Abstract
OATP1B1 (SLCO1B1) is predominantly expressed at the basolateral membrane of hepatocytes and is critically important for the hepatic uptake and clearance of numerous drug substrates and endogenous compounds. In general, the organic anion transporting polypeptides (OATP; SLCO) represent a superfamily of uptake transporters that mediate the sodium‐independent transport of a diverse range of amphipathic organic compounds including bile salts, steroid conjugates, thyroid hormones, anionic peptides, numerous drugs and other xenobiotic substances. OATP1B1 is highly polymorphic and a number of relevant and ethnically dependent polymorphisms have been identified and functionally characterized. In particular, the SLCO1B1 521T>C and 388A>G polymorphisms are commonly occurring variants in ethnically diverse populations and numerous in vitro and clinical studies have evaluated the consequences of these variants to interindividual differences in drug disposition and response. OATP1B1 is particularly important for the disposition of HMG‐CoA reductase inhibitors, or statins, as it is known to efficiently transport most statins to their site of action within hepatocytes. Many studies have focused on the consequences of OATP1B1 variants to statin disposition in vitro and in vivo and would suggest that genetic variability in SLCO1B1 has important implications for statin pharmacokinetics, risk for statin‐induced myopathy, and modulation of statin treatment response. This review describes what is currently known regarding SLCO1B1 genotype, OATP1B1 protein expression and interindividual and interethnic consequences to drug disposition, with particular focus on statin pharmacokinetics and implications for drug response and toxicity.
Keywords: ethnicity, OATP1B1, polymorphism, SLCO1B1, statin, transporter
Tables of Links
| TARGETS | |
|---|---|
| Transporters 2 | OATP (SLCO) |
| ABCG2 (BCRP) | OATP1B1 (SLCO1B1) |
| BSEP (ABCB11) | OATP1B3 (SLCO1B3) |
| MATE | OATP2B1 |
| MDR | OCT |
| MRP | Enzymes 3 |
| NTCP | HMG‐CoA reductase |
| OAT |
| LIGANDS | |
|---|---|
| Atorvastatin | Methotrexate |
| Atrasentan | Pitavastatin |
| Cholesterol | Pravastatin |
| Docetaxel | Rifampicin |
| Estrone‐3‐sulfate | Rosuvastatin |
| Fluvastatin | Simvastatin |
| Lovastatin | Valsartan |
These Tables list key protein targets and ligands in this article that are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY 1, and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 2, 3.
Introduction
Organic anion‐transporting polypeptide 1B1 (OATP1B1; SLCO1B1) is a membrane transport protein of the organic anion‐transporting polypeptide (OATP) transporter superfamily 4, 5. It functions as a sodium‐independent uptake transporter for a broad range of endogenous and xenobiotic compounds with diverse characteristics 4, 6. Substrates tend to be primarily organic anions with a molecular weight greater than 300 Da, but some substrates may be neutral or even positively charged 7. OATP1B1 is a genetically polymorphic transporter and is predominantly expressed on the sinusoidal (basolateral) membrane of human hepatocytes (Figure 1A) 8, 9, 10. Recent studies have demonstrated OATP1B1 and its related counterpart OATP1B3 to be amongst the most highly expressed uptake or efflux membrane transporters expressed in the liver 11, 12. Thus, OATP1B1 plays a major, clinically important role in the hepatic uptake and clearance of many drugs. Functionally relevant and ethnically‐dependent polymorphisms in SLCO1B1 have been identified and characterized. Clinical studies have confirmed OATP1B1 variants to be associated with alterations in the pharmacokinetics of substrate drugs, treatment response, and risk for drug‐induced toxicities. This review provides an update on the role of SLCO1B1 polymorphisms to interindividual and interethnic variability in drug disposition and response.
Figure 1.
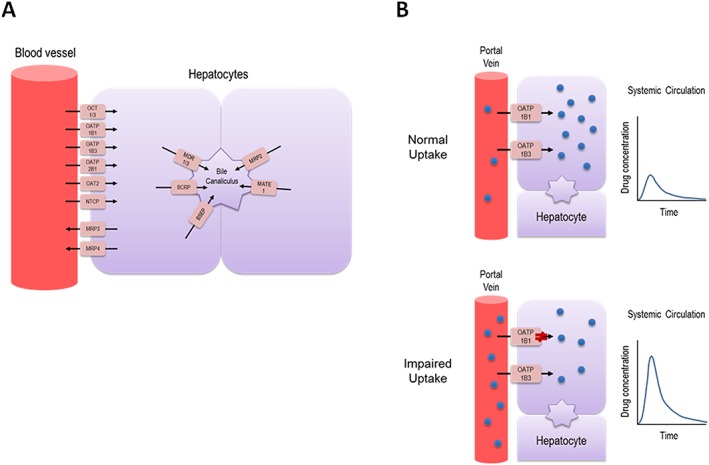
Summary of transporters expressed in human hepatocytes involved in mediating drug uptake and efflux (A), and illustration of the impact of SLCO1B1 genetic variation on drug disposition in vivo (B). Individuals carrying a dysfunctional genetic variant in SLCO1B1 results in impaired hepatocellular uptake of drug substrates as compared to wild type individuals, leading to significantly increased plasma exposure, which may increase risk for drug‐induced adverse effectsBCRP, breast cancer resistance protein; BSEP, bile salt export pump; MATE, multidrug and toxin extrusion; MDR, multidrug resistance protein; MRP, multidrug resistance‐associated protein; NTCP, sodium/taurocholate co‐transporting polypeptide; OAT, organic anion transporter; OATP, organic anion transporting polypeptide; OCT, organic cation transporter
SLCO1B1 genetic variability
