Abstract
A major genomic alteration in prostate cancer (PCa) is frequent loss of chromosome (chr) 8p with a common region of loss of heterozygosity (LOH) at chr8p22 locus. Genomic studies implicate this locus in the initiation of clinically significant PCa and with progression to metastatic disease. However, the genes within this region have not been fully characterized to date. Here we demonstrate for the first time that a microRNA component of this region –miR-383- is frequently downregulated in prostate cancer, plays a critical role in determining tumor initiating potential and is involved in prostate cancer metastasis via direct regulation of CD44, a ubiquitous marker of PCa tumor initiating cells (TICs)/ stem cells. Expression analyses of miR-383 in PCa clinical tissues established that low miR-383 expression is associated with poor prognosis. Functional data suggests that miR-383 regulates PCa tumor initiating/ stem-like cells via CD44 regulation. Ectopic expression of miR-383 inhibited tumor initiating capacity of CD44+ PCa cells. Also, ‘anti-metastatic’ effects of ectopic miR-383 expression were observed in a PCa experimental metastasis model. In view of our results, we propose that frequent loss of miR-383 at chr8p22 region leads to tumor initiation and prostate cancer metastasis. Thus, we have identified a novel finding that associates a long observed genomic alteration to PCa stemness and metastasis. Our data suggests that restoration of miR-383 expression may be an effective therapeutic modality against PCa. Importantly, we identified miR-383 as a novel PCa tissue diagnostic biomarker with a potential that outperforms that of serum PSA.
Keywords: miR-383, chr8p22, genomic deletions, CD44, prostate cancer
Introduction
A critical challenge in prostate cancer (PCa) clinical management is posed by its high rates of recurrence and metastasis, often attributed to the existence of tumor-initiating cells (TIC) or PCa stem cells (CSC) within the bulk of tumor [1, 2]. CSCs have been identified in human prostate tumors and are associated with poor prognosis [1] as these cells are highly tumorigenic, often causing tumor relapse, metastases and therapy resistance [3, 4]. In human PCa, these cells are typically characterized by the expression of cell surface adhesion marker CD44 [1, 5-9] and other markers. The predominant stem cell marker, CD44, is a transmembrane receptor for hyaluronan, that plays critical roles in cellular adhesion, migration, signaling and tumor metastasis [10]. Tumor cells positive for this marker (CD44+) are more proliferative, clonogenic, tumorigenic, and metastatic than the isogenic CD44- cells [7]. Considering the challenges that CD44+ CSCs pose for effective PCa therapy, it is imperative to understand the molecular controls that regulate this subpopulation.
The prostate cancer genome is characterized by a frequent loss of chromosome 8p (chr8p) [11-17] with approximately 30%-50% of cases presenting this genomic alteration. A common region of loss of heterozygosity (LOH) has been mapped to the chr8p22 locus [18, 19] within this region. Genomic studies implicate this region as the site for candidate tumor suppressor genes (TSGs) [20], such as lipoprotein lipase (LPL), though the TSGs within this region has not been fully characterized to date. Studies suggest that chr8p deletion is an important event in both the initiation and metastasis of PCa [21]. High frequency of LOH at chr8p22 is usually observed in high-grade prostate tumors suggesting the existence of a novel putative PCa tumor-suppressor gene [21]. Considering the association of loss of this chromosomal region in PCa with poor prognosis [22], it is crucial to define the role of genetic elements within this region. Thus, the focus of this study was to define the role of microRNA (miRNA) genes located in the frequently deleted chr8p22 region in prostate cancer. MicroRNA genes are often located in fragile chromosomal regions involved in cancers [23]. miRNAs are small non-coding RNA molecules (containing ∼22 nucleotides) that suppress gene expression post transcriptionally via sequence-specific interactions with the 3′- untranslated regions (UTRs) of cognate mRNA targets [24]. In a preliminary screen, we identified that miR-383 is significantly downregulated in prostate cancer. miR-383 is located in the chr8p22 region within intron 3 of the sarcoglycan zeta (SGCZ) gene [25]. Downregulation of microRNA-383 has been associated with male infertility and has been reported to promote testicular embryonal carcinoma cell proliferation by targeting interferon regulatory factor-1 (IRF1) [26]. miR-383 has also been reported to be downregulated in gliomas and medulloblastomas and inhibits cell proliferation, migration, invasion and induces apoptosis via targeting of cyclin D1, GADD45G (growth arrest and DNA-damage-inducible 45 gamma), peroxiredoxin 3 and insulin like growth factor receptor 1 (IGFR1) [27-31]. miR-383 suppresses growth and metastasis of pancreatic carcinomas by directly targeting ROBO3 [32]. These studies point to an important regulatory role of miR-383. However, the role of miR-383 in prostate cancer has never been reported. Here we provide the first evidence that (i) miR-383 is dysregulated in prostate cancer; (ii) miR-383 is a microRNA component of frequently deleted chr8p22 locus that is a critical determinant of PCa TIC/stemness via regulation of CD44, a ubiquitous marker of PCa stem cells; (iii) miR-383 is a potential novel PCa diagnostic biomarker; (iv) miR-383 is an important ‘anti-metastatic’ microRNA in PCa. To our knowledge, this is the first study that associates a long studied frequently lost chromosomal region with PCa TICs via loss of a microRNA component of the region.
Results
MicroRNA-383 located in frequently deleted chr8p22 region is under expressed in prostate cancer
miR-383 is located in a common region of LOH at the chr8p22 locus [18, 19] within the host gene SGCZ (Fig. 1A). We analyzed copy number alterations (CNAs) at this locus in prostate adenocarcinomas in The Cancer Genome Atlas (TCGA) dataset [33, 34]. miR-383 locus was deleted either homozygously or heterozygously in ∼ 48% PCa cases suggesting that prostate tumors are associated with genomic deletion of this miRNA (Fig. 1B). Based on this, we hypothesized that miR-383 expression is lost in PCa and may underlie PCa initiation, progression and metastasis. To test our hypothesis, we analyzed miR-383-5p (major form of miR-383, referred as miR-383) expression in a large cohort of PCa clinical specimens (Fig. 1C-D). miR-383 expression analyses in the TCGA cohort (training cohort) for a set of 187 primary prostate adenocarcinoma cases (Fig. 1C) suggest that ∼72% of PCa cases exhibit miR-383 downregulation. Next we performed miR-383 expression profiling in an independent training-validation cohort of 112 patients seen at SFVAMC (Fig. 1D). miR-383 expression was analyzed in laser capture microdissected (LCM) PCa tissues (n=112) and matched adjacent normals by real-time PCR (Fig. 1D) and was found to be significantly downregulated in ∼76% of PCa cases (P: 0.0003). 9% of PCa cases showed no change and ∼15% of cases exhibited high miR-383 expression. Average miR-383 expression in tumor tissues was ∼4-fold lower than that of normal tissues (P=0.0500) (Fig. 1E). Patients' demographics and clinicopathological characteristics are summarized in Table S1. We also analyzed CNAs within a small subset of SFVAMC cohort (n=22) and found that tumor tissues show a heterozygous or homozygous loss at miR-383 locus in ∼46% of tissues compared to adjacent normals (Fig. 1F). Also, we examined correlation between CNAs at this locus and miR-383 expression within this subset (Fig. 1E) and found a positive correlation (correlation co-efficient= 0.563, P=0.0042). Further, miR-383 expression analyses in prostate cell lines showed that its expression is specifically attenuated in PCa cell lines (LNCaP, DU145, PC3, LAPC9) compared to normal primary prostate epithelial cells (PPEC) (Fig. 1G). Collectively, these data confirm the widespread downregulated expression of miR-383 in PCa.
