Abstract
Cadmium exposure is known to increase lung cancer risk, but the underlying molecular mechanisms in cadmium-stimulated progression of malignancy are unclear. Here, we examined the effects of prolonged cadmium exposure on the malignant progression of A549 human lung adenocarcinoma cells and the roles of Notch1, hypoxia-inducible factor 1α (HIF-1α), and insulin-like growth factor 1 receptor (IGF-1R)/Akt/extracellular signal-regulated kinase (ERK)/p70 S6 kinase 1 (S6K1) signaling pathways. Exposing A549 cells to 10 or 20 μm cadmium chloride (CdCl2) for 9–15 weeks induced a high proliferative potential, the epithelial-mesenchymal transition (EMT), stress fiber formation, high cell motility, and resistance to antitumor drugs. Of note, the CdCl2 exposure increased the levels of the Notch1 intracellular domain and of the downstream Notch1 target genes Snail and Slug. Strikingly, siRNA-mediated Notch1 silencing partially suppressed the CdCl2-induced EMT, stress fiber formation, high cell motility, and antitumor drug resistance. In addition, we found that prolonged CdCl2 exposure induced reduction of E-cadherin in BEAS-2B human bronchial epithelial cells and antitumor drug resistance in H1975 human tumor-derived non-small-cell lung cancer cells depending on Notch1 signaling. Moreover, Notch1, HIF-1α, and IGF-1R/Akt/ERK/S6K1 activated each other to induce EMT in the CdCl2-exposed A549 cells. These results suggest that Notch1, along with HIF-1α and IGF-1R/Akt/ERK/S6K1 signaling pathways, promotes malignant progression stimulated by prolonged cadmium exposure in this lung adenocarcinoma model.
Keywords: drug resistance, epithelial-mesenchymal transition (EMT), lung cancer, metal, Notch pathway, cadmium
Introduction
Cadmium is a widespread pollutant metal with clear carcinogenic potential in humans and animals (1, 2). In humans, its biological half-life is measured in decades (10–30 years) (3). Cadmium inhalation can result from cigarette smoking or via occupational settings (1, 4). Therefore, its carcinogenicity continues to be a major health concern in both environmental and occupational settings.
Lung cancer is the most frequent type of cancer, causing more than one million deaths annually. Association between cadmium exposure and lung cancer is well established (1, 5, 6). The lung absorbs relatively high amounts of cadmium after inhalation (7). Cadmium has been reported to transform human bronchial epithelial cells (8, 9), indicating its role in the first stage of cadmium carcinogenesis. However, little attention has been paid to subsequent events leading to the patients' poor prognosis, namely, malignant progression, including tumor invasiveness and antitumor drug resistance. Tumor invasiveness is caused by cell motility and epithelial-mesenchymal transition (EMT)2 (10, 11). EMT is characterized by the dissolution of cell-cell junctions as well as the loss of apicobasolateral polarity, resulting in the formation of migratory mesenchymal cells with invasive properties (12). During EMT, cells lose the expression of the epithelial marker E-cadherin and gain expression of the mesenchymal markers N-cadherin and vimentin.
The Notch pathway is an evolutionally conserved signaling pathway implicated in various cellular processes, including cell-fate determination, differentiation, proliferation, and cell death (13). In mammals, there are four Notch receptors (Notch1–4). Activation of Notch signaling requires the interaction of the Notch receptors with their ligands such as Jagged1 and -2 and Delta-like 1, 3, and 4 on neighboring cells. Ligand binding leads to sequential cleavages by a disintegrin and metalloprotease (ADAM) and the γ-secretase complex in Notch, resulting in the release of Notch intracellular domain (Notch-ICD) from the membrane. Subsequently, Notch-ICD translocates into the nucleus to modulate Notch-specific target gene expression. Notch signaling regulates gene expression in a highly context- and cell type-dependent manner (14, 15). In epithelial tumor cells, Notch1 expression promotes cell motility, EMT, antitumor drug resistance, and growth of cancer stem cells as well as cell proliferation (10, 16–19). We as well as other groups have previously shown that Notch signaling is activated in response to various stressors such as hypoxia and cadmium exposure (20–22). However, the effects of prolonged cadmium exposure on Notch1 signaling in lung epithelial cells and its possible involvement in cadmium-induced malignant progression have not yet been clarified.
Therefore, we examined whether prolonged cadmium chloride (CdCl2) exposure induces malignant progression in A549 human lung adenocarcinoma cells through the activation of Notch1 signaling. Our results show that cadmium exposure induces Notch1-dependent EMT, stress fiber formation, high cell motility, and antitumor drug resistance in A549 cells. We also found that hypoxia-inducible factor 1α (HIF-1α), a stress-responsive transcription factor, enhanced the transcriptional activity of Notch1 and that insulin-like growth factor 1 receptor (IGF-1R)/Akt/extracellular-signal regulated kinase (ERK)/p70 S6 kinase 1 (S6K1) cascade could interplay with Notch1 signaling and HIF-1α in response to cadmium exposure.
Results
Prolonged cadmium exposure induces EMT, stress fiber formation, and high cell motility in A549 cells
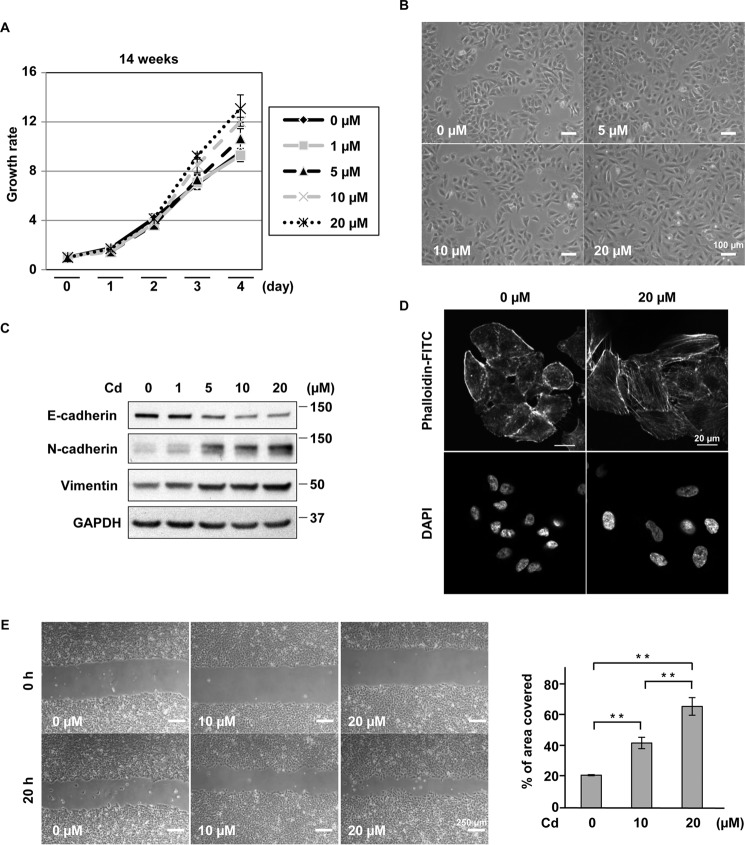
First, we examined the proliferative potential of A549 cells following exposure to varying doses (1–20 μm) of CdCl2 for 1–14 weeks. Cells exposed to 5, 10, or 20 μm CdCl2 for 1 week showed lower proliferation compared with cells without CdCl2 exposure (supplemental Fig. S1a). This lower proliferation became less marked after 8 weeks of exposure (supplemental Fig. S1a). Conversely, cells exposed to 10 or 20 μm CdCl2 for 10 (supplemental Fig. S1a) and 14 weeks (Fig. 1A) showed higher proliferation than cells without CdCl2 exposure. Although A549 cells without CdCl2 exposure maintained a typical epithelial morphology and were organized in compact islets, cells exposed to 10 or 20 μm CdCl2 for 1 week changed in morphology to a rounded shape (supplemental Fig. S1b). In contrast, cells exposed to 10 or 20 μm CdCl2 for 10 weeks showed an EMT-like spindle shape without definite cellular damage (Fig. 1B). This EMT-like change was also observed in cells exposed to 5 μm CdCl2 for 32 weeks (supplemental Fig. S1c). Furthermore, expression of E-cadherin decreased consistently, and that of N-cadherin and vimentin increased in cells exposed to 1–20 μm CdCl2 for 10 weeks in a dose-dependent manner (Fig. 1C). EMT-inducing stressors such as transforming growth factor β (TGF-β) have been reported to induce the formation of stress fibers (23). Exposure to 20 μm CdCl2 for 10 weeks also produced many stress fibers in A549 cells (Fig. 1D). Wound-healing assays showed that exposure to 10 or 20 μm CdCl2 for 10 weeks increased cell motility (Fig. 1E). In addition, the mRNA level of matrix metalloproteinase 2, a type VI collagenase that facilitates tumor invasion (24), was elevated in prolonged CdCl2-exposed cells (supplemental Fig. S2). Taken together, these results indicate that CdCl2 exposure for more than 8 weeks induced EMT, stress fiber formation, and high cell motility in A549 cells. For the subsequent experiments, we used A549 cells that were incubated with 20 μm CdCl2 for 9–15 weeks because cadmium-induced EMT and high cell motility were observed in these cells with prolonged CdCl2 exposure (data not shown).
