Abstract
Cannabinoid receptors (CBR), including CB1 and CB2 have been therapeutic targets for a number of conditions. Recently, splice variants of the CB1R have been identified in humans. The isoforms differ in their N-terminus sequence and pharmacological activity relative to the CB1R, as a result, the differentiation between the CB1 receptor and its isoform is required. As a result, a selected reaction monitoring mass spectrometry (SRM-MS) method was developed for the quantitation of CB1 and the CB1b isoform in CHO cells transduced with CB1 and CB1b. The SRM-MS protocol was assessed with isotopically labeled peptide standards and had high reproducibility of intra-day assay (CVs from 1.9 to 4.3% for CB1 and 0.5 to 5.9% for CB1b) and inter-day assay (CVs from 1.2 to 5.2% for CB1 and 1.2 to 6.1% for CB1b).
Keywords: CB1, CB1b, Cannabinoid receptor isoforms, SRM assay
1. Introduction
The endocannabinoid system consists of the cannabinoid receptors (CBR), endocannabinoids and the enzymes responsible for synthesizing and degrading ECs. CBs are present primarily in brain and in lesser amounts in other parts of the body. The CB is a 45–53 kDa integral membrane G-protein-coupled receptors (GPCRs) containing seven transmembrane domains. To date, three subtypes of cannabinoid receptors have been identified which are CB1R [1], CB2R [2,3] and GPR55 [4]. These GPCRs play an important role in many physiological processes, including metabolic regulation, craving, pain, anxiety, bone growth, and immune function [5,6]. CB1R and CB2Rs mediate their effects via a pertussis toxin-sensitive GTP-binding regulatory protein Gi, while GPR55 does not couple to G(i/o) or G(q) but to Gα13 [7,8]. CB1R are abundant in the central nervous system particularly in regions such as those involved in cognition and short term memory and in motor function and movement [9,10]. They are also expressed in peripheral tissues including immune cells, heart, lung, bone marrow and gastrointestinal tissues [1], where they play major roles in glucose metabolism, cell proliferation, inflammation and apoptosis [3]. In islets of Langerhans the CB1 receptors are reported to be expressed in β cells, and/or α and δ cells [3,5,6,11,12]. As a result, CB1R is a therapeutic target for many conditions including inflammatory bowel disease, depression, metabolic syndrome and cerebral ischemia [3]. More recently, splice variants of the CB1R have also been identified in humans (CB1aR and CB1bR) [9], where they differ in their potency of action [13,14] These isoforms differ in their N-terminus sequence affecting their pharmacologic properties [9,10], therefore the identification and localization of the CB1 receptor and its isoform would be beneficial. Blocking the CB1 receptor in the periphery has been demonstrated to regulate insulin secretion and action [14–16]. While, CB1R is expressed in the central nervous system and peripheral tissue, the isoform CB1a is predominantly expressed in the central nervous system, although its levels are negligible relative to central CB1. CB1b, however, is the main isoform expressed in hepatocytes and is highly expressed in pancreatic β cells, where it regulates adenylyl cyclase activity [14]. Thus it is important for the endocannabinoid field to develop methods to distinguish CB1 and CB1b.
Currently, reverse transcriptase-polymerase chain reaction, western blot analysis, and Immune-histochemistry are the predominant method for the analysis of the cannabinoid receptors. However, the antibodies developed for the CB1 receptor have been fraught with non-specificity. For example, commercial antibodies used in the western blot analysis of CB1 transfected cells, brain lysates and rodent tissues result in multiple bands [17]. Further, we are unable to distinguish between CB1 receptor and its isoforms by western blot analysis. Thus, a targeted proteomic approach for the quantitation of CB1 and CB1b in a cellular matrix was developed and applied to CHO cells transduced with CB1 and CB1b isoforms.