Fig. 1. MicroRNA-383 located in frequently deleted chr8p22 region is under expressed in prostate cancer.
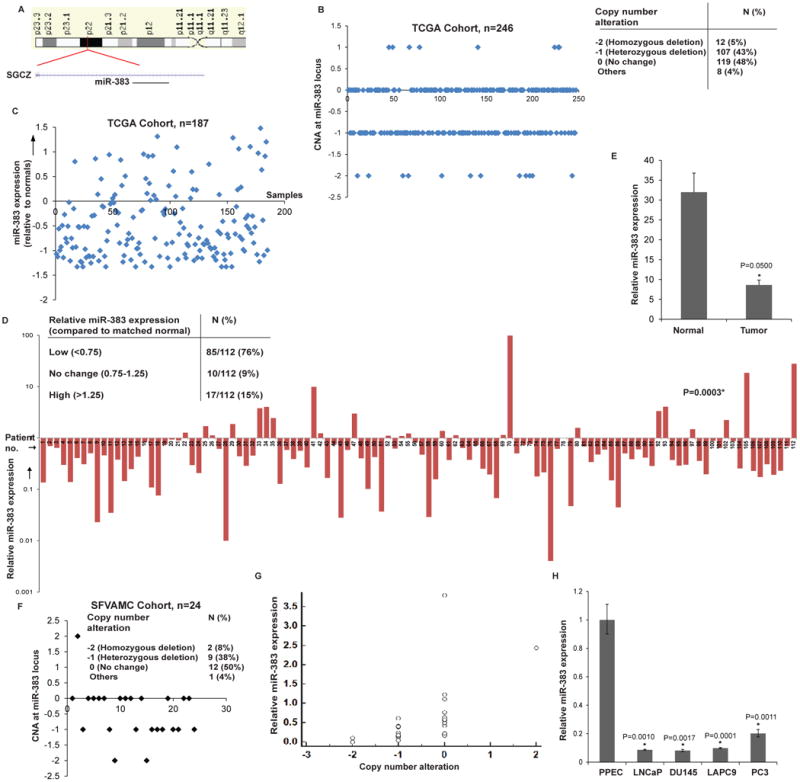
A. Schematic representation of chr8p22 region highlighting the location of miR-383 within the host gene SGCZ.
B. CNAs at miR-383 locus in prostate adenocarcinomas in the TCGA cohort. Table above summarizes the observed CNAs.
C. Relative miR-383 expression levels in the TCGA training cohort (n=187). z-scores were calculated for tumor tissues and plotted along x-axis.
D. Relative miR-383 expression levels in training-validation SFVAMC cohort (n=112). miR-383 expression was analyzed in laser capture microdissected (LCM) PCa tissues (n=112) and matched adjacent normal regions by real-time PCR. Data were normalized to RNU48 control.
E. Average miR-383 expression in LCM microdissected PCa tissues and adjacent normal regions of SFVAMC cohort as assessed by RT-PCR in Fig. 1D. Data are represented as mean ± S.D.
F. CNAs at miR-383 locus in prostate adenocarcinomas in a subset of the SFVAMC cohort (n=24).
G. Correlation between CNAs and miR-383 expression in SFVAMC cohort (n=24).
H. Relative miR-383 expression levels in primary prostate epithelial cells (PPEC) and prostate cancer cell lines as assessed by RT-PCR. Data were normalized to RNU48 control. (* P< .05).
Low miR-383 expression is associated with poor survival outcome in prostate cancer
In view of the observed widespread downregulation of miR-383 expression in PCa clinical specimens, we performed analyses to assess if low miR-383 expression has associated prognostic potential. Kaplan-Meier survival analysis of prostate cancer patients (SFVAMC and TCGA cohort) was performed where patients were stratified into low/high miR-383 expression groups (Fig.2A). Our analyses showed that patients with low miR-383 expression had significantly poor survival outcome (P=0.0102) with the survival probability reduced to ∼81%. These analyses suggest that low miR-383 expression may be associated with poor PCa prognosis. Further, we performed multivariate analyses to determine if miR-383 provides information independent of known prognostic factors such as PSA at diagnosis, Gleason grade and clinical stage (Table S2). Multiple regression analyses for SFVAMC cohort using entry, forward, backward, and stepwise methods (Supplementary Table S2) revealed that miR-383 expression is an independent predictor of overall survival (P < 0.0001).
Fig. 2. Low miR-383 expression is associated with poor survival outcome in prostate cancer.
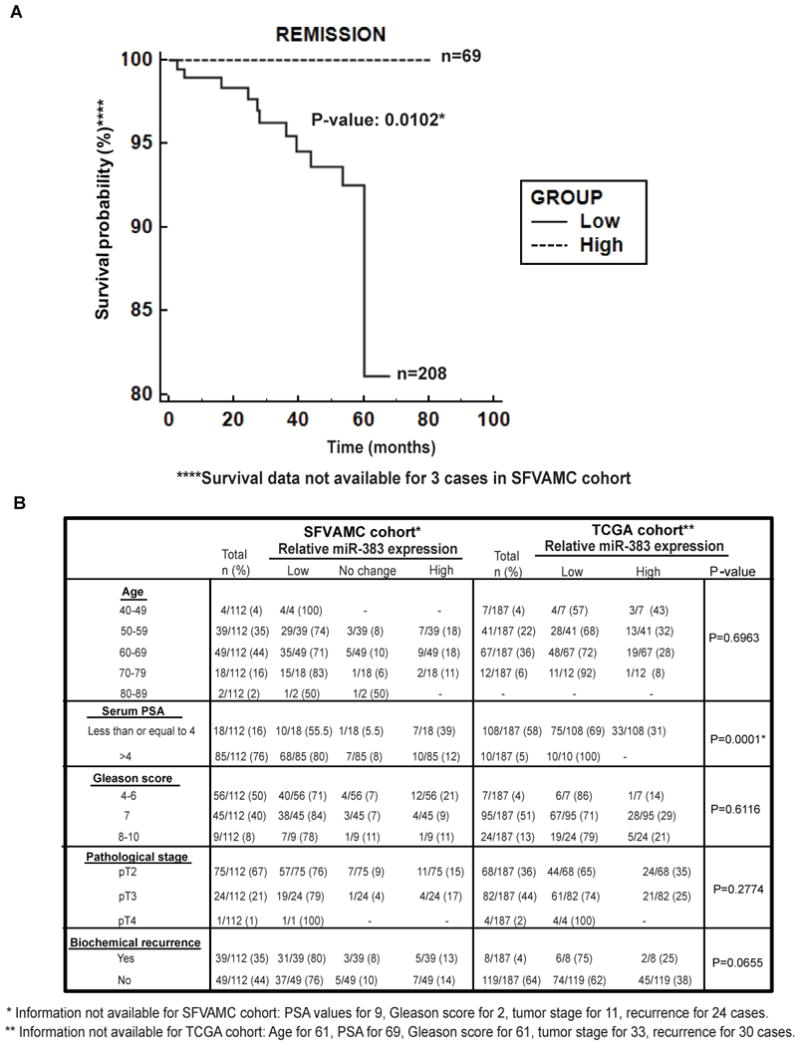
A. Kaplan-Meier survival curve for PCa patients (SFVAMC and TCGA cohorts), stratified based on relative miR-383 levels. P-value based on a log rank test.
B. Correlation of miR-383 expression with clinicopathological parameters in PCa patients. P-values are based on Chi square test.
