Abstract
RecJ/cell division cycle 45 (Cdc45) proteins are widely conserved in the three domains of life, i.e. in bacteria, Eukarya, and Archaea. Bacterial RecJ is a 5′-3′ exonuclease and functions in DNA repair pathways by using its 5′-3′ exonuclease activity. Eukaryotic Cdc45 has no identified enzymatic activity but participates in the CMG complex, so named because it is composed of Cdc45, minichromosome maintenance protein complex (MCM) proteins 2–7, and GINS complex proteins (Sld5, Psf11-3). Eukaryotic Cdc45 and bacterial/archaeal RecJ share similar amino acid sequences and are considered functional counterparts. In Archaea, a RecJ homolog in Thermococcus kodakarensis was shown to associate with GINS and accelerate its nuclease activity and was, therefore, designated GAN (GINS-associated nuclease); however, to date, no archaeal RecJ·MCM·GINS complex has been isolated. The thermophilic archaeon Thermoplasma acidophilum has two RecJ-like proteins, designated TaRecJ1 and TaRecJ2. TaRecJ1 exhibited DNA-specific 5′-3′ exonuclease activity, whereas TaRecJ2 had 3′-5′ exonuclease activity and preferred RNA over DNA. TaRecJ2, but not TaRecJ1, formed a stable complex with TaGINS in a 2:1 molar ratio. Furthermore, the TaRecJ2·TaGINS complex stimulated activity of TaMCM (T. acidophilum MCM) helicase in vitro, and the TaRecJ2·TaMCM·TaGINS complex was also observed in vivo. However, TaRecJ2 did not interact with TaMCM directly and was not required for the helicase activation in vitro. These findings suggest that the function of archaeal RecJ in DNA replication evolved divergently from Cdc45 despite conservation of the CMG-like complex formation between Archaea and Eukarya.
Keywords: archaea, deoxyribonuclease (DNase), DNA helicase, DNA repair, DNA replication
Introduction
DNA replication is essential for all living cells to maintain their genetic information. The molecular basis of DNA replication has been studied in various organisms from the three domains of life, bacteria, Eukarya, and Archaea (1–3). Bacteria and Archaea have small circular genomic DNAs and defined replication origins. However, the DNA replication proteins in Archaea are not similar to those in bacteria but instead resemble their eukaryotic counterparts. Therefore, the bacterial and archaeal/eukaryotic replication systems independently evolved (4). Studies on archaeal DNA replication can contribute to the clarification of the molecular mechanisms of both the archaeal and eukaryotic systems.
During DNA replication, unwinding of dsDNA by a helicase is the essential step for the subsequent nascent DNA strand synthesis. In the eukaryotic DNA replication system, MCM3 (minichromosome maintenance), a heterohexameric complex (Mcm2-7), is the catalytic component of the replicative helicase. However, Mcm2-7 by itself is not the active form, and the formation of the CMG complex, containing Cdc45, MCM, and GINS, activates the helicase activity of MCM (5). An EM study of the CMG complex revealed that Cdc45 and GINS bridge the gap between Mcm2 and Mcm5 in the Mcm2-7 ring (6). In contrast to eukaryotic Mcm2-7, the archaeal MCM hexamer is composed of a single homolog and exhibits the helicase activity by itself in various thermophilic archaea (1).
The eukaryotic GINS complex consists of four different proteins, Sld5, Psf1, Psf2, and Psf3, and is essential for the initiation of DNA replication (7). These four proteins share conserved A- and B-domains, suggesting that they are ancestral paralogs (8). The archaeal GINS tetramer was identified from a cell extract of the crenarchaeon Sulfolobus solfataricus, and the complex contained Gins15 and Gins23, subunits that are more similar to Sld5 and Psf1 and to Psf2 and Psf3, respectively, with a 2:2 molar ratio (9). We have also reported the heterotetrameric GINS, composed of Gins51 (called Gins51 from the order of GINS as 5-1-2-3) and Gins23 with a 2:2 molar ratio (10), and the homotetrameric GINS, composed of only Gins51 (11) from Euryarchaeota. The stimulation of the helicase activity of MCM by the interaction with GINS in vitro has been reported in both Euryarchaeota and Crenarchaeota, although the structural basis for the interaction between archaeal MCM and GINS is unknown (10, 12–15).
Cdc45 was also found to be involved in the initiation of DNA replication in Saccharomyces cerevisiae, and proposed to move with the replication fork (16). The archaeal equivalent of Cdc45 was identified by a bioinformatic analysis. The Cdc45 and RecJ proteins reportedly share a common ancestor and are conserved in all three domains of life (17). Bacterial RecJ has 5′-3′ exonuclease activity and functions in DNA repair pathways, including homologous recombination, base excision repair, and mismatch repair (18–20). Eukaryotic Cdc45 lacks nuclease activity and participates in the CMG complex formation as described above. The archaeal RecJ and eukaryotic Cdc45 are considered to be counterparts in their respective replicative helicases (21). However, the “archaeal CMG-like complex,, containing RecJ, MCM, and GINS, has not been isolated yet.
Different models for the function of RecJ homologs in Archaea have been proposed in several organisms. The RecJ-like protein from S. solfataricus (RecJdbh, RecJ DNA-binding domain homolog) lacks a nuclease domain, and it may function in the detection and signaling of the stalled replication fork (9). Thermococcus kodakarensis RecJ (GAN, GINS-associated nuclease) has the DNA-specific 5′-3′ exonuclease activity, which is stimulated by the interaction with GINS, and GAN may be involved in Okazaki fragment processing (22). Pyrococcus furiosus RecJ (PfRecJ) reportedly cleaved DNA in the 5′-3′ direction and degraded RNA in the 3′-5′ direction (23). PfRecJ was proposed to be involved in the proofreading of mismatched RNA primers in DNA replication.
In this study we focused on the function of the two RecJ homologs, designated as TaRecJ1 and TaRecJ2, from the thermoacidophilic archaeon, Thermoplasma acidophilum. Both proteins are expected to have nuclease activity and to participate in the CMG-like complex. We previously reported that T. acidophilum GINS (TaGINS) is composed of a single homolog (TaGins51), directly interacts with TaMCM, and accelerates the TaMCM ATPase and helicase activities (11, 13). Furthermore, the B-domain of TaGins51 was not required for either the TaGINS tetramer formation or the activation of TaMCM in vitro (24). Our analyses revealed that TaRecJ1 and TaRecJ2 exhibit different nuclease activities. TaRecJ2, but not TaRecJ1, participates in the CMG-like complex formation through the interaction with TaGINS. However, TaRecJ2 did not interact with TaMCM directly, and thus the activation of TaMCM occurs in a TaRecJ2-independent manner.
Results
Preparation of TaRecJ1 and TaRecJ2
The T. acidophilum genome possesses two genes (TA_RS02725 and TA_RS05865) encoding sequences with distinct similarities to the bacterial RecJ and eukaryotic Cdc45 proteins. We cloned these two genes to investigate the functions of the archaeal RecJ-like proteins. However, the nucleotide sequence of TA_RS05865 in the database was different from that of our cloned gene despite several independent PCR and cloning trials. The 1-nt deletion at 556C and the duplication at 622A in the cloned gene caused a difference in 22 amino acid residues (186–207) (supplemental Fig. S1A). The cloned gene product shared high sequence identity with the RecJ homologs from other archaea belonging to Thermoplasmatales (supplemental Fig. S1B), and thus we concluded that the amino acid sequence of our cloned gene product is the original sequence encoded in the T. acidophilum genome. We then aligned the amino acid sequences of these RecJ-like proteins with those of T. kodakarensis (GAN), and Thermus thermophilus (supplemental Fig. S2). All RecJ proteins have seven conserved motifs, which are required for the nuclease activity, and the archaeal RecJs have a long insertion between motifs IV and V as compared with T. thermophilus RecJ. This insertion is present in both the archaeal and eukaryotic RecJ/Cdc45 proteins (25). The crystal structure of human Cdc45 revealed that the insertion plays crucial roles in the CMG formation, and thus the insertion is referred to as CID (CMG-Interaction Domain) (26). Based on the structural similarity to the eukaryotic Cdc45, the TA_RS02725 and TA_RS05865 proteins were both expected to participate in the archaeal CMG-like complex formation. TA_RS02725 also has a short insertion in motif III. In this study the TA_RS02725 and TA_RS05865 proteins were designated as TaRecJ1 and TaRecJ2, respectively, and the recombinant proteins produced in Escherichia coli were purified to homogeneity (supplemental Fig. S3). The aspartic acid residues, Asp-41 and Asp-43 in TaRecJ1 and Asp-34 and Asp-36 in TaRecJ2, were predicted to coordinate a divalent metal ion and to be crucial for the nuclease activity, according to bacterial RecJ (27, 28). Therefore, we also prepared the TaRecJ1-D41A/D43A and TaRecJ2-D34A/D36A mutants as predicted negative controls for the nuclease activity (supplemental Fig. S3).
