Abstract
Although it is well known that the thyroid hormone (T3) is an important positive regulator of cardiac function over a short term and that it also promotes deleterious effects over a long term, the molecular mechanisms for such effects are not yet well understood. Because most alterations in cardiac function are associated with changes in sarcomeric machinery, the present work was undertaken to find novel sarcomeric hot spots driven by T3 in the heart. A microarray analysis indicated that the M-band is a major hot spot, and the structural sarcomeric gene coding for the M-protein is severely down-regulated by T3. Real-time quantitative PCR-based measurements confirmed that T3 (1, 5, 50, and 100 physiological doses for 2 days) sharply decreased the M-protein gene and protein expression in vivo in a dose-dependent manner. Furthermore, the M-protein gene expression was elevated 3.4-fold in hypothyroid rats. Accordingly, T3 was able to rapidly and strongly reduce the M-protein gene expression in neonatal cardiomyocytes. Deletions at the M-protein promoter and bioinformatics approach suggested an area responsive to T3, which was confirmed by chromatin immunoprecipitation assay. Functional assays in cultured neonatal cardiomyocytes revealed that depletion of M-protein (by small interfering RNA) drives a severe decrease in speed of contraction. Interestingly, mRNA and protein levels of other M-band components, myomesin and embryonic-heart myomesin, were not altered by T3. We concluded that the M-protein expression is strongly and rapidly repressed by T3 in cardiomyocytes, which represents an important aspect for the basis of T3-dependent sarcomeric deleterious effects in the heart.
The heart is a major target organ for thyroid hormone (T3) action, and it is well established that hypo- and hyperthyroid patients exhibit changes in cardiac function (1, 2). Acute hyperthyroidism is known to promote a rapid increase in cardiac output, contractility, and ventricular mass mainly due to direct effects (1) and also indirect effects such as sustained volume overload (3, 4) and activation of renin-angiotensin system (5). In addition, damage to the cardiomyocyte-contractile apparatus has been reported in humans with long-term hyperthyroidism (6). Another study has reported that the M-band, an essential element for sarcomere stabilization, is rapidly and severely affected by T3 in the heart, whereas the Z-disc, which anchors actin filaments, endures even when sarcomere is deeply disarrayed (7). Nevertheless, the molecular basis for the T3 deleterious outcome in the myocardium ultrastructure, mediating sarcomeric disarray, remains unclear. In this regard, global gene expression profile assessment of genes related to sarcomere proteins through microarray analysis may produce relevant clues. Indeed, when we addressed global gene expression profile in rat hearts under experimental hyperthyroidism over time, a potentially key sarcomeric hot spot emerged, the M-band. The M-band comprises 4 myomesin proteins, myomesin 1 and its alternative spliced isoform, the embryonic heart myomesin (EH-myomesin) (8), the M-protein (also known as myomesin 2) (9), and the more lately characterized myomesin 3 (10). These closely related proteins are composed mainly of immunoglobulin-like and fibronectin type III domains (11, 12) and were suggested to cross-link the titin and myosin filaments of the M-band. EH-myomesin is preferentially expressed in the embryonic heart of all higher vertebrates and, to a lesser extent, in slow fibers of adult mice (13). Interestingly, EH-myomesin appears to be expressed in a complementary pattern with the M-protein. This differential expression of the M-protein and EH-myomesin suggests that a muscle type can adjust its M-band structure to a determined muscle demand (13). The most recently described myomesin 3 is restricted to the skeletal muscle fibers and therefore is not expressed in the heart (10). In the present study, we employed a microarray analysis to search for highly T3-responsive sarcomeric domains, and the M-protein was identified. We showed that the M-protein expression, but not myomesin 1 and EH-myomesin, is sharply and rapidly down-regulated by T3 in vivo and in vitro. We also showed that the M-protein promoter is responsive to T3, and an area close to the transcription start point is suggested to contain TREs (thyroid hormone elements). Finally, we showed that down-regulation of M-protein in cultured cardiomyocytes drives a severe deficit in contractility, highlighting the important role of M-protein in sarcomere function.
Materials and Methods
Animals
All animals were handled according to an experimental protocol in accordance with ethical principles in animal research adopted by the Brazilian College of Animal Experimentation (COBEA) and approved by Institute of Biomedical Sciences/University of São Paulo Ethical Committee for Animal Utilization (CEUA). Male Wistar rats weighing 200–250 g were obtained from the University of São Paulo, Institute of Biomedical Sciences, in São Paulo, Brazil. The animals were given free access to standard rodent food and water and were housed in a temperature- and light-controlled environment (24°C; 12-hour light/dark cycle).
Microarray
Total RNA from adult rat ventricles was isolated using a Trizol LS reagent (Life Technologies) in accordance with the manufacturer's instructions. Total RNA was further purified with RNAeasy Fibrous Tissue Mini Kit (Qiagen) to reach microarray RNA quality standards. The resulting phenol-free RNA was submitted to spectrophotometric analysis in a Nanodrop spectrophotometer (Uniscience), and RNA quantity was determined by the A260 wavelength method. Total RNA (1 μg) was used in the cDNA first-strand synthesis with One-Cycle Target Labeling and a Control Reagents kit (Affymetrix). At the end of the cycle, samples were repurified using the Cleanup of the Double-Strand cDNA kit (Affymetrix) and requantified in Nanodrop. The biotinylated cRNA synthesis was carried out with the in vitro transcription (IVT) Gene Chip Expression 3′-the Amplification Reagents for IVT labeling kit (Affymetrix) followed by the cRNA fragmentation at a high temperature and a high Mg+2 concentration buffer. For hybridization, we used the Gene Chip Hybridization, Wash and Stain (Affymetrix). Chips were filled in with hybridization buffer, incubated at 45°C for 16 hours and then processed in the washing station (Affymetrix). After all the treatments, the chips were scanned.
Microarray data analysis
Microarray raw data analysis were conducted using “affy” and “simpleaffy” packages by Bioconductor (14), implemented on the R statistical program. MAS5 algorithm was applied. The gene expression values obtained were filtered using a cutoff of 50, allowing us to enrich our samples with genes with relative secure levels of expression above background. Fold changes (fold induction in 12 hours, 24 hours, and 7 days) were calculated for all genes. We decided to adopt a stringent cutoff criterion, only genes down-regulated by ≤ 0.3 and up-regulated by ≥ 3.0-fold were considered acceptable for analysis. Based on Affymetrix annotation files, a list of probes related to sarcomeric proteins was composed and categorized in agreement to its localization in sarcomere.
Animal treatments
For the microarray approach, experimental hyperthyroidism was induced in animals by daily ip injections of T3 (0.07 μg/g of body weight, equivalent to 20 physiological doses) during 12 hours, 24 hours and 7 days. Rats were killed by decapitation. Hearts were then removed and frozen in liquid nitrogen and stored at −80°C for later analysis.