Figure 1.
Prolonged CdCl2 exposure induces EMT, stress fiber formation, and high cell motility in A549 cells. A, cells incubated with 0, 1, 5, 10, or 20 μm CdCl2 for 14 weeks were seeded and allowed to proliferate for 5 days. Total cell numbers were counted each day using a cell counter. Each value is the ratio of the cell numbers at Day 0 and reflects the mean ± S.D. (error bars) of three experiments with duplicate assays in each experiment. B, cells were incubated with 0, 5, 10, or 20 μm CdCl2 for 10 weeks, and their phase-contrast micrographs were taken. Scale bar, 100 μm. C, cells were incubated with 0, 1, 5, 10, or 20 μm CdCl2 (Cd) for 10 weeks. Cell lysates were subjected to Western blotting using the indicated antibodies. Immunoblots shown are representative of at least three independent experiments. D, cells were incubated with 0 or 20 μm CdCl2 for 10 weeks and stained with phalloidin-FITC for F-actin and DAPI for DNA. Scale bar, 20 μm. E, cells were incubated with 0, 10, or 20 μm CdCl2 for 10 weeks and allowed to migrate for 20 h after scratching. Results of quantification of the area covered by cells are also shown (E). Each value (mean ± S.D. of three experiments) was expressed as a percentage of the area covered. Scale bar, 250 μm. **, p < 0.01, significant difference between the samples.
Notch1 is involved in prolonged cadmium exposure-induced EMT, stress fiber formation, and high cell motility in A549 cells
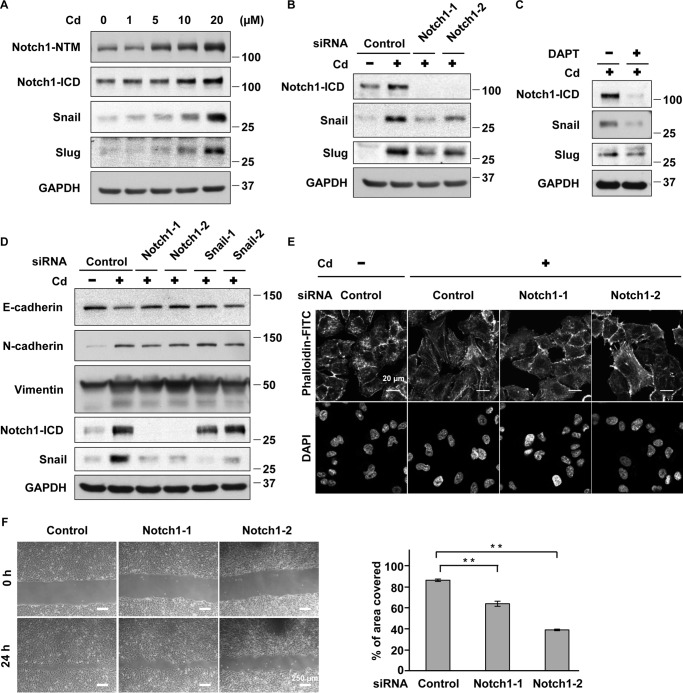
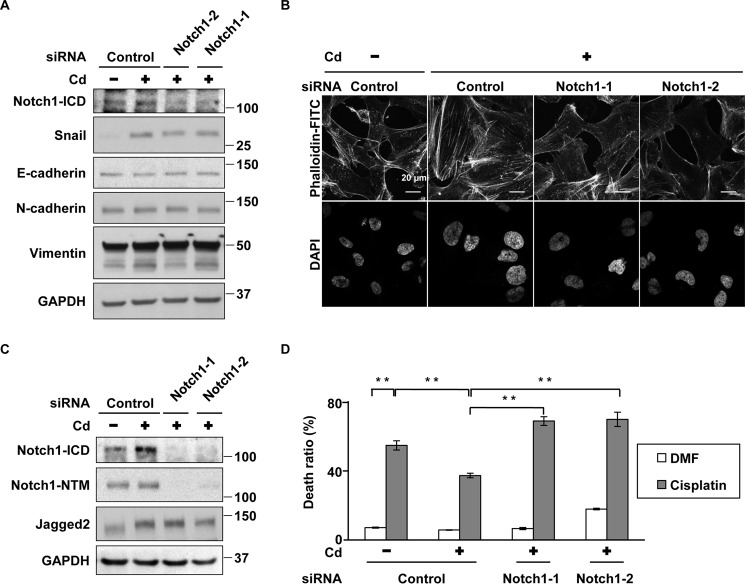
Although Notch3 is highly activated in A549 cells (25), its mRNA expression decreased in cells with prolonged CdCl2 exposure (supplemental Fig. S3a). However, the levels of Notch1 transmembrane subunit (Notch1-NTM), which includes the intracellular region (Fig. 2A), and its mRNA (supplemental Fig. S3b) increased in prolonged CdCl2-exposed cells. In addition, accumulation of Notch1-ICD and its target transcription factors that repress E-cadherin expression, Snail and Slug (26, 27), was found in these cells (Fig. 2A). Notch1 knockdown with siRNAs targeted against the human Notch1 gene (Notch1-1 and Notch1-2) (Fig. 2B) or treatment with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (DAPT) (Fig. 2C) reduced the expression of Notch1-ICD, Snail, and Slug in prolonged CdCl2-exposed cells, indicating the activation of Notch1 signaling by cadmium exposure. Depletion of Notch1 with siRNAs (Notch1-1 and Notch1-2) and that of Snail with siRNAs targeted against the human SNAI1 gene (Snail-1 and Snail-2) suppressed cadmium-induced reduction of E-cadherin expression (Fig. 2D). However, cadmium-induced up-regulation of N-cadherin and vimentin was not altered definitely by either Notch1 or Snail knockdown. In addition, Notch1 depletion with siRNAs (Notch1-1 and Notch1-2) could ameliorate cadmium-induced stress fiber formation (Fig. 2E) and high cell motility (Fig. 2F). These findings indicate that Notch1 signaling reduces E-cadherin expression and induces high cell motility in A549 cells under prolonged CdCl2 exposure.
Figure 2.
