Abstract
The developing tooth offers a model for the study of ectodermal appendage organogenesis. The signaling networks that regulate tooth development have been intensively investigated, but how cell biological responses to signaling pathways regulate dental morphogenesis remains an open question. The increasing use of ex vivo imaging techniques has enabled live tracking of cell behaviors over time in high resolution. While recent studies using these techniques have improved our understanding of tooth morphogenesis, important gaps remain that require additional investigation. In addition, some discrepancies have arisen between recent studies, and resolving these will advance our knowledge of tooth development.
Keywords: tooth development, epithelial dynamics, cell behavior, morphogenesis, molecular signaling
Introduction
Ectodermal appendages, including the hair follicle, mammary gland, nail, and tooth, initiate development through a shared program that involves induction and placode formation (reviewed in Biggs and Mikkola, 2014). The mouse tooth provides an excellent model system to investigate the development of these ectodermally-derived organs, as it is relatively large and can be easily accessed and manipulated. Importantly, the signaling pathways that regulate tooth development are well conserved in different tooth types and between species, as well as during the development of related organs (Fraser et al., 2009; Tummers and Thesleff, 2009).
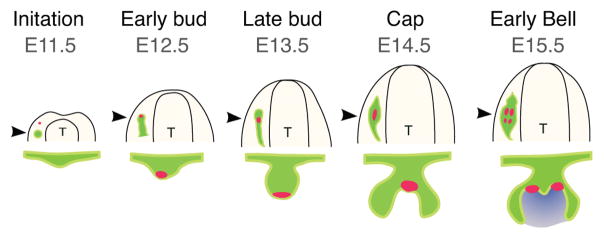
In mice, the first morphological sign of tooth development is observed at embryonic day (E) 11.5, with the appearance of localized thickenings of the oral epithelium known as dental placodes (Fig. 1; reviewed in Tucker and Sharpe, 2004). The instructive information for tooth development resides in the dental epithelium at this stage, and the epithelium has the potential to induce odontogenic fate in any neural crest-derived mesenchyme to form tooth (Mina and Kollar, 1987; Lumsden, 1988). By the early bud stage (E12.5), the placode continues to stratify and invaginate to form the dental lamina, and underlying mesenchyme begins to condense. At this stage, the odontogenic potential shifts to the mesenchyme. Pax9 and Msx1 expression are hallmarks of the dental mesenchyme in the bud and are required for mesenchymal Bmp4 expression, which is important for progression through the subsequent odontogenic stages (Chen et al., 1996; Neubüser et al., 1997; Tucker et al., 1998). By E14.5, the tooth bud extends around the dental mesenchyme (also called the dental papilla) to form a cap-shaped epithelium also known as the enamel organ. A group of post-mitotic cells called the primary enamel knot that forms within the dental epithelium regulates tooth crown development at the cap stage. Signaling molecules secreted by the enamel knot, which include members of the Hh, Wnt, Bmp and Fgf families, play a crucial role in shaping the tooth by maintaining the enamel knot and regulating cell proliferation in adjacent cells that is thought to drive the folding of the epithelium into cusps (Jernvall et al., 1994; Jernvall et al., 2000; Harjunmaa et al., 2014). In addition, the enamel knot expresses p21, Msx2 and Bmp2 to induce growth arrest and subsequently apoptosis of the signaling center (Vaahtokari et al., 1996; Aberg et al., 1997; Jernvall et al., 1998; Thesleff et al., 2001). At E15.5–17.5, the epithelium continues to invaginate, and concurrently, secondary enamel knots form at a distance from the disintegrating primary enamel knot and control the shape of the tooth cusps (Thesleff et al., 2001; Matalova et al., 2005). The shape and size of the tooth is determined throughout these stages, and thus, the early odontogenic stages are critical for determining adult dental morphology. During late odontogenic stages, dental epithelial and mesenchymal cells differentiate into ameloblasts and odontoblasts that lay down enamel and dentin, respectively. Other cell types, such as cementoblasts and cells of the periodontal ligament, differentiate at these stages as well.
Figure 1.
Early stages of molar development. The first morphological sign of molar development is observed at E11.5, with the localized thickening of the oral epithelium to form dental placode. The placode continues to invaginate and grow into distinct 3D shapes during subsequent stages. The shape and size of the tooth is determined throughout these stages, and thus, the early odontogenic stages are critical for determining the adult dental morphology. Each stage is regulated by molecular cues from the signaling center. Top row illustrates the oral surface view of the whole mandible at respective embryonic (E) days. Bottom row illustrates frontal sections of the tooth germ at the locations indicated by arrowheads. T: tongue. Green: dental epithelium. Purple: dental papilla. Red: signaling center.
While the field has accumulated a wealth of knowledge regarding the molecular regulation of tooth development, how individual cells translate these molecular signals into an integrated morphological output remains unclear. One constraint has been that, until recently, the cellular behaviors that drive tooth morphogenesis have been studied using static histological analyses. While such an approach offers us clues about the cellular basis of morphogenesis, it limits our understanding of tissue behaviors, as we are limited to deducing highly dynamic, three-dimensional cellular activities from sequential, often two-dimensional, snapshot analyses. For instance, rigorous histological analysis of the developing incisor placode led to the proposal that an early signaling center located within the incisor field is a rudimentary tooth that subsequently undergoes apoptosis and disintegrates (Hovorakova et al., 2011). However, a recent study using ex vivo imaging suggested that this early signaling center may function as an integral part of the developing incisor (Ahtiainen et al., 2016). In addition, two-dimensional analysis often involves tissue treatments that could introduce artifacts. For example, it was only recently discovered that the molar epithelial progenitor cells initially arrange into a rosette-like structure at the placode stage of tooth development. This is because the structure is sensitive to formaldehyde treatment and thus becomes disrupted upon fixation (Prochazka et al., 2015). For these reasons, the field has moved towards investigating cellular processes under conditions that better resemble in vivo development.
Recently, several groups have proposed different models for how a tooth germ develops using ex vivo imaging techniques on live tissue to link genetics and signaling with the physical morphogenesis of the tooth. Importantly, live explants faithfully recapitulate normal development in terms of tissue morphogenesis, and many gene expression and differentiation markers appear as normal. Below, we review these new findings and propose potential future directions.
