Abstract
Human pluripotent stem cells (hPSCs) can self-renew indefinitely, making them an attractive source for regenerative therapies. This expansion potential has been linked with acquisition of large copy number variants (CNVs) that provide mutant cells with a growth advantage in culture1–3. However, the nature, extent, and functional impact of other acquired genome sequence mutations in cultured hPSCs is not known. Here, we sequenced the protein-coding genes (exomes) of 140 independent human embryonic stem cell (hESC) lines, including 26 lines prepared for potential clinical use4. We then applied computational strategies for identifying mutations present in a subset of cells5. Though such mosaic mutations were generally rare, we identified five unrelated hESC lines that carried six mutations in the TP53 gene that encodes the tumor suppressor P53. Notably, the TP53 mutations we observed are dominant negative and are the mutations most commonly seen in human cancers. We used droplet digital PCR to demonstrate that the TP53 mutant allelic fraction increased with passage number under standard culture conditions, suggesting that P53 mutation confers selective advantage. When we then mined published RNA sequencing data from 117 hPSC lines, we observed another nine TP53 mutations, all resulting in coding changes in the DNA binding domain of P53. Strikingly, in three lines, the allelic fraction exceeded 50%, suggesting additional selective advantage resulting from loss of heterozygosity at the TP53 locus. As the acquisition and favored expansion of cancer-associated mutations in hPSCs may go unnoticed during most applications, we suggest that careful genetic characterization of hPSCs and their differentiated derivatives should be carried out prior to clinical use.
Somatic mutations that arise during cell proliferation and are then subject to positive selection are major causes of cancer and other diseases6. Acquired mutations are often present in a subset of cells in a sample, and can therefore be detected in next generation sequencing data from their presence at allelic fractions less than 50%5,7. We reasoned that similar analysis of sequencing data from a large number of hESCs might reveal previously unappreciated mosaic mutations and mutation-driven expansions acquired during hESC culture at single-nucleotide resolution. This approach would complement previous studies describing culture-derived chromosomal-scale aneuploidies and megabase-scale CNVs in hPSCs1,8,9.
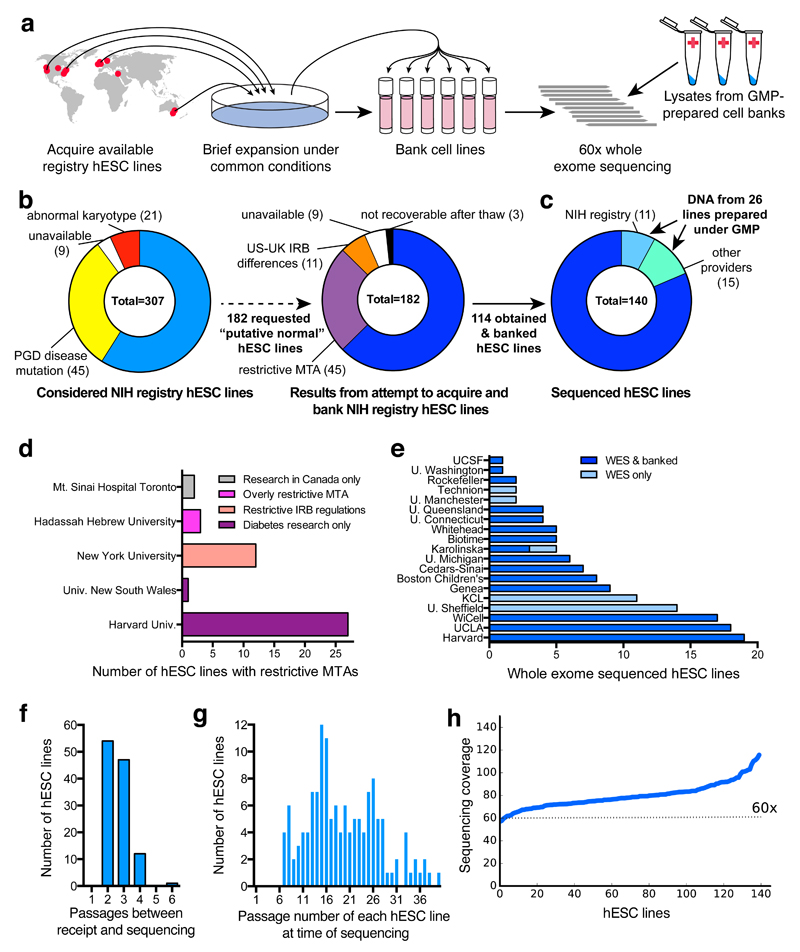
To this end, we sought to collect and perform whole exome sequencing (WES) of hESC lines that were derived under appropriate informed consent and were readily available for distribution (Fig. 1a). We therefore turned to the registry of hESC lines maintained by the US National Institutes of Health (NIH) (Fig. 1b) and were able to obtain, bank, and sequence 114 independent hESC lines (Fig. 1c-e). We selected cell lines at low to moderate passage numbers (mean P18, range P3-P37) and cultured them in a common set of growth conditions for an average of 2.7 ± 0.7 (± STD) passages (range 2-6 passages) prior to banking and sequencing (Fig. 1f,g). Since hESC-derived differentiated cells are currently being studied in clinical trials for their safety and utility in a range of diseases such as macular degeneration10, we also obtained genomic DNA from an additional 26 independent hESC lines that had been prepared under good manufacturing practice (GMP) conditions for potential clinical use (Fig. 1c,e,g). We performed WES of these 140 hESC lines from 19 institutions to a mean read depth of 79.7 ± 0.1 (± SEM) (range 57 for UM4-6 to 115 for UM78-2) (Fig. 1h). Further details on cell line acquisition and selection can be found in Supplementary Table 1 and in Materials and Methods.
Figure 1. Acquisition and WES of 140 hESC lines.
a, Schematic workflow for hESC line acquisition and sequencing. b,c, 114 hESC lines were obtained, banked (b), and analyzed by WES along with 26 GMP-prepared cell lines (c). d, 45 hESC lines were excluded due to use restrictions. e, 140 hESC lines were banked and/or sequenced (see also Supplementary Table 1 and Materials and Methods). f, HESCs were minimally cultured before banking and sequencing. g, Cumulative passage number of hESCs was moderate. h, WES coverage for sequenced hESC lines. IRB, institutional review board; MTA, material transfer agreement; PGD, pre-implantation genetic diagnosis.