2. Materials and method
2.1. Reagents
DMEM/F-12 medium was purchased from Invitrogen, 10% FBS from HyClone, GE Healthcare. Urea, Thiourea, Sodium Chloride, Sodium Carbonate, Silver Nitrate, Tris, Dithiothreitol, Ammonium Bicarbonate, TritonX-100, Protease Inhibitor cocktail, Trichloroacetic acid, Iodoacetamide were purchased from Sigma Aldrich. Sodium Thiosulphate, Potassium Ferricyanide, CHAPs were purchased from Fischer Scientific. LDS sample buffer and Puromycin was purchased from Invitrogen. Trypsin and Protease Max was purchased from Promega. Polybrene was purchased from Santa Cruz (Dallas). Lentivirus was purchased from Genecopia (Rockville, MD). 4–12% “NuPAGE® Novex® 4–12% Bis-Tris” 1 mm gel was purchased from Thermo Fischer Scientific. 2D quant kit was purchased from GE healthcare. Acetonitrile, Ethanol, Acetic Acid, Methanol were purchased from Sigma Aldrich
2.2. CHO cell transduction and culture
CHO cells were maintained in DMEM/F-12 medium (Invitrogen, Grand Island, NY), 10% FBS. CHO cells were transduced with lentiviral particles for each of the CB1 isoforms developed from plasmids containing the appropriate sequence. Vectors for CB1 receptor isoforms were kindly donated by Alex Straiker [13]. Cells were plated on 24-well plates with regular media 24 h before transduction. Transductions were performed with 1 µl of Lentifect™ Lentivirus (Genecopoeia, Rockville, MD) and 8 µg/ml of Polybrene® (Santa Cruz, Dallas, TX) for 24 h. Selection with 10 µg/ml of Puromycin (Invitrogen) was done 72 h after.
2.3. Gel electrophoresis and Western blot analysis
CHO cells stably transduced with human CB1 and CB1b receptor splice variants were lysed in RIPA buffer containing 1 mM sodium orthovanadate, phosphatase and protease inhibitor cocktails, centrifuged at 12,000g for 15 min at 4 °C, and protein quantified using the bicinchoninic acid assay kit (Pierce, ThermoScientific, MA). Protein extracts were prepared with Laemmli sample buffer containing 2, 3, 5 or 6% of SDS and incubated at 37 °C or 95 °C for 30 or 5 min respectively. Protein extracts were resolved on Novex 4%–12% Tris-glycine gels (Invitrogen) under reducing conditions and transferred to polyvinylidene difluoride membranes using the Trans-Blot® SD Semi-Dry Transfer Cell (BioRAd, CA) for 90 min at 25 V. Membranes were blocked with 5% milk (BioRad) in TBS-T. Rabbit polyclonal antibodies generated against CB1 receptor (Rb-Af380-1; Frontier Science, NY) was used 1:1000 in 5% BSA TBS-T overnight at 4 °C. Peroxidase-conjugated secondary antibody was purchased from GE Healthcare (Piscataway, NJ) and ECL reagents from GE Healthcare.
2.4. In-gel digestion
2.4.1. Protein precipitation
CB1 cells were lysed using 600 µl of lysis buffer Tris [50 mM, pH 8.0] containing 8 M urea, 2 M Thiourea, 150 mM NaCl, 4% CHAPs, 1% Triton X, 100 mM DTT and 2 µl protein protease inhibitor. The cells were lysed by sonication on ice (3 s pulse and 15 s breaks) for 5 min followed by passing the cell suspension through 26.5G gauge needle 5 times. The samples were centrifuged at 16,000 × g (Eppendorf Centrifuge 5415D) for 30 min at 4 °C. The supernatant was collected and treated with 10% (w/w) trichloroacetic acid (TCA) in acetone. The solution was kept at 4 °C (1 h for CB1b; 18 h CB1). The solution was centrifuged at 16,000 × g for 15 min at 4 °C, and the precipitate was washed three times with acetone and dried in speed vac for 5 min.