Molar Signaling Centers
The textbook version of the signals governing molar tooth development is that the process is initiated and organized by a single signaling center, the initiation center or primary enamel knot precursor, in the middle of each placode. However, recently more detailed and dynamic analysis has revealed a more complicated picture. The Fibroblast growth factor (Fgf), Sonic Hedgehog (Shh), Wnt and BMP signaling pathways all play multiple roles. In particular, Fgfs, including Fgf8 and 9, are expressed in the dental placode, and Fgf10 is expressed in the mesenchyme from the placode’s earliest appearance. Fgf8 expression appears even earlier in the entire proximal half of the jaw epithelium (Neubüser et al., 1997; Kettunen and Thesleff, 1998; Prochazka et al., 2015; Kettunen et al., 2000). Fgf9- or Fgf10-null mice form mineralized teeth with mild phenotypes, suggesting redundancy of FGF ligands (Yokohama-Tamaki et al., 2006; Haara et al., 2012). However, tissue-specific deletion of Fgf8 arrested tooth development before the early bud stage, indicating that Fgf8 cannot be compensated for by other Fgf family members during early tooth development (Prochazka et al., 2015).
It has been recently reported that the conventionally understood signaling center for the molar placode, as marked by expression of Shh, is at least transiently located distally to the molar placode itself, the latter being defined by cell movements, epithelial thickening and up-regulated Fgf8 expression; this was detected by genetic labeling of the Shh- and Fgf8-expressing populations in a mouse embryo using the double driver strain ShhEgfp/Cre;Fgf8lacZ (Prochazka et al., 2015). Surprisingly, the proximal Fgf8 center also expresses Shh target genes, despite not apparently expressing Shh itself. It still remains unclear at what points the Fgf8-expressing molar placode and the Shh-expressing signaling center are distinct and whether, somehow, these coalesce. Interestingly, expression of Etv4, which is considered to be a direct transcriptional target of the Fgf pathway, is restricted to the molar placode, despite the broad expression of Fgfr2 across the oral epithelium. This suggests selective activation of canonical Fgf signaling in dental epithelium that is under tight temporospatial control. To confirm this, it would be important to analyze the expression of additional Fgf pathway targets, as some of the target genes may differentially respond to signaling inputs (Roehl and Nüsslein-Volhard, 2001). In addition, future experiments involving assessment of temporospatial expression patterns as well as conditional inactivation of Fgf pathway regulators will improve our understanding of the dynamic regulation of Fgf signaling in the molar placode. Several negative feedback regulators of Fgf signaling are expressed in the developing tooth, and these are prime candidates for control of these processes (reviewed in Thisse and Thisse, 2005). For instance, the Sprouty (Spry) gene family has been shown to regulate Fgf signaling in odontogenesis, and loss of Spry2 and Spry4 both result in persistence of Fgf signaling during early tooth development (Klein et al., 2006). Mkp3 is another negative feedback regulator that is expressed adjacent to the Fgf8-expressing domain in the developing limb, at the midbrain/hindbrain boundary, and in the maxillary, mandibular and frontonasal prominence (Klock and Herrmann, 2002). Mkp3 mRNA is also detected in the oral cavity at E11.5, and thus it will be interesting to test if the Mkp3 expression pattern complements Fgf8 expression within the Fgfr2-expressing domain. In addition, a large suite of other negative and positive regulators of the Fgf pathway exists that merit further study.
Molar Progenitor Cell Migration
A very early event in tooth development was studied by Prochazka et al., who proposed that directed cell migration of transiently Fgf8-expressing cells is involved in formation of the tooth germ (Fig. 2A and B). The authors discovered that the expression domain of Fgf8 in the mandibular epithelium is predominantly in the proximal region of the mandible at E11.5, the placode stage of tooth development (Prochazka et al., 2015). Genetic lineage tracing together with ablation of this population revealed that Fgf8-expressing cells are progenitors that give rise to the entire molar epithelium and thus are required for progression through subsequent odontogenic stages. Building on this knowledge, the authors used Fgf8-driven tamoxifen-dependent Cre recombinase to study the cellular dynamics in the molar placode at E11.5. They showed that, at E11.5, the dental epithelial progenitor cells at the proximal region adopt elongated shapes and arrange into a large (perhaps 100–200 μm diameter), rosette-like structure. Such anterior movement would bring these cells towards the above-mentioned Shh-expressing center. Analyses of cells labeled using Fgf8ires-Cre revealed that the cells within the rosette-like structure underwent more directed and dynamic cell migration, compared to cells that are located posteriorly to the rosette-like structure (Fig. 3A and B). Genetic and pharmacological perturbation of the Fgf and Shh pathways resulted in deregulated migration of these dental epithelial cells. However, the two pathways may control different aspects of cell migration. Upon chemical inhibition of the Fgf pathway, using a relatively low dose of the inhibitor SU5402, cell migration was no longer visible; whether this was due to arrest of migration as such, or to the non-formation of a migratory suprabasal layer of cells, remains an open question, as discussed below. Upon chemical inhibition of the Shh pathway, the placodal cells still moved in the general distal-ward direction of the future tooth germ but failed subsequently to converge towards the center of the future tooth. Interestingly, this low-dose pharmacological inhibition of either Fgf or Shh signaling had no apparent effects on cell proliferation and apoptosis. However, as discussed next, the role of these pathways is still somewhat unresolved.
Figure 2.