TaRecJ1 and TaRecJ2 exhibit different nuclease activities
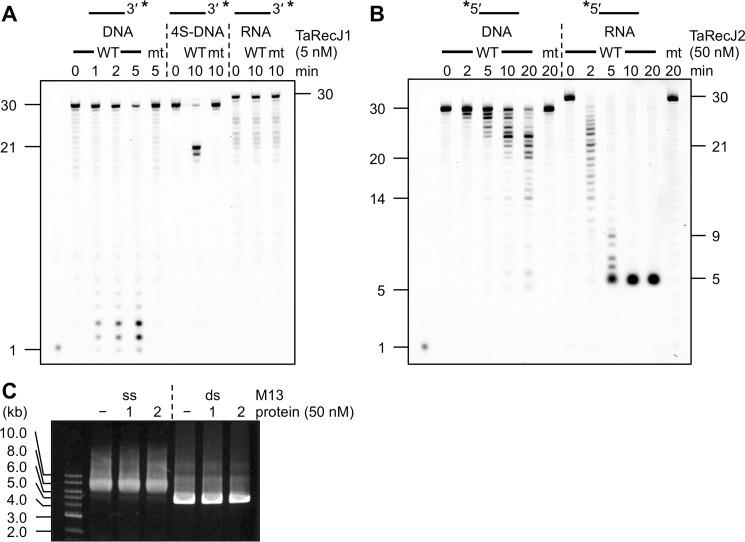
Bacterial RecJ and GAN exhibit 5′-3′ exonuclease activity for single-stranded (ss) DNA. We examined the nuclease activities of TaRecJ1 and TaRecJ2 for ssDNA and RNA. When TaRecJ1 was incubated with a 30-nt ssDNA with an FITC-labeled 3′-terminus (Fig. 1A), short 1–3-nt DNA fragments were observed, suggesting that TaRecJ1 exhibited the exonuclease activity in the 5′-3′ direction. TaRecJ1-D41A/D43A had no detectable activity, as expected. RNA was never degraded by TaRecJ1. To rule out the possibility that TaRecJ1 is an endonuclease that cleaves ssDNA randomly, we introduced four successive phosphorothioate modifications, which generally cause the substrate to be very slowly hydrolyzed by nucleases, into the 10th to 13th bonds in the same oligonucleotide (Table 1, 4S-DNA). TaRecJ1 clearly generated the larger 20- and 21-nt fragments, and thus we concluded that TaRecJ1 exhibits the DNA-specific exonuclease activity in the 5′-3′ direction, similar to bacterial RecJ and GAN. In contrast, when TaRecJ2 was incubated with the 5′-FITC-labeled ssDNA or RNA (Fig. 1B), DNA/RNA ladders were observed, indicating that TaRecJ2 exhibits the exonuclease activity in the 3′-5′ direction for both DNA and RNA. TaRecJ2-D34A/D36A did not generate any detectable product. Although TaRecJ2 did not completely degrade the DNA in 20-min and 60-min reactions, most of the RNA was degraded within 5 min (Figs. 1B and 3A), indicating that TaRecJ2 preferentially degrades RNA over DNA. It should be noted that TaRecJ2 did not generate fragments shorter than 5 nt, although the degradation by TaRecJ1 generated mononucleotides. In addition, neither TaRecJ1 nor TaRecJ2 degraded M13 ssDNA and dsDNA (Fig. 1C), supporting the conclusion that these proteins are exonucleases and not endonucleases.
Figure 1.
Nuclease activities of TaRecJs. A, TaRecJ1 (WT) and its mutant TaRecJ1 D41A/D43A (mt) were incubated at 50 °C with 50 nm 3′-FITC-labeled A30 (DNA), 4S-DNA, or rA30-FITC (RNA) in 20 μl reaction mixtures containing 20 mm Tris-HCl, pH 7.5, 1 mm DTT, 0.1% Triton X-100, and 0.5 mm MnCl2. The reaction products were separated by denaturing 15% PAGE. Asterisks indicate the positions of fluorescent labeling. B, TaRecJ2 (WT) and its mutant TaRecJ2 D34A/D36A (mt) were incubated at 60 °C with 50 nm 5′-FITC labeled A30 (DNA) or FITC-rA30 (RNA) in 20-μl reaction mixtures containing 20 mm Bis-Tris, pH 6.0, 1 mm DTT, 0.1% Triton X-100, and 0.5 mm MnCl2. The reaction products were separated by denaturing 15% PAGE. Asterisks indicate the positions of fluorescent labeling. C, TaRecJ1 (1) and TaRecJ2 (2) were incubated with 1 μg of M13 ssDNA or dsDNA for 30 min. The reaction products were separated by electrophoresis on a 1% agarose gel and visualized by SYBR Gold staining (Invitrogen).
Table 1.
Primers and oligonucleotides used in this study
Asterisks between letters indicate the phosphorothioate modification. Lowercase letters indicate RNA.
| Name | Sequence (5′-3′) |
|---|---|
| TA_RS05395-B-F | dGCCGCGCGGCAGCCATATGAAAGAGGATGGTAGATACGTTCTGG |
| TA_RS05395-R | dCAAGCTTGTCGACGGAGCTCTCATTGATCCAGAACCAGCGCCAC |
| TA_RS02725-F | dATTTTCAGGGCCATATGACCAGCATTCTTGATACATACC |
| TA_RS02725-R | dCAAGCTTGTCGACGGAGCTCTTATTTTATCTCAGTAATGGATG |
| TA_RS05865-F | dAAGGAGATATACATATGATCGAAGATCTCATACCAAAGG |
| TA_RS05865-R | dCAAGCTTGTCGACGGAGCTCCTACCTTGCTTTCGCTTGCACCTTG |
| TaRecJ1-F D41A/S43A | dGCCGCCGCTGGCTTATCCTCATCAGCGATAGC |
| TaRecJ1-R D41A/D43A | dTATATGTGAAACTATTATTATCTTCCTC |
| TaRecJ2-F D34A/D36A | dATAATGCACTATTACCCTTAAAAAATCTGATCC |
| TaRecJ2-R D34A/D36A | dATAATGCACTATTACCCTAAAAAATCTGATCC |
| A30 | dCGAACTGCCTGGAATCCTGACGAACTGTAG |
| 4S-DNA | dCGAACTGCCT*G*G*A*ATCCTGACGAACTGTAG |
| rA30 | cgaacugccuggaauccugacgaacuguag |
| A30–11–30 | dGGAATCCTGACGAACTGTAG |
| A30RC | dCTACAGTTCGTCAGGATTCCAGGCAGTTCG |
| A30RC-11–30 | dTCAGGATTCCAGGCAGTTCG |
| A30temp5 | dTGTCGTGTTCGTTCGCTACAGTTCGTCAGGATTCCAGGCAGTTCG |
| A30temp3 | dCTACAGTTCGTCAGGATTCCAGGCAGTTCGGCTTGCTTGTGCTGT |
| HJ3–54mer | dTCACTCCGCATCTGCCGATTCTGGCTGTGGCGTGTTTCTGGTGGTTCCTAGGTC |
| HJ3–54merRC | dGACCTAGGAACCACCAGAAACACGCCACAGCCAGAATCGGCAGATGCGGAGTGA |
| HJ3RC34 | dCACGCCACAGCCAGAATCGGCAGATGCGGAGTGA |
| HJ-4 | dGACCTAGGAACCACCAGAAACACGCCACAGCCAGGAAGCCGATTGCGAGGCCGTCCTACCATCCTGCAGG |
| Trap DNA | dGACCTAGGAACCACCAGAAACACGCCACAGCCAG |
Figure 3.
