Abstract
We have developed a microfluidic system suitable to be incorporated with a metabolic imaging method to monitor the drug response of cells cultured on a chip. The cells were perfusion-cultured to mimic the blood flow in vivo. Label-free optical measurements and imaging of nicotinamide adenine dinucleotide and flavin adenine dinucleotide fluorescence intensity and morphological changes were evaluated non-invasively. Drug responses calculated using redox ratio imaging were compared with the drug toxicity testing results obtained with a traditional well-plate system. We found that our method can accurately monitor the cell viability and drug response and that the IC50 value obtained from imaging analysis was sensitive and comparable with a commonly used cell viability assay: MTS (3–(4,5-dimethylthiazol-2-yl)–5–(3-carboxymethoxyphenyl)-2–(4-sulfo-phenyl)-2H-tetrazolium) assay. Our method could serve as a fast, non-invasive, and reliable way for drug screening and toxicity testing as well as enabling real-time monitoring of in vitro cultured cells.
I. INTRODUCTION
It is already acknowledged that a highly predictive preclinical drug screening may reduce the high cost of the drug development process.1 Drug failure in the clinical trial is a consequence of the poor predictive power of the existing in vitro models. These models can be grouped into two major categories based on the method used for media refreshing: static and perfusion. The static methods—the most commonly used for in vitro models—are based on multi-well-plates and require robotic systems for drug preparation, administration, and analyses.2 Therefore, the perfusion methods—microengineered cell culture models developed on microfluidic platforms—have become a suitable alternative.2–7 In this direction, the microfluidic setup engages a better control of the cellular microenvironment, improves the cell viability, life span, and metabolic activity, and, moreover, supports different cell culture models.8–12 In microfluidic drug testing platforms, the drug response is usually evaluated with biochemical assays.13–15 One of the most commonly used methods is the MTS (3–(4,5-dimethylthiazol-2-yl)–5–(3-carboxymethoxyphenyl)-2–(4-sulfophenyl)-2H-tetrazolium) assay, a colorimetric method for quantifying the viability of cells. The method is based on the reduction of MTS tetrazolium to a colored formazan product.16 However, these biochemical assays are invasive and can only be performed at experimental endpoints.3,17,18 Therefore, it is essential to design and develop cell viability assays to evaluate the real-time drug response on microfluidic platforms.19
Imaging technologies such as multiphoton microscopy are usually non-invasive and have recently been used for cell and tissue monitoring.20,21 Cellular activities can be evaluated by multiphoton microscopy, through fluorescence signals produced by endogenous cell fluorophores such as nicotinamide adenine dinucleotide (phosphate) (NAD(P)H) and flavin adenine dinucleotide (FAD). NADH and NADPH are often used together as one fluorophore: NAD(P)H due to their similar fluorescence spectrum and their functions as electron carriers in the cellular metabolic function. The two fluorophores in the electron transport chain, NADH and FAD, are coenzymes in mitochondria that contributes to cellular metabolism and oxygen movement.22 NADH, the electron donator to molecular oxygen, has fluorescence excitation and emission maxima at 350 nm and 460 nm; FAD, the electron acceptor, has fluorescence excitation and emission maxima at 450 and 535 nm.23 The redox ratio, defined by the fluorescence intensity of FAD divided by that of NADH, measures the relative changes in the oxidation–reduction state in the cells. The redox ratio responds sensitively to changes in the cellular metabolic rate. The decreased redox ratio usually indicates increased cellular metabolic activity.24,25
Here, in order to achieve non-invasive drug toxicity testing on microfluidic chips, we developed a non-invasive imaging-based assay to evaluate drug toxicity response. We used the optical redox ratio of FAD and NADH to monitor changes in cellular metabolic activity as a result of drug treatment. In our study, we selected HepG2 cells, a human liver cancer cell line due to their high degree of morphological and functional differentiation that resembles polarized human hepatocytes in vitro.26 High resolution (∼400 nm) and label free images of HepG2 cells were acquired by two-photon microscopy. In the imaging process, two photon excitation occurred when a fluorophore was excited by two photons of half the absorption energy of the fluorophore.27 Compared with traditional static cultures, microfluidic chips offer good control of the essential spatiotemporal cues in the microenvironment to maintain organotypic functions close to the physiological context for a longer period of time.2,4,28 The ability to continuously support cell growth and to monitor with real-time imaging favoured this option.29 In this study, we propose a novel two-photon metabolic imaging method for label-free, non-invasive, and real-time drug toxicity testing on a microfluidic chip.
II. MATERIALS AND METHODS
A. Microfluidic chip design and fabrication
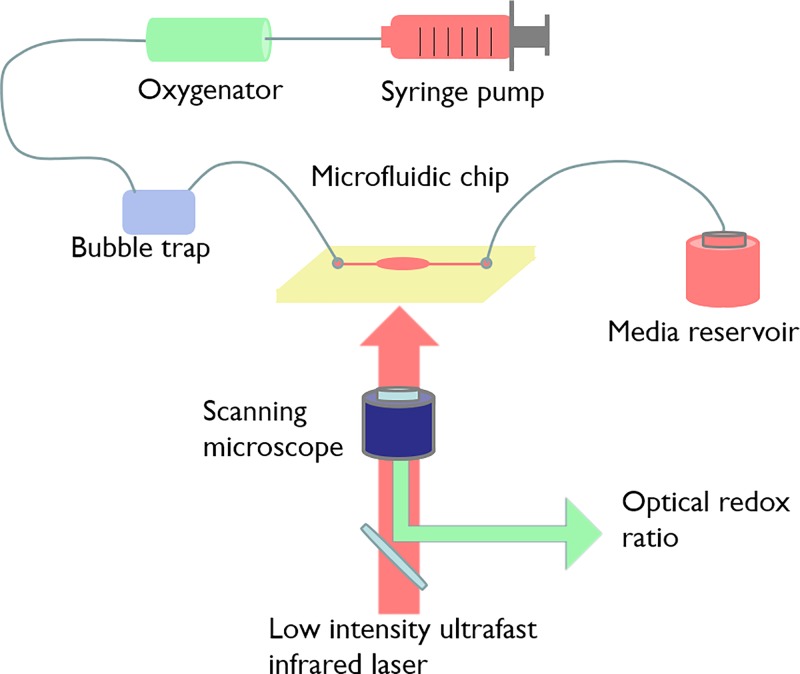
The microfluidic system is a one-pass perfusion system. As shown in Figure 1, it comprised a culture medium reservoir (syringe purchased from Becton Dickinson, USA), a syringe pump (Cole-Parmer, USA), an oxygenator [in house fabricated with Poly(methyl methacrylate) (PMMA)], a bubble trap (in house fabricated with PMMA), and a chip. The oxygenator ensured gas exchange between the culture media and the atmosphere. The bubble trap prevented bubbles from entering the cell culture well. The components were connected by a 1.0 mm interior diameter silicone tubing (New England Small Tubes Corp., USA). The device was primed with ethanol, 0.2% bovine serum albumin (BSA), and 1× PBS (Phosphate Buffer Saline) using a pressure filling technique to remove all the bubbles.13 At the same time, the cells were seeded at the same density of 0.25 × 106 cells/cm2 in 48-well culture plates (Becton Dickinson, USA) containing 2 ml of medium as control.
