SUMMARY
Cell division in Escherichia coli begins with the polymerization of FtsZ into a ring-like structure, the Z-ring, at midcell. All other division proteins are thought to require the Z-ring for recruitment to the future division site. Here, we report that the Z-ring associated proteins ZapA and ZapB form FtsZ-independent structures at midcell. Upon Z-ring disruption by the FtsZ polymerization antagonist SulA, ZapA remained at midcell as a cloud-like accumulation. Using ZapA(N60Y), a variant defective for interaction with FtsZ, it was established that these ZapA structures form without a connection to the Z-ring. Furthermore, midcell accumulations of GFP-ZapA(N60Y) often preceded Z-rings at midcell and required ZapB to assemble, suggesting that ZapB polymers form the foundation of these structures. In the absence of MatP, a DNA-binding protein that links ZapB to the chromosomal terminus region, cloud-like ZapA structures still formed but failed to track with the chromosome terminus and did not consistently precede FtsZ at midcell. Taken together, our results suggest that FtsZ-independent structures of ZapA-ZapB provide additional positional cues for Z-ring formation and may help coordinate its assembly with chromosome replication and segregation.
Keywords: cytokinesis, cell division, FtsZ, cell cycle, divisome
Graphical abstract
We found that E. coli’s FtsZ-ring associated protein ZapA forms FtsZ-independent structures at midcell that are dependent on ZapB. Furthermore, these ZapA-ZapB structures are capable of identifying new sites of division prior to polymerization of the FtsZ-ring and this phenomenon is largely dependent on the ter-binding protein, MatP. Taken together, our results suggest that FtsZ-independent structures of ZapA-ZapB provide additional positional cues for FtsZ-ring formation and may help coordinate its assembly with chromosome replication and segregation.
INTRODUCTION
Bacterial cell division is mediated by a multi-protein complex called the divisome (de Boer, 2010). Assembly of this machine begins with the polymerization of the tubulin homolog FtsZ into a ring-like structure at the future site of division (Bi and Lutkenhaus, 1991). In addition to FtsZ, formation of this so-called Z-ring or proto-ring intermediate requires links between FtsZ and the inner face of the cytoplasmic membrane (Hale and de Boer, 1997; Pichoff and Lutkenhaus, 2002; Pichoff and Lutkenhaus, 2005). In Escherichia coli these connections are provided by the ZipA and FtsA proteins (Hale and de Boer, 1997; Pichoff and Lutkenhaus, 2002; Pichoff and Lutkenhaus, 2005). The Z-ring is also stabilized by a number of FtsZ-associated proteins, including the ZapA-ZapB complex (Gueiros-Filho and Losick, 2002; Ebersbach et al., 2008; Galli and Gerdes, 2010; Durand-Heredia et al., 2011; Hale et al., 2011; Durand-Heredia et al., 2012; Huang et al., 2013; Buss et al., 2013; Buss et al., 2015). Once the Z-ring is assembled, it facilitates the recruitment of over 25 proteins to midcell to form the mature divisome machine capable of promoting cell division (de Boer, 2010).
In E. coli, midcell placement of the Z-ring is principally accomplished by two negative regulators of FtsZ polymerization, the Min system and the nucleoid occlusion factor SlmA (de Boer et al., 1989; Bernhardt and de Boer, 2005). The Min system promotes the pole-to-pole oscillation of the FtsZ antagonist MinC such that its time-averaged concentration is lowest at midcell (Raskin and de Boer, 1999a; Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999b; Meinhardt and de Boer, 2001). FtsZ polymerization thus becomes favored at this site. SlmA is a specific DNA-binding protein that antagonizes FtsZ assembly when it associates with binding sites distributed around the origin-proximal two-thirds of the chromosome (Tonthat et al., 2011; Cho et al., 2011; Cho and Bernhardt, 2013; Tonthat et al., 2013; Du and Lutkenhaus, 2014). It blocks Z-ring assembly over the nucleoid until the later stages of chromosome replication when the SlmA-free chromosome terminus region is positioned at midcell. SlmA is therefore thought to help coordinate cell division with chromosome replication and segregation in addition to functioning as a fail-safe to block division over the nucleoid when replication problems are encountered (Bernhardt and de Boer, 2005; Cho et al., 2011).
Defects in the Min system and SlmA are synthetically lethal (Bernhardt and de Boer, 2005), resulting in a complete division block when both systems are inactivated in cells growing in rich medium. This growth and division defect can be suppressed either by FtsZ overproduction or growth in minimal medium (Bernhardt and de Boer, 2005). It was recently shown that Min− SlmA− cells growing in minimal medium still divide at midcell with surprising precision (Bailey et al., 2014). Midcell localization of the Z-ring in this case was shown to be dependent on the ZapAB complex and the MatP protein that organizes the terminus macrodomain of the nucleoid (Mercier et al., 2008). A direct or indirect interaction between ZapB and MatP has previously been shown to anchor the terminus region of the chromosome to the division site (Espéli et al., 2012). In the case of the Min− SlmA− cells, it appears as though this interaction can also help define the future site of division (Bailey et al., 2014). Consistent with this idea, MatP recruitment to midcell was found to precede that of the Z-ring in Min− SlmA− cells (Bailey et al., 2014). What remains unclear is how the ZapAB-MatP connection might facilitate midcell Z-ring formation and whether its apparent positioning function in Min− SlmA− cells is also used in wild-type cells to promote midcell Z-ring placement.
Here, we identify a ZapAB structure that forms at midcell independent of FtsZ where it likely influences Z-ring formation positively and provides positional information. The structure was initially observed in cells expressing the FtsZ antagonist SulA. When using ZapA-GFP as a marker for Z-ring assembly, we were surprised to find that ZapA persisted at midcell as a cloud-like structure for an extended period of time following dispersal of FtsZ. The independence of this structure from FtsZ was further established using a ZapA variant, ZapA(N60Y), that fails to interact with FtsZ, yet still forms diffusely localized structures at midcell that localize to future sites of division before FtsZ. These ZapA(N60Y) structures require ZapB to form, but not MatP. However, in the absence of MatP the structures fail to track with the chromosome terminus and do not consistently precede FtsZ at midcell. Taken together, our results combined with those described previously (Espéli et al., 2012; Bailey et al., 2014) indicate that ZapAB form an FtsZ-independent structure that can provide a positive signal for midcell Z-ring formation that may also help coordinate its assembly with chromosome replication and segregation.
