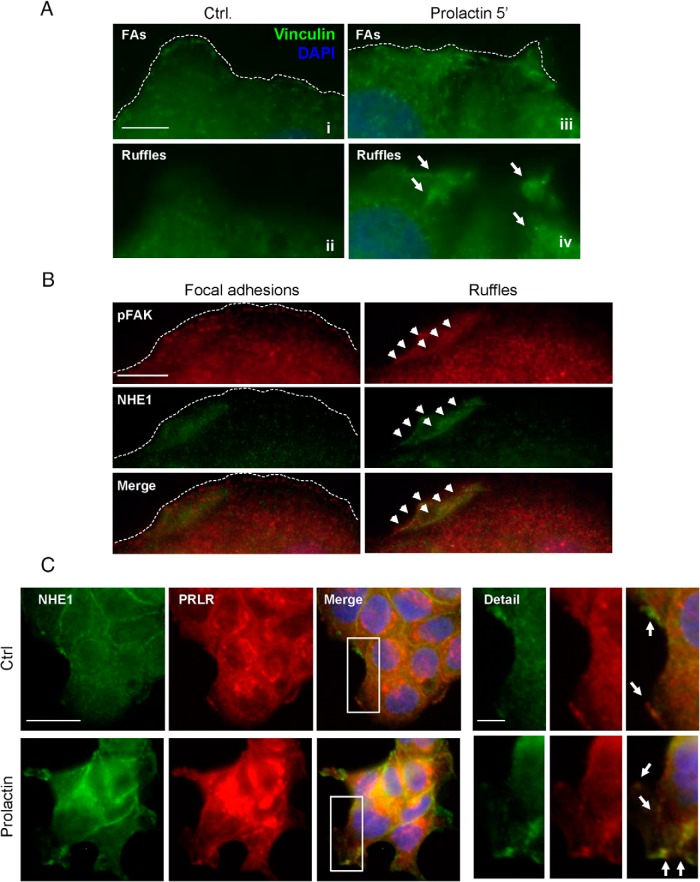
Peripheral membrane ruffling is a phenomenon closely associated with cell motility (46, 47). PRL has previously been proposed to induce membrane ruffles (12, 35) yet without confirmation of their molecular identity. Vinculin, in addition to being a component of focal adhesions is a component of membrane ruffles (70). To study the PRL-induced morphological changes in more detail, T47D cells, a low-invasive human breast cancer cell line, were exposed to PRL (10 nM, 5 min) or control conditions and stained for vinculin to visualize ruffles and focal adhesions by immunofluorescence analysis (IF) (Figure 1A). The PRL-induced peripheral ruffle formation is also illustrated by staining of F-actin in the absence and presence of a 5-minute exposure to PRL (Supplemental Figure 1A). When cells were imaged just above the cell interface with the coverslip, focal adhesions (FAs) were clearly visible, in the absence (i) and presence (iii) of PRL exposure. When imaged 1–2 μm above the level of the coverslip, vinculin labeling revealed the appearance of several-micrometer-long membrane ruffles within a few minutes of PRL exposure (iv), whereas little or no ruffling was observed in the absence of PRL (ii).
Figure 1. Stimulation with PRL induces peripheral membrane ruffling, cytoskeletal reorganization, and NHE1-PRLR colocalization to ruffles in T47D cancer cells.
A, Detail of PRL-induced ruffling in T47D cells. Cells were treated or not with PRL (10 nM, 5 min) and stained for vinculin (green) as a marker of focal adhesions and ruffles. Nuclei are labeled with DAPI (blue), the dotted line indicates the lamellipodial edge, and the arrows the localization of ruffles. Ai, iii, At the cell-coverslip interface, vinculin is predominantly found in FAs; Aii, iv, 1–2 μm above the site of adhesion to the coverslip, vinculin is found in large membrane ruffles induced by PRL. Scale bar, 5 μm. B, T47D cells were treated with PRL (10 nM, 5 min), labeled for pTyr576/577-FAK (red) and NHE1 (green), and imaged at the cell-coverslip interface (FAs) and a few micrometers above (ruffles). The dotted line indicates the lamellipodial edge, and the arrowheads mark the colocalization of NHE1 and p-FAK in ruffles. Note that NHE1 does not appear to localize to FAs but localizes prominently to membrane ruffles. Scale bar, 10 μm. C, Immunofluorescence analysis of NHE1 (green) and the PRLR (red) localization in T47D cells in the absence and presence of PRL (10 nM, 5 min). Where indicated, images are merged with DAPI nuclear staining and a higher-magnification view (×2 zoom) of the indicated square is shown. Arrows mark regions of prominent colocalization. Scale bars (full images), 10 μm; (detail panel), 5 μm. All images are representative of three to four independent experiments per condition. Ctrl, control.
NHE1 is reported to localize to the leading edge lamellipodia (50, 54, 57, 58) and focal adhesions (57–59), but its localization to ruffles is not previously reported and was therefore investigated in further detail. pTyr576/577-FAK was used to label focal adhesions and ruffles after establishing its colocalization with vinculin in both types of structures (Supplemental Figure 1B). In T47D cells exposed to PRL, pTyr576/577-FAK, but not NHE1, clearly localized to focal adhesions, whereas both proteins localized strongly to the PRL-induced ruffles (Figure 1B). The lack of localization of NHE1 to focal adhesions was also seen in the absence of PRL (Supplemental Figure 1C).
To substantiate whether the PRLR and NHE1 might interact in a functionally relevant manner in T47D cells, we first analyzed the expression level and localization of both proteins in T47D cells. Western blotting showed that both NHE1 and the PRLR were expressed in T47D cells at much higher levels than in MCF10A cells, a noncancerous, human breast epithelial cell line (Supplemental Figure 2A). IF analysis showed that NHE1 localized strongly to the plasma membrane in T47D cells, which also exhibited the highest membrane expression of the PRLR (Figure 1C and Supplemental Figure 2B). Both in the presence and absence of PRL (10 nM, 5 minutes), the PRLR and NHE1 colocalized to the plasma membrane (Figure 1C). Notably, NHE1-PRLR colocalization was strong in both the smaller and more sparse ruffles present before PRL and in the larger, more frequent ruffles seen after PRL stimulation (Figure 1C, arrows).
Collectively these data show that PRL induces F-actin- and vinculin-rich membrane ruffles in T47D cells and that NHE1 and the PRLR, which are highly expressed in T47D cells, appear to colocalize in the plasma membrane of unstimulated T47D cells, and more prominently to PRL-induced ruffles.