2.4.2. Protein gel electrophoresis (1D PAGE) and digestion
The precipitate was dissolved in 600 µl of resuspension buffer Tris [50 mM, pH 8.0] containing 8 M urea, 2 M Thiourea, 150 mM NaCl, 100 mM DTT by pipetting at 10 min interval for 30 min at room temperature. The protein concentration was estimated by 2D Quant kit (GE Healthcare). To the solution, 200 µl of Lithium dodecyl sulphate (LDS) sample buffer (Invitrogen) was added and the mixture was warmed at 35 °C for 30 min and kept on ice for 10 min. 600 µg of total protein was loaded in 4–12% “NuPAGE Novex Bis-Tris” 1 mm gel with running buffer as MES buffer. The gel was stained by Silver Stain [18]. After the gel was washed, the protein band was cut into 1cc slices. The gel pieces were destained using 1:1 mixture of 30 mM Potassium ferricyanide and 100 mM Sodium thiosulphates until the gel pieces were no longer brown. They were rinsed with water three times followed by wash with ammonium bicarbonate [25 mM, pH 8] for 10 min. The buffer was discarded and washed with acetonitrile. It was further re-swelled in ammonium bicarbonate [25 mM, pH 8], dehydrated in acetonitrile. The gel pieces were dried in speed vac (savant, SPD111 V) for 20 min. The gel pieces were then incubated in 0.1 M dithiothreitol (DTT) prepared in ammonium bicarbonate [25 mM, pH 8] at 60 °C for 1 h and subsequently, washed with ammonium bicarbonate [25 mM, pH8], followed by incubation with 500 µl of 55 mM iodoacetamide in ammonium bicarbonate [25 mM, pH8] for 30 min in the dark. The gel pieces were then washed twice with ammonium bicarbonate [25 mM, pH 8] and dehydrated by washing with acetonitrile and dried in a speed vac for 30 min. To the dried gel pieces 100 µl of trypsin (12 ng/µl) (Promega) prepared in ammonium bicarbonate [50 mM, pH 8.0] was added and kept overnight at 37 °C. The peptides were collected in a separate vial. To the gel pieces 300 µl of 60% acetonitrile, 0.1% formic acid followed by 90% acetonitrile, 0.1% formic acid. The solution was evaporated under speed vac. The peptides were dissolved in 500 µl water 0.5% trifluroacetic acid (TFA). To the peptides 1 µl of 500 pg/µl of the internal standards was added. The resulting solution was desalted using Oasis HLB cartridge (Waters, Cat No-094225). The peptides were extracted with 80% acetonitrile 0.1% Formic acid. The elute was dried in a speed vac and reconstituted in 5% acetonitrile 0.1% formic acid. Calibration curves were prepared with spiked standards in native CHO-cells (CB1: 2.18–70 ng/ml and 500 ng/ml for the internal standard; CB1b: 0.185–6 µg/ml and 500 ng/ml for the internal standard). The quality control concentrations were as follows: 7, 25 and 40 ng/ml for CB1 and 1, 2 and 6 µg/ml for CB1b. The quantification of CB1 and CB1b was accomplished using area ratios calculated using the corresponding heavy label as the internal standard.
2.5. Immunoprecipitation
The immunoprecipitation of CB1 and CB1b was carried out following a previously published protocol with slight modifications [19]. Briefly, CHO-CB1 or CHO-CB1b cells were lysed in 100 µl Tris HCl buffer [50 mM, pH 7.4] containing 150 mM NaCl, 10 mM iodoacetamide, 2 mM PMSF, 20 mM Na3MoO4, 0.5% NP-40 and protease inhibitor cocktail (SIGMA) for 30 min in ice followed by s sonication bursts 3 times. The solution was centrifuged at 16,000g for 5 min. To the supernatant 2.5 µg Anti-CB1 Polyclonal Antibody (Immuno-Genes, Switzerland) was added and mixed overnight at 4 °C. Then 10 µl slurry of prewashed Protein G agarose beads (Millipore, Billerica, MA) was added and stirred for 2 h at 4 °C. The solution was centrifuged at 500g for 1 min and the supernatant was removed. The beads were washed three times 100 µl of PBS, after which 60 µl of buffer containing 2 M urea, 50 mM ammonium bicarbonate and 5 µg/µl trypsin, 0.1% Protease Max was added for 30 min at 27 °C. The solution was centrifuged for 30 s at 500g, the supernatant was removed and the beads were washed twice with 25 µl of ammonium bicarbonate [50 mM, pH 7.5] containing 2 M urea and 1 mM DTT. The washes and the supernatant were pooled and incubated overnight at room temperature. 20 µl of iodoacetamide (5 mg/ml) was added for 30 min in the dark, then 1 µl of TFA was added. The samples were desalted and reconstituted in 25 µl 95:5 (water: acetonitrile) containing 0.1% formic acid.