FIG. 1.
Schematics of the microfluidic system. It is composed of a culture medium reservoir, a syringe pump, an oxygenator, a bubble trap, and a chip. The system was imaged on a two-photon imaging system.
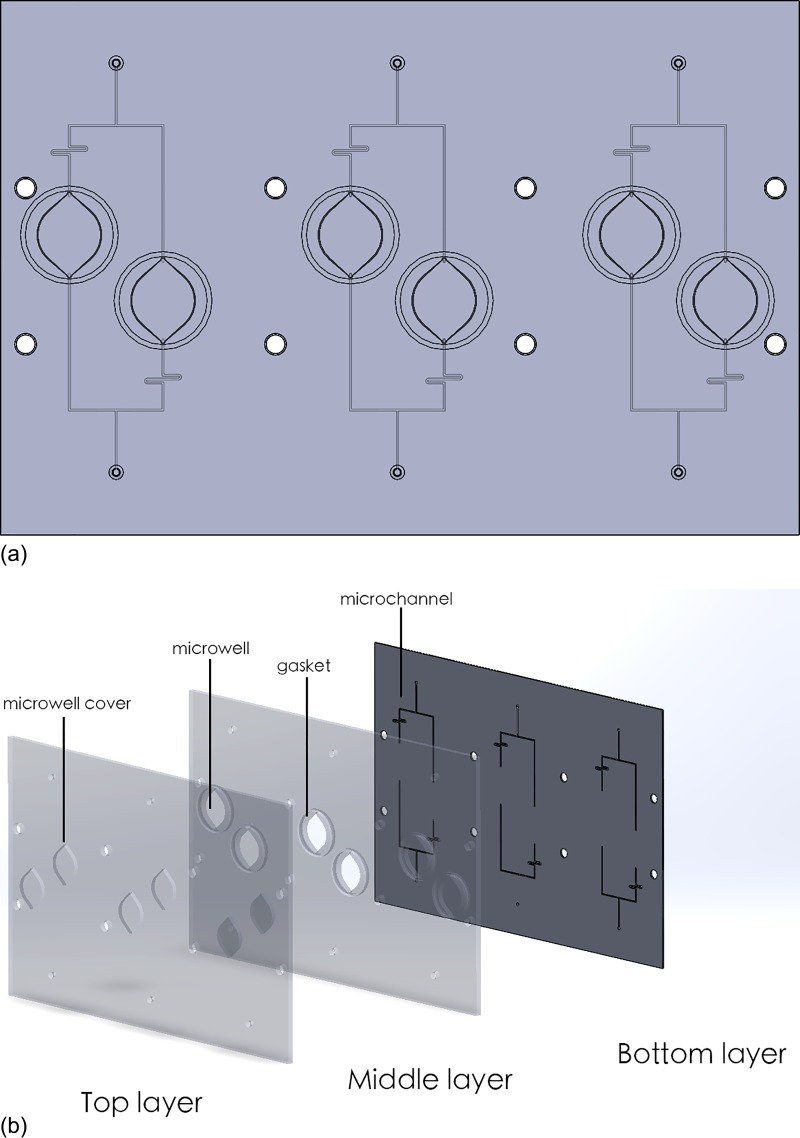
The microfluidic chip was designed in Solidworks 2010 (Dassault Systèmes, France) and fabricated in Poly(methyl methacrylate) (PMMA) by milling (Figure 2). It consists of 4 pairs of microwells for cell culture and microfluidic channels. The wells on-chip have a diameter of 11 mm, the same as the wells in 48-well-plates. This geometry allowed easy comparison of experimental results from a 48-well plate. The inlets and outlets were drilled at two ends of the microchannel and were connected with Nanoports microfluidic connectors (Upchurch, USA). The chip consists of 3 layers: (1) the microchannel was milled onto the bottom layer; (2) through holes were drilled in the middle layer to define the microwells; and (3) the top layer was used to seal the chip. On the middle layer, ring shaped grooves were milled outside the microwells. Rubber gasket rings were placed in these grooves to prevent leaking and cross contamination. The middle and bottom layers were thermally bonded. Prior to bonding, the PMMA surfaces were cleaned with deionized water and Isopropylic alcohol (IPA) followed by N2 drying. The PMMA has a glass transition temperature of approximately 105 °C. The substrates were preheated to 80 °C and clamped by two glass plates. In order to achieve a good bonding, the temperature was increased to 95–100 °C and maintained for 2 h. The final chip was assembled and fastened by bolts and nuts.
FIG. 2.
(a) The layout of the microfluidic chip. There are 3 pairs of microwells, each pair is connected to an inlet and an outlet. (b) The exploded view of the PMMA chip. It consists of 3 layers bonded together using thermo-bonding. Microchannels are fabricated with micro milling.
