Summary
The approximate 30 extant invertebrate phyla have diversified along separate evolutionary trajectories for hundreds of millions of years. Although recent work understandably has emphasized the commonalities of innate defenses, there is also ample evidence, as from completed genome studies, to suggest that even members of the same invertebrate order have taken significantly different approaches to internal defense. These data suggest that novel immune capabilities will be found among the different phyla. Many invertebrates have intimate associations with symbionts that may play more of a role in internal defense than generally appreciated. Some invertebrates that are either long lived or have colonial body plans may diversify components of their defense systems via somatic mutation. Somatic diversification following pathogen exposure, as seen in plants, has been investigated little in invertebrates. Recent molecular studies of sponges, cnidarians, shrimp, mollusks, sea urchins, tunicates, and lancelets have found surprisingly diversified immune molecules, and a model is presented that supports the adaptive value of diversified non-self recognition molecules in invertebrates. Interactions between invertebrates and viruses also remain poorly understood. As we are in the midst of alarming losses of coral reefs, increased pathogen challenge to invertebrate aquaculture, and rampant invertebrate-transmitted parasites of humans and domestic animals, we need a better understanding of invertebrate immunology.
Vive la différence – invertebrates are a heterogeneous lot
One of the most remarkable developments in the recent history of immunology has been the discovery that the internal defense systems of invertebrates, vertebrates, and even plants share striking similarities. The similarities are most notably among signaling receptors of the Toll/interleukin-1 receptor family and in the signaling pathways involved in orchestrating an innate immune response. For very good reasons, this discovery has been big news: it has been important in revealing how a model invertebrate like Drosophila melanogaster defends itself from bacteria and fungi (1, 2); it is pointing the way to the discovery of a previously unknown and important innate defense system in mammals (3, 4), and it is revealing how innate immune responses regulate activation of adaptive immune responses (5). It is no wonder there has been an emphasis on stressing the similarities between vertebrate and invertebrate innate immune systems.
One goal of this article was to accentuate the fact that invertebrates are a heterogeneous lot and in aggregate remain relatively poorly understood from an immunological point of view. All of the 30 or so major lineages of animals that we formally recognize with a phylum name, with one exception, are exclusively invertebrate. The one exception, the Chordata, is also comprised in part of invertebrates. Many of these phyla were recognizably separate by the Cambrian explosion between 520 and 530 million years ago. The ages of the last common ancestors among major animal lineages, such as protostomes and deuterostomes and ecdysozoans and lophotrochozoans, are certainly considerably older, given that the origin of animals is 660 to 720 million years ago (6). Furthermore, invertebrates have proceeded along several disparate evolutionary trajectories: colonial or solitary; benthic or pelagic; short or long lived; and marine, freshwater, or terrestrial. Many invertebrates engage in elaborate and obligatory relationships with symbionts (7–9), and at times, it is hard to know where one partner stops and the other begins. Buchnera, obligate bacterial intracellular mutualists of aphids, are but one example. These bacteria exhibit a pattern of gene loss or reductive evolution that seems to complement similar processes occurring in its hosts (10).
Invertebrates encounter all the usual sorts of challenges to self-integrity. Their habitats are typically laden with infectious agents: viruses, bacteria, fungi, protists, and other animals. Many invertebrate groups have been in existence so long that they harbor their own specialized lineages of pathogens. For example, mollusks (especially gastropods) support the development of approximately 18,000 nominal species of digenetic trematodes that do great damage to these hosts (11). Invertebrates also suffer from neoplasia (12), and in addition, many substrate-inhabiting invertebrates have to compete for space with other members of their own species, a process that may be accompanied by aggressive encounters along lines of contact and by germ or somatic cell parasitism, a process whereby the genetic material of one organism invades and potentially takes over the body of another (13).
So, the term invertebrate embraces a spectacular diversity of body forms and lifestyles, reflective of long, independent evolutionary histories. Invertebrates continually confront an especially broad variety of challenges to self-integrity. It is important that we strive to understand how invertebrate body defenses operate. The hard coral cover of Caribbean reefs has been reduced by 80% in the past three decades (14), and compromised coral defense responses may well be part of the explanation. Invertebrate-transmitted diseases, such as malaria and schistosomiasis, remain common in the developing world (15, 16). Mass-reared invertebrate species, such as shrimps that are proving to be highly vulnerable to virus infection, provide an increasingly important source of human food (17). Although there are certainly important immunological commonalities among the diverse forms of invertebrates, we must retain open minds for differences and for the completely unforeseen. This point is underscored by recent genome studies that show that even relatively closely related invertebrates seem to have adopted quite different approaches to internal defense.
Comparisons among invertebrate defense systems from genome studies
With the completion of a genome sequence, we can begin to glimpse the realms of the possible with respect to an animal’s defense capabilities. The sequence data alone leave much to be deciphered with respect to gene regulation and development, but still some striking points emerge, as shown by the following three brief comparisons.
The first example is of two ecdysozoans, the nematode Caenorhabditis elegans and the dipteran insect D. melanogaster. The study of invertebrate innate defense mechanisms has been greatly invigorated by use of the powerful Drosophila model, which yielded important insights, especially regarding the Toll-signaling pathway (18). The use of C. elegans to dissect immune pathways is in its infancy, but it will no doubt also prove to be a versatile model (19). Although several Toll pathway homologs are present in C. elegans, worms with mutations in several components of the pathway are not significantly affected relative to wildtype worms in response to pathogens (20). At least eight Toll receptors are present in D. melanogaster. Only one Toll-like receptor (TLR) (tol-1) gene, which has not been shown to have a direct role in anti-pathogen responses, has been found in C. elegans (21). In C. elegans, the p38 mitogen-activated protein kinase, programmed cell death and tissue growth factor-β-like pathways are involved in innate immunity (19). The worm also has a large number of C-type lectins that may play a role in pathogen recognition (22, 23), whereas the recognition molecules that have drawn most attention in Drosophila are members of a family of peptidoglycan-recognition proteins (24). Although both are ecdysozoans, it will hardly be surprising if the defense responses of these two model organisms continue to look so different. Their respective phyla have been separate for a long time. Also, a soil dwelling, bacteria-ingesting worm is bound to experience quite different immune challenges from a flying insect that thrives on yeast growing on fermenting fruit.
What is more surprising are the differences that have come to light between two members of the same insect order (Diptera), D. melanogaster and the mosquito Anopheles gambiae (25, 26). The two lineages they represent have been separate for about 250 million years, and the blood-feeding habits of the latter impose unique demands, including exposure to pathogens like malaria parasites and arboviruses imbibed with blood meals. Whereas the Toll signal transduction pathway was found to be conserved relatively, surprisingly large differences were noted between the two insects with respect to the types of putative recognition molecules they possess. In general, a deficit of 1 : 1 orthologs and an over-representation of gene expansions were noted, suggestive of quite different responses to the circumstances in which they live. For example, the Anopheles genome contains a large expansion of genes (about 58 genes) encoding proteins with homology to the C-terminal domain of fibrinogen relative to the Drosophila genome (about 13 genes) (26). Fibrinogen domains have been implicated in non-self recognition in other invertebrates and vertebrates (27–30), and fibrinogen may play a role in hematophagy in mosquitoes by controlling bacteria ingested with blood meals or in defense against Plasmodium parasites (31).
A final example from genome studies is provided by a comparison of immunity-related genes from the ascidian Ciona intestinalis, a non-vertebrate chordate, and other members of the Chordata, namely the vertebrates (32). None of the pivotal genes implicated in the adaptive immune response of vertebrates, such as those encoding T-cell receptors, immuno-globulins, or major histocompatibility complex class I or II molecules, were found in Ciona, although divergent orthologs may still prove to be present. Also lacking in Ciona were immunoproteosome-specific genes, suggesting that this invertebrate lacks an antigen-presenting system for T cells. A variety of genes likely to be involved in innate immunity were present in Ciona, including complement-like, lectin, and TLR genes. Even among putative genes of innate immunity, novelty was found; protein domains expected from such genes were found in unique combinations in Ciona (32). Jiang and Doolittle (33) searched the Ciona genome for 26 proteins involved in clotting or fibrinolysis in vertebrates, and they found no genuine orthologs, although paralogs and constituent domains were found. Gene duplication and shuffling of key modular domains are hypothesized to have occurred, accounting for the appearance of vertebrate-type blood coagulation during the 50–100 million year interval that separates the origins of ascidians and vertebrates.
