Abstract
Diffusion imaging is critical for detecting acute brain injury. However, normal apparent diffusion coefficient (ADC) maps change rapidly in early childhood, making abnormality detection difficult. In this article, we explored clinical PACS and electronic healthcare records (EHR) to create age‐specific ADC atlases for clinical radiology reference. Using the EHR and three rounds of multiexpert reviews, we found ADC maps from 201 children 0–6 years of age scanned between 2006 and 2013 who had brain MRIs with no reported abnormalities and normal clinical evaluations 2+ years later. These images were grouped in 10 age bins, densely sampling the first 1 year of life (5 bins, including neonates and 4 quarters) and representing the 1–6 year age range (an age bin per year). Unbiased group‐wise registration was used to construct ADC atlases for 10 age bins. We used the atlases to quantify (a) cross‐sectional normative ADC variations; (b) spatiotemporal heterogeneous ADC changes; and (c) spatiotemporal heterogeneous volumetric changes. The quantified age‐specific whole‐brain and region‐wise ADC values were compared to those from age‐matched individual subjects in our study and in multiple existing independent studies. The significance of this study is that we have shown that clinically acquired images can be used to construct normative age‐specific atlases. These first of their kind age‐specific normative ADC atlases quantitatively characterize changes of myelination‐related water diffusion in the first 6 years of life. The quantified voxel‐wise spatiotemporal ADC variations provide standard references to assist radiologists toward more objective interpretation of abnormalities in clinical images. Our atlases are available at https://www.nitrc.org/projects/mgh_adcatlases. Hum Brain Mapp 38:3052–3068, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: clinical images, big data informatics, atlas construction, neurodevelopment, diffusion MRI
INTRODUCTION
Apparent diffusion coefficient (ADC) maps are parameter maps derived from diffusion tensor magnetic resonance images. ADC maps provide quantitative measures of water diffusion within brain tissue [Beaulieu, 2002; Le Bihan et al., 2001]. Many early childhood brain disorders cause regional decreases in water diffusion coincident with or before they cause changes on structural MRI [Liauw et al., 2008; Sener, 2001; Wolf et al., 2001]. Therefore, ADC maps are a key component in the diagnosis and prognosis of many early childhood brain disorders (e.g., ischemic insults, metabolic disorders, inflammatory and infectious processes [Counsell et al., 2003; Engelbrecht et al., 2002; Gano et al., 2013; Neil et al., 1998; Padhani et al., 2009]. However, interpreting quantitative ADC maps in early childhood is challenging. For example, in neonatal hypoxic ischemic encephalopathy (HIE), there is 20–50% intra‐/inter‐reader variability even for experienced pediatric neuroradiologists [Goergen et al., 2014; Ozturk, et al., 2008]. Difficulties in interpreting early childhood ADC maps arise because there is considerable individual variation and because the appearance of normal ADC maps changes significantly with age, particularly in the first two years of life [Deoni et al., 2011; Kwan et al., 2015; Neil et al., 1998; Sasaki et al., 2008; Schneider et al., 2009]. Thus, detection of abnormalities in ADC maps, which are often subtle and complex, is confounded by individual variability and rapid regional variations due to normal brain development [Liauw et al., 2008; Wolf et al., 2001]. To objectively and consistently detect abnormalities in ADC maps, ready access to quantitative normal ADC atlases at multiple time points during early childhood brain development would be advantageous [Almli et al., 2007; Mori et al., 2013].
An ideal ADC atlas would provide quantitative information on ADC changes during early brain development. However, existing studies only coarsely sample the ages from birth to 6 years. Some studies report ADC values for four years and up [Morris et al., 2009], or adolescence to young adulthood [Helenius et al., 2002; Naganawa et al., 2003; Sener 2001; Zhai et al., 2003], but did not report ADC changes in early childhood. Other studies report ADC values at birth [Engelbrecht et al., 2002; Kwan et al., 2015; Morriss et al., 1999; Neil et al., 1998; Sadeghi et al., 2013; Zhai et al., 2003], but not months or years after birth. Additional studies report ADC values in the first one [Provenzale et al., 2010], first two [Sener, 2001] or two to four [Engelbrecht et al., 2002] years of life, treating individuals within these intervals as representing one developmental stage. Therefore, prior studies have not captured the fine details of the rapidly changing ADC trajectories within the first 6 years of life. Quantitative ADC atlases providing changing ADC values during early childhood, with densely sampled atlases in the first two years of life, when brain develops most rapidly [Deoni et al., 2011; Kwan et al., 2015; Neil et al., 1998; Sasaki et al., 2008], are lacking.
An ideal ADC atlas would also provide regional as well as temporal ADC variations because different brain regions have different ADC values [Deoni et al., 2011; Watanabe et al., 2013]. However, little information on regional variations exists because: (1) the regions reported in the literature cover only part of the brain (e.g., corpus callosum [Engelbrecht et al., 2002; Morriss et al., 1999; Provenzale et al., 2010; Sadeghi et al., 2013; Zhai et al., 2003], caudate [Neil et al., 1998; Sener, 2001], thalamus [Helenius et al., 2002; Kwan et al., 2015; Naganawa et al., 2003; Neil et al., 1998; Sener, 2001], anterior and/or posterior white matter [Engelbrecht et al., 2002; Helenius et al., 2002; Kwan et al., 2015; Naganawa et al., 2003; Neil et al., 1998; Provenzale et al., 2010; Zhai et al., 2003]); (2) many studies manually annotated a small portion or only several voxels within a region; (3) across‐subject ADC variations were not reported, making it challenging to know if an individual subject's regional ADC value is within or outside normal variation; and (4) reported mean ADC values for large brain regions limits the ability to detect small, subtle changes that are restricted to only a subset of the region. Atlases should ideally quantify normal ADC statistics (means and standard deviations) at every voxel in the brain. However, such atlases do not currently exist for ADC maps in early life.
Existing childhood MRI atlases (Table 1) are either cross‐sectional [Akiyama et al., 2013; Altaye et al., 2008; Fonov et al., 2011; Geng et al., 2012; He and Parikh, 2013; Kazemi et al., 2007; Luo et al., 2014; Nossin‐Manor et al., 2013; Oishi et al., 2011; Prastawa et al., 2005; Shi et al., 2014; Weisenfeld and Warfield, 2009;Weisenfeld et al., 2006a, 2006b; Wilke et al., 2008; Xue et al., 2007; Zhang et al., 2014], or, are often based on structural MRI [Kuklisova‐Murgasova et al., 2011; Sanchez et al., 2012a, 2012b; Serag et al., 2012; Shi et al., 2011; Xie et al., 2015]. The only study that constructed age‐specific diffusion MRI atlases did so at only three time‐points (newborn, 1 year and 2 years of age) [Sadeghi et al., 2013], did not densely sample the first two years of life, and did not provide public access to the atlases. The unmet needs being addressed in our work are: (i) the lack of age‐specific 3D quantitative ADC atlases; (ii) the lack of densely sampled ADC atlases during the first year of life when ADC changes are most rapid (although [Shi et al., 2011] and [Sadeghi et al., 2013] are pioneering studies that constructed structural atlases from subjects 0 to 2 years of age, we extend these studies to include ADC atlases for a larger number of more narrowly defined age bins); and (iii) while existing studies in Table 1 were primarily constructed to enable neuroimaging data analysis (providing atlases for segmentation priors or a standard space for morphometry), there is an emerging demand from clinical radiologists for normative regional ADC values during early childhood to enable more objective identification of abnormalities in clinical ADC maps. To address these unmet needs, we set out to construct a series of whole brain age‐specific ADC atlases to quantify normal ADC variations in every voxel from birth to 6 years of life.
Table 1.
