Abstract
The biosynthesis of many eukaryotic proteins requires accurate targeting to and translocation across the endoplasmic reticulum membrane. Post-translational protein translocation in yeast requires both the Sec61 translocation channel, and a complex of four additional proteins: Sec63, Sec62, Sec71, and Sec72. The structure and function of these proteins are largely unknown. This pathway also requires the cytosolic Hsp70 protein Ssa1, but whether Ssa1 associates with the translocation machinery to target protein substrates to the membrane is unclear. Here, we use a combined structural and biochemical approach to explore the role of Sec71-Sec72 subcomplex in post-translational protein translocation. To this end, we report a crystal structure of the Sec71-Sec72 complex, which revealed that Sec72 contains a tetratricopeptide repeat (TPR) domain that is anchored to the endoplasmic reticulum membrane by Sec71. We also determined the crystal structure of this TPR domain with a C-terminal peptide derived from Ssa1, which suggests how Sec72 interacts with full-length Ssa1. Surprisingly, Ssb1, a cytoplasmic Hsp70 that binds ribosome-associated nascent polypeptide chains, also binds to the TPR domain of Sec72, even though it lacks the TPR-binding C-terminal residues of Ssa1. We demonstrate that Ssb1 binds through its ATPase domain to the TPR domain, an interaction that leads to inhibition of nucleotide exchange. Taken together, our results suggest that translocation substrates can be recruited to the Sec71-Sec72 complex either post-translationally through Ssa1 or co-translationally through Ssb1.
Keywords: 70 kilodalton heat shock protein (Hsp70), endoplasmic reticulum (ER), protein structure, protein translocation, structure-function, Sec71, Sec72, Ssa1, Ssb1, TPR
Introduction
A decisive step in the biosynthesis of many eukaryotic proteins is their accurate targeting to and translocation across the endoplasmic reticulum (ER)3 membrane. In yeast, proteins enter the ER either co- or post-translationally (for review, see Refs. 1, 2). Co-translational translocation is used by secretory proteins with hydrophobic signal sequences and by membrane proteins (3). In co-translational translocation, the translating ribosome is first targeted to the ER membrane by the signal recognition particle (SRP) and its receptor (SR). Subsequently, the ribosome binds to the translocation channel, the Sec61 channel, and the growing polypeptide chain is transferred from the ribosome tunnel into the channel. In the absence of SRP or SR, yeast cells remain viable and the translocation of certain proteins, such as prepro-carboxypeptidase Y (ppCPY) and prepro-α factor (ppαF), is not affected (4). These proteins are translocated only after completion of their synthesis (5–7). Taken together, these observations suggested an SRP-independent ER targeting pathway.
The SRP-independent, post-translational translocation pathway is used by secretory proteins with less hydrophobic signal sequences including many glycosylphosphatidylinositol-anchored membrane proteins (3, 8). Work in Saccharomyces cerevisiae has shown that post-translational translocation is mediated by the Sec complex, an assembly of the Sec61 channel with a tetrameric protein complex consisting of Sec62, Sec63, Sec71 (Sec66), and Sec72 (9, 10). Sec72 is a peripheral membrane protein, whereas Sec71 is a single-spanning membrane protein with a C-terminal cytosolic domain (11, 12). Sec62 and Sec63 have two and three transmembrane regions, respectively, along with cytosolic domains (13, 14). In support of their role in post-translational translocation, mutations or deletions in any one of the genes coding for components of the tetrameric complex results in translocation defects in yeast (3, 11, 12, 15).
The tetrameric complex is required for the initial binding of post-translational substrates (10), as well as for their subsequent translocation across the ER membrane (10). How the individual components of the tetrameric complex participate in substrate recognition and regulate translocation through the Sec61 channel is poorly understood. The only established function of the complex is a role for the J-domain of Sec63; it activates the ATPase activity of Kar2 (BiP), a member of the Hsp70 family of ATPases in the ER lumen (16). Following ATP hydrolysis, Kar2 binds to the translocating polypeptide chain and prevents the backsliding of the chain into the cytosol (17).
Early work has shown that the Ssa proteins, cytoplasmic Hsp70 proteins, are also required for post-translational translocation (18–20). Cross-linking experiments demonstrated that Hsp70 and other cytoplasmic chaperones associate with a translocation substrate as soon as it is released from the ribosome (21). All these proteins are lost when the substrate engages the Sec complex and inserts into the translocon. Photo-crosslinking experiments have shown that the substrate is then in contact with Sec62, Sec71, and Sec72 (21). It is not known whether these proteins can serve as the targeting site for Ssa1. Whether cytoplasmic Hsp70 itself acts as a targeting factor analogous to SRP is also uncertain. It could simply act as a chaperone, keeping substrates in an unfolded or loosely folded conformation, as has been demonstrated previously (18–20), whereas targeting would occur exclusively by an interaction of the signal sequence with the Sec61 channel. Alternatively, Hsp70 could have a specific interaction with the Sec complex components and thus facilitate targeting.
Previous data suggest that the targeting of post-translational substrate to the ER membrane can also occur co-translationally, that is, before completion of polypeptide synthesis (5–7, 22). In addition, using proximity-specific ribosome profiling, it was demonstrated that Sec71 deletion affects the co-translational targeting of certain substrates to the ER membrane (22), even though there is no known mechanism for coordinating translation with recruitment of the translating ribosome to the Sec complex. One possibility is that ribosome-nascent chain complexes bind to the Sec complex through a direct interaction of the ribosome with Sec61. However, this seems unlikely, because the Sec complex, in contrast to the Sec61 channel in isolation, does not interact with ribosomes (10). Alternatively, it is possible that the targeting of ribosome-nascent chain complexes is mediated by another factor. A possible candidate is Ssb1, a cytoplasmic Hsp70 that associates with ribosomes (23) (for review, see Ref. 24) and interacts with nascent chains. Binding of nascent chains to Ssb1 and the ER targeting factor SRP appears to be mutually exclusive (25), raising the possibility that Ssb1, like the Sec complex, participates in the SRP-independent pathway and can serve as an early sorting factor for ribosome-bound nascent chains. However, it is unknown whether Ssb1 has a specific affinity for any of the components of the translocation machinery.
It is likely that structural characterization of the Sec complex or its subcomplexes will be important for unraveling the molecular mechanisms by which these proteins mediate post-translational translocation. However, with the exception of a 21-Å low-resolution cryo-electron microscopy structure of the Sec complex (26), no structural information is currently available.
Here, we report the first structure of a constituent of the Sec complex, the Sec71-Sec72 subcomplex. The structure shows that Sec72 contains a TPR domain that is tethered to the ER membrane by Sec71. We also determined the crystal structure of the TPR domain bound to the C terminus of Ssa1, which suggests how full-length Ssa1 interacts with Sec72. Furthermore, the structure revealed a previously unappreciated variability in the mode of chaperone binding to TPR domains. We demonstrate that the TPR domain of Sec72 also binds the ATPase domain of Ssb1 and inhibits its nucleotide exchange activity. Mutagenesis shows that both binding interfaces, Sec72-Ssa1 and Sec72-Ssb1, play a role in protein translocation. Taken together, our results suggest that translocation substrates can be recruited to the Sec71-Sec72 subcomplex either post-translationally through Ssa1 or co-translationally through Ssb1.