2.6. Targeted proteomic analysis
The tryptic peptides for CB1 and CB1b were selected using NCBI BLAST and UniProt/BLAST searches, with further support for selection of peptides and optimization of transitions through Skyline (Seattle Proteome Center). The CB1 peptide: FPLTSFR and CB1b peptide: TITTDLLGSPFQEK were purchased from New England Peptide (Gardner, MA). For the heavy peptides the C terminal arginine or the lysine was 13C and 15N labelled. The purity of the peptides was greater than 95% as obtained from HPLC. The chromatographic separation was achieved at room temperature on a Waters XBridge C18 column (2.1 mm id × 100 mm, 3.5 µm) protected with a Waters XBridge C18 guard column. The mobile phase consisted of water with 0.1% formic acid as Component A and acetonitrile with 0.1% formic acid as component B. A linear gradient was run as follows: 0 min 5% B; 3 min 5% B; 15 min 15% B; 16 min 15% B; 40 min 50%B; 50 min 80%; 51 min 80%; 53 min 5%B at a flow rate of 0.2 ml/min. The injection volume per sample is 20 µl and the total run time was 60 min per sample. MS/MS analysis was performed using API 5500QTrap system from Applied Biosystems/MDS Sciex equipped with Turbo V electrospray ionization source (TIS)® (Applied Biosystems, Foster City, CA, USA). The data was acquired and analyzed using Analyst version 1.5.1 (Applied Biosystems). Positive electrospray ionization data were acquired using selected reaction monitoring (MRM). The TIS instrumental source settings for temperature, curtain gas, ion source gas 1 (nebulizer), ion source gas 2 (turbo ion spray) and ion spray voltage were 550 °C, 20 psi, 60 psi, 70 psi and 5500 V, respectively. Declustering potential (DP), collision energy (CE) and collision exit potentials (CXP) were optimized for the standard and heavy peptides (the terminal amino acid is 13C labelled). CB1: FPLTSFR: 434.24/360.69 (116 V,23 V,14 V); 434.24/623.351 (45 V,25 V, 10 V); 434.24/510.26 (45 V,25 V, 10 V); 434.24/720.404 (45 V,25 V,10 V); heavy (H2N-FPLTSFR-OH) 439.3/520.3 (91 V, 29 V, 16 V); 439.3/365.4 (117 V, 23 V, 16 V); 439.3/119.8 (91 V, 45 V, 16 V); CB1b: TITTDLLGSPFQEK: 775.408/792.389 (86 V, 33 V,36 V); 775.408/648.335 (86 V, 33 V, 36 V); 775.408/905.473 (120 V, 36 V, 10 V); 775.408/1018.57 (43 V, 38 V, 10 V) and heavy (H2N-TITTDLLGSPFQEK-OH) 779.510/800.430 (86 V,33 V,36 V); 779.510/656.381 (86 V, 47 V, 18 V).
3. Results and discussion
Western blotting has been used in characterizing protein expression levels of CB1 and CB2 receptors with mixed results [14]. For example, with endocrine cells in the islets of Langerhans [1,3,5–8,11,12,16,20,21], there are conflicting data regarding the expression of CB1 or CB2 receptors. In addition, post-translation modifications may result in additional bands. CHO cells were transduced with CB1 full length or with CB1b lentiparticles. Protein extracts from CHO-CB1 were incubated with increasing concentrations of SDS (2, 3, 5 or 6%) at 37 °C or 95 °C for 30 or 5 min respectively. After SDS-PAGE, we found that the best conditions for resolving CB1 receptor on a gel electrophoresis were 5% SDS at 37 °C for 30 min. We then performed an SDS-PAGE in those conditions with protein extracts from CHO-CB1 and CHO-CB1b (data not shown). The expected molecular weights are ~53, and 49 KDa for CB1, and CB1b respectively. Multiple bands were observed at various molecular weights. It is important to note that western blot analysis was not able to distinguish the difference between CB1 and its isoforms. This may be due to post-translational modifications that can alter the run of the proteins in the gel such as glycosylation, as well as to the lack of specificity of the antibodies for the isoforms. Therefore, we sought an alternative approach, targeted proteomics, for the characterization of the CB1 and CB1b isoform.
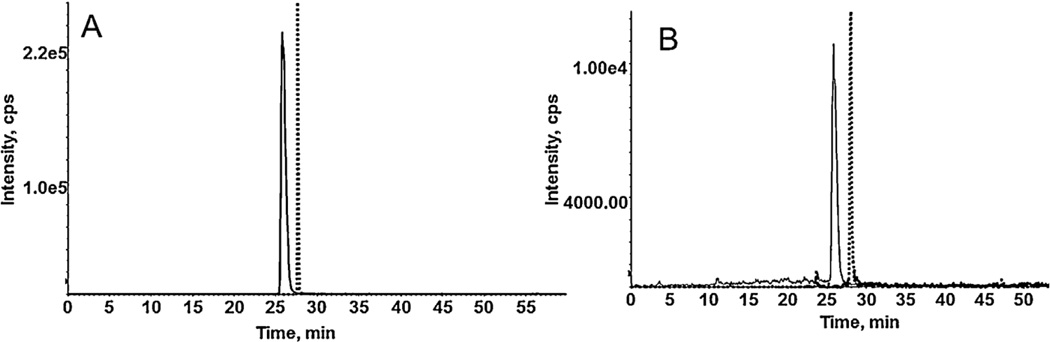
Targeted Proteomics is a quantitative assay for the determination of a targeted protein from a complex matrix using specific peptides. This approach offers advantages over western blot analysis as no antibodies are required and consequently overcomes the limitations of the antibody recognition of highly similar homologous proteins. For CB1 and CB1b receptor, the redundancy in the primary amino acid sequence is greater than 93% [10], with amino acids 22–54 excised in the CB1b isoform. As a result of the high homology, while typically a minimum of three peptides is preferred for a targeted proteomic approach, in this case, only one peptide sequence can be used for each protein (Fig. 1). In silico digestion of the CB1 and CB1b was performed using Protein prospector (University of California, San Francisco). Each peptide for CB1 and CB1b was synthesized to determine the presence of CB1 and CB1b proteins in a cellular matrix. The target peptides: FPLTSFR and TITTDLLGSPFQEK were synthesized for the characterization of the CB1 and CB1b receptors, respectively. For both peptides, four transitions were optimized by MS to achieve the greatest sensitivity. Optimized MS parameters for the respective peptides, with Q1 and Q3, declustering potential (DP), collision energy (CE), entrance potential (EP), and collision cell exit potential (CXP) are reported in Table 1. HPLC optimization results showed a good resolution of peptides (Fig. 2A–B) with a total run time of 60 min using a gradient method with retention times of 25.4 min for CB1 and 27.5 for CB1b.