Low miR-383 expression is correlated with high serum PSA in prostate cancer
We next analyzed if low miR-383 expression is associated with clinicopathological parameters such as age, serum PSA, Gleason grade, pathological stage and biochemical recurrence of the carcinoma (Fig. 2B). Interestingly, we observed a significant inverse correlation between miR-383 and serum PSA levels (P=0.0001). Patients with high serum PSA levels (PSA > 4) exhibited low miR-383 expression. Further, we stratified our clinical cohort based on age and PSA and used age-adjusted PSA ranges to examine the correlation between miR-383 expression and serum PSA (Table S3). We found that miR-383 expression was statistically correlated with age-adjusted PSA levels (P= 0.0007). Low miR-383 expression was observed in 54% of cases with normal age adjusted PSA levels and in 81% of cases with high PSA levels. While there was no significant correlation with age and Gleason score, decreased miR-383 expression was observed in 76% of cases of pathological stage pT2, 79% of cases of pT3 and 100% of pT4 cases (Fig. 2B). Similarly, in the TCGA cohort, low miR-383 expression was observed in 65% of pT2, 74% of pT3 and 100% of pT4 cases. Also, low miR-383 expression was observed in 80% and 75% of cases with biochemical recurrence within the SFVAMC and TCGA cohorts, respectively. However, the correlations between miR-383 expression and pathological stage and biochemical recurrence failed to achieve statistical significance (Fig. 2B).
miR-383 is a potential novel diagnostic biomarker for prostate cancer
In view of the observed correlation between miR-383 expression and serum PSA, we evaluated the diagnostic potential of miR-383 (Fig. 3). Receiver Operating Characteristic (ROC) curve analyses showed that miR-383 expression can be a single significant parameter to discriminate between normal and tumor tissues with an area under the ROC curve (AUC) of 0.94 (95% CI: 0.90-0.98, P=1.696997e-21) (Fig. 3A). The discriminatory ability of miRNA was further characterized using apparent prevalence, true prevalence, sensitivity, specificity, positive predictive value, negative predictive value, positive likelihood ratio (Fig. 3B). These analyses showed that miR-383 as PCa diagnostic biomarker exhibits 100% sensitivity, 72% specificity, 78% positive predictive value and 100% negative predictive value. We also compared the diagnostic ability of miR-383 expression with that of serum PSA, the most widely used PCa biomarker. miR-383 expression outperformed PSA as a diagnostic biomarker in our patient cohort, with PSA showing an AUC of 0.80 (95% CI: 0.71-0.88) (Fig. 3C). The difference between diagnostic abilities of miR-383 vs serum PSA were statistically significant (P = 0.003). Further, a logistic regression model was employed to compare the ability of miRNA and PSA to identify a tumor sample by comparing probability of tumor conditional on miRNA values vs that of PSA values (Fig. 3D). Our analyses suggest that miR-383 is a better diagnostic classifier. As PSA changes (along the x-axis) increasing the probability of tumor, the separation between tumor and normal samples is not apparent (Fig. 3D). On the other hand, when miR-383 expression changes (along the y-axis) increasing the probability of tumor, the majority of tumor samples is at the high probability of tumor, thus showing the advantage of miR-383 as a classifier. This suggests that miR-383 expression is a promising diagnostic biomarker for prostate cancer. In view of these data, we also modelled the combined diagnostic potential of miR-383 and PSA (Fig. 3E) with adjustment for age, Gleason score and grade. Our analyses demonstrates that incorporating miR-383 with PSA increased the AUC of PSA from 0.80 to 0.995 (95% CI: 0.985-0.999, P<0.001).
Fig. 3. miR-383 is a potential novel diagnostic biomarker for prostate cancer.
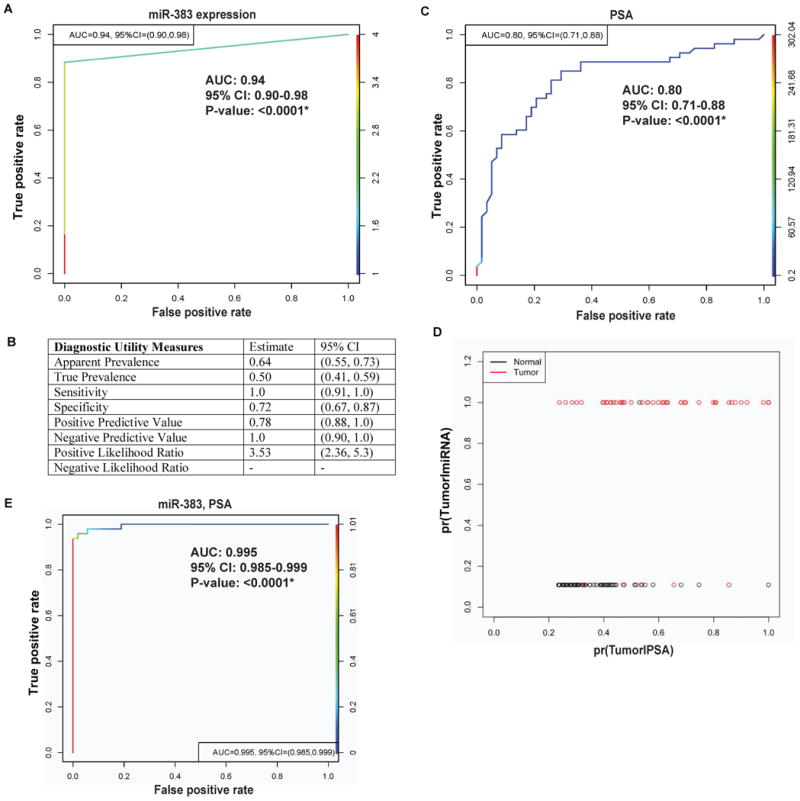
A. ROC curve analysis showing the ability of miR-383 to discriminate between tumor and normal samples.
B. Diagnostic utility measures of miR-383 as a prostate cancer biomarker.
C. ROC curve analysis for PSA to discriminate between tumor and normal samples.
D. Logistic regression analyses to compare the ability of miR-383 and PSA to identify a tumor sample. Probabilities of tumor conditional on miR-383 values are plotted along y-axis while that of PSA along x-axis.
E. ROC curve analysis for combined potential of miR-383 and PSA to distinguish between malignant and normal tissues. (*P< .05).
miR-383 overexpresssion suppresses in vitro attributes of tumorigenicity in PCa cell lines
To assess the functional significance of miR-383 in PCa, we overexpressed miR-383/ control miRNA (miR-CON) in PCa cell lines (Du145, PC3, LNCaP) followed by functional assays (Fig. 4). miR-383 overexpression (Fig. S1) led to a significant increase in the fraction of cells in the G0-G1 cell cycle phase (Fig. 4A) compared to miR-CON transfected cells in Du145, PC3 and LNCaP cells (P = 0.01, 0.04, 0.01 respectively). These data suggest that miR-383 expression leads to G0-G1 arrest in PCa cell lines. Also, miR-383 overexpression led to a significant increase in the apoptotic cell fractions (Early apoptotic + Apoptotic) in Du145/PC3/LNCaP cells as compared to miR-CON treated cells with a concomitant decrease in the viable cell population (Fig. 4B) (P=0.05, 0.04, 0.05 respectively). These analyses demonstrate that miR-383 overexpression induces cell cycle arrest and apoptosis in prostate cancer cell lines. Reduced cellular viabilities upon miR-383 overexpression were also observed by MTS assay (Fig. 4C). Further, miR-383 overexpression led to reduced clonogenicity (Fig. 4D), invasiveness and migration (Fig. 4E) in Du145/PC3/LNCaP cells compared to controls. Collectively, these data suggest that miR-383 suppresses in vitro attributes of tumorigenicity and that miR-383 plays a tumor suppressive role in PCa.
