Abstract
CD8+ T cell immunosurveillance is based on recognizing oligopeptides presented by MHC class I molecules. Despite decades of study, the importance of protein ubiquitylation to peptide generation remains uncertain. Here, we examine the ability of MLN7243, a recently described ubiquitin activating enzyme E1-inhibitor, to block overall cytosolic peptide generation and generation of specific peptides from vaccinia- and influenza A virus-encoded proteins. We show that MLN7243 rapidly inhibits ubiquitylation in a variety of cell lines, and can profoundly reduce generation of cytosolic peptides. Kinetic analysis of specific peptide generation reveals that ubiquitylation of defective ribosomal products (DRiPs) is rate limiting in generating class I peptide complexes. More generally, our findings demonstrate that the requirement for ubiquitylation in MHC class I restricted antigen processing varies with class I allomorph, cell type, source protein, and peptide context. Thus, both ubiquitin-dependent and independent pathways robustly contribute to MHC class I based immunosurveillance.
Introduction
CD8+ T cells play a central role in adaptive immune responses to viruses and other intracellular pathogens, cancers, transplants and autoimmune targets. CD8+ T cells recognize short peptides presented by major histocompatibility complex (MHC) class I molecules. Antigenic peptides typically arise from proteasomal products that are transported by TAP (transporter associated with antigen processing) into the lumen of endoplasmic reticulum (ER), trimmed at their NH2-termini, loaded onto class I molecules, and transported to the cell surface for T cell immunosurveillance.
Two general substrate classes provide antigenic peptides: defective ribosomal products (DRiPs) and retirees (1). Retirees are proteins degraded via the normal process of protein turnover. DRiPs are a substantial subset of nascent gene products degraded more rapidly than their corresponding native retiree pools. DRiPs consist of:
truncated, fragmented or misfolded, proteins
excess subunits of multi-protein assemblies; and
non-canonical translation or mistranslation products, e.g. those resulting from frame shifting, alternative initiation, nuclear translation.
DRiPs are a major source of self-antigenic peptides (2), and appear to account for the vast majority of antigen presentation for at least several viruses (3–5).
For both DRiPs and retirees, proteasome degradation plays an important role in generating cytosolic peptides of between 8 and ~ 17 residues that can be transported into the ER by TAP (6). Classically, proteins are targeted for proteasome degradation by covalent addition of ubiquitin (Ub). Ubiquitylation entails the sequential action of E1, E2 and E3 enzymes. E1s covalently bind Ub via a thioester bond and through the action of E2s, transfer Ub to E3s for covalent modification of substrate amino, thiol, or hydroxyl groups (7). Deubiquitylating enzymes (DUBs), some associated with 26S proteasomes, release and recycle Ub during or prior to protein degradation (8).
While controversial, it appears that a sizeable substrate fraction is targeted for proteasome degradation in a Ub-independent manner (9, 10). This controversy extends to the involvement of ubiquitylation in proteasome mediated-antigen presentation, where evidence supports both Ub dependent- and independent processes (summarized in Table I). Ub-independent presentation is consistent with a large number of reports documenting proteasome-independent, TAP-dependent peptide generation and presentation (22–24), but also likely extends to proteasome mediated degradation (15, 25)
Table I.
Summary of Published Literature on Role of Ubiquitylation in Antigen Presentation
| Peptide | Context | Approaches | Results | Authors’ Conclusions | |
|---|---|---|---|---|---|
| (11) | NY-ESO-1157–165 | NY-ESO-1 | Lysine-less form of NY-ESO-1 | No effect | NY-ESO-1 is governed by non-canonical ubiquitination on non-lysine sites. Atypical ubiquitylation is significant. |
| (12) | IYVLDHLIVV; RAK (BZLF1); YVL (MART-1) | BZLF1 and MART-1 | Knockdown of Ub | Inhibits antigen presentation | Polyubiquitylation of lysine-48 is an essential but indirect signal |
| Expression of a lysine 48 Ub mutant | Inhibits antigen presentation | ||||
| Inhibition of proteasome-associated DUBs | Inhibits antigen presentation | ||||
| Ubiquitinatable amino acid (C,K,S,T)-less form of antigen | No effect | ||||
| (13) | SIINFEKL | SCRAP system | Expression of a dominant negative form of p97 | No effect | Presentation of DRiP-derived peptides is not dependent on the AAA ATPase p97 |
| Inhibition of p97 by DBeQ | No effect | ||||
| (14) | SIINFEKL | SCRAP system and rVV-NPSGFP | Inhibition of DUBs by Eer1 and CI-PGA2 | Inhibits antigen presentation | Polyubiquitin chain disassembly or the actions of deubiquitylating enzymes are important |
| (15) | SIINFEKL | CytoNP; ER-NP; Ub-R-cytoNP; CytoTac; and Tac | Expression of a dominant negative form of ubiquitin | Inhibits presentation from a cytosolic N-rule substrate and ER targeted antigens. No effect on presentation from cytosolic protein | Selective role of ubiquitin |
| Knockdown of the ER-associated Ub ligase Hrd1 | |||||
| (16) | MHC pan Class I surface expression (W6/32) | Knockdown of BAG-6 | Inhibits the cell surface presentation of MHC-I | BAG-6 is necessary for ubiquitin-mediated degradation of newly synthesized defective polypeptides | |
| (17) | SIINFEKL | UbR-NPSGFP; NP-S-GFP; and NP(KEKE)-S-GFP | Temperature-sensitive E1 ts20 cells Knockdown HuS4 to reduce 26S proteasome activity |
No effect | Degradation of full-length DRiPs occurs in a ubiquitin-independent manner |
| (18) | SIINFEKL | OVA | Temperature-sensitive E1 ts85 and ts20 | Inhibits microinjected native OVA but not chemically denatured and alkylated OVA or endogenously synthesized OVA | Modification of the structure of a protein can influence its requirements for ubiquitin conjugation for efficient class I presentation. |
| Methylation of the lysine groups in denatured OVA | Inhibits the presentation of denatured OVA | ||||
| (19) | βgal | Ub-X-βgal and X-βgal | Ub-X-βgal and X-βgal | Enhance presentation when protein is modified with a destabilizing N-terminal residue | Ubiquitin conjugation is a key rate-limiting step in antigen presentation |
| Methylation of amino groups of lysine residues | Inhibits presentation | ||||
| (20) | IAV PB1 | PR8 and VV-PB1 | Temperature-sensitive ubiquitin conjugation mutants ts20 and tsA1S9 | No effect at non permissive temperature | Presentation of endogenous and exogenous antigens is not affected by inactivation of E1 enzyme in ts cell lines. |
| SIINFEKL | OVA and VV-OVA | ||||
| (21) | SIINFEKL | OVA | Temperature-sensitive E1 ts20 cells | Inhibits presentation at non permissive temperature | Ubiquitin-dependent pathway in antigen presentation |
Studies on the Ub-requirement in proteasome-dependent antigen presentation (Table I) are limited by ambiguities associated with the methodologies employed. Studies with temperature sensitive E1s are limited by the incomplete inactivation of E1 and by downstream effects from heat shock conditions needed for E1 inactivation. Studies based on eliminating antigen ubiquitylation sites are limited by discoveries of new types of ubiquitylation sites. Studies adapting genetic knockdowns or dominant negative manipulation of pathway components are limited by residual functions of these generally highly abundant proteins. All approaches suffer from downstream effects of manipulating the Ub-proteasome pathway, which rapidly cascades through many, if not all cellular pathways.
Since E1 activation with Ub is required for all subsequent steps in the ubiquitylation pathway (8), efficiently blocking this step pharmacologically should profoundly block substrate ubiquitylation. Drug-based approaches have the advantage of minimizing the time available for downstream and compensatory alterations in treated cells.
Here, we use MLN7243, a small molecule, cell permeant inhibitor recently described to potently inhibit E1 activity (26), to more conclusively determine the role of ubiquitylation in MHC class I antigen presentation. Our findings reconcile previous conflicting results, and underscore the complexity of generating peptides for CD8+ T cell immunosurveillance.
Materials and Methods
Cells, viruses, and peptides
L-Kb and 293-Kb were maintained in DMEM with 7.5% FBS in a 9% CO2 incubator. DC2.4, P815 and EL4 were maintained in RPMI with 7.5% FBS in a 5% CO2 incubator. rVVs expressing NP-S-GFP, UbR-NP-S-GFP and GFP-Ub-S were previously described (14). B lymphocyte cell lines were generated as previously described (27). Influenza A virus A/Puerto Rico/8/34 (H1N1) (PR8) was grown in 10-day embryonic chicken eggs and used as infectious allantonic fluid. Peptides were synthesized by Mimotopes at >80% purity (Clayton, Melbourne, Australia). MG132 was from EMD Millipore, Darmstadt, Germany. Cychoheximide was from Sigma-Aldrich, St. Louis, MO. MLN7243 was from National Center for Advancing Translational Sciences.