Such comparisons lead us to expect that immune systems will show strong lineage-specific evolution based on processes such as gene duplication and domain shuffling, and that the specific ecological circumstances encountered will have a relatively large effect on the defenses employed and will transcend phylogenetic relatedness to some extent in dictating immune characteristics. Even apparently conserved and homologous systems, such as the nuclear factor-κB-signaling pathway of the innate immune systems of insects and mammals, seem to have evolved to acquire immune function independently in the two groups (34), further indicating the diverse paths taken to achieve internal defense.
How do invertebrates survive without an adaptive immune system?
An oft-asked question is how invertebrates survive in a pathogen-laden world without an anticipatory, specific, and lymphocyte-based immune system capable of clonal expansion, memory, and fine-tuned responses. Some invertebrates, like bivalves or colonial cnidarians, routinely live for decades (35), filtering or trapping all sorts of microorganisms from the water in which they live, including potential pathogens. So why is it that fast-evolving pathogens do not produce variants that eventually overcome the relatively static defenses of such hosts?
Part of the answer surely lies in the fact that innate defense systems, including the use of RNA interference (RNAi), pattern-recognition receptors (PRRs), anti-microbial peptides (AMPs), phagocytic cells, production of toxic oxygen and nitrogen metabolites, and melanization pathways, are brutally effective. The more we learn about these systems, the more we appreciate their capabilities, including the ability to recognize and inactivate viruses (36). The vast majority of multicellular organisms seem to make do with these capabilities, and hence the argument goes that they must be effective. It is further argued that PRRs recognize antigenic features, such as chitin, peptidoglycan, or lipopolysaccharides (LPSs), which are fundamental to microbial existence and simply cannot be modified. Similarly, AMPs must effectively interfere with some vital function or deliver such a lethal hit that invertebrate pathogens simply cannot recover. However, these arguments seem overly simplistic, given the extreme inventiveness of viruses, bacteria, and fungi in acquiring new genetic information, in generating variant surface antigens and novel mechanisms of pathogenicity, and in establishing persistent infections in hosts as diverse as plants and vertebrates (37–39). There is no reason to expect these capabilities would be suspended in invertebrate hosts. As just one example, the ehrlichia Anaplasma marginale establishes persistent infections in ticks just as it does in cattle. A surface antigen known to be varied in cattle is also varied in the ticks, suggesting selective pressures occurring in the environment of the tick (40).
Another part of the answer is that there is likely to be more to invertebrate defenses than readily meets the eye. The development of microarrays will enable investigators to detect more systematically additional components of the innate immune response of which we are presently ignorant. For example, over 230 genes were shown to be induced in adult Drosophila following exposure to microbial infection, many with unknown functions (41). Below, we explore three phenomena not generally considered that might contribute significantly to invertebrate defense: collaboration with symbionts, mosaicism, and production of immune molecules that show a surprising degree of diversity.
If you can’t beat ‘em, join ‘em – collaborating with symbionts in achieving defense
Multicellular animals evolved in a world in which bacteria and other microorganisms were already abundant and hence would have had pervasive interactions with them. Among the gamut of possible associations were some that were mutualistic, either for purposes of nutrition or defense from predators, competitors, or pathogens. Microbial symbionts are hypothesized to be the source of many of the interesting biologically active compounds isolated from marine invertebrates (42). One potential benefit to an animal of mutualistic associations with bacteria is that such bacteria could compete directly for space and resources and thus prevent colonization of the animal’s body by potential pathogens. The adaptability of virulence mechanisms of bacterial pathogens could have been matched by equally adaptable defenses produced by mutualist prokaryotes. Thus, the rather static innate defenses of animals could have acquired a much more dynamic and responsive component. The presence of microflora on both external and internal epithelial surfaces is well known to prevent pathogen colonization (43). The surfaces of marine invertebrates are known to harbor species-specific microbial communities that are distinct from the surrounding environment (44). Another context in which mutualists are enlisted is in the deliberate coating of externally brooded eggs of the shrimp Palaemon macrodactylus with Alteromonas bacteria. The presence of the bacteria protects the eggs from overgrowth and destruction by Lagenidium callinectes, an oomycete crustacean pathogen (45). Experimental removal of bacterial symbionts is followed by rapid pathogen-mediated destruction of egg masses. Reconstitution of the symbiont on egg masses restored resistance to the oomycete. Several species of squid and cuttlefish have a special organ called the accessory nidamental gland associated with the female reproductive system. This gland houses symbiotic bacteria that colonize the layers of the cephalopod egg and protect them from overgrowth (46, 47).
These collaborations suggest that the animal partner can differentiate among different types of microbes and select or favor the mutualists to form a so-called exclusive contract (48), recruit them to specific organs of residence, exclude other microbes from this site, and then prevent the favored symbiont from overrunning the body (9). Although it has nothing to do with defense from pathogens, one system that has been relatively well characterized is the association between Vibrio fischeri and the squid Euprymna scolopes. A virtual monoculture of the bioluminescent bacterium occupies the light-emitting organ of the squid, which is used to project false moonlight downward, thus preventing the formation of a shadow that could be detected by predators (48). Phagocytic defense cells of the squid police the lumen of the light organ and interact with, yet do not eliminate, V. fischeri. Extensive crosstalk between bacteria and tissue is evidenced by changes in the proteome of the squid that only occur in the presence of V. fischeri (49). The tissues that contain bacteria have high levels of a halide peroxidase, which probably functions not only in controlling V. fischeri but also in protecting squid from potential pathogens (50). Similar arrangements are likely to occur in the associations of solemyid clams and vestimentiferan worms with their chemoautotrophic bacterial symbionts (51, 52). The presence of specific, selective, and well-managed bacterial populations living within invertebrates suggests the presence of sophisticated recognition systems that go well beyond the capabilities of PRRs.
Giant bivalves of the genus Tridacna point out an opposite kind of effect with respect to internal defense that might result from an intimate association with mutualist symbionts. Tridacnids harbor photosynthetic dinoflagellate zooxanthellae living in a unique tubular system that arises from the clam’s stomach and extends into the mantle (53). Compared to other bivalves, symbiont-containing tridacnids contain a distinctive type of hemocyte, the morula cell (54), that seems to engage with the symbionts in as yet uncharacterized interactions (55). Furthermore, hemocytes of Tridacna crocea have a diminished ability to produce superoxide anion, nitric oxide, and phenol-oxidase, relative to hemocytes of bivalves lacking zooxanthellae (56). This diminished cytotoxic capability may be critical to allowing the zooxanthellae to establish and persist in tridacnids, but it may make tridacnids vulnerable to parasites that exploit this mutualism. For example, T. crocea is known to be infected with an apicomplexan parasite that lives within the hemocytes (57). In the case of Tridacna, one wonders whether the symbiotic dinoflagellates, in addition to providing photo-synthetic products, might also contribute to defending their hosts from other pathogens. Similar considerations might apply to corals. Do corals that have undergone bleaching (lost their symbiotic zooxanthellae) become more vulnerable to pathogens because the protective contributions of their zooxanthellae have been lost?
Does mosaicism influence invertebrate susceptibility to pathogens?
Long-lived hosts, like bristlecone pine trees or some invertebrates, would seem to be at a profound disadvantage in defending themselves from fast-evolving pathogens. A factor that might improve the odds is the possibility that over time, these initially genetically homogeneous hosts become diversified due to the accumulation of spontaneous somatic mutations, which is called the genetic mosaicism hypothesis (58, 59). Thus, leaves on one branch of a plant might be very susceptible to a particular herbivore, but another branch is not, simply because the genes producing anti-herbivore chemicals have been mutated. The same considerations might apply to a colonial invertebrate with a modular body plan for which mosaic colonies are known to occur in nature (60). Clones produced by asexual reproduction, because they have gone through many cycles of division and have had ample opportunities for spontaneous mutation, may not be as homogeneous as we once thought. Even in non-colonial animals, genes encoding soluble non-self recognition molecules might mutate differently in various parts of the body, and all the resultant gene products might contribute to the organism’s aggregate systemic defense, if they were dispersed throughout the body via the circulatory system. The extent to which somatic mutations might actually influence the ability of animals to resist pathogens is poorly known. As noted by Gill et al. (59), quantitative or qualitative differences in defense may permit the long-lived host to minimize the impact of its destructive enemies. Although many animals follow the so-called Weismann doctrine (61) and have a strict separation of germinal and somatic lines such that mutations in the latter do not affect the former, members of at least 19 animal phyla do not (59). Included among animals for which mutations in somatic tissues could be passed on to progeny are cnidarians, platyhelminths, bryozoans, annelids, and entoprocts.