To date, >45 nonsynonymous (NS) sequence variants have been identified in SLCO1B1 13, 14, some of which are clinically important due to their altered function 15, 16 (Table 1). The first systematic study of SLCO1B1 variants identified 14 NS single‐nucleotide polymorphisms (SNPs) in 15 haplotypes, many of which (217T>C, 245T>C, 467A>G, 521T>C, 1058T>C, 1294A>G, 1463G>C, 1964A>G) were associated with markedly decreased OATP1B1 transport activity in vitro for prototypical substrates such as estrone sulfate and estradiol‐17β‐d‐glucuronide 10. Moreover, genotypic frequencies were dependent on race in a population of European‐ and African‐Americans. In particular, the 521T>C (rs4149056) SNP, resulting in substitution of alanine for valine at amino acid position 174, was common in European‐Americans with an allele frequency of 14% but much less so in African‐Americans at 2%. Conversely, the 388A>G (rs2306283) variant, resulting in an asparagine to aspartic acid amino acid change at amino acid position 130 located in the extracellular region of the transporter, was seen predominantly in African‐Americans with an allele frequency of 74% but significantly less so in European‐Americans at 30%. Additional population genetic studies demonstrate SLCO1B1 variants exhibit marked differences in their ethnically defined allele frequencies amongst major geographical regions. Pasanen et al. 17 investigated the frequencies of 12 SNPs in SLCO1B1, including 5 NS variants and two promoter variants, in 941 individuals from 52 populations including Africa, the Middle East, Asia, Europe, Oceania and the Americas (Amerindians). Consistent with initial reports, the low‐activity haplotypes *5 (388A/521C) and *15 (388G/521C) have a combined frequency of approximately 15–20% in Europeans, 10–15% in Asians and 2% in sub‐Saharan Africans. The *1B (388G) haplotype has a frequency of approximately 26% in Europeans, 39% in South/Central Asians, 63% in East Asians, and as high as 77% in sub‐Saharan Africans.
Table 1.
Selected SLCO1B1 polymorphisms
| Nucleotide change | rs number | Amino acid change | In vitro function | In vivo function | Caucasians | African‐Americans | Asians |
|---|---|---|---|---|---|---|---|
| c.217T>C | rs56101265 | p.F73L | Decreased | 0–2 | 0 | 0 | |
| c.245T>C | rs56061388 | p.V82A | Decreased | 0–2 | 0 | 0 | |
| c.388A>G | rs2306283 | p.N130D | Increased or unchanged | Decreased AUC pravastatin, atorvastatin | 30–45 | 72–83 | 59–86 |
| Increased AUC pitavastatin | |||||||
| c.452A>G | rs2306282 | p.N151S | 0–3 | 0–2 | 0–4 | ||
| c.463C>A | rs11045819 | p.P155T | Unchanged | 13–23 | 2–10 | 0–3 | |
| c.467A>G | rs72559745 | p.E156G | Decreased | 0–2 | 0 | 0 | |
| c.521T>C | rs4149056 | p.V174A | Decreased | Increased AUC statins | 8–20 | 1–8 | 8–16 |
| Increased risk statin‐induced myopathy | |||||||
| Decreased lipid lowering efficacy | |||||||
| c.578T>G | rs72559746 | p.L193R | Decreased | <0.3 | |||
| c.733A>G | rs11045852 | p.I245V | 0 | 0–7 | 0 | ||
| c.1007C>G | rs72559747 | p.P336R | Unchanged | 0 | 1 | ||
| c.1058T>C | rs55901008 | p.I353T | Decreased | 0–2 | 0 | 0 | |
| c.1200C>G | rs59113707 | p.F400L | 0 | 2 | 0 | ||
| c.1294A>G | rs56387224 | p.N432D | Decreased or unchanged | 0–1 | 0 | 0 | |
| c.1385A>G | rs72559748 | p.D462G | Unchanged | 0–1 | 0 | 0 | |
| c.1463G>C | rs59502379 | p.G488A | Decreased | 0 | 3–9 | 0 | |
| c.1495A>G | rs74064213 | p.I499V | 6 | ||||
| c.1929A>C | rs34671512 | p.L643F | Unchanged | 3–9 | 5–13 | 0–1 | |
| c.1964A>G | rs56199088 | p.D655G | Decreased | 0–2 | 0 | 0 | |
| c.2000A>G | rs55737008 | p.E667G | Unchanged | 0–2 | 0–34 | 0 |
The 521T>C variant, located in exon 5, resulted in significantly decreased OATP1B1 membrane expression by Western blot analysis and cell surface biotinylation experiments, resulting in ~75% decreased uptake transport activity toward estrone‐3‐sulfate and estradiol‐17β‐d‐glucuronide in vitro 10. Consistent with its decreased membrane expression, the 521T>C SNP affected mainly the maximum transport velocity in comparison to substrate affinity 10. The deleterious functional consequence of the 521T>C variant was confirmed in subsequent studies with a diverse list of OATP1B1 substrates, including rifampin, pravastatin, atorvastatin, rosuvastatin, atrasentan, ezetimibe glucuronide, methotrexate and docetaxel 9, 13, 18, 19, 20, 21, 22. Another common variant associated with potentially altered transport activity of OATP1B1 is 388A>G (*1b), located in exon 4, but the functional consequences of this variant remain controversial. Quantitative PCR and immunoblotting studies in liver samples have demonstrated the 388A>G variant to be significantly associated with increased OATP1B1 expression, suggesting increased functional activity for this variant 12, 23.