RESULTS
Diffuse ZapA structures persist at midcell upon dissolution of the FtsZ-ring
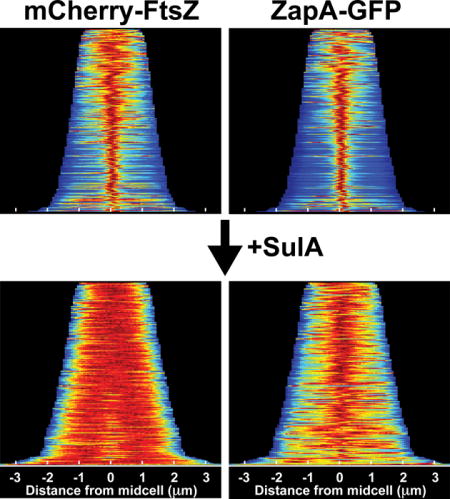
While monitoring Z-ring assembly status following induction of the FtsZ antagonist SulA, we noticed a surprising difference in localization behavior between mCherry-FtsZ and ZapA-GFP markers expressed in the same cell. As expected (Bi and Lutkenhaus, 1993), the band-like fluorescence pattern of mCherry-FtsZ at midcell dissipated rapidly (< 5min) following induction of sulA with arabinose (Fig. 1A, and data not shown). However, a cloud-like accumulation of ZapA-GFP fluorescence persisted at midcell long after the band of mCherry-FtsZ was dissipated by the division inhibitor (Fig. 1A). This phenomenon was observed in the vast majority of cells examined and is readily visualized using population demographs (Fig. 1B–E, G–J, L–O). In these graphics, the fluorescence signal of each cell within the population analyzed was normalized, aligned, and ordered according to increasing cell length (top-to-bottom), which provides a good approximation of cell age. Before induction of sulA (T = 0), a tight distribution of ZapA-GFP fluorescence at midcell was observed throughout the majority of the population (n = 251), with the exception of the smallest newborn cells (Fig. 1C). After 45min of sulA induction (T = 45), a broader distribution of ZapA-GFP signal was observed, but the fluorescence remained strongly biased at midcell within the population (n = 187) (Fig. 1E). This cloud-like localization pattern was not caused by GFP-GFP interactions (Landgraf et al., 2012) as fusions with dimerization defective fluorescent protein variants also displayed this localization following sulA induction (Fig. S1). Additionally, a persistent midcell localization pattern following FtsZ-ring disassembly was not observed for GFP-FtsA or GFP-ZapC fusions (Fig. 1F–O), indicating that localization at midcell independent of FtsZ is not a common property of FtsZ-binding proteins.
Figure 1. ZapA structures persist after FtsZ-ring disassembly.
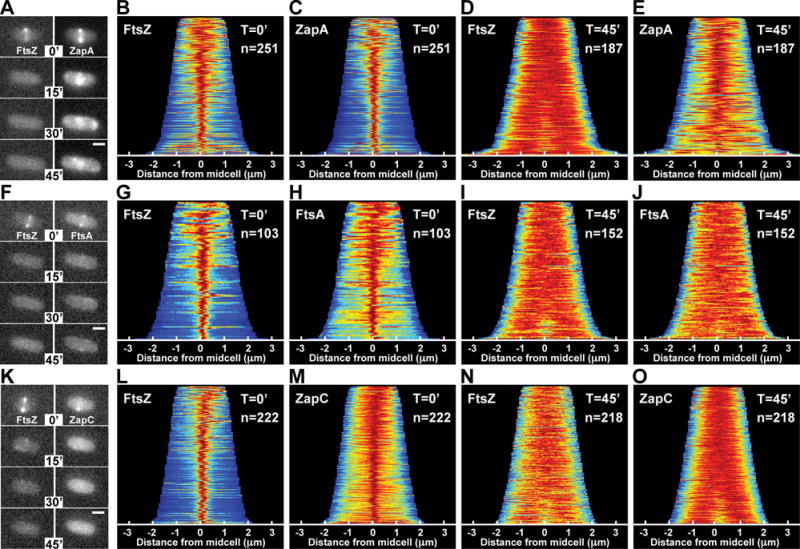
The localization of mCherry-FtsZ (expressed from attλNP145) and either ZapA-GFP (A–E, from its native chromosomal locus), GFP-FtsA (F–J, expressed from attHKHC427) or ZapC-GFP (K–O, expressed from attHKNP162) was followed in wild-type cells during a time-course following sulA induction from pNP146 (0–45min). (A,F,K) Montages of fluorescence images depicting representative single cells in both color channels following sulA induction (0–45min). The protein fusion being monitored is indicated in the top panel (left, red channel; right, green channel). (B–E, G–J, L–O) Population demographs illustrate the localization trend of an indicated fusion protein for a representative population immediately following sulA induction (T=0′) or 45min post-induction (T=45′). The majority of cells at T=0′ display midcell localization for all fluorescence constructs tested (B–C, G–H, L–M). At T=45′, only ZapA-GFP retains midcell localization (D-E, I-J, N-O). Midcell persistence of ZapA-GFP results in an altered appearance transforming from a tight-band (A,C) to a broad, cloud-like structure (A,E). Scale bars = 1μm.
Persistence of ZapA cloud-like structures requires ZapB
ZapB is a coiled-coil protein that self assembles into polymers and interacts directly with ZapA (Ebersbach et al., 2008). We therefore investigated whether ZapB also persists at midcell following the dissolution of Z-rings and whether the formation of the observed cloud-like structures of ZapA-GFP requires ZapB. Indeed, a functional GFP-ZapB fusion (Fig. S2) formed tight bands at midcell in cells prior to sulA induction as expected (Ebersbach et al., 2008), and similar to ZapA-GFP, was retained at midcell in a diffuse localization pattern long after FtsZ-rings were disrupted by SulA (Fig. 2A–C). Consistent with previous results, ZapA-GFP localized to Z-rings in the absence of ZapB with a mild defect (broadening) in its localization pattern (Galli and Gerdes, 2010; Buss et al., 2013) (Fig. 2E). However, following sulA induction in the ΔzapB cells, ZapA-GFP fluorescence rapidly dispersed, converting from its tight band-like pattern to a uniform cytoplasmic signal on a timescale similar to mCherry-FtsZ (Fig. 2D–F). We conclude that ZapA maintains a midcell localized structure following FtsZ-ring disassembly and that this structure is dependent on an underlying foundation of ZapB polymers.
Figure 2. ZapB structures persist after FtsZ-ring disassembly, and are required for ZapA cloud formation.