Plasmids
DNA fragments encoding MYC-SIINFEKL-Venus sequence were amplified from pSLIK-Venus using primer 5′-GAGCTCATGGAGCAGAAGCTCATCTCCGAGGAGGACCTG TCGATCATCAACTTCGAAAAGCTAATGGTGAGCAAGGGCGAGTACTCGAGC-3′ and primer 5′-TACTTGTACAGCTCGTCCATGCCGAGAG-3′. PCR product was then excised with SalI and XhoI restriction enzymes and ligated with similarly digested pCAGGS. DNA fragments encoding ER-targeted SIINFEKL sequence were synthesized by Integrated DNA Technologies (IDT, Coralville, IA) and ligated into pIRES2-EGFP vector (Clontech, Mountain View, CA) excised with NheI and EcoRI restriction enzymes. The signal sequence is MRYMILGLLALAAVCSAA.
Immunoblotting
Immunoblotting was performed as described (28) using rabbit anti-histone H3 (Cell signaling), mouse anti-poly Ub antibody (clone FK1) (Enzo), IRDye 800CW anti-rabbit antibody (LI-COR) and IRDye 680LT anti-mouse antibody (LI-COR).
Immunofluorescence microscopy
L-Kb cells were plated into EZ slide 8-well glass slides (EMD Millipore, Darmstadt, Germany), incubated for overnight at 37°C, and then treated with DMSO or MLN7243 for 1 h at 37°C. Cell were washed twice with cold PBS, fixed with 3.2% paraformaldehyde in PBS for 15 min at room temperature (RT), permeablized in −20°C methanol for 10 min at RT, and rinsed twice in PBS. Block and antibody staining were performed in 5% normal donkey serum in PBS containing 0.02% sodium azide. Cells were blocked for 45 min at RT with gentle shaking, incubated with primary antibodies human ribosomal P antibody (Immunovision) and mouse mono- and polyUb conjugates monoclonal antibody (clone FK2) (Enzo) overnight at 4°C, washed three times with PBS, incubated with secondary antibodies FITC-conjugated AffiniPure donkey anti–mouse IgG (Jackson ImmunoResearch) and Cy5-conjugated AffiniPure donkey anti–human IgG (Jackson ImmunoResearch) for 1 h at RT, washed three times with PBS, rinsed with distilled water, and mounted in Fluoromount-G (SouthernBiotech, Birmingham, AL). Images were acquired using a laser-scanning confocal microscope (TCS SP5; Leica, Mannheim, Germany) with an HC Plan Apo 100×, 1.40 NA oil objective, type FF immersion liquid (Cargille, Cedar Grove, NJ), and LAS AF software (V2.3.1; Leica). Images were processed using Imaris (Bitplane, Zurich, Switzerland).
Fluorescence recovery after photo bleaching (FRAP)
Mel Juso cells stably expressing TAP1-GFP were plated sparsely onto 35mm Ibidi plastic cover-slip bottom dishes (ibidi, Planegg, Germany) and low-intensity cells were chosen for imaging. FRAP was conducted using an inverted Leica TSP SP5 confocal microscope equipped with a CO2 environmental chamber and a Ar-Kr 488 nm laser. A circular ROI with a diameter of 2 μm was placed adjacent to the nuclear envelope and bleaching was performed with 2 scans of the laser at maximum intensity. Effective lateral diffusion of fluorescence into the bleached region was monitored with an attenuated laser. A fluorescence recovery curve was generated by acquiring 50 frames every 250 ms, followed by 40 frames every 1 s after bleaching. The half-time for recovery (t1/2) was extrapolated from each curve after normalization to background and bleaching due to imaging. The diffusion coefficient, D, is calculated from the formula, D = (0.88*w2)/(4 t1/2), where w is the radius of the bleached spot (1 μm in our experiments). D was determined from three independent experiments in which 15–20 cells were imaged per sample and depicted as mean +/−s.e.
Fluorescence correlation spectroscopy (FCS)
Mel Juso cells stably expressing TAP1-GFP were plated on Lab-Tek II chambered coverslips (Nunc, Rochester, NY) one day before the FCS experiment. Cells were treated with 2.5 μM MLN7243, 20 μM MG132, or 25 μg/ml CHX at 37 °C for 30 min prior to the FCS experiment. FCS measurement was performed using a 63x water immersion objective lens (NA 1.2) and a Leica SP8 DMI6000 confocal microscope system (Leica Microsystems, Mannheim, Germany) with a PicoHarp 300 Time-correlated single photon counting module with Single Photon Avalanche diodes detectors (PicoQuant, Berlin, Germany). The TAP1-GFP signal was measured with pulsed white light laser at excitation wavelength of 488 nm. WLL laser power was kept minimum to avoid sample bleaching. A measurement spot was placed on a ER- region and photon counting was performed for 30 s. A total of 12–15 independent points were measured for each condition. Single photon counting data was recorded and stored in pqw format. Correlation analysis was performed using SymPhoTime software (ver.5.3.2.2). Autocorrelation function G(t) was curve fitted with triplet model and calculate relative diffusion coefficient D. Data were exported to Prism 6 (GraphPad Software, La Jolla, CA) for further analysis.
Transfection
293-Kb cells were transfected at 70% confluency using GenJet Ver II (SignaGen, Rockville, MD) according to the manufacturer’s protocol. Medium was changed five hours post-transfection, and cells were acid-stripped 20 hours post-transfection.
TCD8+ cultures
TCD8+ cultures were established as previously described (29). Briefly, murine cultures were established using 1 nM peptide pulsed splenocytes and subsequently cultured in the presence of 10U IL-2/ml. Human cultures were established using 1 mM peptide pulsed PBMCs and subsequently cultured in medium containing 20U IL-2/ml.
In vitro viral infection
For rVV infection, cells were resuspended in 0.1% BSS/BSA and infected with rVV at 10 multiplicity of infection (MOI) at 37°C. For IAV infection, cells were resuspended in FCS-free, acidified RPMI medium and infected with IAV at 10 MOI at 37°C.
Flow cytometry
The kinetics of Kb-SIINFEKL presentation was determined as described (28). To study the kinetics of endogenous peptide presentation, cells were treated in ice-cold citric acid buffer (0.13 M citric acid, 0.061 M Na2HPO4, 0.15 M NaCl, ph 3) at 1 X 107 cells/ml for 120 s, washed three times with PBS, and resuspended in culture media containing drugs specified in figure legends. At indicated time point, an aliquot of cells (generally 5 X 105) was removed and stained with antibodies including mouse anti-HLA A,B,C (W6/32, in house), mouse anti-H-2Kb (HB176, in house), mouse anti-H-2Db (B22.249, in house), FITC anti-mouse H-2Kk (BD Biosciences), and FITC anti-mouse H-2Dk (BD Biosciences). Secondary staining was conducted with Alexa Fluor 647-coupled goat anti-mouse IgG(H+L) (Life Technologies) when necessary. All flow cytometry experiments were conducted using a BD LSRFortessa X-20 flow cytometer (BD Biosciences, San Jose, CA), gated on single cells, and analyzed by FlowJo software (TreeStar, Inc., Ashland, OR).
In vitro activation of antigen-specific TCD8+ and enumeration by intracellular cytokine staining (ICS)
ICS was conducted as described (27, 30). Briefly, PR8 infected APCs were washed and added to monospecific TCD8+ cultures, with BFA added at indicated time points. Following 4 h exposure to BFA, TCD8+ cells were transferred onto ice, fixed with 1% PFA and stained for IFN-γ in the presence of 0.4% saponin.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (GraphPad, San Diego, CA).
Results
MLN7243 blocks protein ubiquitylation
MLN7243 is in phase I clinical trials to treat solid tumors (26). Although it has been described as a potent E1 inhibitor in the brief descriptions published to date, we are unaware of published data demonstrating its ability to inhibit ubiquitylation in cells. Therefore, we tested the capacity of MLN7243 to inhibit protein ubiquitylation in a number of cells lines well suited for studying antigen presentation.
