Abstract
A novel modified nucleic acid nanoparticle harboring an annexin A2 aptamer for ovarian cancer cell targeting and a GC rich sequence for doxorubicin loading is designed and constructed. The system utilizes a highly stable three-way junction (3WJ) motif from phi29 packaging RNA as a core structure. A phosphorothioate-modified DNA aptamer targeting annexin A2, Endo28, was conjugated to one arm of the 3WJ. The pRNA-3WJ motif retains correct folding of attached aptamer, keeping its functions intact. It is of significant utility for aptamer-mediated targeted delivery. The DNA/RNA hybrid nanoparticles remained intact after systemic injection in mice and strongly bound to tumors with little accumulation in healthy organs 6 hours post-injection. The Endo28-3WJ-Sph1/Dox intercalates selectively enhanced toxicity to annexin A2 positive ovarian cancer cells in vitro. The constructed RNA/DNA hybrid nanoparticles can potentially enhance the therapeutic efficiency of doxorubicin at low doses for ovarian cancer treatment through annexin A2 targeted drug delivery.
Keywords: RNA Nanotechnology, aptamer, Annexin A2, Doxorubicin, Ovarian cancer
Graphical Abstract
INTRODUCTION
Ovarian cancer is a highly metastatic and lethal disease with the highest mortality rate of all cancers of the female reproductive system1. Most patients have high-grade disease with metastasis at the time of diagnosis due to vague clinical symptoms at early stages. The 5-year survival rate for patients with advanced disease is very low despite cytoreductive surgery and chemotherapy combination regimens. Therefore, new and innovative therapeutic strategies are needed for the treatment of ovarian cancer to improve the dismal statistics of overall survival of ovarian cancer patients. Targeted delivery of therapeutics to cancer cells, with little collateral damage to healthy cells, remains a promising option for the treatment of ovarian cancer 2.
Doxorubicin, an anthracycline chemotherapy drug, is widely used for treatment of ovarian and other cancers3. It slows or stops the growth of cancer cells by intercalation with DNA and inhibiting macromolecular biosynthesis4. Long-circulating PEGylated liposomal doxorubicin is an FDA approved drug for the treatment of recurrent ovarian cancer5. Given that the delivery system could benefit the therapeutic effect of doxorubicin on ovarian cancers, we explored the possibility of using RNA nanoparticles with a targeting ligand for targeted drug delivery to treat ovarian cancer.
Over the past 30 years, monoclonal antibodies have developed as the affinity ligand of choice for identifying proteins in research, diagnostics, biosensors, imaging, and more recently as therapeutics. The impact of antibodies on biology and medicine has been profound and is an increasingly important component of the biotechnology industry. However, antibodies have significant limitations for in vivo applications due to weak selectivity, batch-to-batch variation, preparation difficulties, immunogenicity and high costs of production and handling. Aptamers are emerging as attractive alternatives for antibodies. Aptamers have been extensively sought and studied as protein-capture reagents, therapeutics, diagnostics, and more recently as biosensors6,7. Unlike monoclonal antibodies, aptamers can be generated against any biomolecules, whole cells8,9 or even tissues with little immunogenicity. Furthermore, aptamers can be easily chemically modified to make them resistant to degradation and to further modulate their affinity and specificity. Thio-DNA aptamers, in which one or both the non-bridging phosphoryl oxygens are replaced by sulfur, are preferred choices, because these substitutions render the thio-DNAs more stable in cellular and plasma environments, mostly due to their enhanced nuclease resistance. Importantly, it has been noted that sulfurization of the phosphoryl oxygens of oligonucleotides often enhances their binding to targeted proteins10. Using Cell-SELEX (Systematic Evolution of Ligands by Exponential Enrichment) on patient-derived ovarian cancer cells, a DNA thioaptamer, Endo28, that specifically binds to human ovarian cancer cells was identified11. The protein target for this aptamer was identified as annexin A2 that is expressed in the vasculature of ovarian tumors11. Annexin A2 is a calcium-binding cytoskeletal protein which is located at the extracellular surface of endothelial cells and multiple types of tumor cells12. The Endo28 aptamer can serve as a targeting module for specific drug delivery to ovarian cancer cells.
Nucleic acid based nanoparticles with variable three-dimensional folding13–15 can be designed to have specific interaction with functional protein, RNA, even small chemicals including ions in the organism16–18. RNA/DNA hybrid nanoparticles have been utilized as multifunctional drug delivery carriers 19–21. The phi29 pRNA three-way junction (3WJ) motif with unusually robust thermostable properties22,23 is used as an platform to construct a new generation of therapeutic nanoparticles23–25. The core structure of pRNA-3WJ can be assembled from three pieces of short RNA oligonucleotides, named 3WJa, 3WJb and 3WJc, with high efficiency23. The rigid pRNA-3WJ scaffold ensures the correct folding of its connected nucleic acid aptamers and other functionalities23,26–29. RNA nanoparticles built with the 3WJ scaffold, while harboring different functional modules, retained the folding and independent functionalities of the modules for specific cell binding, cell entry, gene silencing, catalytic function and cancer targeting, both in vitro and in animal trials27,28,30–32. The pRNA-3WJ nanoparticles are non-toxic and non-immunogenic33. They are also capable of penetrating across heterogeneous biological barriers to selectively target cancer cells in mice and delivering therapeutics after systemic injection with little accumulation in healthy organs and tissues24,27,28,31.
We incorporated the Endo28 aptamer to the 3WJ core and hypothesize that this DNA/RNA hybrid nanoparticle will retain the annexin A2-targeting property in vitro and in vivo. With the addition of fluorescent imaging probe Alexa647 and the chemotherapeutic drug doxorubicin to the pRNA-3WJ scaffold, the nanoparticle can function as a drug carrier to enhance the accumulation of doxorubicin in ovarian cancer cells in vivo, thus reducing the distribution of cargoes to other healthy cells and organs. This would significantly enhance the drug’s therapeutic effects by changing the drug internalization pathway on cancer cells, since free doxorubicin enters cells mainly through passive diffusion, while doxorubicin intercalated into nanoparticles can selectively enter annexin A2 positive cells through receptor mediated endocytosis. An ovarian cancer xenograft mouse model was utilized to evaluate the biodistribution of nanoparticles in vivo.
