Abstract
Old long-lived proteins contain dehydroalanine (Dha) and dehydrobutyrine (Dhb), two amino acids engendered by dehydration of serines and threonines, respectively. Although these residues have a suspected role in protein cross-linking and aggregation, their direct implication has yet to be determined. Here, we have taken advantage of the ability of the enteropathogen Shigella to convert the phosphothreonine residue of the pT-X-pY consensus sequence of ERK and p38 into Dhb and followed the impact of dehydration on the fate of the two MAPKs. To that end, we have generated the first antibodies recognizing Dhb-modified proteins and allowing tracing them as they form. We showed that Dhb modifications accumulate in a long-lasting manner in Shigella-infected cells, causing subsequent formation of covalent cross-links of MAPKs. Moreover, the Dhb signal correlates precisely with the activation of the Shigella type III secretion apparatus, thus evidencing injectisome activity. This observation is the first to document a causal link between Dhb formation and protein cross-linking in live cells. Detection of eliminylation is a new avenue to phosphoproteome regulation in eukaryotes that will be instrumental for the development of type III secretion inhibitors.
Keywords: aging, bacterial protein phosphatase, mitogen-activated protein kinase (MAPK), phosphoproteomics, protein aggregation, type III secretion system (T3SS), Shigella, dehydroalanine, dehydrobutyrine, protein cross-link
Introduction
Upon aging, serines and threonines are occasionally subject to dehydration, resulting in their conversion into dehydroalanine (Dha)3 and dehydrobutyrine (Dhb), respectively. These modified amino acids are alkenes containing unsaturated Cα–β double bonds. This makes them very reactive and subject to nucleophilic attack by cysteines, histidines, and lysines, potentially leading to non-disulfide covalent cross-links.
Such post-translational modifications (PTMs) have been reported on long-lived proteins such as crystallin proteins of the ocular lens, and they are suspected to contribute to the development of age-related lens opacity (1, 2). However, whether Dhb/Dha formation promotes covalent cross-links in a cellular context and thereby nurtures protein aggregation and insolubility remains to be demonstrated. The scarcity of the data is mainly due to the poor reliability of the detection of dehydrated and cross-linked residues, which principally relies on the analysis of acid hydrolyzed samples exposed to high temperature (1, 3, 4). Such extensive heating can artifactually increase formation of Dha from phosphoserine residues, and nucleophilic addition can be catalyzed by acid, raising a concern of artifactual cross-link induced by acid hydrolysis (5, 6). Thus, a clear demonstration of the in vivo implication of Dha/Dhb in protein cross-link is still missing.
The potential instability and the slow emergence of these PTMs have been major obstacles for their detection and the characterization of their impact on protein fate. Interestingly, some bacteria such as Shigella flexneri catalyze the formation of Dhb (7). They use this strategy to irreversibly inhibit the MAP kinase and thereby repress inflammatory gene expression and host immune defense (8). The inactivation of the host MAPK occurs through β-elimination of the phosphate group at the threonine residue within the dually phosphorylated pT-X-pY motif. This reaction leads to the loss of phosphoric acid and H2O and converts the phosphothreonine residue required for MAPK activity into Dhb (see Fig. 1A). The newly formed amino acid lacks the OH group and is no longer phosphorylatable. This reaction, known as eliminylation, causes an irreversible inactivation of the MAPK.
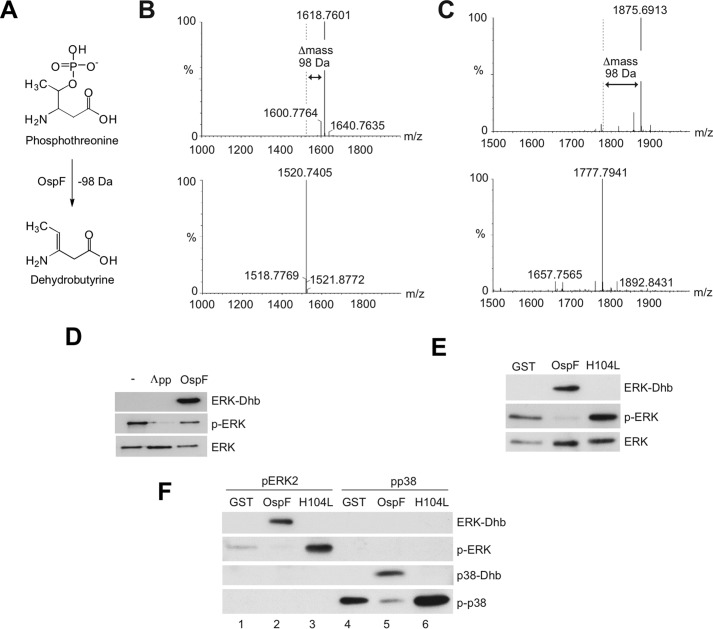
Figure 1.
Generation and characterization of specific anti-Dhb-ERK and anti-Dhb-p38 antibodies. A, diagram of the chemical alteration catalyzed by phosphothreonine lyases. The phosphothreonine residue is converted to Dhb through β-elimination of the phosphate. B, LC-MS spectrum of the molecular mass of the phosphothreonine ERK peptide before and after treatment with a NaOH/dimethyl sulfoxide/ethanol mixture containing barium. A 98-Da difference is observed between the non-treated phospho-ERK peptide ([M + H]+ at m/z 1618.76) and the treated phospho-ERK peptide ([M + H]+ at m/z 1520.74), consistent with the loss of phosphoric acid HPO3 and H2O. C, LC-MS spectrum of the molecular mass of the phosphothreonine p38 peptide before and after treatment with a NaOH/dimethyl sulfoxide/ethanol mixture containing barium. A 98-Da difference is observed between the non-treated phospho-p38 peptide ([M + H]+ at m/z 1875.69) and the treated phospho-p38 peptide ([M + H]+ at m/z 1777.79), consistent with the loss of phosphoric acid HPO3 and H2O. D, immunoblot analysis of in vitro reactions performed with λ phosphatase (Λpp) or OspF, using phospho-ERK2 as substrate. E, immunoblot analysis of in vitro reactions performed with GST, OspF, and the catalytically inactive version of OspF, H104L using phosphoERK2 as substrate. F, in vitro reactions performed with GST, OspF, and the catalytically inactive version of OspF, H104L, using either phospho-ERK2 (pERK2) or phospho-p38α (p-p38) as substrates.
The phosphothreonine lyase activity catalyzing eliminylation has been described for a family of bacterial effectors including OspF from Shigella, SpvC from Salmonella, and HopAI1 from Pseudomonas syringae (7, 9, 10). The effect of these enzymes on MAPK offers a distinctive opportunity to study the impact of Dhb formation on the fate of a protein.
Here, we are using infection by the enteropathogen S. flexneri to monitor Dhb formation in living cells. To trace MAPKs eliminylation in the infected cells, we have engineered the first anti-Dhb antibodies. We demonstrate that eliminylated ERK and p38 form very rapidly after infection and then accumulate. This accumulation can be strictly correlated with the appearance of cross-linked ERK, indicating that the presence of Dhb has a rapid and drastic effect on protein integrity. Our approach also provided valuable information on Shigella, showing that injection of the effector OspF neutralizes the MAPK pathway both by preventing its rephosphorylation and by inducing covalent modifications. In addition, formation of Dhb-modified MAPKs tightly correlated with the activation of the type III secretion apparatus, providing a system to assess Shigella injectisome activity in host cells.