In order to validate the impact of T3 upon the M-band, rats were previously submitted to hypothyroidism by thyroidectomy followed by 0,05% methimazole (Sigma) administrated in drinking water for 20 days. T3 was administered daily to 4 groups (n = 4–6) via ip injection at the following doses: 3.5 ng, 17.5 ng, 0.17 μg, and 0.35 μg/g/ body weight (BW). The dose of T3 to achieve the euthyroid state (3.5 ηg/g BW) is defined as 1× T3 (15), 17.5ng/g BW corresponding to 5× T3, 0.17 μg/g BW to 50× T3, and 0.35 μg/g BW to 100× T3. The other 3 groups (n = 4) were injected with 0.35 μg/g BW T3 for 2 and 8 days. A group of 6 untreated rats was used as control. After the treatments, the animals were killed by decapitation, and their hearts were quickly removed, rinsed in saline, and flash frozen in liquid nitrogen.
Primary culture of cardiomyocytes: preparation and procedures
Cultured neonatal rat cardiomyocytes were prepared as previously described (16). Briefly, myocytes were dispersed from the ventricles of 1- to 2-day-old Wistar rats by digestion with collagenase type II (Worthington) and pancreatin (Invitrogen) at 37°C. Cultures with more than 95% cardiomyocytes were obtained after purification of cell suspensions on a discontinuous Percoll (Pharmacia LKB Biotechnology) gradient. Cell viability was estimated by the Trypan blue method, after which the cells were counted and plated. Myocytes were cultured in DMEM (Invitrogen) containing penicillin-streptomycin (Invitrogen), 10% horse serum (Invitrogen), and 5% newborn calf serum (Invitrogen). The cells were maintained at 37°C under humidified conditions of 95% air and 5% CO2.
Seventy-two hours after plating, the cells were transferred to DMEM containing newborn calf serum 0.5% and maintained overnight. Then, the medium was discarded and the cells were incubated with either serum-free medium (control cells) or serum-free medium containing T3 (10−7 M; Sigma) for 12 hours, 24 hours, and 48 hours. Also cells were transfected with small interfering RNA (siRNA) for M-protein or with a nonspecific siRNA control (Stealth RNAi Negative Control Duplex; Invitrogen) using Lipofectamine 2000 (Invitrogen). The oligo-lipo complex medium was replaced after 6 hours incubation with fresh medium, and the cells were maintained in an incubator for 72 hours. Subsequently, cell contraction properties of cardiomyocytes were evaluated using a video-based sarcomere-spacing acquisition system (SarcLen, IonOptix) as previously described (17). Cardiomyocytes were perfused with normal Tyrode's solution and electrically paced (1–3 Hz via field stimulation). Changes in sarcomere length were recorded and analyzed using IonWizard software (IonOptix).
RNA isolation and real-time quantitative PCR
RNA was isolated from ventricular samples or cardiomyocytes using a Trizol reagent (Invitrogen) followed by cDNA synthesis from 1 μg of total RNA using a SuperScript II Kit (Invitrogen) and oligo-DT primers according to manufacturer's instructions.
A real-time quantitative PCR (qPCR) was performed on the ABI Prism 5700 sequence detection system (Applied Biosystems) using SYBR Green (Applied Biosystems). Cycling conditions were 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 60 seconds. Internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified in separate tubes and data were expressed as the ratio from the target gene over GAPDH. Data obtained previously in our laboratory showed that T3 does not influence cardiac or cardiomyocytic GAPDH expression levels, functioning as an adequate internal control. Data from 5–6 determinations were expressed in all experiments as fold change as compared with control group, arbitrarily set as 1.
Antibodies
Monoclonal mouse antimyomesin antibody (clone B4) (18) and monoclonal mouse anti-M-protein antibody (clone AA259, Ig A) (19) were kindly provided by Dr. Jean Claude Perriard (Zurich University, Switzerland). Antibody against GAPDH was obtained from Abcam. For immunofluorescence, secondary antibodies were fluorescein isothiocyanate-conjugated goat antimouse IgA (Sigma) and Alexa488-conjugated goat antimouse IgG and goat antirabbit (Invitrogen).
Frozen sections of rat heart
Rat ventricles were dissected, snap frozen in liquid nitrogen cooled with isopentane, and stored at −80°C until sectioning. Cross-sections (7 μm thick) through the left ventricle (LV) were cut on a microtome-cryostat (IEC Minotome) at −25°C. The sections were collected on gelatin-coated glass slides, dried for 1 hour at room temperature, and stored at −20°C.
Immunostaining and image acquisition
Cross-sections of rat ventricles were fixed with 4% paraformaldehyde in PB (phosphate buffer, Sigma) for 10 minutes at room temperature, blocked with 0.1 M glycine (Sigma) in PB for 5 minutes, and permeabilized in 0.2% Triton X-100/PB (Sigma) for 10 minutes. Primary antibodies with 3% normal goat serum (Sigma) were incubated overnight at room temperature. After washing with PB, secondary antibodies were added for 2 hours. The specimens were washed in PB and mounted in Vectashield (Vector Laboratories) (20). Slides were evaluated by confocal microscopy (Nikon PCM2000, Nikon Instruments) using appropriate filter combinations of fluorescein isothiocyanate and Alexa488 (excitation filter, 460–490 nm; barrier filter, 515–550 nm). All images were acquired together with the same gain and composed in Adobe Photoshop version 7.0 (Deneba Software).
Cloning of M-protein promoter region
The putative sequence of rat M-protein promoter (accession no. XM_240481.4 GI:109503557) was aligned with murine M-protein (accession no, NM_008664.1 GI:6678995) promoter previously described (12) showing 87% homology using the BLAST tool (21) at NCBI web site (blast.ncbi.nlm.nih.gov). The rat M-protein promoter (−2294/+220bp) construct was generated by amplification of genomic DNA by using sense (5′-TGACATCTTGACCTCTGA-3′) and antisense (5′-TTGGTTTCTCCTACAGGA-3′) primers, both containing MluI restriction site adapters that were used to clone the PCR product into the pGL3-Luc Basic reporter vector (Promega). Fragments comprising the promoter deletions −158/+220bp were generated by digestion with KpnI, and PstI (MBI Fermentas). All construct sequences were checked by an automated sequencing reaction at Human Genome Study Center, University of Sao Paulo, Brazil.
Transfection and luciferase assay
C2C12 myoblasts were cultured in 24-well plates at approximately 50% confluence and transfected using Lipofectamine (Invitrogen) according to manufacturer's instructions. Each plate was cotransfected with 400 ng of the M-protein promoter luciferase reporter gene, 10 ng of control Renilla luciferase vector, 100 ng of thyroid hormone receptor (TR)β or TRα expression vector (generously provided by Dr. Wolfgang Dillmann, University of California, San Diego, CA). Cells were differentiated in horse serum 5% for 2 days and 24 hours before the differentiation endpoint, cells were treated with T3 (0.01; 0.1; 1 and 100 nM) or vehicle. Firefly and Renilla luciferase activity were determined with a Luciferase Assay kit (Promega) in luminometer (Molecular Devices).