NHE1 colocalizes with pAkt and pERK in PRL-induced ruffles in a functionally relevant manner
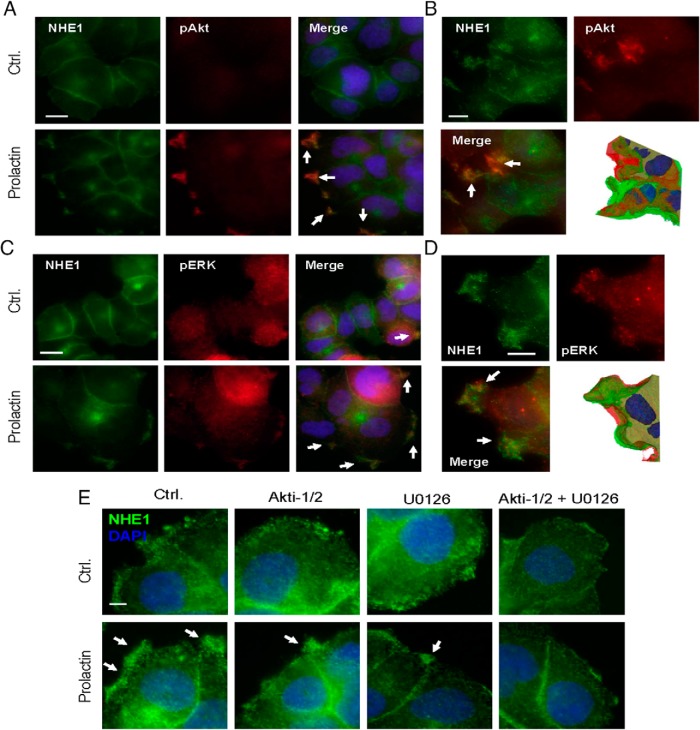
Several PRLR signaling effectors have previously been shown to regulate NHE1, including JAK2-STAT (71), Akt (72), and ERK1/2 (directly or via p90RSK) (68, 73). In accordance with previous reports (74), T47D cells exhibited acute, PRL-induced increases in the relative activity of JAK2, STAT5, Akt, and ERK1/2 (Supplemental Figure 2, C–F). In contrast, and in accordance with their much lower PRLR expression, MCF10A cells exhibited essentially no PRL-induced increases in the activity of these pathways (Supplemental Figure 2, C–F). We therefore next asked whether Akt and ERK colocalize with NHE1 to PRL-induced ruffles. Indeed, in particular, the localization of active (pSer473) Akt to ruffles was specific and dramatic, with essentially all pAkt labeling detected in these structures (Figure 2A). However, also the localization of active (pThr202/Tyr204) ERK1/2 to ruffles was clearly detectable (Figure 2C). Importantly, NHE1 colocalized strongly with pSer473-Akt (Figure 2A) as well as with pThr202/Tyr204-ERK1/2 (Figure 2C) in the PRL-induced membrane ruffles. Given their nature as partially folded membrane sheets, apparent accumulation and colocalization of molecules in ruffles might reflect the imaging of several layers of membrane. Maximal intensity projections and isosurface projections focusing on the ruffled areas were therefore performed, confirming that the colocalization of p-Akt and NHE1 (Figure 2B) and pERK1/2 and NHE1 (Figure 2D) was widely distributed throughout the peripheral ruffles and that especially p-Akt localization was strongly enriched in ruffles compared with other membrane areas.
Figure 2. NHE1 colocalization with pAkt and pERK in PRL-induced ruffles.
Immunofluorescence analysis of T47D cells treated or not with PRL (10 nM, 5 min) and stained for NHE1, pSer473-Akt, pThr202/Tyr204-ERK1/2, and DAPI (nuclear stain, blue), as indicated. Arrows mark regions of prominent colocalization (A–D) or NHE1 localization to ruffles (E). A, NHE1 (green) and pSer473-Akt (red) colocalize in PRL-induced ruffles. Scale bar, 10 μm. B, z-stack imaging, maximal intensity projection, and isosurface projection show details of NHE1-p-Akt colocalization in ruffles. Note the strong p-Akt enrichment in ruffles seen in both panels A and B. Scale bar, 5 μm. C, NHE1 (green) and pThr202/Tyr204-ERK1/2 (red). D, z-stack imaging, maximal intensity projection and isosurface projection show details of NHE1-p-ERK colocalization in ruffles. Scale bar, 5 μm. Scale bar, 10 μm. E, T47D cells were treated with Akti-1/2 or U0126 10 μM for 1 hour, followed by treatment or not with PRL, as indicated, and stained for NHE1 (green) with DAPI nuclear counterstain. Scale bar, 10 μm. All images are representative of at least three independent experiments per condition. Ctrl, control.
Our previous work has implicated Akt and ERK1/2 in membrane localization and activity of NHE1 in murine fibroblasts (51) and shown that NHE1 and ERK2 directly interact (75). We therefore asked whether these kinases play a role in ruffle formation and NHE1 localization to these structures. Treatment with Akti-1/2 and/or U0126 (by preincubation with 10 μM of either compound or both compounds in combination, for 1 h) had the expected inhibitory effect on Akt and ERK1/2 activity in T47D cells under the conditions used (Supplemental Figure 3, A and B). Inhibition of Akt or ERK1/2 and, even more dramatically the combined inhibition of both kinases, resulted in both reduced peripheral ruffle formation and reduced NHE1 localization to the membrane periphery (Figure 2E).
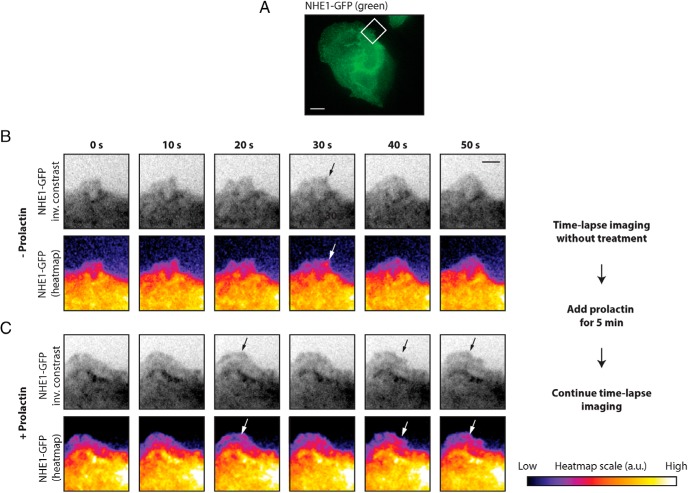
To determine the temporal relationship between PRLR stimulation, membrane ruffle formation, and NHE1 recruitment to ruffles, we carried out time-lapse imaging. Initial attempts to transfect T47D cells with NHE1-GFP and perform time-lapse imaging were not successful because the cells did not tolerate the transfection well and died on the microscope stage. Instead, we used MDCK kidney epithelial cells stably transfected with NHE1-GFP (Figure 3A). MDCK cells have previously been shown to respond to PRL (76), and we confirmed this by Western blot analysis of MDCK-II cells after exposure to 10 nM PRL (Supplemental Figure 4). Similar to the T47D cells, MDCK cells exhibited a basal level of ruffling in the absence of PRL and underwent further membrane ruffling upon addition of PRL (Figure 3 and Supplemental Video 1 [before PRL stimulation] and Supplemental Video 2 [after stimulation with 10 nM PRL]). As seen, NHE1 was already present at the edge of ruffles prior to stimulation with PRL (Figure 3B, arrows, and Supplemental Video 1). Already at time 5 minutes after PRL addition, ruffles were clearly increased in size, their movement appeared more dynamic, and additional NHE1 was recruited to the ruffle edges (Figure 3C, arrows, and Supplemental Video 2).
Figure 3. PRL treatment increases ruffling and NHE-GFP recruitment in MDCK cells.