Fig. 4. miR-383 overexpresssion suppresses in vitro attributes of tumorigenicity in prostate cancer cell lines.
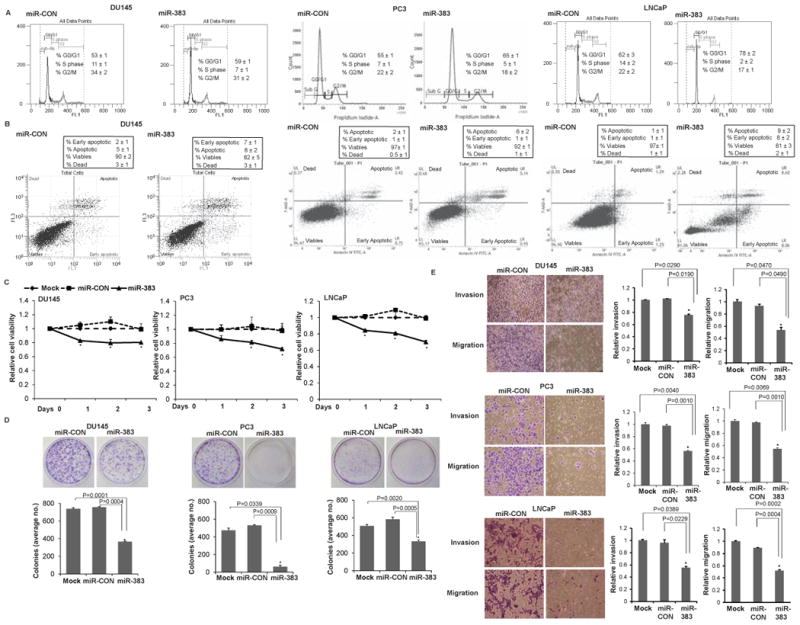
miR-383 mimic/control miR/mock was transfected in Du145/ PC3/ LNCaP cell lines followed by functional assays (performed 72 hrs post-transfection) (*P< .05).
A. Cell cycle assay in Du145/ PC3/ LNCaP cells after miR-CON (left panel) or miR-383 (right panel) transfections.
B. Apoptosis assay in Du145/ PC3/ LNCaP cells after miR-CON (left panel) or miR-383 (right panel) transfections as assessed by ANNEXIN V-FITC /7-AAD staining.
C. MTS cellular viability assays, D. Colony formation assays, and E. Transwell invasion and migration assays in Du145/ PC3/ LNCaP cells transfected with mock/ miR-CON/ miR-383.
miR-383 silencing induces tumorigenicity in normal prostate epithelial cells
In a reciprocal approach, we silenced endogenous miR-383 expression in normal primary prostate epithelial cell line (PPEC) (Fig. S2A) followed by functional assays. miR-383 knockdown led to increased cellular viabilities (Fg. S2B). Further, miR-383 knockdown increased the invasiveness and motility (Fig. S2C-D) of normal prostate epithelial cells. This data suggests that endogenous miR-383 expression is vital to maintain prostate cells in normal untransformed state and miR-383 loss initiates tumorigenicity in these cells.
miR-383 directly regulates prostate cancer stem cell marker CD44
Ongoing studies in our laboratory on microRNA genes located in the frequently deleted chr8p region suggest that these genes are important players in PCa epithelial to mesenchymal transition (EMT), stemness and PCa progression and metastasis (unpublished data). In silico analyses using miRANDA [35] algorithm identified that miR-383 potentially targets an array of stemness related genes including CD44, LIN28A, STAT3 and LEF1 (Table S4). We performed Western blot analyses to validate these targets after miR-383 overexpression (Fig. S3). Our analyses showed that only CD44 levels were subject to regulatory control by miR-383. RT-PCR and immunoblotting analyses of mock/miR-CON/miR-383 overexpressing DU145/PC3/LNCaP cells confirmed that miR-383 expression represses CD44 mRNA and protein expression (Fig. 5A, 5B and S4). This repression was confirmed in bioluminescent PC-3M-luc-C6 cells stably transfected with control miR/miR-383 (Fig. 5A-B). To verify that these effects on CD44 expression are as a result of direct interaction of miR-383 with the corresponding microRNA response element (MRE)/ binding site within CD44 3′ UTR (Fig. 5C), we performed luciferase reporter assays with CD44 3′UTR/Control 3′UTR in miR-CON/miR-383 transfected DU145/PC3 cells (Fig. 5D). A significant decrease in CD44 3′UTR luciferase activity was observed upon miR-383 transfection in both cell lines confirming CD44 as a direct miR-383 target. To validate CD44 as direct miR-383 target, we mutated the putative miR-383 binding site in CD44 3′ UTR (represented in Fig. 5C). Transfection of mutant CD44 3′UTR/control 3′UTR in miR-CON/miR-383 transfected DU145/PC3 cells showed that mutation of the miR-383 binding site prevented the repression of luciferase activity observed upon miR-383 overexpression in DU145 and PC3 cells (Fig. 5D). Since CD44 is a predominant cell surface marker of PCa tumor initiating cells (TICs) [1, 5-9], we next examined the regulation of TICs by miR-383. We purified CD44+ and CD44- cell populations from PCa xenografts generated from DU145, PC3 and LAPC9 cell lines (Fig. S5) and profiled miR-383 expression (Fig. 5E). The average percentage of CD44+ subpopulation observed with Du145 (upper panel), PC3 (middle panel) and LAPC9 (lower panel) are represented in Fig. S5. A consistent significant downregulation of miR-383 was observed in CD44+ cell populations suggesting that miR-383 regulates PCa tumor initiating/ stem-like cells.
Fig. 5. miR-383 directly regulates prostate cancer stem cell marker CD44.
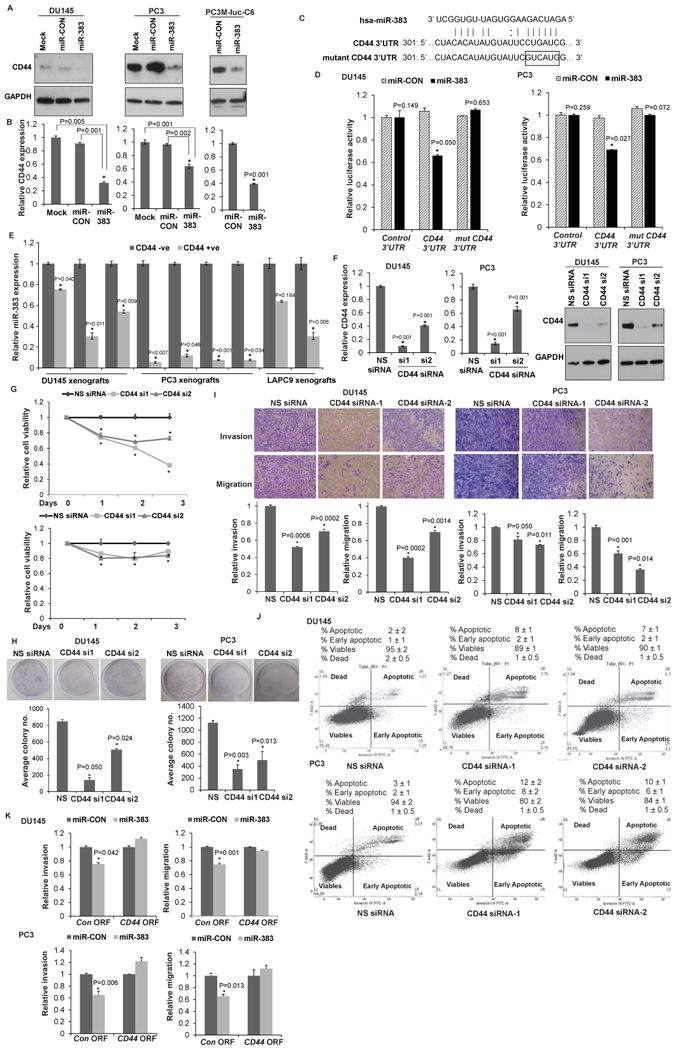
A. Immunoblots of endogenous CD44 in DU145/PC3 cells transfected with mock/miR-CON/miR-383. Right panels show endogenous CD44 in PC-3M-luc-C6 cells stably transfected with control miR/miR-383. GAPDH was used a loading control.