Indeed, 388A>G and 521T>C form four distinct haplotypes: *1A (388A/521T), *1B (388G/521T), *5 (388A/521C) and *15 (388G/521C) 17, 24. In general, the OATP1B1*5 and *15 genotypes confer significantly impaired hepatic uptake 9, 20, 21, 25, resulting in an increased systemic exposure of substrates (Figure 1B), whereas OATP1B1*1B has been associated with decreased systemic exposure due to enhanced hepatic uptake, including statins such as atorvastatin and pravastatin 23, 26, 27. However, additional studies on the functional consequences of the *1B haplotype have yielded conflicting results in its activity as several studies demonstrated no significant differences in *1A versus *1B for evaluated substrates, such as estrone‐3‐sulfate, estradiol‐17β‐d‐glucuronide, and docetaxel 9, 10, 18, and even opposite results with increased plasma exposure of pitavastatin for those with the *1B haplotype, suggesting reduced activity for this variant 28, 29. Additional studies to fully characterize the functional consequences of the 388A>G variant are warranted. Interestingly, coexisting null function mutations in SLCO1B1 and SLCO1B3 which resulted in complete and simultaneous deficiencies of both hepatic OATP1B1 and OATP1B3 have been linked to families with Rotor syndrome, a rare, benign hereditary syndrome characterized by conjugated hyperbilirubinemia, coproporphyrinuria and strongly reduced liver uptake of many diagnostic compounds, including cholescintigraphic tracers 30.
While common OATP1B1 variants have received the most attention, rare SLCO1B1 genotypes may also have functional consequences in vivo. Ramsey et al. 13 identified 93 SNPs in SLCO1B1 in 699 patients, of which 15 were NS SNPs. Three NS SNPs (388A>G, 463C>A and 521T>C) were common, with a minor allele frequency (MAF) >5%, one (1929A>C) had low frequency (MAF 1%–5%), and 11 were rare (MAF < 1%). In examining the impact of rare and common SLCO1B1 variants on drug response, this group noted rare SLCO1B1 NS variants were associated with reduced methotrexate clearance. In a multivariate analysis, SLCO1B1 variants accounted for 10.7% of population variability in methotrexate clearance. The *5 and *15 haplotypes and two rare novel haplotypes, *23 (211G>A) and *31 (388A>G/1463G>C), were associated with low methotrexate clearance. Interestingly, though, while common NS variants accounted for the majority of that variability, rare damaging NS variants comprised 17.8% of SLCO1B1’s effects with larger effect sizes than common variants, suggesting rare variants are likely to have an important effect on pharmacogenetic phenotypes 13.
Ethnic variability in OATP1B1 expression
It has been demonstrated that SLCO1B1 variants are associated with altered OATP1B1 liver expression but until recently, little was known regarding interethnic differences in OATP1B1 expression. Peng et al. 12 utilized a targeted quantitative proteomic approach to determine hepatic protein concentrations of transporters including OATP1B1 in a panel of human livers (n = 141) across ethnic groups, including Caucasians, Asians and African‐Americans. Interestingly, when compared across ethnicity and accounting for genotype, the hepatic expression levels of OATP1B1 were higher in Asians relative to Caucasians, while there were too few African‐American samples to draw any conclusions. Accordingly, additional potential explanations include regulatory or epigenetic modifications that are ethnically‐dependent that result in race‐dependent differences in OATP1B1 expression that are independent of OATP1B1 genotype. Furthermore, the effects of SLCO1B1 polymorphisms on OATP1B1 hepatic abundance were examined by comparing the protein concentrations of different allelic variants. OATP1B1 hepatic protein expression was significantly associated with genotype as 388A>G expression was significantly higher than wild type 388A>A. This is in concordance with a previous study that had noted the 388A>G variant to be associated with increased OATP1B1 hepatic protein expression 23. But, interestingly, there was no difference in protein expression between 521T>C and wild type 521T>T. However, it should be noted that the proposed mechanism of impaired function for the 521T>C variant is a mistrafficking defect that results in significantly decreased cell surface expression as total absolute protein content for the 521T>C compared to the *1a wild type allele was equivalent 10.
Effects of SLCO1B1 polymorphisms on pharmacokinetics and drug disposition
Features of the clinical pharmacokinetics of statins
Statins, 3‐hydroxy‐3‐methylglutaryl coenzyme A reductase (HMG‐CoA reductase) inhibitors, are amongst the most commonly prescribed drugs worldwide and used as antihyperlipidemic agents in the treatment of hypercholesterolemia and the primary and secondary prevention of coronary artery disease 31, 32. However, their use has been associated with adverse drug effects including severe muscle‐related side effects such as rhabdomyolysis and myopathy 33. Statins are partly (pravastatin and rosuvastatin) or rarely (other statins) eliminated into the urine through the kidneys in an unchanged form; they are mainly delivered within hepatocytes to their site of action in inhibiting HMG‐CoA reductase by uptake transporters and eliminated into the bile by efflux transporters with minimal metabolism 33, 34, 35. Many statins are well known to be substrates of hepatic uptake transporters, including OATP1B1, OATP1B3 and OATP2B1 36. However, studies have suggested OATP1B1 to be the major hepatic OATP to be involved in hepatic uptake and clearance. For example, the contribution of OATP1B1 to the overall hepatic uptake of pitavastatin and rosuvastatin was estimated to be 92% 37 and 77% 38, respectively.