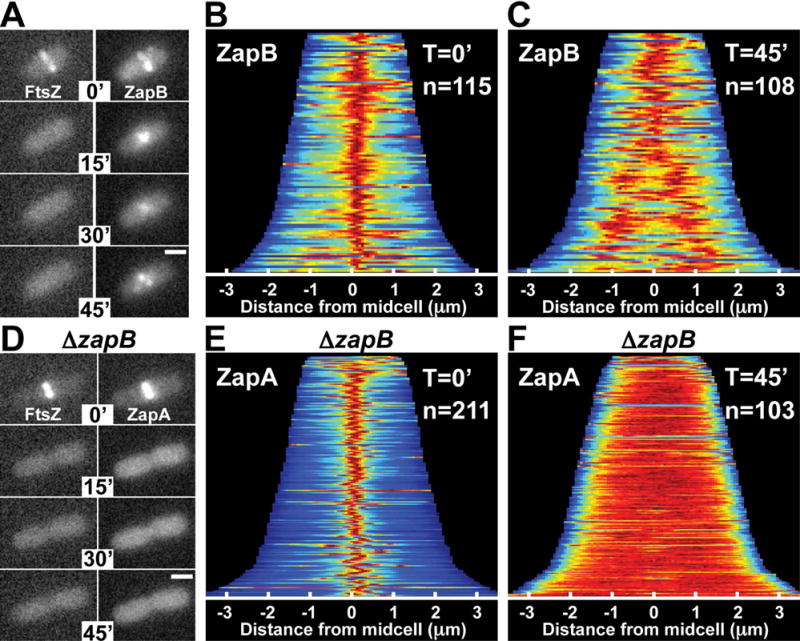
(A) Fluorescence images of mCherry-FtsZ (expressed from attλNP145) and GFP-ZapB (expressed from attHKJAB034) in ΔzapB [DL56] cells during a time-course following sulA induction from pNP146 (0–45min). (B–C) Population demographs for GFP-ZapB in cells from (A) at T=0′ and T=45′ post sulA induction. (D) Fluorescence images of mCherry-FtsZ (expressed from attλNP145) and ZapA-GFP (expressed from its native locus) in ΔzapB cells [NP298] during a time-course following sulA induction (0–45min). (E–F) Population demographs for ZapA-GFP in cells from (D) at T=0′ (E) and T=45′ (F). Scale bars = 1μm.
ZapA structures assemble between segregating nucleoids without prior Z-ring formation
After extended periods of SulA-induced filamentation, additional ZapA-GFP structures became visible and were dispersed at regular intervals along the growing cell filaments (Fig. 3A). This localization pattern is reminiscent of segregating nucleoids. We therefore colocalized the ZapA-GFP structures with a HupA-mCherry fusion expressed from its native locus to label the nucleoids. Following induction of sulA for 120min, nucleoids flanking the original Z-ring showed signs of replication/segregation and adopted a bilobed morphology (Fig. 3B). Additional ZapA-GFP accumulations were observed to localize between these segregating nucleoids (Fig. 3B, magenta arrows). As segregation continued to generate four well-separated nucleoids, the ZapA-GFP accumulations also moved apart and filled the gaps between the nucleoids (Fig. 3B, cyan arrows). The accumulation of ZapA-GFP between replicating nucleoids did not appear to require significant segregation, and demographs indicate that this localization pattern was prevalent within the populations imaged (Fig. S3). This result suggests that ZapA-GFP structures can identify future division sites without prior FtsZ-ring formation.
Figure 3. ZapA structures assemble between segregating nucleoids.
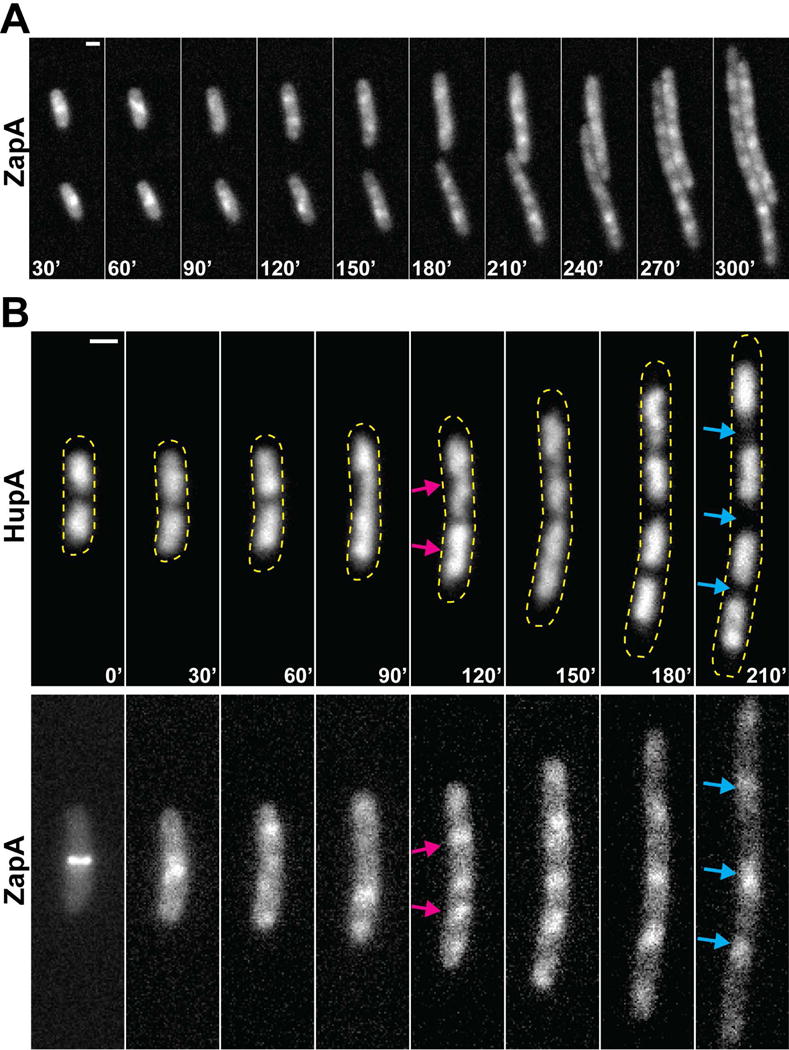
(A) Representative fluorescence time-lapse images of ZapA-GFP (expressed from its native locus in NP1) in two cells 30min after sulA induction from pNP146 (30–300min). (B) Representative fluorescence time-lapse images of a single cell expressing ZapA-GFP and HupA-mCherry (expressed from their native loci in NP127) during a time-course following sulA induction (0–210min). Arrows indicate ZapA-GFP localization between actively segregating (bilobed) nucleoids (magenta) or nucleoid-free regions (cyan). Yellow dashed lines approximate the cellular perimeter. Scale bars = 1μm.