Although background mutation alone could by itself generate considerable somatic diversification in a long-lived animal (62), it is also possible that specialized mechanisms exist to increase the mutation rate in response to pathogens. Following exposure to the oomycete Peronospora parasitica, the vascular plant Arabidopsis thaliana responds by increasing the frequency of somatic recombination. This finding prompts the speculation that possible substrates for somatic recombination might be the large numbers of pathogen-resistance genes known to be present in plants (63). The result could be the generation of new pathogen specificities, some of which might be transmitted to progeny. Kovalchuk et al. (64) similarly noted a threefold increase in homologous recombination frequency in tobacco plants exposed to tobacco mosaic or oilseed rape mosaic virus. The results suggested the presence of a systemic recombination signal that is stimulated by virus infection and that can move through the plant, triggering potentially heritable genomic changes. The utility of such a response could be the generation of new specificities in pathogen-resistance genes.
Production of diverse immune molecules by invertebrates – some theoretical and experimental views
It is not known whether invertebrates have mechanisms of enhanced somatic recombination upon pathogen exposure similar to those coming to light in plants. If such a mechanism exists to generate pathogen-recognition molecules, there is an obvious advantage with respect to increasing the likelihood of binding a rare pathogen epitope or of matching a pathogen that is undergoing antigenic variation. There is also the obvious disadvantage that a recognition molecule that recognizes self could be generated. One possible response would be to eliminate cells producing self-reactive recognition molecules, but perhaps a more realistic scenario would simply be to use a self-recognition system to override any signal provided by a mutated recognition molecule that happened to bind to a self cell. Another reason to wonder whether the ability to produce somatically diversified recognition molecules would be advantageous is simply whether or not a sufficient quantity of the molecules could be produced to be of any value. Without the ability to produce expanded clones of cells that generate unique and relevant recognition capabilities, would somatic diversification be worth the effort?
Under appropriate circumstances, which we explore below with the aid of a mathematical model, the answer is affirmative: diversification, even in the absence of clonal selection, can be advantageous. First, however, we shall re-examine the potential role of diversification in innate immunity with respect to its much more familiar role in adaptive immunity.
In adaptive immunity, diversification is extremely thorough, generating receptors that are essentially random, covering the whole space of potential epitopes. This production is consistent with their role as recognition molecules, which must identify non-self and signal its presence to the rest of the immune system. Holes in the repertoire, under these circumstances, could represent fatal vulnerabilities. Given this state of affairs, the number of cells bearing receptors appropriate for any given epitope will be very small, and amplification by clonal selection will be necessary to accomplish this task. Finally, such expansion could be devastating if directed against host cells. Tolerance to self, another task of selection, becomes another indispensable corollary of diversification.
There is another role of immunoglobulins that may be more relevant in the context of invertebrate immunity. They are effector-enhancement molecules that link effectors of innate immunity, such as phagocytes and complement components, to microbes. In this context, they are not recognition molecules. Indeed, the effectors must already be present and activated. Serum immunoglobulin by itself is not particularly dangerous and is cleared effectively in a short time, so that self-reactivity, if it should arise as a side effect of infection, will not persist.
Suppose there is such a bridge, or effector-enhancement molecule, in an invertebrate. When, if ever, would diversification prove advantageous? Random changes will likely lead to enhancement of effectiveness in a minority of molecules and no change or diminution in the rest. When would this particular tradeoff prove worthwhile? Let us formalize this question and state it mathematically. For the case of a receptor, the on- and off-rates for binding to its ligand are key determinants of its effectiveness. We shall focus attention on the off-rate or its inverse, the mean lifetime of the receptor–ligand complex, τ, but similar arguments can be developed for multiple traits as well. The concentration density of molecules with inverse off-rate τ is designated by c(τ), and the effectiveness of molecules with inverse off-rate τ is designated by f(τ). Effectiveness is intended to be a very general term standing in for any number of possible quantitative measures of immunological competence. The mathematical definition of the effectiveness function will depend on the detailed mechanism of the process in which the molecules participate. We provide one such possible definition below. For the sake of simplicity in the argument, if we assume that the molecules act independently of each other, the total effectiveness is given by the following integral expression:
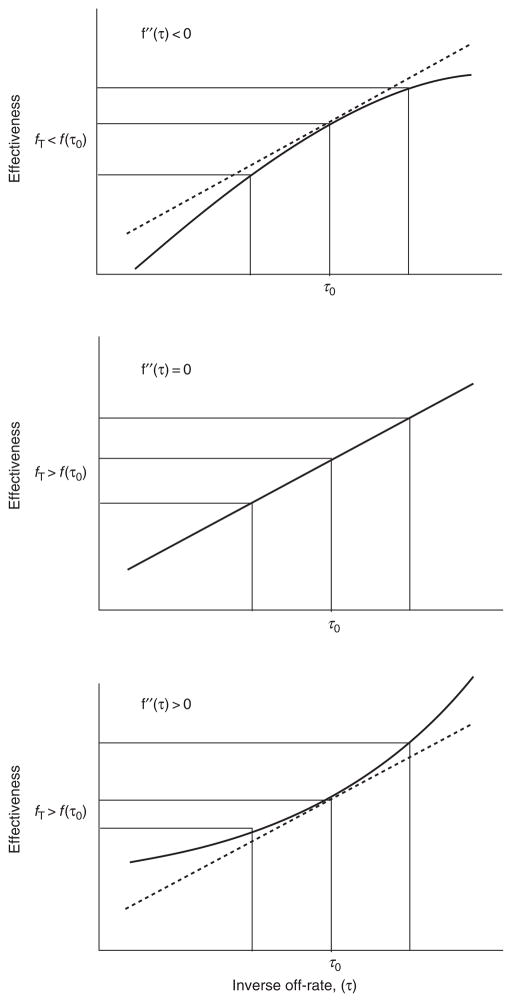
This formula can be regarded as a weighted average effectiveness, with weights being the relative concentrations. Random diversification corresponds to increasing the spread of the distribution c(τ), without changing its location. The variance of τ increases, but the mean remains unchanged. A population with no diversity has variance zero. The question we ask is: when does increasing the variance from zero increase the total effectiveness? That is, when is the derivative of fT with respect to the variance of τ positive? Under very mild assumptions, the answer is that fT increases with the variance, when the second derivative of f with respect to τ (evaluated at the mean value τ0) is positive (Fig. 1).
Fig. 1. An illustration of the conditions under which diversification without subsequent selection is likely to have a positive effect.
The abscissa is the inverse off-rate and the ordinate is the effectiveness of the response. Suppose that τ0 is the current value of the inverse off-rate. The three panels show the different behaviors that can occur in the neighborhood of τ0. The top panel shows the case of a negative second derivative. Note that this condition diminishes the advantage of increasing τ and exacerbates the disadvantage of decreasing τ. Thus, the expected value in the presence of diversification, fT, is less than the current value of the effectiveness f(τ0). The middle panel shows the case where the second derivative is zero, where potential advantage is exactly balanced by potential disadvantage, so that fT = f(τ0). Finally, the bottom panel shows the case of particular interest: positive second derivative, which enhances the advantage of increasing τ and moderates the disadvantage of decreasing τ. Under these circumstances, the expected value under random diversification is greater than the present value, so that diversification becomes a good bet, even without selection.
In many common situations, this second derivative is negative, due to the prevalence of saturating functions in biology and the fact that these functions must have negative second derivatives (at least somewhere and maybe everywhere). There is an important class of processes where the second derivative is generically positive. These are compound processes in which the dissociation of the molecule in question can occur at several places in the chain of reactions. Suppose, for example, that a process involves the following sequence of reactions: (i) the host defense molecule (L) binds/unbinds receptor (R) on the microbial surface; (ii) the host effector (E) binds to the host defense molecule on the surface; and (iii) the host effector neutralizes the microbe. The chemical reaction scheme below illustrates this process:
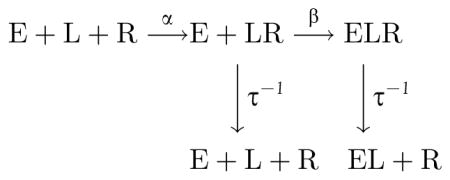
A natural measure of effectiveness for this process is the equilibrium number of complete ELR complexes. A straightforward computation yields the following:
The second derivative is positive for as long as τ is less than some threshold value. That is, if the molecule already binds very well, then diversification alone will not increase the effectiveness. If it binds only moderately well or poorly, random diversification could, even without subsequent selection, improve its performance.