Notch1 is involved in prolonged CdCl2-induced EMT, stress fiber formation, and high cell motility in A549 cells. A, cells were incubated with 0, 1, 5, 10, or 20 μm CdCl2 (Cd) for 10 weeks. Cell lysates were subjected to Western blotting using the indicated antibodies. B, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA, Notch1 siRNA-1, or Notch1 siRNA-2. Cell lysates were subjected to Western blotting using the indicated antibodies. C, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were treated with 0.1% DMSO or 50 μm DAPT for 16 h. Cell lysates were subjected to Western blotting using the indicated antibodies. D, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA, Notch1 siRNA-1, Notch1 siRNA-2, Snail siRNA-1, or Snail siRNA-2. Cell lysates were subjected to Western blotting using the indicated antibodies. Immunoblots shown are representative of at least three independent experiments. e and f, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA, Notch1 siRNA-1, or Notch1 siRNA-2. Transfected cells were stained with phalloidin-FITC for F-actin and DAPI for DNA (E) and used for wound healing assays (F). Scale bar, 20 μm (e), 250 μm (F). Results of quantification of the area covered by cells are also shown (F). Each value (mean ± S.D. (error bars) of three experiments) was expressed as a percentage of the area covered. **, p < 0.01, significant difference between the samples.
Notch1 is involved in prolonged cadmium exposure-induced antitumor drug resistance in A549 cells
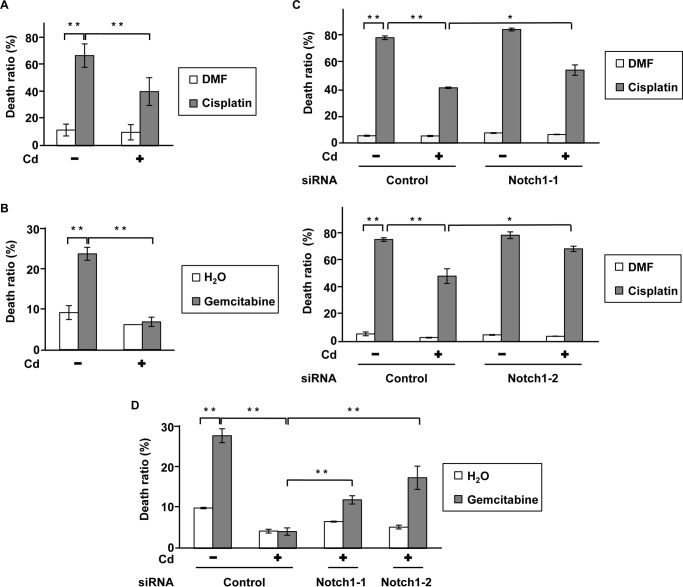
We determined the viability of prolonged CdCl2-exposed A549 cells treated with cisplatin, gemcitabine, and etoposide, antitumor drugs commonly used in lung cancer chemotherapy, using trypan blue exclusion assays. Prolonged CdCl2 exposure resulted in lower cell death induced by treatment with cisplatin (Fig. 3A), gemcitabine (Fig. 3B), and etoposide (data not shown), indicating the development of higher antitumor drug resistance. Deletion of Notch1 expression with siRNAs (Notch1-1 and Notch1-2) partially but significantly increased cell death induced by cisplatin (Fig. 3C) and gemcitabine (Fig. 3D) in prolonged CdCl2-exposed cells. In addition, knockdown of Snail increased cell death induced by cisplatin (supplemental Fig. S4a) and gemcitabine (supplemental Fig. S4b) in these cells. These findings indicate that prolonged CdCl2 exposure induced antitumor drug resistance through Notch1 signaling in A549 cells.
Figure 3.
Notch1 is involved in prolonged CdCl2-induced antitumor drug resistance in A549 cells. A and B, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were treated with 0.1% N,N-dimethylformamide (DMF) or 10 μg/ml cisplatin (A) and 0.1% distilled water or 200 μm gemcitabine (B). The viability of cells was determined by trypan blue exclusion assay. C and D, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA, Notch1 siRNA-1, or Notch1 siRNA-2. Transfected cells were treated with 0.1% N,N-dimethylformamide or 10 μg/ml cisplatin (C) and 0.1% distilled water or 200 μm gemcitabine (D). The viability of cells was determined by trypan blue exclusion assay. Each value is the percentage of trypan blue-positive cells and reflects the mean ± S.D. (error bars) of three experiments with duplicate assays in each experiment. *, p < 0.05; **, p < 0.01, significant difference between the samples.
HIF-1α regulates Notch1 activity in prolonged cadmium-exposed A549 cells
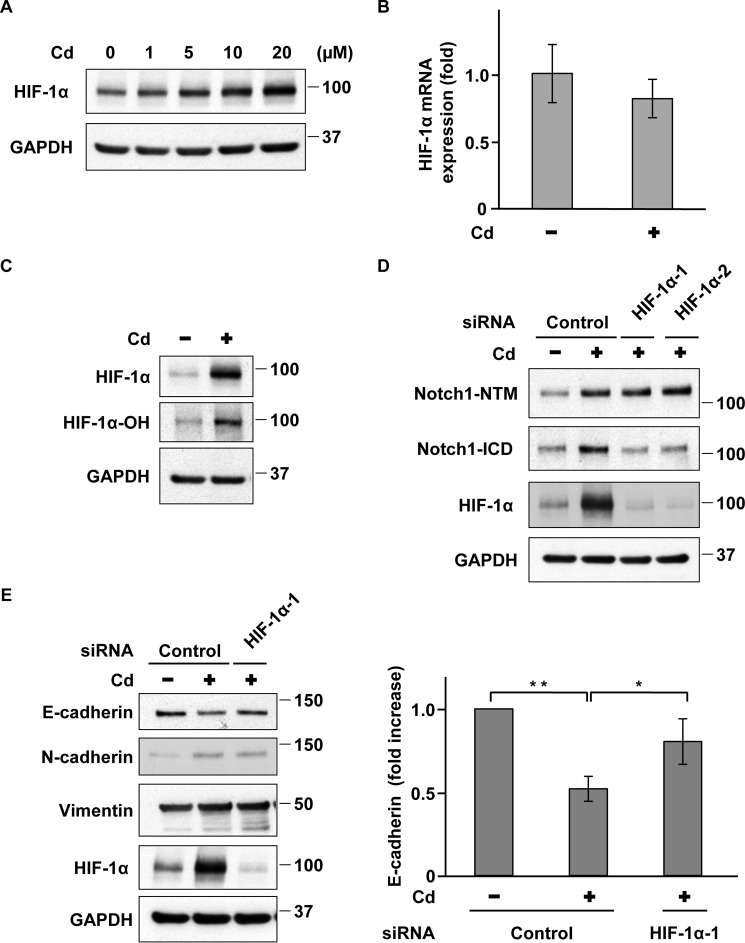
The transcription factor HIF-1α is an important trigger and modulator of EMT and activates Notch1 signaling through a multistep process (28–32). Therefore, we examined whether HIF-1α is involved in Notch1 activation and induces EMT in prolonged CdCl2-exposed A549 cells. HIF-1α protein levels increased in cells exposed to 5–20 μm CdCl2 for 10 weeks (Fig. 4A); however, HIF-1α mRNA levels did not significantly change in prolonged 20 μm CdCl2-exposed cells (Fig. 4B). Although HIF-1α is degraded by the ubiquitin/proteasomal pathway after its hydroxylation at the Pro-564 residue in normoxia (33, 34), prolonged CdCl2 exposure increased the level of HIF-1α protein hydroxylated at Pro-564 (Fig. 4C). The perturbance of proteasomal degradation by an unclear mechanism might induce the stabilization of HIF-1α protein in prolonged CdCl2-exposed A549 cells. HIF-1α knockdown with siRNAs targeted against the human HIF1A gene (HIF-1α-1 and HIF-1α-2) suppressed Notch1-ICD but not Notch1-NTM in prolonged CdCl2-exposed cells (Fig. 4D). Furthermore, HIF-1α depletion suppressed cadmium-induced reduction of E-cadherin expression in prolonged CdCl2-exposed cells, whereas cadmium-induced up-regulation of N-cadherin and vimentin expression was not affected (Fig. 4E). These findings indicate that HIF-1α induces the activation of Notch1 signaling, leading to the down-regulation of E-cadherin expression in prolonged CdCl2-exposed A549 cells.
Figure 4.