Comparison with existing cross‐sectional and age‐specific atlases
| Studies | MRI modality | # Subjects | Age at MRI | # Age bins | Atlases released | Scanner and magnetic field of strength |
|---|---|---|---|---|---|---|
| Cross‐sectional atlases | ||||||
| Prastawa et al., 2005 | T1w | 3 | Neonate | 1 | Siemens 3T | |
| Weisenfeld et al., 2006a | T1w, T2w | 13 | 42 weeks GA | 1 | GE 1.5T | |
| Weisenfeld et al., 2006b | T1w, T2w | 20 | 42 weeks GA | 1 | GE 1.5T | |
| Kazemi et al., 2007 | T1w | 33 | 39–42 weeks GA | 1 | Yes | GE 1.5T/3T, Siemens 3T |
| Xue et al., 2007 | T2w | 20 | 27–44 weeks GA | 1 | Philips 3T | |
| Wilke et al., 2008 | T1w | 404 | 4.8–18.6 years | 1 | GE 1.5T, Siemens 1.5T | |
| Altaye et al., 2008 | T1w | 77 | 9–15 months | 1 | Siemens 3T | |
| Weisenfeld et al., 2009 | T1w, T2w | 15 | 42 weeks GA | 1 | GE 1.5T | |
| Oishi et al., 2011 | T1w, T2w, DTI | 25 | 38–41 weeks PC | 1 | Yes | Philips 3T |
| Geng et al., 2012 | FA, RD, AD | 211 | 0–2 years | 1 | Siemens 3T | |
| He et al., 2013 | T2w | 19 | 23–30 weeks GA | 1 | Philips 3T | |
| Nossin‐Manor et al., 2013 | T1w, T2w, DTI | 54 | 26–34 weeks GA | 1 | GE 1.5T | |
| Akiyama et al., 2013 | T1w | 60 | 6 months | 1 | Yes | GE 1.5T, Siemens 3T |
| Zhang et al., 2014 | T1w, FA | 9 | 2–13 days | 1 | Siemens 3T | |
| Shi et al., 2014 | T2w | 73 | 9–55 days | 1 | Siemens 3T | |
| Luo et al., 2014 | T1w | 53 | 5.93–8.01 years | 1 | Philips 1.5T | |
| Age‐specific atlases | ||||||
| Shi et al., 2011 | T1w, T2w | 95 | 0–2 years | 3 | Yes | Siemens 3T |
| Fonov et al., 2011 | T1w, T2w, PD | 324 | 4.5–18.5 years | 5 | Yes | GE 1.5T, Siemens 1.5T |
| Kuklisova‐Murgasova et al., 2011 | T2w | 142 | 29–44 weeks, GA | 6 | Philips 3T | |
| Sanchez et al., 2012a | T1w, T2w | 154 | 2 weeks – 4 years | 13 | Yes | GE 1.5T, Siemens 1.5T/3T |
| Sanchez et al., 2012b | T1w, T2w | 494 | 4.5–24 years | 32 | Yes | GE 1.5T, Siemens 1.5T/3T |
| Serag et al., 2012 | T1w, T2w | 204 | 26–44 weeks, PC | 9 | Philips 3T | |
| Sadeghi et al., 2013 | FA, MD, RD, AD | 26 | 0–2 years | 3 | Siemens 3T | |
| Xie et al., 2015 | T1w | 138 | 7–16 years | 5 | Siemens 3T, GE 3T | |
| Ours | ADC | 201 | 0–6 years | 10 | Yes | Siemens 3T |
MRI: magnetic resonance imaging; T1w and T2w: T1‐ and T2‐weighted MRI; PD: proton density MRI; DTI: diffusion tensor imaging; GA: gestational age; PC: postconception.
METHODS
Querying, Downloading and Selecting Clinically Acquired Images
This study is HIPAA‐compliant and IRB approved. At our institution, diffusion imaging and generation of ADC maps are part of the pediatric brain MRI protocol for a variety of clinical indications. Our motivation was that, among the vast number of children who had undergone clinical MR scans at the Massachusetts General Hospital (MGH), there were likely to be a small number who were suspected to have brain disorders, but who turned out to be normal after MR evaluation and clinical follow‐up. Those images could provide a normative cohort to guide clinical interpretation of new cases as the diffusion sequence was acquired with our exact clinical protocol. We were aware that clinical data might be highly inconsistent and incomplete, and that children who underwent clinical scans were at risk of minor and subtle abnormalities that were not necessarily captured at the time of scan. Considering these factors, we used three rounds of very strict and conservative reviews to select the final normative cohort.
Query and data download
We used the Research Patient Data Registry (RPDR) to query the EHR and identify the cohort of interest, then, the Medical Imaging Informatics Bench to Bedside (mi2b2, https://www.nmr.mgh.harvard.edu/lab/mi2b2) software [Murphy et al., 2015] to access the identified cases from the PACS at MGH. The criteria for subjects in our initial cohort were: (a) scanned between 2006 and 2013, with scanner‐generated ADC maps; (b) 0 to 6 years of age at the time of the scan; and (c) having radiological reports with short impressions indicating no detected abnormalities [Coakley et al., 2003]. We also excluded images from any neonate with a history of preterm birth noted in the medical record. We found 2,871 term subjects who satisfied all these conditions. Some subjects' images were missing, incomplete or corrupted. So we were only able to obtain complete ADC maps for 1,648 out of those 2,871 subjects.
Selection based on longitudinal clinical reports
Out of the 1,648 subjects whose ADC maps we downloaded, 705 had radiological reports showing no clear abnormality at the time of the scan. To further exclude those who may have abnormalities that were too subtle or too early to tell at the time of the scan, we used RPDR again and pulled the follow‐up clinical and radiological reports of those 705 children up to 2 years after the initial scan. We removed 322 children who were reported with brain abnormalities in later visits. We had 383 children remaining in our dataset.
Selection based on additional expert reviews
These 383 children remaining may still have very minor or subtle abnormalities, which, although not concerning clinically, may contaminate our data. Therefore, we conducted two additional rounds of expert review of the ADC maps at their initial visit. Two experts—a pediatric neuroradiologist (PEG) and a neonatologist (SVB)—reviewed the radiology reports (PEG) and complete clinical records (SVB) of the remaining 383 subjects one by one. If either of the two experts found evidence or strong suspicion of a neurological disorder, the subject was excluded, even if the radiological report did not identify a clear abnormality. With these exclusions, ADC maps of 201 children remained. These 201 subjects were free of neurological disorders at the time of the MRI and at follow‐up visits up to 2 years after the MRI. Those 201 ADC maps became our normative cohort for atlas construction.
Imaging information
All imaging studies were performed on Siemens Trio 3T scanners and our clinical diffusion protocol was as follows: TR = 7,500–9,500 ms, TE = 80–115 ms, b value = 1,000 s/mm2, matrix = 128 × 128 × 60, voxel size = 2 × 2 × 2 mm and number of diffusion directions = 32. ADC maps were automatically generated by the diffusion sequence.
Atlas Construction
Patient grouping
We divided the 201 subjects into 10 age groups. They included: the first two weeks of life, the remaining first quarter, the second, third and fourth quarter of the first year, then, every year from the second to the sixth year of the life. These age divisions were chosen specifically to sample the first two years and, in particular, the first year more densely.
Table 2 lists the numbers of male, female and all subjects in each age group. Figure 1 shows the distributions of the exact ages in each age group. The numbers of males and females were roughly balanced (1.1:1 to be precise).
Table 2.
The numbers of male, female and all subjects in the 10 age bins
| Age | Y1 | Y2 | Y3 | Y4 | Y5 | Y6 | Total | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| W1‐2 | Rest of Q1 | Q2 | Q3 | Q4 | |||||||
| # Subjects | 13 | 13 | 8 | 8 | 13 | 34 | 33 | 25 | 21 | 33 | 201 |
| # Females | 4 | 5 | 4 | 5 | 5 | 17 | 14 | 14 | 10 | 15 | 93 |
“W1‐2” refers to the first 2 weeks of life. “Q2” refers to the second quarter of the first year of life and “Y3” refers to the third year of life. Other abbreviations are similarly defined.
Figure 1.
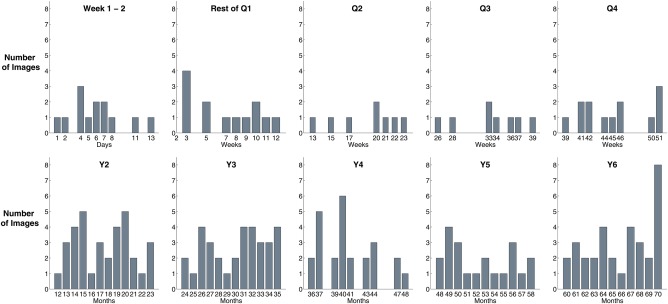
Distribution of children's ages within each atlas age group. [Color figure can be viewed at http://wileyonlinelibrary.com]
Brain extraction
ADC maps were skull stripped by an automated multiatlas brain extraction algorithm [Ou et al., 2015a]. This algorithm achieved consistently high agreement (0.96 ± 0.01 Dice Coefficient) with expert annotations in all age groups [Ou et al., 2015a]. Step 1 in Figure 2 visualizes the automatically computed brain extraction results (red contours) for all subjects in the Q2 age group. We used the skull stripped ADC maps for atlas construction.
Figure 2.