A, MDCK cells stably expressing NHE1-GFP. The box indicates the region shown in panels B and C, Scale bar, 10 μm. B, Montages of NHE1-GFP in ruffle dynamics before addition of PRL. C, Montages of NHE1-GFP in ruffle dynamics of the same region as in panel B after addition of PRL. Panels B and C are shown as inverted-contrast images (upper panels) and with a gradient pseudocolor of GFP intensity (lower panels). The intensity scale is shown in arbitrary units on the right-hand side. The montages are chosen from movies with a total length of 5 minutes (Supplemental Videos 1 and 2, respectively) each, with 10 seconds between each frame. Arrows point to NHE1-GFP in the ruffle edge. Scale bar, 3 μm. Data shown are representative of eight independent experiments. Ctrl, control.
Taken together, these data show that activated Akt and ERK1/2 localize to PRL-induced ruffles, colocalizing extensively with NHE1, and that their inhibition reduces the prevalence of NHE1-containing ruffles. Live imaging analysis showed that NHE1 localizes to existing ruffles in MDCK cells and is further recruited to the more prominent ruffles induced by PRL exposure.
PRL stimulates NHE1 activity and elicits pericellular acidification in T47D cells
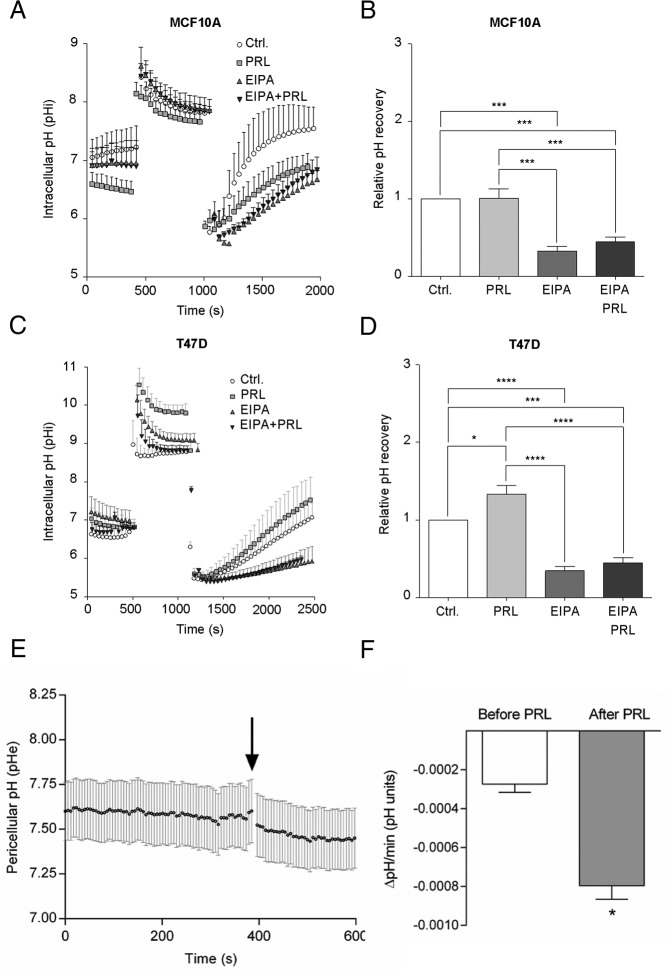
We next asked whether acute exposure to PRL altered NHE1 activity in MCF10A and T47D cells. NHE1 activity was determined as the EIPA-sensitive fraction of the rate of recovery of pHi after an NH4Cl prepulse-induced acid load in the nominal absence of HCO3− (Figure 4). Where indicated, PRL (10 nM) was present from prior to the NH4Cl prepulse, and/or EIPA (10 μM) was present from the point of NH4Cl removal. Figure 4, A and B, show pHi over time and relative pHi recovery rate in MCF10A cells, and Figure 4, C and D, show the corresponding data for T47D cells. MCF10A cells recovered their pHi rapidly after acidification, and recovery was robustly inhibited by EIPA, showing that NHE1 activity plays a dominant role in acidification-induced net acid extrusion in these cells. In agreement with the low PRLR expression level and essential lack of PRL-induced signaling in these cells, pHi recovery in MCF10A cells was unaffected by PRL. Similar to that in MCF10A cells, pHi recovery in T47D cells was highly EIPA sensitive, yet in marked contrast to that in MCF10A cells, the pHi recovery rate in T47D cells was significantly increased by pretreatment with PRL. Treatment with Akti-1/2 or U0126, or both compounds in combination, inhibited the PRL-stimulated NHE1 activity (Supplemental Figure 3C), confirming the known role of these kinases in NHE1 activation fibroblasts (51).
Figure 4. PRL stimulates NHE1 activity and acid extrusion in T47D cells.
A, pHi recovery over time after an NH4Cl prepulse-induced acid load in MCF10A cells ± 10 nM PRL and/or 5 μM EIPA, as indicated. Error bars represent SD from measurements on at least six cells in a single experiment. B, Relative pHi recovery rates in MCF10A cells were calculated from the initial, linear phase of the recovery curve and are shown as mean with SEM error bars of four independent experiments. ***, P ≤ .001, one-way ANOVA with Tukey multiple comparisons posttest. C, As in panel A, except in T47D cells. D, Relative pH recovery rates in T47D cells, calculated from the initial, linear phase of the recovery curve, and are shown as mean with SEM error bars of five independent experiments. *, P ≤ .05, ***, P ≤ .001, ****, P ≤ .0001, one-way ANOVA with Tukey multiple comparisons test. E and F, pHe was measured in T47D cells using Fluorescein-DHPE. E, Sample trace. The arrow shows the time of addition of PRL (10 nM). Error bars are SD from measurements on seven cells in a single experiment. F, Data as in panel E summarized as the change in pHe per minute the last minute before PRL addition and at time 5 minutes after PRL addition. Error bars represent SEM for the four independent experiments. *, Paired, two-way Student's t test. Ctrl, control.
Because we and others have demonstrated that NHE1-dependent acid extrusion can be associated with a pericellular acidification (59, 77), we next asked whether this was also the case after PRL-dependent NHE1 activation. pHe of T47D cells was measured using fluorescein-DHPE, which integrates into the outer leaflet of the cell membrane (59), load in the nominal absence of HCO3−, and in the absence of flow to facilitate detection of pericellular acidification. Consistent with the PRL-induced increase in NHE1 activity, addition of 10 nM PRL elicited a slow but significant increase in the rate of pericellular acidification (Figure 4, E and F).
These results show that PRL stimulates NHE1-dependent pHi recovery after an acid load in T47D cells and that this correlates with a PRL-induced pericellular acidification.