Chromatin immunoprecipitation (ChIP) assay
Cultured neonatal rat myocardial cells were processed using buffers and reagents from EZ ChIP Kit (Upstate Biotechnology) according to manufacturer's instructions. Briefly, cells were fixed in DMEM containing 1% formaldehyde for 10 minutes at room temperature and transferred to lysis buffer. DNA was sheared to fragments of approximately 200–1000 bp by applying 8 bursts of sonication for cells (40% of power; 10 seconds each). Samples were diluted with dilution buffer and precleared for 1 hour at 4°C with protein A-Sepharose (50% slurry) saturated with salmon sperm DNA. An aliquot of 10 μL was collected as “input.” The remaining supernatants were submitted to immunoprecipitation with protein A-Sepharose saturated with salmon sperm DNA and 5 μg of anti-TRα/ß antibody (fl-408; Santa Cruz Biotechnology). In parallel, one sample was incubated with protein A-Sepharose only in order to generate the negative control (no-AB). Sepharose pellets were then washed with buffers provided in the kit and treated with elution buffer. Supernatants were submitted to cross-linking reversal and RNase A treatment. DNA was purified using phenol-chloroform and resuspended in 20 μL of ultrapure H2O. DNA samples were amplified for detection of M-protein and GH genes, the latter used as control gene. A 184-bp fragment of the mouse M-protein promoter was amplified by conventional (40 cycles) and real-time quantitative PCR. The sequences of the primers were: GH sense, 5′-CCCTCGTCCCAGTGAACAAACG-3′; and antisense, 5′-GCTGGAGCCACTGACAGCTTG-3′; M-protein sense, 5′-TCCAGAGCTGGACTGAGGAGGAAA-3′; and antisense, 5′-TTGCTCTGGCAGCATAGGTCCA-3′. The products were visualized as described for RT-PCR. Amplification values were normalized by input.
Putative TREs searching engine
Putative TREs were identified by using a computer script written using Phyton language. Briefly, the consensus sequence A/TGGNC/GA/G/T (sense and antisense) was used to feed the script, allowing identification of putative TREs. This script was developed by Andrei Rozanski.
Statistical analysis
All data were expressed as mean ± SD. Unpaired Student's t test or one-way ANOVA followed by Tukey multiple-comparison test were employed for assessment of significance (GraphPad Prism version 5.0; GraphPad Software, Inc). Differences were considered significant when P < .05.
Results
Thyrotoxicosis-induced heart hypertrophy
T3 treatment efficiency was confirmed by an increase of 51% in LV weight of animals treated with 100× T3 for 8 days, while BW decreased 12.6% as compared with euthyroid animals. LV weights (mg) for the euthyroid and T3-treated were 905.33 ± 150.05 and 1370 ± 134.35, respectively, and LV:BW ratios (mg/g) of 3.8 ± 0.5 and 6.59 ± 0.46; both parameters were significantly different between groups (P < .001; n = 5).
Microarray analysis: identification of T3-dependent sarcomeric hot spots
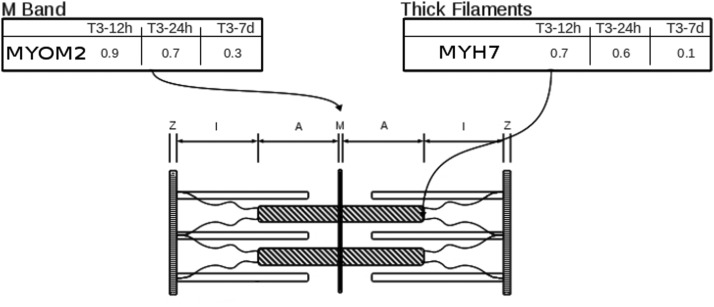
Our first aim was to detect uncovered T3 highly responsive hot spots in the sarcomere. Of 110 sarcomeric genes evaluated, which fitted within 4 distinct categories namely “thick filaments,” “thin filaments,” “M-band,” and “Z-disk,” we found only 2 genes that were considered responsive to T3 (≤0.3- and ≥3.0-fold): Myh7 (0.1-fold, thick filament category) and M-protein (0.3-fold, M-band category) (Figure 1). Previous studies have described the responsiveness of genes related to proteins of thick filaments (1, 3) under experimental hyperthyroidism; nonetheless, no studies have indicated M-band genes as responsive to T3. Therefore we stepped further by analyzing M-band and assuming M-protein as the main finding of our microarray approach under stringent cutoff conditions.
Figure 1.
Identification of hot spots on heart sarcomere under T3 treatment determined by microarray analysis. Fold induction after 12 hours, 24 hours, and 7 days of T3 treatment, along with genes, are shown in boxes described as “M-Band” and “Thick Filaments.”
M-protein gene expression is down-regulated in the heart by T3 treatment and up-regulated in hypothyroid animals.
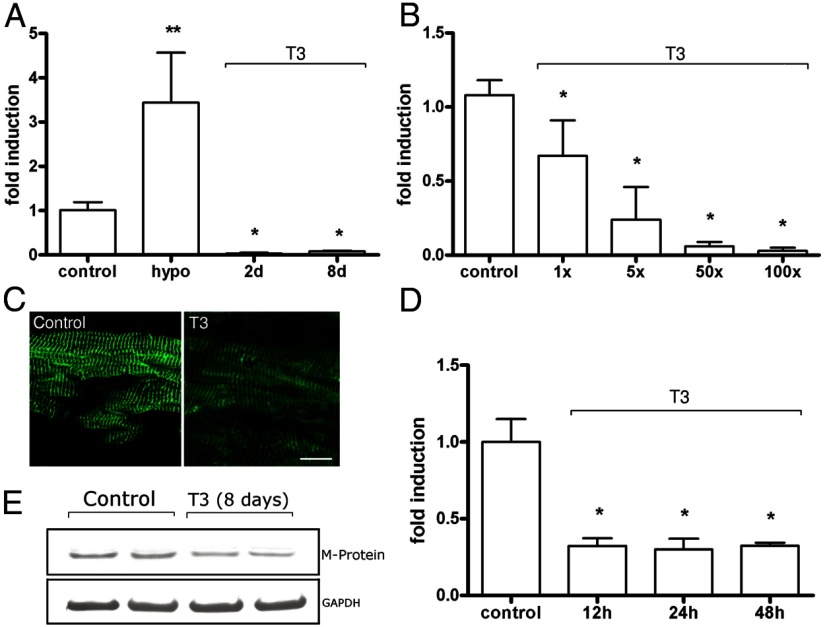
An up-regulation (3.4-fold) was observed on M-protein mRNA levels in hearts from hypothyroid rats, compared with those observed in euthyroid animals (Figure 2A). On the other hand, a down-regulation to virtually undetectable levels was observed on M-protein mRNA levels after 2 or 8 days of treatment (100× T3) (Figure 2A). Increasing doses of T3 for 48 hours elicited a dose-dependent repression profile of M-protein mRNA levels (Figure 2B). The treatment with 1× T3 was enough to decrease M-protein mRNA levels to 67% of control levels (P < .001). Accordingly, increasing doses (5×, 50×. and 100×) further decreased (P < .001) M-protein gene expression (24%, 6%, and 3% of control, respectively) (Figure 2B).
Figure 2.