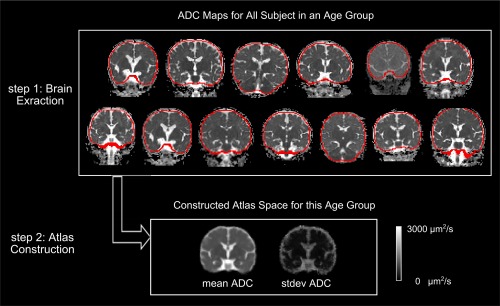
Illustration of atlas construction in one age group (Q2). From all extracted brains (red contours in step 1), we computed an atlas space quantifying mean and standard deviation (stdev) ADC values at every voxel (step 2). [Color figure can be viewed at http://wileyonlinelibrary.com]
Constructing age‐specific atlases
We constructed 10 ADC atlases corresponding to the age groups noted above. Atlas construction was formulated as an unbiased group‐wise deformable registration problem [Fonov et al., 2011], similar to [Fonov et al., 2011; Guimond et al., 2000; Kuklisova‐Murgasova et al., 2011]. Briefly, the atlas construction was unbiased, in the sense that we did not randomly pick any one subject and register every other subject into this subject space, which would introduce bias toward the anatomy and the ADC histogram distributions of this randomly chosen subject. In the unbiased atlas construction, we iteratively computed a virtual space, referred to as the atlas space, which represented the mean geometry and mean anatomy of all images in the group. Mathematically, the virtual atlas space was a space such that the N deformations needed to map the N images in the same age group into this virtual atlas space sum up to zero everywhere within this atlas space [Guimond et al., 2000]. After having computed the atlas space, and after having computed the deformations needed to register images in the same age group to this atlas space, the mean atlas (μ atlas) was computed by directly averaging all mapped ADC values from all subjects at every voxel in the atlas space. The standard deviation map in the atlas space (σ atlas) was similarly computed by computing standard deviations of mapped ADC values from all subjects at every voxel in the atlas space.
The atlas construction was initialized by affine registrations to establish global spatial correspondence between the input images. After that we computed deformable registration to account for regional differences. The resulting atlas space, after affine and deformable processes, became the final atlas space in each age group.
The central part of the atlas construction consisted of pair‐wise registrations. Of the many available registration algorithms [Klein et al., 2009; Ou et al., 2014; Sotiras et al., 2013], we chose the DRAMMS pair‐wise registration software [Ou et al., 2011] (https://www.nitrc.org/projects/dramms) and its group‐wise registration version specifically programmed for unbiased atlas construction (https://www.nitrc.org/projects/popdramms). We chose DRAMMS because of its strong performance compared to more than 10 algorithms in image registration with across‐subject variation [Ou et al., 2014], good performance in ADC modality [Erus et al., 2014; Ou et al., 2015a; Satterthwaite et al., 2014] and good performance in developing brains as rapidly changing as in baboon [Love et al., 2016], mouse [Anderson and Maga, 2015; Ingalhalikar et al., 2015; Young and Maga, 2015] and children as young as neonates [Erus et al., 2014; Ou et al., 2014; Satterthwaite et al., 2014, 2016].
As a conceptual depiction, Step 2 in Figure 2 shows the constructed (unbiased) atlas, with both the mean atlas and the standard deviation map, in the exemplar Q2 age group.
Validation of and Data Extraction from Atlases
We validated the accuracy of our atlases as a representation of the cohort we selected in two ways, corresponding to two hypotheses.
First, we tested the hypothesis that the ADC values in the constructed atlases were representative of the mean ADC values in the collected cohort, in each age group. That was, one atlas could represent multiple individuals. This hypothesis should hold if the unbiased atlas construction served its theory. This means that we can annotate one atlas and arrive at the same exact ADC statistics as if we repeatedly annotate ADC maps from multiple individuals. To test this hypothesis, we compared the brain volume and ADC values in the constructed atlases with the mean values of all individuals in the same age group, for all age groups. A Student's t‐test was calculated to see if the mean of population was statistically significantly different from the values in the atlases.
Second, we tested the hypothesis that once carefully reviewed and processed, clinically acquired ADC maps could generate similar global and regional ADC statistics compared to those from data sets generated from normal subjects recruited into research studies. We tested this hypothesis by comparing the mean ADC values of our atlases, which were based on clinical images, with mean ADC values, both voxel wise and for several regions of interest (ROIs), as reported in the literature for normal subjects.
Quantifying global and regional ADC changes
We summarized whole‐brain mean ADC values in each age‐specific ADC atlas. Additionally, a trained person (KR) annotated representative white and gray matter regions of interests (ROIs, in 3D) in all of our age‐specific ADC atlases. A second person (YO) verified the ROI annotations. We chose ROIs that have been frequently studied in the literature for age‐dependent ADC changes (Table 4). They include bilateral anterior and posterior whiter matter (WM) ROIs, as well as the entire bilateral caudate heads, bilateral thalami and corpus callosum. Anterior and posterior WM ROIs were placed where we had high confidence that only WM was included, and where each ROI spanned at least 3 axial slices and contained at least 50 voxels (in atlases for age groups under 1 year) and 100 voxels (in atlases for 1 year or older ages). The boundaries of caudate head, thalamus and corpus callosum were well defined, so we were able to label them in their entirety using all three planes. We plotted the ROI‐specific mean ADC values across ages to quantitatively summarize neurodevelopmental ADC changes in these ROIs. We further compared our findings with the literature (from individual subjects).
Table 4.
Comparison of the regional ADC values in our constructed atlases with regional ADC values reported in individuals/populations in the literature
| Birth | Q1 | Q2 | Q3 | Q4 | Y2 | Y3 | Y4 | Y5 | Y6 | 20–40 yo | Imaging protocol | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Caudate | ||||||||||||
| Our atlases (L) | 1,235 | 936 | 848 | 814 | 812 | 810 | 804 | 786 | 781 | Siemens 3T, 2 × 2 × 2 mm, b = 1,000 | ||
| Our atlases (R) | 1,226 | 965 | 857 | 840 | 829 | 817 | 800 | 803 | 802 | |||
| Neil et al., 1998 | 1,240 | Siemens 1.5T, 2 × 2 × 5 mm, b = 987/800 | ||||||||||
| Sener et al., 2012 | 820 | Siemens 1.5T, b = 1,000 | ||||||||||
| Thalamus | ||||||||||||
| Our atlases (L) | 1,101 | 899 | 840 | 840 | 818 | 809 | 799 | 783 | 782 | Siemens 3T, 2 × 2 × 2 mm, b = 1,000 | ||
| Our atlases (R) | 1,111 | 904 | 829 | 823 | 800 | 796 | 789 | 768 | 765 | |||
| Neil et al., 1998 | 1,080 | Siemens 1.5T, 2 × 2 × 5 mm, b = 987/800 | ||||||||||
| Sener et al.,2012 | 980 | 830 | Siemens 1.5T, b = 1,000 | |||||||||
| Helenius et al., 2002 | 730 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | ||||||||||
| Naganawa et al., 2003 | 830–910 | Siemens 1.5T, 2 × 2 × 6 mm, b = 1,000 | ||||||||||
| Kwan et al., 2015 | 1,075 | 1,020 | Phillips 3T, 1.8 × 1.8 × 1.8 mm, b = 750 | |||||||||
| Corpus Callosum | ||||||||||||
| Our atlases | 1,524 | 1,359 | 1,204 | 1,145 | 1,057 | 976 | 963 | 960 | 942 | Siemens 3T, 2 × 2 × 2 mm, b = 1,000 | ||
| Morriss et al., 1999 (L genu) | 1,540 | 1,310 | 1,130 | 940 | 1,080 | 869 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | |||||
| Morriss et al., 1999 (R genu) | 1,470 | 1,330 | 1,320 | 950 | 1,060 | 933 | ||||||
| Morriss et al., 1999 (L splenium) | 1,380 | 1,410 | 1,350 | 1,180 | 1,180 | 950 | ||||||
| Morriss et al., 1999 (R splenium) | 1,380 | 1,240 | 1,350 | 1,030 | 1,120 | 960 | ||||||
| Zhai et al., 2003 (genu) | ∼1,200 | ∼780 | Siemens 3T, 1.722 × 5 mm, b = 1,000 | |||||||||
| Zhai et al., 2003 (splenium) | ∼1,150 | ∼850 | ||||||||||
| Engelbrecht et al., 2002 (genu) | 1,280 | 760 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | |||||||||
| Engelbrecht et al., 2002 (splenium) | 1,410 | 770 | ||||||||||
| Provenzale et al., 2007 (genu) | 1,180 | GE 1.5T, 3 × 3 × 5 mm, b = 1,000 | ||||||||||
| Provenzale et al., 2007 (splenium) | 1,110 | |||||||||||
| Sadeghi et al., 2013 | ∼1,350 | ∼1,050 | ∼950 | Siemens 3T, 2 × 2 × 2 mm, b = 1,000 | ||||||||
| White Matter (Anterior) | ||||||||||||
| Our atlases (L) | 1,517 | 1,142 | 1,096 | 1,037 | 998 | 938 | 911 | 880 | 875 | Siemens 3T, 2 × 2 × 2 mm, b = 1,000 | ||
| Our atlases (R) | 1,483 | 1,137 | 1,094 | 1,039 | 980 | 915 | 877 | 850 | 856 | |||
| Neil et al., 1998 | 1,450 | Siemens 1.5T, 2 × 2 × 5 mm, b = 987/800 | ||||||||||
| Zhai et al., 2003 | ∼1,500 | ∼750 | Siemens 3T, 1.722 × 5 mm,b = 1,000 | |||||||||
| Engelbrecht et al., 2002 | 1,500 | 920 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | |||||||||
| Helenius et al., 2002 | 710 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | ||||||||||
| Naganawa et al., 2003 | 780–860 | Siemens 1.5T, 2 × 2 × 6 mm, b = 1,000 | ||||||||||
| Provenzale et al., 2007 | 1,330 | GE 1.5T, 3 × 3 × 5 mm, b = 1,000 | ||||||||||
| Kwan et al., 2015 | 1,760 | 1,508 | Phillips 3T, 1.8 × 1.8 × 1.8 mm, b = 750 | |||||||||
| Whiter Matter (Posterior) | ||||||||||||
| Our atlases (L) | 1,322 | 1,064 | 1,029 | 1,017 | 952 | 915 | 900 | 876 | 865 | Siemens 3T, 2 × 2 × 2 mm, b = 1,000 | ||
| Our atlases (R) | 1,385 | 1,081 | 1,027 | 1,028 | 970 | 921 | 896 | 877 | 863 | |||
| Neil et al., 1998 | 1,500 | Siemens 1.5T, 2 × 2 × 5 mm, b = 987/800 | ||||||||||
| Zhai et al., 2003 | ∼1,550 | ∼880 | Siemens 3T, 1.722 × 5 mm,b = 1,000 | |||||||||
| Engelbrecht et al., 2002 | 1,640 | 900 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | |||||||||
| Helenius et al., 2002 | 710 | Siemens 1.5T, 2 × 2 × 5 mm, b = 1,000 | ||||||||||
| Kwan et al., 2015 | 1,494 | 1,379 | Phillips 3T, 1.8 × 1.8 × 1.8 mm, b = 750 | |||||||||
The unit of ADC values is µm2/s. L is left and R is right. Q1, Q2, Q3 and Q4 are quarters in the first year and Y1, Y2, …, Y6 are years, as defined in Table 1. The unit for b values in the far right column is s/mm2. Bold values represent our data.