One can further elucidate this observation by taking it to its natural limit. Suppose a process absolutely requires an inverse rate constant above some threshold value for it to have any effectiveness at all and that τ0 is below that rate. Then in the non-diversified state, the process is completely ineffective. All diversification has to do is produce a few molecules with τ> τ0 to improve this situation.
This result has salutary implications for the evolutionary origins of diversification mechanisms in immunity. Such mechanisms can arise even in the absence of (and therefore prior to) sophisticated mechanisms for clonal expansion and tolerance induction, which may then have arisen later and co-evolved alongside a progressively more extensive and tightly regulated diversity-generating mechanism.
The existence of active diversification in invertebrates could help resolve an additional puzzle in the evolution of immunity. It seems as if all mechanisms of somatic diversification arose at the same point, some time between the divergence of the vertebrates and of the cartilaginous fishes, as all jawed vertebrates possess V(D)J recombination, TdT, and somatic hypermutation or gene conversion, or both (65). This sudden burst of genetic innovation has been dubbed the big bang of immunology (66, 67). However, if somatic mutation arose prior to and independently of the acquisition of the recombination activation genes (RAG-1 and RAG-2) (68), then the apparent coincidence loses some of its mystery. Perhaps, a pre-existing host-defense mutation mechanism was simply co-opted and enhanced in the novel context of obligatory combinatorial diversification.
Several recent experimental findings suggest that the above may be more than just a theoretical exercise. Now that molecular methods for cloning and sequencing have become more widely available, increasingly detailed analyses have disclosed a diversity of factors involved in the innate immunity of a variety of organisms. Counter to expectations based on the model of pattern recognition in which a limited set of PRRs sufficiently recognizes and responds to groups of pathogens (69), factors that function in the context of innate immunity have been shown to display sequence diversity in vertebrates and even more so in invertebrates. The innate immune system of rainbow trout employs three variants of complement factor C3 (70). Carp and other teleost species also have isoforms of C3 and additional complement factors. These additional iso-forms are hypothesized to increase the range of epitopes that can be recognized (71, 72). Diversity has also been documented from invertebrate deuterostomes. Five different families of diverse V-region-containing chitin-binding proteins that may serve in non-self recognition were described from the proto-chordate Amphioxus (73). Among echinoderms, the genome of the sea urchin Strongylocentrotus purpuratus was predicted to harbor as many as 1200 scavenger receptor cysteine-rich genes, for which expression patterns differ between individuals (74, 75). Additionally, extensive diversity was evident in a group of related sequences that made up 94% of clones recovered from a cDNA library derived from LPS-stimulated coelomocytes of S. purpuratus (76). At the other end of the spectrum of animal phylogeny, extensive diversity of receptors for non-self recognition governing fusion/rejection interactions and internal defense occurs in sponges (Geodia cydonium) (77, 78) and colonial cnidarians (79). As expected, similar diversity occurs also in the other major lineage of animals, the protostomes. The crustacean Penaeus monodon (Ecdysozoa) expresses variant sequences of AMPs such as peneaidins and crustins (80–83). A representative of the Lophotrochozoa, the gastropod Biomphalaria glabrata, expresses diverse parasite-reactive hemolymph lectins in response to infection. This response includes fibrinogen-related proteins (FREPs) that are comprised of N-terminal immunoglobulin superfamily (IgSF) domains and fibrinogen-related sequences at the C-terminus (27). FREP sequences display diversity due to point mutations and alternative splicing. Thirteen different FREP gene subfamilies have been recognized, based on the criterion that members of a subfamily share 86% or higher sequence identity of the N-terminal IgSF sequence (29, 76, 84, 85).
We are at an interesting juncture with respect to interpretation of the diversity revealed by such studies. Genome sequences are not yet available for most of the organisms used, and many of these observations have been based on medium-to-large-scale application of polymerase chain reaction (PCR)-based amplification and sequencing methods. Nucleotide misincorporations occur even when DNA polymerases of the highest fidelity are used (86). Furthermore, amplification of a target sequence that is a member of a family of related genes can potentially generate sequences comprised of portions of the different family members, a PCR artifact known as template switching (87). The observed sequence diversity of PCR amplicons can be falsely increased. These nagging sources of artifact pose a distinct challenge to investigators seeking to document diversity of immune molecules in animals that may otherwise be poorly known. Nonetheless, several laboratories using a variety of methods with quite different experimental subjects are converging on the discovery of surprisingly diverse immune molecules, strongly suggesting that at least some part of the diversity is real. These observations need not be considered inimical to the concept of recognition of pathogen-associated molecular patterns mediated through a set of PRRs of limited diversity. Rather, variable recognition or effector molecules could play an important complementary role in innate defense.
Invertebrates and viruses – a surprising gap in our knowledge
Like all organisms, invertebrates are plagued by viruses that not only cause mortality but also inflict subtle fitness penalties (88). Some of these viruses are of great concern, because they directly threaten the continued vitality of enterprises such as shrimp farming (17, 89) or commercial harvesting of edible bivalves (90). Others, such as the baculoviruses, have been used in the biocontrol of lepidopteran pests (91) or have been developed as important expression vectors of eukaryotic proteins. Over 500 known varieties of arboviruses are exceptionally versatile in that they can infect both cells of their invertebrate vector host and of a vertebrate host (92). One such virus, West Nile virus (93), is currently epidemic in the United States. Although many invertebrate viruses have made themselves conspicuous in one context or another, the overall extent of our knowledge of the diversity and biology of invertebrate viruses is rather scant. We are also surprisingly ignorant about how invertebrates control or eliminate viruses. Vertebrates deal with viruses in multiple ways. Viral infection results in the induction of interferons, which in turn upregulate the transcription of many genes that protect cells from damage and death. Infection may also be countered by the activation of potentially lytic natural killer (NK) cells, production of antibodies to block viral receptors and to promote phagocytosis by opsonization, and use of cytotoxic T cells to kill virally infected cells.
NK cells, antibodies, and cytotoxic T cells are all lacking in invertebrates. No invertebrate has been shown to have obvious homologs of interferon (65, 94), although an important component of a pathway activated by interferon in mammals, the enzyme 2′,5′-oligo A synthetase, has been reported in sponges (95, 96). In mammals, this enzyme activates an RNA-digesting enzyme that can destroy viral RNA and also reduce viral and host cell mRNA levels. The extent to which it functions in protection from viruses in sponges is not known. So how do invertebrates respond to viruses?
Work with model invertebrates and their anti-viral responses has thus far not proceeded from the perspective of basic immunology. These investigations are undertaken mostly from the perspective of trying to understand the basic raison d’être for the phenomenon of RNAi and its role in gene silencing (97). It is not the purpose of this article to review RNAi but rather to try to place it in the general context of invertebrate immunology. RNAi is a sequence-specific response initially triggered by double-stranded RNA (dsRNA) that subsequently targets any cytoplasmic RNA species sharing homology with the triggering sequence. It is conserved across plants, some fungi, and animals, and it is increasingly viewed as a basic trans-eukaryotic genome defense against molecular parasites like viruses and transposons (37, 98, 99). Work with both C. elegans and Drosophila has contributed significantly to unraveling the general properties of RNAi, and an anti-viral context has been shown specifically for Drosophila (100). These authors showed that flock house virus, an RNA virus that can infect both vertebrate and invertebrate hosts, is both an initiator and target of RNAi in Drosophila host cells. It encodes the protein B2 that also operates as an RNA-silencing suppressor, suggesting that RNAi is a potent host defense mechanism that must be circumvented by the virus (100). In general, the role of RNAi as a viral defense mechanism is better understood in plants than animals. Several plant viruses are also known to have elaborated a variety of RNAi counter measures (37).