M-protein mRNA and protein levels are decreased by T3. A, M-protein mRNA levels in euthyroid (control), hypothyroid (hypo) + T3 (100×) for 2 and 8 days (n = 5–6). Data are expressed as fold induction and control is arbitrarily set as 1. Bars represent mean, and the connected vertical lines represent SD. *, P < .05 vs control; **, P < .001 vs control. B, T3 induces repression of M-protein mRNA level in a dose-dependent manner. Rats were treated with increasing doses of T3 (1×, 5×, 50×, and 100× physiological doses) for 48 hours (n = 5–6). *, P < .001 vs control. C, Immunoexpression of M-protein in LV sections from rats treated with 100× of T3 for 8 days. Note decreased intensity of labeling (green) in T3-treated group as compared with control group; Bar, 20 μm. D, M-protein mRNA levels determined by real-time quantitative PCR in cultured cardiomyocytes. Cells were treated with T3 (10−7M) for 12 hours, 24 hours, or 48 hours. *, P < .001 vs control. E, Western blot analysis of T3-treated (8 days) rats show decreased M-protein expression compared with control.
In order to further evaluate the effects of T3 upon the M-band, we used specific antibodies against the M-protein in LV sections. A striated pattern, typical of the M-band localization, was obtained (Figure 2C). Treatment (100× T3) resulted in a decrease in immunolabeling intensity at 8 days of treatment (Figure 2C). Western blot analysis confirmed decreased expression of the M-protein in T3-treated animals (Figure 2E).
M-protein gene expression is down-regulated by T3 treatment in cultured cardiomyocytes
In order to eliminate a possible systemic modulation on the M-protein gene expression, we evaluated the effect of T3 (10−7M) in cultured isolated neonatal cardiomyocytes. Treatment for 12, 24, and 48 hours significantly decreased the M-protein mRNA levels (Figure 2D). Remarkably, as soon as 12 hours of treatment caused a marked reduction (∼70%) on M-protein mRNA levels when compared with control (P < .01). This effect was sustained up to 48 hours (Figure 2D).
TR-mediated repression of M-protein promoter activity
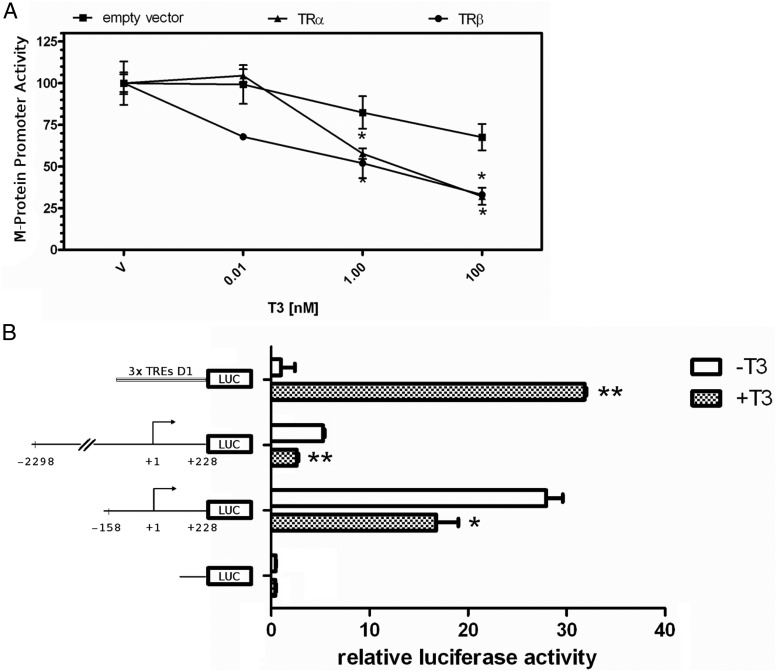
At this point, it was clear that T3 plays a role decreasing the M-protein gene expression. To investigate whether this effect was mediated at the transcriptional level, we used a reporter construct containing the proximal promoter region −2294 to +228 of the M-protein gene driving the firefly luciferase transcription (M-protein-luc) in C2C12 cells. Transfection efficiencies were normalized with Renilla luciferase activity. T3 treatment down-regulated the M-protein promoter activity in the presence of either TRα1 or TRβ1, by using as low as 1 nM T3 (Figure 3A). To decrease the M-protein promoter area responsive to T3, we have deleted the −2294 to +228 fragment down to −158/+228. This shorter construct (pGL3 −158/+228) similarly showed a significant repression response to the T3 treatment when compared with that related to the longer construct (−2294 to +228 bp), although this deletion has substantially increased the basal transcriptional activity (Figure 3B). These results suggest that a T3 response element(s) is/are located between the nucleotides −158 and +228 region.
Figure 3.
M-protein promoter is responsive to T3. A, The rat M-protein promoter driving the firefly luciferase (LUC) reporter construct was cotransfected into C2C12 cells with either TRα, TRβ, or empty expression vector (pCMX). Cells were treated with progressive doses of T3 (0.01, 0.1, 1, and 100 nM) or vehicle (V). *, P < .001 vs V. B, Mean ± SD of transcriptional activity of 3×TRE D1 (positive control), −2298 to +228 bp and −158 to +228 bp promoter, and proximal regions of M-protein gene and promoter less pGL3 plasmid (negative control) transfected in C2C12 cells treated (+T3) or nontreated (−T3) with 50 nM T3. Promoter activity was arbitrarily set as 1 in all nontreated groups, and T3 effect was expressed as fold change. *, P < .01; and **, P < .001 vs respective untreated group. n = 3.
Evidence of physical association of TRs with M-protein promoter
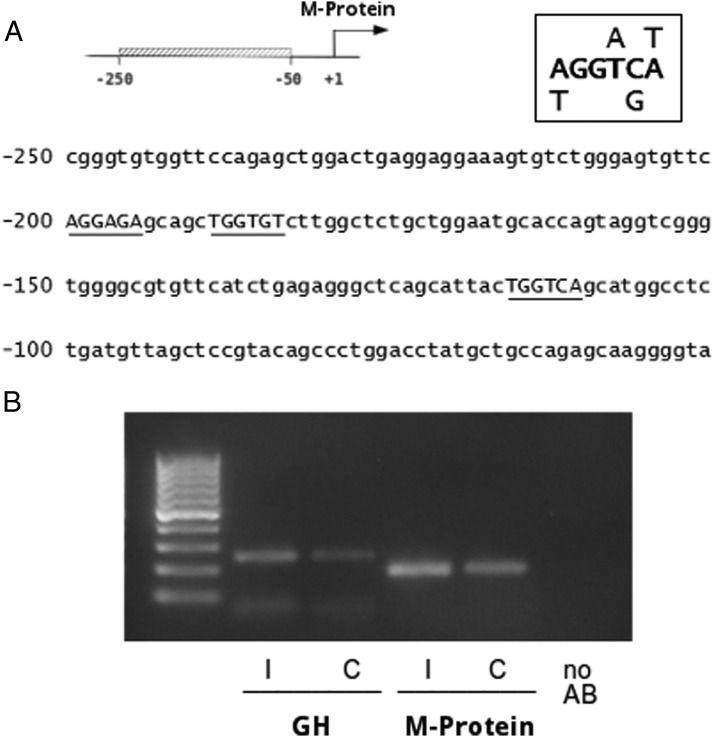
Next, we decided to employ a computer script (developed by Andrei Rozanski; see Material and Methods) designed to identify potential half-sites (negative TREs). By using this tool, we have identified 3 putative negative TREs within the fragment −158/+228, following a quite stringent criterion (A/t G G T/a C/g A/t). All of them are located in the sensus strand and two of them are nearby, spaced by 5 nucleotides (Figure 4A), one at −200 bp and the other at −189 bp relative to the transcriptional start. The third putative negative TRE is located at −116 bp (Figure 4A). In order to obtain experimental evidence of direct binding of TRs to the −158/+228 the M-protein promoter fragment, we have performed a ChIP assay using primary cultures of cardiomyocytes. Amplification of GH promoter was used as a positive control gene because it is well recognized as extremely sensitive to T3 (22). No-antibody (AB) was used as negative control. As expected, TRs was specifically bound to GH promoter. Likewise, TRs were also bound to the −239 to −56 M-protein promoter region (Figure 4B).