METHODS
Construction of pRNA-3WJ nanoparticles harboring Endo28 thioaptamer
DNA/RNA hybrid nanoparticles were constructed using a bottom-up approach, as previously described. Briefly, DNA oligo 3WJc-Endo28-3WJa was synthesized by DNA synthesizer, and Alexa647-3WJb 2′F-RNA, 3WJb-Sph1 2′F-RNA, and Sph1-2′F-RNA were prepared by RNA synthesizer using solid phase synthesis. The sequences are described below (* represents a phosphorothioated bond, lower case letter represents a 2′-fluorine modified base):
3WJc-Endo28-3WJa DNA: 5′-GGA TCA ATC ATG GCA ACG CTC GGA TCG ATA AGC TTC GCT CGT CCC CC*A GGC* AT*A G*AT* ACT CCG CCC CGT C*AC GG*A TCC TCT* AG*A GC*A CTG TTG CCA TGT GTA TGT GGG-3′
Alexa647-3WJb- 2′F RNA: 5′-(Alexa 647) (C6-NH) ccc AcA uAc uuu Guu GAu cc -3′
3WJb-Sph1 2′F RNA: 5′-ccc AcA uAc uuu Guu GAu ccA Auc ccG cGG ccA uGG cGG ccG GGA G -3′
Sph1-2′F RNA: 5′-cuc ccG Gcc Gcc AuG Gcc GcG GGA uu -3′
3WJa-2′F RNA: 5′-uuG ccA uGu GuA uGu GGG -3′
3WJc-2′F RNA: 5′-GGA ucA Auc AuG GcA A -3′
The RNA nanoparticle carrying a scramble aptamer sequence was prepared by in vitro transcription with Y639F T7 polymerase. The DNA template was prepared by two-step PCR using primer 1 and 2 for first step, and primer 3 and 4 for second step PCR. 2′-Fluoro (2′-F)-modified cytosine and uracil were used in the transcription reaction. The transcribed RNA strand was purified by 8 M Urea 8% polyacrylamide gel ran in TBE buffer (89 mM tris-borate, 2 mM EDTA). RNA bands of interest were visualized by UV shadowing, excised from the gel and eluted with elution buffer (0.5 M Ammonium Acetate, 0.1 M EDTA, 0.1% SDS) followed by ethanol precipitation.
Primer1: 5′-TAA TAC GAC TCA CTA TAC CGG ATC AAT CAT GGC AAG TTC GGT TGT GTC GGC GAG TAT AG-3′
Primer 2: 5′-GGA TCA ACA AAG TAT GTG GGA TCG GCA TTA TAC GTA TAG CA-3′
Primer3: 5′-GTA TAA TAC GAC TCA CTA TAG GGC CGG ATC AAT CAT GGC AA-3
Primer4: 5′-CTC CCG GCC ATG GCC GCG GGA TTG GAT CAA CAA AGT ATG TGG-3′
Scramble template: 5′-GTC GGC GAG TAT AGG TGA AGT TGC CAT GTG TAT GTG GGG TGA TGG ATT GCT ATA CGT AT-3′
Nanoparticles were assembled by mixing strands at equal molar concentrations in PBS (w/Ca2+ Mg2+) buffer (0.1 g/L CaCl2, 0.2 g/L KCl, 0.2 g/L KH2PO4, 0.1 g/L MgCl2-6H2O, 8.0 g/L NaCl, 1.15 g/L Na2HPO4). The mixture was heated to 90 °C for 5 minutes then snap cooled on ice.
Loading doxorubicin to Endo28-3WJ-sph1 nanoparticles
Doxorubicin (Sigma) solution (20 μM) was incubated with extended Endo28-3WJ-Sph1 or Scramble-3WJ-sph1 nanoparticles (2 μM) in the intercalation buffer (0.1 M sodium acetate, 0.05 M NaCl, 0.01 M MgCl2) for 1 hour at room temperature. The free doxorubicin was then removed from the system by passing through Sephadex G50 spin column (NucAway™, Ambion). The drug loading efficiency was monitored by measuring the fluorescence intensity of doxorubicin with a fluorescence spectrophotometer (Horiba Jobin Yvon) at excitation wavelength of 480 nm and emission from 500 to 720 nm.
To measure the intercalation constant of Endo28-3WJ-Sph1 nanoparticle with doxorubicin, increasing concentrations of RNA nanoparticles were incubated with 1.4 μM doxorubicin, and the fluorescence intensity of doxorubicin was measured. The fluorescence quenching was plotted as a function of Dox-conjugating aptamer concentration and fitted to Hill equation in Origin to calculate the Kd.
Release of doxorubicin from the nanoparticle-doxorubicin conjugates
Drug release from the nanoparticle-doxorubicin conjugates was monitored using a dialysis bag with 3.5 kDa cutoff under sink condition34. About 400 μL of Endo28-3WJ-Sph1/doxorubicin conjugate containing 1 μM doxorubicin was dialyzed in the intercalation buffer at 37 °C. 100 μL of releasing medium was collected at time points of 0, 0.7, 2.5, 4, 6, 8, 20, and 24 hrs. Free doxorubicin was also dialyzed to test its release profile as a control. The released doxorubicin was measured by fluorescence spectrophotometer (Horiba Jobin Yvon) at excitation wavelength 480 nm and emission from 500 to 720 nm.
Serum stability assay
400 ng of Alexa647-labeled Endo28-3WJ nanoparticle were incubated in PBS buffer containing fetal bovine serum (FBS) at final concentration of 10 %. Samples were taken at multiple time points, including 0, 0.5, 1, 2, 4, 8, 9, and 24 hours after incubation at 37 °C. 8% Native TBM polyacrylamide gel electrophoresis was applied to visualize the RNA. The gel was imaged at Alexa647 channel with Typhoon FLA 7000 (GE Healthcare; Pittsburgh, Pennsylvania).
Nanoparticle size and zeta potential measurement by DLS
Apparent hydrodynamic sizes and zeta potential of assembled Endo28-3WJ nanoparticles were measured by a Zetasizer nano-ZS (Malvern Instruments; Malvern, United Kingdom). RNA nanoparticles were measured at 1 μM in Diethylpyrocarbonate (DEPC)-treated water at 25 °C.
Cell culture
Human ovarian cancer cell line SKOV3 (American Type Culture Collection; Manassas, Massachusetts) was grown and cultured in McCoy’s 5A medium (Life Technologies) containing10% fetal bovine serum (FBS); IGROV-1 cells were cultured in RPMI 1640 medium (Invitrogen; Grand Island, New York) containing 10 % FBS and 1 % gentamicin sulfate (Gibco); and HEK293T cells were cultured in DMEM medium (Life Technologies) containing 10% FBS. All cells were cultured in a 37 °C incubator with a 5 % CO2 and a humidified atmosphere.
Flow cytometry assay
Cells were trypsinized and rinsed with blank medium. Alexa647-labeled Endo28-3WJ and the control 3WJ nanoparticles were each incubated with 2 × 105 SKOV3, IGROV-1 or HEK293T cells at 37 °C for 1 hour, with the final concentration of RNA to be 100 nM. After washing with PBS (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), the cells were resuspended in PBS buffer and subjected to flow cytometry assay. Flow cytometry assay was performed at the UK Flow Cytometry & Cell Sorting Core Facility.
Confocal microscopy imaging
SKOV3, IGROV-1 and HEK293T cells were grown on glass cover slides in their complete medium overnight. Alexa647-labeled Endo28-3WJ and the control 3WJ nanoparticles were each incubated with the cells at 37 °C for 2 h at final concentration of 100 nM. After washing with PBS, the cells were fixed by 4 % paraformaldehyde and stained by Alexa Fluor® 488 phalloidin (Invitrogen; Grand Island, New York) for actin and Prolong® Gold Antifade Reagent with DAPI (Life Technologies) for nucleus. The cells were then assayed for binding and cell entry on an Olympus FV1000 confocal microscope (Olympus Corporation).