Fig. 1.
The amino acid sequence of the CB1 and CB1b. The target peptide chosen for SRM analysis for CB1 and CB1b is in bold.
Table 1.
Parameters for SRM transitions for FPLTSFR and TITTDLLGSPFQEK. Q1, Q3, DP, CE, CXP for the different transitions for both the standards and internal standards are enlisted.
| Q1 | Q3 | DP(V) | CE(V) | CXP(V) | |
|---|---|---|---|---|---|
| FPLTSFR | 434.24 | 360.69 | 116 | 23 | 14 |
| 434.24 | 623.351 | 45 | 25 | 10 | |
| 434.24 | 510.26 | 45 | 25 | 10 | |
| 434.24 | 720.40 | 45 | 25 | 10 | |
| FPLTSFR (HEAVY PEPTIDE) | 439.3 | 520.3 | 91 | 29 | 16 |
| 439.3 | 365.4 | 117 | 23 | 16 | |
| TITTDLLGSPFQEK | 775.408 | 792.389 | 86 | 33 | 36 |
| 775.408 | 648.335 | 86 | 33 | 36 | |
| 775.408 | 905.473 | 120 | 36 | 10 | |
| 775.408 | 1018.57 | 43 | 38 | 10 | |
|
TITTDLLGSPFQEK (HEAVY PEPTIDE) |
779.510 | 800.430 | 86 | 33 | 36 |
| 779.510 | 656.381 | 86 | 47 | 18 |
Fig. 2.
Chromatogram of CB1 (FPLTSFR) (434.2/360.6, solid line) and CB1b (TITTDLLGPFQEK) (775.40/792.38, dashed line) in 95:5 (water: acetonitrile, containing 0.1% formic acid) (A) and spiked in native CHO cells digested by in-gel digestion method (B).
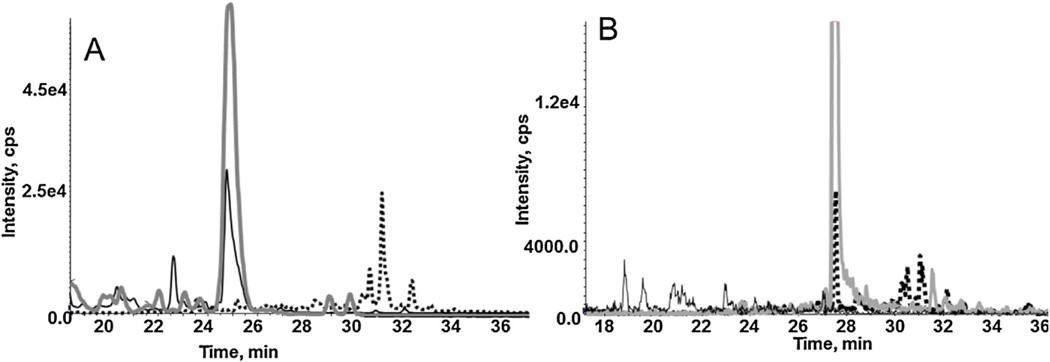
The sample preparation for the transduced CHO cells was optimized in order to enrich the amount of isolated CB1 and CB1b receptors. While several methods exist including gel electrophoresis (1D and 2D), immunoprecipitation, or abundant protein depletion strategy, in this study, only in gel digestion and immunoprecipitation were carried out. Both the transduced and native CHO cells were lysed by sonication followed by passing the cell suspension through needle. Lysis was followed by protein precipitation and resuspension [22], where it was determined that 1 h at 4 °C was sufficient for the CB1b isoform, but 18 h at 4 °C was necessary to have quantitative levels for the CB1 receptor. For protein resuspension, both RIPA and Tris buffer containing urea were used and it was determined that Tris buffer with urea was more effective. While typically, the denaturation step for in-gel digestion is carried out for 10 min at 70 °C, these conditions resulted in non-detectable levels for either CB1 and/or CB1b. As a result, multiple temperatures were assessed and it was determined that 35 °C for 30 min was optimal. For the in-gel digestion of CHO-CB1 cells, only the transitions for CB1 were observed, while for the CHO-CB1b cells, only the CB1b transitions were observed (Fig. 3A–B). While optimizations were carried out for immunoprecipitation [14], the required concentration of the primary antibody was cost-prohibitive and, as a result, the in gel digestion method was preferred.