To identify potentially acquired mutations, we examined the number of sequencing reads at high-quality and high-coverage heterozygous sites across the exome. We observed that the allelic fractions for most variants followed a binomial distribution, reflecting statistical sampling around the 50% level expected of inherited alleles (Fig. 2a), as well as a much smaller set of sites where variant alleles were present at lower fractions (Fig. 2a,b). We applied a statistical test to identify variants for which the observed allelic fractions were unlikely (P<0.01 by binomial test) to have been generated by random sampling of two equally-present alleles. We found 263 candidate mosaic variants, of which 28 were predicted to have a damaging or disruptive effect on gene function (Supplementary Table 2).
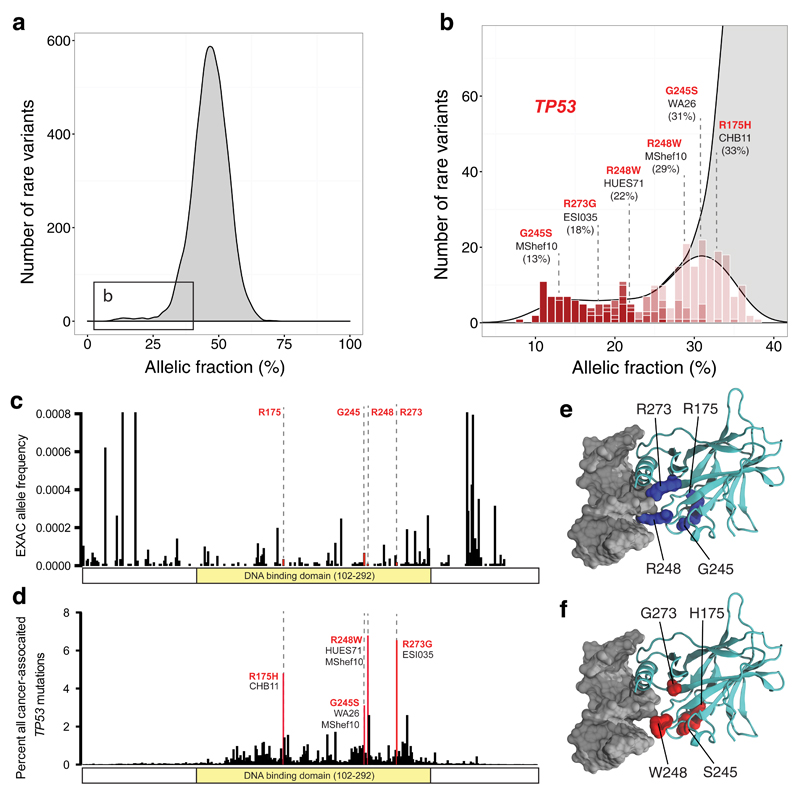
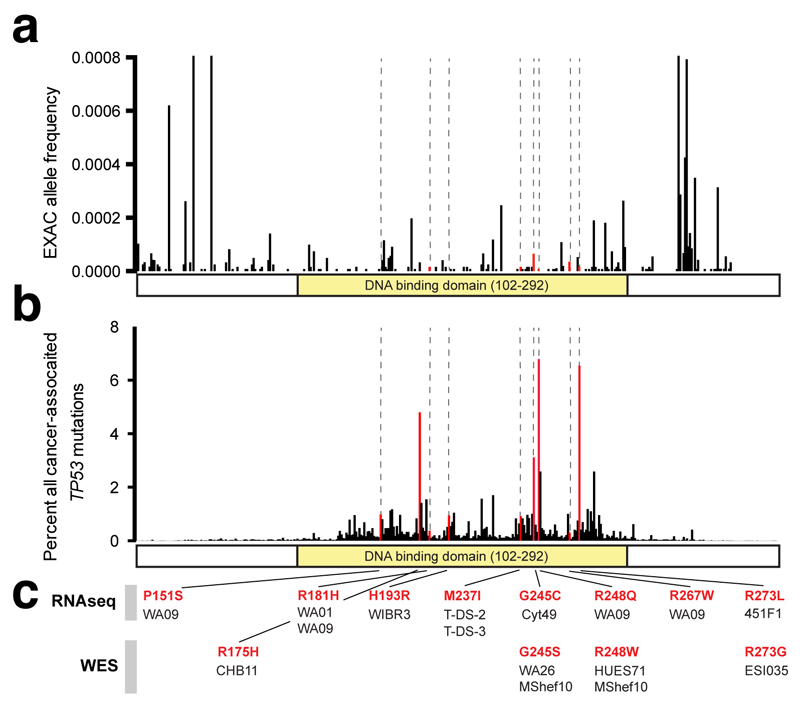
Figure 2. Identification of recurrent, cancer-associated TP53 mutations in hESCs.
a, Some heterozygous variants are present at low allelic fractions (boxed left) in hESCs. b,c Likely mosaic variants (P<0.01, red shading), include six mutations in TP53 (b, Supplementary Table 3) that are rare in ExAC (<0.0001) (c). d, The four affected P53 residues are commonly mutated in human tumors. e, On a crystal structure of P53 bound to DNA, the affected residues map to the DNA binding domain, including to arginine residues that directly interact with DNA. f) The residues mutated in hESCs disrupt DNA binding by P53.
The only gene affected by more than one such mutation was the tumor suppressor gene TP53. TP53 was affected by six of the 28 mutations, found in five unrelated hESC lines (Supplementary Table 3), including a GMP-prepared cell line (MShef10) that carried two distinct TP53 variants (G245S and R248W). These six missense mutations, while rare (<0.01%) in the general population11 (Fig. 2c), mapped to the four residues most frequently disrupted in human cancer (Fig. 2d, Supplementary Table 3)12–14. Since P53 is mutated in approximately 50% of tumors15, coding mutations in these four residues are associated with a substantial fraction of human cancer disease burden. Each of the six mutations involved a cytosine residue of a CpG dinucleotide and may therefore involve a highly mutable site16.
On a crystal structure of the human P53 protein complexed with DNA, each of the mutations mapped to the DNA-binding domain of P5317 (Fig. 2e,f). Mutations at these positions are associated with cancer and often act in a dominant negative fashion to substantially diminish P53’s regulation of apoptosis, cell cycle progression, and genomic stability18. Individuals with germ-line mutations at these residues develop Li-Fraumeni syndrome, an autosomal dominant disease with a lifetime cancer risk of nearly 100%19. In these patients, tumors can arise at any age and can affect most tissues, including the brain, bones, lung, skin, soft tissues, adrenal gland, colon, stomach, and blood20.
