Abstract
The capacity of the liver to regenerate is likely to be encoded as a plasticity of molecular networks within the liver. By applying a combination of comprehensive analyses of the epigenome, transcriptome, and proteome, we herein depict the molecular landscape of liver regeneration. We demonstrated that histone H3 Lys-4 was trimethylated at the promoter regions of many loci, among which only a fraction, including cell-cycle-related genes, were transcriptionally up-regulated. A cistrome analysis guided by the histone methylation patterns and the transcriptome identified FOXM1 as the key transcription factor promoting liver regeneration, which was confirmed in vitro using a hepatocarcinoma cell line. The promoter regions of cell-cycle-related genes and Foxm1 acquired higher levels of trimethylated histone H3 Lys-4, suggesting that epigenetic regulations of these key regulatory genes define quiescence and regeneration of the liver cells. A quantitative proteome analysis of the regenerating liver revealed that conditional protein degradation also mediated regeneration-specific protein expression. These sets of informational resources should be useful for further investigations of liver regeneration.
Keywords: functional genomics, histone methylation, proteomics, regeneration, transcriptomics
Introduction
Recently, an increasing number of patients have been developing primary and metastatic liver cancers (1). Whereas a liver resection is the most curative therapy to improve the prognosis of such patients, the resection of more than half of the liver is often needed for the complete removal of a hepatic tumor. Although both the surgical techniques and the postoperative management have been improved, liver failure is still one of the most severe complications after major hepatectomy (2). Portal vein embolization (PVE),2 a method to stimulate growth of the future remnant liver (not an embolized lobe of the liver), is widely performed preoperatively to prevent postoperative liver failure (3). However, patients who undergo this procedure often fail to achieve a sufficient increase of the remnant liver volume (4). Therefore, it is essential to understand the mechanism of compensatory hypertrophy of the liver.
The liver is known as the metabolic center of the mammalian body and performs many biological functions, such as the metabolism of amino acids, fatty acids, and carbohydrates, and the detoxification of xenobiotics to maintain the homeostasis of the whole body. In addition, the liver is also known as an organ that possesses the capacity for regeneration. Liver regeneration has fascinated the field of regenerative medicine because many other major organs of the human body lack the capacity to undergo a substantial regeneration in the case of a functional deficiency (5). Therefore, the liver has been an attractive organ to investigate the mechanism of tissue regeneration. The method of partial hepatectomy (PH) was described by Higgins and Anderson in 1931 (6) and has been widely applied to the study of liver regeneration. Although more and more knowledge has been accumulated about the liver regeneration after PH (7–9), this model may only clarify the mechanism of postoperative liver regeneration (10). On the other hand, the molecular mechanism by which PVE leads to compensatory hypertrophy of the liver remains largely unclear.
The capacity of the liver to regenerate is likely to be encoded as a plasticity of molecular networks within this organ. Whereas several critical regulators of the liver regeneration, such as NFκB and STAT3, have been reported (11, 12), a bird's eye view of the molecular processes is still lacking. Although several studies have utilized massive molecular profiling approaches (13, 14), little is still known about how changes in one set of molecules would affect another set of molecules. To gain insight into the molecular mechanism of the liver regeneration after PVE, we utilized the portal vein branch ligation (PVBL) technique in mice, which induces a redistribution of the portal blood flow in a manner similar to PVE (15). By applying a combination of comprehensive analyses of the epigenome, transcriptome, and proteome, we herein show the molecular landscape of liver regeneration after PVBL. We demonstrate that histone H3K4 was trimethylated at the promoter regions of many loci, among which cell-cycle-related genes were transcriptionally up-regulated. By a cistrome analysis of genes guided by the transcriptome and epigenome, FOXM1 was found to be the key transcription factor during liver regeneration, whose induction was accompanied by a new acquisition of H3K4me3 at its promoter region. Our results indicate that liver regeneration involves coordinated alterations in both transcriptional and epigenetic regulations.