Results
Crystal structure of the Sec71-Sec72 complex
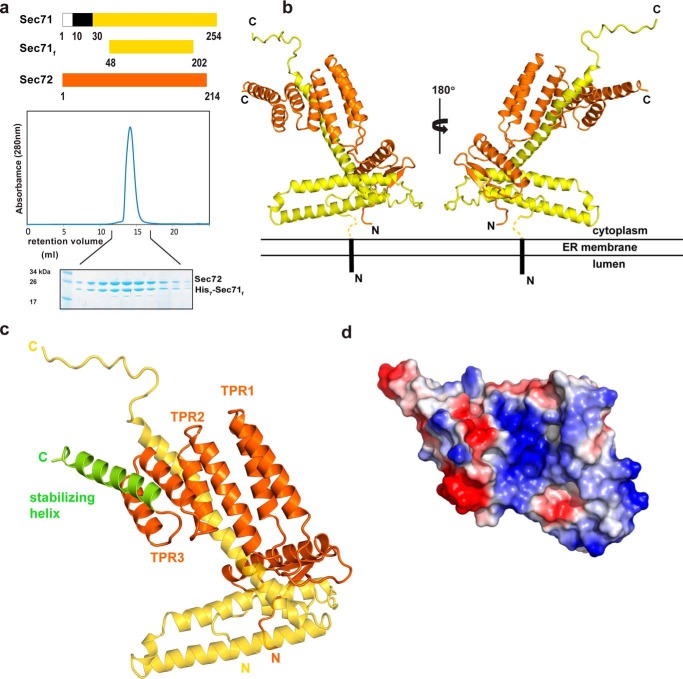
We reasoned that Sec71 and Sec72 may form a subcomplex of the tetrameric complex (Sec63, Sec62, Sec72, and Sec71) because Sec72 is required for the integration of Sec71 into the tetrameric complex (11) and Sec71 is essential for the stability of Sec72 in vivo (11). Given that both Sec71 and Sec72 have domains that are primarily exposed to the cytosol, the N-terminal transmembrane segment of Sec71 is likely not required for the interaction between the two proteins. We therefore expressed Sec71 from Chaetomium thermophilum without its transmembrane segment (Sec71f) together with full-length Sec72 from the same species (Fig. 1a). The complex was expressed in Escherichia coli cells and purified by Ni-NTA chromatography, using an N-terminal His7-tag on Sec71, followed by ion exchange chromatography and gel filtration. The size of the complex in gel filtration and the relative intensity of the two bands in Coomassie Blue-stained SDS gels indicate that the assembly consists of one polypeptide each of Sec71f and Sec72 (Fig. 1a). The data also demonstrate that the cytosolic domain of Sec71 (Sec71f) is sufficient to bind Sec72. The 1:1 stoichiometry is confirmed by the crystal structure of the complex, determined at 3.0-Å resolution (Fig. 1b). Sec71f contains three α-helices, with the N-terminal two helices forming “the base” of Sec71. Given that the N terminus is preceded in the full-length protein by the transmembrane segment, we postulate that these helices are proximal to the membrane. The third helix points away from the base and serves as the anchor for Sec72. This helix is followed by an extended loop that is stabilized by crystal contacts. Sec72 consists of an N-terminal domain that interacts with the base of Sec71 and a C-terminal TPR domain. The TPR domain consists of three sets of helix-loop-helix motifs (TPR1–3) and a C-terminal stabilizing helix (Fig. 1c). The convex surface of the TPR domain interacts with the third helix of Sec71, leaving the concave surface unoccupied. This surface is generally the binding site for ligands in TPR domain-containing proteins. In Sec72, it is positively charged (Fig. 1d), similarly to the TPR domains in Hop, Tom71, and Unc45, which all interact with the negatively charged C termini of chaperones (27–29). These features raise the possibility that the TPR domain of Sec72 also interacts with the negatively charged C terminus of a chaperone.
Figure 1.
Crystal structure of the cytosolic domains of the Sec71-Sec72 complex. a, the cytosolic domains of Chaetomium thermophilum Sec71 and Sec72 indicated in yellow and orange, respectively, were co-expressed in E. coli and purified. Sec71f is a His7-tagged fragment of Sec71 that lacks N-terminal residues, including the TM segment (in black), and some residues at the C terminus, which are not conserved and predicted to be unstructured. The lower panels show the last purification step (gel filtration) with fractions analyzed by SDS-PAGE and Coomassie Blue staining. b, ribbon diagram of the structure of the Sec71-Sec72 complex. Sec71 is shown in yellow and Sec72 in orange. The hypothetical orientation of the complex with respect to the ER membrane is indicated. c, magnified view of the TPR domain of Sec72 bound to Sec71. The three helix-loop-helix motifs are labeled as TPR1–3. The C-terminal, stabilizing helix is shown in green. Sec71 is indicated in yellow. d, space-filling model of the TPR domain, with basic and acidic residues indicated in blue and red, respectively.
The TPR domain of Sec72 binds the C-terminal tail of Ssa1
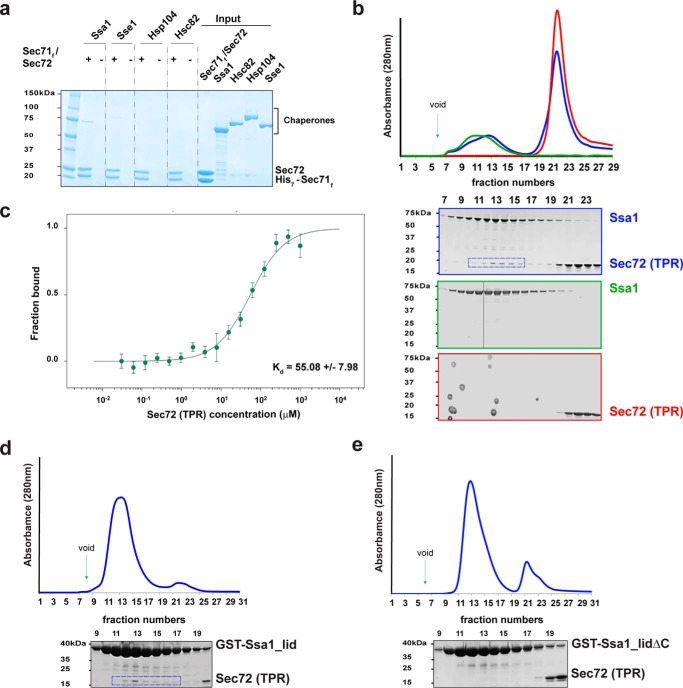
To test whether cytoplasmic chaperones interact with the Sec71-Sec72 complex, we incubated His7-tagged complex with untagged Ssa1, Sse1, Hsc82, or Hsp104. After binding to Ni-NTA resin, the bound material was analyzed by SDS-PAGE and Coomassie Blue staining. Binding was observed for Ssa1, whereas the other chaperones showed little or no interaction (Fig. 2a). Interestingly, Hsc82 (S. cerevisiae Hsp90) has the same four C-terminal amino acids as Ssa1 (EEVD). It is likely that upstream residues of the Hsp90 sequence, in particular a Met residue, interfere with the interaction in a similar manner as observed with the Hsp70-interacting TPR domain of the Hop protein (27).
Figure 2.