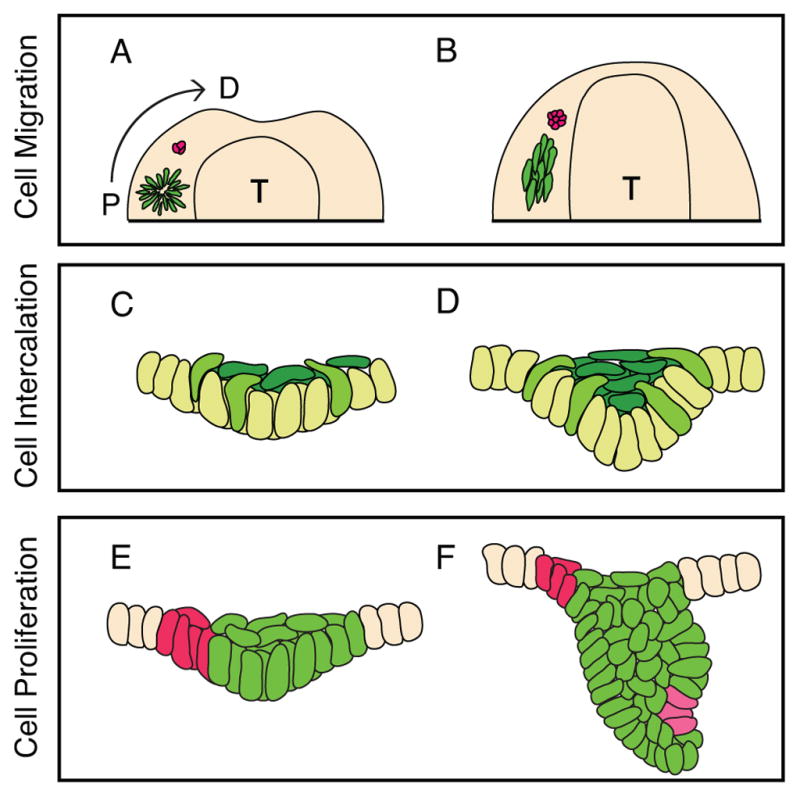
Cell movements in early tooth development. A,B: Prochazka et al. proposed that, viewing the mandible from above, progenitor cells in the molar placode (purple) arrange into a rosette-like structure, then subsequently dissociate to migrate towards a Shh-expressing signaling center (red). C,D: In the model developed by Panousopoulou and Green, shown in frontal aspect, basal cells in the molar placode delaminate towards the center of the placode (yellow-green). The delaminated cells form the suprabasal tissue canopy (dark green), which intercalate along the midline to drive epithelial invagination. E,F: Ahtiainen et al. proposed that, in incisors, a population of non-proliferating cells that co-localizes with morphogen expression (red) instructs the neighboring epithelial cells (green) to proliferate, and this leads to tooth morphogenesis. The enamel knot forms de novo at the tip of the tooth germ (pink). T: tongue. A and C are E11.5. B and D are E12.5. E is E12.0. F is E13.5.
Figure 3.
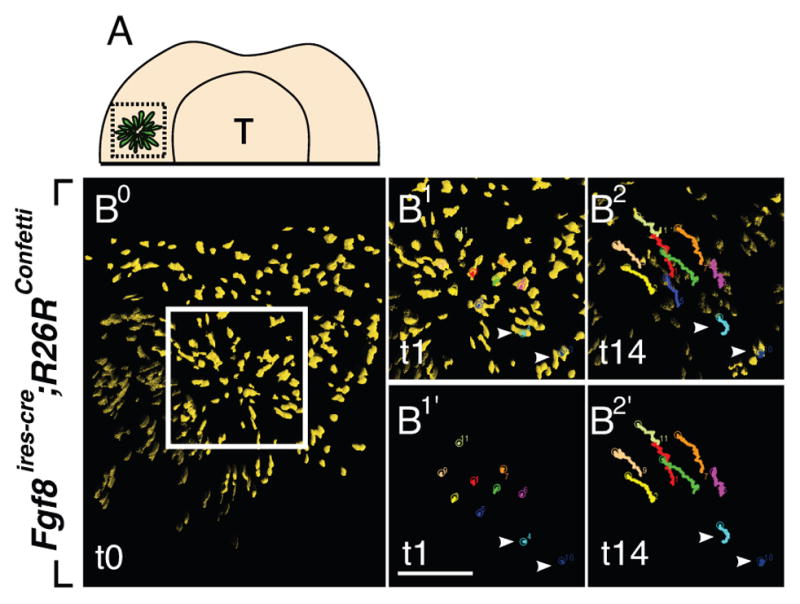
Fgf and Shh signaling instruct molar placodal cells to undergo directed cell migration. A: Schematic presentation of Fgf8-expressing molar placodal cells from oral surface view at E11.5. Dotted square demarcates the molar placode region. B-B2′: Time-lapse imaging via spinning disk confocal microscopy reveals that Fgf8-expressing cells within the rosette-like structure are released and migrate distally. Tracking of the cells within the rosette-like structure (white box) over time (B1-B2 and B1′-B2′). Arrowheads (lower right) indicate posterior cells that do not undergo dynamic migration. Scale bar: 100μm. Reproduced with permission from Elsevier.
Prochazka et al. also observed that genetically hyperactivating Shh signaling, using Fgf8CreER;R26RSmoM2, resulted in distal expansion of the tooth germ into the normally toothless diastema region that separates the incisor from the molars. It has been previously reported that two successive and transient bud-like structures in the diastema may be the rudimentary precursors of the premolars present in ancestral species (Prochazka et al., 2010). These rudimentary buds can develop into a supernumerary tooth in several mutants (Klein et al., 2006; Lochovska et al., 2015). It will be interesting to further investigate if the expanded tooth germ upon increased Shh activity can contribute to supernumerary tooth formation.
Molar Epithelial Stratification and Bending
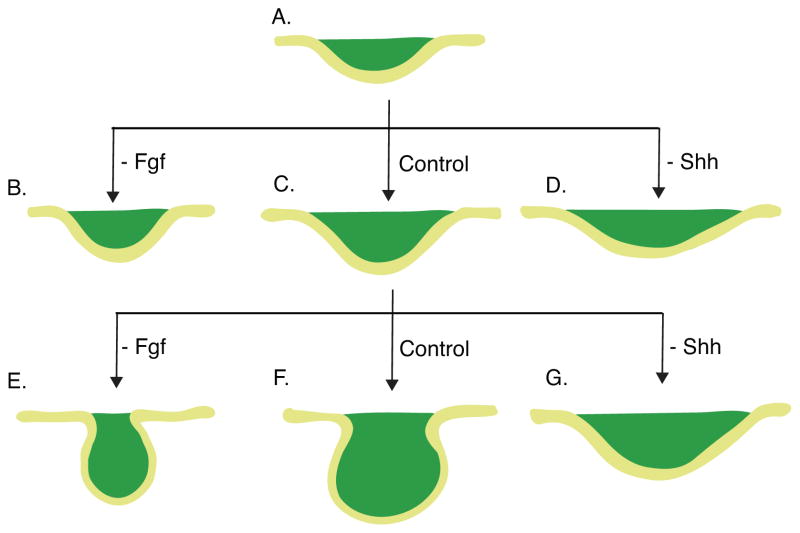
A second recent paper, by Li et al., demonstrates regulatory roles for the Fgf and Shh pathways in early tooth morphogenesis in addition to instructing epithelial cell migration (Li et al., 2016b). In this paper, it was reported that chemical inhibition of the Fgf pathway in E11.5 mandibles significantly impaired the proliferation of the dental epithelial cells, resulting in failure of epithelial stratification of the molar tooth germ (Figs. 4B). However, inhibiting the Fgf pathway after E11.5, when some stratification had already occurred, arrested further proliferation but did not prevent invagination of the developing tooth germ (Fig. 4E). In a complementary experiment, the authors ectopically activated the Fgf pathway by introducing FGF10-soaked beads to single-layered tongue epithelium. Exposure to recombinant FGF10 promoted the stratification of tongue epithelium, suggesting that Fgf signaling is both necessary and sufficient to induce epithelial stratification.