Results
Coordinated transcriptome alterations following PVBL
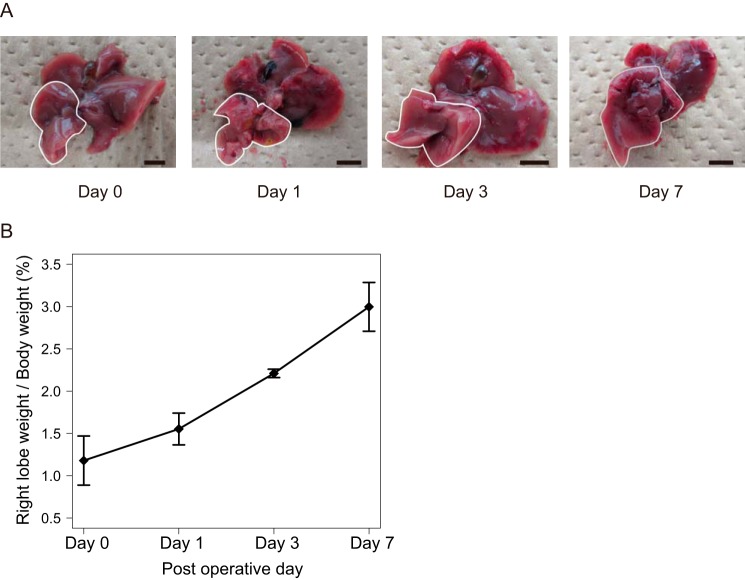
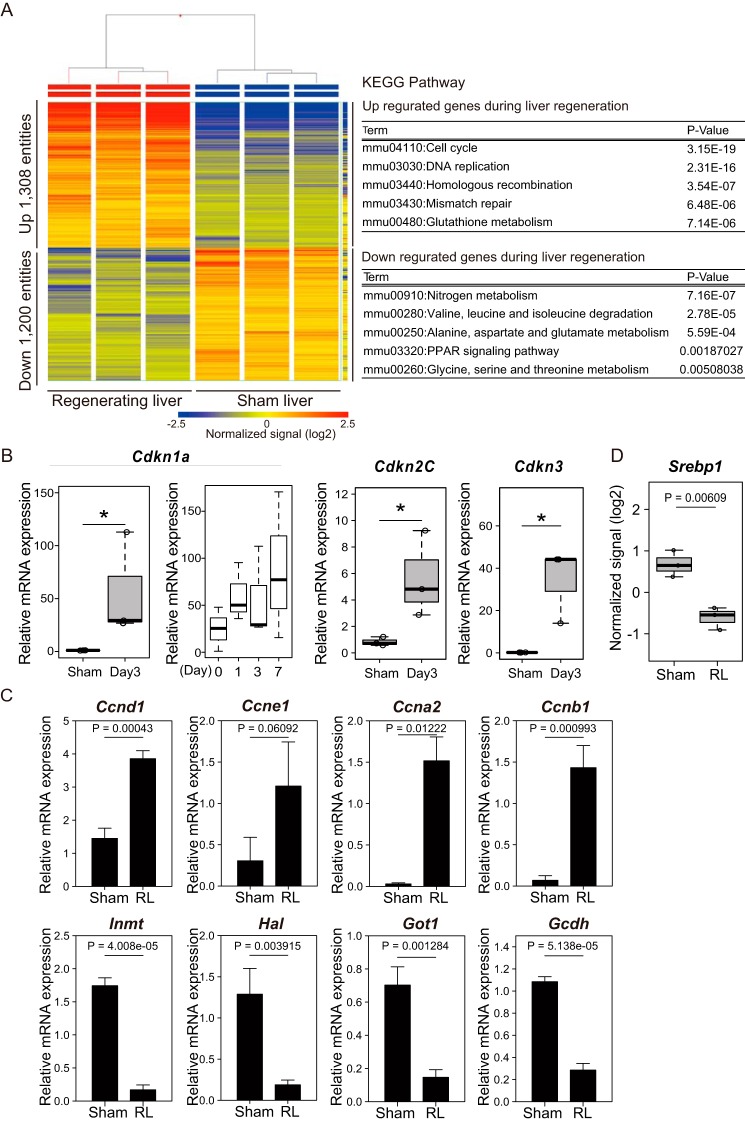
To investigate the mechanism of liver regeneration after PVBL, we performed the PVBL technique on 9–12-week-old male C57BL/6J mice, as described previously (15). Following PVBL, the right lobe of the liver, which was not ligated, gradually enlarged in a time-dependent manner, whereas the ligated lobe became atrophic (Fig. 1). The gene-expression profile during the liver regeneration was determined using microarrays. Because it was reported previously that the cell proliferation state was most activated at day 3 after surgery by this operation in wild-type mice (15), the regenerating liver (RL) at this time point was used. In total, 1,308 and 1,200 entities were significantly up- and down-regulated by > 2-fold, respectively, compared with those in the livers from sham-operated mice (corrected p value < 0.05; Fig. 2A, left).
Figure 1.
Mouse PVBL model. A, macroscopic observation of whole liver. Regenerating liver is indicated with white circles. Scale bars, 5 mm. B, line chart shows the score of right lobe of liver weight/whole body weight (percentage) at the indicated time points following PVBL. The mean values and S.D. (error bars) of the results from 3 mice/group are shown.
Figure 2.
Analysis of transcriptomic changes during liver regeneration. A, the heat map represents the mRNA expression profiles of the regenerating and sham livers at day 3 after the operation (n = 3 mice/group). The tables show the results of KEGG pathway analysis and list the top five pathway terms sorted by p values for up- and down-regulated genes. The color key indicates normalized signal value on a log2 scale. B and C, the expression of the indicated genes was analyzed by RT-qPCR analysis. The mRNA quantities were normalized to Actb (β-actin) mRNA in each sample. Sham, sham liver, respectively. B, the box-and-whisker plots show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. Statistical significance was determined using the Mann-Whitney U test, and asterisks indicate p < 0.05. Expression of Cdkn1a was also analyzed at the indicated time points. C, the mean values and S.D. (error bars) are shown (n = 3/group). Statistical significance was determined using Student's t test, and p values are indicated. D, box plot shows the mRNA expression levels of Srebp1 extracted from the microarray data of A. The box-and-whisker plots show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. Statistical significance was determined using Student's t test, and p values are indicated.
To compare the functions of these up- and down-regulated genes, a KEGG pathway analysis was performed using DAVID bioinformatic resources (16, 17). The terms “Cell cycle” and “DNA replication” emerged significantly often in the analysis of the up-regulated genes (Fig. 2A, right). Importantly, genes involved in not only progression but also cessation of the cell cycle, such as Cdkn1a and Cdkn3 (encoding p21 and KAP, respectively), were induced in RL (Fig. 2B). Interestingly, the pathway terms that were thought to be important for liver functions, such as “nitrogen metabolism,” emerged significantly often in the analysis of the down-regulated genes (Fig. 2A, right). The mRNA expression of genes related to the cell cycle and the metabolic functions of the liver was validated by an RT-qPCR analysis (Fig. 2, B and C). Cyclins important for the G1/S stage (Ccnd1 and Ccne1) and the G2/M stage (Ccna2 and Ccnb1) as well as Cdkn1a were up-regulated, indicating that liver regeneration is programmed to stop proliferation after certain rounds of proliferation. The expression of Cdkn1a was further increased to the later time point (Fig. 2B), suggesting that the level of Cdkn1a expression at day 3 allowed transient proliferation of hepatocytes. Induction of Cdkn1a expression in a partial hepatectomy model was reported previously (18–20). On the other hand, genes encoding enzymes involved in the pathways of amine metabolism (Inmt), amino acid metabolism (Hal), carbon metabolism (Got1), and fatty acid degradation (Gcdh) were down-regulated. One of the key transcription activators of lipid-related genes, SREBP1 (SREBF1), was also reduced in its expression (Fig. 2D). These results indicate that, during the liver regeneration after PVBL, the regenerating liver suppressed some of the metabolic functions and activated genes related to cell proliferation to achieve efficient and rapid regeneration. They also suggest the presence of the coordinated regulation of functionally related genes at the transcriptional level.