PRL treatment elicits ribosomal S kinase activation and NHE1 Ser703 phosphorylation in T47D cells
Stimulation of the GH receptor, which is homologous to PRLR, elicits phosphorylation of human NHE1 at S703 (67), a site previously shown to be phosphorylated by the ERK1/2 effector p90RSK, leading to increased NHE1 activity (68). To determine whether PRL induces S703 phosphorylation of NHE1, we examined S703 phosphorylation of NHE1 using an antibody specific to S703-phosphorylated NHE1 (77, 78). Indeed, stimulation with PRL (10 nM) transiently increased the S703 phosphorylation of NHE1, with maximal phosphorylation induced at 30 minutes of PRL exposure, to a level similar to that induced by exposure to epidermal growth factor (Figure 5A). After 24 hours, NHE1 phosphorylation had returned to control levels. In congruence with this, the activation of p90RSK and ERK1/2 followed a similar time course (Figure 5B). We next asked whether active ribosomal S kinase localized to PRL-induced ruffles. Indeed, both total p90RSK1 (Figure 5C) and active, S380-phosphorylated (Figure 5D) p90RSK were found in PRL-induced ruffles, colocalizing with NHE1, consistent with the notion that p90RSK might elicit the S703 phosphorylation of NHE1 within the confined space of the peripheral ruffles.
We considered that PRL could impact NHE1 activity in other ways than via its phosphorylation, such as increasing NHE1 expression, or by mechanisms involving direct interaction between NHE1 and PRLR. To determine whether long-term PRL exposure increases NHE1 protein expression, T47D cells were exposed to PRL (10 nM) for 24 or 48 hours, followed by Western blotting for NHE1. However, NHE1 protein expression was not altered by long-term PRL exposure (Supplemental Figure 5A). To determine whether NHE1 and PRLR interact directly, NHE1 and PRLR were immunoprecipitated, followed by Western blotting for both proteins. However, despite robust pulldown of both proteins, there was no detectable coimmunoprecipitation, suggesting that either the two proteins do not directly interact or that they do so only relatively weakly/transiently (Supplemental Figure 5B).
Thus, stimulation of T47D cells with PRL elicits rapid and transient phosphorylation of NHE1 on Ser703. NHE1 protein expression is unaltered by long-term PRL exposure, and NHE1 and PRLR do not directly interact in a manner detectable by coimmunoprecipitation.
PRL increases invasiveness of T47D cells in an NHE1-dependent manner
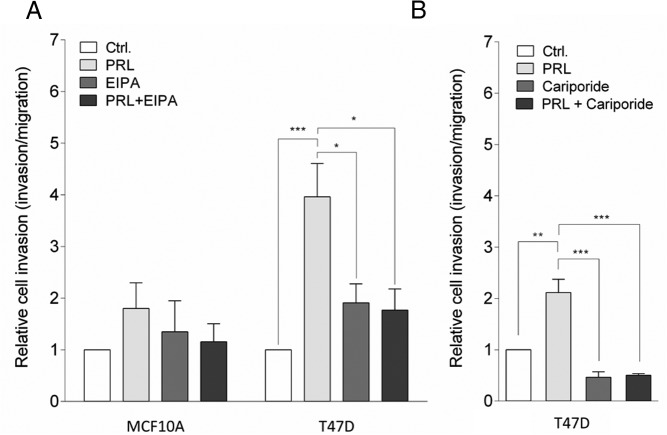
PRL has been reported to act as a chemoattractant for T47D cells (35) and to stimulate invasiveness and/or migration in these cells (12, 32) and in other breast cancer cells (36). Given the known roles of NHE1 in cell motility (54), we therefore next asked whether NHE1 activity contributes to PRL-induced invasion of breast cancer cells. MCF10A and T47D cells were seeded in Boyden chambers with or without PRL as a chemoattractant in the lower chamber and in the absence or presence of EIPA or the more specific, clinically validated NHE1 inhibitor cariporide to inhibit NHE1 (Figure 6). As expected, MCF10A cells exhibited essentially no invasion, neither in the absence nor the presence of PRL as a chemoattractant, and this result was unaffected by EIPA (Figure 6A). T47D cells are also essentially noninvasive under unstimulated conditions (79). Notably, relative invasiveness was more than doubled by PRL in T47D cells, and both unstimulated and PRL-stimulated invasion was essentially abolished by EIPA (Figure 6A) as well as by cariporide (Figure 6B).
Figure 6. PRL induces invasiveness of T47D cells in a NHE1-dependent manner.
Invasiveness of MCF10A (A) and T47D (A and B) cells was assessed in matrigel-coated Boyden chambers. PRL (10 nM) was applied as a chemoattractant to the lower chamber, as indicated, and where indicated, EIPA (5 μM) (A) or cariporide (10 μM) (B) was applied to both chambers to inhibit NHE1. Data are shown as relative cell invasion (invasion relative to migration) in the presence or absence of PRL and/or EIPA or cariporide as indicated. Error bars represent SEM of four (MCF10; A), five (T47D; A), or three (T47D; B) independent experiments. *, P ≤ .05, **, P ≤ .01, ***, P ≤ .001, one-way ANOVA with Tukey posttest. Ctrl, control.
Collectively these results show that NHE1 activity is required for PRL-induced stimulation of the invasiveness of T47D cells.
Discussion
Recent studies have addressed the roles of PRL and the PRLR in breast cancer (3, 4, 25–28, 30). Although there is controversy over the exact roles of PRL signaling, several lines of evidence indicate that PRL signaling contributes to processes such as increased invasiveness and proliferation, major hallmarks of cancer cells (4, 12, 31–33–36). On the other hand, PRL and the PRLR have also been proposed to exert antiinvasive effects (37, 80). The contradictory findings are puzzling and reflect differences in cell types and conditions studied, suggesting that the role of prolactin in cancer is likely highly context dependent. This is underscored by recent findings showing that the outcome of PRL signaling in cancer depends at least in part on STAT5 availability (81) and that the density of the matrix is determining for the effect of prolactin signaling on T47D cell invasiveness (39).
We and others have demonstrated the important role of the NHE1 in cancer development, including breast cancer (55, 56, 82). Specifically, NHE1 has been assigned an important role in cancer cell invasiveness, at least in part because NHE1-mediated acid extrusion favors extracellular protease activity and ECM degradation (53). Because of this and in conjunction with recent phosphoproteomic data revealing that GH, a close relative of PRL, induces human NHE1 phosphorylation on Ser703 (67), we tested the hypothesis that PRL activates breast cancer cell invasion by stimulating NHE1. Our data provide the first demonstration that this is indeed the case: PRL stimulates NHE1 phosphorylation and activation, and PRL-mediated invasiveness of breast cancer cells is NHE1 dependent.
Confirming previous reports (4, 11, 74, 83), exposure to PRL elicited rapid activation of JAK2/STAT5, Akt, and ERK1/2 signaling in T47D cells but not in nontransformed MCF10A mammary epithelial cells, which express much lower levels of PRLR. We demonstrated that both activated Akt and activated ERK1/2 localize to the PRL-induced ruffles. In congruence with this, the localization of Akt to ruffles induced by insulin in hepatocytes and murine embryonic fibroblasts was recently reported (84). In conjunction with the proposed role for PI3K activation in PRL-induced ruffle formation (35), the localization of Akt to the ruffles points to the involvement of a PI3K-Akt signaling axis. Although not studied here, Rac1 may also play a role because its activity is central for ruffle formation (47) and PRL activates Rac signaling (35, 85), apparently in a manner dependent on Nek3 kinase and Vav2 (12). Importantly, NHE1 also strongly localized to the PRL-induced membrane ruffles. Imaging of NHE1-GFP recruitment to ruffles in MDCK cells showed that NHE1 was already present in the smaller, less dynamic ruffles seen before PRL stimulation but was further recruited in parallel with increased ruffle formation upon stimulation with PRL. Thus, NHE1 recruitment is probably not dependent on PRL per se but on the formation of membrane ruffles, possibly in conjunction with the simultaneous phosphorylation and activation of NHE1 (see below).