Figure 4.
Fgf and Shh signaling promote stratification and epithelial bending, respectively. A–D: The molar placode (A) at E11.5 stratifies and bends into early tooth bud (C). However, inhibition of Fgf signaling results in significantly shallower tooth bud (B). Inhibiting the Shh pathway results in a shallower and wider tooth bud (D). E–G: Similar observation is made when the early tooth bud (C, E12.5) is treated with Shh and Fgf inhibitors. Early tooth bud further stratifies and invaginates into late tooth bud (F). However, inhibition of Fgf results in narrower tooth germ (G), while inhibiting Shh results in shallower and wider tooth germ (E). A: E11.5. B–D: E12.5. E–G: E13.5.
The effects of FGF inhibition on proliferation were thus different from those reported by Prochazka et al. One possible explanation for the discrepancies could be the different level of pharmacological treatment. Prochazka et al. exposed the whole mandible explant to 2.5 μM of Fgf pathway inhibitor (SU5402), whereas Li et al. treated mandibular tissue slice culture with 20 μM SU5402. Clearly, the higher dose applied to a smaller piece of tissue could have had different effects, although it was found still to be well below that needed to produce cell death (Li et al., 2016). Several studies have shown that the Fgf pathway can elicit differential cellular responses, including cell proliferation, migration, and differentiation, through activation of different downstream pathways (reviewed in Boilly et al., 2000). For instance, half maximal activity of cell migration required higher FGF concentrations compared to cell proliferation in rat lens epithelial explants (McAvoy and Chamberlain, 1989). It is possible that the dosage used by Prochazka et al. was sufficient to perturb the cell migration, while having a negligible effect on cell proliferation. It is also possible that different durations of drug exposure could have led to different cellular responses, or that differences in experimental setup could explain the distinct outcomes. Further investigation of dose- and duration-dependent response of the dental placode cells to SU5402 through analyzing different downstream pathways would help resolve this discrepancy. Also, the role of Fgf signaling in the dental mesenchyme should be investigated, as Fgfr1IIIc is expressed in the dental mesenchyme at the earliest stages of tooth development (Kettunen and Thesleff, 1998). Reconciling the observations this way would also require that inhibition of the early migration not affect the later cell movements associated with deepening invagination. A recent study showed that the inhibitory effect of SU5402 may not be limited to Fgf signaling, but rather can extend to other tyrosine kinases, and differential dosage and duration of SU5402 treatment may also elicit distinct off-target effects (Gudernova et al., 2016). Therefore, more specific suppression of Fgf signaling, perhaps through genetic deletion of Fgf receptors, may help to address this issue.
Interestingly, the dental placode increased in width at the expense of the depth of invagination upon chemical inhibition of Shh signaling (Figs. 4D and G). The resulting shallower, wider invagination has also been observed in Shh and Hedghog pathway genetic hypomorphs and knockouts, not only in tooth but also in hair follicles, which are thought to be formed at these early stages by much the same signaling and morphogenetic processes (reviewed in Pispa and Thesleff, 2003; St-Jacques et al., 1998; Gritli-Linde, 2002; Prochazka et al., 2015). In the Li et al. study, hyperactivation of the pathway resulted in the opposite phenotype, with accelerated narrowing and deepening of the placode invagination. In both cases, careful quantification showed that there was no significant change in overall cell proliferation rate and apoptosis. These results collectively indicate that the Shh pathway has a regulatory role in epithelial cell rearrangement. The Fgf and Shh pathways together may promote asymmetric cell proliferation that leads to local epithelial stratification and regulation of cell tension and/or motility that leads to epithelial bending. The authors suggest that these processes jointly drive the invagination of the molar epithelium into the underlying mesenchyme.
The Li et al. study demonstrated that Fgf and Shh signaling can promote cell proliferation and epithelial bending, respectively, through gain- and loss-of-function approaches. One important point that could be followed up in the future is that the experimental framework used in the gain-of-function experiments exposed the entire explant to chemical Shh agonist. The lack of a restricted or graded spatial distribution of signal and the degree of signaling activity that may be outside the physiological range may have led to cells not responding in a physiological manner, although a similar effect was seen with SHH protein applied via a protein-soaked bead (Cobourne et al., 2004). It is also worth noting that the experiments could not distinguish between roles of the Shh signal in orienting cell migration versus triggering or maintaining migration as such. Because expression of Shh in the tooth germ is spatially separated from the early molar placode and localized to the distal tip of the early tooth bud, it is even possible that tissue slices, which would normally successfully have undergone invagination, did not contain the endogenous source of Shh (Prochazka et al., 2015). Verifying the endogenous Shh expression within the tissue slice culture would strengthen the relationship between the experimental observations and events that occur in vivo.
Suprabasal Cell Intercalation-Mediated Contraction
One possible cellular mechanism through which Shh signaling promotes epithelial bending is through interaction of cells in the suprabasal layer, as demonstrated by the Li et al. paper described above and another from the same group (Panousopoulou and Green, 2016). Using the molar placode as a model, these authors showed that invagination of the placode is driven by Shh-dependent horizontal intercalation of suprabasal cells (Fig. 2C and D). These suprabasal layers of cells in the tooth germ form a shrinking and thickening canopy over underlying epithelial cells. Experimental incision through the epithelium and mesenchyme that flank the molar placode in jaw slice explants resulted in elastic recoil from the incision site (Fig. 5A). When the incision was introduced specifically in the suprabasal layer, but not the basal layer, the suprabasal layer recoiled laterally from the cut. When an initial incision through the suprabasal cell layer was followed by lateral incision of the flanking tissues, this essentially abolished the recoil of the molar placode (Fig. 5B). These data suggest that the suprabasal cells, but not the basal cell layers, generate a tensile force sufficient and necessary to invaginate the molar placode. The intercalating tissue is, critically, anchored at its edges to the basal lamina by shoulder cells that both contact the lamina basally and intercalate apically. Crucially, Hedgehog pathway inhibition arrested the suprabasal canopy contraction and abolished the intercalation-associated planar cell elongation in both suprabasal and shoulder cells (Li et al., 2016b).