B. Relative CD44 mRNA expression in DU145/PC3/LNCaP cells transfected with mock/miR-CON/miR-383. Data were normalized to GAPDH control.
C. Schematic representation of the CD44 3′-UTR showing the putative miR-383 binding site. Mutant CD44 3′ UTR is represented below.
D. Luciferase reporter assays with the indicated wt and mutated 3′ UTR constructs or control luciferase construct co-tranfected with miR-CON/ miR-383 in DU145 (left panels) and PC3 cells (right panels). Firefly luciferase values were normalized to Renilla luciferase activity and plotted as relative luciferase activity (* P< .05 as compared to miR-CON).
E. Relative miR-383 levels in purified CD44- and CD44+ subpopulations of PCa xenografts (DU145, PC3, LAPC9). Data were normalized to RNU48 control.
F. DU145/PC3 cells were transfected with two sets of siRNAs specific to CD44 (si1 and si2) or a nonspecific (NS) control siRNA/mock transfected for 72 h followed by functional assays. Left panels: Relative CD44 mRNA expression after siRNA transfections as assessed by RT-PCR. p-values are relative to NS siRNA. GAPDH was used as a control. Right panels: Immunoblot analyses for CD44 protein expression after NS/CD44 siRNA transfections. GAPDH was used as a loading control.
G. Cellular viability assay, H. Clonogenicity assay, I. Transwell invasion assay and migration assay after NS/CD44 siRNA-1 and siRNA-2 transfections.
J. Apoptosis assay in DU145 and PC3 cells upon NS siRNA (left panel) or CD44 siRNA-1 (middle panel) or CD44 siRNA-2 (right panel) transfections.
K. Invasion and migration assays in Du145 (upper panels) and PC3 (lower panels) after indicated transfections. (*P< .05).
CD44 is a functionally relevant target of miR-383 in prostate cancer
To determine if CD44 is a functionally relevant miR-383 target in PCa, we performed siRNA mediated knockdown of CD44 in DU145/ PC3 cells followed by functional assays (Fig. 5F-5J). We employed two sets of siRNA, si1-2, against CD44 to achieve efficient knockdown as assessed by RT-PCR (Fig. 5F, left panels) and validated by immunoblot analysis (Fig. 5F, right panels). CD44 knockdown led to decreased cell viability (Fig. 5G), clonogenicity (Fig. 5H), invasiveness and motility (Fig. 5I) and increased apoptosis (Fig. 5J). Also, CD44 knockdown induced G0-G1 cell cycle arrest in Du145 cell line (Fig. S6). Thus, the functional consequences of CD44 knockdown functionally mimicked the effects of miR-383 overexpression. We also performed rescue experiments wherein CD44 ORF clone lacking miR-383 binding site in its 3′ UTR region was overexpressed along with miR-383 overexpression in Du145/PC3 cells. We observed that the effects of miR-383 overexpression on invasion and migration were reversed upon CD44 overexpression as compared to control cells (Fig. 5K) reinforcing CD44 as a functional miR-383 target. However, the effects of miR-383 on cellular proliferation were not significantly abrogated (data not shown).
miR-383 inhibits tumor initiating capacity of CD44+ prostate cancer cells in vitro
To further corroborate miR-383 mediated regulation of TICs via CD44, we asked if modulation of miR-383 expression in the CD44+ subpopulation of xenograft tumors alters their tumor initiating capacity. To test this, we purified CD44+ cells from DU145 mouse xenograft tumors followed by miR-383 transfections and in vitro functional assays (Fig. 6). Overexpression of miR-383 in CD44+ xenograft cells (Fig. 6A) led to markedly inhibited cellular growth (Fig. 6B). The tumor initiating capacity of CD44+ cells was significantly inhibited by miR-383 expression as assessed by clonogenicity (Fig. 6C) and sphere-formation assays (Fig. 6D). To explore the molecular basis for these effects, we performed Western Blot analysis of miR-CON/miR-383 transfected CD44+ xenograft cells (Fig. 6E). Interestingly, ectopic miR-383 expression in CD44+ve PCa TICs led to marked downregulation of CD44 suggesting that miR-383 has remarkable potential to modulate this cell surface marker in PCa TIC/stem cells.
Fig. 6. miR-383 inhibits tumor initiating capacity of CD44+ prostate cancer cells in vitro.
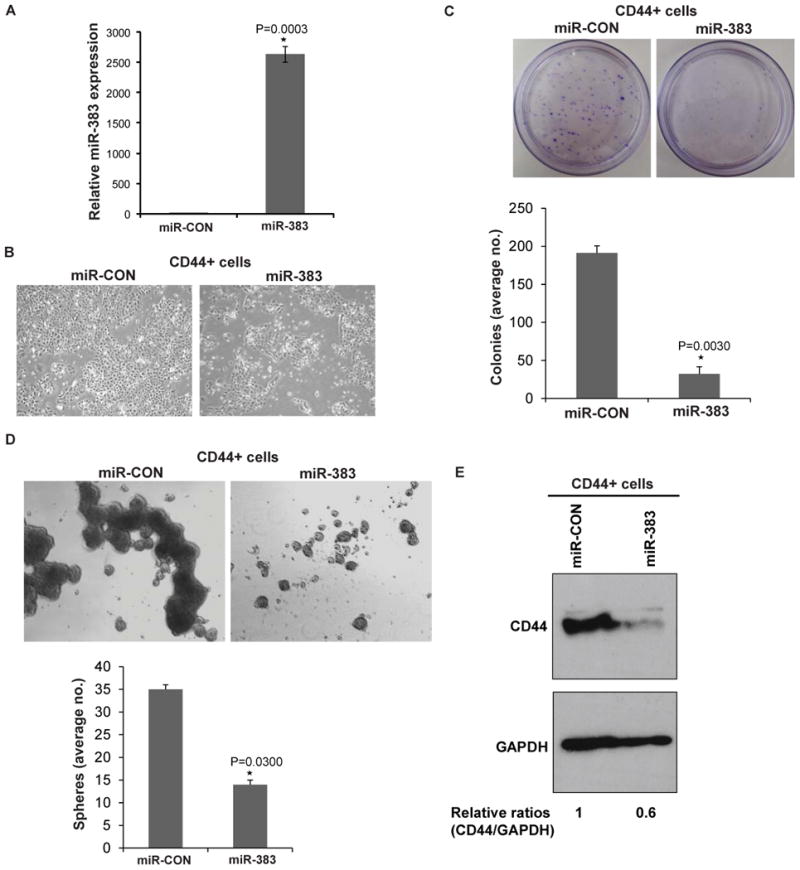
CD44+ subpopulations were purified from DU145 xenograft tumors followed by miR-383 overexpression and in vitro functional assays.