In congruence with this view, both ruffling and NHE1 membrane localization were attenuated by inhibition of Akt, ERK1/2, or both kinases in combination. This agrees well with our previous report that platelet-derived growth factor receptor-α-induced NHE1 leading edge localization in murine fibroblasts is also Akt and ERK1/2 dependent (51), and with the dependence of NHE1 activity on Akt and ERK1/2 (see below), and our recent demonstration of ERK2 scaffolding by NHE1 (75). It may be noted that in contrast to previous reports in other cell types (57–59), NHE1 did not localize detectably to focal adhesions in T47D cells but dramatically to ruffles. Hence, we propose that membrane ruffles are a novel important localization of NHE1.
Ruffles are closely associated with cell motility, and confirming previous reports (12, 35), we found that exposure to PRL stimulated invasion of T47D breast cancer cells through matrigel. Notably, we show here for the first time, using two different NHE1 inhibitors, that PRL-induced invasion is strongly NHE1 dependent. In contrast, as expected, PRL failed to elicit invasive behavior in MCF10A cells, which exhibit much lower PRLR expression and also express less NHE1, compared with T47D cells. Although EIPA also inhibits other transporters, cariporide is highly NHE1 specific (86); hence, the results strongly suggest that NHE1 is required for the PRL-mediated increase in invasiveness. A central proposed role of NHE1 in cancer cell invasiveness is to create a local, acidic pHe conducive to invadopodia formation and function by stimulating the secretion and activity of matrix metalloproteases and other ECM proteases (53, 87). It is therefore very interesting that PRL-induced invasion of breast cancer cells was reported to be associated with increased matrix metalloprotease secretion (33). Our findings that PRL-induced, NHE1-dependent invasiveness was correlated with PRL-induced, NHE1-dependent acid extrusion (both occurring in T47D cells yet not in MCF10A cells) and that PRL elicited pericellular acidification in T47D cells support the notion that PRL-induced invasiveness involves NHE1-dependent pericellular acidification.
Activation of NHE1 is complex and can be induced by multiple stimuli, some of which involve phosphorylation of sites in the C-terminal cytoplasmic tail of NHE1 (see reference 88). One such well-studied site, Ser703 (human NHE1 numbering), is phosphorylated by the ERK1/2 effector p90RSK, leading to NHE1 activation (68). ERK1/2 is the only known upstream activator of p90RSK (except for the constitutively active 3′-phosphoinositide-dependent protein kinase 1 (89)). We show here that inhibition of either Akt or ERK1/2 activity reduces NHE1 localization to the membrane, in agreement with our previous findings in murine fibroblasts (51), as well as NHE1 activity, also in agreement with previous work from us and others (51, 72) (see reference 88). PRL elicits transient phosphorylation of Ser703 of NHE1 with a time course corresponding to the activation of p90RSK by PRL, and active p90RSK colocalizes with NHE1 in PRL-induced ruffles. Given the similarities between PRL and GH signaling, this agrees well with the recent identification in a large phosphoproteomic screen of Ser703 of NHE1 as phosphorylated upon GH exposure of adipocytes (67). In contrast, we detected neither NHE1 up-regulation at the protein expression level after long-term stimulation by PRL nor direct NHE1-PRLR interaction. Obviously this does not exclude that the two proteins interact but, if so, with a low affinity and/or not detectable in the current experimental setting.
Collectively, therefore, our results strongly suggest that the pathway from the PRLR to NHE1 involves PRL-mediated Akt and ERK1/2 activation, the latter in turn activating p90RSK, leading to NHE1 Ser703 phosphorylation. Also consistent with this, both Akt and ERK1/2 inhibition attenuated NHE1-dependent migration of murine fibroblasts (51). Also, FAK has been proposed to regulate NHE1 membrane localization (90) and, given its close proximity to NHE1 in the ruffles, could contribute to its recruitment.
Thus, we envisage that in T47D cells, PRL activates membrane ruffling, at least in part via PI3K-Akt and ERK signaling, and simultaneously activates NHE1, at least in part via ERK1/2-p90RSK-mediated, direct NHE1 phosphorylation. NHE1 is recruited to ruffles, likely in a manner involving PRL-induced ERK1/2, Akt, and/or FAK signaling. This in turn leads to local pericellular acidification around the ruffles as shown for focal adhesions (91) and invadopodia (53). This favors protease secretion and activation and consequently ECM degradation and invasion. NHE1 activity may also further stimulate the activity of FAK, which is pH sensitive in a manner important for focal adhesion formation during cell spreading (65), and the integrin-mediated activation of which has been shown to be NHE1 dependent (92). Collectively our findings support a proinvasive role of PRL-mediated PRLR signaling to NHE1. We speculate that both the pro- (12, 32, 33, 35) and antiinvasive (37, 80) roles of PRL reported in cancer may in part reflect the extent of involvement of NHE1 in invasion in the cancer type and context studied. Finally, it is interesting to note that a role for NHE1 in providing the acidic microenvironment necessary for processing of PRL to its 16-kDa antiangiogenic fragment has been demonstrated (93). How this may contribute, presumably as a potential feedback regulation, to the net aggressiveness of PRLR- and NHE1-expressing tumors is still an open question for future research and could also be a link to understanding the complexity of PRL in regulating breast cancer cell invasion.
In conclusion, we show here that in T47D human breast cancer cells, PRL signaling does the following: 1) elicits the rapid formation of peripheral membrane ruffles to which NHE1 localizes with activated PRLR effectors ERK1/2, Akt, and p90RSK; 2) induces transient NHE1 phosphorylation on Ser703; 3) increases NHE1-dependent pHi recovery and elicits pericellular acidification; and 4) increases cell invasiveness in a manner dependent on NHE1 activity. We propose that PRL-induced NHE1 activation and the resulting NHE1-dependent invasion can contribute to invasive behavior of human breast cancer cells and that the PRL/PRLR-NHE1 signaling axis may be of potential therapeutic interest in breast cancer.
Acknowledgments
We thank Signe A. Sjørup, Katrine Franklin Mark, and Helene S. Müller for their expert technical assistance and G. W. Haxholm and V. Goffin for their discussions and critical reading of the manuscript.
Author contributions include the following: S.F.P. and B.B.K. conceived the study and designed the experiments. J.F., E.P.-C., S.F.P., H.H.J., L.N.N., and A.B. performed the experiments and analyzed the data. S.F.P. and E.P.-C. wrote the manuscript, with input from all the coauthors. All coauthors have seen and approved the final version of the manuscript.
This work was supported by the Danish Council for Independent Research Grants 12–127290 (to S.F.P.) and 12–125862 (to B.B.K.) and support from the Hartmann Foundation (to S.F.P.), the Novo Nordisk Foundation (to B.B.K.), the Harboe Foundation (to S.F.P.); the Lundbeck Foundation (to L.N.N.), the Carlsberg Foundation (to L.N.N.), and MEMBRANES (to L.N.N.) and the Graduate School of Science and Technology (to H.H.J.).