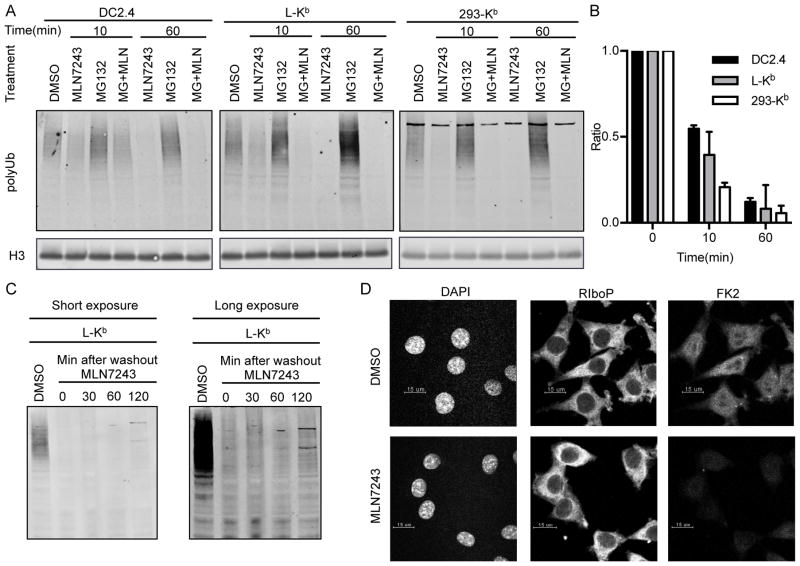
Immunoblotting with the polyubiquitin (polyUb)-conjugate-specific FK1 monoclonal antibody (mAb) (31) revealed that within 10 min of adding to cells, 2.5 μM MLN7243 decreased levels of polyubiquitylated proteins by 45–80% in DC2.4 (mouse dendritic cell like cell line), L-Kb and 293-Kb cells (respectively, mouse L929 and human HEK293 cells expressing the mouse class I molecule H-2Kb from a transgene) (Fig. 1A, and quantification from three independent experiments shown in Fig. 1B). Note that the loss of polyUb following the addition of MLN7243 is expected to result from deubiquitylation mediated by the 19S proteasome subunit as well as myriad other deubquitylases (DUBs) present in cells (8).
Figure 1. MLN7243 blocks protein ubiquitylation.
(A) DC2.4, L-Kb and 293-Kb cells were treated with 2.5 μM MLN7243, 10 μM MG132, or the combination of these two drugs for 10 or 60 min. Whole cell lysates were blotted with FK1 to detect polyubiquitylated proteins. Histone-H3 was used as the loading control. (B) Median intensity of each band was used for quantification. Results from three independent experiments are shown. (C) L-Kb cells were treated with 2.5 μM MLN7243, washed three times with PBS, and cultured in medium without MLN7243 for the indicated times. Whole cell lysates were blotted with FK1 to detect polyubiquitylated proteins. (D) L-Kb cells were cultured in the presence or absence of 2.5 μM MLN7243 for 1 h. DAPI, anti-ribosomal protein P antibody and FK2 were used in immunofluorescence microscopy to detect nuclei (left panel), ribosomes (middle panel) and ubiquitylated proteins (right panel), respectively.
Immunoblotting further revealed that the effects of MLN7243 on polyubiquitylation were irreversible for at least 2 h following its removal from cells (Fig. 1C). This augurs well for the potency of the drug, since it suggests that the MLN7243-E1 complex is highly stable in cells despite being non-covalent.
As expected, incubating cells with the proteasome inhibitor MG132 increased the amount of polyubiquitylated proteins. Importantly, when added together with MG132, MLN7243 abolished MG132-induced accumulation of polyubiquitylated proteins, confirming that MLN7243 functions upstream of MG132 in the ubiquitin proteasome pathway.
Immunofluorescence microscopy further demonstrated the profound inhibition of ubiquitylation exerted by MLN7243 (Fig. 1D). After incubating L-Kb cells with MLN7243 for 1 h, we used DAPI, anti-ribosomal P protein (Ribo-P) antibody, and the Ub-conjugate specific FK2 mAb to visualize nuclear DNA, ribosomes, and ubiquitylated proteins, respectively. MLN7243 reduced the FK2 signal nearly to no primary antibody control values, without diminishing Ribo-P staining, demonstrating its effective inhibition of ubiquitylation.
Taken together, these data demonstrate the efficacy of MLN7243 to block protein ubiquitylation in the cell lines shown. Importantly, we found that HeLa cells are nearly completely resistant to the effects of MLN7243 (Fig. S1A). Thus, while the drug can be highly effective against cultured cells, this must be established for each cell line used, and extrapolating in vivo, for the various cell types present in healthy and diseased tissues.
MLN7243 inhibits overall MHC class I-restricted endogenous antigen processing in an allomorph specific manner
To gauge the effect of MLN7243 on overall class I peptide generation, we acid stripped cell surface class I molecules and measured the recovery of surface class I expression in the presence of MLN7243, vehicle, or the proteasome inhibitor MG132 (Fig. 2) (32, 33). Since peptides are limiting in expression of cell surface class I molecules stable at 37 °C (34, 35), this assay provides a reliable, if indirect, measure of class I peptide ligands generation.
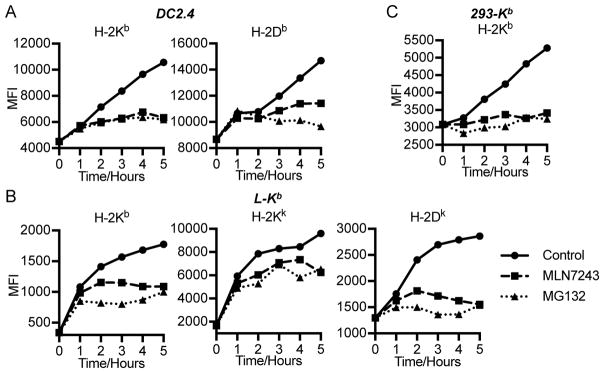
Figure 2. MLN7243 inhibits MHC class I-restricted endogenous antigen surface expression.
DC2.4 (A), L-Kb (B), and 293-Kb (C) cells were washed for 2 min in cold citric acid buffer to destroy native cell surface MHC-I complexes. Cells were then cultured in the presence of 2.5 μM MLN7243 or 10 μM MG132 or equal amount of DMSO, harvested at indicated time points, and analyzed for surface MHC-I complexes by flow cytometry. MFI: medium fluorescent intensity.
For each of the cell lines tested, MLN7243 and MG132 exerted only minor effects in the first hour of recovery (Fig. 2). This is likely due to the cell surface delivery of pre-peptide loaded class I molecules present in the secretory pathway. Over the next 4 h, MLN7243 clearly blocked class I expression, in many cases with similar magnitude as MG132, strongly indicating that blocking ubiquitylation has a major effect on peptide generation. The effect was not always complete, however, and the exceptions are interesting and instructive.
In blocking H-2Kb surface expression, MLN7243 matched MG132 in DC2.4 cells (Fig. 2A), and nearly so in 293-Kb cells (Fig. 2C). By contrast, in L-Kb cells (Fig. 2B), which are more sensitive than DC2.4 cells to MLN7243 inhibition of polyubiquitylation (Fig. 1A and B), MLN7243 was less effective than MG132 at blocking Kb surface expression. This suggests that even for the same class I allomorph, the contribution of ubiquitylation to peptide generation varies between cell lines. For H-2Db in DC2.4 cells (Fig. 2A) and H-2Dk in L-Kb cells (Fig. 2B), MLN7243 was not as effective as MG132, suggesting that in these cells, proteasomes generate a substantial fraction of peptides in a Ub-independent manner. Surprisingly, MLN7243 and MG132 only partially inhibited the recovery of H-2Kk in L-Kb cells, suggesting antigens generated from Ub- and proteasome-independent manner.
These data show that the effects of MLN7243 vary depending on the cell line and MHC class I allomorph, consistent with the idea that a significant fraction of peptides are generated in a Ub-independent manner under a variety of circumstances.
MLN7243 inhibits cytosolic peptide generation
The cell surface class I re-expression assay, while convenient and robust, indirectly measures peptide generation. The most specific assay for the generation of relevant antigenic peptides is the method developed by Reits et al. (36) based on their observation that the mobility of TAP-GFP expressed in the ER membrane of human MelJuSo cells is inversely related to its occupancy by peptide ligands, as measured by fluorescence recovery after photo bleaching (FRAP).
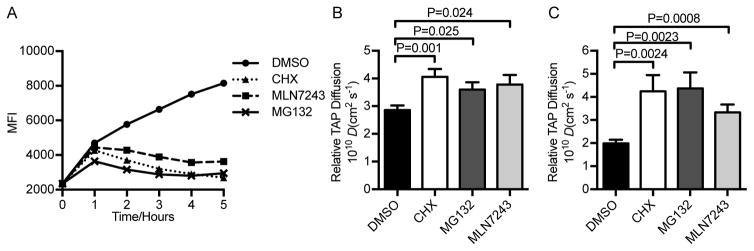
We first repeated the class I stripping experiment on the MelJuSo cells examined in the TAP-FRAP assay using the pan class I specific mAb W6/32 (which binds to most classical and no classical HLA class I molecules) (37, 38) to measure total class I molecule expression (Fig. 3A). This showed that MG132 completely blocks class I surface expression after the bolus of pre-peptide loaded class I molecules is delivered to the cell surface following acid stripping. Cycloheximide (CHX), a protein synthesis inhibitor, was only slightly less effective, consistent with the idea that most peptides are derived from DRiPs, as concluded by Reits et al. (36). MLN7243 was slightly less effective than either drug, consistent with ubiquitylated proteins being a major source of peptides in these cells.