Increased trimethylation of histone H3K4 in the regenerating liver
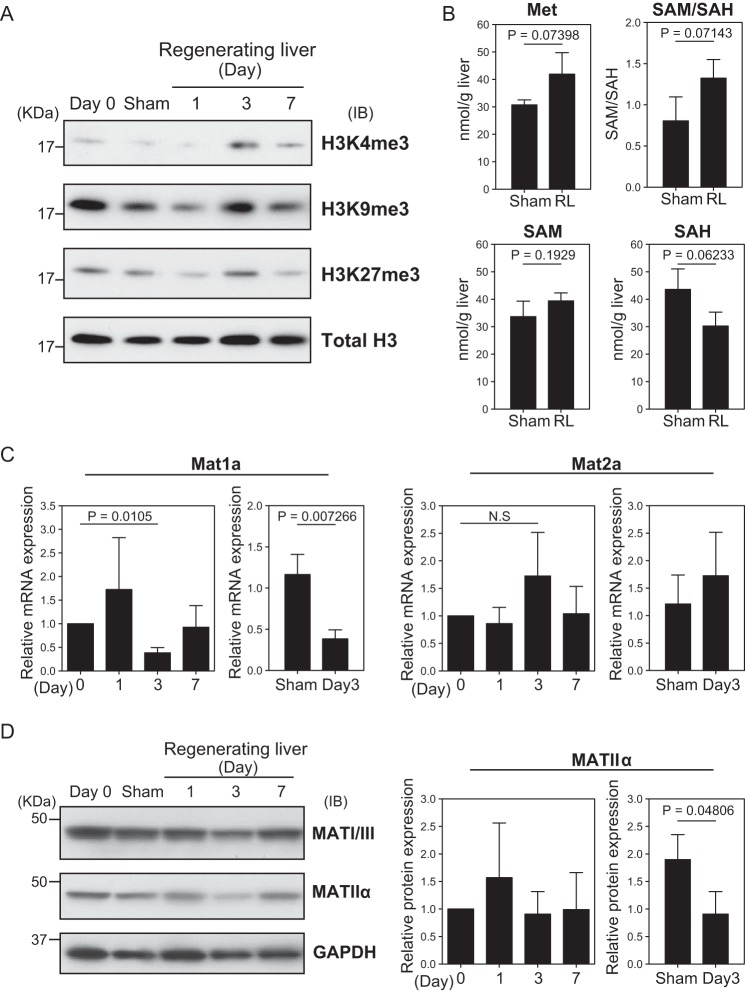
To analyze whether the change of mRNA expression was accompanied by an alteration of histone H3 methylation, histone extracts were prepared from frozen liver tissues at various time points after PVBL, and immunoblotting analysis was performed. We found that the trimethylation of H3K4 (H3K4me3) was strongly induced at day 3 after PVBL compared with that at the other time points (Fig. 3A). The levels of other modifications of histone methylation were not altered as substantially as H3K4me3.
Figure 3.
Analysis of the state of histone H3 methylation and quantification of SAM cycle metabolites. A, immunoblotting analysis of the histone extracts of livers. The livers were isolated from non-treated mice (day 0), those with sham operation (day 3), or those with PVBL at the indicated time points. Immunoblots were performed with antibodies against H3K4me3, H3K9me3, H3K27me3, and total histone H3 as a loading control. Similar results were obtained in three independent experiments. B, the amounts of methionine, S-adenosylmethionine, S-adenosyl-l-homocycteine, and SAM/SAH ratio of the liver. The mean values and S.D. of the results from three mice in the indicated conditions are shown. Statistical significance was determined using Student's t test, and p values are indicated. C, the expression of Mat1a and Mat2a at the indicated time points. The mRNA expression was normalized to Actb mRNA in each sample and is represented as a relative value with regard to that in the day 0 sample. The mean values and S.D. of the results from three mice are shown. Statistical significance between groups was determined using Student's t test, and p values are indicated. N.S, not significant. D, immunoblotting analysis of the whole lysates of regenerating livers. Immunoblots were performed with antibodies against MATI/III, MATIIα, and GAPDH as loading control. The bar plots represent the mean and S.D. (error bars) from the relative intensity of MATIIα protein normalized with GAPDH protein intensity in three mice for each time point. Statistical significance was determined using Student's t test, and the p value is indicated.
Because S-adenosylmethionine (SAM) is the methyl donor of histone methylation, we quantified the concentrations of SAM, methionine, and S-adenosyl-l-homocysteine (SAH) to evaluate the methylation potential of the regenerating liver. The amount of SAM was almost unchanged, but the amounts of methionine and SAH tended to increase and decrease, respectively, in the regenerating liver (Fig. 3B). These alterations resulted in an increase in methylation potential (SAM/SAH ratio; Fig. 3B). These results are consistent with the increased level of global H3K4me3 modification in the regenerating liver, but further studies are required to examine their cause-effect relationship.