To further investigate the propensity of ZapA to form midcell structures independently of FtsZ, we generated ZapA variants defective for interaction with FtsZ. PCR-based mutagenesis was directed to the zapA portion of an expression construct (pJAB132) encoding ZapA-T18 for use in the split adenylate cyclase bacterial two-hybrid (BACTH) assay (Karimova et al., 1998). To assess the ZapA-FtsZ interaction phenotype, the resulting plasmid library was then screened for interaction with FtsZ fused to the other half of adenylate cyclase (FtsZ-T25). A positive interaction in this assay results in increased lacZ expression and the formation of blue colonies on X-gal containing agar. A ZapA-T18 variant with an N60Y substitution was identified that failed to generate a positive interaction signal with FtsZ-T25 (Fig. 4A).
Figure 4. ZapA(N60Y) fails to bind FtsZ, and assembles into ZapB-dependent midcell clouds.
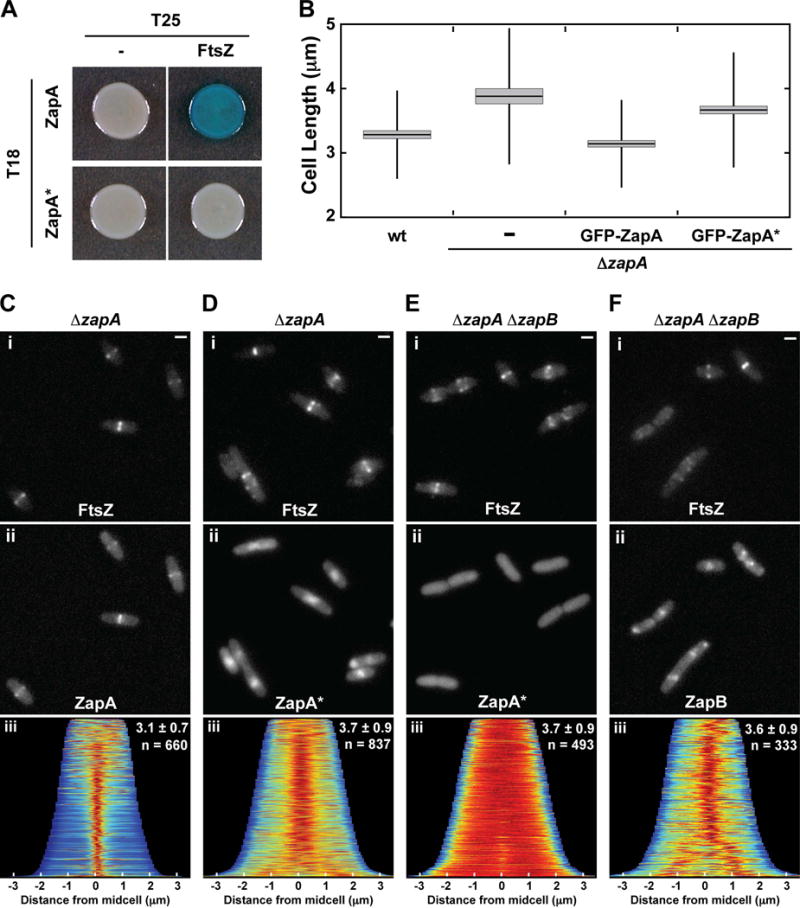
(A) Bacterial two-hybrid results for T18-ZapA (pJB132) and T18-ZapA(N60Y) (ZapA*, pJAB047) paired with unfused T25 (pKT25) and T25-FtsZ (pJB145). (B) Box plots of cell length measurements for wild-type [TB28] and ΔzapA [TB119] cells expressing no fusion protein (−), GFP-ZapA (from attHKJAB032) or GFP-ZapA(N60Y) (GFP-ZapA*, from attHKJAB033). The mean (horizontal line) and standard deviation (vertical line) are illustrated along with the 95% confidence interval (light gray box). Representative micrographs of ΔzapA [TB119] (C–D) or ΔzapA ΔzapB [JAB470] (E–F) cells expressing mCherry-FtsZ (attλNP145, Ci–Fi) and either GFP-ZapA (from attHKJAB032, Cii), GFP-ZapA(N60Y) (attHKJAB033, Dii and Eii), or GFP-ZapB (from attHKJAB034, Fii). Population demographs (Ciii, Diii, and Eiii) illustrate the localization of GFP-ZapA, GFP-ZapA(N60Y), or GFP-ZapB, respectively, in the populations measured in B. Mean length ± standard deviation (μm), along with sample size (n) are indicated in the top right. Scale bars = 1μm
To characterize the ZapA(N60Y) variant further, we tested its ability to correct the mild division phenotype of ZapA-defective cells (Mohammadi et al., 2009; Dajkovic et al., 2010; Buss et al., 2013). Expression of GFP-ZapA from an ectopic locus complements the elongated phenotype of ΔzapA cells, shortening them from 3.9 ± 1.1 μm (n = 304) to 3.1 ± 0.7 μm (n = 664), which is close to the length of wild-type cells [3.3 ± 0.7 μm (n = 471)] (Fig. 4B). As expected, the fusion also co-localized with mCherry-FtsZ and displayed a tight band-like localization pattern at midcell (Fig. 4Ci–iii). GFP-ZapA(N60Y), on the other hand, was unable to correct the ΔzapA division phenotype as ΔzapA cells expressing the fusion had a length distribution of 3.7 ± 0.9 μm (n = 837), which was similar to the ΔzapA control cells (Fig. 4B). Unlike the wild-type fusion, GFP-ZapA(N60Y) did not form tight bands at midcell. Instead, the fusion displayed a diffuse cloud-like localization pattern at the cell center similar to the ZapA-GFP structures observed following FtsZ-inhibition (Fig. 4Di–iii). Also reminiscent of ZapA-GFP following Z-ring disruption, the diffuse midcell structures of GFP-ZapA(N60Y) did not form in actively dividing cells defective for ZapB (Fig. 4Ei–iii). Based on these results, we infer that ZapA(N60Y) is stably produced and functional for its interaction with ZapB. However, GFP-ZapA(N60Y) appears incapable of interacting with FtsZ and forms cloud-like localizations in dividing cells as if FtsZ assembly were inhibited. Similarly, GFP-ZapB also forms cloud-like accumulations at midcell when its connection to FtsZ is severed by deleting zapA (Fig. 4Fi–iii). Importantly, the clouds of GFP-ZapA(N60Y) or GFP-ZapB in ΔzapA cells were often observed to precede Z-ring formation at midcell (Fig. 5A–B, S4), suggesting that these structures are not only capable of forming independently from normal Z-ring assembly but that they also may provide a positive signal promoting Z-ring formation at midcell in otherwise wild-type cells.