Fig. 3.
Chromatogram of CB1 (FPLTSFR) (434.2/360.6, solid line) and CB1b (TITTDLLGPFQEK) (775.40/792.38, dashed line) and their respective internal standards (grey line) in CHO-CB1 (A) and CHO-CB1b (B).
While, in most SRM assays, the addition of the internal standard is typically done prior to analysis, in this study, the internal standard was added at various stages of the sample preparation including pre- and post-desalting and prior to injection with spiked CB1 standard in native CHO cells. A 30% decrease in the calculated concentration was observed when comparing the addition of the internal standard prior to desalting versus prior to analysis (0.30 pg/µg vs 0.21 pg/µg respectively). Further, the addition of the internal standard after desalting resulted in 10% change in calculated concentration of the peptide (0.30 pg/µg vs 0.27 pg/µg, respectively). The results of this study indicate that the addition of the internal standard prior to desalting is necessary and that addition at later stages may result in decreased reported values. The targeted proteomic approach was developed for both peptides in native CHO cells using the 5500 QTRAP, no significant matrix effects were observed for these peptides. The top 3 transitions for both the target peptide of CB1 and in CB1b were observed in the CHO cells transduced with CB1 and CB1b respectively.
The calibration curves were found to be linear over the range of 2.18–70 ng/ml for CB1 (r2 > 0.999) and 0.37–6 µg/ml for CB1b (r2 > 0.994). Quality control (QC) (7, 25, 40) ng/ml for CB1 and (1,2,6) µg/ml for CB1b were spiked in native non-transduced CHO cells and intra and interday accuracy and precision were determined (Table 2). Intraday precision (CV) ranged from 1.9% to 4.3%, and inter-day precision (CV) ranged from 1.2% to 5.2% for CB1. In case of CB1b, the intraday precision ranged from 0.4% to 5.9% while interday precision ranged from 1.2% to 6.1%. The intraday accuracy ranged from 89.7% to 109.8% while inter-day accuracy ranged from 90.6% to 110.3% for CB1. The intraday and interday accuracy for CB1b ranged from 92.01% to 118.5% and 95.2% to 119.05% respectively.
Table 2.
Intraday and interday precision and accuracy for CB1 and CB1b in native CHO cells.
| Concentration CB1 (ng/ml) | Concentration CB1b (µg/ml) | |||||
|---|---|---|---|---|---|---|
| 7 | 25 | 40 | 1 | 2 | 6 | |
| Intra-day CV(%) |
1.9 | 3.3 | 4.3 | 1.1 | 0.4 | 5.9 |
| Inter day CV(%) |
5.2 | 1.2 | 2.8 | 1.9 | 1.2 | 6.1 |
| Intra-day Accuracy |
109.8 | 89.7 | 91.9 | 114.1 | 118.5 | 92.01 |
| Inter-day Accuracy |
110.3 | 90.6 | 90.8 | 115.7 | 119.05 | 95.28 |
The SRM method was applied to determine the concentration of CB1 and CB1b in CHO cells transduced with the CB1 and the CB1b isoform. Based on the developed SRM assay, the concentration of CB1 and CB1b in CHO-CB1 cells and CHO-CB1b cells was determined to be 7.15 ng/ml and 0.87 µg/ml respectively, which fell in the linear range of the developed method. The concentration was further normalized to the total protein content and was determined to be 192.07attomol/µg and 23.39 fmol/µg for target peptide of CB1 and CB1b respectively.