Figure 5.
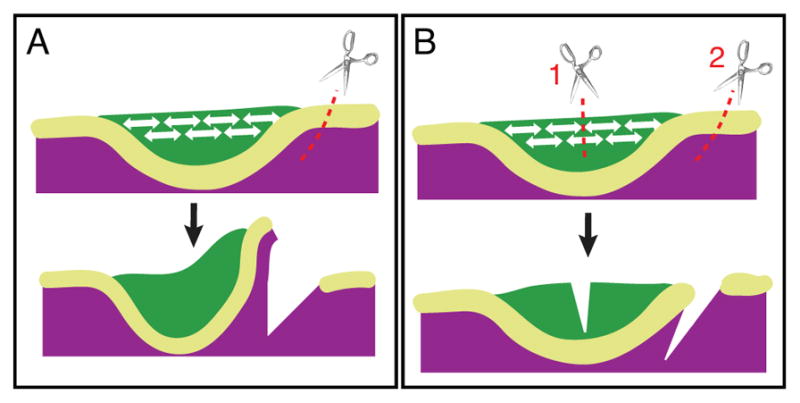
Contractile suprabasal tissue promotes bending of molar placode. A: Single lateral incision in the epithelium that flanks the molar placode at E11.5 allows rapid bending of the molar placode. B: Incision through the suprabasal layer (cut 1) abolishes the lateral cut- (cut 2) induced bending, showing that bending depends on suprabasal tension. Green: suprabasal layer. Yellow-green: basal layer. Purple: mesenchyme.
Given that the above experiments were done in slice explants, effects of the inhibition on long-range planar migration, of the kind observed by Prochazka et al., could not be tested. As with the Fgf inhibition, there might seem to be a discrepancy between the two sets of Shh inhibition experiments. The Prochazka et al. work showed little effect of Hedgehog pathway inhibition on cell motility as such, and the Li et al. treatments in effect arrested cell movement. However, in the both cases there was a definite inhibition of cell convergence, and this may indicate that the Shh signal provides a directional cue rather than a migratory signal as such, and that the later intercalation of cells in the invaginated tooth bud may be more sensitive to its absence than the earlier superficial migration. It would be interesting to see how Shh signaling inhibition affects cell movement and intercalation in real time using time-lapse confocal imaging on pharmacologically treated tissue. Further insights into the cellular response to Shh may be needed, since Gli1 expression is widespread in the tooth and gives few clues as to how it might provide directionality.
Panousopoulou and Green observed enriched actomyosin complex and E-cadherin punctae at the surfaces of the suprabasal cells. They proposed that E-cadherin-based spot adherens junctions enable the cell intercalation, which in turn pulls the tissue towards the midline to hold its morphology. The authors suggested that this process also seals the top of the tooth germ so that cell proliferation and delamination below the intercalating layer further forces the underlying epithelial cells to invaginate into the mesenchyme. The intercalating suprabasal cells are replenished, in part by the incorporation of delaminating basal cells. The authors 3D-rendered the nuclear morphology in the molar placode, using deformation from a spherical shape to reveal the external forces experienced by the cells. They showed that the tension in the suprabasal layer stretched the nuclei from the default sphere shape of unstressed cells into a flattened “lentil” shape, indicating the equal magnitude of force exerted in all directions in the epithelial plane. It is apparent that the cells experience a tensile force along the lingual-buccal axis that results in the most commonly depicted epithelial invagination, but that a similar force in the proximo-distal axes of the jaws sustains invagination in that aspect to form the front and rear of the elongated pit or “boat” shape of the early molar tooth. It will be interesting to further assess which cell behaviors contribute to this proximo-distal force.
Cell Behaviors Along Different Axes
The Prochazka et al. and the Panousopoulou, Li and Green experiments describe two quite different-seeming cell migrations. One involves distal migration of cells distally from the jaw hinge while the other is a convergence of cells into the invaginating tooth bud. One is apparently FGF-dependent, the other FGF-independent. One is apparently Hedgehog pathway oriented, the other, Hedghog pathway dependent. How can these discrepancies be accounted for? One potential explanation involves the different explant culture techniques used to image the tissue. The directed cell migration events proposed by Prochazka et al. are supported by live-tissue imaging of E11.5 hemi-mandible using spinning disk confocal microscopy. The entire hemi-mandible was imaged en face from the oral surface, and the images obtained were processed by maximal intensity projection for cell tracking analysis. This computational method projects the three-dimensional information into two dimensions, and the subsequent analysis does not focus on the tracking information along the rostral-caudal axis. In contrast, Panousopoulou and Green’s suprabasal cell intercalation hypothesis is based on imaging of tooth slices. This method allows observation of the cell behavior that occurs along the vertical (superficial-to-deep) axis without compromising the resolution. However, the tissue slice preparation would likely remove more proximal tissue. Since Prochazka et al. propose that cell migration towards the Shh-signaling center comes from more proximal tissue, this would be largely or entirely absent in the slice cultures. The successful invagination of the placode, which apparently occurs normally in slice cultures, despite the likely absent proximal tissue (near the jaw hinge), suggests that the cell migration and cell intercalation are independent events. It is even possible that slice culture removes critical distal signals or perturbs proximo-distal signals in other ways. Another possibility is that the two events take place in a sequential manner: the dental epithelial cells first migrate distally and define the position and the size of the future tooth germ along the proximo-distal axis, and then they start to proliferate and intercalate to grow and invaginate into the underlying mesenchyme. Various factors can influence the developmental schedule of embryogenesis (e.g. genetic background, transgenic and mutant alleles, and tamoxifen administration) (Naiche and Papaioannou, 2007; Dandekar and Glass, 1987). Therefore, it is possible that the staging of embryos in the two studies were slightly different. One conceptually simple experiment to test this would be to perform live imaging on intact jaw explants (i.e. not slices) in a single genetic background using multiphoton microscopy, which may allow for greater imaging depth compared to traditional spinning disk confocal microscopy, and then analyzing the cell behaviors in four-dimension.