B. Cell culture
HepG2 cells were obtained from the American Type Culture Collection (ATCC, USA). The cells cultured in the biochip were sustained in Minimum Essential Medium (MEM, eagle) supplemented with 1% of HEPES, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1.0 mM sodium pyruvate, and 10% of fetal bovine serum and penicillin-streptomycin (100 U/ml).
HepG2 cells were seeded on glass cover slips in 48-well-plates at the density of 0.25 × 106 cells/cm2 in 0.5 ml of medium (corresponding to 500 000 cells/well). Cells were allowed to attach to the glass surface for 24 h before they were transferred into the microfluidic chip with forceps in a biosafety cabinet. The top cover of the chip sealed the chip for perfusion culture. Circular rubber gaskets were used around the wells for water tight sealing. Subsequently, HepG2 cells were stabilized in the chip for 24 h in perfusion before drug administration. The chip was then kept in a 5% CO2 incubator at 37 °C. MEM culture medium was then perfused at a flow rate of 0.1 ml/h for 72 h.
C. Drug toxicity testing
The HepG2 cells were subjected to chemotherapeutic drugs cisplatin and doxorubicin for toxicity testing. 1 day after culturing on chip, the cells were exposed to 4 concentrations of cisplatin (24–2400 μM) and doxorubicin (1–100 μM) for 24 h under perfusion condition. Dimethyl sulfoxide (DMSO) vehicle controls were established by using medium supplemented with 1% DMSO without drugs. To compare with the results obtained from the redox ratio calculation, cellular viability was tested with MTS assay (Promega, USA). We estimated the drug concentration that produced a 50% inhibitory effect (IC50) by fitting the dose response curve. Its value is determined by non-linear regression curve fitting using GraphPad Prism 6.0 software.
D. Imaging instrumentation
Images were obtained using a confocal microscope with a low intensity ultrafast infrared laser (Carl Zeiss, Germany). The excitation light and emission light were coupled through an inverted microscope. An excitation wavelength of 810 nm was used to excite both fluorophores. Bandpass filters of 409–505 nm and 505–634 nm were used to isolate NADH and FAD emission onto the photomultiplier tube (PMT) detector.30 The calibration of the measurements was done to account for systematic errors such as fluctuations in the throughput and excitation efficiency of the system. Rhodamine B of 5.8 × 10−5 g/ml concentration was used as calibration standard.21 Rhodamine standard images were collected daily in condition identical to those of the cell culture images of that day. In the imaging process, the chip was secured on a temperature-controlled microscope stage, which maintained a constant temperature of 37 °C. HepG2 cells were exposed to steady flow of culture media to maintain a favorable cellular microenvironment. Two-photon images were collected at the NADH excitation wavelength followed by the FAD excitation wavelength.
E. Quantification of the optical redox ratio
The image analysis was completed using ImageJ software (NIH, USA) with methods described in Ref. 31. Each scan produced raw NADH and FAD images with 8 × 8 grids, totaling 64 subimages. We selected and processed 16 out of the 64 based on the cell confluency in the region. The redox ratio was computed for each pixel by dividing the NADH fluorescence intensity with FAD fluorescence intensity. The results were multiplied by a scalar value that accounts for the Rhodamine standard measured for the NADH and FAD images. The mean redox ratio for the image was computed by averaging the redox ratios from all the images. The calculation was done using the formula