Cytotoxicity assay
The cytotoxicity of RNA nanoparticles was evaluated with an MTT assay kit (Promega, Madison, WI) following manufacturer instructions. Briefly, SKOV3 and HEK293T cells were plated in a 96-well plate and cultured at 37 °C in humidified air containing 5 % CO2 overnight. The Endo28-3WJ-sph1/Dox 2′F RNA nanoparticle conjugates, control nanoparticles, and free doxorubicin were incubated with cells at 37 °C for 48 hrs, while keeping the final doxorubicin concentration at 3 μM. Then, 15 μL of dye solution was added to each well and incubated at 37 °C for 4 hrs, and 100 μL of solubilization/stop solution was added to each well and incubated at room temperature for 2hrs for color development. The absorbance at 570 nm was recorded using a microplate reader (Synergy 4, BioTek Instruments, Inc., USA). The cell viability was calculated relative to the absorbance of cells-only control.
In vivo biodistribution and tumor targeting of RNA nanoparticles
SKOV3 cells were cultured in vitro and subcutaneously injected under the skin of 8-week-old female nude mice. A total of 2 × 106 cells were injected in solution. Tumors were grown for 4 weeks until they reached a volume of 200 mm3. Mice were then administered PBS, Endo28-3WJ, or 3WJ each with Alexa647 labels at a dose of 2 μM at 100 μL through the tail vein. Mice were imaged for whole body fluorescence at time points 0, 1, 2, 4, and 6 hours with an In Vivo Imaging System (IVIS) imager (Caliper Life Sciences; Waltham, Massachusetts). Upon the completion of the study, mice were sacrificed; and tumors, hearts, kidneys, livers, and brains were collected and imaged by whole body imager for Alexa647 signal. Additionally, tumors were frozen at −80 °C and sectioned for confocal microscopy as described. Furthermore, tumors were fixed in 4 % paraformaldehyde with 10 % sucrose in 1× PBS buffer at 4 °C overnight. Tumor samples were then placed in Tissue-Tek Optimum Cutting Temperature compound (Sakura Finetek USA; Torrance, California) for frozen sectioning (10 μm thick). Sectioned tissue were then stained with DAPI and mounted with ProLong Gold Antifade Reagent (Life Technologies; Carlsbad, California) overnight. Slides were then imaged by Olympus FV1000-Filter Confocal Microscope System (Olympus; Pittsburgh, Pennsylvania). This animal experiment was done with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of The Ohio State University.
RESULTS
Construction of pRNA-3WJ nanoparticles harboring Endo28 aptamer
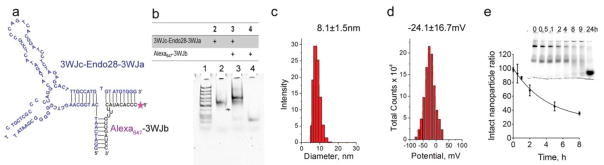
The Endo28 aptamer sequence was incorporated into the pRNA-3WJ 2′F-RNA, creating DNA/RNA hybrid nanoparticles suitable for ovarian cancer targeted drug delivery. The DNA/RNA hybrid nanoparticle with a two-piece design was found to have the highest assembly efficiency, in which the Endo28 thio-DNA aptamer sequence was connected to the 3′ end of 3WJc and 5′ end of 3WJa DNA, forming one DNA strand. The DNA oligo was then assembled with 2′F-modified Alexa647-3WJb 2′F-RNA to form a 2′F RNA/thio-DNA hybrid nanoparticle with 3WJ core structure, named Endo28-3WJ (Figure. 1a). The two-dimensional structure of Endo28 thio-DNA aptamer was predicted by M-Fold 35. The formation of the hybrid nanoparticle was confirmed by native PAGE analysis (Figure. 1b). The hybrid nucleic acid nanoparticles have a mean particle size around 8 nm (Figure. 1c) and negative charge with zeta potential around −24 mV (Figure. 1d) as measured by Dynamic Light Scattering (DLS). They are stable in serum with a half-life of 4 hr in 10 % FBS (Figure. 1e).
Figure 1. Characterization of the Endo38-3WJ nanoparticle construction.
(a). Primary sequence and secondary structure of Endo28-3WJ nanoparticle predicted by M-Fold. (b). Native PAGE test of the assembly of Endo28-3WJ nanoparticles. Lane 1 is ultralow range DNA ladder (Thermo Scientific). (c). Particle size of Endo28-3WJ as determined by DLS. (d). Zeta potential of Endo28-3WJ nanoparticles measured by DLS. (e). Serum stability assay showed the half-life of Endo28-3WJ in 10% FBS is around 4h, which was calculated by quantification of the bands in the gel imaged at Ethidium bromide channel.
Loading doxorubicin into Endo28-3WJ nanoparticles and in vitro release
Doxorubicin can form physically non-covalently conjugate with nucleic acid nanoparticles by preferentially binding to double stranded 5′-GC-3′ or 5′-CG-3′ sequences 4,36,37. To increase the loading ratio of doxorubicin per nanoparticle, we extended the arm at the 3′-end of 3WJb by adding a 26 bp GC rich sequence named Sph1. The extended Endo28-3WJ-Sph1 nanoparticle was assembled from 3 single strands of nucleic acid (Figure. 2a) with high efficiency as detected by native PAGE (Figure. 2b). The size distribution of Endo28-3WJ-Sph1 nanoparticle did not change significantly comparing to that of the Endo28-3WJ nanoparticle, with a mean hydrodynamic diameter around 7 nm (Figure. S1a) and a zeta potential around −24 mV (Figure. S1b) as measured by DLS. Addition of the GC rich 2′F RNA sequence to the nanoparticle did not impact its serum stability significantly either. The Endo28-3WJ-Sph1 hybrid nanoparticles are stable in serum with a half-life of around 4 hr in 10 % FBS (Figure. S1c).
Figure 2. Characterization of the RNA nanoparticle/doxorubicin physical conjugates.