Quantifying growth of mean geometries
We used the 4D‐DRAMMS deformable registration (https://www.nitrc.org/projects/dramms4d, validated in [Ou et al., 2014, 2015b]) to align all constructed ADC atlases. Then, we used tensor‐based morphometry (TBM) to capture how the mean brain geometry changed in space and time. Specifically, we calculated the Jacobian Determinant maps from the series of deformations obtained in the 4D registration for TBM. The non‐negative Jacobian determinant value at a voxel specified fractional volume (volume preservation if = 1, expansion if >1, or shrinkage if <1) needed to transform a voxel from the baseline atlas into the ADC atlas of older ages.
RESULTS
Whole Brain Exploration of Age‐Specific Atlases from Clinically Acquired Images
Figure 3 shows the constructed age‐specific ADC atlases. Figure 4 shows the change of whole‐brain volume and ADC values derived from atlases. As expected, ADC values decline and brain volume expands from birth to 6 years of age. The changes are most rapid in the first 2 years of life and then slow down, reaching a plateau by 5–6 years of age, which is consistent with previous studies [Deoni et al., 2011; Watanabe et al., 2013].
Figure 3.
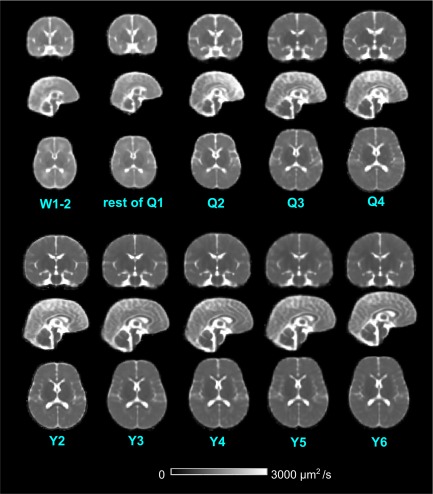
Representative coronal, sagittal and axial views of the constructed age‐specific ADC atlases. [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 4.
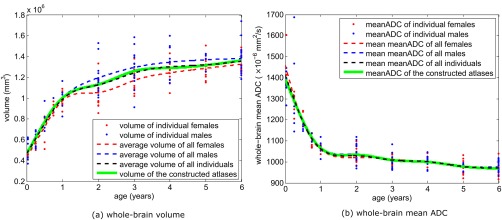
The whole‐brain volume and mean ADC values in individuals and in the constructed atlases. [Color figure can be viewed at http://wileyonlinelibrary.com]
The mean brain volume at birth is around 500 ml and reaches a plateau after 6 years of age, at around 1,300 ml, consistent with existing reports (Table 3). On average, males have bigger brains than the females, but these differences are not always statistically significant. Looking at Tables 2 and 3 together, the differences of male‐versus‐female brain volumes are insignificant (P > 0.05) in those age groups having less than 15 total subjects (the first 5 age groups), and are significant in 4 out of the remaining 5 age groups having more than 15 total subjects. This underscores the need for a replication study with larger cohorts if we want to draw more conclusive observations on gender differences (see Discussion for future work).
Table 3.
Comparison of the whole‐brain volume and the whole‐brain mean ADC values in our atlases and published findings referring to individual subjects
| W1‐2 | Rest Q1 | Q2 | Q3 | Q4 | Y2 | Y3 | Y4 | Y5 | Y6 | Adult (20–40 yo) | Imaging protocol | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole‐brain volume | ||||||||||||
| Male individuals | 506 ± 96 | 609 ± 71 | 799 ± 130 | 922 ± 72 | 1,003 ± 99 | 1,192 ± 143 | 1,297 ± 179 | 1,347 ± 145 | 1,370 ± 62 | 1,383 ± 149 | Siemens 3T, 23 mm | |
| Female individuals | 460 ± 22 | 575 ± 61 | 681 ± 161 | 908 ± 52 | 984 ± 93 | 1,040 ± 123 | 1,176 ± 110 | 1,244 ± 106 | 1,288 ± 124 | 1,328 ± 90 | ||
| P values (male = female) | 0.23 | 0.22 | 0.15 | 0.37 | 0.37 | 0.002 | 0.029 | 0.048 | 0.025 | 0.12 | ||
| Constructed atlases | 496 | 599 | 767 | 929 | 1,006 | 1,120 | 1,267 | 1,289 | 1,317 | 1,364 | ||
| P values (population mean = atlas mean) | 0.52 | 0.52 | 0.70 | 0.79 | 0.65 | 0.52 | 0.75 | 0.33 | 0.44 | 0.62 | ||
| Huang et al., 2006 | 367 ± 18 | 1,258 ± 86 | Phillips 1.5T, 1.83 mm | |||||||||
| Knickmeyer et al., 2008 | 425 | 855 | 983 | Siemens 3T, 13 mm (T1) | ||||||||
| Gilmore et al., 2007 (males) | 525 | Siemens 3T, 13 mm (T1) | ||||||||||
| Gilmore et al., 2007 (females) | 487 | Siemens 3T, 13 mm (T1) | ||||||||||
| Watanabe et al., 2013 (males) | ∼520 | ∼1,350 | ∼1,400 | Phillips 1.5T, 2 × 3 × 5 mm | ||||||||
| Watanabe et al., 2013 (females) | 500 | ∼1,200 | ∼1,200 | Phillips 1.5T, 2 × 3 × 5 mm | ||||||||
| Whole‐brain mean ADC | ||||||||||||
| Male individuals | 1,377 ± 50 | 1,319 ± 177 | 1,163 ± 14 | 1,109 ± 21 | 1,067 ± 29 | 1,043 ± 42 | 1,003 ± 31 | 1,003 ± 26 | 978 ± 18 | 975 ± 26 | Siemens 3T, 23 mm | |
| Female individuals | 1,467 ± 118 | 1,274 ± 58 | 1,204 ± 42 | 1,085 ± 29 | 1,069 ± 18 | 1,025 ± 35 | 1,011 ± 17 | 1,001 ± 23 | 976 ± 35 | 967 ± 36 | ||
| P values (male = female) | 0.06 | 0.32 | 0.06 | 0.14 | 0.45 | 0.10 | 0.22 | 0.41 | 0.45 | 0.22 | ||
| Constructed atlases | 1,385 | 1,290 | 1,202 | 1,096 | 1,068 | 1,039 | 1,006 | 1,001 | 974 | 968 | ||
| P values (population mean = atlas mean) | 0.25 | 0.37 | 0.92 | 0.55 | 0.51 | 0.75 | 0.49 | 0.40 | 0.50 | 0.29 | ||
| Watanabe et al | ∼1,220 | ∼860 | ∼760 | Phillips 1.5T, 2 × 3 × 5 mm | ||||||||
Bold values indicate our result. The unit for b values is s/mm2. In the far right column, the imaging modality is diffusion imaging by default; modalities that are not diffusion imaging are noted in the bracket [e.g., (T1) means T1‐weighted MRI]. For diffusion imaging, b = 1,000 s/mm2 unless otherwise noted. Gray bars are results in our cohort (comparing males versus females, individuals versus atlases). P values indicating statistical significant differences (P < 0.05) are in italics.