Figure 4.
TRs bind to M-protein promoter. A, Sequence of the proximal M-protein is shown (−250 to −50); top right shows the consensus TRE sequence used to search for putative TREs, which are indicated by underlining the sequences. B, Fragments of M-protein (−239/−56) and GH (−200 /−165) promoters were amplified from DNA extracted from the input and ChIP originated from each sample. Values from the amplification of each ChIP sample were normalized by the respective input. AB, antibody.
Impact of M-protein deficiency on contractile properties in neonatal cardiomyocytes
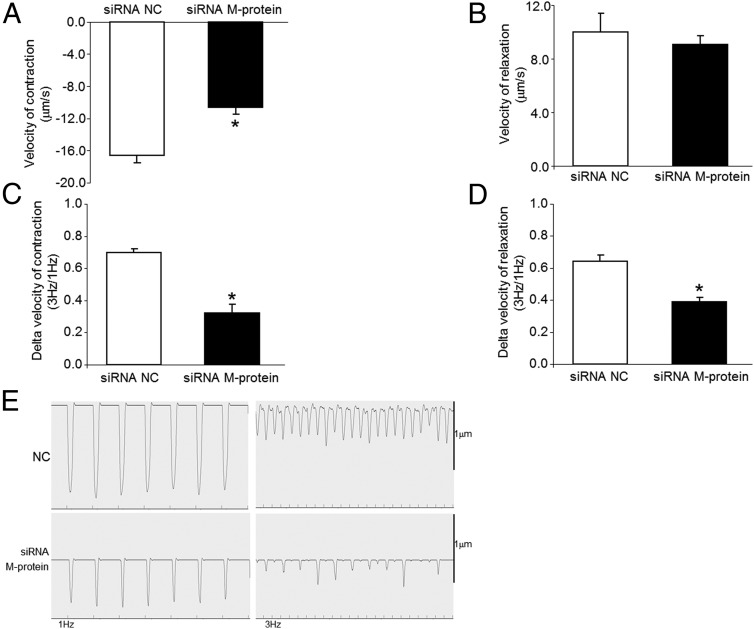
In order to investigate the role of M-protein on cardiomyocyte-contractile function, we siRNA knocked down M-protein and subsequently measured velocity of contraction and velocity of relaxation using a video-based sarcomere-spacing acquisition system (SarcLen, IonOptix) as described elsewhere (17). The results show that M-protein deficiency drives a significant drop in velocity of contraction at 1 Hz stimulation (77% ;P < .05; Figure 5A). No alterations in velocity of relaxation at 1 Hz were detected by silencing M-protein (Figure 5B). Of interest, cardiomyocytes with M-protein deficiency displayed reduced rate of contraction and relaxation upon increasing stimulation frequency compared with control cells (Figure 5, C–E).
Figure 5.
Impact of M-protein deficiency on cardiomyocyte function. Velocity of contraction (A) and relaxation (B) were determined in neonatal siRNA M-protein knocked down cardiomyocytes at 1 Hz, 30 V. Also the delta between velocity of contraction (C) and relaxation (D) at 3Hz/1Hz was determined. Representative acquisitions from negative control (NC; scrambled transfected) and siRNA M-protein knockdown are shown (E). *, P < .05 vs NC.
Effect of T3 upon other M-protein constituents: myomesin and EH-myomesin
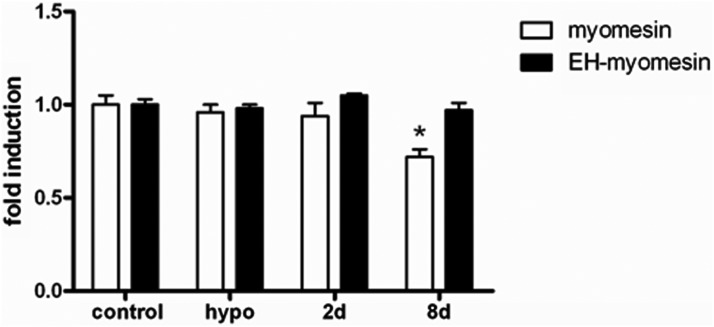
To investigate the response of other structural M-band proteins to T3, we analyzed the mRNA levels of myomesin and EH-myomesin. Myomesin mRNA levels were not altered in hearts from rats treated with 100× T3 for 2 days, whereas those treated for 8 days presented a modest, but significant (P < .05), decrease (∼25%) in myomesin mRNA levels (Figure 6).
Figure 6.
T3 effect upon myomesin and EH-myomesin mRNA levels were determined by real time quantitative PCR. Data is expressed as fold induction (mean ± SD), and euthyroid group (control) is arbitrarily set as 1. Myomesin (open bars) and EH-myomesin (solid bars) mRNA levels in control, hypothyroid (hypo), and euthyroid rats treated with T3 for 2 and 8 days (100×) (n = 5–6). *, P < .001 vs control.
Discussion
In the present study, we demonstrate that supraphysiological doses of T3 have a strong and fast repressive effect upon the M-protein gene and protein expression in the heart and also in isolated cardiomyocytes. This effect was sustained because decreased levels of M-protein gene expression were found after up to 1 week of treatment with supraphysiological doses of T3. Moreover, the molecular mechanism responsible for this effect involves transcription, as evidenced by T3 -dependent down-regulation M-protein promoter activity in transient transfection assays and by ChIP assays.
It is well known that elevated levels of T3 rapidly drive an increase in myocardium speed and strength, resulting in improved functions. These effects are mainly due to alterations in myosin heavy chain transcription (2) and modifications of calcium-regulatory proteins (23, 24). The acute gain of cardiac function driven by T3 excess may result in hypertrophy, which is associated with changes in chamber geometry. On the other hand, sustained T3 elevated levels can, over longer time periods, cause congestive heart failure and atrial arrhythmia, which are accompanied by severe loss of function (2). Due to the acute gain in cardiac function, T3 has been suggested as a therapeutic tool for improving cardiac performance in conditions such as infarct, heart failure, and ischemia reperfusion (25–27). The potential drawback of this strategy is that if supraphysiological doses of T3 are used, T3 can also trigger long-term deleterious effects as stated above. Therefore, characterization of T3-dependent molecular events in the heart will help to develop novel therapeutic strategies, highlighting the beneficial effects of the hormone and minimizing deleterious effects. In fact, it has been shown that a replacement dose of T3 in mice submitted to acute myocardial infarction significantly improves ventricular function and remodeling. On the other hand, a higher dose of T3 increases mortality rate and triggers activation of AKT and ERK, pathways involved in hypertrophy (28).