Formation of Signaling Centers in Early Incisor Development
Many signaling cascades, including the Shh and Fgf pathways, are iteratively activated in both the incisor and the molar placodes. A recent study by Ahtiainen et al. performed ex vivo analysis of the developing incisor placode in a fluorescent ubiquitination–based cell cycle indicator (Fucci) mouse to understand the molecular regulation of cell behaviors in the incisor placode (Ahtiainen et al., 2016). The authors demonstrate that there is a population of non-mitotic cells that resides in the incisor placode near the oral surface. Time-lapse confocal imaging reveals that while these cells actively migrate towards the midline of the tooth bud to condense, the total number of cells remains relatively constant. Once condensed, these cells may act as a signaling center to regulate the early stages of incisor development (Fig. 2E), as the location of the non-mitotic cells coincides with the expression of signaling molecules that regulate tooth development.
The authors define this early population of quiescent, morphogen-secreting cells as the “initiation knot”. Cell quiescence is a common feature of signaling centers in various organs, including the apical ectodermal ridge of the developing limb bud, the developing hair placode, and the enamel knot in the developing tooth germ (e.g., Martin, 1998; Ahtiainen et al., 2014; Jernvall et al., 1994). Notably, the authors reported that the initiation knot, which appears very early, and the enamel knot, which appears later during odontogenesis, are distinct populations. Tracking individual cells in the initiation knot of the mandibular explant revealed that these cells remained within clear boundaries near the oral surface, whereas the enamel knot appeared de novo at the tip of the mature bud (Fig. 1F). This observation is consistent with a previous study that demonstrated, using finely-staged embryos, that the two spatially distinct signaling centers appear sequentially (Hovorakova et al., 2011; Hovorakova et al., 2013).
While the cells in the initiation knot remain quiescent, they may promote proliferation in the neighboring cells to drive tooth morphogenesis. To test this possibility, the authors use Tabby mice, which exhibit smaller tooth and cusp patterning defect due to mutations in the Eda/Edar/NF-κB pathway (Pispa et al., 1999; Harjunmaa et al., 2014). The authors found that the overall number of quiescent cells and the volume of the initiation knot were reduced in Tabby mice. In turn, the smaller initiation knot resulted in reduced expression of genes that are important for progression of tooth development, suggesting a link between the size of the initiation knot and that of the resulting tooth bud. One alternative explanation would be that the smaller initiation knot and tooth bud are independently initiated but sequential events. While it is likely that the tooth bud depends on specific molecular signals from the initiation knot, it could also be that the Eda/Edar/NF-κB pathway determines the size of the dental anlage, and the smaller initiation knot would be a secondary result of a hypomorphic tooth germ. One way to address this would be to perform spatiotemporal ablation of the initiation knot cells. Another remaining question is whether the smaller tooth germ in Tabby mice results from reduced mitogenic signal in the initiation knot. This can be addressed in the future by unbiased molecular characterization of the initiation knot and quantitative analysis of cell proliferation in the tooth germ of Tabby mice.
Cell Behaviors in Molar vs. Incisor Epithelium
Cell migration has been observed in various morphogenetic contexts (reviewed in Aman and Piotrowski, 2010). As described above, molar placode cells migrate and this is essential for formation of the tooth germ; however, in the incisor, as observed by Ahtiainen et al., the migration seems to be limited to the cells in the initiation knot, whereas cell proliferation seems to be the main driving force for subsequent invagination. One possible reason for this discrepancy is the difference in the developmental timing of incisor and molar. Ahtiainen et al. studied similar embryonic stages (E11.5 and E12.0) to those in the molar-focused papers reviewed above. However, because development of the incisor is faster than that of molar, it is likely that Ahtiainen et al. analyzed the cell behaviors at more advanced stages of tooth development. Later stages in molar morphogenesis were not analyzed by either Prochazka et al. or the Green group, and it would be interesting to see if later stages of invagination are, like the incisor, dominated more by proliferation processes than by cell movement. Conversely, an examination of incisor at earlier stages and at higher resolution might reveal similarities to the molar, since Ahtiainen et al. made their observations at relatively low magnification. Another possibility is that signaling centers in incisors and molars have qualitatively different instructive roles. It is important to note that the molar signaling center is spatially distinct from the molar placode, whereas the incisor signaling center (i.e. initiation knot) is located within the incisor field. Moreover, morphogenesis of the incisor and the molar placode differ on a three-dimensional level. The incisor placode maintains its location and adopts a relatively rounder shape as it develops, whereas the molar placode undergoes significant expansion along the proximo-distal axis to adopt its cylindrical shape. Therefore, it is conceivable that cells respond differently to the same signals to contribute to the morphogenesis of the two tooth types.
Conclusions and Future Prospects
The studies discussed in this review have enhanced our understanding of how molecular signaling translates into dental epithelial cell dynamics. However, many mysteries remain, and it is certain that tooth morphogenesis is more complex than our current understanding. Most studies of tooth development have focused on the epithelial contribution due to the availability of genetic tools and the tissue’s well-defined structure. However, it is well known that epithelium and mesenchyme exchange highly dynamic and reciprocal signaling to orchestrate tooth development. The dental mesenchyme takes on an instructive role between E11.5 and E12.5, during which placodal invagination occurs, suggesting that mesenchymal signaling is critical for directing epithelial invagination (Mina and Kollar, 1987). Panousopoulou and Green showed by explantation that the initial invagination forces in the molar were autonomous to the epithelium, but it is theoretically possible that the dental mesenchyme coordinately undergoes remodeling to facilitate invasion by the invaginating epithelium (Panousopoulou and Green, 2016). The presence of abundant F-actin and phosphomyosin in jaw mesenchyme also allows for the possibility of a role of mesenchyme during tooth development. Therefore, investigating dental mesenchymal dynamics would be a logical next step to further our understanding of tooth morphogenesis. Recently, tamoxifen-inducible Cre lines under control of promoters of early dental mesenchymal markers, such as Fgf10 and Msx1, have been generated, and these may be useful for future studies (El Agha et al., 2012; Lallemand et al., 2013). In parallel, the field has also begun to explore dental mesenchymal cell dynamics through combining ex vivo imaging with tissue recombination techniques (Li et al., 2016a; Mammoto et al., 2011).