Many viruses of invertebrates have either single-stranded RNA or dsRNA genomes, and as noted by Ahlquist (101), RNA viruses replicate their genomes through complementary strands, resulting in dsRNA replication intermediates that make them potentially vulnerable to RNAi. For instance, positive sense single-strand RNA viruses from plants have shown to be vulnerable to RNAi (102). Thus, one would expect RNAi to be an important defense mechanism of invertebrates against potentially all RNA viruses. But what about DNA viruses? Some of the important DNA viruses of invertebrates are iridoviruses, baculoviruses, entomopox viruses, and densoviruses. In general, our knowledge of the relationships between RNAi and DNA viruses is still rudimentary. Some DNA viruses of plants have been shown to be silenced by RNAi, possibly because dsRNA was fortuitously produced during their transcription or amplification (37). Other intracellular anti-viral mechanisms, such as the production of RNA-editing enzymes that mutate viral genomes (103), may also be operative in invertebrates.
Given that several arboviruses, like yellow fever, dengue, and West Nile viruses, are important human pathogens, it would be especially valuable to know more about their biology within the cells of their mosquito hosts. Adelman et al. (104) pre-exposed Aedes aegypti to Sindbis virus engineered to contain dengue sequences, and Ae. aegypti were subsequently shown to be highly resistant to challenge with dengue isolates containing homologous sequences, an effect attributed to RNAi. Similar results were obtained by Caplen et al. (105) using Semliki Forest virus and dengue in mosquito cell lines. These studies suggest novel ways to prevent replication of arboviruses in arthropod vectors, such as development of transformed mosquito lines that express viral RNAs that would stimulate RNAi and effectively prevent natural infection with viruses containing homologous sequences (106).
Although it seems very likely that RNAi is an important defense against viruses, do invertebrates have mechanisms other than intracellular ones to protect themselves from viral attack? The state of our very limited knowledge is perhaps best exemplified with respect to anti-viral immunity in marine crustaceans. There are about 20 different viruses that collectively comprise the greatest threat to the world’s shrimp industry (17, 107), yet we know very little about anti-viral mechanisms in shrimps. White spot syndrome virus (WSSV), which has had the greatest impact on shrimp culture, causes a sharp reduction in hemocyte counts of P. monodon, but the underlying reasons for this reaction are unclear (108). Pan et al. (109) found that the tissue extracts obtained from blue crabs (Callinectes sapidus), shrimp (Penaeus setiferus), and crayfish (Procambarus clarkii) possess anti-viral activities that inhibit Sindbis, vaccinia, vesicular stomatitis, mengo, banzi, and poliomyelitis viruses. The blue crab inhibitor prevents viruses from attaching to cells and has a lipid component, but beyond that it remains uncharacterized. Although we are ignorant of the mechanisms, antiviral defenses are clearly present in shrimp and can be enhanced by selection, as shown by the presence of a shrimp strain specifically resistant to taura syndrome virus (110). Also, it has been shown that shrimp can be protected from virulent WSSV by pre-exposure to a generally avirulent infectious hypodermal and hematopoietic necrosis virus, but the underlying mechanism for the interference is not known (111).
Studies of TLRs in mammals have shown that TLR3 recognizes dsRNA and thus could have an anti-viral role (112). TLR9 might also be involved in anti-viral defense (113). There are no data to suggest that invertebrates have comparable receptors capable of recognizing viruses. Also lacking is evidence that defined lectins with anti-viral activity exist in invertebrates. An important mechanism for control of viral infections in invertebrates could be induction of apoptosis before the virus has a chance to complete its development (114), but if so, the molecules involved in detecting viral infection are not known.
Some specific questions
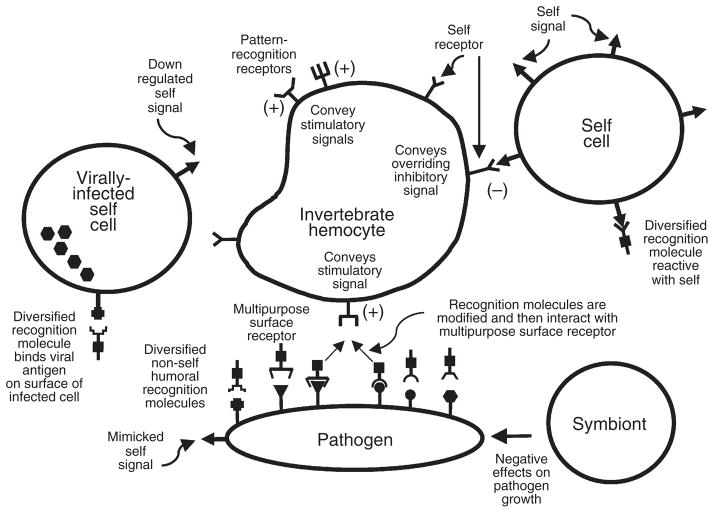
Fig. 2 provides a more extravagant model than normally envisioned for the operation of invertebrate immune systems. This schema is presented more for the purpose of highlighting some of the specific questions that await resolution rather than to promulgate it in preference to simpler and perhaps more fundamentally correct models. As we emphasized above, it is not realistic to expect that all invertebrate immune systems function in the same way, and hence some of the questions we ask below may apply to some groups more than others.
Fig. 2. A general model indicating hypothetical interactions between the major component of the invertebrate defense system, a multipurpose hemocyte, and self, non-self (pathogen), and virally infected self cells.
This model emphasizes the recognition phase of the interaction and does not consider the signaling pathways or effector mechanisms employed by the hemocyte in actually killing a pathogen. Note that the version presented involves a system of dual recognition, featuring both detection of a specific self signal, which is found on self cells, and non-self signals, which are on pathogens or virally infected self cells. Pathogens are assumed to be capable of generating diversified antigens. Receipt of a self signal conveys an inhibitory signal to the hemocyte and downregulates a response, analogous to natural killer (NK) cells, although no strict homology with NK self receptors is implied. In addition to a self receptor, two different kinds of non-self receptors are illustrated, both of which convey stimulatory signals to hemocytes. The response of the hemocyte might depend on the integration of both inhibitory and stimulatory signals. Non-self receptors of the first category are the conventional pattern-recognition receptors such as the surface-associated peptidoglycan-recognition protein receptors of Drosophila (24). These would interact with a repetitive pattern on a bacterium, fungus, or possibly a virus. Also shown is a hemocyte-associated multipurpose receptor that interacts with a set of diversified humoral non-self recognition molecules. These humoral factors might be diversified by processes such as somatic mutation and gene conversion. Once these recognition molecules have bound a pathogen-associated antigen, they are either modified or undergo a conformational change that allows them to interact with the multipurpose hemocyte receptor. Note that some of these diversified recognition molecules might react with components on self cells, creating the possibility of autoreactivity. This tendency could be overridden by simultaneous receipt of the self signal by the self receptor. Note also that the pathogen may bear a mimicked self signal that enables it to escape detection. Another pathogen strategy not shown here is the production of factors that directly harm or interfere with defense cells or molecules. Self cells infected with a virus might either undergo downregulation of self signals or upregulation of membrane-associated viral antigens, tipping the balance in favor of destruction of the infected cell. Depending on the invertebrate involved, symbionts may also play a role in producing anti-microbial peptides or other biologically active compounds that inhibit pathogen growth.
Do invertebrates rely on identification of self as a way to prevent self-reactivity and to help delineate non-self?
NK cells of vertebrates have receptors that transfer inhibitory signals to the cell when a self-signal is received. The model in Fig. 2 illustrates self signals that interact with self receptors to inhibit hemocyte autoreactivity. Although it is clear that invertebrates can clear non-self particles from their blood and react to grafts or to conspecifics competing for space, the extent to which such reactions are dependent on the absence of an inhibitory self signal, as opposed to the recognition of non-self, is not clear. Quesenberry et al. (115) have noted that tunicates express lectins able to recognize self and are therefore potentially able to recognize missing self or induced or altered self. Work with C. elegans or Drosophila has yet to suggest the presence of a self-recognition system. The nature of the molecules that indicate self or of the receptor that interacts with the self signal, if they exist, is unknown for any invertebrate. Progress in identifying such molecules would represent a significant advance in our understanding of invertebrate defenses. They might, for example, hold the key for understanding the basis of host specificity shown by many invertebrate pathogens.
Can invertebrates diversify their non-self recognition molecules?