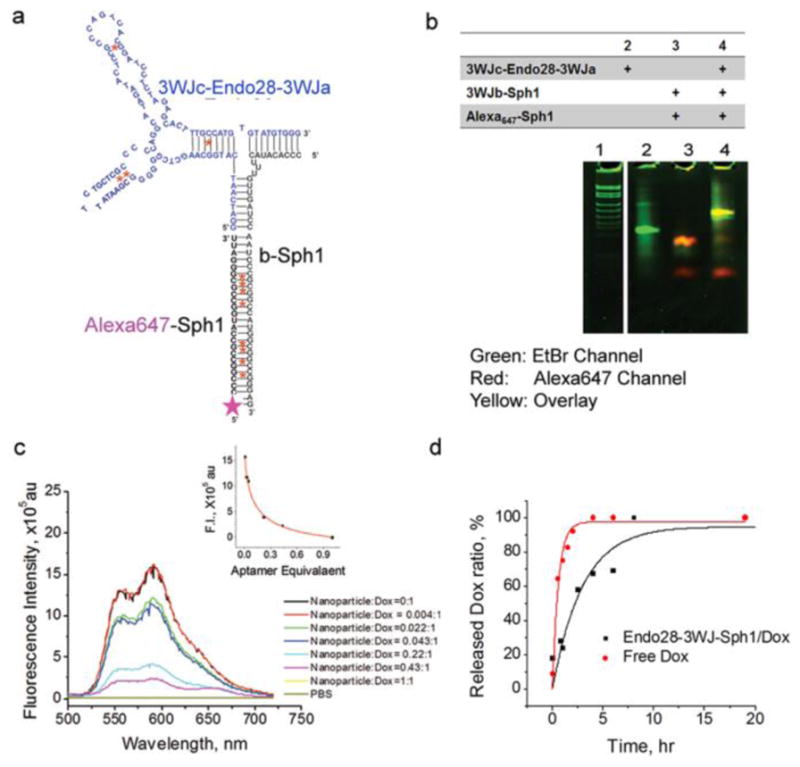
(a) Construct of Endo28-3WJ-sph1 RNA nanoparticle with predicted chelating sites for doxorubicin indicated by red stars. (b) Native PAGE showing the assembly of Endo28-3WJ-sph1 nanoparticles. Lane 1 is 100bp DNA ladder (Thermo Scientific). (c) Fluorescence spectra of doxorubicin solution (1.4μM) with increasing concentration of RNA nanoparticles. Insert: A hill plot for the nanoparticle titration (Kd= 140nM, 0.1 equivalent of the RNA nanoparticles). (d) The percentage of doxorubicin released from the nanoparticle doxorubicin conjugates at 37°C. The doxorubicin concentration was quantified by its fluorescence intensity in release medium.
Doxorubicin (Dox), an anthracycline class drug, has a fluorescence property that can be quenched after interaction with nucleic acid38. The incorporation of Dox to the extended nanoparticle, Endo28-3WJ-Sph1, was examined by fluorescence spectrometry. A decrease in fluorescence intensity was detected when a fixed concentration of Dox was incubated with increasing concentrations of Endo28-3WJ-Sph1 RNA nanoparticles (Figure. 2c). Evaluation of the predicted secondary structure of Endo28-3WJ-Sph1 nanoparticle reveals eleven possible sites for Dox intercalation, as marked by red asterisks in Figure. 2a. The dissociation constant (Kd=140 nM) of the Endo28-3WJ-Sph1/Dox non-vovalent conjugate was derived from the Hill plot (Figure. 2c, insert), while the Dox concentration was kept at 1.4 μM. It suggested that Dox and the nucleic acid nanoparticles spontaneously formed a stable physical conjugate, and that the molar binding ratio of equilibrium Dox to the nucleic acid nanoparticle is around 10:1. The number is consistent with the predicted number of Dox to be intercalated through nearest-neighbor exclusion in each Endo28-3WJ-sph1 RNA nanoparticle (Figure. 2a).
A study of Dox released from the nanoparticle-Dox physical conjugate over time was conducted using a dialysis tube with membrane cutoff of 3 kDa. Upon dialysis, release of more than 80% Dox was observed in 6 hr with first-order kinetics (Figure. 2d). A significantly slower drug release rate at the initial stage suggests that this system is advantageous for in vivo systemically targeted delivery of doxorubicin. Free Dox showed a much faster release profile with more than 80% released within the first hour (Figure. 2d).
Targeting of Endo28-3WJ nanoparticles to cancer cells in vitro
To test the targeting property of Endo28-harboring pRNA-3WJ nanoparticles in cell culture, a fluorescent Alexa647-labeled 3WJb 2′F-RNA strand was assembled to Endo28-3WJ nanoparticles to visualize the RNA nanoparticles in cells. The Endo28-3WJ nanoparticles were incubated with several cell lines, which have different levels of annexin A2 expression. IGROV-110,39 and SKOV340 are tested as annexin A2 positive cells, while HEK293T as annexin A2 negative cells is used as a negative control. Following the RNA incubation and washing steps, cells were analyzed by flow cytometry to confirm the binding of the Endo28-3WJ nanoparticles. The result showed that Endo28-3WJ has stronger binding to IGROV-1 (71.2%) than to SKOV3 (51.7%), while HEK29T showed a very low binding (17.3%), similar to the non-binding 3WJ RNA controls (Figure. 3a). The result agrees with the annexin A2 expression level in cells as reported10,40. The flow cytometry data indicated that the Endo28 aptamer retained its binding property to annexin A2 on cells after incorporation to the pRNA-3WJ, thus providing a branched scaffold to include additional sequences for drug loading.
Figure 3. The binding and internalization of Endo28-3WJ to annexin A2 positive cells.
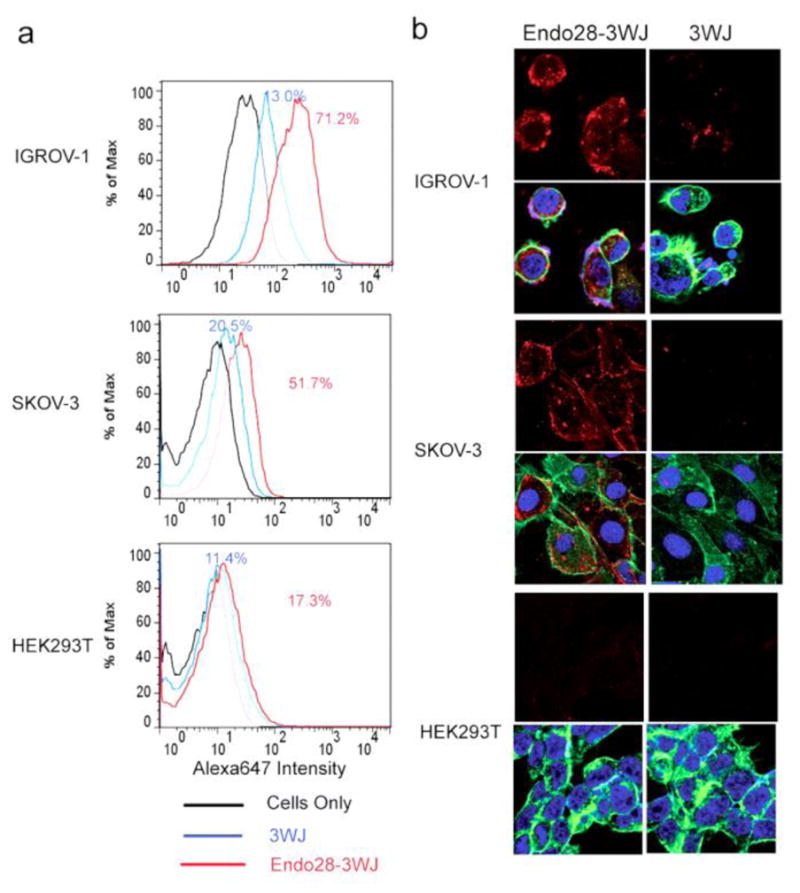
(a) Flow cytometry analysis of the binding of Alexa647-labeled Endo28-3WJ nanoparticles with annexin A2 positive cells (IGROV-1 and SKOV3) and annexin A2 negative cells (HEK293T). (b) Confocal imaging of the internalization of Alexa647-labeled Endo28-3WJ to annexin A2 positive cells (IGROV-1 and SKOV-3) and annexin A2 negative cells (HEK293T).