HIF-1α regulates Notch1 activity in prolonged CdCl2-exposed A549 cells. A, cells were incubated with 0, 1, 5, 10, or 20 μm CdCl2 (Cd) for 10 weeks. Cell lysates were subjected to Western blotting using the indicated antibodies. B, total RNAs from prolonged 20 μm CdCl2-exposed A549 cells (Cd) were subjected to quantitative RT-PCR to determine HIF-1α mRNA levels. Data were normalized to β-actin expression and reflect the mean ± S.D. (error bars) of three experiments. C, cell lysates from prolonged 20 μm CdCl2-exposed A549 cells (Cd) were subjected to Western blotting using the indicated antibodies. D, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA, HIF-1α siRNA-1, or HIF-1α siRNA-2. Cell lysates were subjected to Western blotting using the indicated antibodies. E, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA or HIF-1α siRNA-1. Cell lysates were subjected to Western blotting using the indicated antibodies. Results of densitometric analysis are also shown. Immunoblots shown are representative of at least three independent experiments. Data reflect the mean ± S.D. of three experiments. *, p < 0.05; **, p < 0.01, significant difference between the samples.
HIF-1α transcriptional activity is not required for the activation of Notch1 signaling in prolonged cadmium-exposed A549 cells
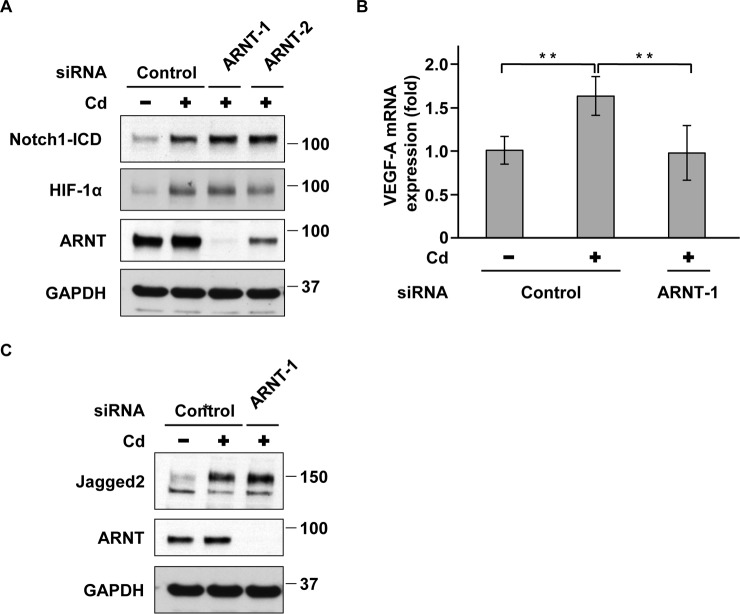
It has been reported that HIF-1α increases the expression of Jagged2 and anterior pharynx-defective 1 (APH-1), a component of the γ-secretase complex, through the binding to their promoters (29, 30), resulting in the activation of Notch1 signaling. To investigate whether the transcriptional activity of HIF-1α is required for Notch1 activation in prolonged CdCl2-exposed cells, we depleted the expression of arylhydrocarbon receptor nuclear translocator (ARNT), an HIF-1α binding partner for DNA binding (28). Transfection with siRNAs targeted against the human ARNT gene (ARNT-1 and ARNT-2) markedly suppressed ARNT expression (Fig. 5A) and almost completely abolished the cadmium-induced increase in mRNA levels of vascular endothelial growth factor A (VEGF-A), an HIF-1α target gene (28) (Fig. 5B). However, there was no significant difference in Notch1-ICD and HIF-1α expression between prolonged CdCl2-exposed cells in the presence and absence of ARNT (Fig. 5A). Although prolonged CdCl2 exposure increased the levels of Jagged2 mRNA (supplemental Fig. S3c) and its protein (Fig. 5C), depletion of ARNT did not change cadmium-induced up-regulation of Jagged2 expression (Fig. 5C). Treatment with 1 nm echinomycin, an HIF-1α transcriptional inhibitor, did not affect the levels of Notch1-ICD and HIF-1α expression in prolonged CdCl2-exposed cells (supplemental Fig. S5), whereas treatment with 0.3 nm echinomycin fully suppressed HIF-1α transcriptional activity (31). Furthermore, there was no significant difference in APH-1a mRNA levels between prolonged CdCl2-exposed cells in the presence and absence of HIF-1α (supplemental Fig. S6). These findings indicate that HIF-1α transcriptional activity is not required for the activation of Notch1 signaling in prolonged cadmium-exposed A549 cells.
Figure 5.
HIF-1α transcriptional activity is not required for activation of Notch1 in prolonged CdCl2-exposed A549 cells. A, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA, ARNT siRNA-1, or ARNT siRNA-2. Cell lysates were subjected to Western blotting using the indicated antibodies. B and C, prolonged 20 μm CdCl2-exposed A549 cells (Cd) were transfected with control siRNA or ARNT siRNA-1. Total RNAs were subjected to quantitative RT-PCR to determine VEGF-A mRNA levels. Data were normalized to β-actin expression and reflect the mean ± S.D. (error bars) of three experiments. *, p < 0.05; **, p < 0.01, significant difference between the samples (B). Cell lysates were subjected to Western blotting using the indicated antibodies (C). Immunoblots shown are representative of at least three independent experiments.
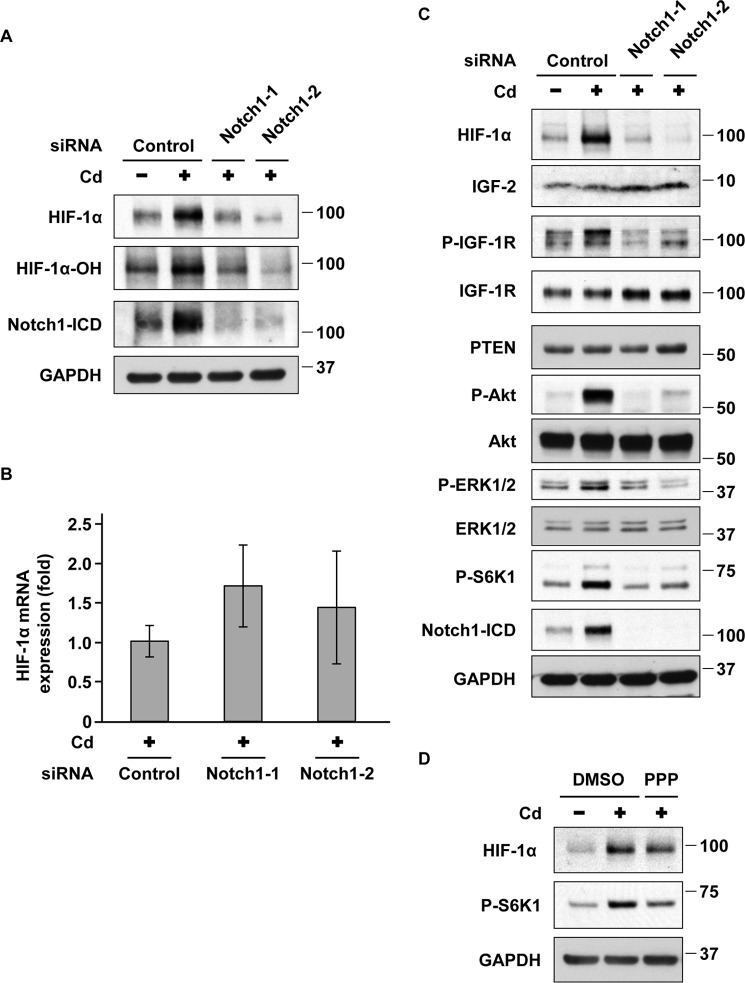
Notch1 regulates HIF-1α expression in prolonged cadmium-exposed A549 cells
Finally, we examined whether Notch1 and HIF-1α form a reciprocal activation loop in prolonged CdCl2-exposed cells. Depletion of Notch1 with siRNAs (Notch1-1 and Notch1-2) suppressed the levels of HIF-1α protein and its hydroxylation at Pro-564 in prolonged CdCl2-exposed cells (Fig. 6A). In contrast, HIF-1α mRNA levels were not changed by the depletion of Notch1 (Fig. 6B), suggesting that Notch1 is involved in the accumulation of HIF-1α protein. Because the translation of HIF-1α is accelerated by the activation of Akt/ERK/S6K1 signaling (35, 36) and Notch1 activates Akt/ERK signaling (30, 37–40), we examined the role of Akt/ERK/S6K1 signaling in the up-regulation of HIF-1α expression in prolonged CdCl2-exposed cells. Depletion of Notch1 with siRNAs (Notch1-1 and Notch1-2) suppressed the phosphorylation of Akt at Thr-308, ERK1/2 at Thr-202 and Tyr-204, and S6K1 at Thr-389 residues in prolonged CdCl2-exposed cells (Fig. 6C), indicating the Notch1-dependent activation of Akt/ERK/S6K1 signaling. It has been reported that Notch1 can activate Akt by increasing the expression of IGF-1R and its ligand IGF-1 (37) or by reducing the expression of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) (40). However, the expression of IGF-1R, IGF-2 (another IGF-1R ligand), and PTEN was not affected by the depletion of Notch1 (Fig. 6C). In contrast, Notch1 silencing suppressed the phosphorylation of IGF-1R at Tyr-1131, which is necessary for IGF-1R activation (41) (Fig. 6C). Treatment with picropodophyllin (PPP), an IGF-1R kinase inhibitor, partially suppressed HIF-1α expression and phosphorylation of S6K1 in prolonged CdCl2-exposed cells (Fig. 6D). We also found that treatment with the Akt inhibitor MK2206, the ERK inhibitor U0126, or the mammalian target of rapamycin (mTOR) inhibitor rapamycin suppressed S6K1 phosphorylation and HIF-1α expression (supplemental Fig. S7). These findings indicate that Notch1 induces HIF-1α expression mainly through the activation of the IGF-1R/Akt/ERK/S6K1 signaling cascade, probably leading to the acceleration of HIF-1α translation, in prolonged CdCl2-exposed A549 cells.