As expected, the brain volumes observed in the atlases are statistically equivalent to those observed in individuals in corresponding age groups (P > 0.05 in all ages, Table 3). This visualizes the mathematical theory that the unbiased construction of atlases do accurately represent the mean whole brain volume in each age group [Fonov et al., 2011].
Males and females in all age groups of our cohort had equivalent whole‐brain mean ADC values (Fig. 4b and Table 3). Moreover, the mean ADC value measures in the atlases are statistically equivalent to that measured from individuals in matched age groups. This ADC result (Fig. 4b), and the results on brain volume (Fig. 4a), both support our hypothesis that the constructed atlas of an age group does faithfully represent the mean of the individuals in the same age group.
Also, the quantitative metrics from our atlases are comparable (given expected variation due to differences in scanners, fields of strength and image resolutions) to reports from individual subjects in published studies [Gilmore et al., 2007; Huang et al., 2006; Knickmeyer et al., 2008; Watanabe et al., 2013].
ROI‐Based Exploration of Age‐Specific Atlases from Clinically Acquired Images
We evaluated ADC values in representative white and gray matter ROIs (Fig. 5). ADC values in white matter at birth are high (1,100–1,500 µm2/s), decrease rapidly in the first two years and reach a relative plateau after 3 years of age (700–900 µm2/s). The mean ADC values in our atlases after 3 years of age are in the same range as those for healthy adults [Helenius et al., 2002; Naganawa et al., 2003; Zhai et al., 2003]. Although we find no evidence of left–right hemispheric differences, our atlases demonstrate the previously reported developmental shift in anterior‐posterior asymmetry. Anterior regions tend to have 100 µm2/s higher ADC values than posterior regions in the first year, but after one year these differences are no longer detected. Finally, the corpus callosum has higher mean ADC values than other ROIs at each stage (except anterior white matter) but this difference diminishes over time, being 200–400 µm2/s higher at birth and only 50–200 µm2/s higher by 3 years of age.
Figure 5.
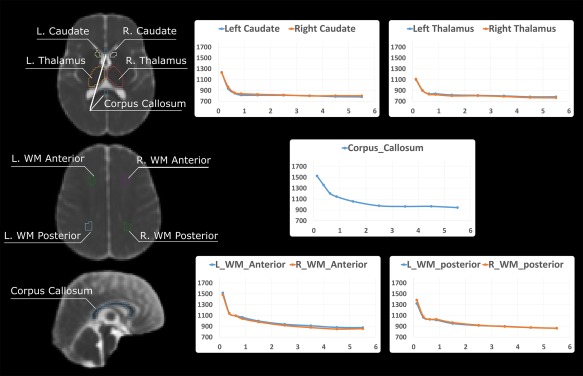
Manual annotations of ROIs in a representative atlas (the Y2 age group), and the change of mean ADC values in these atlas‐based ROIs plotted with respect to age. The ROI annotations were done in 3D in all age‐specific atlases, but only representative 2D planes in the Y2 atlas are displayed here. In all plots, the x axis is age in years and the y axis is the mean ADC value in the ROI (unit: µm2/s). [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 4 further summarizes our findings in the context of existing literature. Prior reports have provided mean ADCs from a sparse set of ROIs in one or in a coarsely sampled set of ages. Each report was based on ROIs drawn on individual subject ADC maps and then averaged together [Engelbrecht et al., 2002; Engelter et al., 2000; Helenius et al., 2002; Morriss et al., 1999; Naganawa et al., 2003; Provenzale et al., 2007; Sasaki et al., 2008]. In contrast, we show results from a sample of representative ROIs placed in each of our 10 atlases from ages 0 to 6 years of age. Our results are similar to published values. This suggests that: (a) regional analysis of one atlas can generate results similar to regional analysis of images from mulitple individuals; (b) with careful review, images acquired clinically produce similar results compared to prospectively gathered normal volunteer cohorts.
Spatiotemporal Patterns of Brain Development
Another application of our atlases is to quantify how the average brain geometry evolves with age. Figure 6 shows the relative volume of each atlas compared to the W1–2 atlas at each voxel. All brain regions underwent their fastest expansion in the first year. The expansion gradually slowed between year 1 and 3, reaching a plateau after year 4. Brain development was also spatially heterogeneous: the frontal lobe and medial temporal areas expanded faster than other regions in agreement with reported findings derived from structural MR modalities [Fonov et al., 2011; Sanchez et al., 2012a; Serag et al., 2012].
Figure 6.
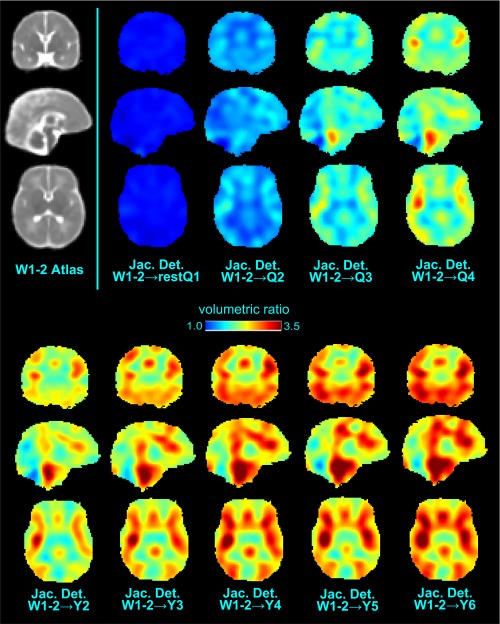
Volumetric changes of age‐specific atlases relative to the W1‐2 atlas. “Jac. Det.” is short for Jacobian Determinant, which measures the volumetric ratio at each voxel. [Color figure can be viewed at http://wileyonlinelibrary.com]
DISCUSSION
Summary and Contributions
In this study, we mined the EHR and PACS to extract a large number of clinically generated ADC maps in normative subjects aged 0 to 6 years of life. We created the largest set of age‐specific normative ADC atlases in this age range, for the first time densely sampling the first two years of age (see Table 1 for comparison with existing atlases). We showed that the resulting age specific ADC atlases have ADC values and volumetric changes that are globally and regionally comparable to those from previously reported studies in prospectively gathered images. In addition, we reported ADC changes not only in a few regions where previous reports exist, but for the first time at every voxel in the brain. Finally, we made these densely sampled quantitative ADC atlases publicly available.
We chose to create quantitative ADC atlases, because of their practical use in clinical diagnoses. In clinical practice, a trainee often learns what is normal by viewing clinical images that have been visually inspected and deemed normal by an experienced neuroradiologist. Experienced neuroradiologists learn what is normal by obtaining follow‐up on clinically acquired cases. Here, we leverage the EHR and clinical PACS to perform these tasks systematically and objectively across a large cohort in greater detail to begin to define age‐specific normative reference values. By making our atlases publicly available we are providing others with the opportunity to understand normal ADC changes during early‐life development. Our future studies will explore the ability of our atlases to decrease variability in clinical reads of ADC maps by providing neuroradiologists a normal reference. In addition, we will explore the ability of our atlases to guide clinical interpretation by detecting statistically significant voxel‐wise ADC changes in a reference study.
Strengths and Significance
This study was possible given recent technical innovations in clinical informatics tools and in clinical pediatric image analysis. First, the ability to use informatics tools to easily access and mine the clinical PACS for clinically acquired images for secondary research use is a recent advance [Murphy et al., 2015]. It allows us to use EHR‐based informatics tools to limit the number of potential cases needing individual vetting by an investigator. Here, started by identifying 2,871 potentially normative cases from the tens of thousands of scans of 0‐ to 6‐year old in our PACS, used our software tools to eliminate all of 383 which needed to be reviewed by an expert to yield our final cohort of 201 ADC maps. In addition, atlas construction was based on exploiting recent advances in image analysis pipelines, which could accurately and consistently skull strip [Doshi et al., 2013; Ou et al., 2015a] and register [Ou et al., 2011, 2014] ADC maps, even though images were from heterogeneous early‐life populations and even though images were clinically acquired.
The resulting atlases (Fig. 3) are with no significant susceptibility distortions that are common in adult diffusion MRI images. This is primarily due to the lack of pneumatization of the frontal sinuses before 5 years of age [Adibelli et al., 2011] and less prominent mastoid air cells compared to adult populations [Cinamon, 2009]. In addition, multiple experts visually reviewed the ADC maps and excluded those with severe motion artifacts and susceptibility distortions; and the atlas construction process was able to compute an average geometry/anatomy, which cancels off any remaining nonsystematic artifacts or distortions from individual ADC maps.