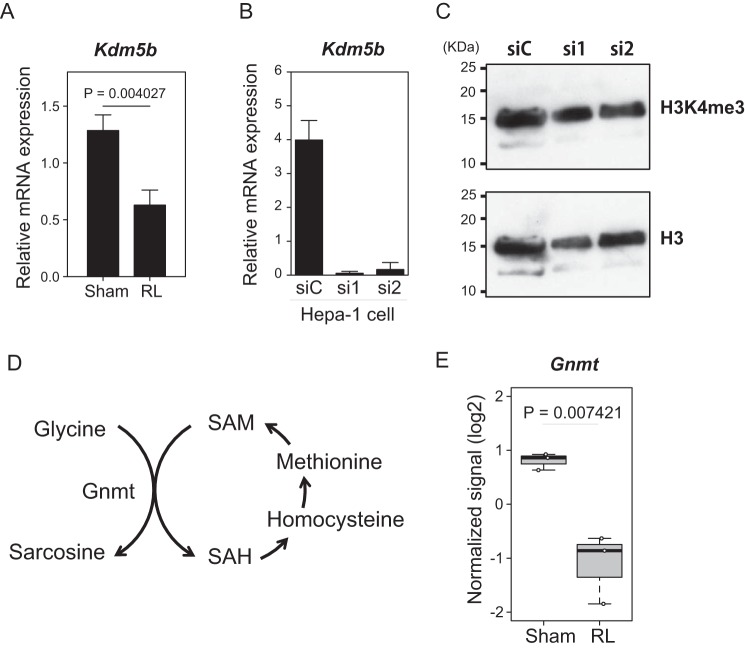
SAM is produced through a reaction catalyzed by methionine adenosyltransferase (MAT). It was reported that liver-specific Mat1a is down-regulated, whereas Mat2a is up-regulated at the transcriptional level during the liver regeneration after PH (21). As expected, the level of Mat1a mRNA was significantly decreased at day 3 after the surgery (Fig. 3C). This observation was confirmed by a comparison with the sham-operated group. On the other hand, Mat2a mRNA did not show any significant alteration. For a further confirmation of these findings, immunoblotting analysis was carried out (Fig. 3D). Consistent with the mRNA levels, MATI/III was transiently decreased at day 3. MATIIα, the catalytic subunit of MATII encoded by Mat2a, was also decreased at day 3 (Fig. 3D). This might be due to an alteration of the stability of the MATIIα protein in the regenerating liver, but the underlying mechanism remains unclear. These observations strongly suggest that post-PVBL regeneration does not involve the switching of MATI/III and MATIIα. Although the level of its protein did not increase, the nuclear localization of MATIIα (22) may be enhanced upon PVBL to support histone methylation. Interestingly, the mRNA expression of Kdm5b, which is a demethylase of H3K4, was decreased in the regenerating liver, as determined by the microarray analysis and an RT-qPCR analysis (Fig. 4A). To examine its function, we depleted Kdm5b in Hepa1 cells (Fig. 4B). However, the level of H3K4me3 was not appreciably affected in immunoblotting analysis (Fig. 4C). It remains an open question whether the suppressed expression of Kdm5b contributed to the induction of H3K4me3.
Figure 4.
Expression of Kdm5b and Gnmt during liver regeneration. A, mRNA expression of Kdm5b. The mRNA quantities were normalized to Actb mRNA in each sample. The mean values and S.D. (error bars) of the results from three mice are shown. Statistical significance was determined using Student's t test, and p values are indicated. B, effects of the indicated siRNAs targeting Kdm5b (si1 and si2) or control siRNA (siC) in Hepa1 cells. C, immunoblotting analysis of the histone extracts of Hepa1 cells treated with the indicated siRNAs using antibodies specific for histone H3K4me3 (top) or bulk H3 (bottom). D, reaction by Gnmt in SAM metabolism. E, box plot shows the mRNA expression levels of Gnmt extracted from the microarray data of Fig. 2 and depicted as in Fig. 2.
A recent study has shown that glycine N-methyltransferase consumes a large fraction of SAM under normal conditions to restrict the amount of SAM in a cell (Fig. 4D) (23). We found that the levels of Gnmt mRNA for this enzyme were reduced in the RL (Fig. 4E). Therefore, the reduction of glycine N-methyltransferase in the RL may promote histone methylation by redirecting SAM away from the damping pathway.
Pervasive H3K4me3 marking for proliferation-related and other categories of genes
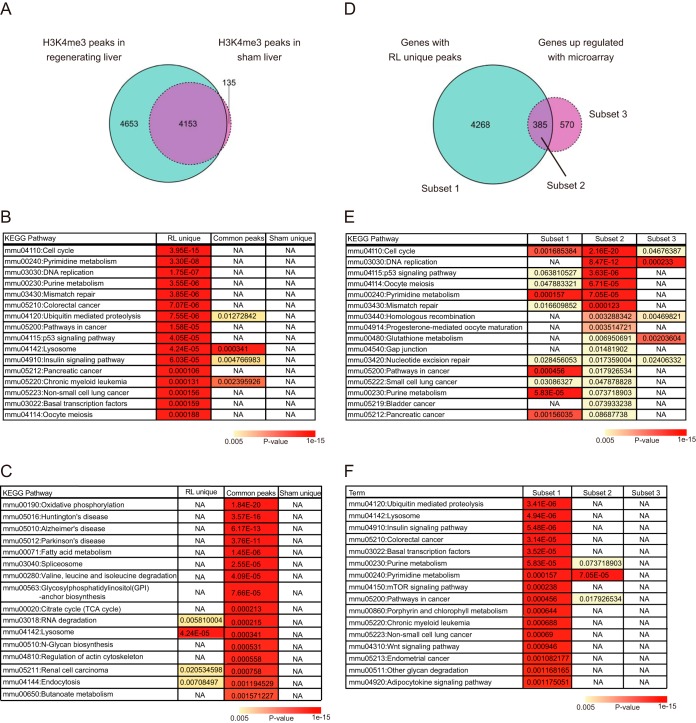
The above immunoblotting analysis revealed that global H3K4me3 was significantly increased during liver regeneration at day 3 after PVBL. To identify the genes that newly acquired the H3K4me3 modification, a ChIP-seq analysis was performed using an anti-H3K4me3 antibody. We identified genes with H3K4me3 peaks at promoters by using the MACS2 peak-calling algorithm, and the union gene sets with called peaks in the biological duplicate were defined as the genes with H3K4me3 peaks. We classified these genes with H3K4me3 peaks into three categories: unaltered under the two conditions (4,153 peaks), specifically detected in the sham condition (135 peaks), or specifically detected after PVBL (4,653 peaks) (Fig. 5A). To evaluate the functional differences between these subsets of peaks, a pathway analysis was carried out using a list of their nearest genes. Compared with the genes with common peaks and with sham unique peaks, the genes with the peaks unique to the RL showed a significant enrichment of pathway terms related to the cell cycle and cell proliferation (Fig. 5B). In contrast, the genes with common peaks showed an enrichment of pathway terms related to various metabolic functions (Fig. 5C). To analyze the relationship between H3K4me3 modification and gene expression, we integrated the ChIP-seq and microarray data sets. Interestingly, among the genes with H3K4me3 peaks unique to RL, only 8.3% (385 of 4,653 genes) were transcriptionally up-regulated (-fold change > 2.0), and most of the genes did not show an alteration in their expression during the liver regeneration (Fig. 5D). These three subsets were used to perform a pathway analysis. The biological pathways contributing to the liver regeneration, such as “cell cycle” and “DNA replication,” were enriched mainly in the analysis of subset 2, which was comprised of the genes with peaks unique to RL and increased mRNA expression (Fig. 5E). In contrast, the genes with peaks unique to RL but that were not induced did not show enrichment of pathway terms related to the cell cycle process (Fig. 5F). These results suggested that many genes acquired the H3K4me3 modification during the liver regeneration, among which those with functions associated with cell proliferation were selectively up-regulated at the level of mRNA transcription. Our data raise the possibility that epigenetic modifications such as H3K4me3 were necessary but not sufficient for the regulation of gene expression during the liver regeneration. Additional mechanisms may be layered upon the pervasive H3K4me3 modification.