Figure 6.
Notch1 regulates HIF-1α expression in prolonged CdCl2-exposed A549 cells. A, B, and C, prolonged 20 μm CdCl2-exposed A549 cells were transfected with control siRNA, Notch1 siRNA-1, or Notch1 siRNA-2. Cell lysates were subjected to Western blotting using the indicated antibodies (A and C). Total RNAs were subjected to quantitative RT-PCR to determine HIF-1α mRNA levels. Data were normalized to β-actin expression and reflect the mean ± S.D. (error bars) of three experiments (B). D, prolonged 20 μm CdCl2-exposed A549 cells were treated with 0.1% DMSO or 200 nm PPP for 16 h. Cell lysates were subjected to Western blotting using the indicated antibodies. Immunoblots shown are representative of at least three independent experiments. P-, phospho-.
Notch1 is involved in prolonged cadmium exposure-induced cellular responses in BEAS-2B and H1975 cells
We examined whether Notch1 signaling regulates prolonged cadmium-induced malignant progression in lung-related cancer cells other than A549 cells, including BEAS-2B human bronchial epithelial cells and H1975 human tumor-derived non-small-cell lung cancer cells. Consistent with the previous report (9), we found that prolonged CdCl2-exposed BEAS-2B cells showed cadmium resistance depending on Nrf2 expression (supplemental Fig. S8, a–c). In addition, Notch1 knockdown partially suppressed cadmium-induced alterations of E-cadherin and Snail levels and stress fiber formation in these resistant BEAS-2B cells (Fig. 7, A and B). Furthermore, we found that prolonged CdCl2-exposed H1975 cells obtained the cisplatin resistance via activation of Notch1 signaling (Fig. 7, C and D).
Figure 7.
Notch1 is involved in prolonged cadmium exposure-induced cellular responses in BEAS-2B and H1975 cells. A and B, prolonged 10 μm CdCl2-exposed BEAS-2B cells (Cd) were transfected with control siRNA, Notch1 siRNA-1, or Notch1 siRNA-2. These cell lysates were subjected to Western blotting using the indicated antibodies (a). These cells were stained with phalloidin-FITC for F-actin and DAPI for DNA (B). Scale bar, 20 μm. c and d, prolonged 30 μm CdCl2-exposed H1975 cells (Cd) were transfected with control siRNA, Notch1 siRNA-1, or Notch1 siRNA-2. These cell lysates were subjected to Western blotting using the indicated antibodies (C). These cells were treated with 0.2% N,N-dimethylformamide (DMF) or 20 μg/ml cisplatin for 34 h. The viability of cells was determined by trypan blue exclusion assay. Each value is the percentage of trypan blue-positive cells and reflects the mean ± S.D. (error bars) of three experiments with duplicate assays in each experiment (D). **, p < 0.01, significant difference between the samples.
Discussion
A549 cells are a human lung adenocarcinoma cell line with properties of type II alveolar epithelial cells (42). We found that exposure to CdCl2 for more than 8 weeks enhanced the proliferative ability of A549 cells. The transcription factor Nrf2 has been reported to be one of the key factors that induce a high proliferative ability in cadmium-transformed BEAS-2B cells (9), and its downstream target, heme oxygenase-1, is involved in the suppression of cadmium toxicity in kidney and pulmonary cells (43, 44). Consistent with these findings, knockdown of Nrf2 (supplemental Fig. S9, a–c) and heme oxygenase-1 (supplemental Fig. S9, d–f) reduced the proliferative potential in prolonged CdCl2-exposed A549 cells. To clarify the regulatory mechanisms of cadmium-induced malignant progression, we used A549 cells that were incubated with 20 μm CdCl2 for 9–15 weeks as the in vitro model of prolonged cadmium-exposed lung epithelial cells.
We found that prolonged CdCl2 exposure induced EMT, stress fiber formation, high cell motility, and antitumor drug resistance in A549 cells. In concordance with the findings that chronic cadmium exposure induces EMT-like characteristics in HPL-1D human peripheral lung epithelial cells (45), cadmium-induced malignant progression was also clearly observed in our model using A549 cells. Furthermore, consistent with our previous findings in HK-2 human renal proximal epithelial cells treated with CdCl2 (22), an increase in the levels of Notch1-ICD and its downstream targets, Snail and Slug, was found in prolonged CdCl2-exposed A549 cells. Notch1 knockdown partially suppressed prolonged CdCl2-induced EMT, stress fiber formation, high cell motility, and antitumor drug resistance. In addition, we also found that prolonged CdCl2 exposure induced reduction of E-cadherin in BEAS-2B cells and antitumor drug resistance in H1975 cells depending on Notch1 signaling. These findings demonstrate for the first time, to our knowledge, that Notch1 signaling is involved in cadmium-induced malignant progression in lung cancer cells, including A549 cells. Because these findings remained in the prolonged CdCl2-exposed A549 cells after removal of CdCl2 from culture medium for 10 weeks, cadmium-induced malignant progression via the Notch1 pathway may be maintained (supplemental Fig. S10).
It has been reported that Notch1 is one of the important components that induce EMT under hypoxia (16), and it is also correlated with antitumor drug resistance in T-cell acute lymphoblastic leukemia cells (16), breast cancer cells (17), and A549 lung cancer cells (18). However, knockdown of Notch1 failed to fully suppress up-regulation of N-cadherin and vimentin in prolonged CdCl2-exposed cells. It has been reported that TGF-β signaling induces EMT (23), and its expression increases in response to cadmium exposure (45, 46). When expression of both TGF-β1 and TGF-β2 was reduced, cadmium-induced up-regulation of N-cadherin and vimentin was found to be only partially suppressed (supplemental Fig. S11). These findings suggest that other pathways might also function in parallel with Notch1 and TGF-β signaling for the induction of EMT in prolonged CdCl2-exposed A549 cells. We found that knockdown of Notch1 did not suppress cadmium-induced up-regulation of Nrf2 expression. Conversely, Nrf2 knockdown suppressed the reduction of E-cadherin and elevation of Snail levels without affecting the Notch1-ICD level in prolonged CdCl2-exposed A549 cells (supplemental Fig. S12). These findings suggest that Nrf2 may be another signaling pathway that regulates E-cadherin expression in response to the prolonged cadmium exposure.