Figure 5. FtsZ-independent ZapA(N60Y) and ZapB structures precede Z-ring assembly at midcell.
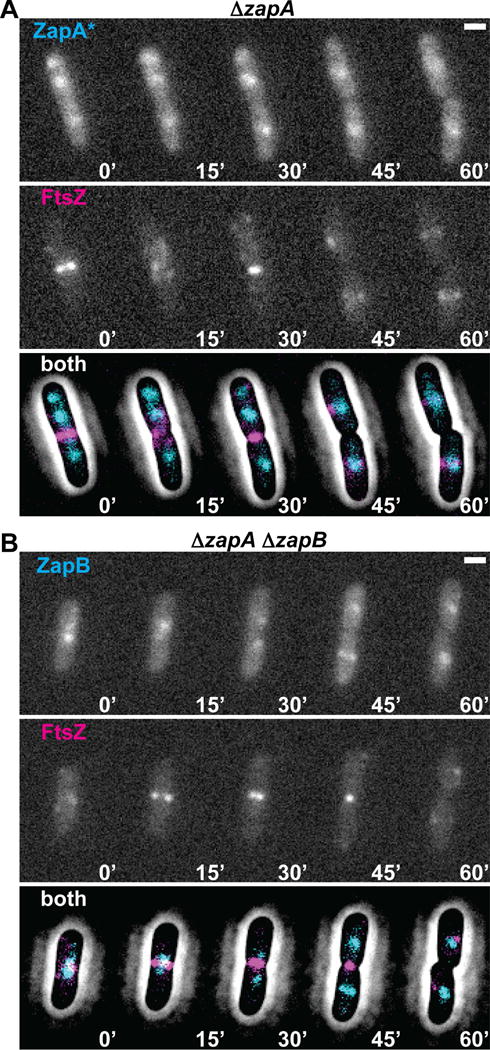
(A) Time-lapse fluorescence images of a single cell expressing GFP-ZapA(N60Y) (ZapA*, from attHKJAB033) and mCherry-FtsZ (from attλNP145) in ΔzapA [TB119] cells. Below the images of individual fluorescence channels is a combined image of GFP-ZapA(N60Y) (cyan) and mCherry-FtsZ (magenta) overlaid with a contrast-adjusted phase image. (B) Similar time-lapse fluorescence images of a single cell expressing GFP-ZapB (from attHKJAB034) and mCherry-FtsZ (from attλNP145) in ΔzapA ΔzapB [JAB470] cells. Images are as in (A). Scale bars = 1μm.
FtsZ-independent ZapA-ZapB structures track with the chromosome terminus
The localization of ZapA-ZapB structures to midcell independent of an FtsZ interaction raises the question of what positional cues are used to guide their assembly. Given the previously described link between ZapB and the chromosome terminus (Espéli et al., 2012), this region of the chromosome was a likely candidate, especially because it is known to be recruited to midcell during the later stages of replication (Wang et al., 2005). To test the potential role of the chromosome terminus (ter) as a positional marker for the ZapAB structures, we tracked its location using GFP-ParB and a parS site integrated ~200kb upstream of the dif site in a strain producing ZapA-mCherry from its native locus. Consistent with previous reports highlighting the apparent coordination between chromosome dynamics and divisome assembly (Espéli et al., 2012; Bailey et al., 2014), the ZapA-mCherry signal largely overlapped with the ter label in the absence of SulA production (T=0′, 89.7% of cells; n = 273) (Fig. 6A). Similarly, following SulA production, the cloud-like ZapA-mCherry structures also predominantly colocalized with the ter label (T=30′, 90.1% of cells, n = 374; T=60′, 87.9% of cells, n = 280; T=90′, 82.8% of cells, n = 470) (Fig. 6B). This co-localization was even observed for ZapA-mCherry structures present outside of the midcell region (Fig. 6C). Co-localization was not 100%, however, and in cases where it was not observed, either a ZapA-mCherry structure never formed, or the structures appeared to persist at the initial division site (i.e. midcell) in the inter-nucleoid space while the ter region segregated to either side of the cloud (Fig. 6D). In cells capable of division, cloud-like structures of ZapA(N60Y)-mCherry were also found to closely colocalize with the ter label (94.7% of cells; n = 619) (Fig. 6E).
Figure 6. ZapA and ZapA(N60Y) largely colocalize with the chromosomal ter region.
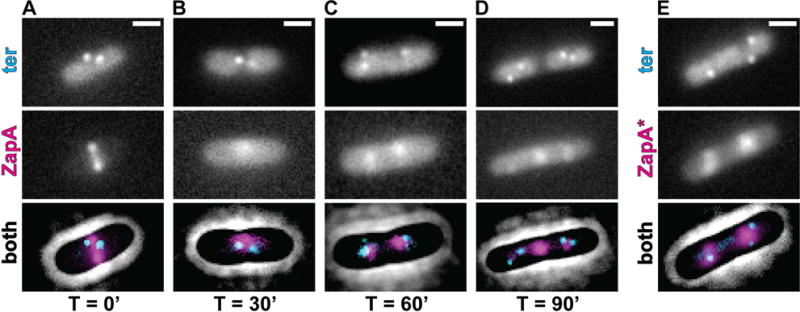
(A–D) Fluorescence micrographs of GFP-ParB (ter, cyan) and ZapA-mCherry (magenta, expressed from its native locus) in strain JAB338 are shown together with a contrast-adjusted phase image during a time course (0–90min) following sulA induction from pNP146. (E) A representative micrograph of GFP-ParB (ter, cyan) and ZapA(N60Y)-mCherry (ZapA*, magenta, expressed from native zapA locus) in strain JAB374 during normal growth. Scale bars = 1μm.