The TPR domain of Sec72 binds to Ssa1. a, purified His7-tagged Chaetomium thermophilum Sec71-Sec72 complex was incubated with untagged Ssa1, Sse1, Hsc82, or Hsp104. After binding to Ni-NTA resin, the bound material was analyzed by SDS-PAGE and Coomassie Blue staining. The right panel shows 10% of the input material for the pulldown experiments. b, purified full-length Ssa1 was mixed with purified TPR domain of Sec72 and subjected to gel filtration. The absorbance at 280 nm was followed (blue curve in upper panel). Controls were performed with Ssa1 or TPR domain alone (green and red curves, respectively). For each experiment, fractions were analyzed by SDS-PAGE and EZBlue staining (lower panels; the colors correspond to the curves in the upper panel). The box indicates fractions containing the TPR domain of Sec72 co-eluting with Ssa1. The vertical line indicates that empty gel lanes were removed from the image of the gel. c, full-length Ssa1 was incubated with increasing concentrations of the TPR domain of Sec72, and binding was assessed by microscale thermophoresis. The dissociation constant was determined from four independent experiments. Error bars, mean ± S.D. d, a GST-fusion of the C-terminal lid domain of Ssa1 was mixed with the TPR domain and subjected to gel filtration. Fractions were analyzed by SDS-PAGE and EZBlue staining. The box indicates fractions containing TPR domain of Sec72 co-eluting with Ssa1. e, as in d, but with a GST-fusion of a lid domain lacking the last 14 amino acids.
To test whether the TPR domain of Sec72 is responsible for the interaction with Ssa1, we performed gel filtration experiments with purified proteins (Fig. 2b). Indeed, a population of the two proteins co-eluted, the low percentage likely reflecting a weak interaction between the two proteins. Determination of the binding constant by microscale thermophoresis gave an apparent binding constant of ∼55 μm (Fig. 2c). We next tested whether Ssa1 binds the TPR domain of Sec72 via its C termini, akin to that seen for Hsp70 binding to other TPR domains such as Hop, Tom71, and Unc45 (27–29). A GST fusion of the C-terminal lid domain of Ssa1 interacted with the TPR domain (Fig. 2d), whereas a construct lacking the last 14 amino acids did not bind (Fig. 2e). These results support the idea that the C terminus of Ssa1 interacts with the TPR domain of Sec72. Weak binding between TPR domains and an Hsp70 partner have also been reported for many other cases (18–230 μm) (27, 29, 30) and may not be surprising, as only a few residues of the Hsp70 interact with the TPR domain.
To further characterize the interaction between the C terminus of Ssa1 and the TPR domain, we attempted co-crystallization of the Sec71-Sec72 complex or of the TPR domain with a synthetic C-terminal Ssa1 peptide, or soaking of Sec71-Sec72 crystals with the peptide. However, the resulting structures lacked bound peptide, likely because the interaction was weak. To facilitate binding, we therefore fused a C-terminal peptide of Ssa1 (HDNDGPTVEEVD) to the TPR domain of Sec72, both derived from C. thermophilum. The fusion protein indeed crystallized, and its structure was determined by molecular replacement and refined to a resolution of 2.87 Å. Eight copies of the fusion protein were located in the asymmetric unit, but they did not differ significantly (root mean standard deviation (r.m.s.d.) 0.35–0.5 Å). In addition, the structure of the TPR domain was essentially the same as in the Sec71-Sec72 structure determined without peptide (r.m.s.d. 0.36Å, supplemental Fig. S1). The only difference is in the orientation of Lys-159. In the Sec71-Sec72 structure, Lys-159 forms a hydrogen bond with Asp-193 of the stabilizing helix, whereas in the Sec72-Ssa1 peptide fusion protein structure, it reorients and contacts the terminal Asp-226 of the Ssa1 peptide.
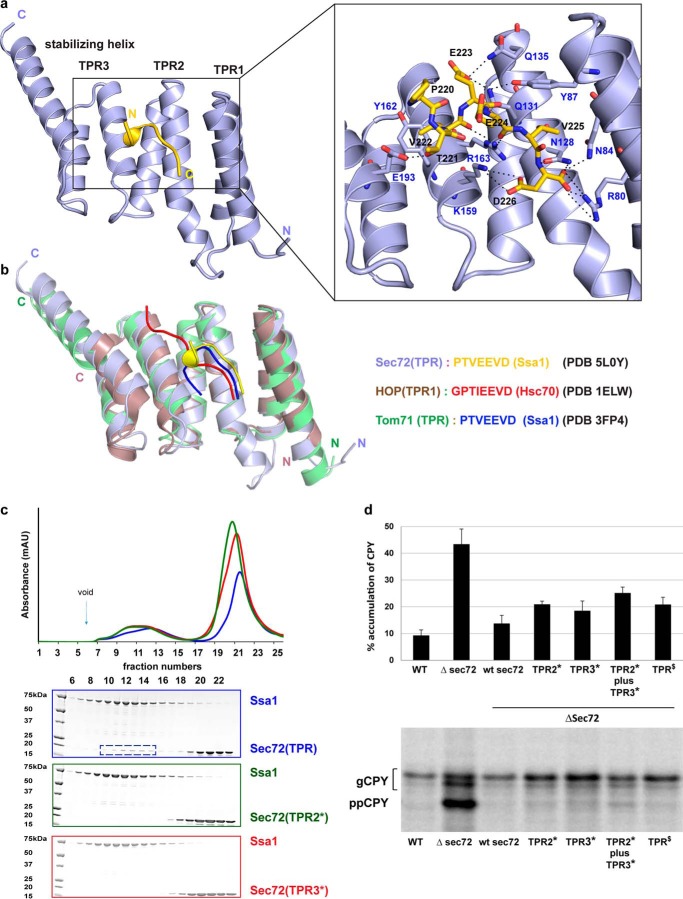
In agreement with our prediction, the negatively charged C terminus of Ssa1 (PTVEEVD) sits in the positively charged pocket formed by the concave surface of the TPR domain. Most of the preceding residues of Ssa1 are invisible, suggesting that they are flexible and do not perturb the binding of the C-terminal tail. The TPR domain forms a two-carboxylate clamp that holds the terminal Asp-226 of the PTVEEVD sequence in place. Asp-226 forms extensive hydrogen bonds and salt bridges with conserved residues in TPR1, TPR2, and TPR3, including Arg-80, Asn-84, Asn-128, and Lys-159 (Fig. 3a). In addition, side chains of Glu-223 and Thr-221 (corresponding to the underlined residues in the PTVEEVD sequence) contact TPR2 through Gln-135 and Gln-131 and the stabilizing helix through Asp-193, respectively (Fig. 3a). The backbone of the Ssa1 peptide also engages in polar contacts with residues Asn-128, Arg-163, Tyr-87, and Arg-80 of the TPR domain (Fig. 3a). In contrast to Hsp70 protein Ssa1, in Hsp90 protein, a Glu residue replaces Thr-221 at the corresponding position, providing another explanation why Sec72 is specific for Hsp70.
Figure 3.