With respect to recognition of non-self, the predominant paradigm is for the presence of both circulating and cell membrane-associated pattern-recognition molecules capable of recognizing determinants characteristic of broad groups of pathogens. Abundant evidence for these molecules now exists, especially in Drosophila, and they could prove to be sufficient to account for all non-self recognition. There have been no reports of the existence of unusual mechanisms to diversify known invertebrate pattern-recognition molecules, but there is a growing body of literature to suggest that invertebrates can produce diversified immune molecules. The extent to which diversified non-self recognition molecules occur in invertebrates, which might be generated by unusual processes such as gene conversion or somatic mutation, is a high priority for study. While being mindful of the possibility of PCR-related artifacts, some of the challenges awaiting such studies are the following: to reveal the mechanisms for generating diversity, to learn whether pathogen exposure provokes diversification, and to determine whether this diversity is relevant in defense. The presence of a self-recognition system would prevent the problem of autoreactivity, which could potentially result from production of diversified receptors.
Fig. 2 suggests one possible way that diversified antigen receptors might work, borrowing from a model proposed by Richards and Renwrantz (116) based on their study of phagocytosis of opsonized particles by hemocytes of the snail Helix pomatia. A hemocyte-associated receptor may be capable of binding to several different humoral recognition factors, but only after the latter have bound non-self structures and have been modified. The modification might be an enzymatic cleavage or conformational change that exposes a ligand site that can be bound by the hemocyte-associated receptor, resulting in phagocytosis of the opsonized structure. The appeal of this model is that a multipurpose membrane-associated receptor could interact with a variety of different humoral recognition molecules.
Do invertebrates have specialized mechanisms to identify and destroy viruses or virally infected cells?
Another purpose for the model provided was to highlight our ignorance about how invertebrates cope with viral infections. We are now rapidly learning about RNAi, and perhaps this and other intracellular mechanisms are all that invertebrates have available for combating viruses. However, it seems premature to conclude that effective means do not exist for blocking viral entry into cells, for promoting their aggregation and phagocytosis, and for identifying and destroying virus-infected cells. Are the mechanisms involved part of the standard invertebrate package of pattern-recognition and phagocytosis, or might more specialized systems for eliminating viruses exist? We have much to learn in this area.
Do invertebrates with abundant symbiont populations use them in internal defense?
For the many invertebrate species that harbor dense symbiont populations, do the symbionts play a role other than in nutrition? Whether symbionts might aid hosts by co-evolving with pathogens and matching virulence factors with anti-virulence factors remains an intriguing subject for future study.
Conclusions
Invertebrates have become popular subjects in the context of ecological immunology, because of the simple mechanisms underpinning their innate immune systems (117). The use of invertebrates as models in all kinds of studies is certainly to be encouraged, but at the same time, it is critically important not to underestimate, over-simplify, or take for granted their immune capabilities. It is not at all clear that studies attempting to measure immune competence by some unidimensional parameter, such as melanin content or hemocyte counts, will conclude anything meaningful. When claims about the immune capabilities of invertebrates are made, such as in the recent study providing evidence of immunological memory in copepod crustaceans (118), they need to be backed up by explicit information about the antigenic nature of the parasites employed. The host responses inferred to be present need be identified and their efficacy ascertained.
The term invertebrate is dangerously broad, so we should expect that the solutions for internal defense in one group would be different from the solutions obtained in other groups. Although invertebrates lack many of the well-known complicated features of vertebrate immunity, they may well employ alternative means to generate diverse and complex responses. In fact, there is much about their immunobiology that we do not fundamentally grasp, and this lack makes the future study of invertebrate immunobiology compelling. As noted above, the impetus for learning more is strongly driven by the urgent need to understand several applied problems that relate to invertebrate defenses.
Acknowledgments
We gratefully acknowledge the support of NIH grants AI24340 (ESL) and AI52363 (CMA). This publication was made possible by NIH grant number IP20RR18754 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources.
References
- 1.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RAB. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 4.Takeda K, Akira S. Toll receptors and pathogen resistance. Cell Microbiol. 2003;5:143–153. doi: 10.1046/j.1462-5822.2003.00264.x. [DOI] [PubMed] [Google Scholar]
- 5.Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 6.Valentine JW. Prelude to the Cambrian explosion. Annu Rev Earth Planet Sci. 1999;30:285–306. [Google Scholar]
- 7.Douglas AE. Symbiotic Interaction. Oxford: Oxford University Press; 1994. [Google Scholar]
- 8.O’Neill SL, Hoffmann AA, Werren JH. Influential Passengers – Inherited Microorganisms and Arthropod Reproduction. Oxford: Oxford University Press; 1997. [Google Scholar]
- 9.McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 10.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 11.Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DTJ. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda) Int J Parasitol. 2003;33:733–755. doi: 10.1016/s0020-7519(03)00049-3. [DOI] [PubMed] [Google Scholar]
- 12.Stephens RE, Walker CW, Reinisch CL. Multiple protein differences distinguish clam leukemia cells from normal hemocytes: evidence for the involvement of p53 homologues. Comp Biochem Physiol C Toxicol Pharmacol. 2001;129:329–338. doi: 10.1016/s1532-0456(01)00208-3. [DOI] [PubMed] [Google Scholar]
- 13.Rinkevich B. Germ cell parasitism as an ecological and evolutionary puzzle: hitchhiking with positively selected genotypes. Oikos. 2002;96:25–30. [Google Scholar]
- 14.Gardner TA, Cote IM, Gill JA, Grant A, Watkinson AR. Long-term region-wide declines in Caribbean corals. Science. 2003;301:958–960. doi: 10.1126/science.1086050. [DOI] [PubMed] [Google Scholar]
- 15.Price RN, Nosten F. Drug resistant falciparum malaria: clinical consequences and strategies for prevention. Drug Resist Update. 2001;4:187–196. doi: 10.1054/drup.2001.0195. [DOI] [PubMed] [Google Scholar]
- 16.Morgan JAT, Dejong RJ, Snyder SD, Mkoji GM, Loker ES. Schistosoma mansoni and Biomphalaria: past history and future trends. Parasitology. 2001;123:S211–S228. doi: 10.1017/s0031182001007703. [DOI] [PubMed] [Google Scholar]
- 17.Lotz JM. Viruses, biosecurity and specific pathogen-free stocks in shrimp aquaculture. World J Microbiol Biotechnol. 1997;13:405–413. [Google Scholar]
- 18.Tzou P, Meister M, Lemaitre B. Methods for studying infection and immunity in Drosophila. Mol Cell Microbiol. 2002;31:507–529. [Google Scholar]
- 19.Alegado RA, Campbell MC, Chen WC, Slutz SS, Tan MW. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell Microbiol. 2003;5:435–444. doi: 10.1046/j.1462-5822.2003.00287.x. [DOI] [PubMed] [Google Scholar]