Localization of cloud-like ZapA-GFP structures in the absence of MatP
Previous results indicate that the ter region of the chromosome is linked to the divisome by a direct or indirect interaction between ZapB and the DNA-binding protein MatP responsible for organizing the ter macrodomain (Mercier et al., 2008; Espéli et al., 2012). We therefore investigated whether MatP was required for the midcell localization of the ZapA cloud structures and their co-localization with the chromosome terminus. To investigate this possibility, we followed ZapA-GFP localization in ΔmatP cells before and after the disruption of Z-rings. Similar to what was observed in wild-type cells, ZapA-GFP displayed its typical tight, band-like localization pattern at midcell in ΔmatP cells prior to sulA induction (Fig. 7A–C). Cloud-like structures of ZapA-GFP were also retained at former division sites following the disruption of FtsZ-rings by SulA (Fig. 7A, D). However, in addition to midcell localization, which was observed in ~60% of the cells, ZapA-GFP also displayed polarly localized “clouds” of fluorescence in a significant portion of ΔmatP cells following SulA production (Fig. 7B, D). As shown in the demograph (Fig. 7D), polar localization of ZapA-GFP clouds was correlated with cell length (Fig. S5). In shorter cells, the ZapA-GFP cloud typically started out at midcell following FtsZ-ring dissolution, but then shifted to the pole (Fig. 7B). The reason for this change in localization is not clear, but it may be due to insufficient nucleoid segregation, which leads to crowding at midcell in these smaller (younger) cells such that the polar location becomes favored. Similarly, GFP-ZapA(N60Y) localized to both midcell and polar sites in ΔmatP cells (Fig. 8A–C). The loss of robust midcell localization of ZapA structures in ΔmatP cells correlated with reduced co-localization of ZapA(N60Y)-mCherry with the ter marker (6.4% of cells; n = 606) (Fig. 8D). These findings support the previously described connection between ZapA and ZapB with the chromosome terminus via MatP as a positional marker for Z-ring assembly (Espéli et al., 2012; Bailey et al., 2014) and further indicate that the connection between these factors occurs independently of the Z-ring.
Figure 7. MatP influences ZapA cloud positioning following sulA induction.
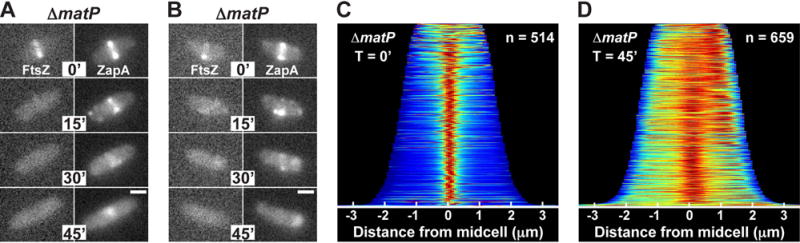
(A–B) Fluorescence images of mCherry-FtsZ (from attλNP145) and ZapA-GFP (native locus) in ΔmatP [NP301] cells during a time course (0–45min) following sulA induction from pNP146 in minimal medium. (C–D) Population demographs for ZapA-GFP immediately following sulA induction (T=0′) or 45min post-induction (T=45′). Scale bars = 1μm.
Figure 8. ZapA(N60Y) localization in ΔmatP cells.
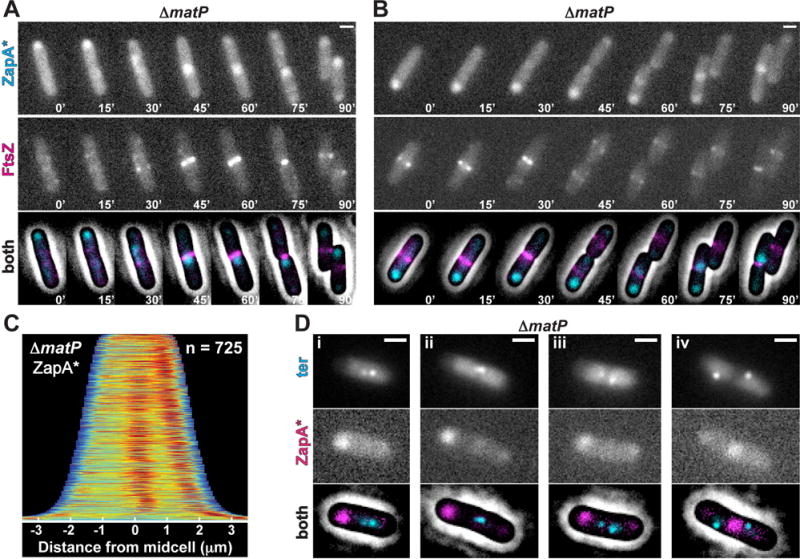
(A–B) Time-lapse fluorescence images of single cells expressing GFP-ZapA(N60Y) (ZapA*, from attHKJAB033) and mCherry-FtsZ (from attλNP145) in ΔzapA ΔmatP [NP314] cells during slow growth. (C) Population demograph illustrating the localization of GFP-ZapA(N60Y) (ZapA*) in a population of ΔzapA ΔmatP [NP314] cells in minimal medium. (Di–iv) Fluorescence images of GFP-ParB (ter, cyan) and ZapA(N60Y)-mCherry (magenta, expressed from its native locus) from JAB378 cells grown in minimal medium. Images are as in Figure 5A. Scale bars = 1μm.
DISCUSSION
The Min system functions as the dominant global regulator responsible for directing Z-ring formation to midcell in E. coli (Sun et al., 1998; Bernhardt and de Boer, 2005). However, the cellular gradient of the MinC division inhibitor established as a result of its pole-to-pole oscillation (Raskin and de Boer, 1999a; Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999b; Meinhardt and de Boer, 2001) is probably only defining a relatively broad area of midcell as permissive for FtsZ polymerization relative to the typical width of a Z-ring (ca. 100 nm) (Fu et al., 2010). Additional determinants are most likely needed to promote FtsZ polymerization within an even tighter zone to stimulate normal Z-ring assembly. Several lines of evidence, including the FtsZ-independent structures we report here, support the notion that ZapA and ZapB represent at least one of the determinants providing this fine-scale positional information for proper Z-ring assembly.