Interaction of the TPR domain of Sec72 with the C terminus of Ssa1. a, crystal structure of a fusion of the TPR domain of Sec72 with a C-terminal Ssa1 peptide, both derived from Chaetomium thermophilum. Shown are ribbon diagrams with Sec72 in light blue and the peptide in yellow. The right panel gives a magnified view with the amino acids of the peptide and interacting TPR residues shown as sticks. Dashed lines indicate salt bridges and hydrogen bonds. b, structures of other TPR domains with bound C-terminal Hsp70 peptides. Note that similar peptides adopt different conformations when bound to different TPR domains. c, purified full-length Ssa1 was mixed with purified wild-type TPR domain of Sec72 and subjected to gel filtration. The absorbance at 280 nm was followed (blue curve in upper panel). Additional experiments were performed with TPR domains that carried mutations in either TPR2 (mutations of Asn-128, Gln-131; mutant TPR2*; green curve) or TPR3 and the stabilizing helix (mutation of Lys-159, Arg-163, and Asp-193; mutant TPR3*; red curve). For each experiment, fractions were analyzed by SDS-PAGE and EZBlue staining (lower panels; the colors correspond to the curves in the upper panel). The box indicates fractions containing the TPR domain of Sec72 co-eluting with Ssa1. d, the translocation of the post-translational substrate carboxypeptidase Y (CPY) was tested by incubating wild-type yeast cells or cells carrying Sec72 mutations with Trans35S-label. After immunoprecipitation, the samples were subjected to SDS-PAGE and autoradiography. The percentage of non-translocated CPY (prepro-CPY (ppCPY)) with respect to total CPY (glycosylated CPY (gCPY) plus ppCPY) was determined by densitometry from two (TPR2*) or four to six (all others) experiments, a representative of which is shown in the lower panel. The bars show mean (TPR2) and standard deviation (all others). TPR2* contains the mutations S111A and D114A in full-length S. cerevisiae Sec72 (equivalent to the TPR2* mutations in C. thermophilum). TPR3* contains the mutations R145A, D141A, and K175A (equivalent to the TPR3* mutations in C. thermophilum), and TPR$ contains D172R and E173K, equivalent to the E190R and E191R mutations in C. thermophilum (Fig. 4).
The TPR domain of Sec72 is structurally similar to the TPR domains of Hop and Tom71 (27, 28), which also bind the negatively charged C terminus of Hsp70 (r.m.s.d. 1.4 Å and 2.0 Å, respectively). Hop and Tom71 also use the two-carboxylate clamp to interact with the C-terminal Asp residue of the Hsp70 peptide. In both cases, the Hsp70 peptide is bound in an extended conformation. In contrast, in the TPR-Ssa1 peptide structure, the Ssa1 peptide forms a 310 helix (Fig. 3b) by virtue of intramolecular hydrogen bonding between Pro-220 and Glu-223 of the Ssa1 peptide. The same Ssa1 peptide forms a β-strand in the structure of the unrelated Sis1 (Hsp40)-Ssa1 complex (31). Taken together, this shows that the C-terminal Ssa1 tail is versatile and can adopt different structures depending on its binding partner.
Next we used site-directed mutagenesis to test whether the TPR-peptide interaction observed in the crystal structure is relevant for complex formation between Ssa1 and Sec72. Residues at the interaction interface of the TPR domain were changed to Ala, and complex formation between purified full-length Ssa1 and the TPR domain of Sec72 was analyzed by gel filtration. Indeed, mutations in either TPR2 (mutations of Asn-128, Gln-131; mutant TPR2*) or TPR3 and the stabilizing helix (mutation of Lys-159, Arg-163, and Asp-193; mutant TPR3*) abolished the interaction (Fig. 3c). These results confirm that the C terminus of Ssa1 interacts with the TPR domain of Sec72.
To test whether the interaction between Sec72 and Ssa1 has a role in protein translocation in vivo, we introduced the TPR mutations that disrupt the interaction of the C. thermophilum components into the interaction surface of full-length S. cerevisiae Sec72. Translocation of the post-translational substrate carboxypeptidase Y (CPY) was tested by incubating yeast cells for a short time period with Trans35S-label and determining the percentage of non-translocated molecules. In wild-type cells, only 9% of CPY remained non-translocated in the cytosol, whereas in cells lacking Sec72 the percentage increased to 43%. Mutants in the Ssa1 interaction surface of Sec72 (TPR2* and TPR3* mutations) showed a moderate translocation defect (19–25%) (Fig. 3d). These results indicate that Ssa1 binding to Sec72 plays a role in post-translational translocation. The stronger translocation defect seen with a sec72 deletion mutant could be because of additional defects in Sec complex function (12) or is suggestive of other factors being involved in Sec72-mediated protein translocation.
Ssb1 interaction with the TPR domain of Sec72
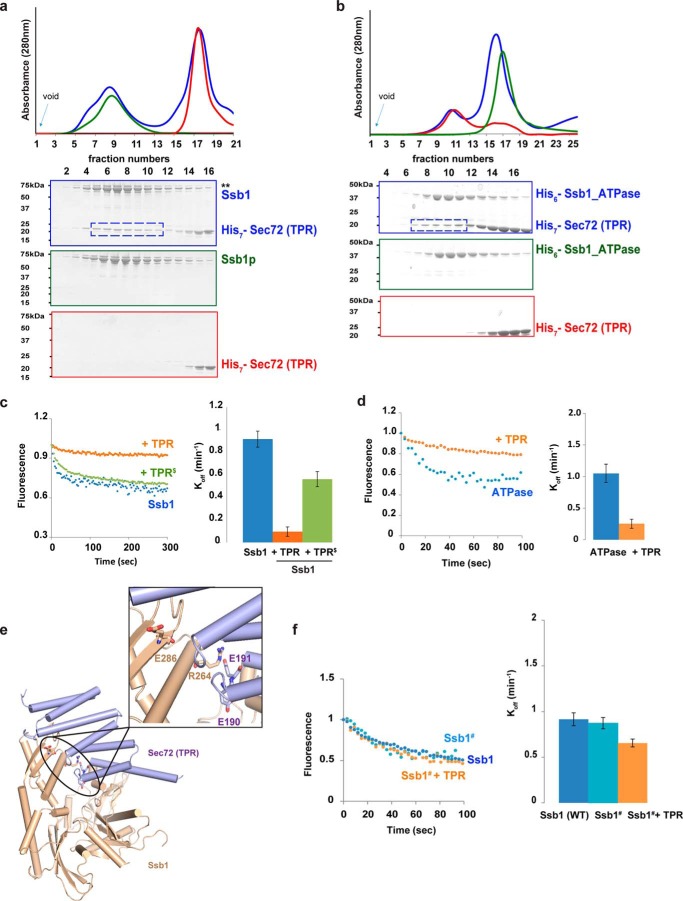
Surprisingly, we found that Ssb1, a cytosolic Hsp70 that lacks the C-terminal EEVD sequence, also interacts with the TPR domain of Sec72 (Fig. 4a). This interaction is mediated by the ATPase domain of Ssb1 (Fig. 4b), an interaction that is reminiscent of the binding of the TPR domain of Hip to the ATPase domain of Hsp70 (32). Because Hip inhibits the nucleotide exchange of the ATPase domain, we tested whether the TPR domain of Sec72 has a similar activity. Full-length Ssb1 was incubated with fluorescently labeled MANT-ADP (2′-(or-3′)-O-(N-methylanthraniloyl)-ADP), and the release of the nucleotide analog was followed by the decrease in fluorescence after addition of ATP. Rapid nucleotide exchange was observed (Fig. 4c, blue dots). When the TPR domain of Sec72 was added, nucleotide exchange was drastically reduced (Fig. 4c, orange dots). A similar effect was observed when the TPR domain was added to the isolated ATPase domain of Ssb1 (Fig. 4d, orange versus blue dots). These results suggest that the TPR domain of Sec72 interacts in a similar way as