Results
Generation and characterization of anti-ERK and anti-p38 Dhb antibodies
To date, detection of eliminylated residues has exclusively relied on mass spectrometry, a resource-demanding and time-consuming technique for which reproducibility and robustness are dependent on the abundance of the PTM. To facilitate the detection of eliminylated residues and also to allow detection in living cells of these inherently instable PTMs, we engineered anti-ERK and anti-p38 Dhb antibodies. The antigens were generated using two synthetic peptides encompassing the conserved Thr(P)-Xaa-Tyr(P) sequence of ERK and p38, respectively, phosphorylated at the threonine residue; within these peptides, the phosphothreonine residues were chemically converted into Dhb. Mass spectrometry confirmed a mass difference of 98 Da between the original phosphopeptide and the derived Dhb-containing peptide (Figs. 1, B and C). Immunization of rabbits with these Dhb-containing peptides generated anti-p38-Dhb and anti-ERK-Dhb polyclonal antibodies. Specificity of the anti-ERK-Dhb antibody toward the Dhb residue was first confirmed in vitro using a phosphorylated ERK2 protein treated with the Shigella phosphothreonine lyase OspF, or as control, with the λ phosphatase. As show in Fig. 1D, anti-ERK-Dhb antibody detected the product of the phosphotheonine lyase reaction, but not that of the phosphatase activity, which dephosphorylated the Thr(P)-Xaa-Tyr(P) sequence of ERK2 (Table 1). The detection was also abolished when WT OspF was substituted by a catalytically inactive OspF carrying a histidine 104 to leucine mutation (OspF H104L; Fig. 1E). Finally, we observed no cross-reaction of the anti-ERK-Dhb antibody with the p38 Dhb signal generated as above by incubating phosphorylated p38 with OspF (Fig. 1F, lane 5). Specificity of the anti-p38Dhb antibody was verified by a similar approach, confirming a strict dependence on OspF catalytic activity (Fig. 1F, lane 5) and with no cross-reactivity with ERK-Dhb (Fig. 1F, lane 2). We concluded from our in vitro analysis that the anti-ERK-Dhb and anti-p38-Dhb antibodies were suitable for detection of MAPK kinase products modified by a phosphothreonine lyase activity.
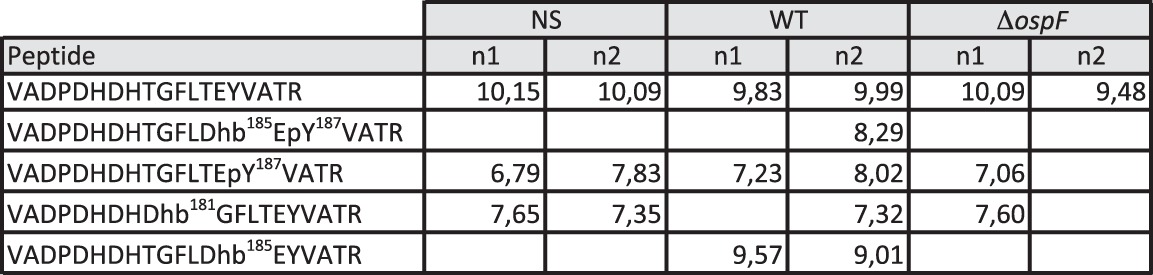
Table 1.
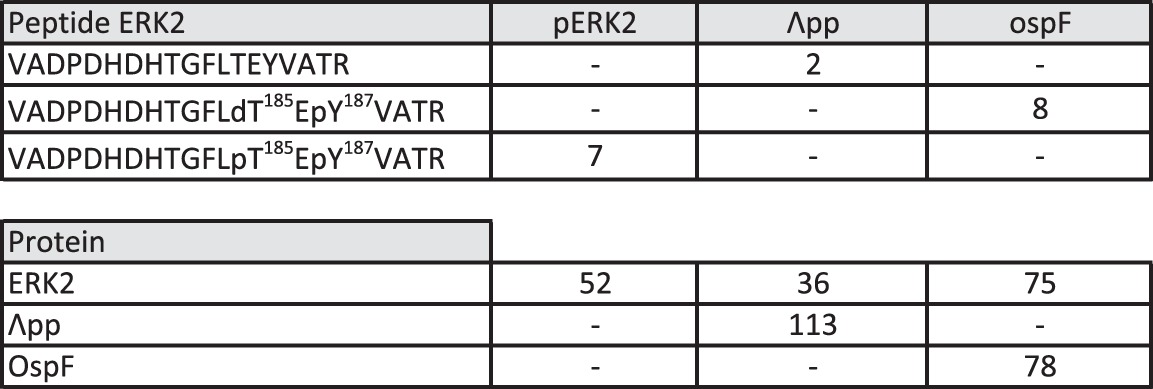
Detection of modified and unmodified TEY-containing peptide
We report the number of peptide spectral matches of each form of the 73VADPDHDHTGFLTEYVATR91 peptide identified in the in vitro ERK2 assay, with OspF and Λpp (λ phosphatase). We also report the number of total peptide spectral matches for ERK2, Λpp, and OspF in each sample.
Dhb-modified MAPKs accumulate rapidly in Shigella-infected cells
To induce the formation of Dhb-modified MAPKs, we infected HeLa cells with the wild-type bacterium S. flexneri serotype 5a (WT); the ospF deficient strain (ΔospF) was used as an internal control. To facilitate detection of the modified proteins, the HeLa cells were expressing a Flag-tagged version of ERK2. We firstly used mass spectrometry to characterize the spectrum of Dhb-modified residues on MAPK in both basal conditions (non-infected) and upon Shigella infection. With the WT strain, the MS/MS spectrum of ERK2 peptide containing the 185TEY187 sequence showed an 18-Da mass decrease compared with an unmodified threonine residue. This mass shift was compatible with a β-elimination reaction of a phosphothreonine converted to Dhb (Fig. 2A). As expected, Dhb at position 185 (ERK2Dhb185) occurred in a strict OspF-dependent manner in the infected cells (Fig. 2B and Table 2). In basal conditions, we also detected a Dhb modification on residue ERK2T181 not modulated by Shigella infection, but the signal intensity of the peptide carrying this modification was low compared with that of ERK2Dhb185 upon Shigella infection (supplemental Fig. S1 and Table 2).
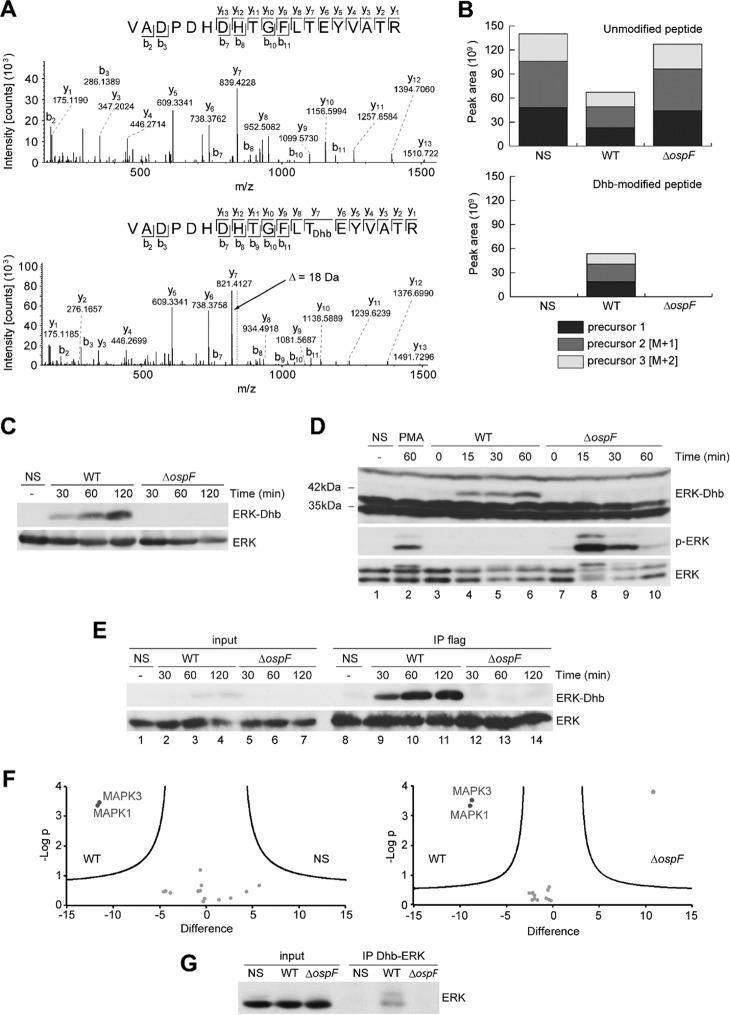
Figure 2.