A. Assessment of miR-383 overexpression in CD44+ cells by real time PCR. Data were normalized to RNU48 control.
B. Phase contrast images of CD44+ cells transfected with miR-CON/miR-383.
C. Clonogenicity assay in miR-CON/miR-383 transfected CD44+ cells.
D. Sphere formation assay in miR-CON/miR-383 transfected CD44+ cells.
E. Western Blot analysis of miR-CON/miR-383 transfected CD44+ xenograft cells. Band intensities were determined by Image J and relative ratios (CD44/GAPDH) were calculated and are shown below the blot.
miR-383 expression suppresses primary prostate xenograft tumor growth in vivo
In view of the in vitro tumor suppressive effects of miR-383, we sought to determine the in vivo effects of miR-383 (Fig. 7). We examined the effects of ectopic expression of miR-383 on both primary prostate xenograft tumor growth (Fig. 7B-C) and metastases (Fig. 7D). Bioluminescent PC-3M-luc-C6 cells were stably transfected with miR-CON/miR-383 (Fig. 7A) and subcutaneously injected into nude mice to generate PCa xenograft tumors (Fig. 7B). Our results show that miR-383 over expression led to significant inhibition of xenograft tumor growth confirming its tumor suppressive effects in PCa. Further, in view of our present results suggesting that miR-383 is an important regulator of CD44, we asked if this regulatory control functions in vivo. To test this, we extracted protein from xenograft tumors followed by Western blotting for CD44 (Fig. 6C). Our data shows that miR-383 expressing tumors (Fig. 7C, lower panels) had lower CD44 protein levels as compared to control tumors (Fig. 7C, upper panels). This further validates that CD44 is a direct miR-383 target in PCa.
Fig. 7. miR-383 overexpression inhibits primary tumor growth and metastasis of prostate cancer in vivo.
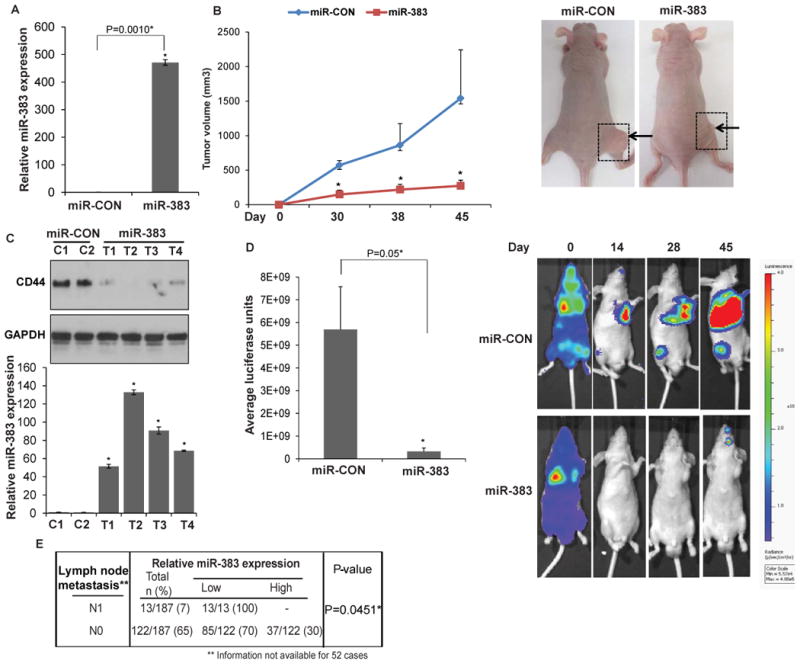
Bioluminescent PC-3M-luc-C6 cells were stably transfected with control miR/miR-383 and were either subcutaneously injected (Fig. 7B-C) or were used to generate mouse PCa experimental metastasis model (Fig. 7D). (* P= .05).
A. Relative miR-383 expression levels in PC-3M-luc-C6 cells stably transfected with control miR/miR-383 as assessed by RT-PCR. Data were normalized to RNU48 control.
B. Tumor volumes of xenograft tumors from miR-CON (blue) and miR-383 (red) groups, at the indicated time points. Data represent the mean of each group ±SD. Right panels: Representative images of mice from miR-CON/ miR-383 group on day 45. Arrows indicate tumors.
C. Upper panels: CD44 protein levels in control (C1-C2) and miR-383 prostate cancer xenografts (T1-T4) as assessed by immunoblot analysis. GAPDH was used as a loading control. Lower panels: Relative miR-383 expression as assessed by real-time PCR in xenograft tumors represented in upper panels. Data were normalized to RNU48 control.
D. miR-CON/miR-383 expressing PC-3M-luc-C6 were injected in the left ventricles of nu/nu mice (day 0) followed by periodic monitoring of metastases by in vivo BLI. Average bioluminescence of mice from miR-CON/ miR-383 groups on day 45 are plotted. Data represent the mean of each group ±SD Right panels: Representative bioluminescence images from miR-CON/miR-383 groups at indicated time-points. The scale bar on the right represents the relative intensity of bioluminescence.
E. Correlation between miR-383 expression and lymph node metastasis in TCGA cohort of prostate adenocarcinomas.
miR-383 expression suppresses prostate cancer metastasis in vivo
Since CD44 plays critical roles in tumor metastasis [10] and in view of our data implicating an important role of miR-383 in PCa invasiveness and migration due to CD44 regulation, we examined the effects of miR-383 overexpression on PCa metastasis. As an experimental metastasis model (Fig. 7D), bioluminescent PC-3M-luc-C6 cells stably expressing control miR/miR-383 were injected into the left ventricles of nude mice followed by periodic monitoring of the in vivo tumor burden by bioluminescent imaging (BLI). Interestingly, miR-383 had a strong inhibitory effect on metastases indicating its important ‘anti-metastatic’ role in PCa. To corroborate this, we examined whether low miR-383 expression is associated with metastasis in PCa clinical specimens (Fig. 7E). The TCGA cohort of prostate adenocarcinoma had 13 cases with lymph node metastasis (N1). 13/13 (100%) of these clinical tissues showed low miR-383 expression suggesting a significant inverse correlation between miR-383 expression and lymph node metastasis (P=0.0451). However, this correlation could not be tested with the SFVAMC cohort as it had no metastatic cases.
Discussion
A major PCa genomic alteration is frequent loss of chromosome 8p [11-17] with a common region of LOH at chr8p22 locus [18, 19]. Here we demonstrate for the first time that a miRNA component of this region is frequently downregulated in PCa and plays a critical role in determining the tumor initiating potential and is involved in PCa metastasis via its direct regulation of CD44. Thus, our study is novel in that it associates a long observed genomic alteration to PCa stemness and metastasis via loss of a miRNA gene. To our knowledge, this is the first study that ascribes this role to this frequently lost genomic region in prostate cancer.
Importantly, our study reports for the first time that miR-383 is dysregulated in prostate carcinomas. Though miR-383 has been reported to be downregulated in other malignancies such as gliomas, medulloblastomas and pancreatic cancer [27-32], its role has never been explored in PCa. Till date, studies focused on microRNA expression profiling in PCa [36, 37] did not report significant alterations of this miRNA. However, our focused analyses of laser capture microdissected PCa clinical tissues compared to matched adjacent normals and reanalysis of TCGA data [36] revealed the widespread downregulation of this miRNA. Further, our analyses suggest that low miR-383 expression is associated with poor prognosis in PCa as clinical specimens with low miR-383 expression had poor survival outcome. Though our clinical samples do not suggest a statistically significant association between Gleason grade or tumor stage, miR-383 expression was significantly inversely correlated with lymph node metastasis. Also, our clinical cohort had a limited number of cases of pathological stage pT4 and metastatic samples. Hence, additional studies with larger datasets are warranted to confirm the prognostic potential of miR-383 in PCa. Previous studies suggest increased chr8p deletions with advanced tumor grade [38] and poor prognosis in PCa [22]. In particular, ch8p22 deletion has been shown to be a strong indicator for predicting disease progression [39] and PCa survival [40].