Figure 3. MLN7243 inhibits cytosolic peptide generation.
(A) TAP1-GFP cells were washed for 2 min in cold citric acid buffer, then cultured in the presence of DMSO, cycloheximide (CHX), MLN7243 or MG132, harvested at indicated time points, and analyzed for surface MHC-I complexes by flow cytometry. (B) Lateral mobility of TAP in TAP1-GFP cells determined by FRAP assay. (C) Diffusion rate of TAP1-GFP measured by fluorescence correlation spectroscopy. GraphPad Prism software was used to calculate P values (Unpaired t test).
TAP-FRAP confirmed that each of the drugs significantly decreases peptide supply to TAP (Fig. 3B). We extended these findings using fluorescence correlation spectroscopy (FCS), which measures diffusion rates by the temporal changes in fluorescence of individual molecules (Fig. 3C). The calculated diffusion rate for TAP-GFP was in reasonable agreement with the value obtained via FRAP. Further, FCS confirmed the effect of CHX, MG132 and MLN7243 on speeding TAP diffusion.
Taken together, these data show that MLN7243 effectively blocks the generation of TAP-transported peptides in MelJuSo cells, consistent with a major role of protein ubiquitylation in targeting the source proteins to proteasomes.
The role of ubiquitylation in MHC-I antigen processing is cell and source-protein dependent
To examine the role of ubiquitylation in generating defined antigenic peptides, we first studied the chicken ovalbumin peptide SIINFEKL, which binds with high affinity to H-2Kb (39). We can precisely quantitate cell surface Kb-SIINFEKL complexes by flow cytometry using the 25D1.16 mAb (40), while simultaneously measuring native forms of the source antigen genetically fused to fluorescent reporter proteins.
We expressed SIINFEKL in the context of a number of fusion proteins encoded by recombinant vaccinia viruses:
NP-S-GFP, influenza A virus nucleoprotein (NP) genetically fused to the SIINFEKL peptide followed by APDPPVAT and terminating with eGFP
UbR-NP-S-GFP, identical to NP-S-GFP except for Ub at its amino terminus and the initiating Met of NP changed to Arg
NP(KEKE)-S-GFP, identical to NP-S-GFP with a 29-residue degradation motif inserted at NP residue 333
GFP-Ub-S
GFP-JAW1-S, SIINFEKL appended to the C-terminus of a type II anchored ER protein
One hour after infecting cells with rVVs, we added vehicle or MLN7243 (0.5 or 2.5 μM) to the cells and then measured GFP fluorescence and Kb-SIINFEKL expression by flow cytometry each hour for four hours.
We initially studied UbR-NP-S-GFP as a model Ub-dependent substrate. Ub is co-translationally cleaved from NP, leaving Arg at the amino terminus of NP-S-GFP as a N-end rule degradation motif (41–43). It is degraded with a half-life of 10 min in a proteasome- and E1-dependent manner (25, 44). To optimize MLN7243 use, we performed a dose titration, adding the drug one hour after infection of DC2.4 cells to enable VV to penetrate cells and initiate infection with an intact Ub-proteasome pathway (Fig. S1C). As expected, after 4 h incubation, MLN7243 increased UbR-NP-S-GFP fluorescence. The drug was effective at the lowest concentration used (0.25 μM), and reached maximal effect at 1 μM. At higher concentrations, there was a slight decrease in UbR-NP-S fluorescence, consistent with a minor effect on viral protein synthesis at these concentrations. The generation of Kb-SIINFEKL complexes at the cell surface demonstrated the opposite effect, with ~67% inhibition achieved at 0.5 μM, and complete inhibition observed at 2.5 μM. In subsequent experiments, we therefore used 0.5 μM MLN7243, where GFP fluorescence is nearly fully rescued, and 2.5 μM MLN7243, where Kb-SIINFEKL expression is completely abolished.
Extending these findings to 293-Kb and L-Kb cells (Fig. 4), MLN7243 increases UbR-NP-S-GFP fluorescence while completely (DC2.4, L-Kb) or nearly completely (293-Kb) inhibiting Kb-SIINFEKL expression (summarized in Table II), consistent with our previous finding that peptide generation from UbR-NP-S-GFP is E1-dependent (25). By contrast, HeLa-Kb cells are completely resistant to the effects of MLN7243 on polyubiquitylated proteins (Fig. S1A), and UbR-NP-S-GFP fluorescence or Kb-SIINFEKL generation in HeLa-Kb cells (Fig. S1D). This is consistent with the idea that the effect of MLN7243 on antigen presentation in cells is based on its E1 antagonism.
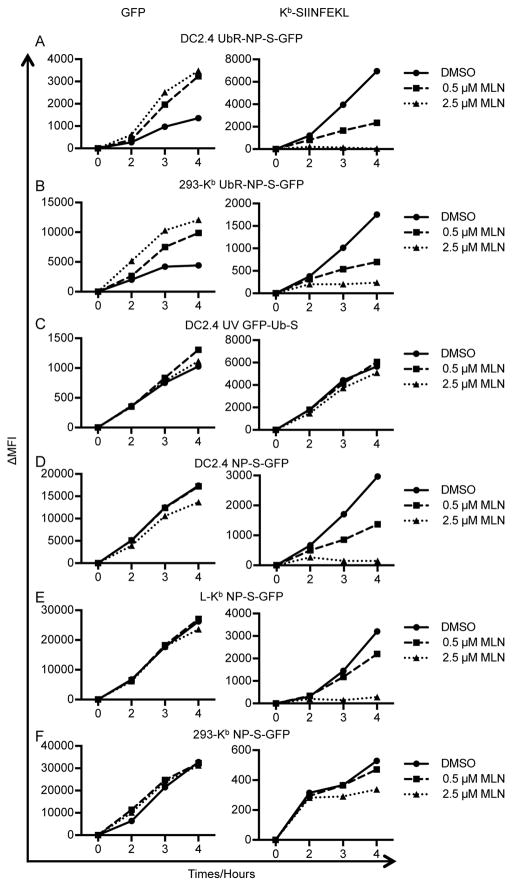
Figure 4. MLN7243 selectively inhibits Kb-SIINFEKL presentation from rVV-expressed proteins.
DC2.4 (A, C and D), L-Kb (E), and 293-Kb (B and F) cells were infected for 1 h with rVV expressing UbR-NP-S-GFP (A and B), partially UV-inactivated rVV expressing GFP-Ub-S (C), or rVV expressing NP-S-GFP (D–F). Cells were then cultured in the presence of DMSO, 0.5 μM MLN7243, or 2.5 μM MLN7243 and harvested at indicated time points. Levels of GFP (left panels) and surface Kb-SIINFEKL (right panels) were determined by flow cytometry. ΔMFI: medium fluorescent intensity after background subtraction.
Table II.
Summary of Kb-SIINFEKL presentation from rVVs
| Cell line | Source Protein | Inhibitor effect |
|---|---|---|
| 293-Kb | NP-S-GFP | + |
| UbR-NP-S-GFP | ++ | |
| NP(KEKE)-S-GFP | ++ | |
| GFP-Ub-S | − | |
| GFP-Jaw1-S | − | |
| L-Kb | NP-S-GFP | +++ |
| UbR-NP-S-GFP | +++ | |
| NP(KEKE)-S-GFP | +++ | |
| GFP-Ub-S | − | |
| GFP-Jaw1-S | − | |
| DC2.4 | NP-S-GFP | +++ |
| UbR-NP-S-GFP | +++ | |
| NP(KEKE)-S-GFP | +++ | |
| GFP-Ub-S | − | |
| GFP-Jaw1-S | − |
The effects of MLN7243, unlike MG132, on UbR-NP-S-GFP fluorescence and peptide generation are irreversible for at least 2 hours (Fig. S1E), consistent with the biochemical findings above (Fig. 1C). NP(KEKE)-S-GFP, which misfolds and is ubiquitylated and degraded by proteasomes with a t½ of ~ 70 min (45), behaves similarly to UbR-NP-S-GFP in all three cells lines, with Kb-SIINFEKL presentation being completely or nearly completely blocked (data summarized in Table II).
By contrast, in each of the cell lines, MLN7243 has essentially no effect on Kb-SIINFEKL generation from GFP-Ub-S (Fig. 4C and Table II), which generates SIINFEKL in a proteasome-independent matter by the rapid action of ubiquitin hydrolases (14). To prevent saturation of the class I pathway by SIINFEKL, which is generated in relatively large amounts compared to its normal liberation by proteasomes, we UV-irradiated virus to limit GFP-Ub-S expression and attained levels of surface Kb-SIINFEKL similar to those expressing UbR-NP-S-GFP. Similarly, MLN7243 did not inhibit Kb-SIINFEKL generation from GFP-Jaw1-S (Table II), a post-translationally targeted ER protein that provides SIINFEKL in a TAP- and proteasome-independent manner (46).