| (1) |
In Equation (1), [Redox] is the final redox ratio image, [FAD] is the FAD intensity image, and [NADH] is the NADH intensity image. RFAD and RNADH are the mean Rhodamine standard intensity scalar values. Redox ratios of the drug-interrogated samples were normalized to redox ratios of the fully viable control samples to get the viability.
III. RESULTS
A. Drug toxicity testing in static culture
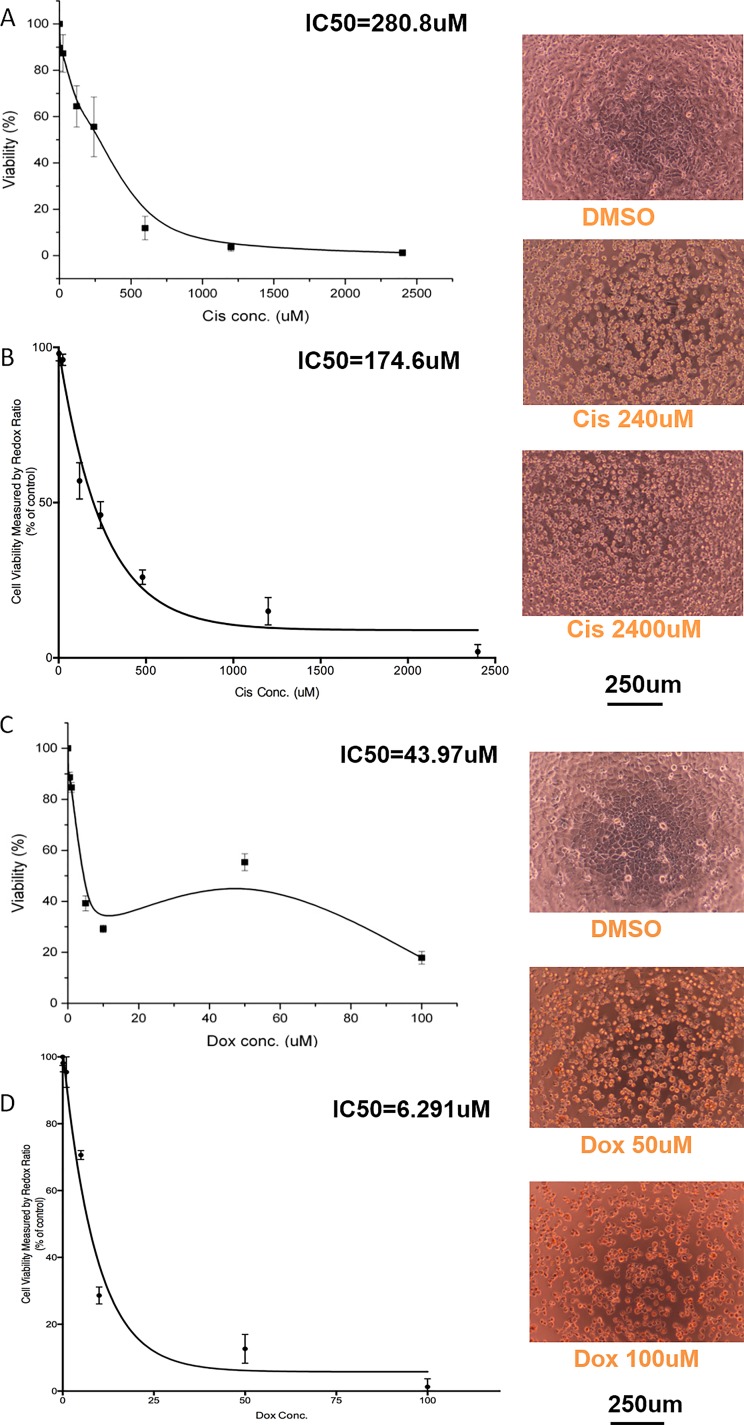
We compared drug toxicity response derived from the MTS assay and redox ratio method to confirm the feasibility of the redox ratio as a drug toxicity assay. We administered two effective chemotherapeutic agents: cisplatin and doxorubicin to HepG2 cells. They demonstrated the inhibitory effect for cancer cells as well as the cell lines derived from cancer cells such as HepG2.32 Traditionally, both the MTS assay and the redox ratio are used to evaluate the cytotoxicity. As presented in Figure 3, the redox ratio method showed a more sensitive drug response compared with MTS assay. Both assay methods were able to show decreased cell viability with increasing drug concentrations. We estimated the drug concentration that produced a 50% inhibitory effect (IC50 value) for each drug. The IC50 values achieved here were comparable to the results from literatures.33–35 For Cisplatin, the IC50 value calculated from the redox ratio method was 174.6 μM, smaller than that acquired from MTS assay: 280.8 μM. For doxorubicin, the IC50 value calculated from the redox ratio method was 6.29 μM, smaller than the IC50 value calculated from MTS assay: 43.97 μM. As depicted in Figure 3(c), when using the MTS method at a doxorubicin concentration of 50 μM, the cell viability response deviated from the dose response curve when the doxorubicin concentration was increased, and when fitting the curve, this resulted in a bump in the curve. This could be possible because of the red colour of the doxorubicin solution interfering with the colorimetric MTS assay reading, increasing the colorimetric reading. The actual cell viability is lower than the value measured by colorimetric assay at the concentration of 50 μM. The MTS assay and redox ratio method results were confirmed by the microscopy images taken for respective concentration groups. For lower drug concentrations (24 μM of cisplatin and 1 μM of doxorubicin), the cellular morphology was similar to the DMSO controls. However, at high drug concentrations (2400 μM of cisplatin and 100 μM of doxorubicin), the cells exhibited apoptotic morphology, such as cellular and nuclear shrinkage and formation of apoptotic bodies from the plasma membrane.36
FIG. 3.
Drug toxicity testing of HepG2 in 2D static culture. The IC50 calculated with MTS assay is compared with that of the redox ratio method to show that the redox ratio method can be used to acquire similar dose response data. Testing of cisplatin using (a) MTS and (b) Redox ratio. Testing of doxorubicin using (c) MTS and (d) Redox ratio. Data are presented as mean ±SD of 3 experiments.
B. Metabolic imaging of HepG2 on chip
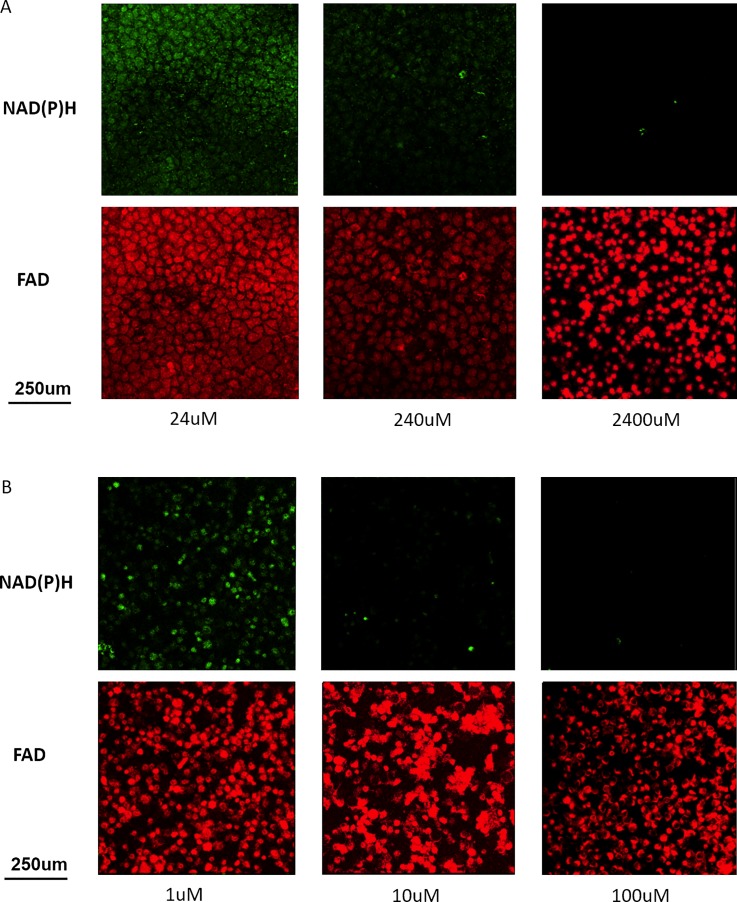
Figure 4 presents the results of two-photon metabolic imaging on microfluidic chips. For both cisplatin and doxorubicin, autofluorescence signals decreased with increasing drug concentrations, corresponding to decreased metabolic activities and decreased cellular viabilities.37 Similar to optical images in Figure 3, morphological changes in apoptotic cells were observed with metabolic imaging.
FIG. 4.