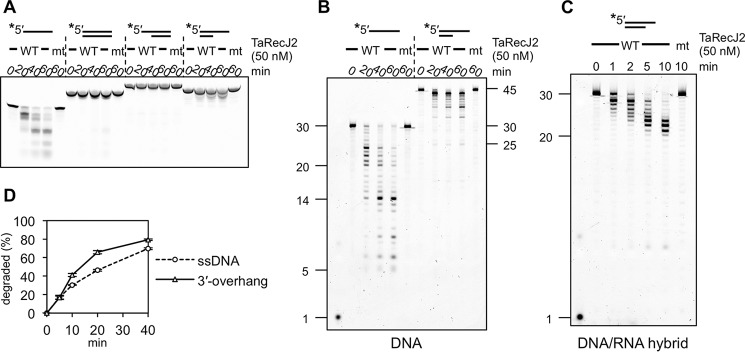
Biochemical properties of TaRecJ2. A–C, TaRecJ2 (WT) and its mutant TaRecJ2 D34A/D36A (mt) were incubated at 50 °C with 50 nm 5′-FITC-labeled 30-nt ssDNA (A30), 30 bp of dsDNA (A30 + A30RC), 30 bp of dsDNA with a 15-nt 5′-overhang (A30temp5 + A30), 20 bp of dsDNA with a 25-nt 3′-overhang (A30temp3 + A30–11-30), or 20 bp of dsDNA/RNA and 10-nt 3′-RNA overhang (rA30 + A30RC-11–30). The reaction products were separated by native 20% PAGE (A) and denaturing 15% PAGE (B and C). Asterisks indicate the positions of fluorescent labeling. D, quantification of the degraded products shown in B. The decreased amount of substrate DNA was quantified and plotted at each time. The averages in three independent experiments with the S.E. are shown.
Biochemical characterization of TaRecJ1 and TaRecJ2
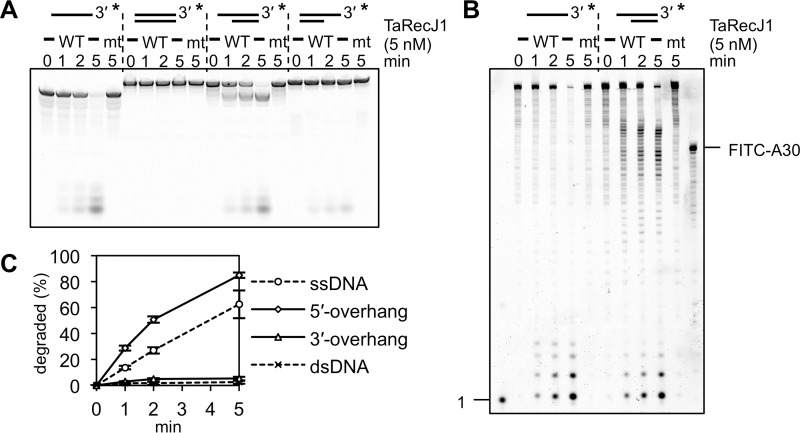
We determined the optimal reaction conditions for the nuclease activities of TaRecJ1 and TaRecJ2 (supplemental Fig. S4). TaRecJ1 exhibited the strongest exonuclease activity for ssDNA with Tris-HCl buffer at pH 7.5 in the presence of 0–50 mm NaCl. The degraded products were observed in the presence of MnCl2, MgCl2, or NiCl2 but not ZnCl2 or CaCl2 (supplemental Fig. S4C), and the strongest nuclease activity was observed in the presence of 1 mm MnCl2. The optimal temperature was 45–55 °C. On the other hand, TaRecJ2 exhibited the strongest nuclease activity with Bis-Tris buffer at pH 6.0–7.0. NaCl strictly inhibited the nuclease activity of TaRecJ2. The degraded product was observed in the presence of MnCl2, with an optimum concentration <0.5 mm. This 3′-5′ exonuclease activity was strictly dependent on manganese, and no cleavage was observed with other metals tested here (supplemental Fig. S4D). The optimal temperature was 55–65 °C. We then examined the substrate specificity using ssDNA, dsDNA, and 5′- and 3′-overhanged DNAs. As shown in Fig. 2A, TaRecJ1 degraded ssDNA and 5′-overhanged DNA, but dsDNA and 3′-overhanged DNA were hardly degraded. The degradation of the 5′-overhanged DNA generated mononucleotides resulting from the resection of dsDNA and longer products, corresponding to the 30-bp dsDNA (Fig. 2B). Therefore, the substrate specificity of TaRecJ1 is highly similar to that of T. kodakarensis GAN (22). This 5′-3′ exonuclease activity was reproducibly observed and was quantified (Fig. 2C). In contrast to TaRecJ1, TaRecJ2 degraded ssDNA and 3′-overhanged DNAs but did not resect dsDNA (Fig. 3, A and B). This 3′-5′ exonuclease activity was also reproducibly observed and was quantified (Fig. 3D). TaRecJ2 was further incubated with a DNA/RNA hybrid with the 3′-extension of RNA. As shown in Fig. 3C, only the 3′-extension was degraded to generate the 20-bp DNA/RNA hybrid, indicating that TaRecJ2 only cleaves ssDNA or RNA.
Figure 2.
Biochemical properties of TaRecJ1. A and B, TaRecJ1 (WT) and its mutant TaRecJ1 D41A/D43A (mt) were incubated at 50 °C with 50 nm 3′-FITC-labeled 54-nt ssDNA (HJ3–54-mer), 54 bp of dsDNA (HJ3–54-mer + HJ3–54-mer RC), 34 bp dsDNA with a 20-nt 5′-overhang (HJ3–54mer + trap DNA), or 34 bp of dsDNA with a 20-nt 3′-overhang (HJ3–54mer + HJ3RC34). The reaction products were separated by native 20% PAGE (A) and denaturing 15% PAGE (B). Asterisks indicate the positions of fluorescent labeling. C, quantification of the degraded products shown in A. The decreased amount of substrate DNA was quantified and plotted at each time. The averages in three independent experiments with the S.E. are shown.
TaRecJ2 forms an archaeal CMG-like complex with TaMCM and TaGINS both in vitro and in vivo
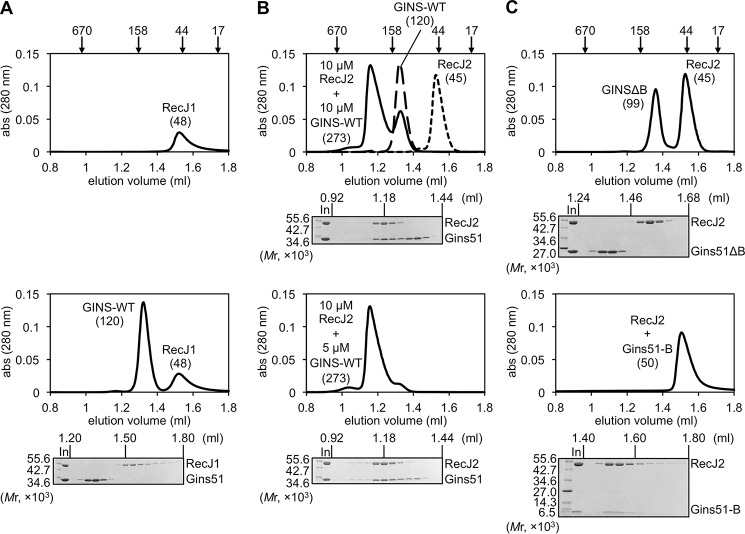
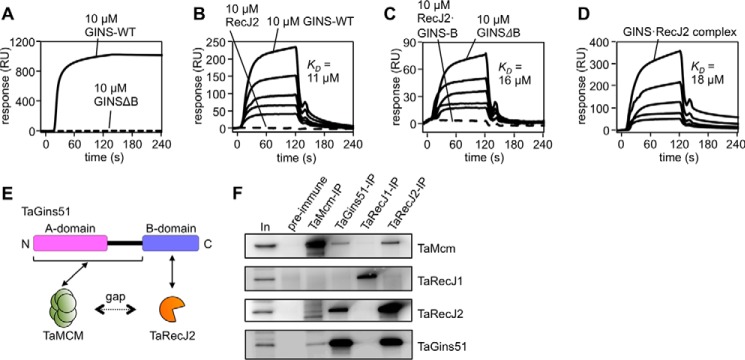
A key question is whether the GAN homolog indeed participates in the archaeal replicative helicase complex. To address this issue, we first investigated the physical interaction of TaRecJs with TaGINS by gel filtration analyses using the recombinant proteins. As shown in Fig. 4, A and B, both TaRecJ1 (Mr 54014.6) and TaRecJ2 (Mr 49647.7) eluted as a single peak corresponding to a monomer, although the molecular weights calculated from the elution positions (TaRecJ1, 48 × 103; TaRecJ2, 45 × 103) were slightly lower than the molecular masses of the proteins because of some structural features. TaGins51-WT was reproducibly eluted as the tetramer, as reported previously (Fig. 4B) (11). In the mixture of TaGins51-WT and TaRecJ1, both proteins eluted separately, indicating that TaRecJ1 does not form a stable complex with TaGINS (Fig. 4A). In contrast, TaRecJ2 eluted with TaGins51-WT in the same fraction, and the estimated molecular weight suggested that this complex is very large (273 × 103) (Fig. 4B). When TaRecJ2 and the TaGins51-WT tetramer were mixed at a 2:1 ratio, the elution profile showed a single peak, suggesting that the TaRecJ2 monomer and the TaGins51 tetramer form a stable complex in a 2:1 ratio. The TaGins51 protein is composed of conserved A- and B-domains, and thus we prepared the B-domain-deleted mutant (TaGins51ΔB), which forms a stable tetramer (24), to examine the interface between TaRecJ2 and TaGINS. As shown in Fig. 4C, TaRecJ2 did not elute with TaGins51ΔB. Although we attempted to prepare the TaGins51-B-domain as a recombinant protein, it was never obtained in the soluble form by itself. Therefore, the TaRecJ2 and TaGins51-B-domain proteins were co-produced in E. coli. The gel filtration analysis demonstrated that the TaRecJ2· TaGins51-B-domain complex was successfully obtained (Fig. 4C). These results indicated that the B-domain of TaGins51 is essential for the interaction with TaRecJ2. We also performed surface plasmon resonance (SPR) analyses to confirm these interactions. Purified TaRecJ2 was immobilized on Biacore CM5 sensor chips by amine coupling. As shown in Fig. 5A, TaGins51-WT strongly associated with TaRecJ2, and TaGins51ΔB did not show the detectable interaction, consistent with the gel filtration chromatography results. We could not evaluate the apparent dissociation constant (KD) value of the TaRecJ2·TaGins51-WT interaction because the interaction was very strong, and the dissociation was hardly detected.