Hip with the ATPase domain of Hsp70 (32). Indeed, a model can be built on the basis of the crystal structure of the Hip-Hsp70 ATPase domain complex, in which the TPR domain of Sec72 replaces that of Hip (Fig. 4e) (overlay shown in supplemental Fig. S2) and the Ssb1 ATPase domain replaces that of Hsp70. A comparison of the structures of the TPR domains shows that five of six Hip residues that interact with the ATPase domain and are required for nucleotide exchange inhibition (32) are also found in Sec72 (Asp-190, Asp-191, Phe-187, Asn-157, Arg-80). A similar analysis showed that only one or none of these residues is found in the TPR domains of mammalian and yeast Hop (TPR1 and TPR2B, respectively), Unc45, CHIP, Sgt2, or Tom71, which all interact with the C terminus of Hsp70, but likely not with the ATPase domain. Based on the homology model for the complex of the Sec72 TPR domain and the Hsp70 ATPase domain (Fig. 4e), we changed two Glu residues to Arg in the loop between TPR3 and the stabilizing helix (Glu-190 and Glu-191), which should interfere with complex formation. When tested in the ADP release assay, this mutant (TPR$) had indeed a reduced activity in inhibiting nucleotide exchange (Fig. 4c, green dots). Importantly, introducing the same mutations into full-length Sec72 reduced post-translational translocation of CPY in S. cerevisiae cells (TPR$) (Fig. 3d), demonstrating the physiological relevance of this interaction. The partial translocation defects seen with each of the mutant classes (TPR2*, TPR3*, and TPR$) (Fig. 3d, upper panel) suggest that they each interfere with only one targeting pathway, Ssa1-dependent or Ssb1-dependent. In strains lacking Sec72, both pathways are likely disrupted, leading to a stronger CPY translocation defect. To further validate that the TPR domain of Sec72 interacts with the ATPase domain of Ssb1, we made a mutant in the ATPase domain of full-length Ssb1 that is expected to affect complex formation (E286A and R264A; Ssb1# mutant) (Fig. 4e). The nucleotide exchange rate of this mutant was similar to that of wild-type Ssb1, but was only moderately inhibited by the TPR domain (Fig. 4f). Taken together, these results suggest that the TPR domain of Sec72 can interact through distinct surfaces not only with the C terminus of Ssa1, but also with the ATPase domain of Ssb1.
Figure 4.
Ssb1 interaction with the TPR domain of Sec72. a, purified full-length Ssb1 was mixed with purified TPR domain of Sec72 and subjected to gel filtration. The absorbance at 280 nm was followed (blue curve in upper panel). Controls were performed with Ssb1 or TPR domain alone (green and red curves, respectively). For each experiment, fractions were analyzed by SDS-PAGE and EZBlue staining (lower panels, the colors correspond to the curves in the upper panel). The box indicates fractions containing the TPR domain of Sec72 co-eluting with Ssb1. b, as in a, but with the purified ATPase domain of Ssb1 instead of the full-length protein. c, full-length Ssb1 was incubated with the fluorescent nucleotide analog MANT-ADP. Excess of ATP was added and the decrease of fluorescence was followed over time (blue dots). Similar experiments were performed in the presence of purified wild-type TPR domain of Sec72 or of TPR$ (E190R and E191R) mutant (orange and green dots, respectively). Error bars, mean ± S.D. d, as in c, but with the ATPase domain of Ssb1, instead of the full-length protein. e, a model of the complex of the TPR domain of Sec72 bound to the ATPase domain of Ssb1, based on a structure of the Hip-ATPase complex (32). The interface residues Glu-190 and Glu-191 in the TPR domain and Arg-264 and Asp-286 in Ssb1 ATPase domain are shown in a magnified view. f, as in c, but with a Ssb1 mutant (Ssb1#) carrying the R264A and D286A mutations in the ATPase domain.
Discussion
Here, we report a high-resolution structure of the Sec71-Sec72 subcomplex, a component of the Sec complex involved in post-translational protein translocation across the yeast ER membrane. The structure demonstrates that Sec71 serves as an ER anchor for Sec72, consistent with observations that deletion mutants of Sec71 or Sec72 have the same phenotype, and that Sec71 deletion leads to destabilization of Sec72 (11, 12). The structure and biochemical experiments show that Sec72 has a TPR domain that interacts with members of two different Hsp70 families, Ssa1 and Ssb1. Ssa1 associates through its C-terminal acidic tail with the TPR domain, whereas Ssb1 binds through the ATPase domain.
The interaction of Sec72 with Ssa1 is consistent with predictions based on the co-evolution of the two binding partners (33). Because Ssa1 interacts through its C terminus with Sec72, its peptide-binding pocket would remain accessible in the complex. We therefore suggest that post-translationally translocated substrates bind to Ssa1 and are then targeted through the Ssa1-Sec72 interaction to the translocation channel (Fig. 5). This is further supported by our in vivo translocation assays, which demonstrate that disruption of the Ssa1 binding interface on Sec72 affects the transport of the post-translational substrate CPY across the ER membrane.
Figure 5.
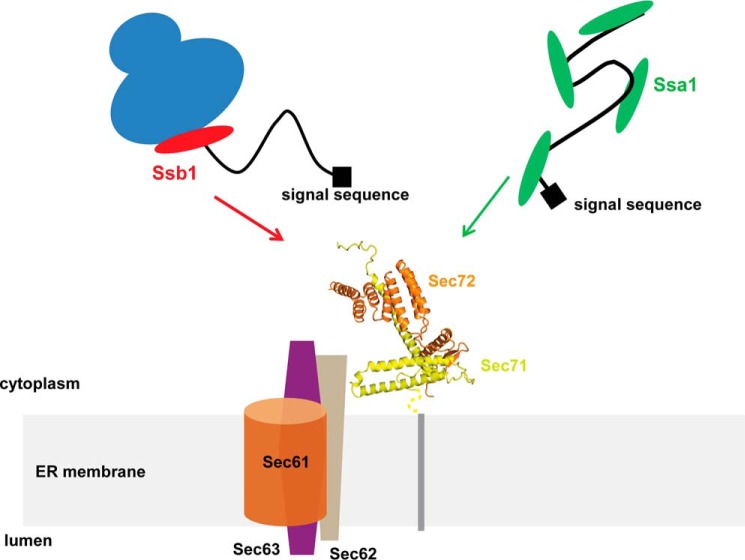
Model for the co- and post-translational targeting of substrates to the Sec complex. Ssb1 would bind to the ribosome-nascent chain complex and then associate through Sec72 with the Sec complex (consisting of the Sec61 channel, the Sec62-Sec63 complex and the Sec71-Sec72 complex). Ssa1 would bind to completed polypeptide chains and mediate their post-translational interaction with the Sec complex. In both cases, translocation would occur through the Sec complex that is dedicated to post-translational translocation and does not require ribosome-Sec61 interaction. It should be noted that, in the case of co-translational targeting, translocation through the Sec complex could commence before the polypeptide is completed.