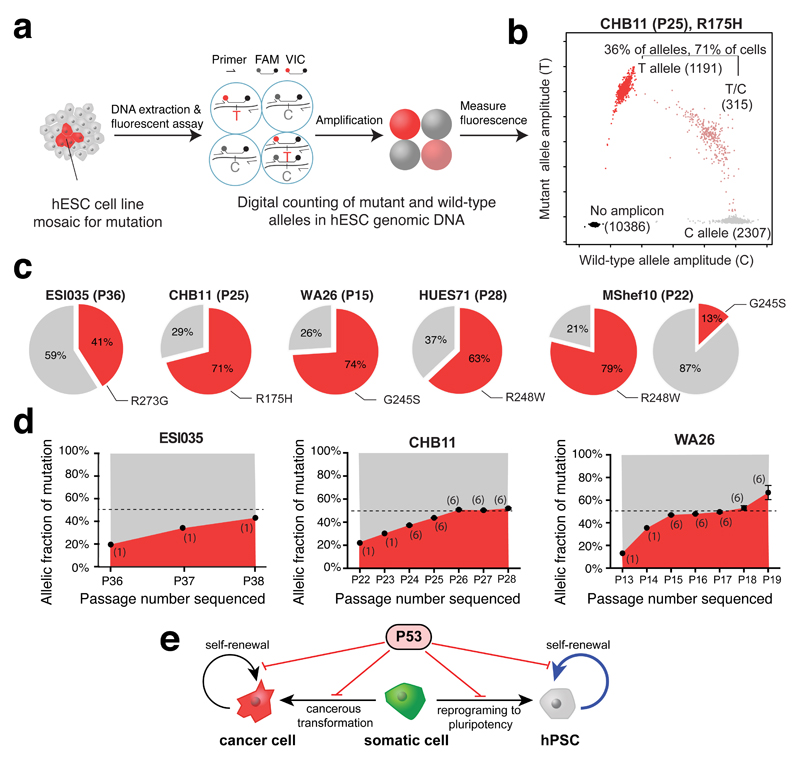
To independently test the hypothesis that the inactivating TP53 mutations were acquired during cell culture, we developed droplet digital PCR (ddPCR) assays to digitally count the abundance of each allele at the four TP53 mutation sites (Fig. 3a,b, Supplementary Table 4). Analysis of genomic DNA derived from the 140 hESC lines confirmed that all six mutations identified by WES were indeed mosaic, with allelic factions ranging from 7-40%, suggesting their presence in 14-80% of cells in culture (Fig. 3c). We did not identify additional cell lines carrying mutations at these positions, suggesting they were either absent or present at allelic fractions below the sensitivity of the assay (approximately 0.1%)21. These findings demonstrate that each of the TP53 mutations identified in hESCs was an acquired mutation and that cells with the mutation had come to represent a significant fraction of cells in affected lines.
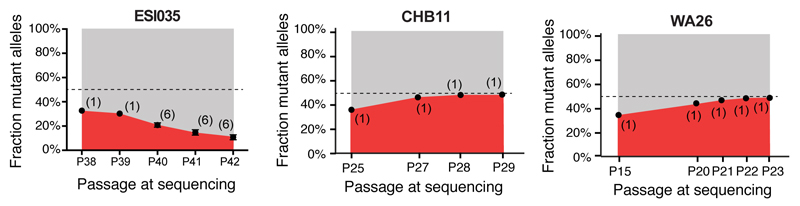
Figure 3. TP53 mutations in hESCs are mosaic and confer strong selective advantage.
a, Droplet digital PCR (ddPCR) assay schematic. b, Representative ddPCR data showing droplets containing the reference allele (gray), mutant allele (red), both alleles (pink), or neither allele (black). c, Estimated fraction of mutant cells (red) in affected hESC lines. d, Mutant allelic fraction rapidly increases during standard hESC culture. Error bars depict SEM and numbers indicate replicate wells. Note further allele-fraction expansion (after P17) for WA26, likely involving LOH. e, Model of P53’s role in both cancer and stem cell biology.
We next asked whether the cells harboring these TP53 mutations expanded their representation within the hESC population across passages. We re-obtained early passage vials for hESC lines that were mosaic for TP53 mutations (CHB11 at P22, and WA26 at P13), thawed a fresh vial of ESI035 at P36, and analyzed the genomic DNA from the frozen vial and at each subsequent passage to test for changes in mutant allelic fraction. In each of the three hESC lines, TP53 mutant alleles increased in representation over passages (Fig. 3d) in all but one experiment, suggesting these mutations conferred a strong selective advantage (approximately 1.9-fold/passage) under standard hESC culture conditions (Fig. 3e, Extended Data Figure 1, Supplementary Table 5). To confirm that this selective advantage was conferred by TP53 mutations and not by CNVs at Chr20q11.211–3, we analyzed all 140 hESC lines using microarrays and found that none of the lines carrying TP53 mutations also carried the Chr20q CNV (Supplementary Table 6). Our overall results are consistent with a model in which positive selective pressure for clonal expansion of mutations that inactivate P53 are present during the routine culture of hPSCs. Indeed, it has previously been reported that loss of P53 activity facilitates the reprogramming of somatic cells to pluripotency22,23 and promotes hPSC survival and proliferation24, suggesting a prominent role for P53 in regulating self-renewal in hPSCs (Fig. 3e).