- 20.Pujol N, et al. Reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 21.Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 22.Dodd RB, Drickamer K. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology. 2001;11:71R–79R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- 23.Mallo GV. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 24.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 25.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 26.Zdobnov EM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 27.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gokudan S, et al. Horseshoe crab acetyl group-recognizing lectins involved in innate immunity are structurally related to fibrinogen. Proc Natl Acad Sci USA. 1999;96:10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S-M, Léonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria glabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. 2001;53:684–694. doi: 10.1007/s00251-001-0386-8. [DOI] [PubMed] [Google Scholar]
- 30.Holmskov U, Thiel S, Jensenius JC. Collectins and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 31.Dimopoulos G, et al. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection. Proc Natl Acad Sci USA. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehal P, et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y, Doolittle RF. The evolution of vertebrate blood coagulation as viewed from a comparison of puffer fish and sea squirt genomes. Proc Natl Acad Sci USA. 2003;100:7527–7532. doi: 10.1073/pnas.0932632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman R, Hughes AL. Molecular evolution of the NF-κB signaling system. Immunogenetics. 2002;53:964–974. doi: 10.1007/s00251-001-0399-3. [DOI] [PubMed] [Google Scholar]
- 35.McMahon RF, Bogan AE. Mollusca. Bivalvia. In: Thorp JH, Covich AP, editors. Ecology and Classification of North American Freshwater Invertebrates. 2. San Diego: Academic Press; 2001. pp. 331–429. [Google Scholar]
- 36.Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Curr Opin Immunol. 2003;15:396–401. doi: 10.1016/s0952-7915(03)00080-3. [DOI] [PubMed] [Google Scholar]
- 37.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 38.Buttner D, Bonas U. Common infection strategies of plant and animal pathogenic bacteria. Curr Opin Plant Biol. 2003;6:312–319. doi: 10.1016/s1369-5266(03)00064-5. [DOI] [PubMed] [Google Scholar]
- 39.Rhen M, Eriksson S, Clements M, Bergstrom S, Normark SJ. The basis of persistent bacterial infections. Trends Microbiol. 2003;11:80–86. doi: 10.1016/s0966-842x(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 40.de la Fuente J, Kocan KM. Expression of Anaplasma marginale major surface protein 2 variants in persistently infected ticks. Infect Immun. 2001;69:5151–5156. doi: 10.1128/IAI.69.8.5151-5156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi J, Ishibashi M. Bioactive metabolites of symbiotic marine microorganisms. Chem Rev. 1993;93:1753–1769. [Google Scholar]
- 43.Boman HG. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 44.Engel S, Jensen PR, Fenical W. Chemical ecology of marine microbial defense. J Chem Ecol. 2002;28:1971–1985. doi: 10.1023/a:1020793726898. [DOI] [PubMed] [Google Scholar]
- 45.Gil-Turnes MS, Hay ME, Fenical W. Symbiotic marine-bacteria chemically defend crustacean embryos from a pathogenic fungus. Science. 1989;246:116–118. doi: 10.1126/science.2781297. [DOI] [PubMed] [Google Scholar]
- 46.Grigioni S, Boucher-Rodoni R, Demarta A, Tonolla M, Peduzzi R. Phylogenetic characterisation of bacterial symbionts in the accessory nidamental glands of the sepioid Sepia officinalis (Cephalopoda: Decapoda) Mar Biol. 2000;136:217–222. [Google Scholar]
- 47.Barbieri E, et al. Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda: Loliginidae) Environ Microbiol. 2001;3:151–167. doi: 10.1046/j.1462-2920.2001.00172.x. [DOI] [PubMed] [Google Scholar]
- 48.Visick KL, McFall-Ngai MJ. An exclusive contract: specificity in the Vibrio fischeri Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemus JD, McFall-Ngai MJ. Alterations in the proteome of the Euprymna scolopes light organ in response to symbiotic Vibrio fischeri. Appl Environ Microbiol. 2000;66:4091–4097. doi: 10.1128/aem.66.9.4091-4097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Small AL, McFall-Ngai MJ. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- 51.Bosch C, Grasse PP. Partial cycle of chemoautotroph symbiotic bacteria and their relations with the bacteriocytes in Riftia pachyptila Jones (Pogonophora, Vestimentifera). 1. The trophosome and the bacteriocytes. C R Acad Sci III. 1984;299:371–376. [Google Scholar]
- 52.Krueger DM, Gustafson RG, Cavanaugh CM. Vertical transmission of chemoautotrophic symbionts in the bivalve Solemya velum (Bivalvia: Protobranchia) Biol Bull. 1996;190:195–202. doi: 10.2307/1542539. [DOI] [PubMed] [Google Scholar]
- 53.Norton JH, Shepherd MA, Long HM, Fitt WK. The zooxanthellal tubular system in the giant clam. Biol Bull. 1992;183:503–506. doi: 10.2307/1542028. [DOI] [PubMed] [Google Scholar]
- 54.Nakayama K, Nishijima M, Maruyama T. Morula-like cells in photo-symbiotic clams harboring zooxanthellae. Zool Sci. 1998;15:339–344. doi: 10.2108/zsj.15.339. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama K, Ishikura M, Maruyama T. Proteins of morula-like cells in hemolymph of the giant clam, Tridacna derasa. Biol Bull. 1997;193:141–146. doi: 10.2307/1542759. [DOI] [PubMed] [Google Scholar]
- 56.Nakayama K, Maruyama T. Differential production of active oxygen species in photo-symbiotic and non-symbiotic bivalves. Dev Comp Immunol. 1998;22:151–159. doi: 10.1016/s0145-305x(97)00060-8. [DOI] [PubMed] [Google Scholar]
- 57.Nakayama K, Nishijima M, Maruyama T. Parasitism by a protozoan in the hemolymph of the giant clam Tridacna crocea. J Invertebr Pathol. 1998;71:193–198. doi: 10.1006/jipa.1997.4747. [DOI] [PubMed] [Google Scholar]
- 58.Whitham TG, Slobodchikoff CN. Evolution by individuals, plant–herbivore interactions, and mosaics of genetic-variability – the adaptive significance of somatic mutations in plants. Oecologia. 1981;49:287–292. doi: 10.1007/BF00347587. [DOI] [PubMed] [Google Scholar]
- 59.Gill DE, Chao I, Perkins SL, Wolf JB. Genetic mosaicism in plants and clonal animals. Annu Rev Ecol Syst. 1995;26:423–444. [Google Scholar]
- 60.Sommerfeldt AD, Bishop JDD, Wood CA. Chimerism following fusion in a clonal ascidian (Urochordata) Zool J Linn Soc. 2003;79:183–192. [Google Scholar]
- 61.Weismann W. The Germ Plasm: A Theory of Heredity. London: Scott; 1893. [Google Scholar]
- 62.Schön I, Martens K. No slave to sex. Proc R Soc Lond B Biol Sci. 2003;270:827–833. doi: 10.1098/rspb.2002.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet. 2002;30:311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- 64.Kovalchuk I, et al. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature. 2003;423:760–762. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- 65.Flajnik MF, Miller K, DuPasquier L. Evolution of the immune system. In: Paul WE, editor. Fundamental Immunology. 5. New York: Raven Press; 2003. pp. 519–570. [Google Scholar]
- 66.Schluter SF, Bernstein RM, Bernstein H, Marchalonis JJ. ‘Big bang’ emergence of the combinatorial immune system. Dev Comp Immunol. 1999;23:107–111. doi: 10.1016/s0145-305x(99)00002-6. [DOI] [PubMed] [Google Scholar]
- 67.Marchalonis JJ, Whitfield GK, Schluter SF. Rapid evolutionary emergence of the combinatorial recognition repertoire. Integr Comp Biol. 2003;43:347–359. doi: 10.1093/icb/43.2.347. [DOI] [PubMed] [Google Scholar]
- 68.Plasterk RV. V(D)J recombination. Ragtime jumping Nature. 1998;394:718–719. doi: 10.1038/29389. [DOI] [PubMed] [Google Scholar]
- 69.Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Zarkadis IK, Sarrias MR, Sfyroera G, Sunyer JO, Lambris JD. Cloning and structure of three rainbow trout C3 molecules: a plausible explanation for their functional diversity. Dev Comp Immunol. 2001;25:11–24. doi: 10.1016/s0145-305x(00)00039-2. [DOI] [PubMed] [Google Scholar]
- 71.Nakao M, Mutsuro J, Nakahara M, Kato Y, Yano T. Expansion of genes encoding complement components in bony fish: biological implications of the complement diversity. Dev Comp Immunol. 2003;27:749–762. doi: 10.1016/s0145-305x(03)00076-4. [DOI] [PubMed] [Google Scholar]
- 72.Sunyer JO, Boshra H, Lorenzo G, Parra D, Freedman B, Bosch N. Evolution of complement as an effector system in innate and adaptive immunity. Immunol Res. 2003;27:549–564. doi: 10.1385/IR:27:2-3:549. [DOI] [PubMed] [Google Scholar]