Functional studies with PCa cell lines demonstrated in vitro and in vivo tumor suppressive role for miR-383 in PCa. miR-383 silencing induced tumorigenicity in normal prostate epithelial cells suggesting a potential role of miR-383 in tumor initiation. Further, observed ‘anti-metastatic’ effects of ectopic miR-383 expression in a PCa experimental metastasis model suggest that miR-383 may play an important ‘anti-metastatic’ role in PCa. These findings are in agreement with earlier genomic studies suggesting that LOH at chr8p22 locus is an important event in the initiation of clinically significant PCa and is associated with subsequent progression to metastatic PCa [21].
Importantly, we demonstrate for the first time that miR-383 is a key negative regulator of PCa TICs via its direct regulation of CD44. Remarkably, ectopic expesssion of miR-383 in CD44+ve PCa TICs led to marked downregulation of CD44 suggesting that miR-383 may regulate this subpopulation of PCa TICs/ stem cells. Further, miR-383 overexpression in PCa cell lines led to direct repression of CD44 in vitro and in vivo, validating it as a direct miR-383 target. Collectively, these data demonstrate that miR-383 is an important regulator of PCa TICs via direct regulation of CD44. CD44 has been previously reported to be regulated by miR-34a [41], miR-708 [42], miR-373 and miR-520c [43] in PCa. Notably, Liu et al demonstrated miR-34a as a key negative regulator of CD44+ PCa cells and miR-34a loss led to PCa development and metastasis [41]. Similarly, we demonstrated miR-708 as another key regulator of CD44+ subpopulation of PCa cells via its direct regulation of CD44 [42]. Yang et. al [43] demonstrated that miR-373 and miR-520c suppress CD44 translation. Thus, CD44 may be combinatorially regulated by several miRNAs lending support to the concept that the miRNAs might cooperate to regulate specific genes by binding to the 3′ UTR of a single gene [44, 45]. Since CD44 has been reported to play critical roles in tumor initiation and metastasis [10], we propose that frequent loss of miR-383 at chr8p22 region leads to tumor initiation and PCa metastasis via its regulation of CD44. Though miR-383 was widely downregulated in PCa clinical specimens, ∼15% of prostate tumors showed upregulated miR-383 levels as compared to matched adjacent normal. In agreement with our hypothesis, these PCa cases were associated with better overall survival outcome.
Our study suggests that miR-383 inhibits PCa proliferation, migration and invasiveness and induces apoptosis. miR-383 expression has been previously reported to inhibit cell proliferation, migration, invasion and induce apoptosis in gliomas and medulloblastomas via targeting of cyclin D1, GADD45G (growth arrest and DNA-damage-inducible 45 gamma), peroxiredoxin 3 and insulin like growth factor receptor 1 (IGFR1) [27-31]. In pancreatic cancer, miR-383 suppresses growth and metastasis by direct targeting of ROBO3 [32]. In skin cells, miR-383 is regulated by STAT3 that suppresses ATR controlling apoptosis [46]. Our study suggests that the effects of miR-383 on PCa cell lines are primarily mediated through its regulation of CD44. Considering the involvement of CD44, a transmembrane receptor for hyaluronan, in cell-cell and cell-matrix adhesion, migration, signaling and tumor metastasis [10], we suggest that miR-383-CD44 regulatory interplay plays an important role in prostate tumorigenesis. Reversal of the effects of miR-383 overexpression on invasion and migration upon CD44 overexpression lends support to our hypothesis. However, since the effects of miR-383 overexpression on cellular proliferation could not be ‘rescued’ by CD44, miR-383 may target additional genes. Future studies in the lab are directed towards identifying additional miR-383 targets in PCa.
Overall, we describe a novel finding that associates the frequently deleted chr8p22 locus to PCa stemness and metastasis via loss of a microRNA gene. Another miRNA component of this region, miR-320, has been similarly reported to be downregulated in PCa and suppress the cancer stem cell-like characteristics of PCa cells by downregulating the Wnt/beta catenin signaling pathway [47]. miR-320 knockdown significantly increased cancer stem-like properties, and conversely, increased miR-320 expression in prostate stem-like TICs significantly suppresses stem cell-like properties of PCa cells. This study lends further support to our hypothesis linking chr8p22 locus with PCa stem cells. Also, miR-320 has been reported to be transactivated by miR-383 in granulosa cells (GC) leading to enhanced miR-320 mediated suppression of GC proliferation via its targeting of E2F1 and SF1 [48]. Though we did not explore if such a regulatory interaction between miR-383 and miR-320 exists in PCa in the present study, future directions will be focused on this aspect.
Though histologic assessment has been used widely to assess PCa from tissue specimens, it has its own limitations as with serum PSA testing [49, 50]. Hence, microRNAs are being explored as alternative PCa biomarkers [51]. Our analyses suggest miR-383 as a potential PCa tissue diagnostic biomarker. Interestingly, serum PSA levels were inversely correlated with miR-383 expression in PCa tissues and factoring in tissue miR-383 expression with serum PSA increased the diagnostic potential of PSA in our clinical cohort. These data suggest that miR-383 expression can be a significant diagnostic parameter that can be used as a supplement to serum PSA testing. In view of our present results, ongoing studies in the lab are focused on testing miR-383 levels in serum from PCa patients and assessing its potential as an alternative biomarker with serum PSA.
Considering the association of chr8p22 loss with poor PCa prognosis [22], our findings are highly significant. Our in vivo data showing a tumor-suppressive role of miR-383 in primary and metastatic PCa mouse models suggest that restoration of miR-383 expression may represent a therapeutic modality in PCa.
Materials and Methods
Cell lines and cell culture
Primary prostate epithelial cells and PCa cell lines (LNCaP, Du145, PC3) were obtained from the American Type Culture Collection (ATCC) and cultured under recommended conditions as described previously [52]. These cell lines were authenticated by DNA short-tandem repeat analysis by ATCC. The experiments with cell lines were performed within 6 months of their procurement/resuscitation.
Tissue samples
Formalin-fixed, paraffin-embedded (FFPE) PCa samples (n=112) were obtained from the SFVAMC. Written informed consent was obtained from all patients and the study was approved by the UCSF Committee on Human Research. All tissue slides were reviewed by a board certified pathologist for the identification of cancer foci as well as adjacent normal glandular epithelium.
Animal studies
All animal care was in accordance with the guidelines of the SFVAMC and the study was approved by the San Francisco VA IACUC. Bioluminescent PC-3M-luc-C6 cells were stably transfected with miR-CON/miR-383 and either subcutaneously injected or were injected into the left ventricles of 6-weeks old male nude mice (strain nu/nu; Simonsen Laboratories, n=8 per group) as detailed in supplemental methods.
Statistics
All experiments were repeated at least three times with similar results each time. All quantified data represents an average of triplicate samples or as indicated. Data are represented as mean ± S.E.M or as indicated. Two-tailed Student's t-test was used for comparisons between groups. Chi square test was used for correlation analyses between miR-383 expression and clinicopathological parameters. Statistical analyses were performed using MedCalc version 10.3.2. Results were considered statistically significant at P ≤ 0.05. Full method descriptions are available in the Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with preparation of the manuscript.