Previous work characterizing the phenotypes of ΔzapA or ΔzapB mutants in otherwise wild-type cells provided clues that ZapA and ZapB might function in part to control Z-ring placement. Without ZapA or ZapB, FtsZ often forms aberrant spiral-like or multiple ring-like structures spread over a wider than normal zone about the cell center (Ebersbach et al., 2008; Mohammadi et al., 2009; Galli and Gerdes, 2010; Dajkovic et al., 2010; Buss et al., 2013). Moreover, approximately 20% of ZapA- or ZapB-defective cells were found to form multiple constriction sites within the midcell region (Buss et al., 2013). These phenotypes have typically been interpreted as representing a defect in Z-ring stability (Huang et al., 2013) resulting from a failure in FtsZ polymer crosslinking or the loss of connections between higher-order FtsZ assemblies mediated by ZapA and ZapB (Dajkovic et al., 2010; Buss et al., 2013). However, ZapB itself does not directly interact with FtsZ or influence its polymerization (Galli and Gerdes, 2012), and ZapA is capable of localizing to the Z-ring in vivo and interacting with and bundling FtsZ polymers in vitro without ZapB (Low et al., 2004; Mohammadi et al., 2009; Galli and Gerdes, 2010; Dajkovic et al., 2010; Galli and Gerdes, 2012). A role for ZapB in bridging FtsZ polymers therefore seems unlikely. Instead, the shared phenotype of ΔzapA and ΔzapB cells involving multiple Z-rings and constriction sites in the midcell area suggests that the local positioning of Z-ring assembly may be the principal function of the ZapA-ZapB complex.
Although a Z-ring positioning function for ZapA and ZapB is suggested by the phenotypes of the single deletion mutants, strong support for this activity was first provided by studies of division placement in Min− SlmA− cells of E. coli grown under permissive conditions (Bailey et al., 2014). Division in these cells was surprisingly found to be biased at midcell instead of displaying a random distribution as expected for cells lacking both known division positioning systems (Bailey et al., 2014). Precise midcell division in the absence of Min and SlmA was further shown to require ZapA, ZapB, and the chromosome terminus organizing protein MatP (Bailey et al., 2014). This dependence as well as the observation that the ter macrodomain arrived at midcell in the Min− SlmA− cells prior to FtsZ led to the proposal that the ZapA-ZapB-MatP connection serves as a positive signal to promote midcell Z-ring assembly (Bailey et al., 2014). Consistent with this possibility, super-resolution microscopy analysis identified a layered ZapA-ZapB-MatP structure at midcell connecting the Z-ring with the chromosome (Buss et al., 2015).
A lingering problem with the model that ZapA and ZapB function to direct midcell Z-ring assembly is that it implies that these proteins possess the ability to identify midcell independently of FtsZ. However, FtsZ is thought to be required for the recruitment of most, if not all, divisome proteins to midcell, especially those directly associating with it (Huang et al., 2013). Here, we resolve this issue by demonstrating that ZapA and ZapB are not entirely subject to this canonical midcell recruitment hierarchy. ZapA not only persisted at midcell following Z-ring disruption by SulA, but was also capable of midcell recruitment when its ability to interact with FtsZ was disrupted. Formation of these cloud-like accumulations of ZapA was found to require ZapB. Thus, ZapB provides the underlying foundation of these structures most likely by forming a polymeric matrix akin to the structures observed in vitro (Ebersbach et al., 2008). ZapB itself is recruited to midcell independently of ZapA and therefore also independent of a connection to FtsZ. Instead of FtsZ, the ultimate positional determinant for these structures was found to be the chromosome ter macrodomain and the associated MatP protein. Without MatP, the structures still correctly localized to midcell on occasion, but also mislocalized to the poles. In this case, localization is probably determined by the polymeric nature of ZapB and the preference for these structures to form in nucleoid-free regions of the cell. This mislocalization phenotype in the absence of MatP correlates with the Z-ring defects observed in ΔmatP cells (Buss et al., 2015) and the loss of precision in midcell division observed in Min− SlmA− cells defective for MatP (Bailey et al., 2014). Finally, as expected for potential Z-ring positioning determinants, the FtsZ-independent clouds of ZapA or ZapB were found to arrive at midcell slightly before the formation of tight Z-rings similar to the early midcell localization of the ter macrodomain in Min− SlmA− cells observed previously (Bailey et al., 2014).
Even though they were capable of identifying midcell, the FtsZ-independent ZapA-ZapB structures we observed were much more diffuse than the tight band-like accumulations of typical divisome proteins. Similar structures were not detected as precursors to Z-rings in wild-type cells where the ZapA-FtsZ interaction was functional. We therefore do not think the FtsZ-independent ZapA-ZapB structures we observe represent true intermediates in Z-ring assembly. Such intermediates are likely to be difficult, if not impossible, to detect under normal circumstances due to cooperativity in Z-ring assembly. Only a relatively small number of ZapA-ZapB complexes recruited to midcell may be needed to kick-off a self-enhancing cycle of FtsZ polymerization at midcell and subsequent Z-ring formation. Nevertheless, the special circumstances employed in this study clearly demonstrate that ZapA-ZapB structures connected to the ter macrodomain can form and identify division sites independent of FtsZ thereby strengthening the emerging model that these proteins provide a positive localization signal promoting proper midcell Z-ring formation. Interestingly, these ZapAB structures may be responsible for the persistent localization of FtsZ at midcell in the absence of its functional membrane tethers, ZipA and FtsA (Pichoff and Lutkenhaus, 2002).
Examples of positive signals promoting proper Z-ring placement have been uncovered or proposed in several other organisms. In Streptomyces coelicolor the SsgA and SsgB proteins define the ladder of division sites in aerial hyphae to promote subsequent Z-ring formation and cell division for spore formation (Willemse et al., 2011). PomZ in Myxococcus xanthus also preceded FtsZ at midcell where it likely serves as a positive regulator of division site placement (Treuner-Lange et al., 2012). Additionally, outgrowing spores of Bacillus subtilis lacking the Min and nucleoid occlusion systems remain capable of accurately identifying the midcell division site, suggesting a positive regulator of Z-ring positioning may be operating in this bacterium as well (Rodrigues and Harry, 2012). Thus far, the mechanism(s) by which these spatial regulators of Z-ring assembly identify the division site are not known. One attractive possibility is that they also use chromosomal loci as positional cues as is the case for the ZapA-ZapB complexes observed here.
EXPERIMENTAL PROCEDURES
Media, bacterial strains and plasmids
Cells were grown with LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or M9 minimal medium (Miller, 1972) supplemented with 0.2% casamino acids and 0.2% glucose (M9+) or no glucose (M9−) at 30°C. Unless indicated otherwise, antibiotic concentrations were as follows: Tetracycline (Tet), 5 μg/mL; Ampicillin (Amp), 25 μg/mL; Chloramphenicol (Cam), 25 μg/mL; Kanamycin (Kan), 25 μg/mL).