The constructed atlases have multiple potential uses:
In radiology reading rooms, our normative ADC atlases can provide valuable quantitative references to assist radiologists in abnormality detection. Our recent preliminary results showed that utilizing our atlases helped reduce inter‐rater variability of radiology reads [Jaims et al., 2015], however, a larger‐scale study is needed to confirm these initial findings.
In radiology classrooms, tutorials can be developed around these atlases for students and radiology residents to help them learn the normative appearance of ADC maps. We believe that our detailed 3D+time atlases will be able to help them explore regional and voxel‐wise normative ADC variations instead of relying on single slice (2D) qualitative exemplar images that are traditionally used.
In computer‐aided detection, our atlases have the potential to provide a means to detect a range of abnormalities as outliers to normal ADC variations. Our preliminary results showed that voxel‐wise comparison to atlases allowed us to automatically identify brain regions with neonatal hypoxic ischemic injury [Ou et al., 2015c], where our accuracy is comparable to that of two independent expert radiologists [Ou et al., 2017]. Again, larger‐scale studies are needed to confirm these initial results.
In neuroscience and brain development studies, our atlases can be used to quantify how water diffusion changes in space and time (see Fig. 6). Additionally, when quantifying regional ADC values (e.g., [Neil et al., 1998; Provenzale et al., 2007; Watanabe et al., 2013; Zhai et al., 2003]), our age‐specific atlases can now be used to annotate the ROIs instead of needing to annotate individual ADC maps, saving both time and effort yet arriving at comparable results (Tables 3 and 4);
In multimodal, longitudinal and/or population imaging studies, our age‐specific ADC atlases offer an additional modality to compare to structural or/and functional acquisitions. They also provide a standard space for spatially normalizing individual ADC atlases in voxel‐based morphometry (VBM) or lesion‐symptom mapping (LSM) studies.
Limitations and Future Work
This study has limitations. Our comparison to published ADC values and growth curves are qualitative and not statistical because prior data is limited (Tables 3 and 4). In addition, for regional analysis we were only able to compare our results to the small set of regions for which results have been previously reported. Also, there were differences in scanners and in imaging sequences between our study and existing studies making exact comparisons difficult. Because of these factors, we could only report that our clinical data and existing data led to similar ADC values but could not carry out a detailed voxel‐wise statistical comparison.
Another potential limitation is the sample size we had available for each template. This is a common issue in other studies, where for example fewer than 10 subjects are in some age groups in [Sadeghi et al., 2014; Sanchez et al., 2012b]. Unfortunately, there is a lack of rigorous studies to suggest an adequate sample size required to capture the true normal variations in a specific population. Nevertheless, our atlases constructed in age groups with fewer subjects (second and third quarter of the first year) fit the longitudinal trend of global and local ADC and volume changes (Tables 3 and 4, Figs. 3, 4, 5, 6). We are continuing to curate our EHR data to identify additional ADC maps from normative subjects and are exploring collaborations with other sites. Larger sample size in future studies may generate atlases with even greater specificity including the ability to stratify by gender and race.
Because all images were from a single Siemens 3T Trio scanner, there is no cross‐scanner ADC variability. However, future studies will demand larger sample sizes and participation of multiple institutions, introducing small cross‐scanner ADC variability if the same vendor [Van Leemput, 2009], or larger cross‐scanner ADC variability if different vendors [Walker et al., 2016; Wu et al., 2011]. Our ultimate goal is to provide atlases to clinicians and researchers irrespective of scanner brand. To achieve this, future prospective studies will include strategies for normalizing diffusion parameter maps across scanners using, for instance, phantoms [Doshi et al., 2016; Iglesias and Sabuncu, 2015] or ideally volunteers as is currently being done in many clinical studies. Fortunately, our analysis pipeline (brain extraction, atlas construction) has been shown to sustain high accuracy and consistency in clinical images from diverse sources [Ou et al., 2015a; Serag et al., 2012].
Due to the rapid ADC changes in the first year of life (Fig. 5), we constructed atlases using incrementally larger age spans beginning with 2‐week spans during the first month and then 3‐month age spans. After one year, the rate of ADC change decreases (Fig. 5) and 12‐month age spans were used. We note that, in all atlases, the standard deviations of ADC values may come from both inter‐individual differences and age‐based ADC variations. This is a limitation of our study. Separating the two is challenging and requires larger data sets. As more data sets become available, each age group can be subdivided more finely to decrease these age‐based variations. For example, we will further divide the Week 0–1 age bin into half‐week intervals, Year 1–2 age bin into quarters or half‐year intervals, and age bins older than year 2 into half‐year intervals. With more subjects, we will also be able to create future sex specific atlases and eventually explore variations with different ethnic groups. The goal is to characterize inter‐individual variation that is distinct from age and sex‐based ADC variations. By doing so, we will better capture subtle and regional ADC development thus increasing the sensitivity of our atlases to identify early‐life and subtle ADC abnormalities.
Moreover, it is crucial to point out that this study is based on cross‐sectional not longitudinal images. Therefore, we cannot determine if age‐dependent regional variations are due to individual differences or due to regional differences in rates of development. To attain this specific goal, research funding will be required as normative clinical subjects are highly unlikely to have longitudinal brain scanned from birth to 6 years of age that would be required [Walker et al., 2016]. In our study, we exploit the fact that MRI may be obtained in young children as an extra step to ensure there is no abnormality when clinical evaluation is equivocal. However, when MRI is normal and follow‐up is normal, additional follow‐up clinical imaging is unlikely to be obtained.
Finally, future efforts will also focus on further improving the atlas quality by additional innovative image analysis techniques. For example, our atlases (e.g., Fig. 3) are blurrier than individual ADC map due to the nature of atlas construction and averaging. More sophisticated atlas construction frameworks (e.g., [Shi et al., 2014; Van Leemput, 2009; Wu et al., 2011]) may lead to sharper edges and higher contrast. Future efforts will also include automatic segmentation of ADC maps into regions of interest [Doshi et al., 2016; Iglesias and Sabuncu, 2015], instead of using the manual annotations used in this article. In addition, when subject sizes are larger we will be able to explore choosing atlas age groups in a more systematic manner [Davis et al., 2007; Serag et al., 2012], and constructing atlases by taking a series of age groups into joint consideration [Davis et al., 2007; Serag et al., 2012]. For instance, the 13 normative ADC maps for week 0‐1 were all from full term neonates without consideration of gender. These neonates have different gestational ages (37–40 weeks) and are a mix of male and female neonates, introducing potential additional variations in the ADC values. Given a larger sample size, we could stratify into subgroups by gender and gestational as well as chronological age to generate more finely tuned atlases.
Conclusion
In conclusion, we have demonstrated that by careful review and selection, the EHR and the clinical PACS can be mined to create a densely sampled normative database. In addition, we showed that robust image analysis techniques can be used to create 10 age specific ADC atlases that quantitatively characterize the mean and standard deviation of ADC values and volumetric change at each voxel over the first 6 years of life. These normative atlases are now publicly available at https://www.nitrc.org/projects/mgh_adcatlases.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Rudolph Pienaar for his assistance in the organization of the medical images, to Nadia Berard and Katie Murphy for their assistance in the review of clinical charts, and to Nathaniel Reynolds for his assistance in the clinical and image data mining.
REFERENCES
- Adibelli ZH, Songu M, Adibelli H (2011): Paranasal sinus development in children: A magnetic resonance imaging analysis. Am J Rhinol. Allergy 25:30–35. [DOI] [PubMed] [Google Scholar]
- Akiyama LF, Richards TR, Imada T, Dager SR, Wroblewski L, Kuhl PK (2013): Age‐specific average head template for typically developing 6‐month‐old infants. PloS One 8:e73821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli CR, Rivkin MJ, McKinstry RC (2007): The NIH MRI study of normal brain development (Objective‐2): Newborns, infants, toddlers, and preschoolers. Neuroimage 35:308–325. [DOI] [PubMed] [Google Scholar]
- Altaye M, Holland SK, Wilke M, Gaser C (2008): Infant brain probability templates for MRI segmentation and normalization. Neuroimage 43:721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Maga AM (2015): A novel procedure for rapid imaging of adult mouse brains with microCT using iodine‐based contrast. PLoS One 10:e0142974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C (2002): The basis of anisotropic water diffusion in the nervous system–a technical review. NMR Biomed 15:435–455. [DOI] [PubMed] [Google Scholar]
- Cinamon U (2009): The growth rate and size of the mastoid air cell system and mastoid bone: A review and reference. Eur Arch Otorhinolaryngol 266:781–786. [DOI] [PubMed] [Google Scholar]
- Coakley FV, Liberman L, Panicek DM (2003): Style guidelines for radiology reporting: A manner of speaking. Am J Roentgenol 180:327–328. [DOI] [PubMed] [Google Scholar]
- Counsell SJ, Allsop JM, Harrison MC, Larkman DJ, Kennea NL, Kapellou O, Cowan FM, Hajnal JV, Edwards AD, Rutherford MA (2003): Diffusion‐weighted imaging of the brain in preterm infants with focal and diffuse white matter abnormality. Pediatrics 112:1–7. [DOI] [PubMed] [Google Scholar]
- Davis BC, Fletcher PT, Bullitt E, Joshi S (2007): Population shape regression from random design data. In: IEEE 11th International Conference on Computer Vision, 2007, ICCV 2007. pp 1–7.