Figure 5.
The ChIP-seq analysis of histone H3K4me3. A–C, the number of genes with H3K4me3 peaks in the indicated samples at day 3 after operation (A) and the results of KEGG pathway analysis (B and C). The Venn diagram represents the number of genes with H3K4me3 peaks. The blue and purple circles indicate genes in regenerating liver and sham liver, respectively. The tables represent the KEGG pathway terms significantly enriched in genes with the RL unique peaks. Significance of enrichment of the pathway terms in genes with common and sham-unique peaks is also shown. NA, pathway terms that are not significantly enriched. D–F, comparison between genes with RL unique H3K4me3 peaks (4,653 genes) and those transcriptionally up-regulated in regenerating livers (both at day 3 after operation). The Venn diagram represents the number of genes in each subset (E). The results of KEGG pathway analysis of subsets 2 and 1 are shown in E and F, respectively, with p values of the enriched pathways in other subsets.
The association of histone methylation patterns with functional gene groups
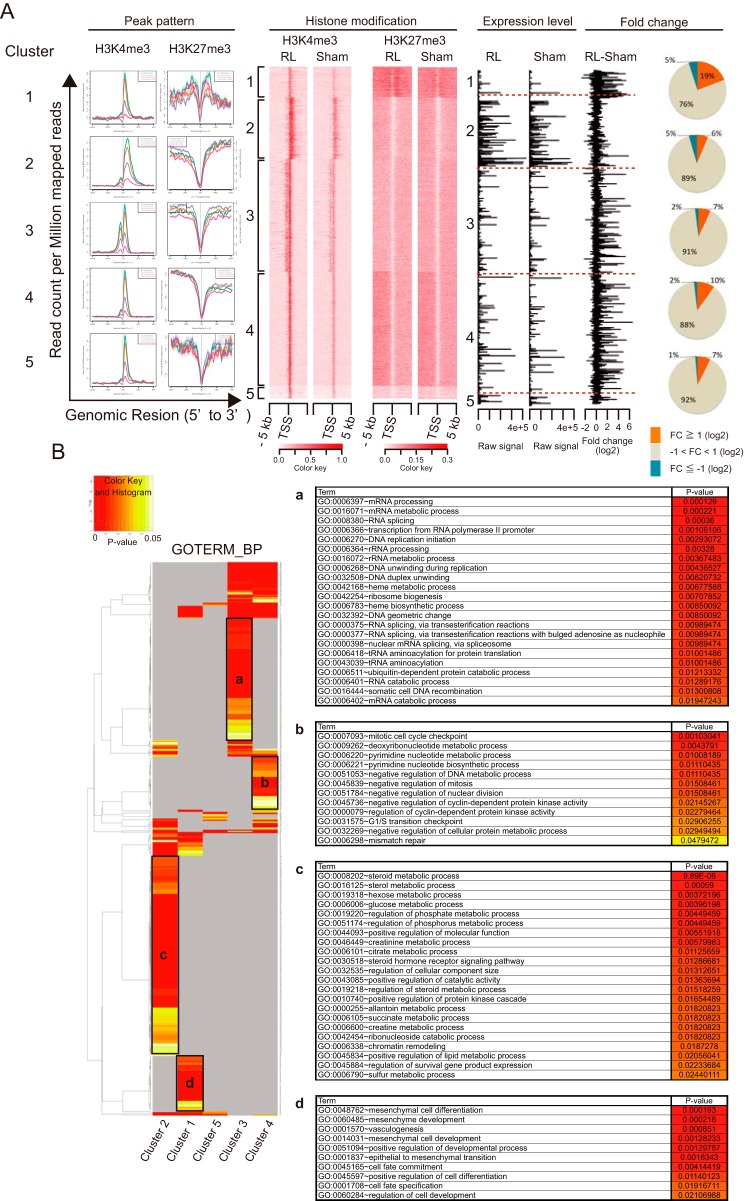
To further classify the genes with H3K4me3 peaks unique to the RL, we added a ChIP-seq analysis using the anti-H3K27me3 antibody. The combined patterns of H3K4me3 and H3K27me3 modifications analyzed by a K-means clustering algorithm classified these genes into five clusters (Fig. 6A). We then analyzed the relationship between the patterns of histone methylation and the mRNA expression profiles. Interestingly, clusters 1 and 2 were clearly distinguished by the patterns of mRNA expression. Cluster 1, which was characterized by positivity for both H3K4me3 and H3K27me3, was relatively enriched with genes for which the expression was significantly increased in the RL compared with that in the sham liver, but their mRNA raw expression values were relatively low (Fig. 6A, Expression level and Fold change). In contrast, cluster 2 was not particularly enriched with up-regulated genes, but their expression levels were considerably higher than those in the other clusters. Whereas there was a slight difference between clusters 3 and 4 in terms of their patterns of mRNA expressions, their patterns of H3K4me3 peaks were clearly different; cluster 3 revealed bimodal patterns of H3K4me3 peaks around transcription start sites, whereas cluster 4 revealed narrow H3K4me3 peaks. These results indicate that there is a distinct correlation between the patterns of histone methylation and of mRNA expression, which may reflect distinct regulatory mechanisms.