HIF-1α has been reported to activate Notch1 signaling in either a transcriptional activity-dependent or -independent manner (28–32). In response to prolonged CdCl2 exposure, HIF-1α protein but not its transcripts was accumulated in A549 cells, likely due to a perturbance of proteasomal degradation of HIF-1α. Silencing of HIF-1α expression suppressed the cadmium-induced increase in Notch1-ICD and reduction of E-cadherin expression. In contrast, inhibition of HIF-1α transcriptional activity with ARNT knockdown or echinomycin treatment failed to suppress Notch1 activation. Although expression of Jagged2, APH-1, and Notch1 has been reported to be up-regulated by HIF-1α (28–30), Jagged2 expression was not affected by the depletion of ARNT. Furthermore, we found that expression of APH-1 and Notch1-NTM was not affected by the depletion of HIF-1α. These findings suggest that HIF-1α induces the activation of Notch1 signaling without depending on its transcriptional activity in prolonged CdCl2-exposed A549 cells. In contrast, it has been reported that a transcriptionally inactive mutant of HIF-1α can stabilize Notch1-ICD in COS-7 cells (32). HIF-1α can also activate Notch1 signaling through a direct interaction with the γ-secretase complex (31). Further studies are needed to examine these transcriptional activity-independent mechanisms in A549 cells exposed to cadmium.
Conversely, the experiments using Notch1 siRNAs showed that Notch1 signaling induces the accumulation of HIF-1α protein in prolonged CdCl2-exposed A549 cells. In addition, activation of the Akt/ERK/S6K1 pathway, which can accelerate the translation of HIF-1α (35, 36), was found to be regulated by Notch1 signaling as has been reported previously (30, 37–40). Notch1 activation increases the expression of IGF-1R and IGF-1 and results in the activation of the downstream Akt pathway in lung adenocarcinoma cells during hypoxia (37). However, depletion of Notch1 had little to no effect on the expression of IGF-1R, IGF-2, and PTEN but did suppress the phosphorylation of IGF-1R and Akt in prolonged CdCl2-exposed A549 cells. The IGF-1R kinase inhibitor PPP at a concentration (200 nm) without detectable cytotoxicity partially suppressed the expression of HIF-1α and the phosphorylation of S6K1. The Akt/ERK pathway has also been reported to induce HIF-1α expression in human bronchial epithelial cells exposed to cadmium (8). We also confirmed that the ERK/Akt/S6K1 pathway was involved in HIF-1α accumulation in prolonged CdCl2-exposed A549 cells. These findings suggest that the activation of the IGF-1R/Akt/ERK/S6K1 pathway is involved in the accumulation of HIF-1α in prolonged CdCl2-exposed A549 cells. Oxidative stressors such as cigarette smoke extract, H2O2, and ozone induce a ligand-independent aberrant activation of a receptor-type tyrosine kinase (47–49). Although the possible involvement of IGF-1 cannot be excluded, a Notch1-mediated mechanism independent of ligand binding remains to be clarified. In addition, cadmium exposure-induced activation of other receptor-type tyrosine kinases, including the epidermal growth factor receptor (22, 50), might contribute to the subsequent activation of the Akt/ERK pathway.
In summary, the present study shows that prolonged CdCl2 exposure activates Notch1 signaling and induces malignant progression in A549 lung adenocarcinoma cells. In addition, the HIF-1α and IGF-1R/Akt/ERK/S6K1 signaling pathways reciprocally activate Notch1 signaling to induce EMT in prolonged CdCl2-exposed A549 cells. Further experiments on Notch1 signaling after exposure to lower concentrations of cadmium, including animal models, will provide clues to understanding the malignant progression of lung cancer caused by cadmium exposure or cadmium from cigarette smoking.
Experimental procedures
Chemicals
CdCl2 was obtained from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). PPP was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). DAPT, cisplatin, etoposide, and gemcitabine hydrochloride were obtained from Wako Pure Chemical Industries, Ltd. Antibodies against phospho-IGF-1R (Tyr-1131), total IGF-1Rβ (D23H3) XP®, phospho-Akt (Thr-308) (C31E5E), total Akt (pan) (C67E7), phospho-p70 S6 kinase (Thr-389) (108D2), cleaved Notch1 (Val-1744) (D3B8), Notch1 (D1E11) XP, Jagged2 (C23D2), Snail (C15D3), Slug (C19G7), E-cadherin (24E10), PTEN (D4.3) XP, ARNT (D28F3) XP, phospho-p44/42 (ERK1/2), p44/42 MAPK (ERK1/2), and hydroxy-HIF-1α (Pro-564) (D43B5) XP were obtained from Cell Signaling Technology, Inc. (Beverly, MA). IGF-2 antibody was obtained from Abcam plc (Cambridge, England). N-cadherin, vimentin, HIF-1α, and GAPDH (GT239) antibodies were obtained from Genetex Inc. (Irvine, CA). The siRNAs targeted against the human Notch1 (siRNA-1, Hs_NOTCH1_3 FlexiTube siRNA, SI00119028; siRNA-2, Hs_NOTCH1_4 FlexiTube siRNA, SI00119035), SNAI1 (siRNA-1, Hs_SNAI1_1 FlexiTube siRNA, SI00083398; siRNA-2, Hs_SNAI1_5 FlexiTube siRNA, SI02636424), HIF1A (siRNA-1, Hs_HIF1A_5 FlexiTube siRNA, SI02664053; siRNA-2, Hs_ HIF1A_10 FlexiTube siRNA, SI04249308), and ARNT (siRNA-1, Hs_ARNT_5 FlexiTube siRNA, SI03020913; siRNA-2, Hs_ ARNT_2 FlexiTube siRNA, SI00304220) and non-target siRNA (AllStars Negative Control siRNA) were purchased from Qiagen (Hilden, Germany). See supplemental “Materials and Methods” for more information.
Cell culture and treatments
A549 cells were obtained from the Health Science Research Resources Bank (Japan Health Sciences Foundation, Osaka, Japan) and grown in Earle's minimum essential medium (MEM) with non-essential amino acids supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco, Invitrogen) in a humidified atmosphere of 5% CO2, 95% air at 37 °C. BEAS-2B cells were obtained from DS Pharma Biomedical Corp. (Osaka, Japan) and grown in Dulbecco's modified Eagle's medium (DMEM) with non-essential amino acids supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2, 95% air at 37 °C. H1975 cells were kindly provided from Dr. Kenji Tanabe and grown in RPMI 1640 medium, HEPES with non-essential amino acids supplemented with 1 mm sodium pyruvate, 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2, 95% air at 37 °C. A549 cells were exposed continuously to CdCl2 (0, 1, 5, 10, and 20 μm) for 1–15 weeks. For each experiment, exponentially growing A549 cells were seeded at 3 × 105 or 5 × 105 cells/well in 6-well culture plates and cultured for 1 day before the experiments. DAPT and PPP were dissolved in dimethyl sulfoxide (DMSO). Cells were incubated with DMSO (0.1%) or each compound for 16 h. BEAS-2B and H1975 cells were exposed continuously to 10 and 30 μm CdCl2 for 10 weeks, respectively. For each experiment, exponentially growing cells were seeded at 3 × 105 (for BEAS-2B cells) or 4 × 105 cells/well in 6-well culture plates (for H1975 cells).
Preparation of whole cell lysates
After incubation, cells were washed with phosphate-buffered saline and lysed with sodium dodecyl sulfate-polyacrylamide gel Laemmli sample buffer. Cell lysates were collected, sonicated, and boiled for 5 min. Protein concentrations were determined using the RC DC Protein Assay (Bio-Rad).