OATP1B1 is thus considered the major transporter involved in the hepatic uptake of statins. However, several OATP1B1 genetic variants have been found to be associated with impaired uptake activity of numerous drug substrates, including statins, most notably the commonly occurring 521T>C variant. In a number of clinical studies, subjects with the SLCO1B1 521T>C polymorphism have demonstrated reduced nonrenal (hepatic) clearance of pravastatin 24, 39, 40, 41, rosuvastatin 42, 43, 44, 45, atorvastatin 42, 45, 46, simvastatin acid 45, 47, 48, 49 and pitavastatin 50, as compared with subjects with the SLCO1B1 wild‐type alleles. The clinical consequences of individuals carrying loss of function SLCO1B1 alleles have also been noted with regard to statin‐induced adverse effects. A genome‐wide association study (GWAS) identified an intronic variant in SLCO1B1 in near complete linkage disequilibrium (r2 = 0.97) with the SLCO1B1 521T>C allele to be the single most important predictor of simvastatin‐associated myopathy risk in the SEARCH trial in patients who received high‐dose 80 mg simvastatin 51. The odds ratio for myopathy risk due to SLCO1B1 521T>C was 4.5 for heterozygous carriers and 16.9 for homozygous carriers. These results were replicated in 10 000 patients who received 40 mg simvastatin in the Heart Protection Study, demonstrating a myopathy odds ratio of 2.6 for each copy of the 521T>C allele. The vast majority of the subjects in this study were Caucasian and the study conclusions may have more important implications for Caucasian and Asian populations as the genotypic frequency of the 521T>C allele approaches 15–20% in these ethnic populations, whereas they are significantly less common in African‐American and Hispanic populations. The association between the 521T>C SNP and statin adverse events was subsequently confirmed for simvastatin and also demonstrated for atorvastatin, but not associated in subjects taking pravastatin, suggesting the risk for statin‐induced adverse events modulated by OATP1B1 genotype is influenced by the choice of statin being administered 52. Furthermore, in the setting of SLCO1B1 521T>C genotype, gender has been reported to also contribute to interindividual variability in pharmacokinetics and treatment efficacy of statins 53, 54.
The functional consequences of the SLCO1B1 388A>G polymorphism (SLCO1B1*1b) has also been evaluated. Maeda et al. 27 showed a good correlation with the *1b allele among the area under the plasma concentration–time curve (AUC) values of pravastatin, valsartan and temocapril in a three‐period crossover pharmacokinetic study. When oral administration of these drugs was performed separately in each period in 23 healthy subjects with carriers of SLCO1B1 388A>G and/or 521T>C variants, the subjects with 388A>G revealed higher nonrenal clearance, representing the hepatic uptake clearance of pravastatin, and increased transport activity compared to the *1a allele. This would be consistent with studies demonstrating increased protein expression for those who carry the *1b allele 12, 23. While this higher nonrenal (hepatic) clearance in subjects with 388A>G was also observed when pravastatin or lovastatin was used in Caucasians 26, 41, 55, and pitavastatin in Koreans 56, other conflicting studies have demonstrated either no differences or even opposite differences to the *1a allele such as with rosuvastatin in Caucasians 44, Koreans 57 and Chinese patients 43, pitavastatin in Chinese 29, or simvastatin acid in Koreans 49. Further adequately powered studies should be conducted to assess the influence of SLCO1B1 388A>G genotype on statin pharmacokinetics and determine whether this has any clinical implications for statin response and risk for adverse effects.
There is also data regarding the influence of OATP1B1 genotype to statin response. In short‐term clinical studies of less than 3 months, when compared to those who are wild‐type, the SLCO1B1 521T>C variant was associated with modest increases in LDL‐cholesterol for patients receiving pravastatin, but not rosuvastatin 58, 59, 60, 61. This is consistent with the impaired function of this variant resulting in reduced hepatic uptake and thus decreased inhibition of HMG‐CoA reductase within hepatocytes. A more recent population‐based study confirmed a statistically significant association with lower statin‐induced total cholesterol reduction in those with SLCO1B1 521T>C genotype and receiving either simvastatin, pravastatin, lovastatin or fluvastatin 40. No effect was noted for atorvastatin. Assessment of achievement of treatment goals according to published cardiovascular guidelines showed a lower rate of successful treatment for those harbouring the SLCO1B1 521T>C variant and receiving either simvastatin or pravastatin. The SLCO1B1 388A>G variant has generally not been associated with significant changes in LDL or total cholesterol response 40, 62, 63.
Ethnic variability in the pharmacokinetics of statins
Ethnic variability has been observed in the plasma exposures of statins between Caucasians and Asians or between Caucasians and African‐American populations. For example, a higher systemic plasma exposure of pravastatin was observed in Caucasians as compared with African‐Americans 41 and plasma exposure of rosuvastatin was higher in Asians than that in Caucasians living in the same environments, Singapore 64 or the United States 44. When Caucasians and African‐Americans were compared after a single oral 40 mg dose of pravastatin was administrated, a higher plasma exposure of pravastatin was observed in Caucasians as compared with African‐Americans 41. The CL/F (oral clearance) in African‐Americans was 1.4 times higher than in Caucasians. This significant ethnic variability was still observed even after adjusting for SLCO1B1 521T>C and 388A>G genotypes, gender, and BSA, and assay sensitivity as covariates, suggesting other genetic or non‐genetic factors may contribute to the interethnic and interindividual variability in pravastatin disposition 41.
Systemic exposure to rosuvastatin was observed to be approximately two‐fold higher in Japanese subjects living in Japan compared with white subjects in Western Europe or the United States 65, 66, 67. A population pharmacokinetic analysis revealed that the apparent oral clearance of rosuvastatin was reduced by 44% in Asian subjects, mainly Japanese subjects living in Japan, compared to Caucasian subjects living in the UK, Europe or the USA 68. Moreover, clinical rosuvastatin pharmacokinetic studies have been conducted with subjects living in the same environment, either Singapore or the US 44, 64. When the pharmacokinetics of rosuvastatin in Asians were compared with those in Caucasian subjects living in the same environment, a remarkable difference in plasma AUC values between Caucasians and Asians was confirmed. Rosuvastatin plasma exposure was significantly higher in Asians and similar to that in Japanese populations and SLCO1B1 genotypes (388A>G or 521T>C) did not account for the observed pharmacokinetic differences. Rosuvastatin exposure was higher in subjects carrying the SLCO1B1 521C allele compared with that in non‐carriers of this allele, but since the allele frequency of this variant is similar in Asians and Caucasians, interethnic differences in statin disposition cannot be explained by SLCO1B1 genotype alone 44. Based on the data from these collective studies, in the US, the recommended initial dose of rosuvastatin for Asians is 5 mg, which is half of the dose recommended for Caucasians.