The bacterial strains and plasmids used in this study are listed in Tables S1 and S2, respectively. A description of their construction is included in the Supplementary Text along with a list of employed primers (Table S3). All strains used for fluorescence imaging are derivates of MG1655 and all deletions were either obtained or designed to be identical to those from the Keio knockout collection (Baba et al., 2006). All plasmids used for integration at phage attachment sites (HK022 or λ) via the helper vectors pTB102 (Bernhardt and de Boer, 2005) and pInt-ts (Haldimann and Wanner, 2001), were derivates of the CRIM plasmids of Wanner and co-workers (Haldimann and Wanner, 2001). Fluorescent fusions and deletion constructs were transferred between strains using P1 transduction (Miller, 1972) and resistance markers flanked by frt sites were cured using pCP20 as described previously (Datsenko and Wanner, 2000).
Microscopy and image analysis
Cultures were grown overnight in LB at 30°C. The following day cultures were diluted 1:1000 in M9+ in the presence of the indicated inducer and grown at 30°C to an OD600 between 0.2 and 0.4. We induced the production of GFP-FtsA (attHKHC427), ZapC-GFP (attHKNP162), GFP-ZapA (attHKJAB032) and GFP-ZapA(N60Y) (attHKJAB033) with 1000μM Isopropyl-B-D-thiogalactopyranoside (IPTG) throughout growth. GFP-ZapB (attHKJAB034) was typically induced with 250μM IPTG, however a range of concentrations (250 – 1000μM) rescued the length defect of ΔzapB cells (Fig. S2). We induced sulA from pNP146 by addition of 0.2% arabinose to the media or agarose pad (see below).
Fluorescence microscopy was performed as described previously (Meisner et al., 2013) with the following exceptions. Prior to image acquisition, cultures were centrifuged at 6,000 × g for 1.5min and concentrated 10-fold in M9+ media. If induction of Para::sulA was desired, pelleted cells were resuspended in M9− and imaged on M9− pads supplemented with 0.2% glycerol and 0.2% arabinose. Imaging was performed at room temperature. For time-lapse imaging, the coverslip was sealed and images were acquired every 15min as specified.
Image analysis, including cell length measurements, was performed with Oufti (Paintdakhi et al., 2015) and MATLAB. Population demographs were constructed using Oufti and formatted in MATLAB and Adobe Illustrator. The normalized fluorescence intensity of each cell is displayed using the ‘jet’ colormap from MATLAB, where red signifies high intensity and blue indicates low intensity. Raw images were cropped with ImageJ and labeled with Adobe Illustrator. Both the colocalization of ZapA-mCherry or ZapA(N60Y)-mCherry with ter (i.e. ParB-GFP), and the polar or midcell localization of GFP-ZapA or GFP-ZapA(N60Y) in ΔmatP were determined by eye.
Bacterial two-hybrid complementation assay
We used the bacterial adenylate cyclase two-hybrid system (Karimova et al., 1998) in the E. coli cya strain BTH101 ΔzapA (JAB321). Random mutagenesis was performed on zapA from pJB132 with primers using the GeneMorph II kit (Agilent). We selected for transformants containing the BACTH plasmids at 30°C on LB containing 100 μg/mL Amp, 75 μg/mL Kan, 500 μM IPTG, and 40 μg/mL 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-gal).
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Bernhardt and Xiao labs for advice and helpful discussions. This work was supported by the National Institutes of Health (R01 AI083365 to TGB, and RO1 GM086447 to JX), a National Science Foundation EAGER award (MCB-1019000 to JX), and a Hamilton Smith Innovative Research Award to JX.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J. Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in Escherichia coli. PLoS Genet. 2014;10:e1004504. doi: 10.1371/journal.pgen.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, Hatem C, Xiao J. In vivoorganization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Molecular Microbiology. 2013;89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J. A Multi-layered Protein Network Stabilizes the Escherichia coli FtsZ-ring and Modulates Constriction Dynamics. PLoS Genet. 2015;11:e1005128. doi: 10.1371/journal.pgen.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Bernhardt TG. Identification of the SlmA active site responsible for blocking bacterial cytokinetic ring assembly over the chromosome. PLoS Genet. 2013;9:e1003304. doi: 10.1371/journal.pgen.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Pichoff S, Lutkenhaus J, Wirtz D. Cross-linking FtsZ polymers into coherent Z rings. Molecular Microbiology. 2010;78:651–668. doi: 10.1111/j.1365-2958.2010.07352.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Lutkenhaus J. SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD. PLoS Genet. 2014;10:e1004460. doi: 10.1371/journal.pgen.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a Cell Division Factor That Promotes the Assembly of FtsZ in Escherichia coli. J Bacteriol. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol. 2011;193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Molecular Microbiology. 2008;68:720–735. doi: 10.1111/j.1365-2958.2008.06190.x. [DOI] [PubMed] [Google Scholar]
- Espéli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 2012;31:3198–3211. doi: 10.1038/emboj.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS ONE. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Molecular Microbiology. 2010;76:1514–1526. doi: 10.1111/j.1365-2958.2010.07183.x. [DOI] [PubMed] [Google Scholar]
- Galli E, Gerdes K. FtsZ-ZapA-ZapB interactome of Escherichia coli. J Bacteriol. 2012;194:292–302. doi: 10.1128/JB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PAJ. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol. 2011;193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Molecular Microbiology. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Huang KH, Durand-Heredia J, Janakiraman A. FtsZ Ring Stability: of Bundles, Tubules, Crosslinks, and Curves. J Bacteriol. 2013;195:1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat Methods. 2012;9:480–482. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Moncrieffe MC, Löwe J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J Mol Biol. 2004;341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, de Boer PA. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci USA. 2001;98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Molecular Microbiology. 2013;89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espéli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; New York: 1972. [Google Scholar]
- Mohammadi T, Ploeger GEJ, Verheul J, Comvalius AD, Martos A, Alfonso C, et al. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry. 2009;48:11056–11066. doi: 10.1021/bi901461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. - PubMed - NCBI. Molecular Microbiology. 2015;99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Molecular Microbiology. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999a;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CDA, Harry EJ. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 2012;8:e1002561. doi: 10.1371/journal.pgen.1002561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Yu XC, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Molecular Microbiology. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, et al. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci USA. 2013;110:10586–10591. doi: 10.1073/pnas.1221036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuner-Lange A, Aguiluz K, van der Does C, Gómez-Santos N, Harms A, Schumacher D, et al. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Molecular Microbiology. 2012;87:235–253. doi: 10.1111/mmi.12094. [DOI] [PubMed] [Google Scholar]
- Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