RNA nanoparticles suitable for targeted therapeutics delivery need to be internalized into the target cells for proper release of therapeutic agents. The entry of the DNA/RNA hybrid nanoparticles harboring the Endo28 into annexin A2 positive cells including IGROV-1 and SKOV3 cells was examined by confocal microscopy, and the annexin A2 low expression HEK293T cells were used as negative controls. After incubating Alexa647 labeled Endo28-3WJ nanoparticles with the cells, Endo28-3WJ showed clear internalization to IGROV-1 and SKOV3 cells, but very little signal was observed on HEK293T cells (Figure. 3b). Additionally, low Alexa647 signal was observed around all cells for the 3WJ nanoparticles without the annexin A2 aptamers (Figure. 3b). These results suggest that Endo28-3WJ nanoparticles entered the cells in an annexin A2-dependent fashion.
Intracellular delivery of doxorubicin to ovarian cancer cells in vitro
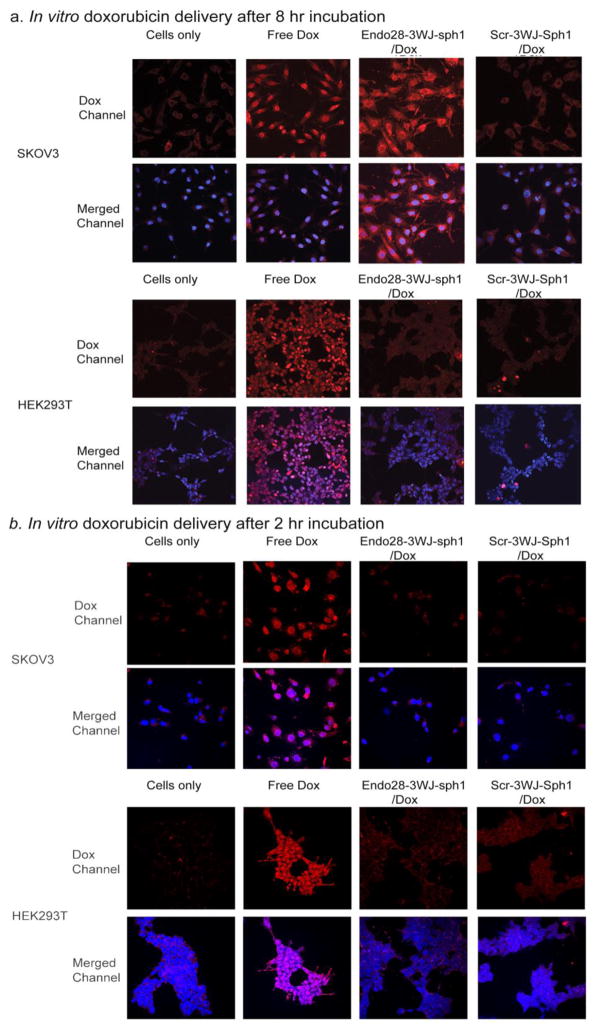
To determine if the Endo28-3WJ-Sph1 nanoparticle could deliver Dox to annexin A2-overexpressing ovarian cancer cells, confocal microscopy imaging was performed. Both SKOV3 (annexin A2 positive) and HEK293T (annexin A2 negative) cells were incubated with free Dox, Endo28-3WJ-Sph1/Dox, or Scr-3WJ-Sph1/Dox RNA nanoparticle conjugates for 8 hrs and then analyzed by confocal imaging. Scr-3WJ-Sph1RNA nanoparticle with the pRNA-3WJ core structure and a similar Dox loading efficiency but a scrambled aptamer sequence was used as a negative control. The SKOV3 cells treated with Endo28-3WJ-Sph1/Dox intercalates exhibited strong Dox fluorescence signal, similar to free Dox in the confocal microscopy assay. However, when treated with the Scr-3WJ-Sph1/Dox intercalates, the cells showed much weaker cellular uptake of Dox (Figure. 4a). The released Dox from Endo28-3WJ-sph1/Dox complex was found mostly located in the cytoplasm and nuclei (Figure. 4a); while in contrast free Dox was located in cell nuclei after the same treatment (Figure. 4a). When incubated with annexin A2 negative HEK293T cells, both Endo28-3WJ-Sph1/Dox and Scr-3WJ-Sph1/Dox intercalates showed weak uptake (Figure. 4a). The cellular localization of doxorubicin after 2 hr incubation was also imaged. Free doxorubicin was found in nuclei and cytoplasm, while less doxorubicin were detected in Endo28-3WJ-Sph1/Dox treated SKOV3 cells. This is possibly due to the sustained release profile of Endo28-3WJ-Sph1/Dox in vitro; only around 20% dox was released from the Endo28-3WJ-Sph1/Dox after 2hrs, comparing to 80% in free dox (Figure. 4b). The distribution of doxorubicin in SKOV3 cells after 8 hr treatment with Endo28-3WJ-Sph1/Dox was similar to 2 hr free dox treated cells. It suggests that Dox can enter cell cytoplasm and intercalated to nucleus DNA later. Thus free Dox released from Endo28-3WJ-Sph1/Dox could eventually intercalate into cell nucleus. These results suggest that Endo28-3WJ-Sph1/Dox non-covalent conjugates can specifically deliver Dox into annexin A2 positive cell lines.
Figure 4. In vitro delivery of doxorubicin by Endo28-3WJ-Sph1 nanoparticles to cells.
Both annexin A2 positive SKOV3 and annexin A2 negative HEK293T cells were tested. Nanoparticles harboring doxorubicin conjugated with scramble aptamer sequences were tested as negative controls. The cells were treated with nanoparticles for (a) 8 hrs or (b) 2hrs, and imaged. The doxorubicin channel is shown as red; cell nuclei stained with DAPI is shown as blue.
Effects of Endo28-3WJ-sph1/Dox conjugate on cell cytotoxicity
Cell cytotoxicity was evaluated with an MTT assay, which monitors cell metabolic activity41. The cytotoxicity effect of Endo28-3WJ-Sph1/Dox conjugates was tested with both annexin A2 positive SKOV3 cells and annexin A2 negative HEK293T cells. Various concentrations (0.625 μM, 1.250 μM, 3.000 μM) of Dox from Endo28-3WJ-Sph1/Dox and controls including Scr-3WJ-Sph1/Dox and free Dox were incubated with cells. Endo28-3WJ-sph1/Dox showed significantly higher toxicity on SKOV3 cells than the controls in all three tested concentrations (Figure. 5a). Such a difference in cytotoxicity was not detected with HEK293T cells (Figure. 5b). The results suggest that the Endo28-3WJ-Sph1 nanoparticles are able to deliver Dox selectively to annexin A2 positive cells and exert a higher cytotoxicity effect on the targeted cells.
Figure 5. Cell cytotoxicity assay for Endo28-3WJ-Sph1/Dox intercalates in vitro.