Determination of OspF lyase activity in infected cells. A, fragmentation mass spectrum of an unmodified 173–191 ERK peptide, and fragmentation mass spectrum of the same peptide carrying the Dhb modification on Thr185, both found in HeLa cells transfected with pcDNA-Flag-ERK2 construct and then infected 2 h by the WT S. flexneri. The site attribution is determined by the masses of the fragment ions: only the ions y7 to y13 contain the Dhb modification because they are 18 Da lighter than the non-modified fragment ions. The alignment showing this mass difference is represented with a gray dashed line for the y7 ion. B, MS1 area extraction of the signal of the precursor ion for the non-modified 173–191 peptide (first three isotopes, z = 3), and MS1 area extraction of the signal of the precursor ion for the peptide 173–191 carrying the Dhb modification on Thr185 (first three isotopes, z = 3). C, HeLa cells transfected with a pcDNA-Flag-ERK2 construct and then untreated (NS) or infected by the WT Shigella or the mutant for the ospF gene (ΔospF) for different durations. D, HeLa cells untreated (NS), stimulated with 1 μg/ml of PMA, or infected by the WT Shigella or the mutant for the ospF gene (ΔospF) for different durations. E, immunoprecipitation of the Dhb-ERK-with a Flag antibody in HeLa cells transfected with pcDNA-ERK2-Flag and then non-treated (NS) or infected with the WT Shigella or the ospF mutant (ΔospF) for different durations. F, volcano plot representations of proteins immunoprecipitated with the anti-ERK-Dhb antibody and identified by mass spectrometry in three independent experiments. Left panel, proteins immunoprecipitated in HeLa cells infected by the WT Shigella versus non-infected HeLa cells. Right panel, HeLa cells infected by the WT Shigella versus the ospF mutant. The data were determined by significance test (FDR < 0.05%, delimited by the black lines). The x axis shows the difference of the average of the logarithm of label free quantification intensities between NS and WT (B) or ospF and WT (C). The y axis shows the negative logarithm of t test p value. G, immunoprecipitation of ERK with the anti-ERK-Dhb antibody from lysates of HeLa cells non-treated (NS), infected with the WT Shigella, or the ospF mutant (ΔospF) for 60 min.
Table 2.
Detection of modified and unmodified TEY-containing peptide
We report the logarithms of the intensity of each form of the 73VADPDHDHTGFLTEYVATR91 peptide identified in the anti-Flag ERK2 immunoprecipitation in HeLa cells non-infected (NS), infected with WT Shigella, or the mutant for the ospF gene (ΔospF). n1 and n2 are two biological replicates.
We next used the anti-Dhb antibodies to trace MAP kinase Dhb modifications in live cells upon Shigella infection. ERK-Dhb signal appeared early (30 min) after WT Shigella infection of Hela cells expressing Flag-tagged ERK2, and the signal was not observed when infecting with the ΔospF strain (Fig. 2C; see also supplemental Fig. S8 for the visualization of the full blot). We also examined modification of endogenous ERK1/2 proteins in HeLa cells infected with WT or ΔospF strains or stimulated by phorbol myristate acetate (PMA) as a positive control for MAPK activation. In these experiments and consistent with our earlier studies, only the ΔospF strain allowed for ERK phosphorylation (Fig. 2D, lanes 7–10; see also supplemental Fig. S8 for the visualization of the full blot). Using the anti-ERK-Dhb antibody, we found again that the WT but not the ΔospF strain induced the formation of a 42-kDa species as early as 15 min post-infection (Fig. 2D, lanes 4–6). Similar results were obtained in other cell types, including the mouse embryonic fibroblasts (MEFs) and the enterocytic TC7 cells (supplemental Fig. S2). Finally, transcomplementation of the ΔospF mutant with a plasmid encoding OspF (ΔospF pOspF) restored Dhb signal, whereas complementation with catalytically inactive OspF (ΔospF pH104L) did not. This further documented the requirement for OspF catalytic activity to acquire Dhb signal (supplemental Fig. S3, compare lanes 5 and 6). To ascertain that the 42-kDa Dhb product was ERK2, we performed immunoprecipitation studies. Endogenous ERK or Flag-tagged ERK2 immunoprecipitates were probed with the Dhb-ERK antibody, thereby allowing the detection of a 42-kDa signal accumulating upon infection with the WT but not the ospF-deficient strain (Fig. 2E, lanes 9–11, and supplemental Fig. S4). We could achieve similar results by immunoprecipitating p38 MAPK using the Dhb p38 antibody (supplemental Fig. S5). We next investigate the ability of the Dhb-ERK antibody to immunoprecipitate the modified kinases upon infection. We firstly characterized the quality of the Dhb immunoprecipitate by mass spectrometry. The results confirmed that both ERK1 and ERK2 were specifically detected in the immunoprecipitates and only upon OspF bacterial injection (Fig. 2F, left and right panels). Remarkably, all the ERK peptides carrying the TEY sequences were Dhb-modified at Thr185 for ERK2 and Thr202 for ERK1, indicating that the antibody immunoprecipitated exclusively the modified ERK-Dhb MAPKs isoforms (Table 3; see also supplemental Table S1 for a complete set of data). Only few additional proteins were immunoprecipitated by the antibody in an OspF-independent manner and without any detectable Dhb residue (Table 4; see also supplemental Table S2 for a complete set of data). Finally, immunoblot analysis confirmed that both endogenous ERK1 and ERK2 were immunoprecipitated with the anti-Erk-Dhb antibody, showing that each of the two ERK isoforms are subject to Dhb modifications in a strict OspF-dependent manner (Fig. 2G).
Table 3.
Detailed list of MAPK1 and MAPK3 peptides identified in the anti-Dhb-ERK immunoprecipitates
We report the peptide sequence, the number of trypsin missing cleavages the peptide identification score, the log(2) of the intensity of the signal obtained for each peptide attributed to MAPK1 and MAPK3 (confident protein identification requires at least two unique peptides fragmented per protein per sample). Common peptides to MAPK1 and MAPK3 are reported as non-unique to the protein (Unique (Proteins) = no). Modified sites reports the Dhb sites identified. n1, n2, and n3 are three biological replicates.