- 73.Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- 74.Pancer Z. Dynamic expression of multiple scavenger receptor cysteine-rich genes in coelomocytes of the purple sea urchin. Proc Natl Acad Sci USA. 2000;97:13156–13161. doi: 10.1073/pnas.230096397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pancer Z. Individual-specific repertoires of immune cells SRCR receptors in the purple sea urchin (S. purpuratus) In: Beck G, Sugumaran M, Cooper EL, editors. Phylogenetic Perspectives on the Vertebrate Immune System. New York: Kluwer Academic/Plenum Publications; 2001. pp. 31–40. [DOI] [PubMed] [Google Scholar]
- 76.Du Pasquier L, Smith LC. Workshop report: evolutionary immunobiology – new approaches, new paradigms. Dev Comp Immunol. 2003;27:263–271. doi: 10.1016/s0145-305x(02)00069-1. [DOI] [PubMed] [Google Scholar]
- 77.Pancer Z, et al. Polymorphism in the immunoglobulin-like domains of the receptor tyrosine kinase from the sponge Geodia cydonium. Cell Adhes Commun. 1996;4:327–333. doi: 10.3109/15419069609010776. [DOI] [PubMed] [Google Scholar]
- 78.Pancer Z, Skorokhod A, Blumbach B, Muller WEG. Multiple Ig-like featuring genes divergent within and among individuals of the marine sponge Geodia cydonium. Gene. 1998;207:227–233. doi: 10.1016/s0378-1119(97)00631-8. [DOI] [PubMed] [Google Scholar]
- 79.Cadavid LF, Powell AE, Nicotra ML, Moreno M, Buss LW. An invertebrate histocompatibility complex. Genetics. doi: 10.1534/genetics.167.1.357. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Destoumieux D, Munoz M, Bulet P, Bachere E. Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda) Cell Mol Life Sci. 2000;57:1260–1271. doi: 10.1007/PL00000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gross PS, Bartlett TC, Browdy CL, Chapman RW, Warr GW. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus. Dev Comp Immunol. 2001;25:565–577. doi: 10.1016/s0145-305x(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 82.Cuthbertson BJ, Shepard EF, Chapman RW, Gross PS. Diversity of the penaeidin antimicrobial peptides in two shrimp species. Immunogenetics. 2002;54:442–445. doi: 10.1007/s00251-002-0487-z. [DOI] [PubMed] [Google Scholar]
- 83.Bartlett TC, Cuthbertson BJ, Shepard EF, Chapman RW, Gross PS, Warr GW. Crustins, homologues of an 11.5-kDa antibacterial peptide, from two species of penaeid shrimp. Litopenaeus vannamei and Litopenaeus setiferus. Mar Biotechnol (NY) 2002;4:278–293. doi: 10.1007/s10126-002-0020-2. [DOI] [PubMed] [Google Scholar]
- 84.Léonard PM, Adema CM, Zhang S-M, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- 85.Zhang S-M, Loker ES. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Dev Comp Immunol. 2003;27:175–187. doi: 10.1016/s0145-305x(02)00091-5. [DOI] [PubMed] [Google Scholar]
- 86.Cline J, Braman JC, Hogrefe HH. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rothman LD, Myers JH. Debilitating effects of viral diseases on host Lepidoptera. J Invertebr Pathol. 1996;67:1–10. [Google Scholar]
- 89.Spann KM, Lester RJG. Viral diseases of penaeid shrimp with particular reference to four viruses recently found in shrimp from Queensland. World J Microbiol Biotechnol. 1997;13:419–426. [Google Scholar]
- 90.Elston R. Bivalve mollusc viruses. World J Microbiol Biotechnol. 1997;13:393–403. [Google Scholar]
- 91.Moscardi F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu Rev Entomol. 1999;44:257–289. doi: 10.1146/annurev.ento.44.1.257. [DOI] [PubMed] [Google Scholar]
- 92.Borucki MK, Kempf BJ, Blitvich BJ, Blair CD, Beaty BJ. La Crosse virus: replication in vertebrate and invertebrate hosts. Microbes Infect. 2002;4:341–350. doi: 10.1016/s1286-4579(02)01547-2. [DOI] [PubMed] [Google Scholar]
- 93.Garmendia AE, Van Kruiningen HJ, French RA. The West Nile virus: its recent emergence in North America. Microbes Infect. 2001;3:223–229. doi: 10.1016/s1286-4579(01)01374-0. [DOI] [PubMed] [Google Scholar]
- 94.Beschin A, Bilej M, Torreele E, De Baetselier P. On the existence of cytokines in invertebrates. Cell Mol Life Sci. 2001;58:801–814. doi: 10.1007/PL00000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuusksalu A, Pihlak A, Muller WEG, Kelve M. The (2′–5′) oligoadenylate synthetase is present in the lowest multicellular organisms, the marine sponges – demonstration of the existence and identification of its reaction-products. Eur J Biochem. 1995;232:351–357. [PubMed] [Google Scholar]
- 96.Lopp A, Kuusksalu A, Reintamm T, Muller WEG, Kelve M. 2′,5′-oligoadenylate synthetase from a lower invertebrate, the marine sponge Geodia cydonium, does not need dsRNA for its enzymatic activity. Biochem Biophys Acta. 2002;1590:140–149. doi: 10.1016/s0167-4889(02)00207-0. [DOI] [PubMed] [Google Scholar]
- 97.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 98.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 99.Plasterk RHA. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 100.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 101.Ahlquist P. RNA-dependent RNA polymerases, viruses, RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 102.Li WX, Ding SW. Viral suppressors of RNA silencing. Curr Opin Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 103.Kewal Ramani VN, Coffin JM. Weapons of mutational destruction. Science. 2003;301:923–925. doi: 10.1126/science.1088965. [DOI] [PubMed] [Google Scholar]
- 104.Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol Biol. 2001;10:265–273. doi: 10.1046/j.1365-2583.2001.00267.x. [DOI] [PubMed] [Google Scholar]
- 105.Caplen NJ, Zheng ZL, Falgout B, Morgan RA. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol Ther. 2002;6:243–251. doi: 10.1006/mthe.2002.0652. [DOI] [PubMed] [Google Scholar]
- 106.Olson KE, Adelman ZN, Travanty EA, Sanchez-Vargas I, Beaty BJ, Blair CD. Developing arbovirus resistance in mosquitoes. Insect Biochem Mol Biol. 2002;32:1333–1343. doi: 10.1016/s0965-1748(02)00096-6. [DOI] [PubMed] [Google Scholar]
- 107.Loh PC, Tapay LM, Lu YN, Nadala ECB. Viral pathogens of the penaeid shrimp. Adv Virus Res. 1997;48:263–312. doi: 10.1016/S0065-3527(08)60290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van de Braak CBT, Botterblom MHA, Huisman EA, Rombout JHWM, van der Knaap WPW. Preliminary study on haemocyte responses to white spot syndrome virus infection in black tiger shrimp Penaeus nonodon. Dis Aquat Organ. 2002;51:149–155. doi: 10.3354/dao051149. [DOI] [PubMed] [Google Scholar]
- 109.Pan J, et al. Broad antiviral activity in tissues of crustaceans. Antiviral Res. 2000;48:39–47. doi: 10.1016/s0166-3542(00)00117-0. [DOI] [PubMed] [Google Scholar]
- 110.Hizer SE, Dhar AK, Klimpel KR, Garcia DK. RAPD markers as predicators of infectious hypodermal and hematopoietic necrosis virus (IHHNV), resistance in shrimp (Litopenaeus stylirostris) Genome. 2002;45:1–7. doi: 10.1139/g01-117. [DOI] [PubMed] [Google Scholar]
- 111.Tang KFJ, Durand SV, White BL, Redman RM, Mohney LL, Lightner DV. Induced resistance to white spot syndrome virus infection in Penaeus stylirostris through pre-infection with infectious hypodermal and hematopoietic necrosis virus – a preliminary study. Aquaculture. 2003;216:19–29. [Google Scholar]
- 112.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 113.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Narayanan K. Apoptosis: its role in microbial control of insect pests. Curr Sci. 1998;75:114–122. [Google Scholar]
- 115.Quesenberry MS, Ahmed H, Elola MT, O’Leary N, Vasta GR. Diverse lectin repertoires in tunicates mediate broad recognition and effector innate immune responses. Integr Comp Biol. 2003;43:323–330. doi: 10.1093/icb/43.2.323. [DOI] [PubMed] [Google Scholar]
- 116.Richards EH, Renwrantz LR. 2 lectins on the surface of Helix pomatia hemocytes – a Ca2+-dependent, Galnac-specific lectin and a Ca2+-independent, mannose 6-phosphate specific lectin which recognizes activated homologous opsonins. J Comp Physiol B Biochem Syst Environ Physiol. 1991;161:43–54. [Google Scholar]
- 117.Rolff J, Siva-Jothy MT. Invertebrate ecological immunology. Science. 2001;301:472–475. doi: 10.1126/science.1080623. [DOI] [PubMed] [Google Scholar]
- 118.Kurtz J, Franz K. Evidence for memory in invertebrate immunology. Nature. 2003;425:37–38. doi: 10.1038/425037a. [DOI] [PubMed] [Google Scholar]