We acknowledge UCSF CTSI (Dr. Iryna V. Lobach) for help with statistical analysis. This work was supported by the National Cancer Institute at the National Institutes of Health (Grant Number RO1CA177984, RO1CA138642), VA program project on PCa (BX001604).
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose.
References
- 1.Tang DG, Patrawala L, Calhoun T, Bhatia B, Choy G, Schneider-Broussard R, et al. Prostate cancer stem/progenitor cells: identification, characterization, and implications. Mol Carcinog. 2007;46:1–14. doi: 10.1002/mc.20255. [DOI] [PubMed] [Google Scholar]
- 2.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes & development. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Tang DG. Prostate cancer stem cells and their potential roles in metastasis. J Surg Oncol. 2011;103:558–562. doi: 10.1002/jso.21806. [DOI] [PubMed] [Google Scholar]
- 4.Wicha MS. Stemming a tumor with a little miR. Nat Med. 2011;17:162–164. doi: 10.1038/nm0211-162. [DOI] [PubMed] [Google Scholar]
- 5.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res. 2008;68:1820–1825. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- 7.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 8.Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 9.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796–6805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 10.Iczkowski KA. Cell adhesion molecule CD44: its functional roles in prostate cancer. Am J Transl Res. 2010;3:1–7. [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer research. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- 12.Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer research. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 13.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberfarb ME, Lin M, Lechpammer M, Li C, Tanenbaum DM, Febbo PG, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer research. 2003;63:4781–4785. [PubMed] [Google Scholar]
- 15.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer research. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 16.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 17.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bova GS, Carter BS, Bussemakers MJ, Emi M, Fujiwara Y, Kyprianou N, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer research. 1993;53:3869–3873. [PubMed] [Google Scholar]
- 19.Macoska JA, Trybus TM, Benson PD, Sakr WA, Grignon DJ, Wojno KD, et al. Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer research. 1995;55:5390–5395. [PubMed] [Google Scholar]
- 20.Kagan J, Stein J, Babaian RJ, Joe YS, Pisters LL, Glassman AB, et al. Homozygous deletions at 8p22 and 8p21 in prostate cancer implicate these regions as the sites for candidate tumor suppressor genes. Oncogene. 1995;11:2121–2126. [PubMed] [Google Scholar]
- 21.Lu W, Takahashi H, Furusato B, Maekawa S, Ikegami M, Sudo A, et al. Allelotyping analysis at chromosome arm 8p of high-grade prostatic intraepithelial neoplasia and incidental, latent, and clinical prostate cancers. Genes, chromosomes & cancer. 2006;45:509–515. doi: 10.1002/gcc.20314. [DOI] [PubMed] [Google Scholar]
- 22.El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Kollermann J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piovani G, Savio G, Traversa M, Pilotta A, De Petro G, Barlati S, et al. De novo 1Mb interstitial deletion of 8p22 in a patient with slight mental retardation and speech delay. Molecular cytogenetics. 2014;7:25. doi: 10.1186/1755-8166-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lian J, Tian H, Liu L, Zhang XS, Li WQ, Deng YM, et al. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell death & disease. 2010;1:e94. doi: 10.1038/cddis.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z, Cen D, Luo X, Li D, Li P, Liang L, et al. Downregulation of miR-383 promotes glioma cell invasion by targeting insulin-like growth factor 1 receptor. Med Oncol. 2013;30:557. doi: 10.1007/s12032-013-0557-0. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, Tian H, Duan Z, Cao Y, Zhang XS, Sun F. microRNA-383 impairs phosphorylation of H2AX by targeting PNUTS and inducing cell cycle arrest in testicular embryonal carcinoma cells. Cellular signalling. 2014;26:903–911. doi: 10.1016/j.cellsig.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Xu D, Ma P, Gao G, Gui Y, Niu X, Jin B. MicroRNA-383 expression regulates proliferation, migration, invasion, and apoptosis in human glioma cells. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-3378-2. [DOI] [PubMed] [Google Scholar]
- 30.Xu Z, Zeng X, Tian D, Xu H, Cai Q, Wang J, et al. MicroRNA-383 inhibits anchorage-independent growth and induces cell cycle arrest of glioma cells by targeting CCND1. Biochemical and biophysical research communications. 2014;453:833–838. doi: 10.1016/j.bbrc.2014.10.047. [DOI] [PubMed] [Google Scholar]
- 31.Zhao L, Gu H, Chang J, Wu J, Wang D, Chen S, et al. MicroRNA-383 regulates the apoptosis of tumor cells through targeting Gadd45g. PloS one. 2014;9:e110472. doi: 10.1371/journal.pone.0110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Cao C, Tang T, Lu C, Xu J, Wang S, et al. ROBO3 promotes growth and metastasis of pancreatic carcinoma. Cancer letters. 2015;366:61–70. doi: 10.1016/j.canlet.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer research. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 38.Matsuyama H, Pan Y, Yoshihiro S, Kudren D, Naito K, Bergerheim US, et al. Clinical significance of chromosome 8p, 10q, and 16q deletions in prostate cancer. The Prostate. 2003;54:103–111. doi: 10.1002/pros.10173. [DOI] [PubMed] [Google Scholar]
- 39.Matsuyama H, Pan Y, Oba K, Yoshihiro S, Matsuda K, Hagarth L, et al. The role of chromosome 8p22 deletion for predicting disease progression and pathological staging in prostate cancer. Aktuelle Urologie. 2003;34:247–249. doi: 10.1055/s-2003-41607. [DOI] [PubMed] [Google Scholar]
- 40.Matsuyama H, Oba K, Matsuda K, Yoshihiro S, Tsukamoto M, Kinjo M, et al. Haploinsufficiency of 8p22 may influence cancer-specific survival in prostate cancer. Cancer genetics and cytogenetics. 2007;174:24–34. doi: 10.1016/j.cancergencyto.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature medicine. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini S, Majid S, Shahryari V, Arora S, Yamamura S, Chang I, et al. miRNA-708 control of CD44(+) prostate cancer-initiating cells. Cancer research. 2012;72:3618–3630. doi: 10.1158/0008-5472.CAN-12-0540. [DOI] [PubMed] [Google Scholar]
- 43.Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol. 2009;2:361–369. [PMC free article] [PubMed] [Google Scholar]
- 44.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 45.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 46.Liao XH, Zheng L, He HP, Zheng DL, Wei ZQ, Wang N, et al. STAT3 regulated ATR via microRNA-383 to control DNA damage to affect apoptosis in A431 cells. Cellular signalling. 2015;27:2285–2295. doi: 10.1016/j.cellsig.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, et al. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 48.Yin M, Wang X, Yao G, Lu M, Liang M, Sun Y, et al. Transactivation of micrornA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. The Journal of biological chemistry. 2014;289:18239–18257. doi: 10.1074/jbc.M113.546044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooks DD, Wolf A, Smith RA, Dash C, Guessous I. Prostate cancer screening 2010: updated recommendations from the American Cancer Society. J Natl Med Assoc. 2010;102:423–429. doi: 10.1016/s0027-9684(15)30578-2. [DOI] [PubMed] [Google Scholar]
- 50.Wolf AM, Wender RC, Etzioni RB, Thompson IM, D'Amico AV, Volk RJ, et al. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA Cancer J Clin. 2010;60:70–98. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
- 51.Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, et al. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