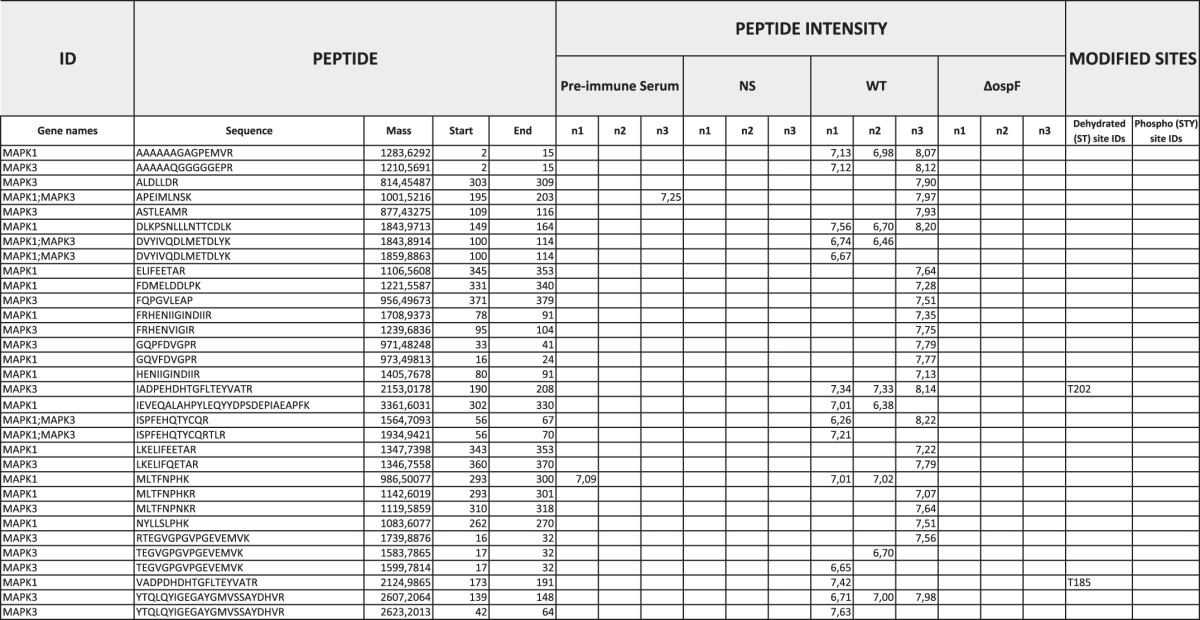
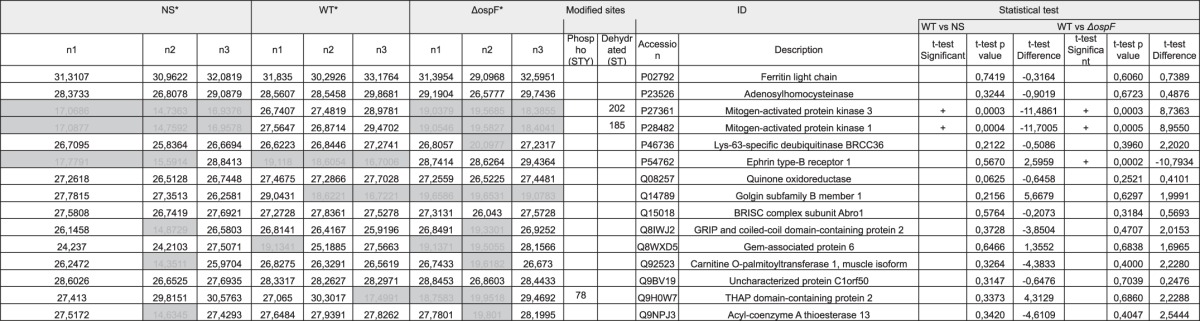
Table 4.
Detailed list of proteins immunoprecipited by the anti-ERK-Dhb antibody
We report the log(2)LFQ (label-free intensity) of the proteins identified by high resolution mass spectrometry in the immunoprecipitation. Gray cells represent non-detected intensities (missing values). Only proteins non-detected in the in the preimmune serum were selected. For statistical analysis, missing values were imputed using a Gaussian distribution of values (width 0.3 and downshift of 6 S.D. of the distribution of the intensity of all the proteins of each sample). Identification parameters include the identification score, the total protein intensity, and the total number of peptides used for the identification (MS/MS count). Modified sites reports the amino acid position for the Dhb and phosphorylation sites identified for each protein. Statistical test reports the p value and the difference of the averages for WT Shigella versus non-stimulated (NS) and for WT versus ΔospF. * The proteins listed in these columns were never identified in preimmune serum. n1, n2, and n3 are three biological replicates.
Shigella induces cross-linking of ERK proteins
Dhb intermediates are predicted to be unstable and subject to nucleophilic attack, which might promote cross-links. Therefore, we searched for the formation of ERK cross-links induced by the catalytic activity of OspF. As described above, ERK-Dhb accumulated early; later time points revealed that the species persisted at least 24 h post-infection and was detectable both in the nuclear and cytosolic extracts with the anti-ERKDhb185 antibody (Figs. 2E, lanes 9–11, and 3a and supplemental Fig. S6). Concomitantly with the formation of Dhb, a species of ∼170 kDa immunoreactive to the ERK antibody accumulated in a strict OspF-dependent manner (Fig. 3B). Like ERKDhb185, this 170-kDa species was detected in both the cytoplasmic and the nuclear compartments (Fig. 3C). Harsh denaturing conditions such as boiling in 2.5% β-mercaptoethanol, 8 m urea, or diethylenetriaminepentaacetic acid did not result in any decrease in the intensity of the slow-migrating species, suggesting that a highly stable molecular complex containing ERK forms in the presence of OspF (supplemental Fig. S7).
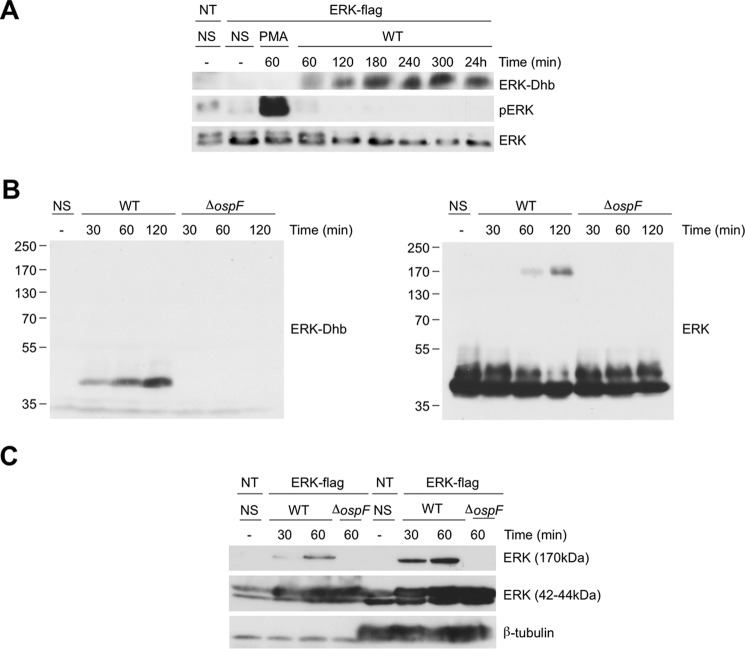
Figure 3.