Interestingly, a common nonsynonymous variant, 421C>A (rs2231142; p. Gln141Lys) resulting in significantly decreased transport activity, in ABCG2 (BCRP), an efflux transporter localized to the apical membranes of hepatocytes and intestinal enterocytes and capable of statin transport, may partially explain interethnic differences in rosuvastatin exposure between Asians and Caucasians 44, 69, 70. The frequency of 421C>A in ABCG2 is lower in Caucasians (~9–14%) than in Asians (~35%) 70, 71. Birmingham et al. 45 recently assessed plasma exposure of atorvastatin, rosuvastatin and simvastatin in Caucasian and Asian subjects. Polymorphisms in SLCO1B1 521T>C or ABCG2 421C>A were associated with higher exposure to rosuvastatin, atorvastatin and simvastatin acid across ethnic populations. However, in individuals carrying wild‐type alleles for both SLCO1B1 and ABCG2, plasma AUC still appeared to be higher for rosuvastatin, atorvastatin and simvastatin acid in Chinese and Japanese subjects compared with Caucasians, respectively. Therefore, despite the differences in the frequency of the ABCG2 421C>A allele amongst Asians and Caucasians, this alone does not explain the increased plasma exposure of statins in Asians when compared to Caucasians. Tomita et al. 72 conducted quantitative pharmacokinetic analyses of published studies to date evaluating statin plasma exposure in Asian and Caucasian populations. They suggest that OATP1B1‐mediated hepatic intrinsic uptake clearance, where the ratio of OATP1B1 activity in Japanese/Caucasians is 0.584 and is independent of SLCO1B1 genotype, may explain the ethnic variability in plasma AUCs of statins. However, the reasons underlying ethnic differences in OATP1B1 activity are not understood. An analysis of cell surface protein expression levels of OATP1B1 in liver samples from Caucasians and Asians may help to support this hypothesis regarding differences in intrinsic activity.
Conclusion
OATP1B1 is an important hepatic drug uptake transporter that can mediate the uptake and clearance of numerous endogenous compounds and drugs. It is also highly polymorphic and a number of functionally relevant and ethnically dependent polymorphisms have been identified and characterized. In this review, we describe what is currently known regarding SLCO1B1 genotype, OATP1B1 protein expression, and the resulting interindividual and interethnic consequences to drug disposition, focusing particularly on statin pharmacokinetics and drug response in populations of Caucasian, African‐American and Asian subjects. SLCO1B1 genotype significantly influences statin pharmacokinetics, risk for statin‐induced adverse effects such as myopathy, and the treatment efficacy of certain statins. However, SLCO1B1 genotype alone does not entirely account for observed significant differences in plasma statin exposure amongst ethnic groups. Additional studies focused on delineating the membrane‐specific expression of OATP1B1 amongst these groups may help to clarify these differences.
Competing Interests
The authors have no competing interests to declare.
This work was supported by grants from a Hyundai Scholar Award and NIH R01 GM099924 (to R.H. Ho).
Lee, H. H. , and Ho, R. H. (2017) Interindividual and interethnic variability in drug disposition: polymorphisms in organic anion transporting polypeptide 1B1 (OATP1B1; SLCO1B1). Br J Clin Pharmacol, 83: 1176–1184. doi: 10.1111/bcp.13207.
References
- 1. Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP, et al The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucl Acids Res 2016; 44: D1054–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander SPH, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E, et al The Concise Guide to PHARMACOLOGY 2015/16: Transporters. Br J Pharmacol 2015; 172: 6110–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE, et al The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 2015; 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta 1609; 2003: 1–18. [DOI] [PubMed] [Google Scholar]
- 5. Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch 2004; 447: 653–665. [DOI] [PubMed] [Google Scholar]
- 6. Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica 2008; 38: 778–801. [DOI] [PubMed] [Google Scholar]
- 7. Hagenbuch B, Stieger B. The SLCO (former SLC21) superfamily of transporters. Mol Aspects Med 2013; 34: 396–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Konig J, Cui Y, Nies AT, Keppler D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am J Physiol Gastrointest Liver Physiol 2000; 278: G156–G164. [DOI] [PubMed] [Google Scholar]
- 9. Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, et al. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology 2006; 130: 1793–1806. [DOI] [PubMed] [Google Scholar]