On-chip metabolic imaging of HepG2 incubated with (a) cisplatin and (b) doxorubicin. Each scan produced raw NADH and FAD images with 8 × 8 grids, totaling 64 subimages. 16 of the 64 subimages were selected and processed based on the cell confluency in the region. The figure shows individual subimages. The redox ratio was computed for each pixel by dividing the NADH fluorescence intensity with FAD fluorescence intensity.
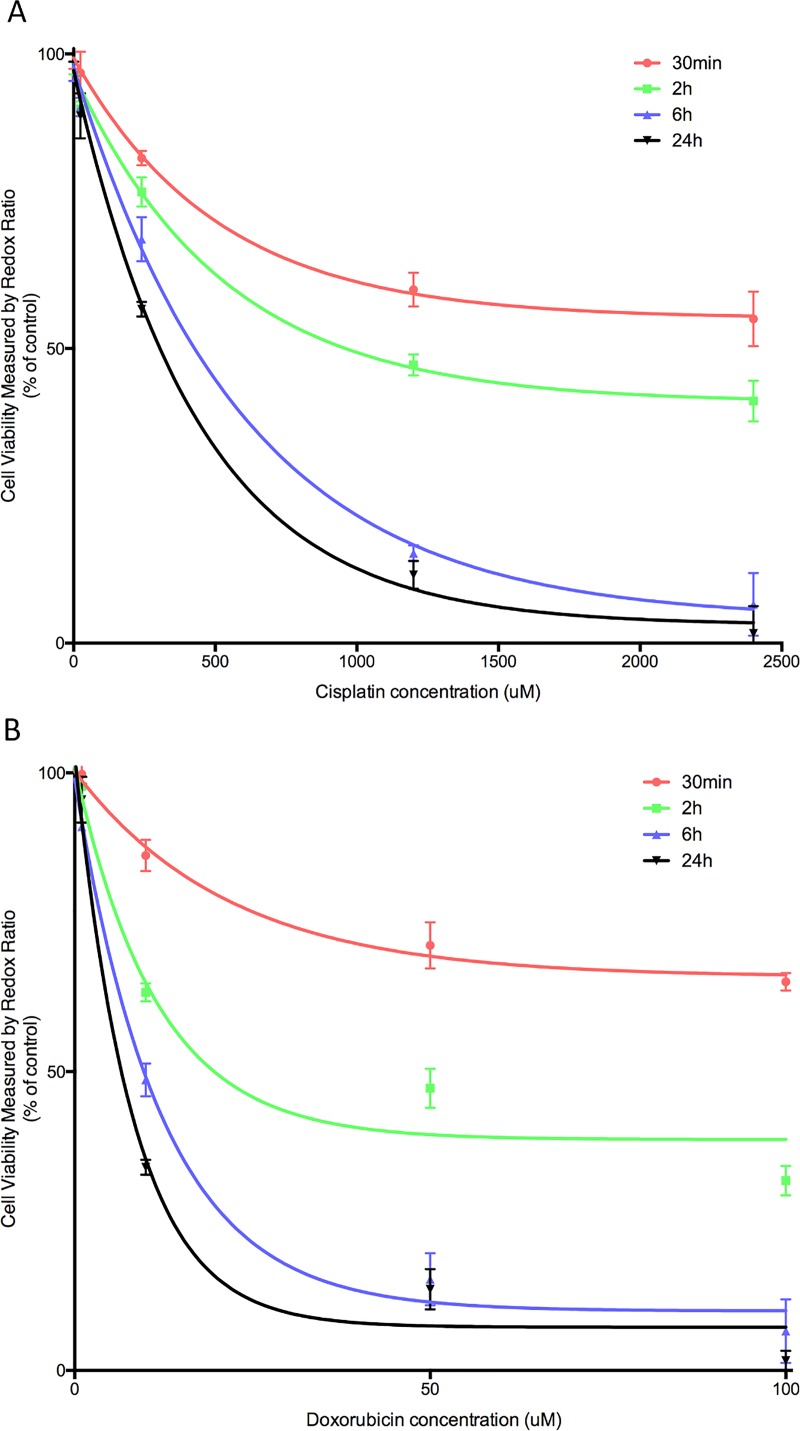
C. Time dependent dose response on chip
After demonstrating the feasibility of using the redox ratio method to evaluate drug toxicity responses in static cultures, we conducted a time dependent drug dose response study on chip, using the redox ratio method (Figure 5). Real-time redox ratio images were acquired on chip during perfusion culture to calculate the time dependent change in cellular viability. After administering the drugs, a trend of decreasing cells' viability was observed in both cisplatin and doxorubicin administered to HepG2s from 30 min to 24 h. The control group, in which only DMSO was added, remained viable during the observation period. The cells responded rapidly to the addition of drugs: a decrease in metabolic functions was detected at 30 min. At 24 h, the same experimental time point as for the static cultures, HepG2s cultured on chips exhibited similar dose response curves compared with the curve obtained from cultures in the well plate. IC50 values estimated at 24 h for cisplatin and doxorubicin were 303.7 and 5.659 μM, respectively. The higher IC50 value for cisplatin on the microfluidic chip compared with the static culture (174.6 μM) could be due to the favourable culture condition, leading to better cellular viability and metabolic function under perfusion.38,39
FIG. 5.
Real-time monitoring of the dose response of HepG2 to (a) cisplatin and (b) doxorubicin using the redox ratio method. Images taken at 30 min, 2 h, 6 h, and 24 h after drug administration. IC50 values estimated at 24 h for cisplatin and doxorubicin are 303.7 and 5.659 μM respectively. Data are presented as mean ±SD of 3 experiments.