Figure 4.
Physical interactions between TaGINS and TaRecJs. The physical interactions between TaGINS and TaRecJs were analyzed by gel filtration using a Superdex 200 3.2/30 column pre-equilibrated with 10 mm HEPES-NaOH, pH 7.5, and 0.15 m NaCl. The loading volume was 20 μl. The elution profiles, monitored by the absorbance at 280 nm, are shown. The peak positions of the marker proteins are indicated on the top. Aliquots (10 μl) of each fraction from the eluates were subjected to 10–20% SDS-PAGE followed by Coomassie Brilliant Blue staining. A, TaRecJ1 (10 μm) or the mixture of TaRecJ1 and TaGINS-WT (10 μm each) was applied to the column, and the elution profiles were shown in the upper and lower panels, respectively. B, the elution profiles of TaRecJ2 (10 μm), TaGINS-WT (10 μm), and the mixture of TaRecJ2 (10 μm) and TaGINS-WT (10 μm) are shown in the upper panel, and that of the mixture of TaRecJ2 (10 μm) and TaGINS-WT (5 μm) was shown in the lower column. C, the elution profiles of the mixture of TaRecJ2 (10 μm) and TaGINSΔB (10 μm), and the mixture of TaRecJ2 (10 μm) and TaGINS-B-domain (10 μm) is shown in the upper and lower panels, respectively.
Figure 5.
Physical interactions of TaRecJ2 with TaGINS and TaMCM. A--D, SPR analyses were performed. Purified TaRecJ2 (A) and TaMCM (B--D) were each immobilized on CM5 Sensor Chips. The loaded proteins are shown at the top of the sensorgrams. A, TaGINS-WT and TaGINSΔB (10 μm each) were loaded onto the chip for 120 s. RU, resonance units. B, various concentrations (10, 5, 2.5, 1.25, and 0.63 μm) of TaGINS-WT and TaRecJ2 (10 μm) were loaded onto the chip for 120 s. C, Various concentrations (10, 5, 2.5, 1.25, and 0.63 μm) of TaGINSΔB and the TaRecJ2·TaGINS-B-domain complex (10 μm) were loaded onto the chip for 120 s. D, various concentrations (10, 5, 2.5, 1.25, and 0.63 μm as a 2:1 complex) of the TaRecJ2·TaGINS complex were loaded onto the chip for 120 s. The calculated KD values from these SPR analyses are shown in each panel. E, the scheme for interactions among TaGins51, TaMCM, and TaRecJ2. Formation of the CMG-like complex depends on bridging the gap between TaMCM and TaRecJ2 by TaGins51. F, Immunoprecipitation analyses were performed to confirm the formation of a complex including TaMCM, TaGINS, TaRecJ1, and TaRecJ2 in the T. acidophilum cell extract. The immunocomplexes were captured with anti-TaMcm, anti-TaGins51, anti-TaRecJ1, and anti-TaRecJ2 antibodies, respectively, from the whole cell extract (as shown on the top) and were subjected to 10–20% SDS-PAGE followed by Western blot analyses using these antibodies (shown on the right side). The whole cell extracts without immunoprecipitation (In) or precipitated with DynaBeads Protein G (Novex) treated with pre-immune serum were also loaded as positive and negative controls, respectively.
We then investigated whether TaRecJ2 forms a complex with TaMCM, immobilized on Biacore CM5 sensor chips by amine coupling. However, TaRecJ2 did not associate with TaMCM directly, although TaGINS and TaGINSΔB did with KD values of 11 μm and 16 μm, respectively (Fig. 5, B and C). Interestingly, the TaRecJ2·TaGINS-WT complex (2:1 complex) successfully associated with TaMCM, indicating that the archaeal CMG-like complex could be formed in vitro. TaRecJ2 had minimal effects on the affinity between TaGINS and TaMCM, from the KD value of 18 μm between TaRecJ2·TaGINS and TaMCM (Fig. 5D). We also investigated the interaction between the TaRecJ2·TaGINS-B-domain complex and TaMCM. However, no interaction was detected (Fig. 5C), and thus the formation of the TaRecJ2·TaMCM·TaGINS complex depends on the interaction between TaMCM and the TaGINS-A-domain (Fig. 5E). To investigate whether the CMG-like complex is formed in the T. acidophilum cells, an immunoprecipitation assay was performed using extracts from exponentially growing cells. As shown in Fig. 5F, TaRecJ1 was not detected in the precipitations using either an anti-TaMcm, anti-TaGins51, or anti-TaRecJ2 antibody. In contrast, TaRecJ2 was co-precipitated with each of the anti-TaMcm and anti-TaGins51 antibodies. These results suggest that the TaRecJ2·TaMCM·TaGINS-containing complex is also formed in vivo.
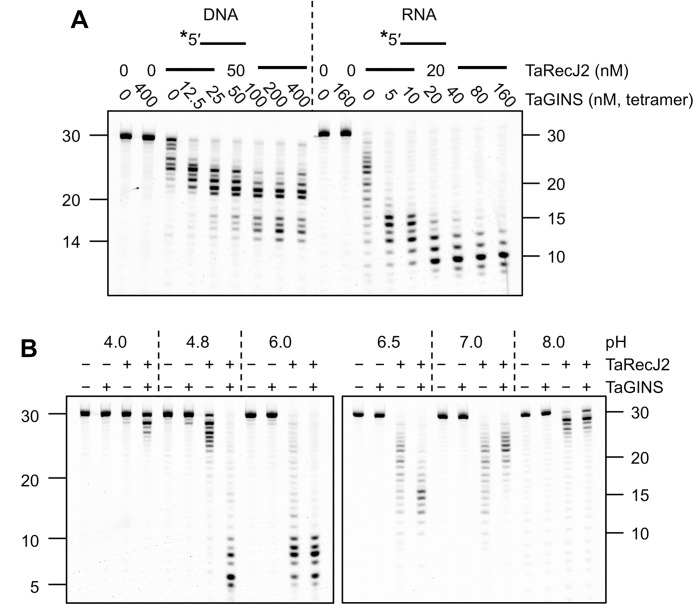
TaGINS stimulates the nuclease activity of TaRecJ2 at an acidic pH
GINS stimulates the exonuclease activity of GAN from T. kodakarensis in vitro (22). We evaluated the exonuclease activities of TaRecJ1 and TaRecJ2 in the presence of TaGINS. The exonuclease activity of TaRecJ1 was the same in the presence and absence of TaGINS (supplemental Fig. S5). In contrast, the 3′-5′ exonuclease activity of TaRecJ2 was stimulated by TaGINS for both DNA and RNA, as shown in Fig. 6A. Although we detected a stable 2:1 complex of TaRecJ2 and TaGINS by gel filtration chromatography, an excess amount of TaGINS was required for the saturation of the TaRecJ2 nuclease activity, perhaps due to the reassembly inefficiency at the low protein concentrations in the reaction conditions. This concentration dependence, in addition to a very fast and successive nucleolytic reaction of the exonucleolytic activity, made the quantification of stimulation not easy. Intriguingly, the stimulation of the nuclease activity of TaRecJ2 by TaGINS was observed at pH values <7.0 (Fig. 6B). In contrast, TaGINS inhibited the nuclease activity at pH 7.0 and 8.0.