OspF induced a long-lasting accumulation of Erk-Dhb with concomitant formation of a 170-kDa Erk molecular complex. A, HeLa cells transfected with a pcDNA-Flag-ERK2 construct, then untreated (NS), and stimulated with 1 μg/ml of PMA, infected by the WT Shigella or the mutant for the ospF gene (ΔospF) for different durations. The data are representative of two separate experiments. B, ERK-Dhb and ERK immunoblot of lysates of HeLa cells transfected with a pcDNA-Flag-ERK2 construct, then untreated (NS), and infected by the WT Shigella or the ospF mutant (ΔospF) for different durations. Molecular weight markers are on the left. The data are representative of three separate experiments. C, nuclear or cytoplasmic lysates of HeLa cells transfected with a pcDNA-Flag-ERK2 construct, then non-stimulated (NS), and infected with the WT Shigella or the ospF mutant (ΔospF) for different durations. The data are representative of two separate experiments.
We next sought to determine the molecular identity of the 170-kDa complex. ERK2 immunoprecipitates were separated in a SDS gel, and the 170-kDa band was excised and submitted to mass spectrometry analysis. The results confirmed the presence of ERK peptides in the complex upon infection with the WT strain, whereas no peptide from other proteins species was significantly detected (Table 5). Overall, our data are the first demonstration that emergence of Dhb residues rapidly results in covalent protein cross-linking in live cells.
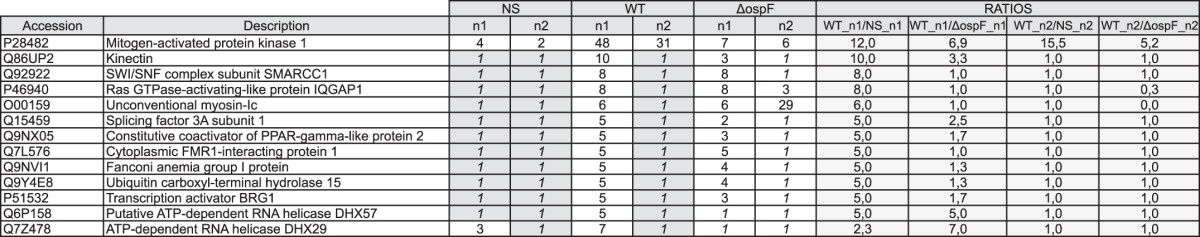
Table 5.
Detailed list of proteins detected at the 170-kDa Erk complex
Ratios were calculated using peptide spectral matches of the WT condition compared to the non-stimulated and ΔospF conditions to show the more abundant proteins detected in the WT condition. n1 and n2 are two biological replicates. To calculate the ratios, missing values were replaced by 1 and reported in italics. We report only the proteins with a ratio of ≥5.
Coupling of MAPK-Dhb formation with type III secretion activity
The phosphothreonine lyase OspF is injected upon activation of the Shigella type III secretion system (T3SS) (8). Thus, OspF injection and consequently Dhb formation should be an indicator of T3SS activity. At bacterial entry, T3SS activation is triggered by its contact with the host cell membrane, leading to effector protein injection into host cytosol allowing bacterial internalization into a phagocytic vacuole (Fig. 4A). Following vacuole rupture, bacteria gain access to the host cytoplasm, and the T3SS activity is down. Reactivation of the T3SS is strictly dependent on actin-based motility and formation of plasma membrane protrusions during cell-to-cell spread (11). We therefore examined whether accumulation of Erk-Dhb could be used as a marker of T3SS activity at both invasion and spreading steps. As shown in Fig. 4B, a Dhb signal was observed at a multiplicity of infection (MOI) as low as 10, and signal intensity correlated with the MOI, indicating that Dhb accumulation correlated with the size of the bacterial inoculum and thus with the initial T3SS activity. To determine whether Dhb formation also accounted for T3SS reactivation within the intracellular epithelial niche, we used HeLa cells stably expressing Connexin 26 (HeLa Cx26), a component of the gap junctions necessary for intercellular bacterial spreading. We firstly investigated the pattern of T3SS activity upon infection of HeLa Cx26 using a previously characterized transcription-based secretion activity reporter (TSAR) (11). This reporter expresses a fast-maturing GFP under the control of the promoter of one of the effectors injected by T3SS. Bacteria harboring this reporter fluoresce green when the T3SS is active. HeLa Cx26 cells were infected with WT, ΔospF, and ΔicsA strains. The latter remains invasive but is unable to spread because it does not produce the protein IcsA, an activator of the actin polymerization machinery that drives the bacterium within the host cytosol. As expected, intracellular bacteria with high TSAR activity were detected with both WT and ΔospF Shigella (green bacteria), whereas TSAR signal remained undetectable with the non-motile ΔicsA mutant, in agreement with its inability to spread (Fig. 4C). A Western blot with the anti-ERK-Dhb antibody allowed recapitulating the outcome of the TSAR assay. Indeed, a strong enrichment of the Dhb signal in the WT but not the ΔicsA mutant was detected in an OspF-dependent manner (Fig. 4D, compare lanes 2 and 3). Thus, the formation of an ERK-Dhb signal tightly correlated with the T3SS activity at both invasion and cell-to cell spreading steps.
Figure 4.
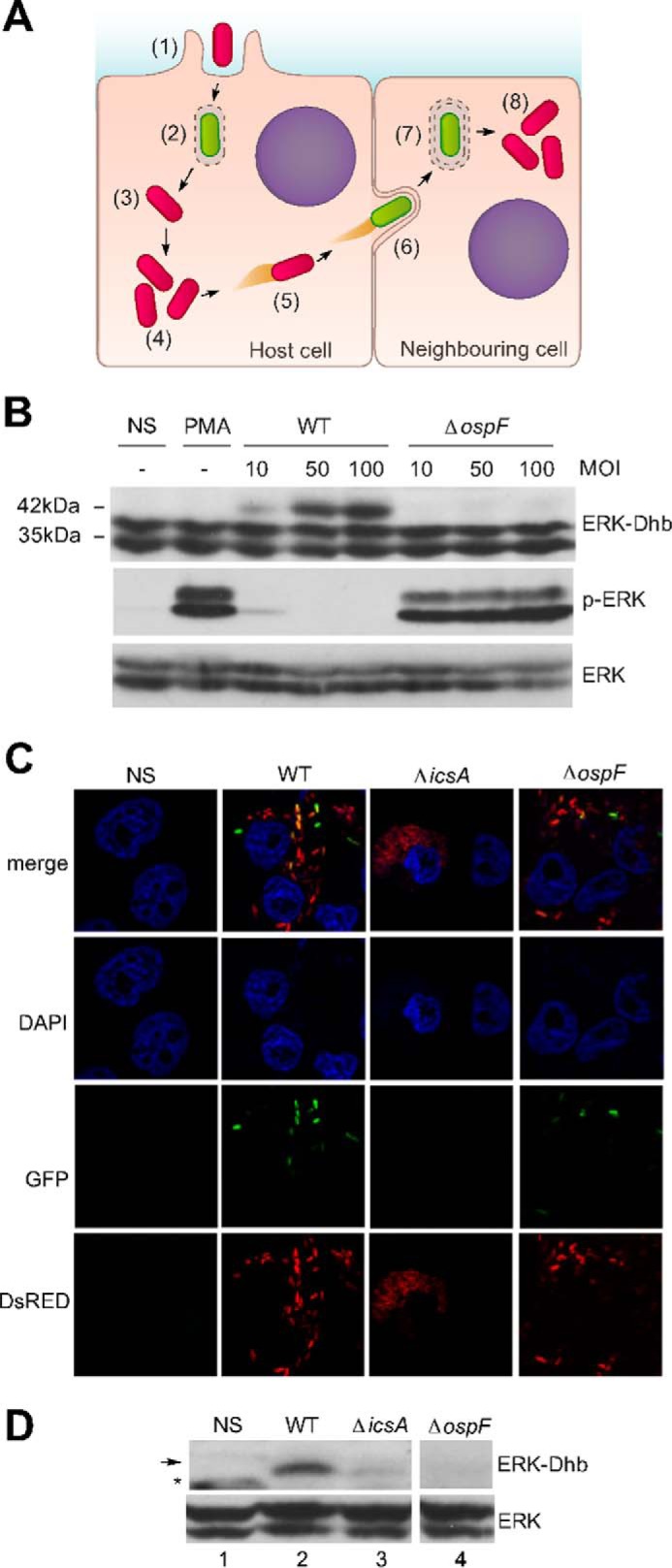
Erk-Dhb is a marker of type III secretion system activity. A, diagram showing the waves of T3SS activity of S. flexneri. Shigella is internalized (1) in a vacuole, and the T3SS is active (2). The vacuole is quickly disrupted (3), leaving Shigella free in the cytosol (4). Shigella is motile in the cytoplasm by forming actin tails, thanks to the IcsA virulence protein (5). When Shigella encounters the cell membrane, the bacterium forms protrusions and reactivates its T3SS (6), is internalized in a vacuole (7), and upon vacuole disruption is again free to multiply and disseminate (8). B, HeLa cells untreated (NS), stimulated with 1 μg/ml of PMA, or infected by the WT Shigella or the ospF mutant (ΔospF) with different multiplicities of infection. C and D, confluent HeLa cells constitutively expressing the connexin 26 were infected with S. flexneri (WT), the mutant for the icsa gene (ΔicsA) or the ospF mutant (ΔospF) at a MOI of 20. The experiment was analyzed by confocal microscopy for a qualitative analysis, and Western blotting was performed with anti-ERK-Dhb and ERK antibodies. Asterisks indicate nonspecific bands (not OspF-dependent). The arrow designates the specific band.