So far, it has been assumed that a specific targeting process exists only for co-translational translocation, where SRP and its membrane receptor mediate the binding of a translating ribosome to the translocon. Although the cytosolic Hsp70 molecules Ssa1–4 have been implicated in post-translational translocation (18–20), it was thought that they would merely keep polypeptides in a loosely folded, translocation-competent state. Our results now suggest that Ssa1 is a targeting factor, as it has a specific interaction with the translocon.
A similar targeting mechanism is seen in post-translational transport into mitochondria, where the TPR domain-containing protein Tom70 functions as a receptor for mitochondrial precursor proteins (34). Similar to the ER protein Sec72, Tom70 is a constituent of the mitochondrial translocation machinery, interacts with the C terminus of Ssa1 (28), and recruits polypeptides associated with Ssa1 to the mitochondrial membrane. Because Ssa1 likely does not discriminate between proteins destined to the ER or mitochondria, it is likely that the targeting specificity comes from the signal sequence. Nevertheless, Ssa1-mediated interaction with the translocons in the two organelles could increase the local concentration of transport substrates and thus enhance targeting efficiency. Because only a few C-terminal amino acids of Ssa are involved in the interaction with Sec72 or Tom70, the binding constants are low and the interaction is likely transient. Indeed, as in the case of Sec72, Tom70 interacts only weakly with Ssa1 (34). A weak interaction of Ssa1 with receptor proteins is expected, as the targeting systems are dynamic, with Ssa1 likely associating and dissociating both with the translocation substrate and with the receptors in the organelles. A TPR domain-containing protein, Pex5, is also involved in the targeting of proteins to peroxisomes (for review, see Ref. 35). In this case, the TPR domain interacts with the C-terminal targeting sequence SKL of proteins destined to the peroxisomal matrix (36). Again, the binding constant is low, but still allows specific targeting. Taken together, TPR domains appear to be a common theme in substrate targeting to different organelles, either directly as in the case of Pex5, or indirectly through Ssa1, as in the case of Tom70 and Sec72.
We made the surprising observation that Ssb1, a cytosolic Hsp70 lacking an acidic C terminus, also binds to the TPR domain of Sec72. In this case, the binding is mediated by the ATPase domain. As in the precedent case of Hip (32), the interaction with the TPR domain leads to inhibition of nucleotide exchange. The interaction surface of Ssb1 for the TPR domain is also found in the ATPase domains of Ssa1 and Sse1, but our data indicate that Ssa1 only binds through its C-terminal tail (Fig. 3c) and Sse1 binds weakly or not at all (Fig. 2a). Perhaps, the binding to the N-terminal ATPase domains is somehow inhibited in the full-length Ssa1 and Sse1 proteins.
Based on the interaction between Ssb1 and Sec72, we propose that substrates can be co-translationally recruited to the ER membrane, even when they are translocated by the Sec complex that is dedicated to post-translational transport. In this model, Ssb1 would mediate the interaction of a ribosome-nascent chain complex with the Sec71-Sec72 components of the Sec complex (Fig. 5). A population of Ssb1 is indeed bound to ribosome-nascent chain complexes, and could therefore potentially target proteins to the translocon (25). Based on an ER proximity assay to analyze ribosome targeting, there is also evidence that Sec71-Sec72 is involved in the recruitment of translating ribosomes (22). CPY was one of the Sec71-dependent, post-translational substrates that was synthesized by ribosomes targeted to the ER membrane (22). Whereas the ribosome binds directly to the Sec61 channel during co-translational translocation, in this model it would bind through the nascent chain and Ssb1 to Sec72. The binding of the TPR domain of Sec72 to the ATPase domain of Ssb1 would lead to inhibition of nucleotide exchange, which in turn would prolong the interaction of the nascent chain with Ssb1 until translocation commences. Further experiments should allow this and other models to be tested directly, including the mechanisms by which exchange of Ssb1 for Ssa1, as well as release of the Hsp70s and subsequent protein translocation, are coordinated.
Experimental procedures
Protein purification
Sec71 (residues 48–202, referred to as Sec71f) and full-length Sec72 of C. thermophilum were co-expressed in E. coli. The TPR domain of Sec72 (68–214) or a Sec72-Ssa1 peptide fusion (residues 68–214 of Sec72 fused to C. thermophilum Ssa1 C-terminal sequence HDNDGPTVEEVD) were expressed individually. These constructs were cloned into expression plasmids based on pQLink (Addgene plasmid, accession numbers 13670 and 13667) (37). GST-Ssa_lid (524–642) and GST-Ssa1lid_ΔC (524–627) from S. cerevisiae were expressed after cloning into the plasmid pGEX-4T1 (GE Healthcare). Ssa1 (1–642), Ssb1 (1–613), the ATPase domain of Ssb1 (1–384) from S. cerevisiae were expressed after cloning into the plasmid pProEx HTb (Gibco). Mutations were introduced using QuikChange Mutagenesis (Stratagene). All constructs were confirmed by DNA sequencing.
N-terminally His-tagged proteins were produced in BL21 E. coli (New England Biolabs) grown in 2x YT medium at 37 °C to an A600 of 1.0–1.4. Protein expression was induced by the addition of 0.3–0.5 mm isopropyl β-d-thiogalactopyranoside (IPTG). After growth overnight at 18 °C (Ssb1_ATPase domain, 1–384) or 24 °C (all others), the cells were harvested and the tagged proteins were purified from cell lysates by Ni2+-affinity chromatography followed by removal of the His-tag by digestion with tobacco etch virus protease. When necessary, the proteins were further purified by anion exchange (MonoQ, GE Healthcare) and size exclusion (S200, GE Healthcare) chromatography. For Ssa1 and Ssb1, the proteins were treated with EDTA overnight to strip off nucleotide prior to anion exchange (MonoQ, GE Healthcare) and size exclusion (S200, GE Healthcare) chromatography (38). Ssb1_ATPase domain (1–384) was purified as described for the ATPase domain of Hsp70 (32). Purified proteins were flash frozen in liquid nitrogen and stored at −80 °C. For the preparation of selenomethionine (Se-Met)-labeled Sec71 (residues 48–202, Sec71f) and full-length Sec72 (residues 1–214), methionine synthesis was suppressed by metabolic inhibition (39). Sec71 (48–202) and Sec72 (1–214) were co-expressed in BL21 E. coli cells grown in M9 media supplemented with 5% (w/v) dextrose and 0.7% (w/v) yeast nitrogen base without amino acids (Difco) to an A600 of ∼0.8. l-selenomethionine (Acros Organics) was added to a final concentration of 50 mg/liter (lysine, phenylalanine, threonine, arginine, isoleucine, leucine, valine) (final concentrations, 50 mg/liter) (Sigma). After 20 min, protein expression was induced by adding 1 mm IPTG and shaking overnight at 24 °C. Se-Met proteins were purified as above, with 5 mm DTT present throughout.
N-terminally GST-tagged proteins were produced in BL21 E. coli (New England Biolabs) cells grown in 2x YT media at 37 °C to an A600 of 1.0–1.4. Protein expression was induced by the addition of 0.3–0.5 mm IPTG. Cells were harvested after growing overnight at 24 °C, and the tagged proteins were purified from cell lysates by glutathione-affinity chromatography followed by size exclusion (S200, GE Healthcare) chromatography. Purified proteins were flash frozen in liquid nitrogen and stored at −80 °C. Hsc82, Sse1, and Hsp104 were purified as reported (40).