A targeted proteomic SRM assay was developed for the determination of CB1 and CB1b in a cellular matrix. All QCs had precision less than <6% and accuracy within 20% of the nominal concentration. The developed assay may be used in determination of CB1 and CB1b in other matrices including tissue.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute on Aging. We thank Alex Straiker for the plasmids.
References
- 1.Bermudez-Silva FJ, Suarez J, Baixeras E, Cobo N, Bautista D, Cuesta-Munoz AL, Fuentes E, Juan-Pico P, Castro MJ, Milman G, Mechoulam R, Nadal A, Rodriguez de Fonseca F. Presence of functional cannabinoid receptors in human endocrine pancreas. Diabetologia. 2008;51:476–487. doi: 10.1007/s00125-007-0890-y. http://dx.doi.org/10.1007/s00125-007-0890-y. [DOI] [PubMed] [Google Scholar]
- 2.Bouaboula M, Poinot-Chazel C, Bourrie B, Canat X, Calandra B, Rinaldi-Carmona M, Le Fur G, Casellas P. Activation of mitogen-activated protein kinases by stimulation of the central cannabinoid receptor CB1. Biochem. J. 1995;312(Pt. 2):637–641. doi: 10.1042/bj3120637. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8526880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakata M, Yada T. Cannabinoids inhibit insulin secretion and cytosolic Ca2+ oscillation in islet beta-cells via CB1 receptors. Regul. Pept. 2008;145:49–53. doi: 10.1016/j.regpep.2007.08.009. http://dx.doi.org/10.1016/j.regpep.2007.08.009, S0167-0115(07)00157-7 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Lauckner JE, Jensen JB, Chen HY, Lu HC, Hille B, Mackie K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2699–2704. doi: 10.1073/pnas.0711278105. http://dx.doi.org/10.1073/pnas.0711278105 (0711278105 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malenczyk K, Jazurek M, Keimpema E, Silvestri C, Janikiewicz J, Mackie K, Di Marzo V, Redowicz MJ, Harkany T, Dobrzyn A. CB1 cannabinoid receptors couple to focal adhesion kinase to control insulin release. J. Biol. Chem. 2013;288:32685–32699. doi: 10.1074/jbc.M113.478354. http://dx.doi.org/10.1074/jbc.m113.478354, M113.478354 [pii] (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starowicz KM, Cristino L, Matias I, Capasso R, Racioppi A, Izzo AA, Di Marzo V. Endocannabinoid dysregulation in the pancreas and adipose tissue of mice fed with a high-fat diet. Obesity (Silver Spring) 2008;16:553–565. doi: 10.1038/oby.2007.106. http://dx.doi.org/10.1038/oby.2007.106 (oby2007106 [pii]) [DOI] [PubMed] [Google Scholar]
- 7.Kargl J, Balenga N, Parzmair GP, Brown AJ, Heinemann A, Waldhoer M. The cannabinoid receptor CB1 modulates the signaling properties of the lysophosphatidylinositol receptor GPR55. J. Biol. Chem. 2012;287:44234–44248. doi: 10.1074/jbc.M112.364109. http://dx.doi.org/10.1074/jbc.m112.364109, M112.364109 [pii] (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKillop AM, Moran BM, Abdel-Wahab YH, Flatt PR. Evaluation of the insulin releasing and antihyperglycaemic activities of GPR55 lipid agonists using clonal beta-cells, isolated pancreatic islets and mice. Br. J. Pharmacol. 2013;170:978–990. doi: 10.1111/bph.12356. http://dx.doi.org/10.1111/bph.12356 (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shire D, Carillon C, Kaghad M, Calandra B, Rinaldi-Carmona M, Le Fur G, Caput D, Ferrara P. An amino-terminal variant of the central cannabinoid receptor resulting from alternative splicing. J. Biol. Chem. 1995;270:3726–3731. doi: 10.1074/jbc.270.8.3726. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7876112. [DOI] [PubMed] [Google Scholar]