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by the Danish Council for Independent Research Grants 12–127290 (to S.F.P.) and 12–125862 (to B.B.K.) and support from the Hartmann Foundation (to S.F.P.), the Novo Nordisk Foundation (to B.B.K.), the Harboe Foundation (to S.F.P.); the Lundbeck Foundation (to L.N.N.), the Carlsberg Foundation (to L.N.N.), and MEMBRANES (to L.N.N.) and the Graduate School of Science and Technology (to H.H.J.).
Footnotes
- BCECF
- 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein
- DAPI
- 4′,6-diamidino-2-phenylindole
- ECM
- extracellular matrix
- EIPA
- 5-(N-ethyl-isopropyl)amiloride
- FA
- focal adhesion
- FAK
- focal adhesion kinase
- GFP
- green fluorescent protein
- IF
- immunofluorescence analysis
- IR
- isotonic Ringer
- JAK
- Janus kinase
- NHE1
- Na+/H+ exchanger
- NP40
- Nonidet P-40
- pHe
- pericellular pH
- Fluorescein-DHPE
- N-(fluorescein-5-thiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine
- pHi
- intracellular pH
- PI3K
- phosphatidyl-inositol-3 kinase
- PRL
- prolactin
- PRLR
- PRL receptor
- p90RSK
- p90 ribosomal S kinase
- STAT
- signal transducer and activator of transcription
- TBST
- TBS + Tween 20.
References
- 1. Teilum K, Hoch JC, Goffin V, Kinet S, Martial JA, Kragelund BB. Solution structure of human prolactin. J Mol Biol. 2005;351:810–823. [DOI] [PubMed] [Google Scholar]
- 2. Marano RJ, Ben-Jonathan N. Minireview: extrapituitary prolactin: an update on the distribution, regulation, and functions. Mol Endocrinol. 2014;28:622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clevenger CV, Furth PA, Hankinson SE, Schuler LA. The role of prolactin in mammary carcinoma. Endocr Rev. 2003;24:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clevenger CV, Gadd SL, Zheng J. New mechanisms for PRLr action in breast cancer. Trends Endocrinol Metab. 2009;20:223–229. [DOI] [PubMed] [Google Scholar]
- 5. Brooks CL. Molecular mechanisms of prolactin and its receptor. Endocr Rev. 2012;33:504–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gadd SL, Clevenger CV. Ligand-independent dimerization of the human prolactin receptor isoforms: functional implications. Mol Endocrinol. 2006;20:2734–2746. [DOI] [PubMed] [Google Scholar]
- 7. Brown RJ, Adams JJ, Pelekanos RA, et al. . Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol. 2005;12:814–821. [DOI] [PubMed] [Google Scholar]
- 8. Tallet E, Fernandez I, Zhang C, et al. . Investigation of prolactin receptor activation and blockade using time-resolved fluorescence resonance energy transfer. Front Endocrinol (Lausanne). 2011;2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broutin I, Jomain JB, Tallet E, et al. . Crystal structure of an affinity-matured prolactin complexed to its dimerized receptor reveals the topology of hormone binding site 2. J Biol Chem. 2010;285:8422–8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clevenger CV, Medaglia MV. The protein tyrosine kinase P59fyn is associated with prolactin (PRL) receptor and is activated by PRL stimulation of T-lymphocytes. Mol Endocrinol. 1994;8:674–681. [DOI] [PubMed] [Google Scholar]
- 11. Acosta JJ, Munoz RM, Gonzalez L, et al. . Src mediates prolactin-dependent proliferation of T47D and MCF7 cells via the activation of focal adhesion kinase/Erk1/2 and phosphatidylinositol 3-kinase pathways. Mol Endocrinol. 2003;17:2268–2282. [DOI] [PubMed] [Google Scholar]
- 12. Miller SL, Antico G, Raghunath PN, Tomaszewski JE, Clevenger CV. Nek3 kinase regulates prolactin-mediated cytoskeletal reorganization and motility of breast cancer cells. Oncogene. 2007;26:4668–4678. [DOI] [PubMed] [Google Scholar]
- 13. Srivastava S, Matsuda M, Hou Z, et al. . Receptor activator of NF-κB ligand induction via Jak2 and Stat5a in mammary epithelial cells. J Biol Chem. 2003;278:46171–46178. [DOI] [PubMed] [Google Scholar]
- 14. Barclay JL, Anderson ST, Waters MJ, Curlewis JD. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int J Cancer. 2009;124:1756–1766. [DOI] [PubMed] [Google Scholar]
- 15. Rasmussen LM, Frederiksen KS, Din N, et al. . Prolactin and oestrogen synergistically regulate gene expression and proliferation of breast cancer cells. Endocr Relat Cancer. 2010;17:809–822. [DOI] [PubMed] [Google Scholar]
- 16. Damiano JS, Wasserman E. Molecular pathways: blockade of the PRLR signaling pathway as a novel antihormonal approach for the treatment of breast and prostate cancer. Clin Cancer Res. 2013;19:1644–1650. [DOI] [PubMed] [Google Scholar]
- 17. Muraoka-Cook RS, Sandahl M, Hunter D, Miraglia L, Earp HS III. Prolactin and ErbB4/HER4 signaling interact via Janus kinase 2 to induce mammary epithelial cell gene expression differentiation. Mol Endocrinol. 2008;22:2307–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamauchi T, Yamauchi N, Ueki K, et al. . Constitutive tyrosine phosphorylation of ErbB-2 via Jak2 by autocrine secretion of prolactin in human breast cancer. J Biol Chem. 2000;275:33937–33944. [DOI] [PubMed] [Google Scholar]
- 19. Oakes SR, Rogers RL, Naylor MJ, Ormandy CJ. Prolactin regulation of mammary gland development. J Mammary Gland Biol Neoplasia. 2008;13:13–28. [DOI] [PubMed] [Google Scholar]
- 20. Horseman ND, Gregerson KA. Prolactin actions. J Mol Endocrinol. 2014;52:R95–R106. [DOI] [PubMed] [Google Scholar]
- 21. Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiol Rev. 2000;80:1523–1631. [DOI] [PubMed] [Google Scholar]
- 22. Nitze LM, Galsgaard ED, Din N, et al. . Reevaluation of the proposed autocrine proliferative function of prolactin in breast cancer. Breast Cancer Res Treat. 2013;142:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wennbo H, Gebre-Medhin M, Gritli-Linde A, Ohlsson C, Isaksson OG, Tornell J. Activation of the prolactin receptor but not the growth hormone receptor is important for induction of mammary tumors in transgenic mice. J Clin Invest. 1997;100:2744–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ginsburg E, Vonderhaar BK. Prolactin synthesis and secretion by human breast cancer cells. Cancer Res. 1995;55:2591–2595. [PubMed] [Google Scholar]
- 25. Tworoger SS, Eliassen AH, Zhang X, et al. . A 20-year prospective study of plasma prolactin as a risk marker of breast cancer development. Cancer Res. 2013;73:4810–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bogorad RL, Courtillot C, Mestayer C, et al. . Identification of a gain-of-function mutation of the prolactin receptor in women with benign breast tumors. Proc Natl Acad Sci USA. 2008;105:14533–14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Courtillot C, Chakhtoura Z, Bogorad R, et al. . Characterization of two constitutively active prolactin receptor variants in a cohort of 95 women with multiple breast fibroadenomas. J Clin Endocrinol Metab. 2010;95:271–279. [DOI] [PubMed] [Google Scholar]