These positive controls (UbR-NP-S-GFP, NP(KEKE)-S-GFP) demonstrate that MLN7243 effectively inhibits Kb-SIINFEKL generation from Ub-dependent substrates in each cell line tested, while the negative controls (GFP-Ub-S, GFP-Jaw1-S) show that inhibition is not based on blocking antigen synthesis, TAP transport, class I synthesis, peptide loading, transport to the cell surface or stability of cell surface complexes. Would MLN7243 block Kb-SIINFEKL generation from DRiPs generated from NP-S-GFP, a metabolically stable viral protein (44, 45)?
MLN7243 had little to no effect on NP-S-GFP fluorescence in each of the three cell lines tested (Fig. 4D–F, left panels), indicating that there is not a pool of fluorescent DRiPs degraded in a Ub-dependent manner that is detectable by flow cytometry. In DC2.4 or L-Kb cells, 2.5 μM MLN7243 completely inhibited Kb-SIINFEKL generation. By contrast, although MLN7243 was most effective in 293-Kb cells in blocking polyubiquitylation (Fig. 1A) and surface Kb re-expression following acid stripping (Fig. 2C), it was least effective in blocking Kb-SIINFEKL generation from NP-S-GFP. There was no effect in the first two hours post-infection, where presentation should be dominated by the DRiP pool, given the extremely limited size of the retiree pool and the long half-life of NP-S-GFP (estimated at 69 h, see below).
Extending these findings to antigens synthesized from cellular mRNAs, we found that after acid stripping, MLN7243 clearly blocked cell surface Kb-SIINFEKL re-expression from cells transfected with a plasmid encoding SIINFEKL inserted between Myc epitope and Venus (Myc-S-Venus), but not from cells transfected with a plasmid encoding SIINFEKL at the C-terminus of an ER targeting sequence (ER-S), which is presented in TAP and proteasome independent manner (Fig. S1F).
These findings demonstrate that MLN7243 can be used to specifically measure the involvement of ubiquitylation in generation of class I peptide complexes, and further, that the requirement for ubiquitylation in generating a given peptide varies with the context of peptide in a source protein and the cell line expressing the source protein.
Ubiquitylation is rate limiting for NP-S-GFP DRiP peptide generation
We next used MLN7243 in conjunction with CHX and MG132 to examine the NP-S-GFP and UbR-NP-S-GFP DRiP substrate pool size and degradation kinetics in L-Kb cells, where generation of Kb-SIINFEKL from both substrates is strictly Ub-dependent. We added inhibitors three hour post-rVV infection, when antigen synthesis and presentation rates reach their Vmax, and measured their effects on substrate fluorescence and Kb-SIINFEKL surface expression at five 30 min intervals (Fig. 5).
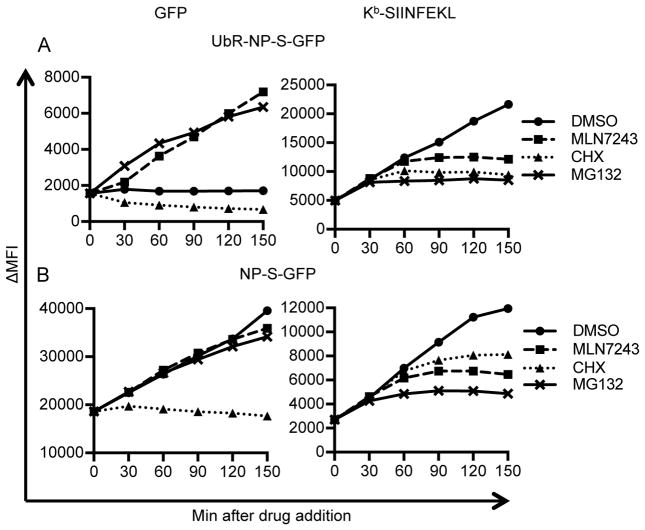
Figure 5. Fine kinetics of inhibitor blockade.
L-Kb cells were infected with rVVs expressing UbR-NP-S-GFP (A) or NP-S-GFP (B) for 1 h. At 3 h, DMSO, MLN7243, CHX or MG132 were added into cell cultures. Cells were harvested at indicated time points after drug addition. Levels of GFP (left panels) and surface Kb-SIINFEKL (right panels) were determined by flow cytometry. ΔMFI: medium fluorescent intensity after background subtraction.
Looking first at UbR-NP-S-GFP fluorescence (Fig. 5A, left panel), in the presence of DMSO, GFP levels are steady, indicating that the rate of degradation matches the rate of synthesis. CHX immediately shutdown GFP synthesis, while MLN7243, after a slightly less than 30 min lag, increased GFP fluorescence to the same or even greater extent as MG132 (compare the slopes of MLN7243 after 30 min and of MG132 in Fig. 5A, left panel). This is consistent with a complete functional blockade of ubiquitylation and complete dependence of UbR-NP-S-GFP degradation on ubiquitylation. We interpret the 30 min delay between MLN7243 and MG132 action to represent the time (t½ ~10 min) required to deplete Ub from existing E1, E2 and E3 complexes involved in UbR-NP-S-GFP degradation (though we cannot eliminate a contribution from a lag in MLN7234 blockade of E1 after adding it to cells).
Neither MG132 nor MLN7243 modified the linear increase in NP-S-GFP fluorescence, demonstrating that they do no act by blocking protein synthesis (Fig. 5B, left panel). Following CHX addition, NP-S-GFP fluorescence decreases by 2% in 2h, consistent with a 69 h t½. for fluorescent molecules. UbR-NP-S-GFP fluorescence decays much more rapidly, but still this is an overestimate of the metabolic stability, since non-fluorescent molecules, which make up the bulk of the population, are degraded much more rapidly, with the entire population exhibiting a biochemical t½ of ~ 10 min (44).
Turning to antigenic peptide generation (Fig. 5A and B, right panels), each of the inhibitors completely blocked Kb-SIINFEKL surface expression after various intervals. As with UbR-NP-S-GFP fluorescence, MG132 acted more rapidly than MLN7243. Interestingly, CHX shutdown of Kb-SIINFEKL expression was faster than MLN7243 for UbR-NP-S-GFP, but slower for NP-S-GFP. This can be most clearly seen by normalizing the data to the maximum expression of Kb-SIINFEKL achieved after the shutdown, where the kinetics of shutdown with MLN7243 and MG132 are nearly identical between UbR-NP-S-GFP and NP-S-GFP (Fig. S2A and B), while CHX is clearly slower for NP-S-GFP (Fig. S2C).
For both NP-S-GFP and UbR-NP-S-GFP, after adding MG132, Kb-SIINFEKL surface complexes reach near maximal values within 60 min with t½ around 20 min (Fig. S2A, t½s are shown by dotted lines in Fig. S2A–C). This is consistent with an <30 min t½ for delivery of proteasome generated-peptides to the cell surface via Kb molecules (Fig. S2A). By contrast, after adding MLN7243, an additional 10 min is required to reach half maximal values (t½ ~30 min, Fig. S2B), consistent with a ~10 min t½ for discharging of the relevant E3-Ub complexes. For both NP-S-GFP and UbR-NP-S-GFP, considerably more complexes are generated post addition of MLN7243 vs. MG132 (Fig. 5), implying that a large fraction of antigen that is ubiquitylated is committed to proteasome degradation and not subject to deubiquitylation that would preclude proteasome degradation.
Notably, the kinetic behavior of Kb-SIINFEKL complexes generated from NP-S-GFP and UbR-NP-S-GFP differ following CHX addition, with t½s of UbR-NP-S-GFP <30 min and NP-S-GFP ~ 40 min (Fig. S2C). Taking into account the time required for Kb-SIINFEKL surface delivery post-proteasomal processing, we can estimate that the relevant pools of UbR-NP-S-GFP and NP-S-GFP for antigen presentation are degraded, respectively, with t½s of < 5 min and ~ 10 min (in the latter case ~400-fold faster than NP-S-GFP fluorescence).
We can also analyze the numbers of Kb-SIINFEKL complexes delivered to the cell surface after drug treatment. By equating this cohort relative to minutes of expression required to observe similar increase in complex numbers in untreated cells, we can normalize for the greater Kb-SIINFEKL complex generation from UbR-NP-S-GFP vs. NP-S-GFP (Table III and Fig. S2D and E). After MG132 treatment, the relative pool sizes are similar, at 30–40 expression min. The post-MLN7243 pool size again is equivalent, with both at 60 expression min, consistent as above that after depleting existing E3-Ub conjugates, 20–30 min is required for proteasomal processing of Ub-antigen conjugate. By contrast, the post-CHX pools are not equivalent. For UbR-NP-S-GFP the post-CHX pool is smaller than the post- MLN7243 pool size (45 vs. 60 expression min, due to the lag in MLN7243 action), while the opposite is true with NP-S-GFP (80 vs. 60 expression min). This is completely consistent with pool sizes and degradation kinetics of UbR-NP-S-GFP vs. NP-S-GFP. With their slower targeting for ubiquitylation, NP-S-GFP DRiPs are more slowly degraded once protein synthesis is abrogated.