- 10.Ryberg E, Vu HK, Larsson N, Groblewski T, Hjorth S, Elebring T, Sjogren S, Greasley PJ. Identification and characterisation of a novel splice variant of the human CB1 receptor. FEBS Lett. 2005;579:259–264. doi: 10.1016/j.febslet.2004.11.085. http://dx.doi.org/10.1016/j.febslet.2004.11.085, S0014-5793(04)01482-6 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Tharp WG, Lee YH, Maple RL, Pratley RE. The cannabinoid CB1 receptor is expressed in pancreatic delta-cells. Biochem. Biophys. Res. Commun. 2008;372:595–600. doi: 10.1016/j.bbrc.2008.05.077. http://dx.doi.org/10.1016/j.bbrc.2008.05.077, S0006-291X(08)00983-2 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Kim W, Doyle ME, Liu Z, Lao Q, Shin YK, Carlson OD, Kim HS, Thomas S, Napora JK, Lee EK, Moaddel R, Wang Y, Maudsley S, Martin B, Kulkarni RN, Egan JM. Cannabinoids inhibit insulin receptor signaling in pancreatic beta-cells. Diabetes. 2011;60:1198–1209. doi: 10.2337/db10-1550. http://dx.doi.org/10.2337/db10-1550, db10-1550 [pii] (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straiker A, Wager-Miller J, Hutchens J, Mackie K. Differential signalling in human cannabinoid CB 1 receptors and their splice variants in autaptic hippocampal neurones. Br. J. Pharmacol. 2012;165:2660–2671. doi: 10.1111/j.1476-5381.2011.01744.x. http://dx.doi.org/10.1111/j.1476-5381.2011.01744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Mariscal I, Krzysik-Walker SM, Doyle ME, Liu Q-R, Cimbro R, Santa-Cruz Calvo S, Ghosh S, Cieśla Ł, Moaddel R, Carlson OD, Witek RP, O’Connell JF, Egan JM. Human CB1 receptor isoforms, present in hepatocytes and β-cells, are involved in regulating metabolism. Sci. Rep. 2016;6:33302. doi: 10.1038/srep33302. http://dx.doi.org/10.1038/srep33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tam J, Vemuri VK, Liu J, Bátkai S, Mukhopadhyay B, Godlewski G, Osei-Hyiaman D, Ohnuma S, Ambudkar SV, Pickel J, Makriyannis A, Kunos G. Peripheral CB1 cannabinoid receptor blockade improves cardiometabolic risk in mouse models of obesity. J. Clin. Invest. 2010;120:2953–2966. doi: 10.1172/JCI42551. http://dx.doi.org/10.1172/JCI42551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juan-Picó P, Fuentes E, Bermúdez-Silva FJ, Javier Díaz-Molina F, Ripoll C, Rodríguez de Fonseca F, Nadal A. Cannabinoid receptors regulate Ca(2+) signals and insulin secretion in pancreatic beta-cell. Cell Calcium. 2006;39:155–162. doi: 10.1016/j.ceca.2005.10.005. http://dx.doi.org/10.1016/j.ceca.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Grimsey NL, Goodfellow CE, Scotter EL, Dowie MJ, Glass M, Graham ES. Specific detection of CB1 receptors; cannabinoid CB1 receptor antibodies are not all created equal! J. Neurosci. Methods. 2008;171:78–86. doi: 10.1016/j.jneumeth.2008.02.014. http://dx.doi.org/10.1016/j.jneumeth.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Chevallet M, Luche S, Rabilloud T. Silver staining of proteins in polyacrylamide gels. Nat Protoc. 2006;1:1852–1858. doi: 10.1038/nprot.2006.288. http://dx.doi.org/10.1038/nprot.2006.288 (nprot.2006.288 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turriziani B, Garcia-Munoz A, Pilkington R, Raso C, Kolch W, von Kriegsheim A. On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology (Basel) 2014;3:320–332. doi: 10.3390/biology3020320. http://dx.doi.org/10.3390/biology3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bermdez-Silva FJ, Surez Prez J, Nadal A, Rodrguez de Fonseca F. The role of the pancreatic endocannabinoid system in glucose metabolism. Best Pract. Res. Clin. Endocrinol. Metab. 2009;23:87–102. doi: 10.1016/j.beem.2008.10.012. http://dx.doi.org/10.1016/j.beem.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Romero-Zerbo SY, Rafacho A, Díaz-Arteaga A, Suárez J, Quesada I, Imbernon M, Ross RA, Dieguez C, de Fonseca FR, Nogueiras R, Nadal A, Bermúdez-Silva FJ. Role for the putative cannabinoid receptor GPR55 in the islets of Langerhans. J. Endocrinol. 2011;211:177–185. doi: 10.1530/JOE-11-0166. http://dx.doi.org/10.1530/JOE-11-0166. [DOI] [PubMed] [Google Scholar]
- 22.Fic E, Kedracka-Krok S, Jankowska U, Pirog A, Dziedzicka-Wasylewska M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 2010;31:3573–3579. doi: 10.1002/elps.201000197. http://dx.doi.org/10.1002/elps.201000197 (n.d) [DOI] [PubMed] [Google Scholar]