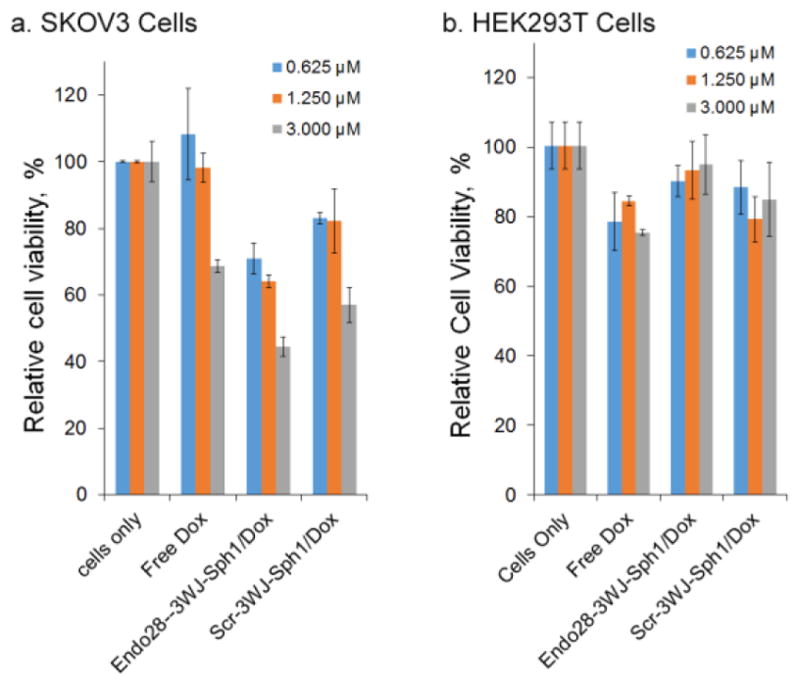
(a). Cell cytotoxicity assay with annexin A2 positive SKOV3 cells. With n=3 biological replicates, statistics were calculated using a two-sided t-test with center values presented as averages and errors as s.d. p =1.2e-2, 3e-4, 1e-4 comparing Endo28-3WJ-Sph1/Dox to free doxorubicin; and1.3e-2, 3.2e-2, 1.5e-3 comparing Endo28-3WJ-Sph1/Dox toScr-3WJ-Sph1/Dox at concentrations of 0.625, 1.250, and 3.000 μM respectively. (b) Cell cytotoxicity assay with annexin A2 negative HEK293T cells.
Targeting of Endo28-3WJ nanoparticles to cancer cells in vivo
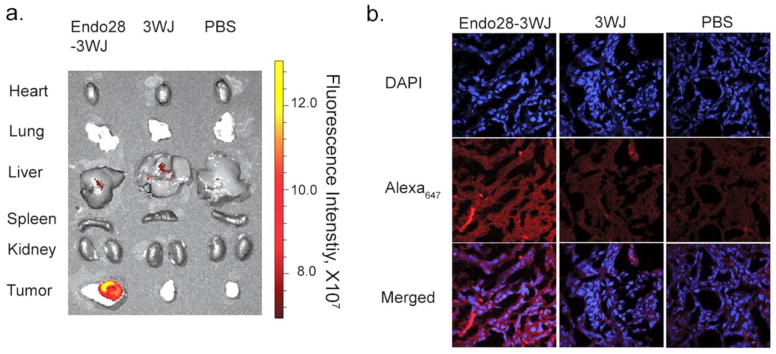
Ovarian cancer xenograft mice models were developed through subcutaneous injection of SKOV3 cells to female nude mice. Tumors were fully developed after 4 weeks. 100 μL of 10 μM Alexa647-labeled Endo28-3WJ nanoparticles were administered to the mice through tail-vein injection. Mice were whole body imaged to monitor the biodistribution of nanoparticles in vivo at designated time points. The mice were sacrificed after 6 hrs, and nanoparticle accumulation in organs was imaged. Alexa647 was detected throughout the whole body of the mice after 30 min of injection, indicating that nanoparticles successfully circulated through the mice. During the early time points, Alexa647 signal was detected in the tumor, liver, and bladder of the mice. After 6 hrs, fluorescence signal remained only in the xenograft tumor, but was not detectable in all healthy organs (Figure. 6a). Confocal microscopy imaging of the tumor sections shows specific targeting and accumulation of the Endo28-3WJ nanoparticles to the SKOV3 xenograft tumor, while the control group, including 3WJ nanoparticles and PBS, showed much lower fluorescent intensity in tumor cells (Figure. 6b). The results demonstrated that the 3WJ RNA nanoparticles harboring annexin A2 aptamer are suitable for targeted in vivo drug delivery to cancer cells.
Figure 6. Evaluation of the targeting effect of Endo28-3WJ nanoparticles using SKOV3 xenograft mice model.
(a) Fluorescence image showing the specific targeting of Alexa647-labeled Endo28-3WJ nanoparticles in ovarian tumors 6 hr post systemic administration. (b) Histological assay of the ovarian tumor frozen cross sections (10 μm) by confocal microscopy showing the binding and entry of Endo28-3WJ nanoparticles to tumor cells. Blue: nuclei; Red: RNA nanoparticles.
DISCUSSION
Nanoparticles are attractive drug delivery systems. Although liposomes are the most commonly used nanoparticles, they have some limitations. Liposomes are known to accumulate in the liver, and the drug-loaded liposomes can cause significant liver toxicity42. Nucleic acids, beyond the native function for RNA interference43–45, enzyme-like activity46, DNA repair47, and other genome editing48, can also be designed and constructed with defined shape and structure49,50. Nucleic acid nanoparticles have great potential to overcome the liver accumulation limitations for in vivo application23,24,27,28,31,51, with the nature of negative surface charge and well defined particle shape and size. The multivalent nature of the pRNA-3WJ allows us to conjugate multiple molecules with different functions. The stoichiometry of the different functional molecules can be carefully controlled. By conjugating the DNA thioaptamer that specifically binds to annexin A2 positive cells to the pRNA-3WJ loaded with doxorubicin, we created a multi-functional nanoparticle that specifically targets ovarian cancer cells and the tumor vasculature.
Nanoparticles with Endo28 aptamer not only specifically bind to annexin A2 positive cells, but also enter through receptor mediated endocytosis as expected. The polyvalency of the pRNA-3WJ scaffold also allowed for harboring of imaging probes for tracking and therapeutics for treatment. By incorporating a GC-rich sequence specifically designed for the binding of a chemotherapy drug, doxorubicin, efficient loading of the drug was achieved at around 10 molecules per nanoparticle. The nanoparticles showed a sustained release profile of doxorubicin, with the drug release reaching 80 % in 6 hrs, compared to the naked doxorubicin that reached 80% releasing within only 1 hr. Such sustained release will ensure that majority of the loaded drug will be released after the carrier nanoparticle reaches the targeted tumor site, thus reducing the distribution of toxic chemical drugs to healthy organs and minimizing side effects.
Upon confirmation of the annexin A2-targeting nanoparticles entering into ovarian cancer cells and proper loading/releasing profiling of doxorubicin, in vitro cancer cell-targeted delivery of doxorubicin was further tested. SKOV3 cells are known to overexpress annexin A2 and are also resistant to multiple drugs including doxorubicin, thus serving as a suitable model. The Endo28-3WJ-Sph1/Dox complex showed significantly higher toxicity towards SKOV3 cells than the scramble control and the free Dox, suggesting a possible change in the intracellular trafficking pathway of the drug when an aptamer is used to deliver the drug. Confocal imaging showed that the doxorubicin released from the Endo28-3WJ-Sph1/Dox nanoparticles mainly accumulated in cell cytoplasm, as opposed to free doxorubicin that was located mostly in cell nuclei. The difference can be attributed to the difference in doxorubicin cell entry pathways. Free doxorubicin enters cells mainly through passive diffusion, while the Endo28-3WJ-Sph1/Dox enters cells through annexin A2 receptor-mediated endocytosis.