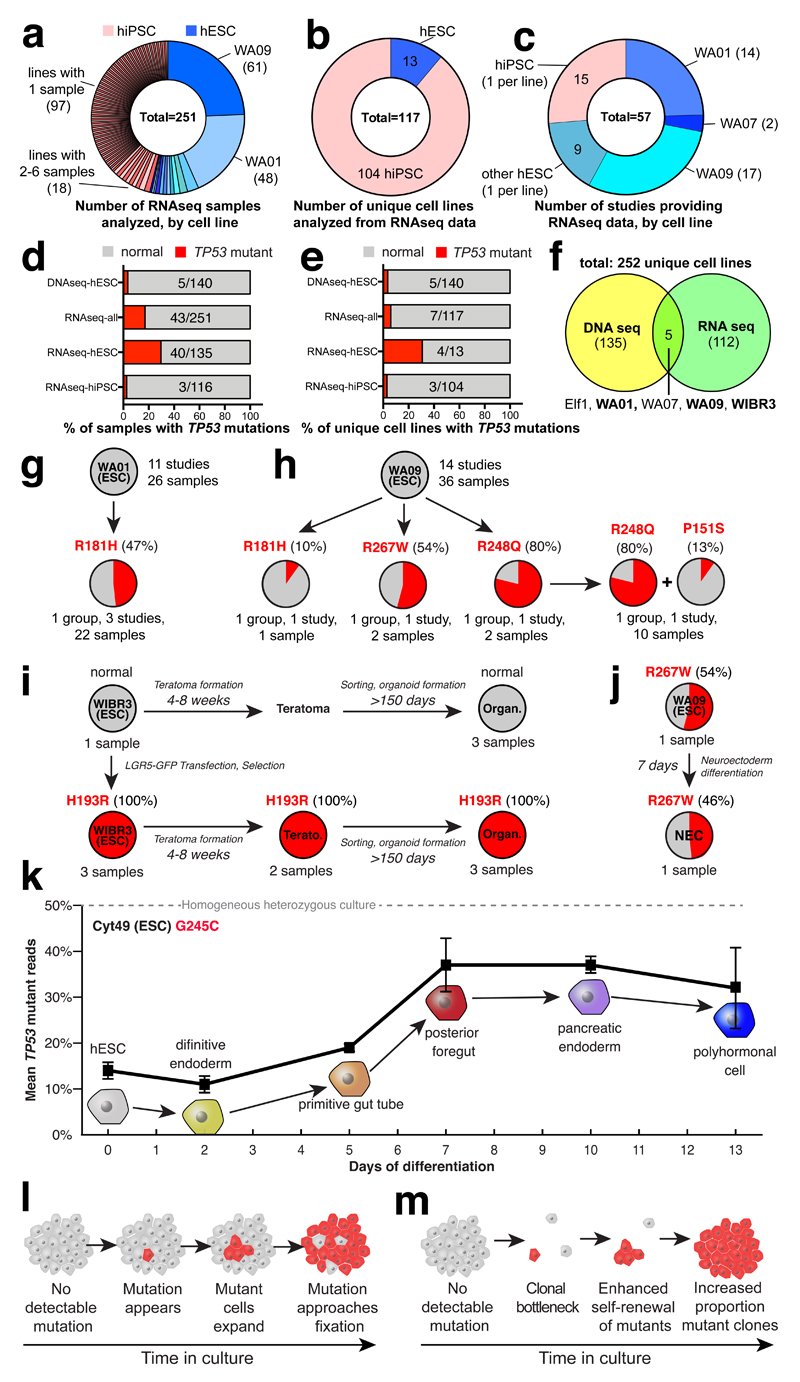
To test the reproducibility of our observations and to explore the effects of P53 mutations in additional contexts, we screened for TP53 mutations in publically available RNA sequencing data from 251 hPSC samples in 57 published studies, corresponding to 13 hESC and 104 hiPSC lines (Fig. 4a-c, Supplementary Table 7). The relatively high expression of TP53 in stem cells provided sufficient read depth for allelic counting and allowed us to identify nine instances of eight distinct point mutations in TP53, three of which we had independently seen by WES. Like the mutations ascertained by WES, each of these eight mutations led to missense substitutions in the DNA-binding domain of P53 (Fig. 4d-f, Supplementary Table 3, Extended Data Figure 2). When we considered both WES and RNAseq data sets, we identified four codons that were recurrently mutated in hPSCs: R181, G245, R248, and R273. Notably, we identified four distinct TP53 mutations in the commonly used WA09 (H9) hESC line grown in different laboratories (P151S, R181H, R248Q, and R267W) indicating that the mutations arose during cell culture (Fig. 4h).
Figure 4. A substantial fraction of hPSCs in published studies harbor TP53 mutations.
a-e, Published RNAseq data show that 7/117 (6%) unique hPSC lines harbor P53 mutations. f, Combined DNAseq and RNAseq analysis reveals 12/252 (5%) distinct cell lines affected by 15 TP53 mutations (Supplementary Table 3). g,h,i, P53 mutant WA01 was seen in three (g), WA09 acquired four distinct TP53 mutations in three groups (h), and WIBR3 lost all normal copies of TP53 after gene editing (i). j,k, TP53 mutant cells could be differentiated (j,k), and expanded relative to WT cells (k). Error bars depict SEM. l,m, Model of TP53 mutation enrichment during hPSC culture (l) or during clonal bottlenecks (m).
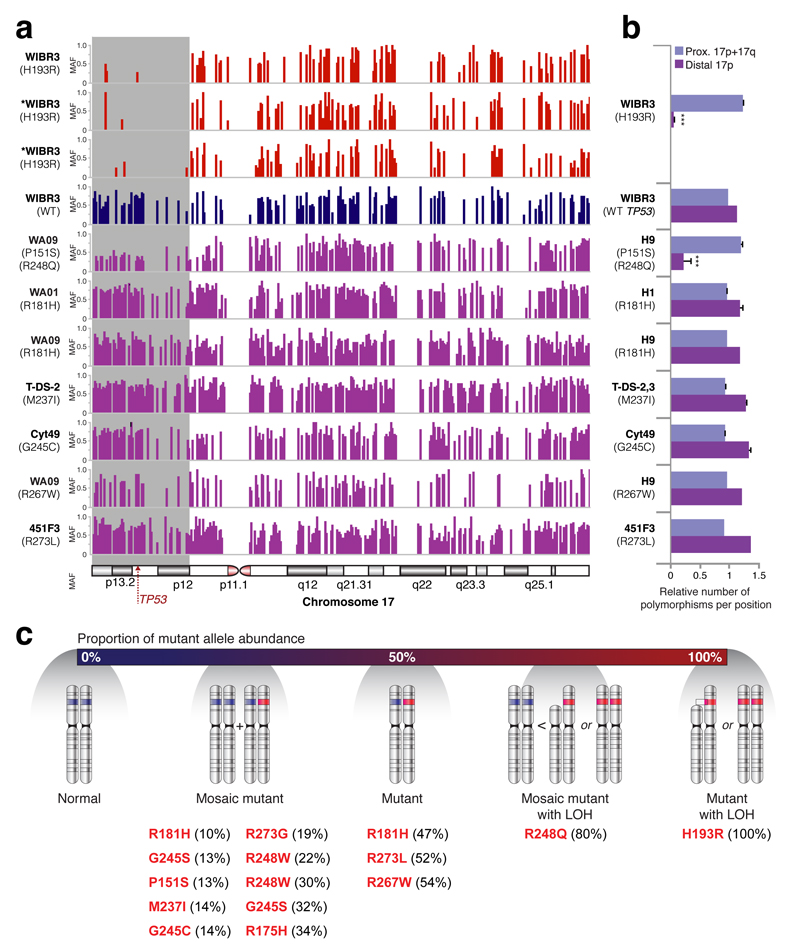
Of the 15 instances of TP53 mutations observed by either WES or RNAseq, the percentage of mutant reads suggested that 10 were mosaic and that three had reached fixation (50% of allelic fraction). Surprisingly, two cell lines, WA09 (R248Q) and WIBR3 (H193R) had 80% ± 3% and 100% mutant reads, respectively (Supplementary Table 3). These findings were consistent with the excess allelic fraction observed during the culture of WA26 (Fig. 3d) and suggested the presence of additional mutational mechanisms affecting mutant TP53 allelic fraction. Indeed, we observed loss of heterozygosity (LOH) of a large telomeric domain including the TP53 locus along chromosome 17 (Extended Data Figure 3) that was almost complete in a gene-targeted derivative of WIBR3 (Fig. 4i), and was partial in WA09, consistent with the observed high fraction (80%) of mutant TP53 reads. These results suggest that follow-on LOH after an initial TP53 point mutation likely confers additional selective advantage.