Figure 6.
Effect of TaGINS on the nuclease activity of TaRecJ2. A, TaRecJ2 was incubated at 50 °C with 50 nm 5′-FITC labeled A30 (DNA) or rA30 (RNA) in 20-μl reaction mixtures containing 20 mm Bis-Tris, pH 6.0, 1 mm DTT, 0.1% Triton X-100, and 0.5 mm MnCl2, with increasing amounts of TaGINS for 30 min for DNA and 2 min for RNA. The reaction products were separated by denaturing 15% PAGE. B, TaRecJ2 (50 nm) was incubated at 50 °C with 5′-FITC-labeled rA30 (RNA) in 20-μl reaction mixtures containing 20 mm sodium acetate, Bis-Tris, and Tris-HCl in the pH ranges of 4.0–4.8, 6.0–7.0, and 8.0, respectively, in the presence or absence of 50 nm TaGINS for 2 min. The reaction products were separated by denaturing 15% PAGE. Asterisks indicate the positions of fluorescent labeling.
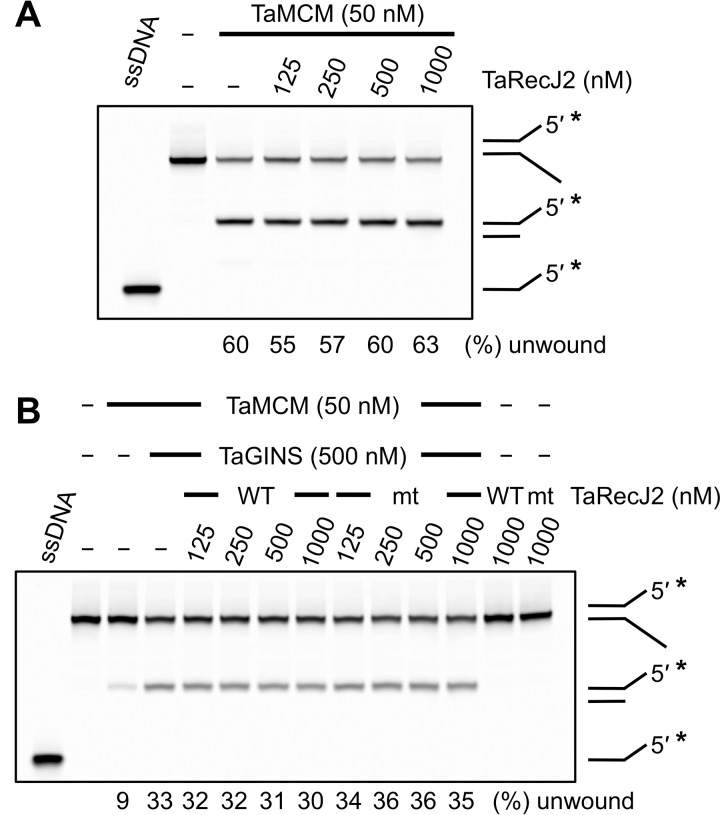
The TaRecJ2·TaGINS complex stimulates the TaMCM helicase activity
Finally, we investigated the effect of the CMG-like complex formation on the helicase activity of TaMCM. Although we used reaction mixtures containing Tris-HCl, pH 8.0, in the previous studies to evaluate the in vitro helicase activity of TaMCM (13, 24), the functional interaction between TaRecJ2 and TaGINS was observed only at acidic pH values, as described above. Therefore, Bis-Tris, pH 6.0, was employed to examine the helicase activity of the CMG-like complex. We could not evaluate the helicase activity of TaMCM at pH 4.8, where the TaRecJ2·TaGINS complex exhibits the strongest nuclease activity, because TaMCM became insoluble at this pH (data not shown). As shown in Fig. 7A, TaRecJ2 had no effect on the helicase activity of TaMCM. The degradation of a splayed-arm substrate by TaRecJ2 was not detected because TaRecJ2 does not cleave DNA in coordination with Mg2+. TaGINS stimulated the TaMCM helicase activity reproducibly, as we previously reported (13) (Fig. 7B). The activation of TaMCM by TaGINS was also observed in the presence of TaRecJ2, indicating that the TaRecJ2·TaGINS complex can stimulate the TaMCM helicase activity despite the lack of stimulation with TaRecJ2 by itself. The increased unwinding efficiency in the presence of TaGINS was not changed by the addition of TaRecJ2. TaRecJ2 does not seem to work actively for the activation of the TaMCM helicase directly, although TaRecJ2 is involved in the helicase complex by the distinctive tight binding between TaGINS and TaRecJ2.
Figure 7.
Effect of the TaGINS·TaRecJ2 complex on the helicase activity of TaMCM. A, the Cy5-labeled splayed-arm DNA substrate (50 nm) was incubated with TaMCM and increasing amounts of TaRecJ2. The reaction was performed at 60 °C for 15 min. B, the same substrate was incubated with 50 nm TaMCM, 500 nm TaGINS, and increasing amounts of TaRecJ2 (WT) or TaRecJ2-D34A/D36A (mt). The reaction was performed at 50 °C for 15 min. The helicase activity is expressed at the bottom of the panel as the relative amount of unwound DNA (%) in each reaction condition. ssDNA was loaded in parallel as controls for the unwinding reaction. Asterisks indicate the positions of fluorescent labeling.
Discussion
The GAN protein from T. kodakarensis is included in arCOG00427. T. acidophilum has two RecJ homologs, which both belong to arCOG00427 (21), and therefore, these two proteins may have similar functions to GAN. It is also interesting that many euryarchaeal organisms have two or more GAN homologs. We investigated the biochemical properties of TaRecJ1 and TaRecJ2 to reveal the functions of the RecJ homologs in Archaea and found that only TaRecJ2 forms a stable complex with TaGINS. TaRecJ1 exhibited nuclease activity but not in a “GINS-associated” manner. These results indicated that the GAN homologs belonging to arCOG00427 have diverse functions.
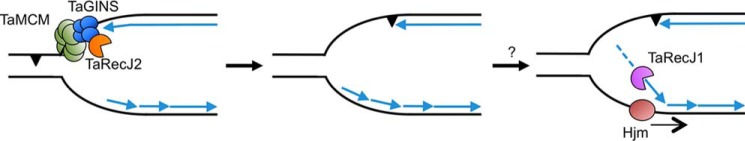
TaRecJ1 exhibited the DNA-specific exonuclease activity in the 5′-3′ direction, similar to bacterial RecJ, and did not form a complex with TaMCM and TaGINS in vivo. Therefore, we speculate that TaRecJ1 is involved in the stalled replication repair and recombinational repair pathways, similar to the bacterial RecJ functions. Actually, RecJ homologs from Methanocaldococcus jannaschii, a methanogenic archaeon, can partially complement the recJ mutant phenotype (UV sensitivity) of E. coli (29). Although the degradation of 5′-overhanged DNA by TaRecJ1 generated mononucleotides in vitro, distinct amounts of dsDNA were also observed. E. coli RecJ was reported to resect the duplex region efficiently, in coordination with the RecQ helicase in vitro (30). TaRecJ1 may also require a RecQ-like helicase for the efficient resection of dsDNA in T. acidophilum cells, and perhaps it is Hjm, the functional counterpart in Archaea (31). In contrast, TaRecJ2 showed quite different properties from TaRecJ1. TaRecJ2 formed a stable complex with GINS, as reported in S. solfataricus, T. kodakarensis, and P. furiosus (9, 22, 23), and exhibited 3′-5′ exonuclease activity for both DNA and RNA. TaGINS stimulated the exonuclease activity of TaRecJ2 only at acidic pH values <pH 7.0. A similar situation was recently reported for Picrophilus torridus, in which GINS stimulates the MCM helicase activity only under acidic conditions at pH 3.0 and pH 4.0 (14). The intracellular pH values of T. acidophilum and P. torridus are reportedly 5.5 and 4.6, respectively (33, 34), which are consistent with the finding that GINS efficiently functions only at acidic pH values to stimulate its partners in Thermoplasmatales. The possible models for the functions of TaRecJ1 and TaRecJ2 in T. acidophilum cells are summarized in Fig. 8. The two RecJ/Cdc45 family proteins in T. acidophilum may function in different processes for precise replication fork progression. TaRecJ1 may also work in double-strand break repair to start homologous recombination in the bacterial RecFOR pathway.