Discussion
Our work provided the first evidence for the traceability of Dhb-modified kinases in living cells. Dhb modifications irreversibly affect the phospho-state of the molecule, and so far, the implication of this PTM in the regulation of the cellular phosphoproteome remains scarcely investigated. Eliminylated residues from phosphothreonine or phosphoserine (the latter leading to Dha) were predicted to be unstable and therefore unlikely to be immunogenic. Our work brings the proof of concept that this type of antibodies can be engineered to trace this modification in live cells. Our antibody-based approach will allow exploring tracks completely out of reach for mass spectrometry and opens the way to the making of an entirely new toolbox for the study of irreversible protein modifications, in the context of bacterial infections and also in the context of aging. Eliminylation is catalyzed by the virulence effector OspF that belongs to a small family of bacterial effectors with phospholyase activity (7). In contrast, the molecular mechanisms causing eliminylation in the context of aging remain elusive. Dha and Dhb can be detected already at an early age in crystallin, a transparent structural protein of the lens. However, high levels of covalent cross-linking, aggregation, and insolubility of crystallin is seen only with age (2, 12). Proteins of tooth dentin also contain harboring Dha and Dhb residues (4). These two proteins belong to the longest-lived protein in the body, which might facilitate the detection of age-related post-translational modifications. However, our report demonstrates that the phenomenon can be extended to the ERK MAPK that exhibits a protein half-life of about 24 h (13). Importantly, OspF phospholyase activity specifically induced the accumulation of ERK-Dhb and ERK protein cross-links at both cytoplasmic and nuclear compartments, thereby establishing a causal relationship between Dhb formation and covalent modifications. Interestingly, also in basal conditions, we could detect a Dhb modification on residue ERK2T181, a phosphosite with unknown function. This observation suggests that, as in human lens, eliminylated residues form naturally in ERK and that physiological mechanism(s) preventing cross-links may exist to avoid protein aggregation. The current model established from the in vivo data obtained in human lens points out a protective function of the antioxidant system through the nucleophilic attack of the Dhb/Dha residues by glutathione, resulting in the formation of thiol metabolite adducts that may prevent protein cross-linking and aggregation (12). Whether chronic inflammation and/or fall in anti-oxidant defense, as seen in aging, promotes these modifications in various tissues and especially in long-lived cells such as neurons remains to be investigated.
Finally, we have showed that the anti-ERK-Dhb antibody can be used as a tool to assess Shigella injectisome activity. Monitoring T3SS activity has proved to be technically challenging because tracer dyes such as biarsenic in FlAsH or CCF4 in FRET are often toxic to eukaryotic cells (14, 15). Anti-Dhb antibodies provide a simple tool to track activation of the injectisome without the need of artificial reporter systems. This will be useful to detect infected host cells in the absence of bacterial invasion, as shown for T lymphocytes (16). T3SS has emerged as an attractive target for antimicrobial therapeutics by reducing selection pressures on pathogens to develop drug-resistant mutations, and our antibody-based approach can potentially provide a screening system for the identification of small molecules inhibiting the type III secretion activity.
In conclusion, we document for the first time that Dhb modifications result in non-disulfide covalent protein cross-links in living cells. Thus, our work showed evidence that eliminylated residues, once formed, can rapidly change protein fate. Finally, we believe that the mere possibility of producing Dhb antibodies opens the way for thorough characterization of eliminylated residues in physiopathological situations, such as diseases involving protein aggregation.
Experimental procedures
Generation of anti-ERK-Dhb and anti-p38-Dhb antibodies
Two peptides with the following sequences were purchased from GenScrip: Ac-H-T-G-F-L-pT-E-Y-V-A-T-R-C-CONH2 (ERK), Ac-R-H-T-D-D-E-M-pT-G-Y-V-A-T-R-C-amide (p38). According to Mattila et al. (17), the peptides (11 mg) were dissolved in 11 ml of 0.2 m NaOH/dimethyl sulfoxide/ethanol mixture (8:3:1) containing barium nitrate (40 mm). Phosphate eliminylation was followed by LC-MS. After 1.5 h at 37 °C, the Thr(P) peptides were entirely converted to β-methyldehydroalanine peptides (Dhb). After treatment with pure acetic acid (1 ml), the compounds were isolated by C18 reverse-phase HPLC using a linear gradient of acetonitrile in 0.08% (v/v) aqueous TFA (1%/min). Synthesis yields were 48% (Dhb-ERK) and 43% (Dhb-p38). A polyclonal antibody was generated by immunization of rabbits with this purified peptide (Eurogentec). The anti-ERK-Dhb antibody was affinity purified on Dhb-ERK peptide, as previously described (18).
Cell culture and bacterial strains
HeLa cells (American Type Culture Collection, CCL-2), HeLa cells stably expressing Connexin 26, MEF cells, and TC7/Caco-2 cells (derived from a human colon carcinoma) were used in this study. HeLa, HeLa Cx26, and MEF cells were cultivated at 37 °C, 10% CO2 in DMEM supplemented with 1 g/liter glucose (Gibco) and 10% (v/v) fetal bovine serum. TC7 cells were cultivated at 37 °C, 5% CO2 in DMEM supplemented with 1 g/liter glucose (Gibco), 20% (v/v) fetal bovine serum, and 1% (v/v) of non-essential amino acids (Life Technologies Inc.). The wild-type invasive strain of S. flexneri serotype 5a, the mutant for the ospF gene, and the ospF recomplemented strain (8) were used in this study. To obtain a mutant expressing a catalytically inactive form of OspF (OspF H104L), a pUC18 plasmid containing the ospF gene was modified to insert a double point mutation on the 311th and 312th nucleotides of the ORF sequence of the ospF gene, using a QuikChange site-directed mutagenesis kit (Stratagene). The sequences of the primers introducing the mutation were as follows: GTTGGGGACAAGTTTCTAATTAGTATAGCTAGG (forward primer) and CCTAGCTATACTAATTAGAAACTTGTCCCCAAC (reverse primer). The ospF deficient strain was transformed with this plasmid, allowing the expression of OspF H104L protein. The M90T WT strain and ospF and icsA mutants carrying a T3SS secretion activity reporter plasmid (TSAR 2.4s) were used in this study (11). All the strains were grown at 30 °C overnight in trypticase soy broth supplemented with the appropriate antibiotics when necessary. On the day of infection, bacterial cultures were diluted 40× in trypticase soy broth and cultured at 37 °C until A600 nm = 0.7.