In addition, there are a number of exciting studies that investigate cellular behaviors underlying different processes of tooth morphogenesis which have not been highlighted in this review. While this review focuses on dental placodal invagination, it is equally interesting to understand the process through which placodes are formed. Li et al. suggest that the vertical mitotic angle of placodal cells relative to the basement membrane may promote the thickening, suggesting that perturbation of genes known to be involved in regulating mitotic orientation could further this line of investigation (Li et al., 2016b). Another group explored the mechanism through which bud-to-cap transition occurs. The tooth germ at E12.5, which is essentially an invaginated bud of epithelium, deforms into a distinct shape that resembles a cap by E14.5 (Fig. 1). To understand this process, Morita et al. combined long-term live imaging with Fucci cell cycle analysis, correlating the tissue growth pattern with mitotic orientation and frequency in the tooth germ at E14.5 (Morita et al., 2016). The authors reinforced previous studies showing that differential cell proliferation is one of the driving forces of tooth morphogenesis (Jernvall et al., 1994). In addition, through quantitative analysis of fluorescence recovery of YFP-actin after photobleaching, the authors showed that dynamic actin remodeling might play a role in tooth germ morphogenesis.
Another interesting question is the mechanism through which the second and third molars develop. Both humans and mice develop three molars per jaw quadrant, with each subsequent molar forming at the posterior of the previous tooth germ. Gaete et al. explored this phenomenon at tissue and cellular level. Through combining explant culture with DiI labeling and tissue truncation, they found that second and third molars are derived from progenitors that reside in the “tail” of the first molar tooth germ, a structure more superficial than the bulk of the tooth germ (Gaete et al., 2015). This finding complements previous work examining development of molars (Juuri et al., 2013). Investigating different stages of tooth morphogenesis will provide us with an understanding of how the dental organ derives its characteristic 3D shape from a flat epithelial sheet.
The tooth has long been an important model for developmental biologists. A deeper understanding of the morphogenetic events in tooth development will be of broad significance to the field of organogenesis, as many signaling pathways and cellular events in tooth development are not restricted to odontogenesis, but rather are deployed in various developmental contexts.
Acknowledgments
The authors were funded by NIDCR grants R35-DE026602 to O.D.K. and F30-DE025160 to R.K
We are grateful to Drs. Jimmy Hu, Amnon Sharir, Jan Prochazka, and members of the Klein laboratory for helpful discussions and critical reading of the manuscript, and to Dr. Adriane Joo for assistance with the figures. The authors were funded by NIDCR grants R35-DE026602 to O.D.K. and F30-DE025160 to R.K.
References
- Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210:383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Ahtiainen L, Lefebvre S, Lindfors PH, Renvoise E, Shirokova V, Vartiainen MK, Thesleff I, Mikkola ML. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev Cell. 2014;28:588–602. doi: 10.1016/j.devcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Ahtiainen L, Uski I, Thesleff I, Mikkola ML. Early epithelial signaling center governs tooth budding morphogenesis. The Journal of Cell Biology. 2016;214:753–767. doi: 10.1083/jcb.201512074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman A, Piotrowski T. Cell migration during morphogenesis. Developmental Biology. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Mikkola ML. Early inductive events in ectodermal appendage morphogenesis. Seminars in Cell & Developmental Biology. 2014;25–26:11–21. doi: 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Boilly B, Vercoutter-Edouart AS, Hondermarck H, Nurcombe V, Le Bourhis X. FGF signals for cell proliferation and migration through different pathways. Cytokine & Growth Factor Reviews. 2000;11:295–302. doi: 10.1016/s1359-6101(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Miletich I, Sharpe PT. Restriction of sonic hedgehog signalling during early tooth development. Development. 2004;131:2875–2885. doi: 10.1242/dev.01163. [DOI] [PubMed] [Google Scholar]
- Dandekar PV, Glass RH. Development of mouse embryos in vitro is affected by strain and culture medium. Gamete Res. 1987;17:279–285. doi: 10.1002/mrd.1120170402. [DOI] [PubMed] [Google Scholar]
- El Agha E, Al Alam D, Carraro G, MacKenzie B, Goth K, De Langhe SP, Voswinckel R, Hajihosseini MK, Rehan VK, Bellusci S. Characterization of a novel fibroblast growth factor 10 (Fgf10) knock-in mouse line to target mesenchymal progenitors during embryonic development. PLoS One. 2012;7:e38452. doi: 10.1371/journal.pone.0038452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An Ancient Gene Network Is Co-opted for Teeth on Old and New Jaws. Plos Biology. 2009;7:233–247. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaete M, Fons JM, Popa EM, Chatzeli L, Tucker AS. Epithelial topography for repetitive tooth formation. Biol Open. 2015;4:1625–1634. doi: 10.1242/bio.013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde ABM, Maas R, Zhang XM, Linde A, McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- Gudernova I, Vesela I, Balek L, Buchtova M, Dosedelova H, Kunova M, Pivnicka J, Jelinkova I, Roubalova L, Kozubik A, Krejci P. Multikinase activity of fibroblast growth factor receptor (FGFR) inhibitors SU5402, PD173074, AZD1480, AZD4547 and BGJ398 compromises the use of small chemicals targeting FGFR catalytic activity for therapy of short-stature syndromes. Hum Mol Genet. 2016;25:9–23. doi: 10.1093/hmg/ddv441. [DOI] [PubMed] [Google Scholar]
- Haara O, Harjunmaa E, Lindfors PH, Huh SH, Fliniaux I, Aberg T, Jernvall J, Ornitz DM, Mikkola ML, Thesleff I. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development. 2012;139:3189–3199. doi: 10.1242/dev.079558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjunmaa E, Seidel K, Hakkinen T, Renvoise E, Corfe IJ, Kallonen A, Zhang ZQ, Evans AR, Mikkola ML, Salazar-Ciudad I, Klein OD, Jernvall J. Replaying evolutionary transitions from the dental fossil record. Nature. 2014;512:44–48. doi: 10.1038/nature13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovorakova M, Prochazka J, Lesot H, Smrckova L, Churava S, Boran T, Kozmik Z, Klein O, Peterkova R, Peterka M. Shh expression in a rudimentary tooth offers new insights into development of the mouse incisor. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2011;316B:347–358. doi: 10.1002/jez.b.21408. [DOI] [PubMed] [Google Scholar]