- 10. Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP‐C: identification of multiple allelic variants associated with altered transport activity among European‐ and African‐Americans. J Biol Chem 2001; 276: 35669–35675. [DOI] [PubMed] [Google Scholar]
- 11. Burt HJ, Riedmaier AE, Harwood MD, Crewe HK, Gill KL, Neuhoff S. Abundance of hepatic transporters in Caucasians: a meta‐analysis. Drug Metab Dispos 2016; 44: 1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng KW, Bacon J, Zheng M, Guo Y, Wang MZ. Ethnic variability in the expression of hepatic drug transporters: absolute quantification by an optimized targeted quantitative proteomic approach. Drug Metab Dispos 2015; 43: 1045–1055. [DOI] [PubMed] [Google Scholar]
- 13. Ramsey LB, Bruun GH, Yang W, Trevino LR, Vattathil S, Scheet P, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res 2012; 22: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev 2011; 63: 157–181. [DOI] [PubMed] [Google Scholar]
- 15. Konig J, Seithel A, Gradhand U, Fromm MF. Pharmacogenomics of human OATP transporters. Naunyn Schmiedebergs Arch Pharmacol 2006; 372: 432–443. [DOI] [PubMed] [Google Scholar]
- 16. Maeda K, Sugiyama Y. Impact of genetic polymorphisms of transporters on the pharmacokinetic, pharmacodynamic and toxicological properties of anionic drugs. Drug Metab Pharmacokinet 2008; 23: 223–235. [DOI] [PubMed] [Google Scholar]
- 17. Pasanen MK, Neuvonen PJ, Niemi M. Global analysis of genetic variation in SLCO1B1. Pharmacogenomics 2008; 9: 19–33. [DOI] [PubMed] [Google Scholar]
- 18. Lee HH, Leake BF, Teft W, Tirona RG, Kim RB, Ho RH. Contribution of hepatic organic anion‐transporting polypeptides to docetaxel uptake and clearance. Mol Cancer Ther 2015; 14: 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tirona RG, Leake BF, Wolkoff AW, Kim RB. Human organic anion transporting polypeptide‐C (SLC21A6) is a major determinant of rifampin‐mediated pregnane X receptor activation. J Pharmacol Exp Ther 2003; 304: 223–228. [DOI] [PubMed] [Google Scholar]
- 20. Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP‐C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15 + C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 2005; 15: 513–522. [DOI] [PubMed] [Google Scholar]
- 21. Katz DA, Carr R, Grimm DR, Xiong H, Holley‐Shanks R, Mueller T, et al. Organic anion transporting polypeptide 1B1 activity classified by SLCO1B1 genotype influences atrasentan pharmacokinetics. Clin Pharmacol Ther 2006; 79: 186–196. [DOI] [PubMed] [Google Scholar]
- 22. Oswald S, Konig J, Lutjohann D, Giessmann T, Kroemer HK, Rimmbach C, et al. Disposition of ezetimibe is influenced by polymorphisms of the hepatic uptake carrier OATP1B1. Pharmacogenet Genomics 2008; 18: 559–568. [DOI] [PubMed] [Google Scholar]
- 23. Nies AT, Niemi M, Burk O, Winter S, Zanger UM, Stieger B, et al. Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med 2013; 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishizato Y, Ieiri I, Suzuki H, Kimura M, Kawabata K, Hirota T, et al. Polymorphisms of OATP‐C (SLC21A6) and OAT3 (SLC22A8) genes: consequences for pravastatin pharmacokinetics. Clin Pharmacol Ther 2003; 73: 554–565. [DOI] [PubMed] [Google Scholar]
- 25. Deng JW, Song IS, Shin HJ, Yeo CW, Cho DY, Shon JH, et al. The effect of SLCO1B1*15 on the disposition of pravastatin and pitavastatin is substrate dependent: the contribution of transporting activity changes by SLCO1B1*15. Pharmacogenet Genomics 2008; 18: 424–433. [DOI] [PubMed] [Google Scholar]
- 26. Mwinyi J, Johne A, Bauer S, Roots I, Gerloff T. Evidence for inverse effects of OATP‐C (SLC21A6) 5 and 1b haplotypes on pravastatin kinetics. Clin Pharmacol Ther 2004; 75: 415–421. [DOI] [PubMed] [Google Scholar]
- 27. Maeda K, Ieiri I, Yasuda K, Fujino A, Fujiwara H, Otsubo K, et al. Effects of organic anion transporting polypeptide 1B1 haplotype on pharmacokinetics of pravastatin, valsartan, and temocapril. Clin Pharmacol Ther 2006; 79: 427–439. [DOI] [PubMed] [Google Scholar]
- 28. Hu M, Mak VW, Yin OQ, Chu TT, Tomlinson B. Effects of grapefruit juice and SLCO1B1 388A>G polymorphism on the pharmacokinetics of pitavastatin. Drug Metab Pharmacokinet 2013; 28: 104–108. [DOI] [PubMed] [Google Scholar]
- 29. Wen J, Xiong Y. OATP1B1 388A>G polymorphism and pharmacokinetics of pitavastatin in Chinese healthy volunteers. J Clin Pharm Ther 2010; 35: 99–104. [DOI] [PubMed] [Google Scholar]
- 30. van de Steeg E, Stranecky V, Hartmannova H, Noskova L, Hrebicek M, Wagenaar E, et al. Complete OATP1B1 and OATP1B3 deficiency causes human Rotor syndrome by interrupting conjugated bilirubin reuptake into the liver. J Clin Invest 2012; 122: 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaughan CJ, Gotto AM Jr, Basson CT. The evolving role of statins in the management of atherosclerosis. J Am Coll Cardiol 2000; 35: 1–10. [DOI] [PubMed] [Google Scholar]
- 32. Farmer JA. Aggressive lipid therapy in the statin era. Prog Cardiovasc Dis 1998; 41: 71–94. [DOI] [PubMed] [Google Scholar]