Table III.
Precursor SIINFEKL pool size: minutes of normal expression
| NP-S-GFP | UbR-NP-S-GFP | |
|---|---|---|
| CHX | 80 | 45 |
| MLN7243 | 60 | 60 |
| MG132 | 40 | 30 |
Ubiquitylation requirement for antigen presentation varies widely among influenza A virus (IAV) peptides
We next examined the requirement for ubiquitylation in generating seven distinct peptides generated from two different IAV proteins expressed in the context of IAV infection of mouse or human cells (Fig. 6). Since we do not have Abs specific for the relevant peptide class I complexes, we measured class I peptide expression by activation of in vitro propagated CD8+ T cell lines (27, 30). To quantitate the effect of MLN7243 on the generation of cognate cell surface class I peptide complexes, we treated infected cells with BFA to ensure that we were measuring presentation under complex limiting conditions. We then related complexes generated in the presence and absence of MLN7243 to peptide titration curves performed in parallel on the same day of the assay (Figs. S3A and C). In this way, we could determine the relative number of complexes presented by infected cells in the presence and absence of MLN7243 (quantitated in Supplementary Table). We also measured viral protein expression by infected cells, to account for potential effects of MLN7243 on viral gene expression (Fig. S3B and D).
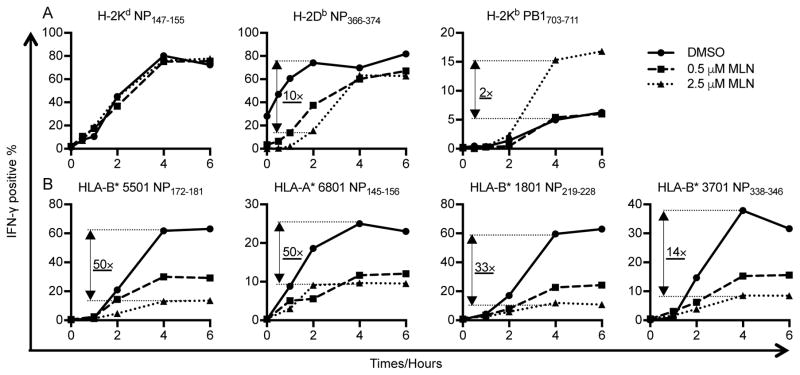
Figure 6. MLN7243 differentially affects mouse and human MHC-I presentation of IAV antigens.
Antigen presenting cells were infected with IAV PR8 for 60 min, cultured in the presence of DMSO, 0.5 μM MLN7243, or 2.5 μM MLN7243, washed, and used to stimulate T cell lines with BFA addition at indicated time points. Antigen presentation of mouse H-2Kd restricted NP147–155, H-2Db restricted NP366–374 and H-2Kb restricted PB1703–711 (A) and human HLA-B* 5501 restricted NP172–181, HLA-A* 6801 restricted NP145–156, HLA-B* 3701 NP338–346 and HLA-B* 1801 restricted NP219–228 (B) was assessed by specific TCD8+ cultures by IFN-γ ICS.
We first examined antigen presentation by mouse P815 (H-2d) and EL4 (H-2b) cells. MLN7243 effectively blocked ubiquitylation in these cells (Fig. S1B), and also moderately decreased viral gene expression as assessed by flow cytometric measurement of intracellular NP (Fig. S3B). Interestingly, of three peptides examined (Fig. 6A, quantification in Supplementary Table), only the Db-restricted peptide NP366–374, was inhibited by MLN7243, with an approximate 10-fold reduction in peptide presentation early in infection. By contrast, presentation of the Kb restricted PB1703–711 peptide was enhanced two-fold by 2.5 μM MLN7243 and unchanged by 0.5 μM MLN7243. MLN7243 had no effect on presentation of H-2Kd restricted NP147–155, consistent with the findings of Huang et al. (15).
MLN7243 inhibited all HLA-restricted IAV NP peptides we examined using IAV infected autologous EBV-transformed B lymphocyte cell lines as APCs. DMSO treated cells generated 14- to 50-fold more complexes than 2.5 μM MLN7243 treated cells at 4 h post infection (Fig. 6B, quantification in Supplementary Table). The dramatic inhibition cannot be explained by the slight decrease of NP signal in the corresponding antigen presenting cells (Fig. S3D). Taken together, these findings reinforce our conclusion that the requirement for ubiquitylation varies widely for presentation of any given antigenic peptide.
Discussion
Our findings reveal a varied role of ubiquitylation in generating MHC class I peptide ligand, depending on cell type, class I allomorph, source protein and precise peptide examined. Our results reconcile previous reports that used varied strategies to examine presentation of individual defined antigenic peptides with conflicting results (Table 1).
We show that MLN7243 rapidly blocks ubiquitylation in cultured cells, as clearly shown by blocking MG132 induced increases in polyubiquitylated proteins, apparent after 10 min (Fig. 1). The MLN7243 E1 blockade lasts at least two hours after removing the drug from the media, suggesting that it irreversibly inactivates E1 in cells (Fig. 1 and Fig. S1E). While we cannot conclude that MLN7243 completely blocks ubiquitylation in the cells we examined, our data strongly suggests that the block is at least nearly complete.
In addition to E1 (UBA1), MLN7243 inhibits ubiquitin-activating enzyme E1-like protein 2 (UBA6) and thus blocks FAT10yation of proteins. We believe it is unlikely, however, that inhibition of FAT10ylation makes a major contribution to our findings because UBA6 is present at ~10% of the levels of UBA1 in most cells (8) and the evidence to date does not support a major role for FAT10 in antigen presentation (47).
Because of the complexity and broad influence of ubibuitylation in cellular functions, it is impossible to perfectly isolate the participation of Ub in peptide generation from other pathways can could potentially modulate peptide generation or class I biogenesis. By limiting the duration and concentration of MLN7243, however, we can minimize the downstream effects of cell stress pathways that are activated by the inhibition of ubiquitylation.
MLN7243 nearly completely blocks generation of Kb-SIINFEKL complexes from two rVV encoded misfolding forms of NP that are degraded in a proteasome dependent manner (44, 45), providing a clear example of Ub-dependent peptide presentation (Fig. 4). MLN7243 had no significant effect on presentation of proteasome independent substrates, include ER-targeted SIINFEKL from GFP-Jaw1-S and cytosolic liberated SIINFEKL from Ub-GFP-S, demonstrating that ongoing ubiquitylation is not required for TAP-mediated peptide transport, peptide loading in the ER, transport of class I peptide complexes to the cell surface, or stable surface class I surface expression (Fig. 4).
Things get interesting with a more natural DRiP substrate, NP-S-GFP (Fig. 4). Here, generation of surface Kb-SIINFEKL complexes is Ub-dependent in DC2.4 and L-Kb cells, but largely Ub-independent in 293-Kb cells, despite highly effective blockade of ubiquitylation shown by immunoblotting (Fig. 1), and also by a nearly complete blockade of Kb re-expression after acid stripping of the same cells (Fig. 2). Although the auto-catalytic cleavage of GFP in NP-S-GFP (28), responsible for ~50% of its DRiP dependent Kb-SIINFEKL generation, makes it a highly unusual substrate, we also observe MLN7243-resistant (or even enhanced) presentation of a two IAV peptides generated from unmodified viral proteins in IAV infected-cells, NP147–155, previously described as proteasome dependent (48), and PB1703–711, whose proteasome-dependence can be inferred by its dependence on immunoproteasome subunit expression (4).
Indeed, the effect of MLN7243 on class I re-expression clearly demonstrates its varied effect on peptide generation. In DC2.4 and 293-Kb cells, the effects of MLN7243 on Kb surface expression are nearly identical to MG132 (proteasome inhibitor), while in L-Kb cells MLN7243 is less effective than MG132, suggesting a considerable contribution of Ub-independent, proteasome dependent peptide generation. We see similar discrepancies with Db in DC2.4 cells and Dk in L-Kb cells. Strikingly, for KK in L-Kb cells, the bulk of peptides appear to be generated in a Ub- and proteasome-independent manner.
Such allomorph dependent proteasome inhibitor resistant bulk peptide-presentation has been previously reported (24, 49, 50). Specifically, HLA-A3, HLA-A11, and HLA-B35 appear to be loaded with peptides normally while peptide loading of many other HLA class I allomorphs are inhibited by the proteasome inhibitor lactacystin (50). As it is difficult to establish that proteasome inhibitors completely block all proteasome active sites the parallel effects of MLN7243 and MG132 we observe, strongly support the conclusion that at least some of these previous observations provide an accurate assessment of a major contribution of proteasome independent-peptide generation to immunosurveillance.