The in vivo biodistribution study showed a promising profile with accumulation of the nanoparticles in tumors, but not healthy organs. Thus, using the Endo28-3WJ nanoparticle for cancer cell-targeted drug delivery will benefit ovarian cancer patients by reducing the side effects of cancer chemotherapeutics and increasing its local concentration in the tumor microenvironment after systemic administration. Incorporating doxorubicin into annexin A2 aptamer-harboring nanoparticles possibly changed the cell internalization pathway of the drug. Free doxorubicin enters cell mainly through nonspecific passive diffusion; while the doxorubicin complexed in nanoparticles mainly enters cells through receptor mediated endocytosis. Loading doxorubicin into nucleic acid nanoparticles would not only increase its targeting efficiency to cancer cells, but also potentially reduce its drug resistance. P-glycoprotein is one of the main mediators for doxorubicin resistance52. Complexing doxorubicin into nucleic acid nanoparticles changes its cell internalization pathway from passive diffusion to receptor mediated endocytosis, which can reduce its chance to be recognized by p-glycoproteins, potentially reducing the drug resistance. The action mechanism of doxorubicin in cells would not change however, as it only forms non covalent complex with nucleic acid nanoparticles and can be released from nanoparticles eventually.
Overall, the results demonstrated that stable thio-DNA/2′F-RNA hybrid nanoparticles harboring annexin A2 aptamer are suitable as nanocarriers for targeted delivery of doxorubicin to ovarian cancer cells. The DNA/RNA hybrid nanoparticles were proven to remain chemically and thermodynamically stable for in vivo application. The annexin A2-specific nanoparticles produce good binding profiles at 50 nM RNA concentrations and later proved to provide specific delivery of doxorubicin to SKOV3 cells, with significant higher toxicity than targeted-scramble controls. The Endo28 thioaptamer-harboring nanoparticles showed specific targeting to ovarian cancer after systemic administration in vivo. Large-scale preparation of this thio-DNA/2′F-RNA hybrid nanoparticle should be pursued for further moving this new technology to animal therapy and clinical trials.
Supplementary Material
We report here the novel design and construction of a chemical modified DNA/RNA hybrid nanoparticle for ovarian cancer targeted drug delivery. A thio-DNA aptamer targeting annexin A2 was conjugated to a highly thermodynamically stable three way junction (3WJ) core motif derived from pRNA of phi29 bacteriophage. The other arm of pRNA-3WJ is extended with GC rich sequences for doxorubicin loading. The constructed nanoparticles retained the annexin A2 targeting property, can significantly enhance the toxicity of doxorubicin towards ovarian cancer cells and specifically target annexin A2 overexpression cancer cells in vivo. This novel cancer cell targeted drug delivery system is promising for enhancing chemical drug efficiency in ovarian cancer treatment in a targeted manner.
Acknowledgments
This research was supported by grants from NIH R01EB003730 and U01CA151648 to PG U54CA151668 to DGG and from Rivkin Center for Ovarian Cancer Research to VT. The University of Kentucky Flow Cytometry & Cell Sorting Core Facility is supported in part by the Office of the Vice President for Research, the Markey Cancer Center and an NCI Center Core Support Grant (P30 CA177558) to the University of Kentucky Markey Cancer Center. Funding to Peixuan Guo’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is by the C. M. Chen Foundation. Other support includes grants from the NIH (U54 CA151668, UH3 TR000943, P50 CA083639), the RGK Foundation, the American Cancer Society Professor Award and the Frank McGraw Memorial Chair in Cancer Research.
Footnotes
Conflict of Interest: PG is a consultant of Oxford Nanopore and RNA Nanobio, Ltd.. The companies licensed his patents from the University of Kentucky, University of Cincinnati and Purdue University. He is a confounder of P&Z Biological Medicine Ltd. DGG is a current member of AM Biotechnology, a company that has licensed various aptamer technologies from his UT laboratories.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Gullotti E, Yeo Y. Extracellularly activated nanocarriers: a new paradigm of tumor targeted drug delivery. Mol Pharm. 2009;6:1041–51. doi: 10.1021/mp900090z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A’hern RP, Gore ME. Impact of doxorubicin on survival in advanced ovarian cancer. J Clin Oncol. 1995;13:726–32. doi: 10.1200/JCO.1995.13.3.726. [DOI] [PubMed] [Google Scholar]
- 4.Frederick CA, Williams LD, Ughetto G, van der Marel GA, van Boom JH, Rich A, et al. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29:2538–49. [PubMed] [Google Scholar]
- 5.Rose PG. Pegylated liposomal doxorubicin: optimizing the dosing schedule in ovarian cancer. Oncologist. 2005;10:205–14. doi: 10.1634/theoncologist.10-3-205. [DOI] [PubMed] [Google Scholar]
- 6.Brody E, Gold L. Aptamers as therapeutic and diagnostic agents. J Biotechnol. 2000;74(1):5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 7.Leonard M, Zhang Y, Zhang X. Small non-coding RNAs and aptamers in diagnostics and therapeutics. Methods Mol Biol. 2015;1296:225–33. doi: 10.1007/978-1-4939-2547-6_21. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Rossi JJ. Evolution of Cell-Type-Specific RNA Aptamers Via Live Cell-Based SELEX. Methods Mol Biol. 2016;1421:191–214. doi: 10.1007/978-1-4939-3591-8_16. [DOI] [PubMed] [Google Scholar]
- 9.Lyu Y, Chen G, Shangguan D, Zhang L, Wan S, Wu Y, et al. Generating Cell Targeting Aptamers for Nanotheranostics Using Cell-SELEX. Theranostics. 2016;6:1440–52. doi: 10.7150/thno.15666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somasunderam A, Thiviyanathan V, Tanaka T, Li X, Neerathilingam M, Lokesh GL, et al. Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of human CD44. Biochemistry. 2010;49:9106–12. doi: 10.1021/bi1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangala L, Wang H, Jiang D, Wu S, Somasunderam A, VolK D, et al. Improving tumor vascular maturation using non-coding RNAs increases anti-tumor effect of chemotherapy. J Clin Invest. 2016;1(17):e87754. doi: 10.1172/jci.insight.87754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lokman NA, Ween MP, Oehler MK, Ricciardelli C. The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenviron. 2011;4:199–208. doi: 10.1007/s12307-011-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashida S, Inoue T, Saito H. Three-dimensionally designed protein-responsive RNA devices for cell signaling regulation. Nucleic Acids Res. 2012;40:9369–78. doi: 10.1093/nar/gks668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q, Yoo SY, Chung YH, Lee JY, Min J, Choi JW. Control of electrochemical signals from quantum dots conjugated to organic materials by using DNA structure in an analog logic gate. Bioelectrochemistry. 2016;111:1–6. doi: 10.1016/j.bioelechem.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Lee T, Yagati AK, Pi F, Sharma A, Choi J-W, Guo P. Construction of RNA-quantum dot chimera for nanoscale resistive biomemory application. ACS Nano. 2015;9:6675–82. doi: 10.1021/acsnano.5b03269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLong RK, Hurst MN, Aryal S, Inchun NK. Unique Boron Carbide Nanoparticle Nanobio Interface: Effects on Protein-RNA Interactions and 3-D Spheroid Metastatic Phenotype. Anticancer Res. 2016;36:2097–103. [PubMed] [Google Scholar]