Figure 8.
Possible models for the functions of TaRecJ1 and TaRecJ2 in the T. acidophilum cells. TaRecJ2 is present in the CMG-like replicative helicase complex through the interaction with the TaGins51-B-domain, although there is no interaction between TaRecJ2 and TaMCM (left). TaRecJ1 may function as the counterpart of bacterial RecJ (middle and right) in the stalled replication fork repair process. If DNA polymerase encounters a lesion on the leading strand template, for example, the replication fork is halted with the extended lagging strand (middle). TaRecJ1 may degrade Okazaki fragments in coordination with Hjm (right) followed by fork regression to process the repair.
Interestingly, all GINS-associated nucleases characterized to date have exhibited different nuclease activities. The GAN proteins identified in T. kodakarensis show 5′-3′ exonuclease activity, which is stimulated by GINS (22). We recently solved the crystal structure of GAN complexed with the B-domain of Gins51 and proposed a conserved interaction between GAN/Cdc45 and GINS in Archaea and Eukarya (35). In this work we presented the GAN (RecJ2)·MCM·GINS complex formation in Archaea for the first time and confirmed that the CMG-like complex is actually formed in Archaea. However, in Thermoplasma, GAN is TaRecJ2, a 3′-5′ exonuclease, and it exhibited much stronger activity for RNA than DNA (degradation of ssDNA was not complete even after a 60-min incubation). Therefore, the functions of the RecJ family proteins in Archaea seem to be significantly diversified. Further analyses are required to know whether TaRecJ2 function as an RNase and/or DNase in T. acidophilum cells. A RecJ-like protein in P. furiosus showed the 3′-5′ exonuclease activity for RNA strands and was suggested to remove mismatched RNA primers in DNA replication, although no direct evidence was shown (23). It is important to investigate whether the nuclease activity of TaRecJ2 actually functions in the replisome. It is also possible that TaRecJ2 may participate in an RNA metabolic pathway as well as in DNA replication.
We demonstrated that the B-domain of TaGins51 was essential for the TaRecJ2·TaGINS interaction and that the TaRecJ2·TaGINS complex associates with TaMCM to stimulate the helicase activity. However, we also found that TaRecJ2 does not interact with TaMCM, although TaRecJ2 has a CID-like insertion between motifs IV and V. Furthermore, TaGINS and the TaRecJ2·TaGINS complex stimulated the TaMCM helicase activity in vitro, with comparable efficiencies. There is a gap between RecJ and MCM in the archaeal CMG-like complex. In the EM structure of the eukaryotic CMG complex, CID of Cdc45 was suggested to bind to the N-terminal extension of Mcm2 (26, 36, 37). The phosphorylation of the N-terminal extension of Mcm2 by Dbf4-dependent kinase (DDK) reportedly promotes the Cdc45·Mcm2-7 interaction (39). These are possibly the crucial differences between the archaeal and eukaryotic replicative helicases, because there is no evidence for the phosphorylation-mediated regulation of helicase components in Archaea. Furthermore, the archaeal Mcm proteins lack the N-terminal extension. We speculate that eukaryotic Cdc45 acquired the essential function in the MCM activation, which is accomplished by the direct Cdc45·Mcm2 interaction, after the division into Eukarya. Considering that TaGINSΔB stimulates the TaMCM helicase activity with a comparable efficiency to TaGINS-WT (24), TaMCM could be activated in a TaGins51-B-domain/TaRecJ2-independent manner in T. acidophilum cells. The genes encoding GAN were indeed deleted in other Euryarchaeota, including Haloferax volcanii (40) and T. kodakarensis.4 However, it is also possible that other modifications have been introduced into archaeal replication factors and facilitate the TaRecJ2·MCM interaction. Lysine methylation of S. solfataricus MCM reportedly enhanced the helicase activity at high temperature (41). Other protein modifications besides phosphorylation should be investigated to elucidate the regulation of archaeal MCM. Furthermore, an unidentified protein may bridge the gap between the CID of archaeal RecJ and MCM. It should also be noted that the successful deletion of the recJ genes in archaeal organisms does not mean that RecJ is not involved in DNA replication, because DNA replication may occur by a backup pathway in the absence of RecJ. We are now searching for the interacting partners of TaRecJ1 and TaRecJ2 by yeast two-hybrid assays using a T. acidophilum genome prey library. This work will help to elucidate the diverged functions of the archaeal RecJ homologs in DNA replication. Further studies on the RecJ proteins from Archaea will contribute to revealing the common and diverse functions in the DNA replication machinery of Archaea and Eukarya.
During the review process of this work, a complex formation of MCM, GINS, and Cdc45 from S. solfataricus, a crenarchaeal hyperthermophiles, has been demonstrated by Bell and co-workers (42). No effect of GINS on the helicase activity of MCM was consistent with their early work (9). However, the association of Cdc45/RecJ with GINS and MCM robustly stimulates the helicase activity. Cdc45/RecJ by itself did not show any effect on MCM helicase activity. This report indicated that the CMG complex is the central component of the replicative helicase in S. solfataricus. In the case of S. solfataricus, another group reported that GINS stimulated the DNA binding and processivity of MCM helicase in vitro (15). Further studies will elucidate the structure and functions of the replicative helicase complex more precisely for the efficient replication fork progression in Archaea.
Experimental procedures
Cloning of the Tagins51-B-domain and the TarecJ1 and TarecJ2 genes
The DNA fragments encoding the Tagins51-B-domain, TarecJ1, and TarecJ2 were amplified by PCR using the primer sets TA_RS05395-B-F/TA_RS05395-R, TA_RS02725-F/TA_RS02725-R, and TA_RS05865-F/TA_RS05865-R (Table 1), respectively, with T. acidophilum genomic DNA as the template. The Tagins51-B-domain, TarecJ1, and TarecJ2 fragments were inserted into the pET-28a(+) (Novagen), pCDF-643, and pCDF-644 vectors, respectively, using an In-Fusion HD Cloning kit (Clontech), according to the manufacturer's instructions, and their nucleotide sequences were confirmed. The pCDF-643 and pCDF-644 vectors are modified pCDF1-b (Novagen) vectors in which the cloning region is replaced with that from the pET-21a(+) (Novagen) vector. The pCDF-644 vector also carries an N-terminal His-tag sequence and the recognition sequence for TEV protease inserted upstream of the NdeI site. The resultant plasmids were designated as pET28a-TaGins51-B-domain, pCDF-His-TaRecJ1, and pCDF-TaRecJ2, respectively. Amino acid substitutions were introduced into the TA_RS02725 and TA_RS05865 genes on the pCDF-His-TaRecJ1 and pCDF-TaRecJ2 plasmids by PCR-mediated mutagenesis (KOD-Plus-mutagenesis Kit; TOYOBO) using the primers listed in Table 1.