Crystallization and structure determination
Crystals of Se-Met-substituted Sec71-Sec72 complex were obtained by vapor diffusion at 4 °C using a 1:1 ratio of protein (11 mg/ml) and well buffer (0.1 m Tris-HCl, pH 8.5, 1.4 m ammonium tartrate dibasic). Crystals grew over a period of 2 days and were subsequently cryo-protected using well buffer supplemented with sequentially increasing concentrations of glycerol (up to 15% (v/v)) before flash freezing in liquid nitrogen. Crystals of the Sec72-Ssa1 fusion construct were obtained by vapor diffusion at 23 °C using a 1:1 ratio of protein (15 mg/ml) and well buffer (0.1 m ammonium citrate, pH 7.0, 12% PEG 3350). Streak seeding was required to produce high-quality crystals.
Crystals were cryo-protected by a brief soak in buffer (14% PEG 3350, 0.1 m ammonium citrate, pH 7.0) supplemented with 15% (v/v) glycerol and flash frozen in liquid nitrogen. All data were collected at NE-CAT of the Advanced Photon Source (Argonne National Laboratory) and processed using X-ray Detector Software (XDS) (41).
The His7-Sec71-Sec72 complex crystallized in space group P6522, with one molecule in the asymmetric unit. The crystals diffracted to a maximum resolution of 3.0 Å. The structure was determined by SAD phasing using Se-Met-substituted protein. To improve the anomalous signal for phasing, Se-Met SAD datasets from two different crystals were merged using XDS. The PHENIX suite of programs was used to find the Se-Met sites, calculate the initial electron density maps, improve the phases, and generate the initial model (42). The final model was obtained after iterative cycles of model building using COOT (43) with refinement in PHENIX. The final model includes all residues of Sec72, except amino acids 1–4 and 214 (residues 5–213). The model also includes essentially all residues of the Sec71 construct (residues 30–184 of Sec71 and the sequence NLYFQGS preceding the Sec71 sequence), except the His-tag and the linker region between the His-tag and the tobacco etch virus cleavage site. 96.0% of the residues are in the most favored regions of the Ramachandran plot, whereas 4.0% fall in allowed regions, as judged using MolProbity (44).
The Sec72-peptide fusion protein crystallized in space group P21212, with eight molecules in the asymmetric unit. The structure was solved by molecular replacement using PHASER (45), with Sec72 TPR domain (68–213) as the search model. The final model, built using COOT and refined using PHENIX, includes Sec72 residues 68–214 and the Ssa1 residues PTVEEVD. 96.3% of the residues are in the most favored regions of the Ramachandran plot, with the remaining 3.7% falling in allowed regions as judged using MolProbity (44). Data collection and structure refinement statistics for both structures are summarized in supplemental Table 1.
Crystal contacts were analyzed with PISA (46), and structural homologs identified with DALI (47). Structure-based sequence alignment was guided by DaliLite (48), and molecular illustrations were prepared with PyMOL (49).
Binding experiments
Analytical size exclusion chromatography to monitor complex formation between Sec72 (TPR) and Ssa1p or Ssb1p was performed at 4 °C. 100 μm Ssa1 or Ssb1 was incubated with 3-fold excess of wild-type Sec72 (TPR) or mutant Sec72 (TPR2*, TPR3*) for 30 min on ice, and samples were loaded onto a Superdex 5/150 column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH8.0. Fractions were analyzed by SDS-PAGE and stained with EZBlue (Invitrogen). For measuring binding to Ssb1_ATPase, 200 μm of the ATPase domain was incubated with 10-fold excess of Sec72 (TPR) and loaded onto a Superdex 5/150 column as above. For measuring binding to GST-fusion proteins (GST-Ssa1_lid, GST-Ssa1_lid ΔC), 250 μm fusion protein was incubated with a 3-fold excess of Sec72 (TPR), and binding was tested as above.
The interaction of Ssa1, Sse1, Hsc82, or Hsp104 with His7-Sec71-Sec72 complex was tested in 50 μl of 20 mm Hepes/KOH, pH7.5, 100 mm KCl, 10 mm imidazole, 5 mm MgCl2, 5 mm ADP, 5 mm 2-mercaptoethanol. 50 μm His-tagged complex was incubated with 5 μm chaperone on ice for 2 h and then added to Ni-NTA agarose beads. After 10 min incubation, the resin was washed twice with binding buffer and the bound material was eluted with binding buffer containing 300 mm imidazole.
Microscale thermophoresis
Assays were carried out with a Monolith NT.115 instrument (Nano Temper, Munich, Germany) at 22 °C. Purified full-length Ssa1 was labeled with L001 Monolith NT.115 Protein Labeling Kit RED-NHS (Amine Reactive) dye according to the manufacturer's instructions. All components were in 20 mm Hepes/KOH pH 8.0, 50 mm KCl, 0.05% (v/v) Tween20, 0.5 mm tris(2-carboxyethyl)phosphine (TCEP). 10 nm of labeled Ssa1 was added to serial dilutions of unlabeled, Sec72 TPR domain (0.03–1000 μm), incubated for 5 min and loaded into capillaries. Data evaluation was performed with the Monolith software. The binding constant reported was determined from four independent experiments.
Nucleotide release measurements
Nucleotide release kinetics of the fluorescent nucleotide analog MANT-ADP was followed with a Synergy H1M multi-mode reader (Biotek). All components were in 15 mm Tris, pH 8.0, or 15 mm Hepes, pH 7.0, 60 mm KCl, and 5 mm MgCl2 buffer (buffer A) containing 1 mm 2-mercaptoethanol. MANT-ADP complexes were formed by mixing 150 μm Ssb1, Ssb1 ATPase domain or Ssb1# mutant (E286A, R264A), in the ratio of 0.9:1 for 20 min at 27 °C and then incubating at 4 °C overnight. At the beginning of the experiment, the ATPases were diluted to a concentration of 5 μm in buffer A. The nucleotide exchange solutions contained 2.5 mm ATP and protein factors (used in 90-fold excess), as indicated in Fig. 4. To determine the nucleotide release rates, equal volumes of the solutions were mixed at 30 °C and release of labeled nucleotide was monitored by the decrease in MANT-ADP fluorescence (excitation 350 nm, emission cutoff filter 448 nm). The curves were analyzed assuming first-order kinetics and a constant drift from bleaching. Rates reported were determined from at least three independent experiments.
Strain construction and immunoprecipitation of radiolabeled proteins
The SEC72 gene in strain MAT a, trp1–1, ade2, leu2–1,112, ura3, his3–11, can1 was disrupted using a kanamycin-resistant cassette as described previously (50) to obtain RGY700. Gene disruption was confirmed by PCR.
RGY700 strain was transformed with LEU2-marked plasmid (pRS315) encoding wild type or mutated SEC72, as detailed in the text. Cells were incubated with Trans 35S-label (Perkin Elmer) and radiolabeled carboxypeptidase (CPY) was immunoprecipitated, as described previously (50).
Accession numbers
Coordinates and structure factors for the Sec71-Sec72 complex and Sec72-Ssa1 peptide fusion protein have been deposited in the Protein Data Bank under the accession numbers 5L0W and 5L0Y, respectively.