Figure 6.
Classification of genes according to the patterns of histone H3 methylation. A, the genes with H3K4me3 peaks unique to regenerating liver were classified by the patterns of H3K4me3 and H3K27me3 methylation. Line charts represent the pattern of H3K4me3 and H3K27me3 peaks of each cluster. A heat map also represents the patterns of histone methylation, and bar plots represent the raw expression values and -fold change of mRNA expression as revealed by microarray analysis. Pie charts show the proportion of up- and down-regulated genes in each cluster. B, GO analysis of genes in each cluster. The heat map represents p values in the color key of the GO terms enriched in each cluster, and the black boxes indicate the GO terms uniquely enriched in each cluster. A portion of such GO terms characterizing each cluster are shown in the tables on the right (a–d) with p values.
To analyze the functional similarities and differences among the above clusters of genes, a GO analysis was performed (Fig. 6B). This analysis revealed differences in gene functions between clusters 1 and 2. The GO terms enriched in cluster 1 included the terms related to the development of various organs and pluripotency, such as “mesenchymal cell differentiation,” “vasculogenesis,” and “cell fate commitment.” It was reported that bivalent domains, consisting of large regions of H3K27me3 harboring smaller regions of H3K4me3, frequently overlay developmental genes in embryonic stem cells (24). The results from the analysis of cluster 1 suggested that genes within such bivalent domains were activated upon the shifting of the liver to a dedifferentiated state to regenerate. A GO analysis of cluster 2 revealed the enrichment of terms related to metabolic function. The presence of broad H3K4me3 peaks toward the 3′ end may be important for the expression of these genes at higher levels compared with the genes in other clusters. Specifically enriched GO terms in cluster 3 pointed to processes related to nucleotide metabolism, such as “mRNA processing” and “mRNA metabolic process,” and those in cluster 4 were terms related to cell cycle checkpoints, such as “mitotic cell cycle checkpoint” and “G1/S transition checkpoint.” The GO terms related to a regenerating state, such as “cell cycle” and “cell cycle process,” were enriched in the analyses of clusters 1, 3, and 4, suggesting that the genes classified into these clusters would contribute to the proliferation of liver cells. By combining the ChIP-seq and microarray analyses, we succeeded in revealing that, during the liver regeneration, the genes of multiple regulatory pathways were activated by distinct mechanisms, which can be stratified according to the patterns of histone methylation.
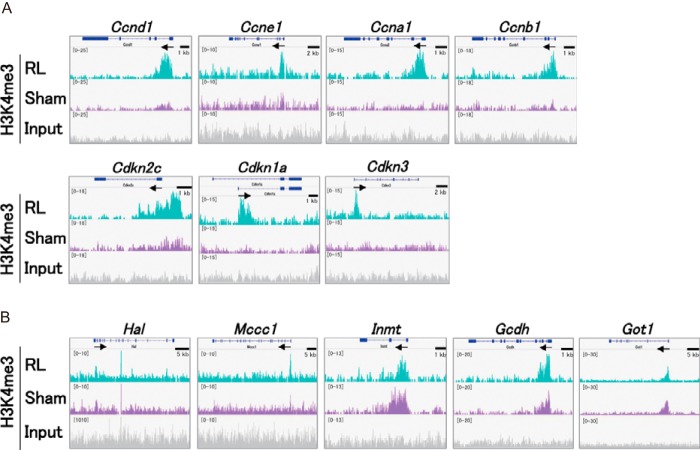
This idea was further validated by analyzing individual genes that were identified in the GO analysis. Cell cycle regulator genes acquired high levels of H3K4me3 around their promoter regions, consistent with their induction upon regeneration (Fig. 7A). In contrast, metabolism-related genes retained H3K4me3 although their expression was reduced upon regeneration (Fig. 7B). These observations suggest two mechanisms in the liver. First, the absence of H3K4me3 at the cell cycle-related genes ensures that liver cells will not start proliferation under normal conditions. Second, the maintenance of H3K4me3 at the metabolism-related genes during regeneration ensures a rapid re-expression of the liver-specific functions once regeneration stops.
Figure 7.
Patterns of H3K4me3 in regenerating liver. A and B, genomic track display for H3K4me3 across the cell-cycle gene loci (A) and metabolism-related gene loci (B). Sham, sham liver. The RefSeq gene tracks are shown above.