Overproduction and purification of recombinant proteins
The recombinant TaGins51-WT and TaGins51ΔB proteins were prepared as described previously (24). To obtain the recombinant TaRecJ1 and TaRecJ2 proteins, E. coli BL21-CodonPlus (DE3)-RIL cells (Agilent Technologies) bearing the pCDF-His-TaRecJ1 or pCDF-TaRecJ2 plasmid were cultured at 37 °C in 1 liter of LB medium containing 50 μg/ml streptomycin and 34 μg/ml chloramphenicol. When the cell density reached an A600 of 0.50, the gene expression was induced by adding isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm followed by further cultivation for 16 h at 18 °C. The cells were harvested by centrifugation (10 min, 5,200 × g) and were disrupted by sonication for 10 min in buffer A (50 mm Tris-HCl, pH 8.0, and 10% glycerol) containing 0.5 m NaCl and 20 mm imidazole for TaRecJ1 and buffer A for TaRecJ2. For TaRecJ1, the soluble extracts obtained by centrifugation (10 min, 22,000 × g) were subjected to chromatography on a 1-ml HisTrap HP column (GE Healthcare), which was developed with a linear gradient of 20–300 mm imidazole. The eluted protein fractions were mixed with 200 μg of TEV protease to cleave the N-terminal His tag and then dialyzed against buffer A containing 0.1 m NaCl and 1 mm DTT for 16 h at 4 °C. To remove the TEV protease and the uncleaved proteins, the protein fraction was incubated with 1 ml of Ni-NTA Superflow resin (Qiagen) for 30 min at 4 °C on a rotary shaker, and the flow-through fraction was pooled. EDTA was then added to a final concentration of 1 mm, and the protein fraction was loaded on a Mono Q 5/50 column (GE Healthcare), which was developed with a linear gradient of 0.1–0.3 m NaCl. The eluted protein fractions were loaded on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare), which was equilibrated with buffer A containing 0.15 m NaCl and 1 mm DTT. The eluted protein fractions were pooled, concentrated with an Amicon Ultra filter (Millipore), and stored at −25 °C. For TaRecJ2, the soluble cell extract obtained by centrifugation (10 min, 22,000 × g) was heated at 55 °C for 20 min. The heat-resistant fraction, obtained by centrifugation (10 min, 22,000 × g), was combined with buffer A containing 2 m (NH4)2SO4 to a final concentration of 1 m (NH4)2SO4 and then was subjected to chromatography on a 5-ml HiTrap Butyl-S FF column (GE Healthcare), which was developed with a linear gradient of 1–0 m (NH4)2SO4. The eluted protein fractions were pooled, dialyzed against buffer A containing 1 mm EDTA, and loaded on a Mono Q 5/50 column, which was developed with a linear gradient of 0–1 m NaCl. The eluted protein fractions were loaded on a HiLoad 16/600 Superdex 200 pg column, which was equilibrated with buffer A containing 0.15 m NaCl. The eluted protein fractions were pooled, concentrated with an Amicon Ultra filter, and stored at −25 °C. The TaRecJ2·TaGins51-B-domain complex was also purified from BL21-CodonPlus (DE3)-RIL cells bearing the pCDF-TaRecJ2 and pET28a-TaGins51-B-domain plasmids and cultured at 37 °C in 1 liter of LB medium containing 50 μg/ml streptomycin, 50 μg/ml kanamycin, and 34 μg/ml chloramphenicol by the same procedures used for TaRecJ2, except that the complex was purified using the N-terminal His-tag of the TaGins51-B-domain, before loading on the 5-ml HiTrap Butyl-S FF column. The protein concentrations were determined using their absorbances at 280 nm and the extinction coefficients of 11,920 m−1 cm−1 for TaGins51-WT, 7,450 m−1 cm−1 for TaGins51ΔB, 23,380 m−1 cm−1 for TaRecJ1, 39,310 m−1 cm−1 for TaRecJ2, and 43,780 m−1 cm−1 for the TaRecJ2·TaGins51-B-domain complex as a monomer. The extinction coefficients were calculated for each protein by the method described earlier (38).
Nuclease assay
The nuclease activities of TaRecJ1 and TaRecJ2 were measured in 20-μl reaction mixtures containing 20 mm Tris-HCl, pH 7.5 (TaRecJ1) or 20 mm Bis-Tris, pH 6.0 (TaRecJ2), 0.5 mm MnCl2, 1 mm DTT, 0.1% Triton X-100, 50 nm DNA or RNA substrate (Table 1) and the proteins described in each figure legend. After an incubation at 50 °C for TaRecJ1 and 60 °C for TaRecJ2, the samples were immediately transferred to ice, and 80 μl of stop solution (formamide, containing 0.1% bromphenol blue and 0.1% xylene cyanol) or 5 μl of 4× stop buffer (100 mm EDTA, 4% SDS, 10% Ficoll, and 0.1% Orange G) was added. An aliquot (3 μl) was separated by electrophoresis on an 8 m urea-containing 15% acrylamide gel in 1× TBE (90 mm Tris borate, pH 8.5, and 1 mm EDTA) or a 20% gel in 1× TBE. The gel image was obtained with an image analyzer, Typhoon Trio+ (GE Healthcare), and the nuclease activity was quantified using the ImageQuant TL software (GE Healthcare).
Gel filtration chromatography
Gel filtration chromatography was performed using the SMART system (GE Healthcare). The purified recombinant TaGins51-WT, TaGins51ΔB, TaRecJ1, TaRecJ2, and TaRecJ2·TaGins51-B-domain complex were applied to a Superdex 200 3.2/30 column (GE Healthcare) pre-equilibrated with 10 mm HEPES-NaOH, pH 7.5, and 0.15 m NaCl. To analyze the interaction between TaGins51 and TaRecJs, TaGins51-WT or TaGins51ΔB was mixed with TaRecJ1 or TaRecJ2 and applied to the same column. The molecular masses of the proteins were estimated from the elution profiles of standard marker proteins, including thyroglobulin (Mr 670,000), γ-globulin (Mr 158,000), ovalbumin (44,000), and myoglobin (Mr 17,000).
SPR analysis
The SPR analysis was performed using a BIACORE J system (GE Healthcare). The purified recombinant TaRecJ2 and TaMCM were each immobilized on a CM5 Sensor Chip by amine coupling according to the manufacturer's protocol (GE Healthcare). The BIACORE analyses were performed at 25 °C in 10 mm HEPES-NaOH, pH 7.5, containing 150 mm NaCl and 0.05% Tween 20 for the TaRecJ2-immobilized chip and in 10 mm HEPES-NaOH, pH 7.5, containing 0.05% Tween 20 for TaRecJ2-immobilized chip at a flow rate of 30 μl/min. To measure the kinetic parameters, various concentrations of proteins (10, 5, 2.5, 1.25, and 0.63 μm as the tetramer) were loaded, and the apparent KD was calculated from the association and dissociation curves of the sensorgrams using the BIAevaluation program (GE Healthcare).
Immunoprecipitation assay
T. acidophilum cells were cultured in 200 ml of medium at 56 °C with shaking as described previously (32) and were harvested at the exponential growth phase (A600 = 0.25) by centrifugation (10 min, 6,000 × g). The cells (1 × 1011) were suspended and disrupted in lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 0.5% Triton X-100) by sonication for 1 min (5 s on/5 s off). A portion (10 μl) of Dynabeads Protein G was washed twice with PBS-T (10 mm sodium phosphate, pH 7.5, 150 mm NaCl, 0.1% Tween 20), mixed with PBS-T containing 10 μl of anti-TaMcm, anti-TaGins51, anti-TaRecJ1, or anti-TaRecJ2 antiserum (prepared by injecting the purified recombinant proteins into rabbits), and incubated at room temperature for 1 h on a rotary shaker. Each mixture was washed twice with TBS-T and then twice with 0.2 m triethanolamine, pH 8.0. The antibody was cross-linked to the Dynabeads Protein G with dimethyl pimelimidate dihydrochloride (DMP, Thermo Scientific Pierce) according to the manufacturer's protocol. Preimmune serum was used for negative control experiments. After equilibration of the antibody-conjugated Dynabeads Protein G with lysis buffer, an aliquot of the cell extract (500 μl, 1.7 × 1010 cells) was added, and the mixture was incubated at room temperature for 1 h on the rotary shaker. The precipitates were washed twice with lysis buffer, and the immunoprecipitated proteins were eluted from the beads with 40 μl of gel loading solution (50 mm Tris-HCl, pH 6.8, 10% glycerol, 100 mm DTT, 0.2 mg/ml bromphenol blue, 2% SDS) at 98 °C for 3 min. Five-microliter portions of the eluates were subjected to 10–20% SDS-PAGE followed by Western blot analysis.
Helicase assay
The 54-mer oligonucleotide HJ-3–54-mer, with a Cy5-labeled 5′-terminus (Table 1), was annealed with HJ-4 (Table 1) and purified as described previously. The helicase activity of TaMCM was measured in 20-μl reaction mixtures containing 20 mm Bis-Tris, pH 6.0, 10 mm Mg(CH3COO)2, 1 mm DTT, 0.1% Triton X-100, 2.5 mm ATP, 50 nm DNA substrate, 50 nm trap DNA to prevent re-annealing of the unwound DNA, and 50 nm TaMCM, with increasing amounts of TaGins51 and/or TaRecJ2 proteins. After an incubation at 50 °C or 60 °C for 15 min, the samples were immediately transferred to ice, and 7 μl of 4× stop buffer (100 mm EDTA, 4% SDS, 10% Ficoll, and 0.1% Orange G) was added. An aliquot (3 μl) was loaded onto a 10% polyacrylamide gel in 1× TBE and electrophoresed at 15 mA for 40 min. The gel image was obtained with an image analyzer, Typhoon Trio+, and the helicase activity was quantified using the ImageQuant TL software.