Protein purification and in vitro reactions
Escherichia coli DH5α were transformed with a pGEX4T2-OspF plasmid encoding the OspF protein fused to a GST tag (8) GST-OspF recombinant protein was purified from a lysate of E. coli transformed with pGEX4T2-OspF using a glutathione affinity column. In vitro reactions were performed for 2 h at 30 °C in 50 mm Tris-HCl, pH 7.5, and 100 mm sodium acetate, with 5 μg of GST-OspF or OspF H104L (catalytically inactive form of OspF), and 100 ng of MAPK kinase (phospho-ERK2 (14-173) and phospho-p38α (14-251); Millipore).
In vitro bacterial infections
For Western blotting analysis, 6 × 106 cells were plated in 100-mm Petri dishes and used for experiments 16 h later. For immunofluorescence experiments, 7 × 105 cells were plated on sterilized cover slips in 6-well plates. The cells were incubated for 1 h in DMEM supplemented with 1 g/liter glucose (Gibco) and infected with S. flexneri (A600 nm = 0.7) at a multiplicity of infection of 40. Bacteria and cells were incubated at room temperature for 10 min or centrifuged for 10 min at 1200 rpm for bacteria not expressing AfaE adhesin and then incubated at 37 °C, 5% CO2 for 20 min. The cells were washed with Dulbecco's modified Eagle's medium supplemented with 1 g/liter glucose (Gibco) and incubated at 37 °C, 10% CO2 with complete medium supplemented with gentamycin (50 μg/ml) to kill extracellular bacteria for different durations. When mentioned, PMA (100 μm) or MG132 (100 μm) was added to the medium.
Immunoblot analysis
The cells were washed in PBS and then lysed in urea buffer (8 m urea, 10 mm Tris, pH 7.5, 1 mm EDTA, 1 mm dithiothreitol, 25 mm β-glycerophosphate, 1 mm orthovanadate, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride, and proteases inhibitors (Roche)). Laemmli sample buffer (92016; Bio-Rad) supplemented with 10% β-mercaptoethanol was added to the lysates, which were analyzed by SDS-PAGE. For nuclear and cytosolic fractioning, the cells were lysed in sucrose buffer (70 mm sucrose, 220 mm mannitol, 5 mm HEPES, 10 mm KCl, 2 mm MgCl2, 5 mm EDTA, 5 mm EGTA) for 5 min. The lysates were centrifuged for 2 min at 16,000 × g at 4 °C, and the supernatant (cytosolic fraction) and pellet (nuclear fraction) were collected. The nuclear fraction was lysed in urea buffer. Laemmli sample buffer (92016; Bio-Rad) supplemented with 10% β-mercaptoethanol was added to supernatant and cytosolic fractions and analyzed by SDS-PAGE. The following antibodies were used: the polyclonal antibodies anti-ERK-Dhb (1:2000) and anti-p38-Dhb (1:2000), polyclonal anti-OspF antibody, anti-ERK1/2 C terminus (1:5000, 06-182; Millipore), anti-ERK1/2 L34F12 preferentially recognizing ERK2 (1:3000, catalog no. 4696; Cell Signaling Technology), anti-phospho-ERK1/2 (1:3000, catalog no. 9101; Cell Signaling Technology), anti-β-tubulin (1:5000, T8535; Sigma), anti-histone H3 (1:5000, ab1791; Abcam), and anti-Flag M2 (10 μg/ml F3165; Sigma). The secondary antibodies used were the following: horseradish peroxidase-conjugated goat IgG to rabbit IgG (1:5000; Nordic Immunology) and peroxidase-conjugated sheep IgG anti-mouse IgG (1:5000, NXA931; GE Healthcare).
Immunoprecipitation studies
The cells were washed in PBS and then lysed in radioimmunoprecipitation assay buffer (150 mm NaCl, 25 mm Tris, pH 7.5, 0,1% SDS, 1% Nonidet P-40, 0.5% sodium desoxycholate, 25 mm β-glycerophosphate, 1 mm orthovanadate, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride, and proteases inhibitors (Roche)). 4 mg of the cell lysate was incubated overnight at 4 °C with 3 μg of the antibody used for immunoprecipitation. Then lysates were incubated for 1 h with 40 μl of protein A-Sepharose beads (Sigma) and analyzed by SDS-PAGE or mass spectrometry.
Mass spectrometry analysis
For anti-Flag immunoprecipitation, eluates were separated by 10% SDS-PAGE, and proteins were stained using silver nitrate (19). The bands corresponding to Flag-ERK were excised. For anti-Dhb-ERK immunoprecipitation, the eluates were concentrated on top of a 10% SDS-PAGE, and the band containing all the proteins in the samples was excised. The gel bands were processed digested in-gel with a 1:50 trypsin-to-protein ratio solution (20). Peptides were dried in a SpeedVac and resuspended in 10% acetonitrile, 0.1% TFA prior nanoRSLC-Q exactive Orbitrap Plus MS (Dionex RSLC Ultimate 3000; Thermo Fisher Scientific) analysis. Peptides were separated on a reverse-phase liquid-chromatographic 50-cm column (Pepmap C18; Dionex). Chromatography solvents were (A) 0.1% formic acid in water, and (B) 80% acetonitrile, 0.08% formic acid. Peptides were eluted from the column with using a 38-min gradient or a 120-min gradient (for Flag-ERK single band analysis and total IP anti-Dhb-ERK, respectively). Two blanks, each with two 25-min linear gradient, were run between biological replicates to prevent sample carryover. Peptides eluting from the column were analyzed by data-dependent MS/MS, using top-10 acquisition method. Briefly, the instrument settings were as follows: resolution was set to 70,000 for MS scans and 17,500 for the data-dependent MS/MS scans to increase speed. The MS AGC target was set to 3 × 106counts, whereas MS/MS AGC target was set to 1 × 105. The MS scan range was from 400 to 2000 m/z. MS and MS/MS scans were recorded in profile mode. Dynamic exclusion was set to 30-s duration.
Raw files corresponding to Flag-ERK bands were processed using the Proteome Discoverer 1.4 software (Thermo Scientific, San Jose, CA) and searched against the Human Uniprot KB/Swiss-Prot database 2014-11 release integrated with sequence of FLAG-ERK. Search parameters included fixed modification: carbamidomethyl (Cys), and variable modification: oxidation (Met), dehydratation (Ser, Thr, and Tyr), phosphorylation (Ser, Thr, and Tyr), and two missed cleavage. Enzyme was trypsin, monoisotopic peptide mass tolerance was ± 3 ppm, and fragment mass tolerance was ± 0.02 Da. The false discovery rate (FDR) was set to 1%. Annotated MS/MS spectra were extracted using Proteome Discoverer 1.4 software. The extracted ion chromatograms and the histogram reporting the intensities of the unmodified and Dhb-modified peptide were obtained using Skyline v2.6.0.