The integration of the cistrome and the transcriptome points to Foxm1 as the regulator of regeneration
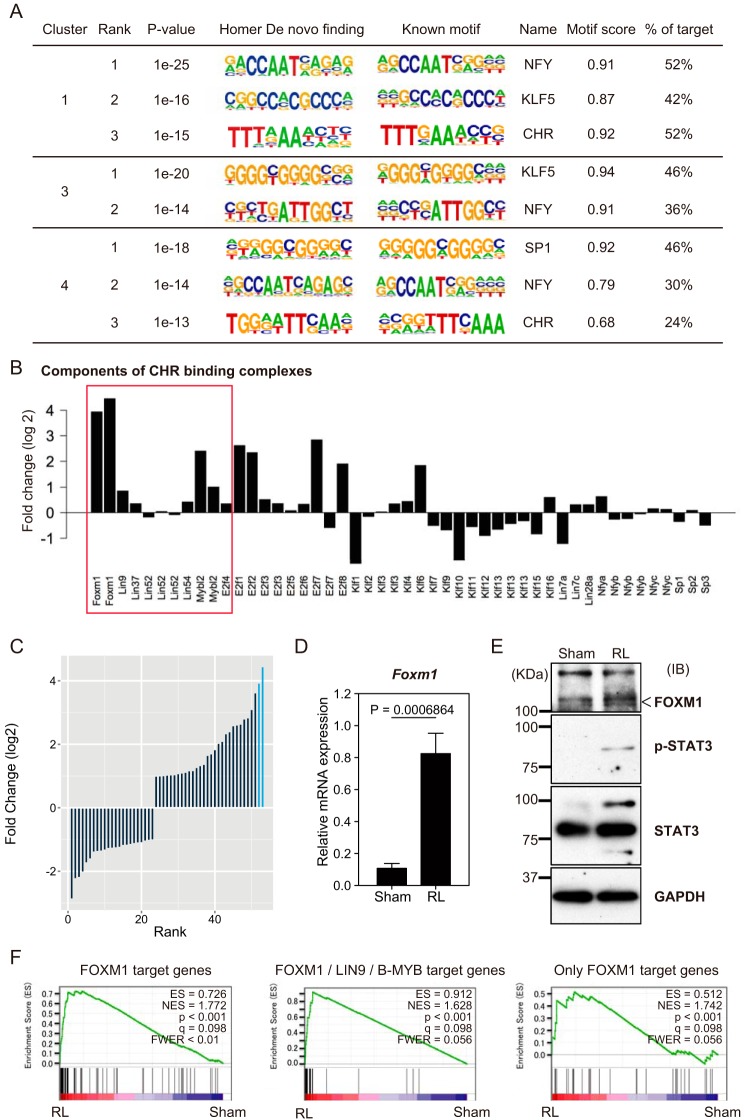
The results of the GO analysis (Fig. 6B) suggested that the clusters 1, 3, and 4 contained genes related to the cell cycle and might be central to the proliferation of liver cells. To investigate whether specific regulatory DNA sequences were enriched at the promoter regions of the genes with H3K4me3 peaks unique to the RL, a motif analysis of the promoter regions (i.e. cistrome) was performed using the HOMER algorithm (25). We identified the NFY, KLF5, cell cycle gene homology region (CHR), and SP1 motifs by separately analyzing these clusters (Fig. 8A). It was previously reported that the CHR is bound by a complex of FOXM1, B-MYB, and MuvB (FOXM1-MMB), resulting in the expression of genes important for the progression of the G2/M phase of the cell cycle (26). Interestingly, by analyzing the microarray data, we found that Mybl2 (which encodes B-MYB), Lin9 (which is a component of the MuvB complex), and Foxm1 were transcriptionally up-regulated in the regenerating liver (Fig. 8B). Moreover, among the DNA-binding factors with significant changes in their transcriptional level as determined by the microarray, Foxm1 showed the highest degree of induction (Fig. 8C, blue bars). We confirmed the mRNA expression of Foxm1 by an RT-qPCR analysis (Fig. 8D). An immunoblotting analysis showed an increase of Foxm1 protein (Fig. 8E). A recent report has shown that Foxm1 is the target of the STAT3 transcriptional activator in K562 cells (27). We found that the amounts of STAT3 and its phosphorylated form were increased in the regenerating liver (Fig. 8E). These results suggested the possibility that STAT3 enhanced the expression and activation of FOXM1, which activated the target genes in the regenerating liver by forming the FOXM1-MMB complex. Using the gene lists from the ChIP-seq experiments of LIN9 and B-MYB performed in proliferating HeLa cells (28) and FOXM1 in U2OS cells (26), a gene set enrichment analysis was performed to investigate whether the FOXM1 target genes were activated transcriptionally during the liver regeneration following PVBL. We found that these gene sets were indeed activated in the regenerating liver (Fig. 8F).
Figure 8.
Motif analysis of promoter regions with H3K4me3 peaks unique to regenerating liver. A, motif analysis of promoter regions of genes acquiring H3K4me3 peaks during liver regeneration using HOMER. Each cluster indicates the gene subset classified in Fig. 5, and the top two or three de novo findings are shown. Known motifs and their names are also indicated. B, bar plots represent the transcriptional changes of DNA-binding transcription factors related to the motifs enriched in Fig. 6A. The red box indicates the components of CHR binding complexes. C, bar plots represent the transcriptional changes of DNA binding transcription factors that were significantly altered by the microarray analysis. Blue bars, alterations of Foxm1. D, mRNA expression of Foxm1 by RT-qPCR analysis. The mRNA quantities were normalized to Actb mRNA in each sample. The mean values and S.D. (error bars) of the results from three mice are shown. Statistical significance was determined using Student's t test, and p values are indicated. E, immunoblotting analysis of the whole lysates of regenerating liver and sham liver. Immunoblots were performed with antibodies against Foxm1, STAT3, phospho-STAT3 (Tyr-705), and GAPDH as loading control. F, gene set enrichment analysis to test whether the FOXM1 target genes were randomly distributed or tended to occur toward the extremes of the list of genes that were differentially expressed during liver regeneration. The left panel shows the analysis using all FOXM1 target genes as a gene set; the middle shows the analysis using the genes with ChIP-seq peaks of FOXM1, LIN9, and B-MYB; and the right shows the analysis using the genes with only FOXM1 peaks (26). Enrichment score (ES), normalized enrichment score (NES), nominal p value (p), false discovery rate (q), and familywise error rate (FWER) are indicated.