In an attempt to better understand the global impact of elevated levels of T3 in sarcomeric genes, we have employed the microarray approach focused on genes related to sarcomeric proteins. From 330 sarcomeric genes selected for analysis, only 2 were found to be responsive under the criterion of ≤0.3- and ≥3.0-fold. By using this relatively rigorous criterion, it was possible to recognize that the effects of T3 in the sarcomere are rather confined. Actually, it is striking that T3 can achieve such powerful functional modifications in the sarcomere by acting in a relatively small set of genes. In addition, this approach enables the identification of hot spots, which can greatly increase our understanding of sarcomeric responsiveness to excess levels of T3. Out of those 2 genes, one codes for β-myosin heavy chain, which is well known to be negatively responsive to T3, and the other codes for M-protein. M-protein gene caught our attention because the degree of negative response to T3 was similar to β-myosin heavy chain (0.3- and 0.1-fold change, respectively) and also because it codes for a protein thought to play an important role in M-band function, which includes increased stability of the thick filament lattice (29).This increased stability seems to allow proper anchoring and, at the same time, allows for certain flexibility. It is believed that these characteristics could promote subtle imbalances between the 2 thick filament halves, caused by a different proportion of the activated cross-bridges, allowing thick-thin filament coupling to function optimally (30). We have actually, in a previous study, foreseen that the M-band could be a target of elevated levels of T3 (7). In this study, we have described a severe M-band disarray driven by T3, whereas the other anchoring domain of the sarcomere, the Z-disc, is essentially not affected (7). These previous ultrastructural findings are in line with the present study in which we have found the M-band as a T3 target in contrast to the Z-disk.
It is clear that an excess of T3, over short term, strongly overexpresses the α-myosin heavy-chain gene, while repressing the β-myosin heavy-chain gene (1, 3) in agreement with our findings in the microarray assay (Figure 1). This effect drives increased speed of contraction, supporting the gain in chronotropism induced by T3. The sarcoendoplasmic reticulum Ca+2 ATPase-2 gene is also induced by T3 over a short term, being therefore another main molecular event supporting increase in relaxation speed (31, 32). It is of great interest to better understand, at the molecular level, why the T3-stimulated heart evolves from the compensated to the uncompensated state. α-Myosin heavy chain (33) and sarcoendoplasmic reticulum Ca+2 ATPase-2 (34) are decreased when uncompensated hypertrophy takes place, indicating that these genes are causative factors for transition from compensated to uncompensated heart hypertrophy. Another possibility regarding candidate genes for T3-dependent transition to cardiac failure may involve structural sarcomeric components. It is noteworthy to highlight that, despite a wide range of studies approaching T3 and contraction-related gene expression, little is known regarding T3 influence upon structural sarcomeric genes. Structural sarcomeric proteins are crucial for sarcomere architecture maintenance and are also important regulators of signaling pathways involved in cell size control. For example, it is well known that Z-disc contains calcineurin, an important phosphatase involved in the pathogenesis of cardiac hypertrophy (35, 36). Moreover, protein kinase Cϵ, an important kinase controlling contractility and hypertrophy, is a Z-disc component (37). Accordingly, the M-band portion of titin contains an activated serine-threonine kinase domain which is involved in important signaling cascade (38). The M-band components, myomesin and M-protein, are known to provide the binding platform for muscle creatine kinase, an important enzyme involved in the restoring of a reservoir of energy (39). Enolase, phosphofructokinase, and adenylate kinase also localized in the M-band, suggesting an additional role of the M-band as a structure linked to regulation of energy metabolism (40, 41). Overall, it is clear that sarcomeric anchoring elements can also be important signaling stations; therefore a better understanding of structural sarcomeric genes will be of use to increase our knowledge of heart plasticity.
To our knowledge, this is the first study that systematically addresses M-band structural proteins under T3 treatment in the heart. A previous study has shown a role of T3 in fetal to adult titin isoform switching via a nongenomic pathway (42). Nonetheless, no studies were conducted in an adult rat heart. Our results obtained from T3-treated rats show that M-protein gene is particularly sensitive to the hormone. A single physiological dose evoked significant reduction in M-protein mRNA levels in a dose-dependent manner. The high sensitivity of this gene to T3 was also confirmed in hypothyroid rats in which M-protein gene expression was increased by approximately 3.5-fold. The negative effect of T3 upon M-protein mRNA expression parallels protein levels, as evidenced by Western blot and immunolabeling experiments. The level of M-protein expression is clearly decreased 8 days after T3 treatment.
The cell specificity of T3 action was assessed by in vitro cardiomyocyte experiments. The results clearly showed that T3 acts directly on cardiomyocytes to drive M-protein repression and does not depend on interactions with other systems such as other hormones, ie, GH, the vascular bed, and autonomic regulation.
The fact that both M-protein mRNA and protein levels were rapidly and intensely down-regulated, raises the possibility of a transcriptional control. Although, specific fast proteolysis such as triggered by the ubiquitin/proteasome system, could also be involved in T3-dependent M-protein cell clearance. We have not approached mRNA and protein stability in this study; however, we show that the M-protein promoter is strongly responsive to T3. M-protein promoter behaves similarly to TRH (43) and the pituitary TSH β-subunit gene (44), which all contain negative TREs. Our promoter deletion experiments pointed the fragment −158 and +228 bp as responsive to T3. Subsequently, by using a computer script designed to pinpoint putative half-sites (negative TREs), we have found 3 potential sequences that could bind TRs. All of them are located at the sensus strand, and the first and second putative half-sites are spaced by 5 nucleotides (−200 and −189bp). The third putative half-site is located at −116 bp. Evidence for binding of TRs to those half-sites comes from ChIP assay experiments, in which we have shown that TR binds directly to M-protein promoter −239/−56 fragment (which includes those putative half-sites). It will be of interest in future studies to provide further insight on the putative negative TREs in M-protein promoter, using additional approaches such as point mutations and footprinting. These results are in accordance with other studies showing the presence of negative TREs in the promoters very close to the TATA box (45–47). Thus, our results suggest that TR-DNA binding activity is required for down-regulation of the M-protein promoter.