Raw files corresponding to the proteins immunoprecipitated using anti-Dhb-ERK antibody were analyzed using MaxQuant 1.5.3.30 software against the human Uniprot KB/Swiss-Prot database 2016-01 (21). To search parent mass and fragment ions, we set a mass deviation of 3 and 20 ppm, respectively, with no match between runs allowed. Carbamidomethylation (Cys) was set as fixed modification, whereas oxidation (Met), N-terminal acetylation, dehydratation (Ser and Thr), and phosphorylation (Ser, Thr, Tyr) were set as variable modifications. The FDRs at the protein and peptide level were set to 1%. The scores were calculated in MaxQuant as described previously (21). The peptides were quantified according to the MaxQuant MS1 signal intensities (21, 22). Statistical and bioinformatic analysis, including volcano plot, were performed with Perseus software, version 1.5.0.31. For statistical comparison, we set four groups, each containing three biological replicates: antibody control (preimmune serum), non-stimulated (NS), WT Shigella, and ospF deficient Shigella (ΔospF) strains. We filtered out all proteins identified in the preimmune serum condition. We then retained only proteins that were quantified three times in at least one group. Next, the data were imputed to fill missing data points by creating a Gaussian distribution of random numbers with a standard deviation of 33% relative to the standard deviation of the measured values and 6 S.D. downshift of the mean to simulate the distribution of low signal values. We performed a t test and represented the data on a volcano plot (FDR < 0.05, S0 = 3, 20 randomizations).
Author contributions
All the authors contributed to the discussion of this work. B. M. M., S. M. J., and C. R. designed and performed the experiments. For proteomic analysis, samples were processed and analyzed by I. G., C. C., and J. L. The production of the Erk and p38 Dhb peptides was done by F. B., while L. A. and C. M. conceived the project, and L. A. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank G. Tran Van Nhieu for providing the Cx26 HeLa cell line and F. X. Campbell-Valois for providing the TSAR 2.4s plasmid and strains and for useful discussions.
This work was supported by grants from the Agence National de la Recherche and INSERM and by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche and ENS Cachan (to B. M. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2 and Figs. S1–S8.
- Dha
- dehydroalanine
- Dhb
- dehydrobutyrine
- PTM
- post-translational modification
- PMA
- phorbol myristate acetate
- MEF
- mouse embryonic fibroblast
- T3SS
- type III secretion system
- MOI
- multiplicity of infection
- TSAR
- transcription-based secretion activity reporter
- FDR
- false discovery rate
- NS
- non-stimulated.
References
- 1. Linetsky M., Hill J. M., LeGrand R. D., and Hu F. (2004) Dehydroalanine crosslinks in human lens. Exp. Eye Res. 79, 499–512 [DOI] [PubMed] [Google Scholar]
- 2. Srivastava K., Chaves J. M., Srivastava O. P., and Kirk M. (2008) Multi-crystallin complexes exist in the water-soluble high molecular weight protein fractions of aging normal and cataractous human lenses. Exp. Eye Res. 87, 356–366 [DOI] [PubMed] [Google Scholar]
- 3. Bessems G. J., Rennen H. J., and Hoenders H. J. (1987) Lanthionine, a protein cross-link in cataractous human lenses. Exp. Eye Res. 44, 691–695 [DOI] [PubMed] [Google Scholar]
- 4. Cloos P. A., and Jensen A. L. (2000) Age-related de-phosphorylation of proteins in dentin: a biological tool for assessment of protein age. Biogerontology 1, 341–356 [DOI] [PubMed] [Google Scholar]
- 5. Goswami S., and Singh D. K. (2009) Biodegradation of α and β endosulfan in broth medium and soil microcosm by bacterial strain Bordetella sp. B9. Biodegradation 20, 199–207 [DOI] [PubMed] [Google Scholar]
- 6. Samuel D., and Silver B. L. (1963) Elimination reactions and hydrolysis of serine phosphate. J. Chem. Soc. 289–296 [Google Scholar]
- 7. Li H., Li H., Xu H., Xu H., Zhou Y., Zhou Y., Zhang J., Zhang J., Long C., Long C., Li S., Li S., Chen S., Chen S., Zhou J.-M., Zhou J.-M., Shao F., and Shao F. (2007) The phosphothreonine lyase activity of a bacterial type III effector family. Science 315, 1000–1003 [DOI] [PubMed] [Google Scholar]
- 8. Arbibe L., Kim D. W., Batsche E., Pedron T., Mateescu B., Muchardt C., Parsot C., and Sansonetti P. J. (2007) An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8, 47–56 [DOI] [PubMed] [Google Scholar]
- 9. Mazurkiewicz P., Thomas J., Thompson J. A., Liu M., Arbibe L., Sansonetti P., and Holden D. W. (2008) SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol. Microbiol. 67, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J., Shao F., Li Y., Cui H., Chen L., Li H., Zou Y., Long C., Lan L., Chai J., Chen S., Tang X., and Zhou J.-M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185 [DOI] [PubMed] [Google Scholar]
- 11. Campbell-Valois F.-X., Schnupf P., Nigro G., Sachse M., Sansonetti P. J., and Parsot C. (2014) A fluorescent reporter reveals on/off regulation of the Shigella type III secretion apparatus during entry and cell-to-cell spread. Cell Host Microbe 15, 177–189 [DOI] [PubMed] [Google Scholar]
- 12. Wang Z., Lyons B., Truscott R. J., and Schey K. L. (2014) Human protein aging: modification and crosslinking through dehydroalanine and dehydrobutyrine intermediates. Aging Cell 13, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pagès G., Lenormand P., L'Allemain G., Chambard J. C., Meloche S., and Pouysségur J. (1993) Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc. Natl. Acad. Sci. U.S.A. 90, 8319–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Enninga J., Mounier J., Sansonetti P., and Tran Van Nhieu G. (2005) Secretion of type III effectors into host cells in real time. Nat. Methods 2, 959–965 [DOI] [PubMed] [Google Scholar]
- 15. Charpentier X., and Oswald E. (2004) Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186, 5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konradt C., Frigimelica E., Nothelfer K., Puhar A., Salgado-Pabon W., di Bartolo V., Scott-Algara D., Rodrigues C. D., Sansonetti P. J., and Phalipon A. (2011) The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe 9, 263–272 [DOI] [PubMed] [Google Scholar]
- 17. Mattila K., Siltainsuu J., Balaspiri L., Ora M., and Lönnberg H. (2005) Derivatization of phosphopeptides with mercapto- and amino-functionalized conjugate groups by phosphate elimination and subsequent Michael addition. Org. Biomol. Chem. 3, 3039–3044 [DOI] [PubMed] [Google Scholar]
- 18. LeGouy E., Thompson E. M., Muchardt C., and Renard J. P. (1998) Differential preimplantation regulation of two mouse homologues of the yeast SWI2 protein. Dev. Dyn. 212, 38–48 [DOI] [PubMed] [Google Scholar]
- 19. Shevchenko A., Wilm M., Vorm O., and Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 20. Andrzejewska Z., Nevo N., Thomas L., Chhuon C., Bailleux A., Chauvet V., Courtoy P. J., Chol M., Guerrera I. C., and Antignac C. (2016) Cystinosin is a component of the vacuolar H+-ATPase-ragulator-rag complex controlling mammalian target of rapamycin complex 1 signaling. J. Am. Soc. Nephrol. 27, 1678–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 22. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., and Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.