Author contributions
A. T. prepared and crystallized the proteins, performed biochemical experiments, and determined the crystal structures. E. C. M. and R. G. conducted experiments in yeast. T. A. R. and A. T. wrote the paper, and T. A. R. supervised the project.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of the staff of NE-CAT of the Advanced Photon Source (Argonne National Laboratory) with X-ray data collection and the SBGrid consortium at Harvard Medical School for support with computer programs. We thank William Clemons Jr. for sending vectors for expression of Hsc82, Hsp104, and Sse1.
Note added in proof
There were several errors in the version of this article that was published as a Paper in Press on March 12, 2017. The gels in Fig. 2, a and b, did not conform to JBC policy regarding the presentation of gel data. These errors have now been corrected and do not affect the results or conclusions of this work.
This work was supported by National Institutes of Health Grants RO1 GM35687 (to R. G.) and RO1 GM 052586 (to T. A. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1 and S2 and supplemental Table 1.
The atomic coordinates and structure factors (codes 5L0W and 5L0Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ER
- endoplasmic reticulum
- TPR
- tetratricopeptide repeat
- SRP
- signal recognition particle
- CPY
- carboxypeptidase
- MANT-ADP
- 2′-(or-3′)-O-(N-methylanthraniloyl)-ADP
- r.m.s.d.
- root mean standard deviation
- IPTG
- isopropyl β-d-thiogalactopyranoside
- Se-Met
- selenomethionine
- SAD
- single-wavelength anomalous diffraction
- XDS
- X-ray Detector Software
- COOT
- Crystallographic Object-Oriented Toolkit.
References
- 1. Park E., and Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 [DOI] [PubMed] [Google Scholar]
- 2. Mandon E. C., Trueman S. F., and Gilmore R. (2013) Protein translocation across the rough endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ng D. T., Brown J. D., and Walter P. (1996) Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hann B. C., and Walter P. (1991) The signal recognition particle in S. cerevisiae. Cell 67, 131–144 [DOI] [PubMed] [Google Scholar]
- 5. Hansen W., and Walter P. (1988) Prepro-carboxypeptidase Y and a truncated form of pre-invertase, but not full-length pre-invertase, can be posttranslationally translocated across microsomal vesicle membranes from Saccharomyces cerevisiae. J. Cell Biol. 106, 1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rothblatt J. A., and Meyer D. I. (1986) Secretion in yeast: translocation and glycosylation of prepro-α-factor in vitro can occur via an ATP-dependent post-translational mechanism. EMBO J. 5, 1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waters M. G., and Blobel G. (1986) Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. J. Cell Biol. 102, 1543–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ast T., Cohen G., and Schuldiner M. (2013) A network of cytosolic factors targets SRP- independent proteins to the endoplasmic reticulum. Cell 152, 1134–1145 [DOI] [PubMed] [Google Scholar]
- 9. Deshaies R. J., Sanders S. L., Feldheim D. A., and Schekman R. (1991) Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349, 806–808 [DOI] [PubMed] [Google Scholar]
- 10. Panzner S., Dreier L., Hartmann E., Kostka S., and Rapoport T. A. (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570 [DOI] [PubMed] [Google Scholar]
- 11. Feldheim D., and Schekman R. (1994) Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J. Cell Biol. 126, 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feldheim D., Yoshimura K., Admon A., and Schekman R. (1993) Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol. Biol. Cell 4, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deshaies R. J., and Schekman R. (1990) Structural and functional dissection of Sec62p, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol. Cell. Biol. 10, 6024–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feldheim D., Rothblatt J., and Schekman R. (1992) Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12, 3288–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rothblatt J. A., Deshaies R. J., Sanders S. L., Daum G., and Schekman R. (1989) Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 109, 2641–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Misselwitz B., Staeck O., and Rapoport T. A. (1998) J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603 [DOI] [PubMed] [Google Scholar]
- 17. Matlack K. E., Misselwitz B., Plath K., and Rapoport T. A. (1999) BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97, 553–564 [DOI] [PubMed] [Google Scholar]
- 18. Becker J., Walter W., Yan W., and Craig E. A. (1996) Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16, 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., and Schekman R. (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332, 800–805 [DOI] [PubMed] [Google Scholar]
- 20. Chirico W. J., Waters M. G., and Blobel G. (1988) 70K heat shock related proteins stimulate protein translocation into microsomes. Nature 332, 805–810 [DOI] [PubMed] [Google Scholar]
- 21. Plath K., and Rapoport T. A. (2000) Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J. Cell Biol. 151, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jan C. H., Williams C. C., and Weissman J. S. (2014) Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nelson R. J., Ziegelhoffer T., Nicolet C., Werner-Washburne M., and Craig E. A. (1992) The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71, 97–105 [DOI] [PubMed] [Google Scholar]
- 24. Peisker K., Chiabudini M., and Rospert S. (2010) The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803, 662–672 [DOI] [PubMed] [Google Scholar]
- 25. Willmund F., del Alamo M., Pechmann S., Chen T., Albanèse V., Dammer E. B., Peng J., and Frydman J. (2013) The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harada Y., Li H., Wall J. S., Li H., and Lennarz W. J. (2011) Structural studies and the assembly of the heptameric post-translational translocon complex. J. Biol. Chem. 286, 2956–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., and Moarefi I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 28. Li J., Qian X., Hu J., and Sha B. (2009) Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284, 23852–23859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gazda L., Pokrzywa W., Hellerschmied D., Löwe T., Forné I., Mueller-Planitz F., Hoppe T., and Clausen T. (2013) The myosin chaperone UNC-45 is organized in tandem modules to support myofilament formation in C. elegans. Cell 152, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panigrahi R., Adina-Zada A., Whelan J., and Vrielink A. (2013) Ligand recognition by the TPR domain of the import factor Toc64 from Arabidopsis thaliana. PLoS One 8, e83461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J., Wu Y., Qian X., and Sha B. (2006) Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem. J. 398, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z., Hartl F. U., and Bracher A. (2013) Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 20, 929–935 [DOI] [PubMed] [Google Scholar]
- 33. Schlegel T., Mirus O., von Haeseler A., and Schleiff E. (2007) The tetratricopeptide repeats of receptors involved in protein translocation across membranes. Mol. Biol. Evol. 24, 2763–2774 [DOI] [PubMed] [Google Scholar]
- 34. Young J. C., Hoogenraad N. J., and Hartl F. U. (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 35. Rucktäschel R., Girzalsky W., and Erdmann R. (2011) Protein import machineries of peroxisomes. Biochim. Biophys. Acta 1808, 892–900 [DOI] [PubMed] [Google Scholar]
- 36. Gatto G. J. Jr., Geisbrecht B. V., Gould S. J., and Berg J. M. (2000) Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 [DOI] [PubMed] [Google Scholar]
- 37. Scheich C., Kümmel D., Soumailakakis D., Heinemann U., and Büssow K. (2007) Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 35, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Polier S., Dragovic Z., Hartl F. U., and Bracher A. (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 39. Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 40. Chartron J. W., Gonzalez G. M., and Clemons W. M. Jr. (2011) A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J. Biol. Chem. 286, 34325–34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kabsch W. (2010) XDS. Acta Crystallogr. D, Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D, Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen V. B., Arendall W.B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D, Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 47. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holm L., and Park J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 49. DeLano W. L. (2002) The Pymol Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- 50. Cheng Z., Jiang Y., Mandon E. C., and Gilmore R. (2005) Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 168, 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.