What non-proteasomal processes might be responsible for generating such peptides? An exciting report attributing a significant role to TPPII in cellular proteostasis (51) based on TPPII up regulation in cells propagated with the proteasome inhibitor NLVS was contradicted by subsequent findings that such cells were more sensitive to other proteasome inhibitors than untreated cells (52). Indeed, it now appears that cells lack endoproteases that can replace the protean activity of proteasomes in degrading large proteins.
Cells do, however, express abundant that exopeptidases and endopeptidases that can potentially participate in peptide generation (reviewed in (53)), particularly if presented with already misfolded or fragmented polypeptides as might arise as DRiPs. Such presentation would presumably be enhanced when proteasome function is impaired, allowing lower affinity peptides or peptides generated from kinetically inferior pathways to predominate.
A possible explanation for proteasome-independent antigen presentation is that some class I allomorphs specialize in presenting the oligopeptides synthesized as alternative translation products, perhaps generated by immunoribosomes, proposed ribosomes specialized for antigen presentation (54, 55). Such ribosomes might generate short polypeptides from non-standard open reading frames (56), such as introns (57, 58) and other mRNAs translated in the nucleus (59–62), alternative initiation codons (63–66), frame shifted (67) (68) or prematurely terminated translation products. Such translation product peptides could be short enough for immediate TAP transport, or might require trimming by cytosolic aminopeptidases (69–72).
Our findings with bulk peptide generation (Fig. 2) also support a role for Ub-independent, proteasome dependent-peptide generation. This was first described for ornithine decarboxylase (73), which is targeted to the 19S regulatory subunit of 26S proteasomes by antizyme (74), rather than polyUb. More broadly, 20S proteasome lacking 19S regulators appear to be specialized for degrading oxidatively damaged non-ubiquitylated proteins (10), and may play an important role in antigen presentation.
In summary, MLN7243 enabled a new and powerful approach to understanding the role of protein ubiquitylation in the generation of class I peptide ligands. Our findings conclusively demonstrate a varied role of ubiquitylation in class I antigenic peptide generation, underscoring the use of multiple pathways to generate peptides for CD8+ cell immunosurveillance.
Supplementary Material
Acknowledgments
Glennys Reynoso provided outstanding technical assistance.
Footnotes
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbriavation used: DRiPs, defective ribosomal products; ER, endoplasmic reticulum; DUB, deubiquitylating enzyme; Ub, ubiquitin; rVV, recombinant vaccinia virus; RT, room temperature; IAV, influenza A virus; ICS, intracellular cytokine staining; FRAP, fluorescence recovery after photo bleaching; CHX, cycloheximide.
References
- 1.Anton LC, Yewdell JW. Translating DRiPs: MHC class I immunosurveillance of pathogens and tumors. J Leukoc Biol. 2014;95:551–562. doi: 10.1189/jlb.1113599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourdetsky D, Schmelzer CE, Admon A. The nature and extent of contributions by defective ribosome products to the HLA peptidome. Proc Natl Acad Sci USA. 2014;111:E1591–1599. doi: 10.1073/pnas.1321902111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purcell AW, Croft NP, Tscharke DC. Immunology by numbers: quantitation of antigen presentation completes the quantitative milieu of systems immunology! Curr Opin Immunol. 2016;40:88–95. doi: 10.1016/j.coi.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Zanker D, Waithman J, Yewdell JW, Chen W. Mixed Proteasomes Function To Increase Viral Peptide Diversity and Broaden Antiviral CD8+ T Cell Responses. J Immunol. 2013;191:52–59. doi: 10.4049/jimmunol.1300802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croft NP, Smith SA, Wong YC, Tan CT, Dudek NL, Flesch IEA, Lin LCW, Tscharke DC, Purcell AW. Kinetics of Antigen Expression and Epitope Presentation during Virus Infection. PLoS Pathog. 2013;9:e1003129. doi: 10.1371/journal.ppat.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggensperger S, Tampe R. The transporter associated with antigen processing: a key player in adaptive immunity. Biol Chem. 2015;396:1059–1072. doi: 10.1515/hsz-2014-0320. [DOI] [PubMed] [Google Scholar]
- 7.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 8.Clague MJ, Heride C, Urbé S. The demographics of the ubiquitin system. Trends Cell biol. 2015;25:417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Raynes R, Pomatto LC, Davies KJ. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol Aspects Med. 2016;50:41–55. doi: 10.1016/j.mam.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golnik R, Lehmann A, Kloetzel PM, Ebstein F. Major Histocompatibility Complex (MHC) Class I Processing of the NY-ESO-1 Antigen Is Regulated by Rpn10 and Rpn13 Proteins and Immunoproteasomes following Non-lysine Ubiquitination. J Biol Chem. 2016;291:8805–8815. doi: 10.1074/jbc.M115.705178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiebiger BM, Pfister H, Behrends U, Mautner J. Polyubiquitination of lysine-48 is an essential but indirect signal for MHC class I antigen processing. Eur J Immunol. 2015;45:716–727. doi: 10.1002/eji.201444830. [DOI] [PubMed] [Google Scholar]
- 13.Palmer AL, Dolan BP. MHC class I antigen presentation of DRiP-derived peptides from a model antigen is not dependent on the AAA ATPase p97. PloS one. 2013;8:e67796. doi: 10.1371/journal.pone.0067796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan BP, Li L, Veltri CA, Ireland CM, Bennink JR, Yewdell JW. Distinct Pathways Generate Peptides from Defective Ribosomal Products for CD8+ T Cell Immunosurveillance. J Immunol. 2011;186:2065–2072. doi: 10.4049/jimmunol.1003096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, Marvin JM, Tatsis N, Eisenlohr LC. Cutting Edge: Selective role of ubiquitin in MHC class I antigen presentation. J Immunol. 2011;186:1904–1908. doi: 10.4049/jimmunol.1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 2010;190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian SB, Bennink JR, Yewdell JW. Quantitating defective ribosome products. Methods Mol Biol. 2005;301:271–281. doi: 10.1385/1-59259-895-1:271. [DOI] [PubMed] [Google Scholar]
- 18.Michalek MT, Grant EP, Rock KL. Chemical denaturation and modification of ovalbumin alters its dependence on ubiquitin conjugation for class I antigen presentation. J Immunol. 1996;157:617–624. [PubMed] [Google Scholar]
- 19.Grant EP, Michalek MT, Goldberg AL, Rock KL. Rate of antigen degradation by the ubiquitin-proteasome pathway influences MHC class I presentation. J Immunol. 1995;155:3750–3758. [PubMed] [Google Scholar]
- 20.Cox JH, Galardy P, Bennink JR, Yewdell JW. Presentation of endogenous and exogenous antigens is not affected by inactivation of E1 ubiquitin-activating enzyme in temperature-sensitive cell lines. J Immunol. 1995;154:511–519. [PubMed] [Google Scholar]
- 21.Michalek MT, Grant EP, Gramm C, Goldberg AL, Rock KL. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. 1993;363:552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 22.Engelhard VH, Brickner AG, Zarling AL. Insights into antigen processing gained by direct analysis of the naturally processed class I MHC associated peptide repertoire. Mol Immunol. 2002;39:127–137. doi: 10.1016/s0161-5890(02)00096-2. [DOI] [PubMed] [Google Scholar]
- 23.Gromme M, Neefjes J. Antigen degradation or presentation by MHC class I molecules via classical and non-classical pathways. Mol Immunol. 2002;39:181–202. doi: 10.1016/s0161-5890(02)00101-3. [DOI] [PubMed] [Google Scholar]
- 24.Vinitsky A, Anton LC, Snyder HL, Orlowski M, Bennink JR, Yewdell JW. The generation of MHC class I-associated peptides is only partially inhibited by proteasome inhibitors: involvement of nonproteasomal cytosolic proteases in antigen processing? J Immunol. 1997;159:554–564. [PubMed] [Google Scholar]
- 25.Qian SB, Princiotta MF, Bennink JR, Yewdell JW. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem. 2006;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- 26.Milhollen M, Sappal D, Duffy J, Hoar K, Huck J, Sha P, Koenig E, Hyer M, Ciavarri J, Bence N. 577 Characterization of the cellular mechanism of action of the first in class investigational inhibitor of the Ubiquitin Activating Enzyme, MLN7243. Eur J Cancer. 2014;50:186. [Google Scholar]