IV. DISCUSSIONS
Cell viability assays are commonly used to determine the cytotoxicity of certain substances. These assays are widely used for drug screening in the pharmaceutical industry.21 Scientists can either look for cytotoxic compounds to develop drugs that target specific cell types such as cancer cells or they can screen through a wide range of drug candidates for unwanted toxicity before further developing the drugs. Viability can normally be assessed with a number of biochemical assays. For example, MTS assay measures the activity of NAD(P)H-dependent enzyme activities,16 lactate dehydrogenase (LDH) assay measures the release of LDH from the cytoplasmic membrane in the presence of damage and toxicity,40 and ATP assay quantifies the amount of adenosine triphosphate (ATP) by measuring the light produced through its reaction with luciferase.41 In this study, we aim to avoid using any biochemical marker to assess cell viability and drug response. We showed the feasibility of the using redox ratio acquired from two-photon metabolic imaging for the evaluation of the drug toxicity response. This method is a non-invasive, fast, and reliable alternative to MTS assay. The microfluidic chip is a good platform for the maintenance of cell culture during real-time imaging. Time dependent dose responses could be obtained, which is otherwise hard to achieve with traditional colorimetric assays. Moreover, more sensitive drug responses could be obtained with redox ratio assay compared with MTS assay.
Two-photon imaging have been used in many applications, including monitoring of metabolic changes in vivo,31 detection of cancer related biomarkers,37,42 monitoring of stem cell differentiation status,43 real-time characterization of tissue engineered constructs,44,45 and ex vivo metabolic measurements of isolated tissue.46 This study is the first to demonstrate the feasibility of using the redox ratio as a potential method to evaluate in vitro drug toxicity responses. Metabolic imaging was often used for the in vivo measurement of metabolism for various tissue types due to its good imaging depth, high resolution, and label-free nature.20,47–49 Redox ratio measurements enhanced the sensitivity detection of metabolic variations while reducing intensity-based artifacts, such as signal variations due to optical loss, making it ideal for quantification studies. Therefore, label-free technologies such as metabolic imaging allowing real-time, non-invasive evaluation on the cellular and molecular levels is a useful way to provide a more complete and accurate method to test drug response in vitro.
Microfluidic technologies were coupled with imaging modalities as a way to do on-line monitoring of metabolic activities: using mass spectroscopy to analyse tumour metabolism,50 using radioassay to image the glycolysis process,51 and using the oxygen imaging approach to evaluate cellular respiratory activity.52 These studies, which are invasive, require fluorescent or radioactive dyes, to be added to the cell. However, by using autofluorescence signals, we achieved real-time imaging of cellular metabolism non-invasively. We demonstrated the chip's capability of performing real-time imaging without the use of any label, allowing pharmaco-kinetics/pharmaco-dynamics (PK/PD) profile of drugs to be investigated easily. The future developments will integrate this system with automated imaging platforms to achieve high-throughput screening of drug candidates.
V. CONCLUSIONS
We incorporated a microfluidic perfusion cell culture chip with an optical metabolic imaging system to achieve label-free and non-invasive testing of anti-cancer drug toxicity. Drug toxicity responses calculated with the redox ratio measurement were comparable and more sensitive compared with MTS assay. The microenvironment in the chip supported the real-time imaging assessment of drug responses of cellular viability without the need for any fluorescent dyes. The on-chip metabolic imaging system could be impactful for the pharmaceutical industry as a cost-effective and non-invasive way for drug toxicity studies.
ACKNOWLEDGMENTS
This work was supported in part by the Institute of Bioengineering and Nanotechnology, Singapore Institute of Manufacturing Technology, and grants from JCO, ASTAR; MBI (through NRF and MOE), NMRC-CBRG, SMART-BioSyM, NUHS to H.Y.; F.Y. was a NGS scholar.
References
- 1. Dambach D. M., Andrews B. A., and Moulin F., “ New technologies and screening strategies for hepatotoxicity: Use of in vitro models,” Toxicol. Pathol. 33, 17–26 (2005). 10.1080/01926230590522284 [DOI] [PubMed] [Google Scholar]
- 2. Dittrich P. S. and Manz A., “ Lab-on-a-chip: Microfluidics in drug discovery,” Nat. Rev. Drug Discov. 5, 210–218 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- 3. Toh Y. C., Lim T. C., Tai D., Xiao G., Van Noort D., and Yu H., “ A microfluidic 3D hepatocyte chip for drug toxicity testing,” Lab Chip 9, 2026–2035 (2009). 10.1039/b900912d [DOI] [PubMed] [Google Scholar]
- 4. Neužil P., Giselbrecht S., Länge K., Huang T. J., and Manz A., “ Revisiting lab-on-a-chip technology for drug discovery,” Nat. Rev. Drug Discov. 11, 620–632 (2012). 10.1038/nrd3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El-Ali J., Sorger P. K., and Jensen K. F., “ Cells on chips,” Nature 442, 403–411 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- 6. Yu F., Iliescu F. S., and Iliescu C., “ A comprehensive review on perfusion cell culture systems,” Informacije MIDEM 46, 163–175 (2016); available at http://www.midem-drustvo.si/Journal%20papers/MIDEM_46(2016)4p163.pdf. [Google Scholar]
- 7. Choudhury D., van Noort D., Iliescu C., Zheng B., Poon K.-L., Korzh S., Korzh V., and Yu H., “ Fish and Chips: A microfluidic perfusion platform for monitoring zebrafish development,” Lab Chip 12, 892–900 (2012). 10.1039/C1LC20351G [DOI] [PubMed] [Google Scholar]