- 33. Shitara Y, Sugiyama Y. Pharmacokinetic and pharmacodynamic alterations of 3‐hydroxy‐3‐methylglutaryl coenzyme A (HMG‐CoA) reductase inhibitors: drug–drug interactions and interindividual differences in transporter and metabolic enzyme functions. Pharmacol Ther 2006; 112: 71–105. [DOI] [PubMed] [Google Scholar]
- 34. Singhvi SM, Pan HY, Morrison RA, Willard DA. Disposition of pravastatin sodium, a tissue‐selective HMG‐CoA reductase inhibitor, in healthy subjects. Br J Clin Pharmacol 1990; 29: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin PD, Warwick MJ, Dane AL, Hill SJ, Giles PB, Phillips PJ, et al. Metabolism, excretion, and pharmacokinetics of rosuvastatin in healthy adult male volunteers. Clin Ther 2003; 25: 2822–2835. [DOI] [PubMed] [Google Scholar]
- 36. Shitara Y, Maeda K, Ikejiri K, Yoshida K, Horie T, Sugiyama Y. Clinical significance of organic anion transporting polypeptides (OATPs) in drug disposition: their roles in hepatic clearance and intestinal absorption. Biopharm Drug Dispos 2013; 34: 45–78. [DOI] [PubMed] [Google Scholar]
- 37. Hirano M, Maeda K, Shitara Y, Sugiyama Y. Contribution of OATP2 (OATP1B1) and OATP8 (OATP1B3) to the hepatic uptake of pitavastatin in humans. J Pharmacol Exp Ther 2004; 311: 139–146. [DOI] [PubMed] [Google Scholar]
- 38. Kitamura S, Maeda K, Wang Y, Sugiyama Y. Involvement of multiple transporters in the hepatobiliary transport of rosuvastatin. Drug Metab Dispos 2008; 36: 2014–2023. [DOI] [PubMed] [Google Scholar]
- 39. Niemi M, Schaeffeler E, Lang T, Fromm MF, Neuvonen M, Kyrklund C, et al. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide‐C (OATP‐C, SLCO1B1). Pharmacogenetics 2004; 14: 429–440. [DOI] [PubMed] [Google Scholar]
- 40. Meyer zu Schwabedissen HE, Albers M, Baumeister SE, Rimmbach C, Nauck M, Wallaschofski H, et al. Function‐impairing polymorphisms of the hepatic uptake transporter SLCO1B1 modify the therapeutic efficacy of statins in a population‐based cohort. Pharmacogenet Genomics 2015; 25: 8–18. [DOI] [PubMed] [Google Scholar]
- 41. Ho RH, Choi L, Lee W, Mayo G, Schwarz UI, Tirona RG, et al. Effect of drug transporter genotypes on pravastatin disposition in European‐ and African‐American participants. Pharmacogenet Genomics 2007; 17: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M. Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther 2007; 82: 726–733. [DOI] [PubMed] [Google Scholar]
- 43. Lee HK, Hu M, Lui S, Ho CS, Wong CK, Tomlinson B. Effects of polymorphisms in ABCG2, SLCO1B1, SLC10A1 and CYP2C9/19 on plasma concentrations of rosuvastatin and lipid response in Chinese patients. Pharmacogenomics 2013; 14: 1283–1294. [DOI] [PubMed] [Google Scholar]
- 44. Birmingham BK, Bujac SR, Elsby R, Azumaya CT, Zalikowski J, Chen Y, et al. Rosuvastatin pharmacokinetics and pharmacogenetics in Caucasian and Asian subjects residing in the United States. Eur J Clin Pharmacol 2015; 71: 329–340. [DOI] [PubMed] [Google Scholar]
- 45. Birmingham BK, Bujac SR, Elsby R, Azumaya CT, Wei C, Chen Y, et al. Impact of ABCG2 and SLCO1B1 polymorphisms on pharmacokinetics of rosuvastatin, atorvastatin and simvastatin acid in Caucasian and Asian subjects: a class effect? Eur J Clin Pharmacol 2015; 71: 341–355. [DOI] [PubMed] [Google Scholar]
- 46. Daka A, Dimovski A, Kapedanovska A, Vavlukis M, Eftimov A, Labachevski N, et al. Effects of single nucleotide polymorphisms and haplotypes of the SLCO1B1 gene on the pharmacokinetic profile of atorvastatin in healthy Macedonian volunteers. Pharmazie 2015; 70: 480–488. [PubMed] [Google Scholar]
- 47. Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics 2006; 16: 873–879. [DOI] [PubMed] [Google Scholar]
- 48. Ramsey LB, Johnson SG, Caudle KE, Haidar CE, Voora D, Wilke RA, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin‐induced myopathy: 2014 update. Clin Pharmacol Ther 2014; 96: 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi HY, Bae KS, Cho SH, Ghim JL, Choe S, Jung JA, et al. Impact of CYP2D6, CYP3A5, CYP2C19, CYP2A6, SLCO1B1, ABCB1, and ABCG2 gene polymorphisms on the pharmacokinetics of simvastatin and simvastatin acid. Pharmacogenet Genomics 2015; 25: 595–608. [DOI] [PubMed] [Google Scholar]
- 50. Ieiri I, Suwannakul S, Maeda K, Uchimaru H, Hashimoto K, Kimura M, et al. SLCO1B1 (OATP1B1, an uptake transporter) and ABCG2 (BCRP, an efflux transporter) variant alleles and pharmacokinetics of pitavastatin in healthy volunteers. Clin Pharmacol Ther 2007; 82: 541–547. [DOI] [PubMed] [Google Scholar]
- 51. Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, et al. SLCO1B1 variants and statin‐induced myopathy – a genomewide study. N Engl J Med 2008; 359: 789–799. [DOI] [PubMed] [Google Scholar]