We next asked if TP53 mutations might affect cell differentiation or affect the survival of differentiated cells. To this end, we examined studies for which there was RNA sequencing data for both hESCs and their differentiated progeny. Cell lines with substantial factions of TP53 mutant cells could readily form teratomas, gut epithelial cells25 (Fig. 4i), neuroepithelial cells26 (Fig. 4j), and pancreatic polyhormonal cells27 (Fig. 4k). Notably, a hESC line harboring a mosaic G245C mutation expanded its mutant allelic fraction over the course of differentiation27, suggesting a continued selective advantage in differentiating mutant cells.
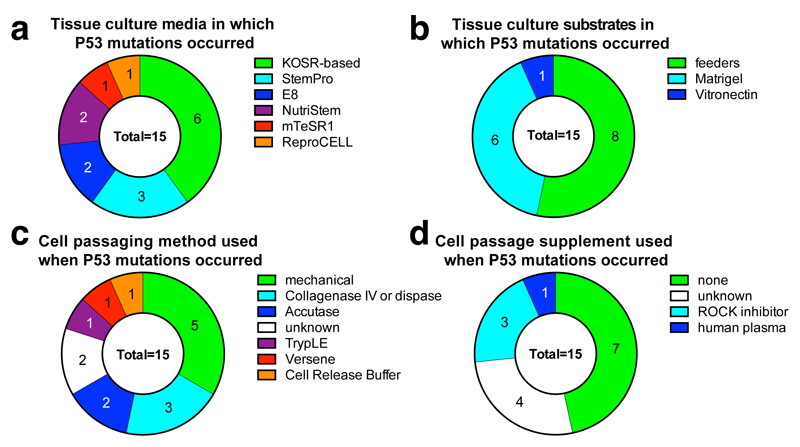
Together, our analyses indicate that researchers have unknowingly and routinely used hPSCs that harbor cancer-related missense mutations in TP53, sometimes accompanied with LOH. These findings have practical implications for the use of hESCs in disease modeling and transplantation medicine. The fact that we observed TP53 mutations among both hESC and hiPSCs cultured with a wide variety of media, substrates, and passaging methods (Extended Data Figure 4) suggests that new culture conditions should be explored to reduce the selective pressure for TP53 mutations. We also suggest that hPSCs should be regularly subjected to genetic testing, particularly before and after stressful interventions such as gene editing or single-cell cloning that force hPSC populations through bottlenecks (Fig. 4l,m). Our specific findings here suggest that the P53 pathway should be an immediate focus for these genetic tests. However, a comprehensive ascertainment of recurrent culture-acquired mutations will require the analysis of still-larger collections of stem cell lines by both exome and whole genome sequencing.
Our findings also demonstrate that sequencing tests provide an opportunity to detect potentially harmful mutations in differentiated cell preparations derived from hPSCs, thereby increasing the safety of cell replacement therapies for conditions ranging from diabetes to Parkinson’s disease. In support of this notion, clinical trials with hPSC-derived materials have recently been halted due to the discovery of undisclosed mutations28, but have since resumed. We suggest that hPSCs and their derivatives be subjected to genome-wide analyses at several key steps: during initial cell line selection, as part of the characterization of a master bank of hPSCs, and as an end-stage release criterion prior to the transplantation of the hPSC-derived cellular product. Importantly, although TP53 mutations recurred at detectable fractions in several cell lines, most lines (~95%) were free of detectable TP53 mutations despite having spent extensive time in culture. Regenerative medicine remains a viable and exciting goal that is more likely to succeed as potential pitfalls, like the one we report here, are identified and addressed.
Materials and Methods
hESC acquisition
As a source of hESCs for this study, we focused on those that had been voluntarily listed by research institutions on the registry of hESC lines maintained by the US National Institutes of Health (NIH) (http://grants.nih.gov/stem_cells/registry/current.htm). As of July 8, 2015, a total of 307 hESC lines were listed on this registry. Of these, we requested viable frozen stocks of the 182 lines annotated to be available for distribution and to lack known karyotypic abnormalities or disease-causing mutations. During our effort to obtain these cell lines, we found that 45 were subject to overly restrictive material transfer agreements that precluded their use in our studies and 11 could not be readily obtained as frozen stocks due to differences in human subjects research regulations between the U.S. and the U.K. Nine cell lines were unavailable upon request or were overly difficult to import, and three could not be cultured despite repeated attempts. Further details on the availability of cell lines can be found in Supplementary Table 1.
The generation of hESCs used in this study was previously approved by the institutional review boards (IRBs) of all providing institutions and reviewed by either the United States National Institutes of Health (US-NIH) or the United Kingdom Stem Cell Steering Committee (UK-SCSC) as all cell lines used in this study were listed on the US-NIH Embryonic Stem Cell Registry or UK Stem Cell Registry. Use of the hESCs for sequencing at Harvard was further approved and determined not to constitute Human Subjects Research by the Committee on the Use of Human Subjects in Research at Harvard University.
hESC culture
In a separate document, we describe in detail a protocol for the adaptation of hESC lines from diverse culture conditions (xxxxxx Reference to Nature Protocol Exchange paper once this DOI is generated). Briefly, we considered that different laboratories employ different methods to culture hESCs, raising the question of how best to thaw and culture the cell lines we obtained from multiple sources. Traditionally, hESCs are maintained on gelatinized plates and co-cultured with replication-incompetent mouse embryonic fibroblast (MEF) feeder cells in tissue culture medium containing knockout serum replacement (KOSR). More recently, hESCs have been cultured on a substrate of cell line-derived basal membrane proteins known by the trade names of Matrigel (BD Biosciences) or Geltrex (Life Technologies), in mTeSR129, E830, or similar in the absence of feeder cells. In previous work, we found that a medium containing an equal volume of KOSR-based hESC medium (KSR) and mTeSR1 (STEMCELL Technologies) (KSR:mTeSR1) robustly supports the pluripotency of hESCs undergoing antibiotic selection during the course of gene targeting experiments under feeder-free conditions31. To minimize stress to hESCs previously cultured and frozen under diverse conditions, cell lines were thawed in the presence of 10 μM Y-27632 (DNSK International) into two wells of a 6-well plate, one of which contained KSR:mTeSR1 on a substrate of Matrigel, and the other containing KOSR-based hESC medium on a monolayer of irradiated MEFs. After 24 hours, Y-27632 was removed and cells were fed daily with the aforementioned media in the absence of any antibiotics. All cultures were tested for the presence of mycoplasma and cultured in a humidified 37°C tissue culture incubator in the presence of 5% CO2 and 20% O2.