- 17.Zhu Y, Chen SJ. Many-body effect in ion binding to RNA. J Chem Phys. 2014;141:055101. doi: 10.1063/1.4890656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Qing Z, Liu C, Tang Q, Li J, Yang S, et al. Direct Fluorescent Detection of Blood Potassium by Ion-Selective Formation of Intermolecular G-Quadruplex and Ligand Binding. Anal Chem. 2016;88:9285–92. doi: 10.1021/acs.analchem.6b02667. [DOI] [PubMed] [Google Scholar]
- 19.Afonin KA, Viard M, Tedbury P, Bindewald E, Parlea L, Howington M, et al. The Use of Minimal RNA Toeholds to Trigger the Activation of Multiple Functionalities. Nano Lett. 2016;16:1746–53. doi: 10.1021/acs.nanolett.5b04676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Afonin KA, Viard M, Kagiampakis I, Case CL, Dobrovolskaia MA, Hofmann J, et al. Triggering of RNA Interference with RNA–RNA, RNA–DNA, and DNA–RNA Nanoparticles. ACS Nano. 2014;9(1):251–9. doi: 10.1021/nn504508s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park Y, Kim H, Lee JB. Self-assembled DNA-Guided RNA Nanovector via Step-wise Dual Enzyme Polymerization (SDEP) for Carrier-free siRNA Delivery. ACS Biomaterials Science & Engineering. 2016;2(4):616–24. doi: 10.1021/acsbiomaterials.5b00554. [DOI] [PubMed] [Google Scholar]
- 22.Binzel DW, Khisamutdinov EF, Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry. 2014;53:2221–31. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nature Nanotechnology. 2011;6:658–67. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, et al. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–57. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, et al. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev. 2014;66C:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu D, Khisamutdinov E, Zhang L, Guo P. Programmable folding of fusion RNA complex driven by the 3WJ motif of phi29 motor pRNA. Nucleic Acids Res. 2013;42:e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, et al. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–40. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binzel D, Shu Y, Li H, Sun M, Zhang Q, Shu D, et al. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Molecular Therapy. 2016;24(7):1267–77. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo P, Shu Y, Binzel D, Cinier M. Synthesis, Conjugation, and Labeling of Multifunctional pRNA Nanoparticles for Specific Delivery of siRNA, Drugs and Other Therapeutics to Target Cells. Methods in Molecular Biology. 2012;928:197–219. doi: 10.1007/978-1-62703-008-3_16. [DOI] [PubMed] [Google Scholar]
- 30.Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, et al. Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano. 2015;9:1108–16. doi: 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee TJ, Haque F, Vieweger M, Yoo JY, Kaur B, Guo P, et al. Functional assays for specific targeting and delivery of RNA nanoparticles to brain tumor. Methods Mol Biol. 2015;1297:137–52. doi: 10.1007/978-1-4939-2562-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui D, Zhang C, Liu B, Shu Y, Du T, Shu D, et al. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Scientific reports. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelmawla S, Guo S, Zhang L, Pulukuri S, Patankar P, Conley P, et al. Pharmacological characterization of chemically synthesized monomeric pRNA nanoparticles for systemic delivery. Molecular Therapy. 2011;19:1312–22. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Souza SS, Deluca PP. Methods to assess in vitro drug release from injectable polymeric particulate systems. Pharm Res. 2006;23:460–74. doi: 10.1007/s11095-005-9397-8. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaires JB, Herrera JE, Waring MJ. Preferential binding of daunomycin to 5′ATCG and 5′ATGC sequences revealed by footprinting titration experiments. Biochemistry. 1990;29:6145–53. doi: 10.1021/bi00478a006. [DOI] [PubMed] [Google Scholar]
- 37.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45:8149–52. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 38.Valentini L, Nicolella V, Vannini E, Menozzi M, Penco S, Arcamone F. Association of anthracycline derivatives with DNA: a fluorescence study. Farmaco Sci. 1985;40:377–90. [PubMed] [Google Scholar]
- 39.Zhang N, Wu ZM, McGowan E, Shi J, Hong ZB, Ding CW, et al. Arsenic trioxide and cisplatin synergism increase cytotoxicity in human ovarian cancer cells: therapeutic potential for ovarian cancer. Cancer Sci. 2009;100:2459–64. doi: 10.1111/j.1349-7006.2009.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lokman NA, Elder AS, Ween MP, Pyragius CE, Hoffmann P, Oehler MK, et al. Annexin A2 is regulated by ovarian cancer-peritoneal cell interactions and promotes metastasis. Oncotarget. 2013;4:1199–211. doi: 10.18632/oncotarget.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciapetti G, Cenni E, Pratelli L, Pizzoferrato A. In vitro evaluation of cell/biomaterial interaction by MTT assay. Biomaterials. 1993;14:359–64. doi: 10.1016/0142-9612(93)90055-7. [DOI] [PubMed] [Google Scholar]
- 42.Yingchoncharoen P, Kalinowski DS, Richardson DR. Lipid-Based Drug Delivery Systems in Cancer Therapy: What Is Available and What Is Yet to Come. Pharmacol Rev. 2016;68:701–87. doi: 10.1124/pr.115.012070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Hsu SH, Wang X, Kutay H, Bid HK, Yu J, et al. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59:555–66. doi: 10.1002/hep.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palli SR. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci. 2014;6:1–8. doi: 10.1016/j.cois.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C, et al. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145:853–64. doi: 10.1053/j.gastro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao L, Yan X. Nanozymes: an emerging field bridging nanotechnology and biology. Sci China Life Sci. 2016;59:400–2. doi: 10.1007/s11427-016-5044-3. [DOI] [PubMed] [Google Scholar]
- 47.Meers C, Keskin H, Storici F. DNA repair by RNA: Templated, or not templated, that is the question. DNA Repair (Amst) 2016;44:17–21. doi: 10.1016/j.dnarep.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu L, Park KH, Zhao L, Xu J, El RM, Gao Y, et al. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Mol Ther. 2016;24:564–9. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boerneke MA, Dibrov SM, Hermann T. Crystal-Structure-Guided Design of Self-Assembling RNA Nanotriangles. Angew Chem Int Ed Engl. 2016;55:4097–100. doi: 10.1002/anie.201600233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han D, Park Y, Kim H, Lee JB. Self-assembly of free-standing RNA membranes. Nature Communications. 2014;5:4367. doi: 10.1038/ncomms5367. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Zhang K, Pi F, Guo S, Shlyakhtenko L, Chiu W, et al. Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting. Adv Mater. 2016 doi: 10.1002/adma.201601976. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.