- 52. Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, et al. The SLCO1B1*5 genetic variant is associated with statin‐induced side effects. J Am Coll Cardiol 2009; 54: 1609–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Q, Chen QX, Ruan ZR, Yuan H, Xu HM, Zeng S. CYP2C9*3(1075A>C), ABCB1 and SLCO1B1 genetic polymorphisms and gender are determinants of inter‐subject variability in pitavastatin pharmacokinetics. Pharmazie 2013; 68: 187–194. [PubMed] [Google Scholar]
- 54. Hubacek JA, Dlouha D, Adamkova V, Lanska V, Ceska R, Vrablik M. Possible gene–gender interaction between the SLCO1B1 polymorphism and statin treatment efficacy. Neuro Endocrinol Lett 2012; 33 (Suppl 2): 22–25. [PubMed] [Google Scholar]
- 55. Tornio A, Vakkilainen J, Neuvonen M, Backman JT, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of lovastatin acid. Pharmacogenet Genomics 2015; 25: 382–387. [DOI] [PubMed] [Google Scholar]
- 56. Chung JY, Cho JY, Yu KS, Kim JR, Oh DS, Jung HR, et al. Effect of OATP1B1 (SLCO1B1) variant alleles on the pharmacokinetics of pitavastatin in healthy volunteers. Clin Pharmacol Ther 2005; 78: 342–350. [DOI] [PubMed] [Google Scholar]
- 57. Choi JH, Lee MG, Cho JY, Lee JE, Kim KH, Park K. Influence of OATP1B1 genotype on the pharmacokinetics of rosuvastatin in Koreans. Clin Pharmacol Ther 2008; 83: 251–257. [DOI] [PubMed] [Google Scholar]
- 58. Igel M, Arnold KA, Niemi M, Hofmann U, Schwab M, Lutjohann D, et al. Impact of the SLCO1B1 polymorphism on the pharmacokinetics and lipid‐lowering efficacy of multiple‐dose pravastatin. Clin Pharmacol Ther 2006; 79: 419–426. [DOI] [PubMed] [Google Scholar]
- 59. Takane H, Miyata M, Burioka N, Shigemasa C, Shimizu E, Otsubo K, et al. Pharmacogenetic determinants of variability in lipid‐lowering response to pravastatin therapy. J Hum Genet 2006; 51: 822–826. [DOI] [PubMed] [Google Scholar]
- 60. Zhang W, Chen BL, Ozdemir V, He YJ, Zhou G, Peng DD, et al. SLCO1B1 521T‐‐>C functional genetic polymorphism and lipid‐lowering efficacy of multiple‐dose pravastatin in Chinese coronary heart disease patients. Br J Clin Pharmacol 2007; 64: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bailey KM, Romaine SP, Jackson BM, Farrin AJ, Efthymiou M, Barth JH, et al. Hepatic metabolism and transporter gene variants enhance response to rosuvastatin in patients with acute myocardial infarction: the GEOSTAT‐1 Study. Circ Cardiovasc Genet 2010; 3: 276–285. [DOI] [PubMed] [Google Scholar]
- 62. Couvert P, Giral P, Dejager S, Gu J, Huby T, Chapman MJ, et al. Association between a frequent allele of the gene encoding OATP1B1 and enhanced LDL‐lowering response to fluvastatin therapy. Pharmacogenomics 2008; 9: 1217–1227. [DOI] [PubMed] [Google Scholar]
- 63. Yang GP, Yuan H, Tang B, Zhang W, Wang LS, Huang ZJ. et al. Lack of effect of genetic polymorphisms of SLCO1B1 on the lipid‐lowering response to pitavastatin in Chinese patients. Acta Pharmacol Sin 2010; 31: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee E, Ryan S, Birmingham B, Zalikowski J, March R, Ambrose H, et al. Rosuvastatin pharmacokinetics and pharmacogenetics in white and Asian subjects residing in the same environment. Clin Pharmacol Ther 2005; 78: 330–341. [DOI] [PubMed] [Google Scholar]
- 65. Warwick MJ, Dane AL, Raza A, Schneck DW. Single‐ and multiple‐dose pharmacokinetics and safety of the new HMG‐CoA reductase inhibitor ZD4522. Atherosclerosis 2000; 151: 39. [Google Scholar]
- 66. Martin PD, Mitchell PD, Schneck DW. Pharmacodynamic effects and pharmacokinetics of a new HMG‐CoA reductase inhibitor, rosuvastatin, after morning or evening administration in healthy volunteers. Br J Clin Pharmacol 2002; 54: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sekino H, Onishi T. Phase I study of ZD4522 (rosuvastatin), a new HMG‐CoA reductase inhibitor – evaluation of tolerance and pharmacokinetics in healthy adult male volunteers after single and repeated oral administration. J Clin Ther Med 2005; 21: 187–203. [Google Scholar]
- 68. Tzeng TB, Schneck DW, Birmingham BK, Mitchell PD, Zhang H, Martin PD, et al. Population pharmacokinetics of rosuvastatin: implications of renal impairment, race, and dyslipidaemia. Curr Med Res Opin 2008; 24: 2575–2585. [DOI] [PubMed] [Google Scholar]
- 69. Kondo C, Suzuki H, Itoda M, Ozawa S, Sawada J, Kobayashi D, et al. Functional analysis of SNPs variants of BCRP/ABCG2. Pharm Res 2004; 21: 1895–1903. [DOI] [PubMed] [Google Scholar]
- 70. Keskitalo JE, Zolk O, Fromm MF, Kurkinen KJ, Neuvonen PJ, Niemi M. ABCG2 polymorphism markedly affects the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther 2009; 86: 197–203. [DOI] [PubMed] [Google Scholar]
- 71. Zhang W, Yu BN, He YJ, Fan L, Li Q, Liu ZQ, et al. Role of BCRP 421C>A polymorphism on rosuvastatin pharmacokinetics in healthy Chinese males. Clin Chim Acta 2006; 373: 99–103. [DOI] [PubMed] [Google Scholar]
- 72. Tomita Y, Maeda K, Sugiyama Y. Ethnic variability in the plasma exposures of OATP1B1 substrates such as HMG‐CoA reductase inhibitors: a kinetic consideration of its mechanism. Clin Pharmacol Ther 2013; 94: 37–51. [DOI] [PubMed] [Google Scholar]