Western blotting
Equal amounts of protein (20 μg) were subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane (Hybond-ECL, Amersham Biosciences). The membrane was blocked with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 for 1 h at room temperature. The membrane was then incubated overnight at 4 °C with the primary antibody, and protein was detected with a Phototope-HRP Western blot detection kit (Cell Signaling Technology, Inc.). The bands on the developed film were quantified with ImageJ 1.42 (National Institutes of Health, Bethesda, MD). The density of each band was normalized to that of GAPDH.
RNA isolation and reverse transcription-PCR
Total RNA was isolated with RNeasy (Qiagen), and first-strand cDNA was synthesized using the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). Quantitative reverse transcription-PCR (RT-PCR) analysis was performed using the THUNDERBIRD® SYBR® qPCR Mix (Toyobo) in the StepOneTM Real-Time PCR system (Life Technologies Japan Ltd.). The expression level was normalized to that of glyceraldehyde-3-phosphate dehydrogenase or β-actin. PCR primers are listed in supplemental Table S1.
Proliferation assay
4 × 104 cells were seeded on 24-well culture plates for 5 days. Each day, the total cell number was counted using a TC10TM automated cell counter (Bio-Rad).
Fluorescence microscopy
For staining with phalloidin, cells were grown on chamber slides, then fixed in 4% paraformaldehyde at room temperature, permeabilized in 0.5% Triton X-100, and incubated with phalloidin-FITC at 1:1000. Fluorescence was examined by a confocal laser-scanning microscope (Carl Zeiss, Oberkochen, Germany).
Gene knockdown of Notch1, SNAI1, HIF1A, ARNT, NFE2L2, HMOX1, TGFB1, and TGFB2 by siRNA
Transfection of siRNAs against human Notch1, SNAI1, HIF1A, ARNT, NFE2L2, HMOX1, TGFB1, and TGFB2 and non-target siRNA into cells was performed using Lipofectamine® 2000 (Invitrogen) (for A549 and H1975 cells) or Lipofectamine RNAiMAX (Invitrogen) (for BEAS-2B cells) according to the manufacturer's instructions with some adjustments. The siRNAs were dissolved in nuclease-free water and diluted to 0.2 μm with 250 μl of Opti-MEM (Invitrogen). 5 μl of Lipofectamine 2000 was also diluted 50-fold with Opti-MEM. Equal volumes of these two solutions were mixed (500 μl total) and immediately added to 2 ml of culture medium at the time of cell plating. After incubation for 24 h, cells were washed with medium and used for experiments.
Wound-healing assay
Cells were grown in MEM under appropriate conditions until a monolayer was formed. A wound was generated by creating a scratch using a sterile 1-ml pipette tip. Images were acquired immediately after the scratch at 0 h and again after 20- or 24-h incubation at 37 °C.
Trypan blue exclusion assay
Culture medium was aspirated and reserved. After trypsinization, cells were suspended in MEM, and the culture medium was returned. The mixture was centrifuged to pellet the cells. Cellular suspension and 0.4% trypan blue in Hanks' balanced salt solution were mixed, and the number of viable cells was counted using a TC10 automated cell counter.
Statistical analysis
Results are expressed as the mean ± S.D. Statistical significance was determined by Student's t test. A value of p < 0.05 was considered to be statistically significant.
Author contributions
K. F. and M. M. designed the studies. K. F., H. I., and T. M. performed the experiments and analyzed the data. K. F. and M. M. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Ryosuke Fujiki, Yuta Komoike, Kenji Tanabe, Kouki Ishikawa, Koji Morimoto, Motoyuki Itoh, Tomoki Yano, Maho Hamasaki, Hiroshi Kimura, Seishiro Hirano, and Taisuke Tomita for helpful discussions.
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grants15K18910 and 26460175 and a grant-in-aid for medicine from the Takeda Science Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Materials and Methods, Figs. S1–S12, and Table S1.
- EMT
- epithelial-mesenchymal transition
- APH-1
- anterior pharynx-defective 1
- ARNT
- arylhydrocarbon receptor nuclear translocator
- DAPT
- N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester
- HIF-1α
- hypoxia-inducible factor 1α
- IGF
- insulin-like growth factor
- IGF-1R
- insulin-like growth factor 1 receptor
- PTEN
- phosphatase and tensin homologue deleted on chromosome 10
- Notch-ICD
- Notch intracellular domain
- S6K1
- p70 S6 kinase 1
- Notch1-NTM
- Notch1 transmembrane subunit
- MEM
- minimum essential medium
- PPP
- picropodophyllin.
References
- 1. International Agency for Research on Cancer (IARC) (2012) A review of human carcinogens: arsenic, metals, fibres, and dusts, in IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 100C, pp 121–141, IARC, Lyon, France: [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization (WHO) (2008) World Health Organization guidelines for drinking water quality: Incorporating 1st and 2nd Addenda, Vol. 1, Recommendations, 3rd Ed., pp. 317–319,World Health Organization, Geneva [Google Scholar]
- 3. Järup L., Berglund M., Elinder C. G., Nordberg G., and Vahter M. (1998) Health effects of cadmium exposure: a review of the literature and a risk estimate. Scand. J. Work Environ. Health 24, Suppl. 1, 1–51 [PubMed] [Google Scholar]
- 4. Huff J., Lunn R. M., Waalkes M. P., Tomatis L., and Infante P. F. (2007) Cadmium-induced cancers in animals and in humans. Int. J. Occup. Environ. Health 13, 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. International Agency for Research on Cancer (IARC) (1993) Cadmium and cadmium compounds, in IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 58, pp. 119–237, IARC, Lyon, France: [PMC free article] [PubMed] [Google Scholar]
- 6. National Toxicology Program (NTP) (2014) 13th Report on Carcinogens, Cadmium and Cadmium Compounds, pp. 1–4, Department of Health and Human Services, Research Triangle Park, NC [Google Scholar]
- 7. Beveridge R., Pintos J., Parent M. E., Asselin J., and Siemiatycki J. (2010) Lung cancer risk associated with occupational exposure to nickel, chromium VI, and cadmium in two population-based case-control studies in Montreal. Am. J. Ind. Med. 53, 476–485 [DOI] [PubMed] [Google Scholar]
- 8. Jing Y., Liu L. Z., Jiang Y., Zhu Y., Guo N. L., Barnett J., Rojanasakul Y., Agani F., and Jiang B. H. (2012) Cadmium increases HIF-1 and VEGF expression through ROS, ERK, and AKT signaling pathways and induces malignant transformation of human bronchial epithelial cells. Toxicol. Sci. 125, 10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Son Y. O., Pratheeshkumar P., Roy R. V., Hitron J. A., Wang L., Zhang Z., and Shi X. (2014) Nrf2/p62 signaling in apoptosis resistance and its role in cadmium-induced carcinogenesis. J. Biol. Chem. 289, 28660–28675 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Iwatsuki M., Mimori K., Yokobori T., Ishi H., Beppu T., Nakamori S., Baba H., and Mori M. (2010) Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 101, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wan L., Pantel K., and Kang Y. (2013) Tumor metastasis: moving new biological insights into the clinic. Nat. Med. 19, 1450–1464 [DOI] [PubMed] [Google Scholar]
- 12. Singh A., and Settleman J. (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin L., Velazquez O. C., and Liu Z.-J. (2010) Notch signaling: emerging molecular targets for cancer therapy. Biochem. Pharmacol. 80, 690–701 [DOI] [PubMed] [Google Scholar]
- 14. Capaccione K. M., and Pine S. R. (2013) The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis 34, 1420–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwanbeck R., Martini S., Bernoth K., and Just U. (2011) The Notch signaling pathway: molecular basis of cell context dependency. Eur. J. Cell Biol. 90, 572–581 [DOI] [PubMed] [Google Scholar]