As mentioned above, we have previously shown by electron microscopy that the M-band is firstly and primarily affected by T3 (7), an effect that precedes cardiac failure, suggesting that degeneration of the M-band could trigger sarcomere disassembling. At the ultrastructural level, it has been shown that alcohol consumption disturbs the normal M-band structure in fetal myocardium (48). Interestingly, this effect seems to be specific to the M-band because other sarcomeric structures are well preserved, ie, I and A bands and Z-discs (48). Subsequent studies have addressed myomesin abundance in heart diseases. For instance, Hein et al (49) reported that ischemia in the myocardium results in decreased myomesin immunoexpression. On the other hand, Wang et al (50) found no alteration in myomesin immunoexpression in hearts submitted to pressure overload. Finally, immunoexpression was used to evaluate myomesin abundance in dystrophic hamsters and no alteration was found (51). There are no studies demonstrating M-protein regulation in the heart through cardiac overload or other cardiomyopathies. In addition no studies have directly addressed M-protein function. In this study we have siRNA knocked down M-protein in cardiomyocytes and evaluated contractile function and noted a 77% reduction in contraction velocity in the M-protein deficient cells, which is in line with an important role of this protein in sarcomere function. It is currently not clear whether M-protein, in addition to having a structural role, could also affect/control the contractility process; nonetheless, our functional results raises the possibility that M-protein could regulate the activity of proteins such as myosin heavy chain, myosin light chain, and Troponin C. It is interesting to note that in other pathophysiological conditions, such as heart hypertrophy and failure driven by pressure overload, the Z-disc seems to be the preferential target rather than the M-band (52–54), which leads to the possibility that anchoring proteins in the sarcomere are differentially affected under different demands.
Because myomesin and EH-myomesin are M-band components as the M-protein, we also investigated its expression. Interestingly, we noticed no alterations on the EH-myomesin expression pattern under T3 treatment and hypothyroidism. Myomesin showed similar results, differing only by an approximately 25% reduction of fold induction under T3 treatment for 8 days. These findings support M-protein as the main gene sensitive to T3 at M-band level.
In summary, this study reveals that M-protein can be selectively regulated by an important modulator of cardiac function, and further studies using other models of cardiac plasticity, such as hypertension, diabetes, and genetic cardiomyopathies, are necessary to further explore the role of this structural sarcomeric element. It would be interesting, in the future; to address the role of M-band proteins using different experimental models/strategies such as myocardial infarction and heart failure in order to further address the important role of this sarcomeric protein in other cardiovascular diseases.
Acknowledgments
This work was supported by Grants 03/02401–0 and 00/12037–6, both from Fundação de Amparo à Pesquisa do Estado de São Paulo, a Sao Paulo State Research Foundation, Brazil.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by Grants 03/02401–0 and 00/12037–6, both from Fundação de Amparo à Pesquisa do Estado de São Paulo, a Sao Paulo State Research Foundation, Brazil.
Footnotes
- BW
- body weight
- ChIP
- chromatin immunoprecipitation
- EH-myomesin
- embryonic heart myomesin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- LV
- left ventricle
- PB
- phosphate buffer
- siRNA
- small interfering RNA
- TR
- thyroid hormone receptor
- TRE
- thyroid hormone element.
References
- 1. Klein I, Ojamaa K. Thyroid hormone and the cardiovascular system. N Engl J Med. 2001;344:501–509. [DOI] [PubMed] [Google Scholar]
- 2. Kahaly GJ, Dillmann WH. Thyroid hormone action in the heart. Endocr Rev. 2005;26:704–728. [DOI] [PubMed] [Google Scholar]
- 3. Dillmann WH. Biochemical basis of thyroid hormone action in the heart. Am J Med. 1990;88:626–630. [DOI] [PubMed] [Google Scholar]
- 4. Klein I, Hong C. Effects of thyroid hormone on cardiac size and myosin content of the heterotopically transplanted rat heart. J Clin Invest. 1986;77:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu LW, Benvenuti LA, Liberti EA, Carneiro-Ramos MS, Barreto-Chaves ML. Thyroxine-induced cardiac hypertrophy: influence of adrenergic nervous system versus renin-angiotensin system on myocyte remodeling. Am J Physiol Regul Integr Comp Physiol. 2003;285:R1473–R1480. [DOI] [PubMed] [Google Scholar]
- 6. Abinder AA, Paukov VS, Rom-Bugoslavskaia ES. Contractile function of the myocardium of the right ventricle in thyrotoxicosis. Probl Endokrinol (Mosk). 1975;21:22–27. [PubMed] [Google Scholar]
- 7. Ferreira PJ, L'Abbate C, Abrahamsohn PA, Gouveia CA, Moriscot AS. Temporal and topographic ultrastructural alterations of rat heart myofibrils caused by thyroid hormone. Microsc Res Tech. 2003;62:451–459. [DOI] [PubMed] [Google Scholar]
- 8. Agarkova I, Auerbach D, Ehler E, Perriard JC. A novel marker for vertebrate embryonic heart, the EH-myomesin isoform. J Biol Chem. 2000;275:10256–10264. [DOI] [PubMed] [Google Scholar]
- 9. Masaki T, Takaiti O. M-protein. J Biochem. 1974;75:367–380. [DOI] [PubMed] [Google Scholar]
- 10. Schoenauer R, Lange S, Hirschy A, Ehler E, Perriard JC, Agarkova I. Myomesin 3, a novel structural component of the M-band in striated muscle. J Mol Biol. 2008;376:338–351. [DOI] [PubMed] [Google Scholar]
- 11. Bantle S, Keller S, Haussmann I, Auerbach D, Perriard E, Mühlebach S, Perriard JC. Tissue-specific isoforms of chicken myomesin are generated by alternative splicing. J Biol Chem. 1996;271:19042–19052. [DOI] [PubMed] [Google Scholar]
- 12. Steiner F, Weber K, Fürst DO. M band proteins myomesin and skelemin are encoded by the same gene: analysis of its organization and expression. Genomics. 1999;56:78–89. [DOI] [PubMed] [Google Scholar]
- 13. Agarkova I, Schoenauer R, Ehler E, et al. . The molecular composition of the sarcomeric M-band correlates with muscle fiber type. Eur J Cell Biol. 2004;83:193–204. [DOI] [PubMed] [Google Scholar]
- 14. Gentleman RC, Carey VJ, Bates DM, et al. . Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miyabara EH, Aoki MS, Soares AG, et al. . Thyroid hormone receptor-β-selective agonist GC-24 spares skeletal muscle type I to II fiber shift. Cell Tissue Res. 2005;321:233–241. [DOI] [PubMed] [Google Scholar]
- 16. Barreto-Chaves ML, Heimann A, Krieger JE. Stimulatory effect of dexamethasone on angiotensin-converting enzyme in neonatal rat cardiac myocytes. Braz J Med Biol Res. 2000;33:661–664. [DOI] [PubMed] [Google Scholar]
- 17. Fajardo G, Zhao M, Urashima T, et al. . Deletion of the β2-adrenergic receptor prevents the development of cardiomyopathy in mice. J Mol Cell Cardiol. 2013;63:155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grove BK, Kurer V, Lehner C, Doetschman TC, Perriard JC, Eppenberger HM. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J Cell Biol. 1984;98:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vinkemeier U, Obermann W, Weber K, Fürst DO. 1993 The globular head domain of titin extends into the center of the sarcomeric M band. cDNA cloning, epitope mapping and immunoelectron microscopy of two titin-associated proteins. J Cell Sci. 106(Pt 1):319–330. [DOI] [PubMed] [Google Scholar]