- 27.Wu C, Zanker D, Valkenburg S, Tan B, Kedzierska K, Zou QM, Doherty PC, Chen W. Systematic identification of immunodominant CD8+ T-cell responses to influenza A virus in HLA-A2 individuals. Proc Natl Acad Sci U S A. 2011;108:9178–9183. doi: 10.1073/pnas.1105624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei J, Gibbs JS, Hickman HD, Cush SS, Bennink JR, Yewdell JW. Ubiquitous Autofragmentation of Fluorescent Proteins Creates Abundant Defective Ribosomal Products (DRiPs) for Immunosurveillance. J Biol Chem. 2015;290:16431–16439. doi: 10.1074/jbc.M115.658062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanker D, Xiao K, Oveissi S, Guillaume P, Luescher IF, Chen W. An optimized method for establishing high purity murine CD8(+) T cell cultures. J Immunol Methods. 2013;387:173–180. doi: 10.1016/j.jim.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Grant E, Wu C, Chan KF, Eckle S, Bharadwaj M, Zou QM, Kedzierska K, Chen W. Nucleoprotein of influenza A virus is a major target of immunodominant CD8 + T-cell responses. Immunol Cell Biol. 2013;91:184–194. doi: 10.1038/icb.2012.78. [DOI] [PubMed] [Google Scholar]
- 31.Fujimuro M, Sadada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Letters. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 32.Sugawara S, Abo T, Kumagai K. A simple method to eliminate the antigenicity of surface class I MHC molecules from the membrane of viable cells by acid treatment at pH 3. J Immunol Methods. 1987;100:83–90. doi: 10.1016/0022-1759(87)90175-x. [DOI] [PubMed] [Google Scholar]
- 33.Polakova K, Karpatova M, Russ G. Dissociation of β2-microglobulin is responsible for selective reduction of HLA class I antigenicity following acid treatment of cells. Mol Immunol. 1993;30:1223–1240. doi: 10.1016/0161-5890(93)90037-c. [DOI] [PubMed] [Google Scholar]
- 34.Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heinen E, Bastin J, Schumacher TNM, Townsend A, Karre K, Ploegh HL. Empty MHC class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 35.Day PM, Esquivel F, Lukszo J, Bennink JR, Yewdell JW. Effect of TAP on the generation and intracellular trafficking of peptide-receptive major histocompatibility complex class I molecules. Immunity. 1995;2:137–147. doi: 10.1016/s1074-7613(95)80014-x. [DOI] [PubMed] [Google Scholar]
- 36.Reits EA, Vos JC, Gromme M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- 37.Maryanski JL, Casanova JL, Falk K, Gournier H, Jaulin C, Kourilsky P, Lemonnier FA, Luthy R, Rammensee HG, Rotzschke O, Servis C, Lopez JA. The diversity of antigen-specific TCR repertoires reflects the relative complexity of epitopes recognized. Hum Immunol. 1997;54:117–128. doi: 10.1016/s0198-8859(97)00082-7. [DOI] [PubMed] [Google Scholar]
- 38.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens--new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 39.Rotzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee HG. Exact prediction of a natural T cell epitope. Eur J Immunol. 1991;21:2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]
- 40.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 41.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 42.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townsend A, Bastin J, Gould K, Brownlee G, Andrew M, Coupar B, Boyle D, Chan S, Smith G. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J Exp Med. 1988;168:1211–1224. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 45.Anton LC, Schubert U, Bacik I, Princiotta MF, Wearsch PA, Gibbs J, Day PM, Realini C, Rechsteiner MC, Bennink JR, Yewdell JW. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snyder HL, Bacik I, Bennink JR, Kearns G, Behrens TW, Bachi T, Orlowski M, Yewdell JW. Two novel routes of transporter associated with antigen processing (TAP)-independent major histocompatibility complex class I antigen processing. J Exp Med. 1997;186:1087–1098. doi: 10.1084/jem.186.7.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basler M, Buerger S, Groettrup M. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Mol Immunol. 2015;68:129–132. doi: 10.1016/j.molimm.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Wherry EJ, Golovina TN, Morrison SE, Sinnathamby G, McElhaugh MJ, Shockey DC, Eisenlohr LC. Re-evaluating the generation of a “proteasome-independent” MHC class I-restricted CD8 T cell epitope. J Immunol. 2006;176:2249–2261. doi: 10.4049/jimmunol.176.4.2249. [DOI] [PubMed] [Google Scholar]
- 49.Luckey CJ, King GM, Marto JA, Venketeswaran S, Maier BF, Crotzer VL, Colella TA, Shabanowitz J, Hunt DF, Engelhard VH. Proteasomes can either generate or destroy MHC class I epitopes: evidence for nonproteasomal epitope generation in the cytosol. J Immunol. 1998;161:112–121. [PubMed] [Google Scholar]
- 50.Benham AM, Gromme M, Neefjes J. Allelic differences in the relationship between proteasome activity and MHC class I peptide loading. J Immunol. 1998;161:83–89. [PubMed] [Google Scholar]
- 51.Glas R, Bogyo M, McMaster JS, Gaczynska M, Ploegh HL. A proteolytic sysytem that compensates for loss of proteasome function. Nature. 1998;392:618–622. doi: 10.1038/33443. [DOI] [PubMed] [Google Scholar]
- 52.Princiotta MF, Schubert U, Chen W, Bennink JR, Myung J, Crews CM, Yewdell JW. Cells adapted to the proteasome inhibitor 4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc Natl Acad Sci USA. 2001;98:513–518. doi: 10.1073/pnas.021132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazaro S, Gamarra D, Del Val M. Proteolytic enzymes involved in MHC class I antigen processing: A guerrilla army that partners with the proteasome. Mol Immunol. 2015;68:72–76. doi: 10.1016/j.molimm.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Yewdell J. To DRiP or not to DRiP: generating peptide ligands for MHC class I molecules from biosynthesized proteins. Mol Immunol. 2002;39:139–146. doi: 10.1016/s0161-5890(02)00097-4. [DOI] [PubMed] [Google Scholar]
- 55.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Van Pel A, Boon T. T cell-recognized antigenic peptides derived from the cellular genome are not protein degradation products but can be generated directly by transcription and translation of short subgenic regions. A hypothesis. Immunogenetics. 1989;29:75–79. doi: 10.1007/BF00395854. [DOI] [PubMed] [Google Scholar]
- 57.Apcher S, Millot G, Daskalogianni C, Scherl A, Manoury B, Fahraeus R. Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci USA. 2013;110:17951–17956. doi: 10.1073/pnas.1309956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robbins PF, El-Gamil M, Li YF, Fitzgerald EB, Kawakami Y, Rosenberg SA. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J Immunol. 1997;159:303–308. [PubMed] [Google Scholar]
- 59.Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells 4. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 60.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolan BP, Knowlton JJ, David A, Bennink JR, Yewdell JW. RNA polymerase II inhibitors dissociate antigenic peptide generation from normal viral protein synthesis: a role for nuclear translation in defective ribosomal product synthesis? J Immunol. 2010;185:6728–6733. doi: 10.4049/jimmunol.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Apcher S, Daskalogianni C, Lejeune F, Manoury B, Imhoos G, Heslop L, Fåhraeus R. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc Natl Acad Sci USA. 2011;108:11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- 64.Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. Unanticipated antigens: Translation initiation at CUG with leucine. PLoS Biol. 2004;2:1774–1784. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bullock TN, Patterson AE, Franlin LL, Notidis E, Eisenlohr LC. Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression. J Exp Med. 1997;186:1051–1058. doi: 10.1084/jem.186.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bullock TNJ, Eisenlohr LC. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J Exp Med. 1996;184:1319–1329. doi: 10.1084/jem.184.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fetten JV, Roy N, Giboa E. A frameshift mutation at the NH2 terminus of the nucleoprotein gene does not affect generation of cytotoxic T lymphocyte epitopes. J Immunol. 1991;147:2697–2705. [PubMed] [Google Scholar]
- 68.Zook MB, Howard MT, Sinnathamby G, Atkins JF, Eisenlohr LC. Epitopes derived by incidental translational frameshifting give rise to a protective CTL response. J Immunol. 2006;176:6928–6934. doi: 10.4049/jimmunol.176.11.6928. [DOI] [PubMed] [Google Scholar]
- 69.Lopez D, Gil-Torregrosa BC, Bergmann C, del Val M. Sequential cleavage by metallopeptidases and proteasomes is involved in processing HIV-1 ENV epitope for endogenous MHC class I antigen presentation. J Immunol. 2000;164:5070–5077. doi: 10.4049/jimmunol.164.10.5070. [DOI] [PubMed] [Google Scholar]
- 70.Saric T, Beninga J, Dax C, Akopian TN, Rock KL, Goldberg AL. MHC class I-presented peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J Biol Chem. 2001;276:36474–36481. doi: 10.1074/jbc.M105517200. [DOI] [PubMed] [Google Scholar]
- 71.Beninga J, Rock KL, Goldberg AL. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 72.Stoltze L, Schirle M, Schwarz G, Schroter C, Thompson MW, Hersh LB, Kalbacher H, Stevanovic S, Rammensee HG, Schild H. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 73.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 74.Murakami Y, Matsufuji S, Kameji T, Hayashi Si, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitin. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.