- 28. Rose-Hellekant TA, Arendt LM, Schroeder MD, Gilchrist K, Sandgren EP, Schuler LA. Prolactin induces ERα-positive and ERα-negative mammary cancer in transgenic mice. Oncogene. 2003;22:4664–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robertson FG, Harris J, Naylor MJ, et al. . Prostate development and carcinogenesis in prolactin receptor knockout mice. Endocrinology. 2003;144:3196–3205. [DOI] [PubMed] [Google Scholar]
- 30. Vomachka AJ, Pratt SL, Lockefeer JA, Horseman ND. Prolactin gene-disruption arrests mammary gland development and retards T-antigen-induced tumor growth. Oncogene. 2000;19:1077–1084. [DOI] [PubMed] [Google Scholar]
- 31. Wen Y, Zand B, Ozpolat B, et al. . Antagonism of tumoral prolactin receptor promotes autophagy-related cell death. Cell Rep. 2014;7:488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hammer A, Rider L, Oladimeji P, et al. . Tyrosyl phosphorylated PAK1 regulates breast cancer cell motility in response to prolactin through filamin A. Mol Endocrinol. 2013;27:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rider L, Oladimeji P, Diakonova M. PAK1 regulates breast cancer cell invasion through secretion of matrix metalloproteinases in response to prolactin and three-dimensional collagen IV. Mol Endocrinol. 2013;27:1048–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Howell SJ, Anderson E, Hunter T, Farnie G, Clarke RB. Prolactin receptor antagonism reduces the clonogenic capacity of breast cancer cells and potentiates doxorubicin and paclitaxel cytotoxicity. Breast Cancer Res. 2008;10:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maus MV, Reilly SC, Clevenger CV. Prolactin as a chemoattractant for human breast carcinoma. Endocrinology. 1999;140:5447–5450. [DOI] [PubMed] [Google Scholar]
- 36. Doll F, Pfeilschifter J, Huwiler A. Prolactin upregulates sphingosine kinase-1 expression and activity in the human breast cancer cell line MCF7 and triggers enhanced proliferation and migration. Endocr Relat Cancer. 2007;14:325–335. [DOI] [PubMed] [Google Scholar]
- 37. Nouhi Z, Chughtai N, Hartley S, Cocolakis E, Lebrun JJ, Ali S. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res. 2006;66:1824–1832. [DOI] [PubMed] [Google Scholar]
- 38. Galbaugh T, Feeney YB, Clevenger CV. Prolactin receptor-integrin cross-talk mediated by SIRPα in breast cancer cells. Mol Cancer Res. 2010;8:1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. Stiff collagen matrices increase tumorigenic prolactin signaling in breast cancer cells. J Biol Chem. 2013;288:12722–12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barcus CE, Holt EC, Keely PJ, Eliceiri KW, Schuler LA. Dense collagen-I matrices enhance pro-tumorigenic estrogen-prolactin crosstalk in MCF-7 and T47D breast cancer cells. PLoS One 2015;10:e0116891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang N, Liu C, Peck AR, et al. . Prolactin-Stat5 signaling in breast cancer is potently disrupted by acidosis within the tumor microenvironment. Breast Cancer Res. 2013;15:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kulkarni MV, Tettamanzi MC, Murphy JW, et al. . Two independent histidines, one in human prolactin and one in its receptor, are critical for pH-dependent receptor recognition and activation. J Biol Chem. 2010;285:38524–38533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen MJ, Olsen JG, Bernichtein S, et al. . Development of prolactin receptor antagonists with reduced pH-dependence of receptor binding. J Mol Recognit. 2011;24:533–547. [DOI] [PubMed] [Google Scholar]
- 44. Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. [DOI] [PubMed] [Google Scholar]
- 45. Abercrombie M, Heaysman JE, Pegrum SM. The locomotion of fibroblasts in culture. II. “ruffling.” Exp Cell Res. 1970;60:437–444. [DOI] [PubMed] [Google Scholar]
- 46. Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–657. [DOI] [PubMed] [Google Scholar]
- 47. Borm B, Requardt RP, Herzog V, Kirfel G. Membrane ruffles in cell migration: indicators of inefficient lamellipodia adhesion and compartments of actin filament reorganization. Exp Cell Res. 2005;302:83–95. [DOI] [PubMed] [Google Scholar]
- 48. Skalski M, Yi Q, Kean MJ, et al. . Lamellipodium extension and membrane ruffling require different SNARE-mediated trafficking pathways. BMC Cell Biol. 2010;11:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. [DOI] [PubMed] [Google Scholar]
- 50. Schneider L, Stock CM, Dieterich P, et al. . The Na+/H+ exchanger NHE1 is required for directional migration stimulated via PDGFR-α in the primary cilium. J Cell Biol. 2009;185:163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clement DL, Mally S, Stock C, et al. . PDGFRα signaling in the primary cilium regulates NHE1-dependent fibroblast migration via coordinated differential activity of MEK1/2-ERK1/2-p90RSK and AKT signaling pathways. J Cell Sci. 2013;126:953–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stock C, Gassner B, Hauck CR, et al. . Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J Physiol. 2005;567:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Busco G, Cardone RA, Greco MR, et al. . NHE1 promotes invadopodial ECM proteolysis through acidification of the peri-invadopodial space. FASEB J. 2010;24:3903–3915. [DOI] [PubMed] [Google Scholar]
- 54. Schwab A, Fabian A, Hanley PJ, Stock C. Role of ion channels and transporters in cell migration. Physiol Rev. 2012;92:1865–1913. [DOI] [PubMed] [Google Scholar]
- 55. Boedtkjer E, Bunch L, Pedersen SF. Physiology, pharmacology and pathophysiology of the pH regulatory transport proteins NHE1 and NBCn1: similarities, differences, and implications for cancer therapy. Curr Pharm Des. 2012;18:1345–1371. [DOI] [PubMed] [Google Scholar]
- 56. Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–677. [DOI] [PubMed] [Google Scholar]
- 57. Martin C, Pedersen SF, Schwab A, Stock C. Intracellular pH gradients in migrating cells. Am J Physiol Cell Physiol. 2011;300:C490–C495. [DOI] [PubMed] [Google Scholar]
- 58. Lagana A, Vadnais J, Le PU, et al. . Regulation of the formation of tumor cell pseudopodia by the Na+/H+ exchanger NHE1. J Cell Sci. 2000;113(Pt 20):3649–3662. [DOI] [PubMed] [Google Scholar]
- 59. Stock C, Mueller M, Kraehling H, et al. . pH nanoenvironment at the surface of single melanoma cells. Cell Physiol Biochem. 2007;20:679–686. [DOI] [PubMed] [Google Scholar]
- 60. Krahling H, Mally S, Eble JA, Noel J, Schwab A, Stock C. The glycocalyx maintains a cell surface pH nanoenvironment crucial for integrin-mediated migration of human melanoma cells. Pflugers Arch. 2009;458:1069–1083. [DOI] [PubMed] [Google Scholar]