- 20. Moriscot AS, Baptista IL, Bogomolovas J, et al. . MuRF1 is a muscle fiber-type II associated factor and together with MuRF2 regulates type-II fiber trophicity and maintenance. J Struct Biol. 2010;170:344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. [DOI] [PubMed] [Google Scholar]
- 22. Petty KJ, Desvergne B, Mitsuhashi T, Nikodem VM. Identification of a thyroid hormone response element in the malic enzyme gene. J Biol Chem. 1990;265:7395–7400. [PubMed] [Google Scholar]
- 23. Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res. 2012;110:1646–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ojamaa K, Petrie JF, Balkman C, Hong C, Klein I. Posttranscriptional modification of myosin heavy-chain gene expression in the hypertrophied rat myocardium. Proc Natl Acad Sci USA. 1994;91:3468–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicolini G, Pitto L, Kusmic C, et al. . New insights into mechanisms of cardioprotection mediated by thyroid hormones. J Thyroid Res. 2013;2013:264387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pingitore A, Chen Y, Gerdes AM, Iervasi G. Acute myocardial infarction and thyroid function: new pathophysiological and therapeutic perspectives. Ann Med. 2012;44:745–757. [DOI] [PubMed] [Google Scholar]
- 27. Henderson KK, Danzi S, Paul JT, Leya G, Klein I, Samarel AM. Physiological replacement of T3 improves left ventricular function in an animal model of myocardial infarction-induced congestive heart failure. Circ Heart Fail. 2009;2:243–252. [DOI] [PubMed] [Google Scholar]
- 28. Mourouzis I, Mantzouratou P, Galanopoulos G, et al. . Dose-dependent effects of thyroid hormone on post-ischemic cardiac performance: potential involvement of Akt and ERK signalings. Mol Cell Biochem. 2012;363:235–243. [DOI] [PubMed] [Google Scholar]
- 29. Pask HT, Jones KL, Luther PK, Squire JM. M-band structure, M-bridge interactions and contraction speed in vertebrate cardiac muscles. J Muscle Res Cell Motil. 1994;15:633–645. [DOI] [PubMed] [Google Scholar]
- 30. Agarkova I, Perriard J. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. [DOI] [PubMed] [Google Scholar]
- 31. Muller A, Zuidwijk MJ, Simonides WS, van Hardeveld C. Modulation of SERCA2 expression by thyroid hormone and norepinephrine in cardiocytes: role of contractility. Am J Physiol. 1997;272:H1876–H1885. [DOI] [PubMed] [Google Scholar]
- 32. Takeuchi K, Minakawa M, Otaki M, et al. . Hyperthyroidism causes mechanical insufficiency of myocardium with possibly increased SR Ca2+-ATPase activity. Jpn J Physiol. 2003;53:411–416. [DOI] [PubMed] [Google Scholar]
- 33. Lowes BD, Minobe W, Abraham WT, et al. . Changes in gene expression in the intact human heart. Downregulation of α-myosin heavy chain in hypertrophied, failing ventricular myocardium. J Clin Invest. 1997;100:2315–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mercadier JJ, Lompré AM, Duc P, et al. . Altered sarcoplasmic reticulum Ca2(+)-ATPase gene expression in the human ventricle during end-stage heart failure. J Clin Invest. 1990;85:305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frey N, Barrientos T, Shelton JM, et al. . Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat Med. 2004;10:1336–1343. [DOI] [PubMed] [Google Scholar]
- 36. Molkentin JD, Lu JR, Antos CL, et al. . A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dorn GW 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest. 2005;115:527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lange S, Xiang F, Yakovenko A, et al. . The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. [DOI] [PubMed] [Google Scholar]
- 39. Hornemann T, Kempa S, Himmel M, Hayess K, Fürst DO, Wallimann T. Muscle-type creatine kinase interacts with central domains of the M-band proteins myomesin and M-protein. J Mol Biol. 2003;332:877–887. [DOI] [PubMed] [Google Scholar]
- 40. Keller A, Demeurie J, Merkulova T, et al. . Fibre-type distribution and subcellular localisation of α and β enolase in mouse striated muscle. Biol Cell. 2000;92:527–535. [DOI] [PubMed] [Google Scholar]
- 41. Lange S. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci. 2002;115:4925–4936. [DOI] [PubMed] [Google Scholar]
- 42. Krüger M, Sachse C, Zimmermann WH, Eschenhagen T, Klede S, Linke WA. Thyroid hormone regulates developmental titin isoform transitions via the phosphatidylinositol-3-kinase/ AKT pathway. Circ Res. 2008;102:439–447. [DOI] [PubMed] [Google Scholar]
- 43. Feng P, Li QL, Satoh T, Wilber JF. Ligand (T3) dependent and independent effects of thyroid hormone receptors upon human TRH gene transcription in neuroblastoma cells. Biochem Biophys Res Commun. 1994;200:171–177. [DOI] [PubMed] [Google Scholar]
- 44. Bodenner DL, Mroczynski MA, Weintraub BD, Radovick S, Wondisford FE. A detailed functional and structural analysis of a major thyroid hormone inhibitory element in the human thyrotropin β-subunit gene. J Biol Chem. 1991;266:21666–21673. [PubMed] [Google Scholar]
- 45. Belandia B, Latasa MJ, Villa A, Pascual A. Thyroid hormone negatively regulates the transcriptional activity of the β-amyloid precursor protein gene. J Biol Chem. 1998;273:30366–30371. [DOI] [PubMed] [Google Scholar]
- 46. Pérez-Juste G, García-Silva S, Aranda A. An element in the region responsible for premature termination of transcription mediates repression of c-myc gene expression by thyroid hormone in neuroblastoma cells. J Biol Chem. 2000;275:1307–1314. [DOI] [PubMed] [Google Scholar]
- 47. Santos GM, Afonso V, Barra GB, et al. . Negative regulation of superoxide dismutase-1 promoter by thyroid hormone. Mol Pharmacol. 2006;70:793–800. [DOI] [PubMed] [Google Scholar]
- 48. Syslak PH, Nathaniel EJ, Novak C, Burton L. Fetal alcohol effects on the postnatal development of the rat myocardium: an ultrastructural and morphometric analysis. Exp Mol Pathol. 1994;60:158–172. [DOI] [PubMed] [Google Scholar]
- 49. Hein S, Scheffold T, Schaper J. Ischemia induces early changes to cytoskeletal and contractile proteins in diseased human myocardium. J Thorac Cardiovasc Surg. 1995;110:89–98. [DOI] [PubMed] [Google Scholar]
- 50. Wang X, Li F, Campbell SE, Gerdes AM. Chronic pressure overload cardiac hypertrophy and failure in guinea pigs. II. Cytoskeletal remodeling. J Mol Cell Cardiol. 1999;31:319–331. [DOI] [PubMed] [Google Scholar]
- 51. Eppenberger ME, Schoenenberger R, Eppenberger HM. Myofibrillar M-line structure in normal and dystrophic hamster muscle. Muscle Nerve. 1984;7:304–311. [DOI] [PubMed] [Google Scholar]
- 52. Knöll R, Hoshijima M, Hoffman HM, et al. . The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. [DOI] [PubMed] [Google Scholar]
- 53. Pyle WG, Solaro RJ. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res. 2004;94:296–305. [DOI] [PubMed] [Google Scholar]
- 54. Solaro RJ. Remote control of A-band cardiac thin filaments by the I-Z-I protein network of cardiac sarcomeres. Trends Cardiovasc Med. 2005;15:148–152. [DOI] [PubMed] [Google Scholar]