- Deoni SC, Mercure E, Blasi A, Gasston D, Thomson A, Johnson M, Williams SC, Murphy DG (2011): Mapping infant brain myelination with magnetic resonance imaging. J Neurosci 31:784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C (2013): Multi‐atlas skull‐stripping. Acad Radiol 20:1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Resnick SM, Gur RC, Gur RE, Satterthwaite TD, Furth S, Davatzikos C, and Alzheimer's Neuroimaging Initiative. (2016): MUSE: MUlti‐atlas region segmentation utilizing ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 127:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht V, Scherer A, Rassek M, Witsack HJ, Mödder U (2002): Diffusion‐weighted MR imaging in the brain in children: Findings in the normal brain and in the brain with white matter diseases. Radiology 222:410–418. [DOI] [PubMed] [Google Scholar]
- Engelter ST, Provenzale JM, Petrella JR, DeLong DM, MacFall JR (2000): The effect of aging on the apparent diffusion coefficient of normal‐appearing white matter. Am J Roentgenol 175:425–430. [DOI] [PubMed] [Google Scholar]
- Erus G, Battapady H, Satterthwaite TD, Hakonarson H, Gur RE, Davatzikos C, Gur RC (2014): Imaging patterns of brain development and their relationship to cognition. Cereb. Cortex bht425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL (2011): Unbiased average age‐appropriate atlases for pediatric studies. Neuroimage 54:313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gano D, Chau V, Poskitt KJ, Hill A, Roland E, Brant R, Chalmers M, Miller SP (2013): Evolution of pattern of injury and quantitative MRI on days 1 and 3 in term newborns with hypoxic‐ischemic encephalopathy. Pediatr Res 74:82–87. [DOI] [PubMed] [Google Scholar]
- Geng X, Gouttard S, Sharma A, Gu H, Styner M, Lin W, Gerig G, Gilmore JH (2012): Quantitative tract‐based white matter development from birth to age 2 years. Neuroimage 61:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Lin W, Prastawa MW, Looney CB, Vetsa YSK, Knickmeyer RC, Evans DD, Smith JK, Hamer RM, Lieberman JA, Gerig G (2007): Regional gray matter growth, sexual dimorphism, and cerebral asymmetry in the neonatal brain. J Neurosci 27:1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goergen SK, Ang H, Wong F, Carse EA, Charlton M, Evans R, Whiteley G, Clark J, Shipp D, Jolley D, Paul E (2014): Early MRI in term infants with perinatal hypoxic–ischaemic brain injury: Interobserver agreement and MRI predictors of outcome at 2 years. Clin Radiol 69:72–81. [DOI] [PubMed] [Google Scholar]
- Guimond A, Meunier J, Thirion J‐P (2000): Average brain models: A convergence study. Comput Vis Image Underst 77:192–210. [Google Scholar]
- He L, Parikh NA (2013): Atlas‐guided quantification of white matter signal abnormalities on term‐equivalent age MRI in very preterm infants: Findings predict language and cognitive development at two years of age. PLoS One 8:e85475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J, Soinne L, Perkiö J, Salonen O, Kangasmäki A, Kaste M, Carano RA, Aronen HJ, Tatlisumak T (2002): Diffusion‐weighted MR imaging in normal human brains in various age groups. Am J Neuroradiol 23:194–199. [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Wakana S, Zhang W, Ren T, Richards LJ, Yarowsky P, Donohue P, Graham E, van Zijl PC, Mori S (2006): White and gray matter development in human fetal, newborn and pediatric brains. Neuroimage 33:27–38. [DOI] [PubMed] [Google Scholar]
- Iglesias JE, Sabuncu MR (2015): Multi‐atlas segmentation of biomedical images: A survey. Med Image Anal 24:205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M, Parker D, Ghanbari Y, Smith A, Hua K, Mori S, Abel T, Davatzikos C, Verma R (2015): Connectome and maturation profiles of the developing mouse brain using diffusion tensor imaging. Cereb. Cortex 25:2696–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaims C, Ou Y, Shih J, Bates S, O'Reilly D, Soul J, Gollub R, Grant PE, Zöllei L (2015): Apparent diffusion coefficient Z‐score maps compared to normative atlas in hypoxic ischemic encephalopathy. In: ASNR 53rd Annual Meeting & The Foundation of the ASNR Symposium 2015. Chicago, IL.
- Kazemi K, Moghaddam HA, Grebe R, Gondry‐Jouet C, Wallois F (2007): A neonatal atlas template for spatial normalization of whole‐brain magnetic resonance images of newborns: Preliminary results. Neuroimage 37:463–473. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH (2009): Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46:786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knickmeyer RC, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, Hamer RM, Lin W, Gerig G, Gilmore JH (2008): A structural MRI study of human brain development from birth to 2 years. J Neurosci 28:12176–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklisova‐Murgasova M, Aljabar P, Srinivasan L, Counsell SJ, Doria V, Serag A, Gousias IS, Boardman JP, Rutherford MA, Edwards AD, Hajnal JV (2011): A dynamic 4D probabilistic atlas of the developing brain. Neuroimage 54:2750–2763. [DOI] [PubMed] [Google Scholar]
- Kwan S, Boudes E, Benseler A, Gilbert G, Saint‐Martin C, Shevell M, Wintermark P (2015): Evolution of apparent diffusion coefficient and fractional anisotropy in the cerebrum of asphyxiated newborns treated with hypothermia over the first month of life. Neural Plast 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan D, Mangin JF, Poupon C, Clark CA, Pappata S, Molko N, Chabriat H (2001): Diffusion tensor imaging: Concepts and applications. J Magn Reson Imaging 13:534–546. [DOI] [PubMed] [Google Scholar]
- Liauw L, van Wezel‐Meijler G, Veen S, van Buchem MA, van der Grond J (2008): Do apparent diffusion coefficient measurements predict outcome in children with neonatal hypoxic‐ischemic encephalopathy? Am J Neuroradiol 30:264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love SA, Marie D, Roth M, Lacoste R, Nazarian B, Bertello A, Coulon O, Anton JL, Meguerditchian A (2016): The average baboon brain: MRI templates and tissue probability maps from 89 individuals. Neuroimage 132:526–533. [DOI] [PubMed] [Google Scholar]
- Luo Y, Shi L, Weng J, He H, Chu WC, Chen F, Wang D (2014): Intensity and sulci landmark combined brain atlas construction for Chinese pediatric population: Intensity and sulci landmark combined brain atlas construction. Hum Brain Mapp 35:3880–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Oishi K, Faria AV, Miller MI (2013): Atlas‐based neuroinformatics via MRI: Harnessing information from past clinical cases and quantitative image analysis for patient care. Annu Rev Biomed Eng 15:71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EB, Phillips NS, Laningham FH, Patay Z, Gajjar A, Wallace D, Boop F, Sanford R, Ness KK, Ogg RJ (2009): Proximal dentatothalamocortical tract involvement in posterior fossa syndrome. Brain 132:3087–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriss MC, Zimmerman RA, Bilaniuk LT, Hunter JV, Haselgrove JC (1999): Changes in brain water diffusion during childhood. Neuroradiology 41:929–934. [DOI] [PubMed] [Google Scholar]
- Murphy SN, Herrick C, Wang Y, Wang TD, Sack D, Andriole KP, Wei J, Reynolds N, Plesniak W, Rosen BR, Pieper S (2015): High throughput tools to access images from clinical archives for research. J Digit Imaging 28:194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganawa S, Sato K, Katagiri K, Mimura T, Ishigaki T (2003): Regional ADC values of the normal brain: Differences due to age, gender, and laterality. Eur Radiol 13:6–11. [DOI] [PubMed] [Google Scholar]
- Neil JJ, Shiran SI, McKinstry RC, Schefft GL, Snyder AZ, Almli CR, Akbudak E, Aronovitz JA, Miller JP, Lee BC, Conturo TE (1998): Normal brain in human newborns: Apparent diffusion coefficient and diffusion anisotropy measured by using diffusion tensor MR imaging. Radiology 209:57–66. [DOI] [PubMed] [Google Scholar]
- Nossin‐Manor R, Card D, Morris D, Noormohamed S, Shroff MM, Whyte HE, Taylor MJ, Sled JG (2013): Quantitative MRI in the very preterm brain: Assessing tissue organization and myelination using magnetization transfer, diffusion tensor and T1 imaging. Neuroimage 64:505–516. [DOI] [PubMed] [Google Scholar]
- Oishi K, Mori S, Donohue PK, Ernst T, Anderson L, Buchthal S, Faria A, Jiang H, Li X, Miller MI, van Zijl PC (2011): Multi‐contrast human neonatal brain atlas: Application to normal neonate development analysis. Neuroimage 56:8–20. May [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Sotiras A, Paragios N, Davatzikos C (2011): DRAMMS: Deformable registration via attribute matching and mutual‐saliency weighting. Med. Image Anal 15:622–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Akbari H, Bilello M, Da X, Davatzikos C (2014): Comparative evaluation of registration algorithms in different brain databases with varying difficulty: Results and insights. IEEE Trans Med Imaging 33:2039–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Gollub RL, Retzepi K, Reynolds N, Pienaar R, Pieper S, Murphy SN, Grant PE, Zöllei L (2015a): Brain extraction in pediatric ADC maps, toward characterizing neuro‐development in multi‐platform and multi‐institution clinical images. Neuroimage 122:246–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Weinstein SP, Conant EF, Englander S, Da X, Gaonkar B, Hsieh MK, Rosen M, DeMichele A, Davatzikos C, Kontos D (2015b): Deformable registration for quantifying longitudinal tumor changes during neoadjuvant chemotherapy. Magn Reson Med 73:2343–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Jaims C, Gollub RL, Retzepis K, Bates S, Murphy S, Grant PE, Zöllei L (2015c): Neonatal Brain Injury Detection in MRI: An Atlas‐based Fully‐Automatic Approach. In: Organization of Human Brain Mapping (OHBM) Annual Meeting. Honolulu, HI.