Colonies of cells with hESC morphology and with a diameter of approximately 400-1000 micrometers were transferred into KSR:mTeSR1 medium containing 10 μM Y-27632 on a substrate of Matrigel by manual picking under a dissecting microscope. Cells with differentiated morphology were removed from plates by aspiration during feeding. Once cultures consisting of cells with homogeneous pluripotent stem cell morphology had been established, they were passaged by brief (2-10 minute) incubation in 0.5 mM EDTA in PBS followed by gentle trituration in KSR:mTeSR1 medium containing 10 μM Y-27632 and re-plating. Once cultures had reached approximately 90% confluence in one well of a 6 well plate, they were passaged with ETDA onto a Matrigel-coated 10 cm plate. Upon reaching approximately 90% confluence, cell lines were dissociated with EDTA as described above and banked for later use in cryoprotective medium containing 50% KSR:mTeSR1, 10 μM Y-27632, 10% DMSO, and 40% fetal bovine serum (HyClone). A subset of hESC lines (Supplementary Table 1) were passaged enzymatically with TrypLE Express (Life Technologies), expanded onto two 15 cm plates, and frozen down in 25 cryovials.
Whole exome sequencing and genotyping
Cell pellets of approximately 1-5 million cells were generated from banked cryovials of research-grade hESC lines, or were obtained directly from institutions providing GMP-grade hESC lines. Cell pellets were digested overnight at 50°C in 500 μl lysis buffer containing 100 μg/ml proteinase K (Roche), 10 mM Tris (pH 8.0), 200 mM NaCl, 5% w/v SDS, 10 mM EDTA, followed by Phenol:Chloroform precipitation, ethanol washes, and resuspension in 10 mM Tris buffer (pH 8.0). Genomic DNA was then transferred to the Genomics Platform at the Broad Institute of MIT and Harvard for Illumina Nextera library preparation, quality control, and sequencing on the Illumina HiSeq 2500 platform. Sequencing reads (150 bp, paired-end) were aligned to the hg19 reference genome using the BWA alignment program. Genotypes from WES data for the cell lines were computed using best practices from GATK software32 compiled July 31, 2015. Sequencing quality and coverage were analyzed using Picard tool metrics. Cross sample contamination was estimated using VerifyBamID (v1.1.2)33. Data from each cell line was independently processed with the HaplotypeCaller walker and further aggregated with the CombineGVCFs and GenotypeGVCFs walkers to generate a combined variant call format (VCF) file. Genotyped sites were finally filtered using the ApplyRecalibration walker.
To determine whether lines with or without acquired TP53 mutations show other chromosomal aberrations or smaller regional changes in copy number, additional genotyping of the 140 hESC lines was performed using a custom high density SNP array (“Human Psych array”) that contains more than half a million SNPs across the genome. CNVs larger than 500 kb were identified using the PennCNV (v1.0.0)34 tool (penncnv.openbioinformatics.org). All CNVs were manually reviewed and are shown in Supplementary Table 6.
Mosaic variant analysis
To identify candidate mosaic variants, a table of heterozygous variants was generated from the VCF (Supplementary Table 2). To limit the frequency of false positive calls due to sequencing artifacts and PCR errors, variants were included if they had a variant read depth (DP) of at least 10, if they were either flagged as a “PASS” site or were not reported in the Exome Aggregation Consortium’s (ExAC) database11, and if they were not located in regions of the genome with low sequence complexity, common large insertions and segmental duplications, as described by Genovese and colleagues5. Multiallelic sites were split, left-aligned, and normalized. The resulting list of 2.1 million “high-quality heterozygous variants” was further refined to include sites that were covered by at least 60 unique reads and had a high confidence variant score (“PASS”) as ascertained by GATK’s Variant Quality Score Recalibration software (840,222 variants). To exclude common inherited variants we selected variants present in less than 0.01% of the (ExAC) control population and restricted our analysis to only singleton or doubleton variants (9490 variants present in 1-2 of the 140 samples). Coverage was calculated by summing reference and alternate allele counts for each variant. Allelic fraction (AF) was calculated by dividing the alternate allele count by the total read coverage (both alleles) of the site.
Although the allelic fraction of inherited heterozygous variants is expected to be 50%, reference capture bias (a tendency of hybrid selection to capture the reference allele more efficiently than alternative alleles) causes the actual expected allele fraction for SNPs and indels to be closer to 45% and 35% respectively5. To account for these technical biases, we used a binomial test with a null model centered at 45% allelic fraction for inherited SNPs and 35% for inherited indels. Variants for which this binomial test was nominally significant (P < 0.01) were deemed to be candidate mosaic variants. The nominal P-value threshold of 0.01 was chosen as an inclusive threshold in order to screen sensitively for potentially mosaic variants, at the expense of also capturing false positives for which low allelic fractions represented statistical sampling fluctuations. For this reason, we considered it important to further evaluate putative mosaic variants by independent molecular methods that deeply sample alleles at the nominated sites (Figure 3). A much more stringent computational screen based on a P-value threshold of 10-7 identified three of the six TP53 variants, and TP53 was also the only gene with multiple putatively mosaic variants in this screen.
We also identified all high quality heterozygous variants that passed the inclusive statistical threshold of (P<0.01) in our binomial test and could potentially be mosaic (n=36,396). These data are included in Supplementary Table 2.
Variant annotation was performed using SnpEff with GRCh37.75 Ensembl gene models. Variants with moderate impact were classified as damaging by a consensus model based on seven in silico prediction algorithms35.
Assessment of TP53 mutation frequency in cancer
We turned to the ExAC database11 that complies the whole exome sequences of over 60,000 individuals to assess the frequency at which the amino acid residues we observed to be mutated in some hESCs were affected in the general population. We then consulted the COSMIC (http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=TP53)12, ICGC (https://cbioportal.gdc.cancer.gov/cbioportal/)13, and IARC P53 (http://p53.iarc.fr/TP53SomaticMutations.aspx)14 databases and plotted the percent of tumors carrying a mutation in each codon (Fig. 2d, Extended Data Figure 2b).