- 8. Alhasan L., Qi A., Al-Abboodi A., Rezk A., Chan P. P. Y., Iliescu C., and Yeo L. Y., “ Rapid enhancement of cellular spheroid assembly by acoustically driven microcentrifugation,” ACS Biomater. Sci. Eng. 2, 1013–1022 (2016). 10.1021/acsbiomaterials.6b00144 [DOI] [PubMed] [Google Scholar]
- 9. Tong W. H., Fang Y., Yan J., Hong X., Hari Singh N., Wang S. R., Nugraha B., Xia L., Fong E. L., Iliescu C., and Yu H., “ Constrained spheroids for prolonged hepatocyte culture,” Biomaterials 80, 106–120 (2016). 10.1016/j.biomaterials.2015.11.036 [DOI] [PubMed] [Google Scholar]
- 10. Xia L., Ng S., Han R., Tuo X., Xiao G., Leo H. L., Cheng T., and Yu H., “ Laminar-flow immediate-overlay hepatocyte sandwich perfusion system for drug hepatotoxicity testing,” Biomaterials 30, 5927–5936 (2009). 10.1016/j.biomaterials.2009.07.022 [DOI] [PubMed] [Google Scholar]
- 11. Zhang S., Tong W., Zheng B., Susanto T. A., Xia L., Zhang C., Ananthanarayanan A., Tuo X., Sakban R. B., and Jia R., “ A robust high-throughput sandwich cell-based drug screening platform,” Biomaterials 32, 1229–1241 (2011). 10.1016/j.biomaterials.2010.09.064 [DOI] [PubMed] [Google Scholar]
- 12. Choudhury D., Mo X., Iliescu C., Tan L. L., Tong W. H., and Yu H., “ Exploitation of physical and chemical constraints for three-dimensional microtissue construction in microfluidics,” Biomicrofluidics 5, 022203 (2011). 10.1063/1.3593407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toh Y.-C., Zhang C., Zhang J., Khong Y. M., Chang S., Samper V. D., van Noort D., Hutmacher D. W., and Yu H., “ A novel 3D mammalian cell perfusion-culture system in microfluidic channels,” Lab Chip 7, 302–309 (2007). 10.1039/b614872g [DOI] [PubMed] [Google Scholar]
- 14. Yu L., Chen M. C., and Cheung K. C., “ Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing,” Lab Chip 10, 2424–2432 (2010). 10.1039/c004590j [DOI] [PubMed] [Google Scholar]
- 15. Ma B., Zhang G., Qin J., and Lin B., “ Characterization of drug metabolites and cytotoxicity assay simultaneously using an integrated microfluidic device,” Lab Chip 9, 232–238 (2009). 10.1039/B809117J [DOI] [PubMed] [Google Scholar]
- 16. Malich G., Markovic B., and Winder C., “ The sensitivity and specificity of the MTS tetrazolium assay for detecting the in vitro cytotoxicity of 20 chemicals using human cell lines,” Toxicology 124, 179–192 (1997). 10.1016/S0300-483X(97)00151-0 [DOI] [PubMed] [Google Scholar]
- 17. Huh D., Leslie D. C., Matthews B. D., Fraser J. P., Jurek S., Hamilton G. A., Thorneloe K. S., McAlexander M. A., and Ingber D. E., “ A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice,” Sci. Transl. Med. 4, 159ra147 (2012). 10.1126/scitranslmed.3004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. B. and Sung J. H., “ Organ-on-a-chip technology and microfluidic whole-body models for pharmacokinetic drug toxicity screening,” Biotechnol. J. 8, 1258–1266 (2013). 10.1002/biot.201300086 [DOI] [PubMed] [Google Scholar]
- 19. Haycock J. W., “ 3D cell culture: A review of current approaches and techniques,” Methods Mol. Biol. 695, 1–15 (2011). 10.1007/978-1-60761-984-0 [DOI] [PubMed] [Google Scholar]
- 20. Zipfel W. R., Williams R. M., and Webb W. W., “ Nonlinear magic: Multiphoton microscopy in the biosciences,” Nat. Biotechnol. 21, 1369–1377 (2003). 10.1038/nbt899 [DOI] [PubMed] [Google Scholar]
- 21. Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., and Ramanujam N., “ In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia,” Proc. Nat. Acad. Sci. U.S.A. 104, 19494–19499 (2007). 10.1073/pnas.0708425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vander Heiden M. G., Cantley L. C., and Thompson C. B., “ Understanding the Warburg effect: The metabolic requirements of cell proliferation,” Science 324, 1029–1033 (2009). 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramanujam N., “ Fluorescence spectroscopy of neoplastic and non-neoplastic tissues,” Neoplasia 2, 89–117 (2000). 10.1038/sj.neo.7900077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramanujam N., Richards-Kortum R., Thomsen S., Mahadevan-Jansen A., Follen M., and Chance B., “ Low temperature fluorescence imaging of freeze-trapped human cervical tissues,” Opt Express 8, 335–343 (2001). 10.1364/OE.8.000335 [DOI] [PubMed] [Google Scholar]
- 25. Drezek R., Brookner C., Pavlova I., Boiko I., Malpica A., Lotan R., Follen M., and Richards-Kortum R., “ Autofluorescence microscopy of fresh cervical-tissue sections reveals alterations in tissue biochemistry with dysplasia,” Photochem. Photobiol. 73, 636–641 (2001). 10.1562/0031-8655(2001)0730636AMOFCT2.0.CO2 [DOI] [PubMed] [Google Scholar]
- 26. Wilkening S., Stahl F., and Bader A., “ Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties,” Drug Metab. Dispos. 31, 1035–1042 (2003). 10.1124/dmd.31.8.1035 [DOI] [PubMed] [Google Scholar]
- 27. Denk W., Strickler J. H., and Webb W. W., “ Two-photon laser scanning fluorescence microscopy,” Science 248, 73–76 (1990). 10.1126/science.2321027 [DOI] [PubMed] [Google Scholar]
- 28. Huh D., Hamilton G. A., and Ingber D. E., “ From 3D cell culture to organs-on-chips,” Trends Cell Biol. 21, 745–754 (2011). 10.1016/j.tcb.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bhatia S. N. and Ingber D. E., “ Microfluidic organs-on-chips,” Nat. Biotechnol. 32, 760–772 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 30. Huang S., Heikal A. A., and Webb W. W., “ Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein,” Biophys. J. 82, 2811–2825 (2002). 10.1016/S0006-3495(02)75621-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skala M. and Ramanujam N., “ Multiphoton redox ratio imaging for metabolic monitoring in vivo,” Methods Mol. Biol. 594, 155–162 (2010). 10.1007/978-1-60761-411-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. T. H. Guthrie, Jr. , McElveen L. J., Porubsky E. S., and Harmon J. D., “ Cisplatin and doxorubicin. An effective chemotherapy combination in the treatment of advanced basal cell and squamous carcinoma of the skin,” Cancer 55, 1629–1632 (1985). [DOI] [PubMed] [Google Scholar]