- Ou Y, Gollub R, Wang J, Fan Q, Bates SV, Chou J, Weiss R, Retzepis K, Pieper S, Jaimes C, Murphy SN, Zöllei L, Grant PE (2017): MRI Detection of Neonatal Hypoxic Ischemic Encephalopathy: Machine v.s. Radiologists. In: Organization for Human Brain Mapping (OHBM). Vancouver, BC, Canada.
- Ozturk A, Sasson AD, Farrell JAD, Landman BA, da Motta ACBS, Aralasmak A, Yousem DM (2008): Regional differences in diffusion tensor imaging measurements: Assessment of intrarater and interrater variability. Am J Neuroradiol 29:1124–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhani AR, Liu G, Mu‐Koh D, Chenevert TL, Thoeny HC, Takahara T, Dzik‐Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA (2009): Diffusion‐weighted magnetic resonance imaging as a cancer biomarker: Consensus and recommendations. Neoplasia 11:102–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M, Gilmore JH, Lin W, Gerig G (2005): Automatic segmentation of MR images of the developing newborn brain. Med Image Anal 9:457–466. [DOI] [PubMed] [Google Scholar]
- Provenzale JM, Liang L, DeLong D, White LE (2007): Diffusion tensor imaging assessment of brain white matter maturation during the first postnatal year. Am J Roentgenol 189:476–486. [DOI] [PubMed] [Google Scholar]
- Provenzale JM, Isaacson J, Chen S, Stinnett S, Liu C (2010): Correlation of apparent diffusion coefficient and fractional anisotropy values in the developing infant brain. Am J Roentgenol 195:W456–W462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi N, Prastawa M, Fletcher PT, Wolff J, Gilmore JH, Gerig G (2013): Regional characterization of longitudinal DT‐MRI to study white matter maturation of the early developing brain. Neuroimage 68:236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CE, Richards JE, Almli CR (2012a): Age‐specific MRI templates for pediatric neuroimaging. Dev Neuropsychol 37:379–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CE, Richards JE, Almli CR (2012b): Neurodevelopmental MRI brain templates for children from 2 weeks to 4 years of age. Dev Psychobiol 54:77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Yamada K, Watanabe Y, Matsui M, Ida M, Fujiwara S, Shibata E (2008): Variability in absolute apparent diffusion coefficient values across different platforms may be substantial: A multivendor, multi‐institutional comparison study. Radiology 249:624–630. [DOI] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Ruparel K, Loughead J, Prabhakaran K, Calkins ME, Hopson R, Jackson C, Keefe J, Riley M, Mentch FD (2014): "Neuroimaging of the Philadelphia neurodevelopmental cohort. Neuroimage 86:544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Calkins ME, Vandekar SN, Erus G, Ruparel K, Roalf DR, Linn KA, Elliott MA, Moore TM, Hakonarson H (2016): Structural brain abnormalities in youth with psychosis spectrum symptoms. JAMA Psychiatry 73:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MM, Berman JI, Baumer FM, Glass HC, Jeng S, Jeremy RJ, Esch M, Biran V, Barkovich AJ, Studholme C, Xu D (2009): Normative apparent diffusion coefficient values in the developing fetal brain. Am J Neuroradiol 30:1799–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener RN (2001): Diffusion MRI: Apparent diffusion coefficient (ADC) values in the normal brain and a classification of brain disorders based on ADC values. Comput Med Imaging Graph 25:299–326. [DOI] [PubMed] [Google Scholar]
- Serag A, Aljabar P, Ball G, Counsell SJ, Boardman JP, Rutherford MA, Edwards AD, Hajnal JV, Rueckert D (2012): Construction of a consistent high‐definition spatio‐temporal atlas of the developing brain using adaptive kernel regression. Neuroimage 59:2255–2265. [DOI] [PubMed] [Google Scholar]
- Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D (2011): Infant brain atlases from neonates to 1‐and 2‐year‐olds. PLoS One 6:e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Wang L, Wu G, Li G, Gilmore JH, Lin W, Shen D (2014): Neonatal atlas construction using sparse representation: Neonatal Atlas Construction. Hum Brain Mapp 35:4663–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiras A, Davatzikos C, Paragios N (2013): Deformable medical image registration: A survey. IEEE Trans Med Imaging 32:1153–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K (2009): Encoding probabilistic brain atlases using Bayesian inference. IEEE Trans Med Imaging 28:822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L, Chang LC, Nayak A, Irfanoglu MO, Botteron KN, McCracken J, McKinstry RC, Rivkin MJ, Wang DJ, Rumsey J, Pierpaoli C (2016): The diffusion tensor imaging (DTI) component of the NIH MRI study of normal brain development (PedsDTI). Neuroimage 124:1125–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Sakai O, Ozonoff A, Kussman S, Jara H (2013): Age‐related apparent diffusion coefficient changes in the normal brain. Radiology 266:575–582. [DOI] [PubMed] [Google Scholar]
- Weisenfeld NI, Warfield SK (2009): Automatic segmentation of newborn brain MRI. Neuroimage 47:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Mewes AU, Warfield SK (2006a): Highly accurate segmentation of brain tissue and subcortical gray matter from newborn MRI In: Medical Image Computing and Computer‐Assisted Intervention–MICCAI 2006. Springer; pp 199–206. [DOI] [PubMed] [Google Scholar]
- Weisenfeld NI, Mewes AUJ, Warfield SK (2006b): Segmentation of newborn brain MRI. In: 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro. pp 766–769.
- Wilke M, Holland SK, Altaye M, Gaser C (2008): Template‐O‐Matic: A toolbox for creating customized pediatric templates. Neuroimage 41:903–913. [DOI] [PubMed] [Google Scholar]
- Wolf RL, Zimmerman RA, Clancy R, Haselgrove JH (2001): Quantitative apparent diffusion coefficient measurements in term neonates for early detection of hypoxic‐ischemic brain injury: Initial experience 1. Radiology 218:825–833. [DOI] [PubMed] [Google Scholar]
- Wu G, Jia H, Wang Q, Shen D (2011): SharpMean: Groupwise registration guided by sharp mean image and tree‐based registration. Neuroimage 56:1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Richards JE, Lei D, Zhu H, Lee K, Gong Q (2015): The construction of MRI brain/head templates for Chinese children from 7 to 16 years of age. Dev Cogn Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Srinivasan L, Jiang S, Rutherford M, Edwards AD, Rueckert D, Hajnal JV (2007): Automatic segmentation and reconstruction of the cortex from neonatal MRI. Neuroimage 38:461–477. [DOI] [PubMed] [Google Scholar]
- Young R, Maga AM (2015): Performance of single and multi‐atlas based automated landmarking methods compared to expert annotations in volumetric microCT datasets of mouse mandibles. Front Zool 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai G, Lin W, Wilber KP, Gerig G, Gilmore JH (2003): Comparisons of regional white matter diffusion in healthy neonates and adults performed with a 3.0‐t head‐only MR imaging unit1. Radiology 229:673–681. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chang L, Ceritoglu C, Skranes J, Ernst T, Mori S, Miller MI, Oishi K (2014): A Bayesian approach to the creation of a study‐customized neonatal brain atlas. Neuroimage 101:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]