Molecular modeling of P53 protein
To visualize the spatial location of the amino acid residues affected by TP53 mutations observed in hESCs by WES on the P53 protein, we downloaded the 1.85 Angstrom X-ray diffraction based structure file from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (file 2AHI) and built the model protein/DNA system (Chain IDs D, G, and H) to visualise the secondary structure of a P53 monomer complexed to DNA as a ribbon diagram. DNA was illustrated as a space-filling model. Water molecules were discarded when building the wild-type model and minimized in two steps using the AMBER 16 package36. Affected residues were indicated as space-filling model superimposed on the ribbon diagram of P53 and highlighted in blue (wild-type) or red (mutated) without consideration of how the mutations might affect the secondary or tertiary structure of the protein.
Measurement of TP53 variant allele fraction by ddPCR
We assayed the allelic fraction of the four distinct TP53 mutations identified by WES (Supplementary Table 3) in the 140 hESC lines by droplet digital PCR (ddPCR). Each ddPCR analysis incorporated a custom TaqMan assay (IDT). Assays were designed with Primer3Plus and consisted of a primer pair and a 5’ fluorescently labeled probe (HEX or FAM) with 3’ quencher (Iowa Black with Zen) for either the control (reference) or mutant (alternative) base for each identified P53 variant (Supplementary Table 4). Genomic DNA from each hESC line was analyzed by ddPCR according to the manufacturer’s protocol (BioRad). The frequency of each allele for a given sample was estimated first by Poisson correction of the endpoint fluorescence reads21. These corrected counts were then converted to fractional abundance estimates of the mutant allele and multiplied by two to determine the fraction of cells carrying the variant allele.
Longitudinal hESC culture and calculation of TP53 mutation expansion
To assess how the allelic fraction of TP53 mutations might change over time in culture, hESC lines CHB11 (passage 22 or 25), WA26 (passage 13 or 15), and ESI035 (passage 36 in two separate experiments) were serially passaged in mTeSR1 media (STEMCELL Technologies) at a density of approximately 30,000 cells/cm2 in the presence of 10 μM Y-27632. Cells were fed daily with mTeSR1 and passaged with Accutase (Innovative Cell Technologies Inc.) at approximately 90% confluence. To monitor changes in allelic fractions, genomic DNA from cells at the indicated passages were analyzed by ddPCR. To calculate the relative expansion rate of mutant relative to wild-type cells, we applied the following formula:
where R0 is defined as the ratio of (variant positive cells)/(variant negative cells) after some number of starting passages and R1 and R2 represent the aforementioned ratios measured on the same sample at T1 and T2>T1 passages respectively. From this equation, the estimation of variant positive cells after t passages from starting ratio R0 can be defined as
Note that this equation estimates the relative growth rate of cells carrying the variant allele with a round of passaging as unit of time, with both relative survival and growth being incorporated. These data are included in Supplementary Table 5. For the subsequent calculation of the earliest passage at which these mutations might have become detectable, the detection thresholds (R0) for WES and ddPCR was assumed to be 0.1 (10/100 reads) and 0.001 (1 per 1000 droplets), respectively.
RNA sequencing analysis
In order to identify P53 mutations in hPSCs, we analyzed 256 publicly available high-throughput RNA sequencing samples of hPSCs from the SRA database (http://www.ncbi.nlm.nih.gov/sra)37. Data accession numbers for SRA (and GEO, where applicable) are provided in Supplementary Table 7. Five of these 256 samples were not considered further as they were from single cells rather than cell lines. Following sequence alignment to the hg19 reference genome with Tophat238, single nucleotides divergent from the reference genome were identified using GATK HaplotypeCaller32. As sufficient sequencing-depth is required to deduce sequence mutation, a threshold of 25 reads per nucleotide was set. Under this criterion, 43 samples (40 hESC and 3 hiPSC) had a missense mutation in TP53. 10 of the 40 hESC samples (WA09) carried two separate mutations (Supplementary Table 7). Upon the identification of cell lines carrying mutant reads, RNA sequencing data from studies containing differentiated samples were included for analysis.
Loss of heterozygosity analysis
In order to evaluate TP53 alleles, we assessed the level of polymorphism by calculating the ratio between the minor and major alleles across chromosome 17. In order to minimize sequencing noise and errors, we included SNPs covered by more than 10 reads and that are located in the dbSNP build 142 database39. The resulted wig files were then plotted using Integrative Genomics Viewer (IGV)40 (Extended Data Figure 4). In order to quantify the difference in polymorphism between samples, we converted the wig files to BigWig using UCSC Genome Browser utilities41 and summed the allelic ratios between the distal part of the short arm of chromosome 17 (17p), the proximal side of this arm and the long arm of chromosome 17 (17q). The allelic ratio sum was then divided by the region's length (bp), which resulted in the proportion of SNPs, followed by one-sided Z score test for two population proportion to compare between the chromosome 17 areas within each sample. While most samples with mutations in TP53 showed a comparable, non-significant rate of polymorphic sites along the chromosome, WIBR3 samples with H193R mutations and WA09 samples with both P151S and R248Q mutations had a significantly different proportion (p<0.001) of polymorphic sites, in the distal part of the short arm of the chromosome (first 16x106 base pairs), including TP53 site. Unlike the three mutant WIBR3 samples, the wild-type WIBR3 sample had a normal distribution of polymorphic sites with no significant difference between the short and long arms.
Extended Data
Extended Data Figure 1. Replicates of cell competition assays carried out at earlier starting passages.
Note that while the mutant allelic fractions for lines CHB11 and WA26 approach fixation, that the fraction of mutant cells unexpectedly decreases for ESI035 over several passages, indicating a potential selective disadvantage that co-segregates with the TP53 mutation in this experiment. The number of replicate wells is indicated in each graph. Error bars depict SEM.
Extended Data Figure 2. Summary of all observed P53 mutations.
a-b, Graphical representation of each of the 9 mutated bases in P53 observed across the 252 whole exome sequenced (WES) and RNA sequenced (RNAseq) hPSC lines depicting their allele frequency in ExAC (a) and the incidence with which the relevant codons are mutated in human cancer (b). c, The 15 instances of these mutations in 12 distinct cell lines is represented along with whether the mutation was seen by WES or RNAseq. Although the M237I event is seen in two distinct iPSC lines, it is conservatively counted here as a single event since the hiPSC clones may be related.