- 61. Paradise RK, Lauffenburger DA, Van Vliet KJ. Acidic extracellular pH promotes activation of integrin α(v)β(3). PLoS one 2011;6:e15746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Srivastava J, Barreiro G, Groscurth S, et al. . Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc Natl Acad Sci USA. 2008;105:14436–14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frantz C, Karydis A, Nalbant P, Hahn KM, Barber DL. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J Cell Biol. 2007;179:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Frantz C, Barreiro G, Dominguez L, et al. . Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol. 2008;183(5):865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choi CH, Webb BA, Chimenti MS, Jacobson MP, Barber DL. pH sensing by FAK-His58 regulates focal adhesion remodeling. J Cell Biol. 2013;202:849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Magalhaes MA, Larson DR, Mader CC, et al. . Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. 2011;195:903–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ray BN, Kweon HK, Argetsinger LS, Fingar DC, Andrews PC, Carter-Su C. Research resource: identification of novel growth hormone-regulated phosphorylation sites by quantitative phosphoproteomics. Mol Endocrinol. 2012;26:1056–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Takahashi E, Abe J, Gallis B, et al. . p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem. 1999;274:20206–20214. [DOI] [PubMed] [Google Scholar]
- 69. Goffin V, Kelly PA. The prolactin/growth hormone receptor family: structure/function relationships. J Mammary Gland Biol Neoplasia. 1997;2:7–17. [DOI] [PubMed] [Google Scholar]
- 70. Rinnerthaler G, Geiger B, Small JV. Contact formation during fibroblast locomotion: involvement of membrane ruffles and microtubules. J Cell Biol. 1988;106:747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Garnovskaya MN, Mukhin YV, Turner JH, Vlasova TM, Ullian ME, Raymond JR. Mitogen-induced activation of Na+/H+ exchange in vascular smooth muscle cells involves Janus kinase 2 and Ca2+/calmodulin. Biochemistry. 2003;42:7178–7187. [DOI] [PubMed] [Google Scholar]
- 72. Meima ME, Webb BA, Witkowska HE, Barber D. The Na-H exchanger NHE1 is an Akt substrate necessary for actin filament reorganization by growth factors. J Biol Chem. 2009;284(39):26666–26675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Malo ME, Li L, Fliegel L. Mitogen-activated protein kinase-dependent activation of the Na+/H+ exchanger is mediated through phosphorylation of amino acids Ser770 and Ser771. J Biol Chem. 2007;282:6292–6299. [DOI] [PubMed] [Google Scholar]
- 74. Aksamitiene E, Achanta S, Kolch W, Kholodenko BN, Hoek JB, Kiyatkin A. Prolactin-stimulated activation of ERK1/2 mitogen-activated protein kinases is controlled by PI3-kinase/Rac/PAK signaling pathway in breast cancer cells. Cell Signal. 2011;23:1794–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hendus-Altenburger R, Pedraz-Cuesta E, Olesen CW, et al. . The human Na+/H+ exchanger 1 is a membrane scaffold protein for extracellular signal regulated kinase. BMC Biol 2016;14:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Peixoto EB, Collares-Buzato CB. Modulation of the epithelial barrier by dexamethasone and prolactin in cultured Madin-Darby canine kidney (MDCK) cells. Cell Biol Int. 2006;30:101–113. [DOI] [PubMed] [Google Scholar]
- 77. Lauritzen G, Stock CM, Lemaire J, et al. . The Na+/H+ exchanger NHE1, but not the Na+, HCO3− cotransporter NBCn1, regulates motility of MCF7 breast cancer cells expressing constitutively active ErbB2. Cancer Lett. 2012;317:172–183. [DOI] [PubMed] [Google Scholar]
- 78. Chen S, Mackintosh C. Differential regulation of NHE1 phosphorylation and glucose uptake by inhibitors of the ERK pathway and p90RSK in 3T3-L1 adipocytes. Cell Signal. 2009;21:1984–1993. [DOI] [PubMed] [Google Scholar]
- 79. Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL. Breast cell invasive potential relates to the myoepithelial phenotype. Int J Cancer. 2003;106:8–16. [DOI] [PubMed] [Google Scholar]
- 80. Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–760. [DOI] [PubMed] [Google Scholar]
- 81. Gutzman JH, Rugowski DE, Nikolai SE, Schuler LA. Stat5 activation inhibits prolactin-induced AP-1 activity: distinct prolactin-initiated signals in tumorigenesis dependent on cell context. Oncogene. 2007;26:6341–6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lauritzen G, Jensen MB, Boedtkjer E, et al. . NBCn1 and NHE1 expression and activity in DeltaNErbB2 receptor-expressing MCF-7 breast cancer cells: contributions to pHi regulation and chemotherapy resistance. Exp Cell Res. 2010;316:2538–2553. [DOI] [PubMed] [Google Scholar]
- 83. Canbay E, Norman M, Kilic E, Goffin V, Zachary I. Prolactin stimulates the JAK2 and focal adhesion kinase pathways in human breast carcinoma T47-D cells. Biochem J. 1997;324(Pt 1):231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhao Y, Lin Y, Zhang H, et al. . Ubl4A is required for insulin-induced Akt plasma membrane translocation through promotion of Arp2/3-dependent actin branching. Proc Natl Acad Sci USA. 2015;112:9644–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. James J, Murry DJ, Treston AM, et al. . Phase I safety, pharmacokinetic and pharmacodynamic studies of 2-methoxyestradiol alone or in combination with docetaxel in patients with locally recurrent or metastatic breast cancer. Invest New Drugs. 2007;25:41–48. [DOI] [PubMed] [Google Scholar]
- 86. Masereel B, Pochet L, Laeckmann D. An overview of inhibitors of Na+/H+ exchanger. Eur J Med Chem. 2003;38:547–554. [DOI] [PubMed] [Google Scholar]
- 87. Stock C, Cardone RA, Busco G, Krahling H, Schwab A, Reshkin SJ. Protons extruded by NHE1: digestive or glue? Eur J Cell Biol. 2008;87(8–9):591–599. [DOI] [PubMed] [Google Scholar]
- 88. Hendus-Altenburger R, Kragelund BB, Pedersen SF. Structural dynamics and regulation of the mammalian SLC9A family of Na+/H+ exchangers. Curr Top Membr. 2014;73:69–148. [DOI] [PubMed] [Google Scholar]
- 89. Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012;441:553–569. [DOI] [PubMed] [Google Scholar]
- 90. Ilic D, Mao-Qiang M, Crumrine D, et al. . Focal adhesion kinase controls pH-dependent epidermal barrier homeostasis by regulating actin-directed Na+/H+ exchanger 1 plasma membrane localization. Am J Pathol. 2007;170:2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ludwig FT, Schwab A, Stock C. The Na+/H+ exchanger (NHE1) generates pH nanodomains at focal adhesions. J Cell Physiol. 2013;228:1351–1358. [DOI] [PubMed] [Google Scholar]
- 92. Tominaga T, Barber DL. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol Biol Cell. 1998;9:2287–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Piwnica D, Fernandez I, Binart N, Touraine P, Kelly PA, Goffin V. A new mechanism for prolactin processing into 16K PRL by secreted cathepsin D. Mol Endocrinol. 2006;20:3263–3278. [DOI] [PubMed] [Google Scholar]