Author contributions
H. O., S. I., and Y. I. conceived the project. H. O. and S. I. designed the experiments. H. O. performed the experiments with help from S. I. H. O., S. I., D. K., and Y. I. analyzed the data and contributed to manuscript preparation. H. O., S. I., and Y. I. wrote the manuscript.
Supplementary Material
Acknowledgment
We thank Dr. Nils-Kåre Birkeland for critical discussion.
This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grant 26242075 (to Y. I.) and in part by the Institute for Fermentation, Osaka (IFO). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5.
Nagata, M., Ishino, S.,Yamagami, T., Ogino, H., Simons, J.-R., Kanai, T., Atomi, H., and Ishino, Y., unpublished data.
- MCM
- minichromosome maintenance
- TaMCM
- T. acidophilum MCM
- CMG
- Cdc45, MCM, and GINS
- GAN
- GINS-associated nuclease
- TaGINS
- T. acidophilum GINS
- nt
- nucleotide(s)
- CID
- CMG interaction domain
- ssDNA
- single-stranded DNA
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SPR
- surface plasmon resonance
- TEV
- tobacco etch virus.
References
- 1. Ishino Y., and Ishino S. (2012) Rapid progress of DNA replication studies in Archaea, the third domain of life. Sci. China Life Sci. 55, 386–403 [DOI] [PubMed] [Google Scholar]
- 2. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., and Oda M. (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79, 89–130 [DOI] [PubMed] [Google Scholar]
- 3. Mott M. L., and Berger J. M. (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 4. Leipe D. D., Aravind L., and Koonin E. V. (1999) Did DNA replication evolve twice independently? Nucleic Acids Res. 27, 3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ilves I., Petojevic T., Pesavento J. J., and Botchan M. R. (2010) Activation of the Mcm2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M. R., and Berger J. M. (2011) The structural basis for Mcm2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takayama Y., Kamimura Y., Okawa M., Muramatsu S., Sugino A., and Araki H. (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makarova K. S., Wolf Y. I., Mekhedov S. L., Mirkin B. G., and Koonin E. V. (2005) Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 33, 4626–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marinsek N., Barry E. R., Makarova K. S., Dionne I., Koonin E. V., and Bell S. D. (2006) GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 7, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshimochi T., Fujikane R., Kawanami M., Matsunaga F., and Ishino Y. (2008) The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J. Biol. Chem. 283, 1601–1609 [DOI] [PubMed] [Google Scholar]
- 11. Ogino H., Ishino S., Mayanagi K., Haugland G. T., Birkeland N. K., Yamagishi A., and Ishino Y. (2011) The GINS complex from the thermoacidophilic archaeon, Thermoplasma acidophilum may function as a homotetramer in DNA replication. Extremophiles 15, 529–539 [DOI] [PubMed] [Google Scholar]
- 12. Ishino S., Fujino S., Tomita H., Ogino H., Takao K., Daiyasu H., Kanai T., Atomi H., and Ishino Y. (2011) Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells 16, 1176–1189 [DOI] [PubMed] [Google Scholar]
- 13. Ogino H., Ishino S., Haugland G. T., Birkeland N. K., Kohda D., and Ishino Y. (2014) Activation of the MCM helicase from the thermophilic archaeon, Thermoplasma acidophilum, by interactions with GINS and Cdc6–2. Extremophiles 18, 915–924 [DOI] [PubMed] [Google Scholar]
- 14. Goswami K., Arora J., and Saha S. (2015) Characterization of the MCM homohexamer from the thermoacidophilic euryarchaeon Picrophilus torridus. Sci. Rep. 5, 9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lang S., and Huang L. (2015) The Sulfolobus solfataricus GINS complex stimulates DNA binding and processive dna unwinding by minichromosome maintenance helicase. J. Bacteriol. 197, 3409–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aparicio O. M., Weinstein D. M., and Bell S. P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69 [DOI] [PubMed] [Google Scholar]
- 17. Sanchez-Pulido L., and Ponting C. P. (2011) Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics 27, 1885–1888 [DOI] [PubMed] [Google Scholar]
- 18. Persky N. S., and Lovett S. T. (2008) Mechanisms of recombination: lessons from E. coli. Crit. Rev. Biochem. Mol. Biol. 43, 347–370 [DOI] [PubMed] [Google Scholar]
- 19. Dianov G., and Lindahl T. (1994) Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 4, 1069–1076 [DOI] [PubMed] [Google Scholar]
- 20. Burdett V., Baitinger C., Viswanathan M., Lovett S. T., and Modrich P. (2001) In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 98, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makarova K. S., Koonin E. V., and Kelman Z. (2012) The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol. Direct 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z., Pan M., Santangelo T. J., Chemnitz W., Yuan W., Edwards J. L., Hurwitz J., Reeve J. N., and Kelman Z. (2011) A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 39, 6114–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan H., Liu X. P., Han Z., Allers T., Hou J. L., and Liu J. H. (2013), RecJ-like protein from Pyrococcus furiosus has 3′-5′ exonuclease activity on RNA: implications for proofreading of 3′-mismatched RNA primers in DNA replication. Nucleic Acids Res. 41, 5817–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogino H., Ishino S., Oyama T., Kohda D., and Ishino Y. (2015) Disordered interdomain region of Gins is important for functional tetramer formation to stimulate MCM helicase in Thermoplasma acidophilum. Biosci. Biotechnol. Biochem. 79, 432–438 [DOI] [PubMed] [Google Scholar]
- 25. Krastanova I., Sannino V., Amenitsch H., Gileadi O., Pisani F. M., and Onesti S. (2012) Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 287, 4121–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simon A. C., Sannino V., Costanzo V., and Pellegrini L. (2016) Structure of human Cdc45 and implications for CMG helicase function. Nat. Commun. 7, 11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutera V. A. Jr., Han E. S., Rajman L. A., and Lovett S. T. (1999) Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J. Bacteriol. 181, 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamagata A., Kakuta Y., Masui R., and Fukuyama K. (2002) The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 99, 5908–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajman L. A., and Lovett S. T. (2000) A thermostable single-strand DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. J. Bacteriol. 182, 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morimatsu K., and Kowalczykowski S. C. (2014) RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 111, E5133–E5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujikane R., Shinagawa H., and Ishino Y. (2006) The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells 11, 99–110 [DOI] [PubMed] [Google Scholar]
- 32. Yasuda M., Oyaizu H., Yamagishi A., and Oshima T. (1995) Morphological variation of new Thermoplasma acidophilum isolates from Japanese hot springs. Appl. Environ. Microbiol. 61, 3482–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Searcy D. G. (1976) Thermoplasma acidophilum: Intracellular pH and potassium concentration. Biochim. Biophys. Acta 451, 278–286 [DOI] [PubMed] [Google Scholar]
- 34. Ciaramella M., Napoli A., and Rossi M. (2005) Another extreme genome: how to live at pH 0. Trends Microbiol. 13, 49–51 [DOI] [PubMed] [Google Scholar]
- 35. Oyama T., Ishino S., Shirai T., Yamagami T., Nagata M., Ogino H., Kusunoki M., and Ishino Y. (2016) Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Res. 44, 9505–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Costa A., Renault L., Swuec P., Petojevic T., Pesavento J. J., Ilves I., MacLellan-Gibson K., Fleck R. A., Botchan M. R., and Berger J. M. (2014) DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife 3, e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuan Z., Bai L., Sun J., Georgescu R., Liu J., O'Donnell M. E., and Li H. (2016) Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 23, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., and Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, (Walker J. M., ed) pp. 571–607, Humana Press, New York [Google Scholar]
- 39. Perez-Arnaiz P., Bruck I., and Kaplan D. L. (2016) Mcm10 coordinates the timely assembly and activation of the replication fork helicase. Nucleic Acids Res. 44, 315–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giroux X., and MacNeill S. A. (2015) Molecular genetic methods to study DNA replication protein function in Haloferax volcanii, a model archaeal organism. Methods Mol. Biol. 1300, 187–218 [DOI] [PubMed] [Google Scholar]
- 41. Xia Y., Niu Y., Cui J., Fu Y., Chen X. S., Lou H., and Cao Q. (2015) The helicase activity of hyperthermophilic archaeal MCM is enhanced at high temperatures by lysine methylation. Front. Microbiol. 6, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu Y., Gristwood T., Hodgson B., Trinidad J. C., Albers S.-V., and Bell S. D. (2016) Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc. Natl. Acad. Sci. U.S.A. 113, 13390–13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.