- 16. Yuan X., Wu H., Han N., Xu H., Chu Q., Yu S., Chen Y., and Wu K. (2014) Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J. Hematol. Oncol. 7, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martz C. A., Ottina K. A., Singleton K. R., Jasper J. S., Wardell S. E., Peraza-Penton A., Anderson G. R., Winter P. S., Wang T., Alley H. M., Kwong L. N., Cooper Z. A., Tetzlaff M., Chen P. L., Rathmell J. C., et al. (2014) Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci. Signal. 7, ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J., Mao Z., Huang J., Xie S., Liu T., and Mao Z. (2014) Blocking the NOTCH pathway can inhibit the growth of CD133-positive A549 cells and sensitize to chemotherapy. Biochem. Biophys. Res. Commun. 444, 670–675 [DOI] [PubMed] [Google Scholar]
- 19. Wael H., Yoshida R., Kudoh S., Hasegawa K., Niimori-Kita K., and Ito T. (2014) Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer 85, 131–140 [DOI] [PubMed] [Google Scholar]
- 20. Ding X., Zhu F., Li T., Zhou Q., Hou F. F., and Nie J. (2011) Numb protects renal proximal tubular cells from puromycin aminonucleoside-induced apoptosis through inhibiting Notch signaling pathway. Int. J. Biol. Sci. 7, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., De Marco M. A., Graziani I., Gazdar A. F., Strack P. R., Miele L., and Bocchetta M. (2007) Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Res. 67, 7954–7959 [DOI] [PubMed] [Google Scholar]
- 22. Fujiki K., Inamura H., and Matsuoka M. (2014) Detrimental effects of Notch1 signaling activated by cadmium in renal proximal tubular epithelial cells. Cell Death Dis. 5, e1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu J., Lamouille S., and Derynck R. (2009) TGF-β-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bauvois B. (2012) New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim. Biophys. Acta 1825, 29–36 [DOI] [PubMed] [Google Scholar]
- 25. Konishi J., Kawaguchi K. S., Vo H., Haruki N., Gonzalez A., Carbone D. P., and Dang T. P. (2007) γ-Secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 67, 8051–8057 [DOI] [PubMed] [Google Scholar]
- 26. Saad S., Stanners S. R., Yong R., Tang O., and Pollock C. A. (2010) Notch mediated epithelial to mesenchymal transformation is associated with increased expression of the Snail transcription factor. Int. J. Biochem. Cell Biol. 42, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 27. Shao S., Zhao X., Zhang X., Luo M., Zuo X., Huang S., Wang Y., Gu S., and Zhao X. (2015) Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol. Cancer 14, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haase V. H. (2009) Oxygen regulates epithelial-to-mesenchymal transition: insights into molecular mechanisms and relevance to disease. Kidney Int. 76, 492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang R., Zhang Y. W., Zhang X., Liu R., Zhang X., Hong S., Xia K., Xia J., Zhang Z., and Xu H. (2006) Transcriptional regulation of APH-1A and increased γ-secretase cleavage of APP and Notch by HIF-1 and hypoxia. FASEB J. 20, 1275–1277 [DOI] [PubMed] [Google Scholar]
- 30. Díaz B., Yuen A., Iizuka S., Higashiyama S., and Courtneidge S. A. (2013) Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J. Cell Biol. 201, 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Villa J. C., Chiu D., Brandes A. H., Escorcia F. E., Villa C. H., Maguire W. F., Hu C. J., de Stanchina E., Simon M. C., Sisodia S. S., Scheinberg D. A., and Li Y. M. (2014) Nontranscriptional role of Hif-1α in activation of γ-secretase and notch signaling in breast cancer. Cell Rep. 8, 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U., and Bondesson M. (2005) Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617–628 [DOI] [PubMed] [Google Scholar]
- 33. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., and Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 34. Tian Y. M., Yeoh K. K., Lee M. K., Eriksson T., Kessler B. M., Kramer H. B., Edelmann M. J., Willam C., Pugh C. W., Schofield C. J., and Ratcliffe P. J. (2011) Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J. Biol. Chem. 286, 13041–13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Semenza G. L. (2003) Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 36. Zhou J., Callapina M., Goodall G. J., and Brüne B. (2004) Functional integrity of nuclear factor κB, phosphatidylinositol 3′-kinase, and mitogen-activated protein kinase signaling allows tumor necrosis factor tumor necrosis factor α-evoked Bcl-2 expression to provoke internal ribosome entry site-dependent translation of hypoxia-inducible factor 1α. Cancer Res. 64, 9041–9048 [DOI] [PubMed] [Google Scholar]
- 37. Eliasz S., Liang S., Chen Y., De Marco M. A., Machek O., Skucha S., Miele L., and Bocchetta M. (2010) Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene 29, 2488–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sriuranpong V., Borges M. W., Ravi R. K., Arnold D. R., Nelkin B. D., Baylin S. B., and Ball D. W. (2001) Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 61, 3200–3205 [PubMed] [Google Scholar]
- 39. Kim T. H., Woo J. S., Kim Y. K., and Kim K. H. (2014) Silibinin induces cell death through reactive oxygen species-dependent downregulation of notch-1/ERK/Akt signaling in human breast cancer cells. J. Pharmacol. Exp. Ther. 349, 268–278 [DOI] [PubMed] [Google Scholar]
- 40. Hales E. C., Taub J. W., and Matherly L. H. (2014) New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell. Signal. 26, 149–161 [DOI] [PubMed] [Google Scholar]
- 41. Hernández-Sánchez C., Blakesley V., Kalebic T., Helman L., and LeRoith D. (1995) The role of the tyrosine kinase domain of the insulin-like growth factor-I receptor in intracellular signaling, cellular proliferation, and tumorigenesis. J. Biol. Chem. 270, 29176–29181 [DOI] [PubMed] [Google Scholar]
- 42. Lieber M., Smith B., Szakal A., Nelson-Rees W., and Todaro G. (1976) A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int. J. Cancer 17, 62–70 [DOI] [PubMed] [Google Scholar]
- 43. Satarug S., Nishijo M., Lasker J. M., Edwards R. J., and Moore M. R. (2006) Kidney dysfunction and hypertension: role for cadmium, p450 and heme oxygenases? Tohoku J. Exp. Med. 208, 179–202 [DOI] [PubMed] [Google Scholar]
- 44. Surolia R., Karki S., Kim H., Yu Z., Kulkarni T., Mirov S. B., Carter A. B., Rowe S. M., Matalon S., Thannickal V. J., Agarwal A., and Antony V. B. (2015) Heme oxygenase-1-mediated autophagy protects against pulmonary endothelial cell death and development of emphysema in cadmium-treated mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L280–L292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Person R. J., Tokar E. J., Xu Y., Orihuela R., Ngalame N. N., and Waalkes M. P. (2013) Chronic cadmium exposure in vitro induces cancer cell characteristics in human lung cells. Toxicol. Appl. Pharmacol. 273, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang B., Li Y., Tan Y., Miao X., Liu X. D., Shao C., Yang X. H., Turdi S., Ma L. J., Ren J., and Cai L. (2012) Low-dose Cd induces hepatic gene hypermethylation, along with the persistent reduction of cell death and increase of cell proliferation in rats and mice. PLoS One 7, e33853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khan E. M., Lanir R., Danielson A. R., and Goldkorn T. (2008) Epidermal growth factor receptor exposed to cigarette smoke is aberrantly activated and undergoes perinuclear trafficking. FASEB J. 22, 910–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Filosto S., Khan E. M., Tognon E., Becker C., Ashfaq M., Ravid T., and Goldkorn T. (2011) EGF receptor exposed to oxidative stress acquires abnormal phosphorylation and aberrant activated conformation that impairs canonical dimerization. PLoS One 6, e23240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu W., Wages P. A., Devlin R. B., Diaz-Sanchez D., Peden D. B., and Samet J. M. (2015) Src-mediated EGF receptor activation regulates ozone-induced interleukin 8 expression in human bronchial epithelial cells. Environ. Health Perspect. 123, 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gao X., Yu L., Moore A. B., Kissling G. E., Waalkes M. P., and Dixon D. (2015) Cadmium and proliferation in human uterine leiomyoma cells: evidence of a role for EGFR/MAPK pathways but not classical estrogen receptor pathways. Environ. Health Perspect. 123, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.