- Hovorakova M, Smrckova L, Lesot H, Lochovska K, Peterka M, Peterkova R. Sequential Shh expression in the development of the mouse upper functional incisor. J Exp Zool B Mol Dev Evol. 2013;320:455–464. doi: 10.1002/jez.b.22522. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Keranen SV, Thesleff I. Evolutionary modification of development in mammalian teeth: quantifying gene expression patterns and topography. Proc Natl Acad Sci U S A. 2000;97:14444–14448. doi: 10.1073/pnas.97.26.14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the Role of the Enamel Knot as a Control Center in Mammalian Tooth Cusp Formation - Nondividing Cells Express Growth-Stimulating Fgf-4 Gene. International Journal of Developmental Biology. 1994;38:463–469. [PubMed] [Google Scholar]
- Juuri E, Jussila M, Seidel K, Holmes S, Wu P, Richman J, Heikinheimo K, Chuong CM, Arnold K, Hochedlinger K, Klein O, Michon F, Thesleff I. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development. 2013;140:1424–1432. doi: 10.1242/dev.089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen P, Laurikkala J, Itaranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock A, Herrmann BG. Cloning and expression of the mouse dual-specificity mitogen-activated protein (MAP) kinase phosphatase Mkp3 during mouse embryogenesis. Mechanisms of Development. 2002;116:243–247. doi: 10.1016/s0925-4773(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Lallemand Y, Moreau J, Cloment CS, Vives FL, Robert B. Generation and characterization of a tamoxifen inducible Msx1(CreERT2) knock-in allele. Genesis. 2013;51:110–119. doi: 10.1002/dvg.22350. [DOI] [PubMed] [Google Scholar]
- Li CY, Hu J, Lu H, Lan J, Du W, Galicia N, Klein OD. alphaE-catenin inhibits YAP/TAZ activity to regulate signalling centre formation during tooth development. Nat Commun. 2016a;7:12133. doi: 10.1038/ncomms12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chatzeli L, Panousopoulou E, Tucker AS, Green JBA. Epithelial stratification and placode invagination are separable functions in early morphogenesis of the molar tooth. Development. 2016b doi: 10.1242/dev.130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochovska K, Peterkova R, Pavlikova Z, Hovorakova M. Sprouty gene dosage influences temporal-spatial dynamics of primary enamel knot formation. BMC Dev Biol. 2015;15:21. doi: 10.1186/s12861-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103(Suppl):155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- Mammoto T, Mammoto A, Torisawa YS, Tat T, Gibbs A, Derda R, Mannix R, de Bruijn M, Yung CW, Huh D, Ingber DE. Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev Cell. 2011;21:758–769. doi: 10.1016/j.devcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes & Development. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Matalova E, Antonarakis GS, Sharpe PT, Tucker AS. Cell lineage of primary and secondary enamel knots. Dev Dyn. 2005;233:754–759. doi: 10.1002/dvdy.20396. [DOI] [PubMed] [Google Scholar]
- McAvoy JW, Chamberlain CG. Fibroblast growth factor (FGF) induces different responses in lens epithelial cells depending on its concentration. Development. 1989;107:221–228. doi: 10.1242/dev.107.2.221. [DOI] [PubMed] [Google Scholar]
- Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- Morita R, Kihira M, Nakatsu Y, Nomoto Y, Ogawa M, Ohashi K, Mizuno K, Tachikawa T, Ishimoto Y, Morishita Y, Tsuji T. Coordination of Cellular Dynamics Contributes to Tooth Epithelium Deformations. PLoS One. 2016;11:e0161336. doi: 10.1371/journal.pone.0161336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–775. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- Neubüser A, Peters H, Balling R, Martin GR. Antagonistic Interactions between FGF and BMP Signaling Pathways: A Mechanism for Positioning the Sites of Tooth Formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Panousopoulou E, Green JB. Invagination of Ectodermal Placodes Is Driven by Cell Intercalation-Mediated Contraction of the Suprabasal Tissue Canopy. PLoS Biol. 2016;14:e1002405. doi: 10.1371/journal.pbio.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J, Jung HS, Jernvall J, Kettunen P, Mustonen T, Tabata MJ, Kere J, Thesleff I. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Developmental Biology. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Prochazka J, Pantalacci S, Churava S, Rothova M, Lambert A, Lesot H, Klein O, Peterka M, Laudet V, Peterkova R. Patterning by heritage in mouse molar row development. Proc Natl Acad Sci U S A. 2010;107:15497–15502. doi: 10.1073/pnas.1002784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazka J, Prochazkova M, Du W, Spoutil F, Tureckova J, Hoch R, Shimogori T, Sedlacek R, Rubenstein JL, Wittmann T, Klein OD. Migration of Founder Epithelial Cells Drives Proper Molar Tooth Positioning and Morphogenesis. Dev Cell. 2015;35:713–724. doi: 10.1016/j.devcel.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl H, Nüsslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Current Biology. 2001;11:503–507. doi: 10.1016/s0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J, Danielian PS, McMahon JA, Lewis PM, Paus R, McMahon AP. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Keranen S, Jernvall J. Enamel Knots as Signaling Centers Linking Tooth Morphogenesis and Odontoblast Differentiation. Advances in Dental Research. 2001;15:14–18. doi: 10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Developmental Biology. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Sharpe PT. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Developmental Dynamics. 1998;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. The importance of signal pathway modulation in all aspects of tooth development. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2009;312B:309–319. doi: 10.1002/jez.b.21280. [DOI] [PubMed] [Google Scholar]
- Vaahtokari A, Aberg T, Jernvall J, Keranen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mechanisms of Development. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Voutilainen M, Lindfors PH, Lefebvre S, Ahtiainen L, Fliniaux I, Rysti E, Murtoniemi M, Schneider P, Schmidt-Ullrich R, Mikkola ML. Ectodysplasin regulates hormone-independent mammary ductal morphogenesis via NF-kappaB. Proc Natl Acad Sci U S A. 2012;109:5744–5749. doi: 10.1073/pnas.1110627109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokohama-Tamaki T, Ohshima H, Fujiwara N, Takada Y, Ichimori Y, Wakisaka S, Ohuchi H, Harada H. Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359–1366. doi: 10.1242/dev.02307. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev Cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]