Extended Data Figure 3. Analysis of loss of heterozygosity in RNA sequencing samples.
a, Polymorphic sites on chromosome 17 in different hPSCs with mutations in TP53. WIBR3 cells with H193R mutation and H9 cells with both P151S and R248Q mutations show less polymorphism in the distal part of chromosome 17p compared to the proximal part of 17p and 17q. *samples with less than 25 reads. b, Summation of the polymorphic sites in the distal part of chromosome 17p compared to the proximal part of 17p and 17q, divided by the overall frequency of polymorphic sites along chromosome 17. WIBR3 cells with H193R mutation and H9 cells with both P151S and R248Q mutations have a significantly different proportion between the two parts of the chromosome, implying loss of heterozygosity (LOH). Error bars depict SEM, ***p < 0.001. c, A schematic representation of possible allele states of TP53 in cultured hPSCs with all observed mutations depicted. Depending on the percentage of mutant reads in a culture, one can deduce if the culture is homogenous or mosaic for a mutation, and whether, in addition to a point mutation, LOH has occurred in the TP53 locus. MAF, minor allele frequency.
Extended Data Figure 4. Culture and passaging method employed for samples bearing P53 mutations.
a, P53 mutations were observed in hPSCs grown in a broad array of culture media including home-made medium supplemented with knockout serum replacement (KOSR), and defined, commercial media such as E8. b, Similar numbers of P53 mutations were observed from cells grown with feeder cells or under feeder-free conditions. c, Since passaging hPSCs can introduce stresses or clonal bottlenecks, we examined whether P53 mutations were consistently seen when a particular passaging method was used, but we observed a wide variety of passaging methods associated with these mutations. Note that the interpretation of these data are complicated by the fact that the culture methods employed in the final published study may not reflect the previous culture history of that cell line, which may have previously passed through multiple laboratories, as well as by the lack of detail about culture methods present in some published studies. d, The addition of supplements such as rock inhibitor at passages does not appear to be sufficient to prevent P53 mutations in hPSCs.
Supplementary Material
Acknowledgements
We thank the many institutions and investigators world-wide that provided their cell lines and supported the publication of the results. We are indebted to Diane Santos, Melissa Smith, Kristen Elwell, Mary Anna Yram, Stacey Ellender, Liz Bevilacqua, and Diane Gage for their assistance with the regulatory and logistical efforts required to acquire and sequence hESC lines. We also thank Kiki Lilliehook for her comments and Ilyas Yildirim for his assistance with the molecular modeling of P53 mutations. We regret the omission of any relevant references or discussion due to space limitations. The Genomics Platform at the Broad Institute performed sample preparation, sequencing, and data storage. YA is a Clore Fellow. NB is the Herbert Cohn Chair in Cancer Research and was partially supported by The Rosetrees Trust and The Azrieli Foundation. Costs associated with acquiring and sequencing hESC lines were supported by HHMI and the Stanley Center for Psychiatric Research. FTM, SAM, and KE were supported by grants from the NIH (HL109525, 5P01GM099117, 5K99NS08371). KE was supported by the Miller consortium of the HSCI and FTM is currently supported by funds from the Wellcome Trust, the Medical Research Council (MR/P501967/1), and the Academy of Medical Sciences (SBF001\1016).
Footnotes
Data Availability Statement
Sequence data that support the findings of this study have been deposited in EGA (https://www.ebi.ac.uk/ega/home) under controlled access with the accession code EGAS00001002400 and in dbGaP (https://www.ncbi.nlm.nih.gov/gap) under controlled access with the accession code xxxxxxx. Computer code relevant to data analysis will be made available upon request.
Author contributions
FTM, SG, SAM, and KE conceived the project. FTM and KE acquired hESC lines with the assistance of MC and GS, who also assisted with regulatory issues pertaining to sequencing and data distribution. FTM cultured and banked hESC lines, prepared them for sequencing, and coordinated efforts to interpret and visualize sequencing data with the assistance of SG. SG performed computational data analysis and visualization with the help of GG, BH and SK. YA preformed the analysis of TP53 mutations in the RNA-seq database with the assistance of SB and NB. Data were interpreted by FTM, SG, NK, GG, YA, SB, NB, SAM, and KE. NK, JM and CM designed, performed and analyzed experiments to measure the mosaic nature and competitive expansion of TP53 mutations. SM derived HUES 68, 69, 70, 74, 75, and DI provided the KCL lines. FTM, SG, SAM, and KE prepared drafts of the manuscript text and figures with contributions and comments from all authors.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.ISCI. screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nature Biotechnology. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery S, et al. BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Reports. 2013;1:379–386. doi: 10.1016/j.stemcr.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen HT, et al. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol Hum Reprod. 2014;20:168–177. doi: 10.1093/molehr/gat077. [DOI] [PubMed] [Google Scholar]
- 4.Unger C, Skottman H, Blomberg P, Dilber MS, Hovatta O. Good manufacturing practice and clinical-grade human embryonic stem cell lines. Human Molecular Genetics. 2008;17:R48–53. doi: 10.1093/hmg/ddn079. [DOI] [PubMed] [Google Scholar]
- 5.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 7.Jaiswal S, et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nature Biotechnology. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 9.Baker D, et al. Detecting Genetic Mosaicism in Cultures of Human Pluripotent Stem Cells. Stem Cell Reports. 2016;7:998–1012. doi: 10.1016/j.stemcr.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz SD, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt's macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015;385:509–516. doi: 10.1016/S0140-6736(14)61376-3. [DOI] [PubMed] [Google Scholar]
- 11.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Research. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database. 2011;2011:bar026–bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouaoun L, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 16.Rideout WM, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 17.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 18.Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene. 2004;23:2330–2338. doi: 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- 19.Malkin D. Li-fraumeni syndrome. Genes & Cancer. 2011;2:475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, et al. Heterogeneity of Li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep. 2014;4:4223. doi: 10.1038/srep04223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Amir H, et al. Spontaneous Single-Copy Loss of TP53 in Human Embryonic Stem Cells Markedly Increases Cell Proliferation and Survival. STEM CELLS. 2016 doi: 10.1002/stem.2550. [DOI] [PubMed] [Google Scholar]
- 25.Forster R, et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Reports. 2014;2:838–852. doi: 10.1016/j.stemcr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie R, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber K. RIKEN suspends first clinical trial involving induced pluripotent stem cells. Nature Biotechnology. 2015;33:890–891. doi: 10.1038/nbt0915-890. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nature Biotechnology. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merkle FT, et al. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11:875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganna A, et al. Ultra-rare disruptive and damaging mutations influence educational attainment in the general population. 2016 doi: 10.1101/050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Case DA, et al. AMBER. 2016 [Google Scholar]
- 37.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 38.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT, et al. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.