- 33. Chen Z., Ke L. D., Yuan X. H., and Adler-Storthz K., “ Correlation of cisplatin sensitivity with differential alteration of EGFR expression in head and neck cancer cells,” Anticancer Res. 20, 899–902 (2000). [PubMed] [Google Scholar]
- 34. Zhang R., Niu Y., and Zhou Y., “ Increase the cisplatin cytotoxicity and cisplatin-induced DNA damage in HepG2 cells by XRCC1 abrogation related mechanisms,” Toxicol. Lett. 192, 108–114 (2010). 10.1016/j.toxlet.2009.10.012 [DOI] [PubMed] [Google Scholar]
- 35. Al-Qubaisi M., Rozita R., Yeap S. K., Omar A. R., Ali A. M., and Alitheen N. B., “ Selective cytotoxicity of goniothalamin against hepatoblastoma HepG2 cells,” Molecules 16, 2944–2959 (2011). 10.3390/molecules16042944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elmore S., “ Apoptosis: A review of programmed cell death,” Toxicol. Pathol. 35, 495–516 (2007). 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walsh A. J., Cook R. S., Manning H. C., Hicks D. J., Lafontant A., Arteaga C. L., and Skala M. C., “ Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer,” Cancer Res. 73, 6164–6174 (2013). 10.1158/0008-5472.CAN-13-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sung J. H., Kam C., and Shuler M. L., “ A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip,” Lab Chip 10, 446–455 (2010). 10.1039/b917763a [DOI] [PubMed] [Google Scholar]
- 39. Prot J. M., Aninat C., Griscom L., Razan F., Brochot C., Guillouzo C. G., Legallais C., Corlu A., and Leclerc E., “ Improvement of HepG2/C3a cell functions in a microfluidic biochip,” Biotechnol. Bioeng. 108, 1704–1715 (2011). 10.1002/bit.23104 [DOI] [PubMed] [Google Scholar]
- 40. Lobner D., “ Comparison of the LDH and MTT assays for quantifying cell death: Validity for neuronal apoptosis?,” J. Neurosci. Methods 96, 147–152 (2000). 10.1016/S0165-0270(99)00193-4 [DOI] [PubMed] [Google Scholar]
- 41. Kurbacher C. M., Cree I. A., Bruckner H. W., Brenne U., Kurbacher J. A., Müller K., Ackermann T., Gilster T. J., Wilhelm L. M., and Engel H., “ Use of an ex vivo ATP luminescence assay to direct chemotherapy for recurrent ovarian cancer,” Anti-Cancer Drugs 9, 51–57 (1998). 10.1097/00001813-199801000-00006 [DOI] [PubMed] [Google Scholar]
- 42. Ostrander J. H., McMahon C. M., Lem S., Millon S. R., Brown J. Q., Seewaldt V. L., and Ramanujam N., “ Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status,” Cancer Res. 70, 4759–4766 (2010). 10.1158/0008-5472.CAN-09-2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rice W. L., Kaplan D. L., and Georgakoudi I., “ Quantitative biomarkers of stem cell differentiation based on intrinsic two-photon excited fluorescence,” J. Biomed. Opt. 12, 060504 (2007). 10.1117/1.2823019 [DOI] [PubMed] [Google Scholar]
- 44. Quinn K. P., Bellas E., Fourligas N., Lee K., Kaplan D. L., and Georgakoudi I., “ Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios,” Biomaterials 33, 5341–5348 (2012). 10.1016/j.biomaterials.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen L. C., Lloyd W. R., Kuo S., Kim H. M., Marcelo C. L., Feinberg S. E., and Mycek M. A., “ The potential of label-free nonlinear optical molecular microscopy to non-invasively characterize the viability of engineered human tissue constructs,” Biomaterials 35, 6667–6676 (2014). 10.1016/j.biomaterials.2014.04.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walsh A. J., Poole K. M., Duvall C. L., and Skala M. C., “ Ex vivo optical metabolic measurements from cultured tissue reflect in vivo tissue status,” J. Biomed. Opt. 17, 116015 (2012). 10.1117/1.JBO.17.11.116015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhuo S., Yan J., Kang Y., Xu S., Peng Q., So P. T. C., and Yu H., “ In vivo, label-free, three-dimensional quantitative imaging of liver surface using multi-photon microscopy,” Appl. Phys. Lett. 105, 023701 (2014). 10.1063/1.4890593 [DOI] [Google Scholar]
- 48. D'Amore J. D., Kajdasz S. T., McLellan M. E., Bacskai B. J., Stern E. A., and Hyman B. T., “ In vivo multiphoton imaging of a transgenic mouse model of Alzheimer disease reveals marked thioflavine-S-associated alterations in neurite trajectories,” J. Neuropathol. Exp. Neurol. 62, 137–145 (2003). 10.1093/jnen/62.2.137 [DOI] [PubMed] [Google Scholar]
- 49. Levene M. J., Dombeck D. A., Kasischke K. A., Molloy R. P., and Webb W. W., “ In vivo multiphoton microscopy of deep brain tissue,” J. Neurophysiol. 91, 1908–1912 (2004). 10.1152/jn.01007.2003 [DOI] [PubMed] [Google Scholar]
- 50. Chen Q., Wu J., Zhang Y., and Lin J. M., “ Qualitative and quantitative analysis of tumor cell metabolism via stable isotope labeling assisted microfluidic chip electrospray ionization mass spectrometry,” Anal. Chem. 84, 1695–1701 (2012). 10.1021/ac300003k [DOI] [PubMed] [Google Scholar]
- 51. Vu N. T., Yu Z. T., Comin-Anduix B., Sondergaard J. N., Silverman R. W., Chang C. Y., Ribas A., Tseng H. R., and Chatziioannou A. F., “ A beta-camera integrated with a microfluidic chip for radioassays based on real-time imaging of glycolysis in small cell populations,” J. Nucl. Med. 52, 815–821 (2011). 10.2967/jnumed.110.078725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ungerbock B., Charwat V., Ertl P., and Mayr T., “ Microfluidic oxygen imaging using integrated optical sensor layers and a color camera,” Lab Chip 13, 1593–1601 (2013). 10.1039/c3lc41315b [DOI] [PubMed] [Google Scholar]