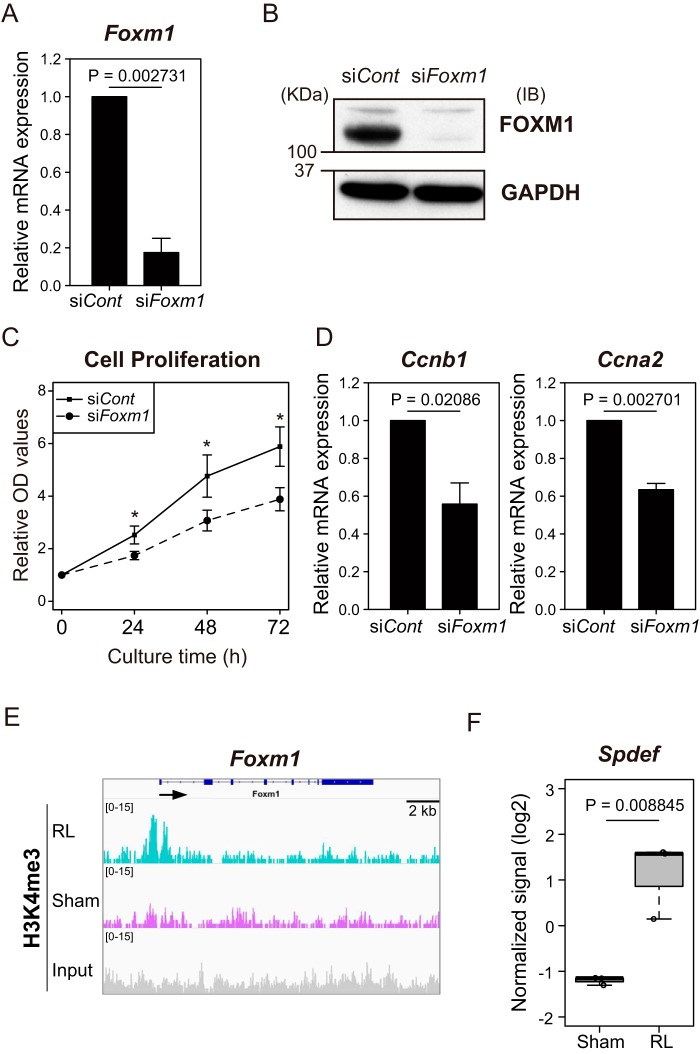
To examine the function of FOXM1 in liver cell proliferation, we performed the knockdown of Foxm1 using Hepa1 cells. Using the Foxm1 siRNA, we could achieve a roughly 80% reduction of Foxm1 mRNA expression (Fig. 9A). We also confirmed the decrease of the level of FOXM1 protein amount by immunoblotting analysis (Fig. 9B). We then carried out a cell proliferation assay to determine whether knocking down Foxm1 would affect the cell proliferation. The growth curves showed that the cell proliferation of Hepa1 cells transfected with siFoxm1 was inhibited significantly compared with that of the cells transfected with siControl (Fig. 9C). Furthermore, we found that the amounts of mRNA of FOXM1 target genes Ccnb1 and Ccna2 were decreased by knockdown of Foxm1 (Fig. 9D), suggesting the role of FOXM1 in the activation of these genes. It has been reported that the overexpression of the FOXM1 protein is associated with aggressive tumor features and a poor prognosis of hepatocellular carcinoma (29) and that the silencing of FOXM1 inhibits hepatocellular carcinoma growth (30). Furthermore, FOXM1 is required for DNA replication and mitosis after partial hepatectomy (31). Together with these previous reports on FOXM1, our observations strongly suggest that FOXM1 promotes cell proliferation by activating its target genes related to the cell cycle progression in hepatocytes.
Figure 9.
The effect of Foxm1 knockdown on hepatoma cell proliferation. A, the relative mRNA expression of Foxm1 was analyzed by RT-qPCR analysis using Hepa1 cells transfected with siControl or siFoxm1 at 24 h following transfection. The mRNA expression was normalized to Actb mRNA in each sample and represented as relative value with regard to that in the siControl sample. The mean values and S.D. (error bars) of the results from three independent experiments are shown. Statistical significance was determined using Student's t test, and p values are indicated. B, immunoblotting analysis of the whole-cell lysates of Hepa1 cells transfected with the indicated siRNAs at 24 h following transfection. Immunoblots were performed with antibodies against FOXM1 and GAPDH as loading control. C, growth curves were measured with Hepa1 cells transfected with the indicated siRNAs. A total of 5,000 cells were seeded in each well (in quadruplicate) of a 96-well culture plate. The absorbance value at 450 nm was measured at the indicated time points and represented as relative value with regard to that in the 0 h sample. The mean values and S.D. are shown, and statistical significance was determined using Student's t test; *, p < 0.05, relative to the siControl at the same time points. D, the relative mRNA expression of the indicated genes was analyzed by RT-qPCR analysis using Hepa1 cells transfected with siControl or siFoxm1 at 24 h following transfection. The mRNA expression was normalized to Actb mRNA in each sample and represented as relative value with regard to that in the siControl sample. The mean values and S.D. of the results from three independent experiments are shown. Statistical significance was determined using Student's t test, and p values are indicated. E, genomic track display for H3K4me3 across around the Foxm1 gene. Sham, sham liver. The RefSeq gene track is shown above the profiles. F, the data for Spdef expression level are from the microarray data of Fig. 1A and are shown as box plots. The box-and-whisker plots show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. Statistical significance was determined using Student's t test, and p values are indicated.
The above multiomics analyses allowed further insights into the mechanisms for the regulation of Foxm1. The ChIP-seq data showed that the promoter region of Foxm1 acquired H3K4me3 during regeneration (Fig. 9E). The absence of H3K4me3 in the normal liver may ensure that the liver cells would not enter into cell cycle under normal conditions. Whereas FOXM1 activates its own expression, the Ets-related transcription factor SPDEF inhibits the Foxm1 promoter activity (32). Interestingly, the mRNA expression of SPDEF was induced in the RL (Fig. 9F), suggesting that Foxm1 expression is eventually silenced by SPDEF upon completion of the liver regeneration.
The liver regeneration involved post-transcriptional regulation of the proteome
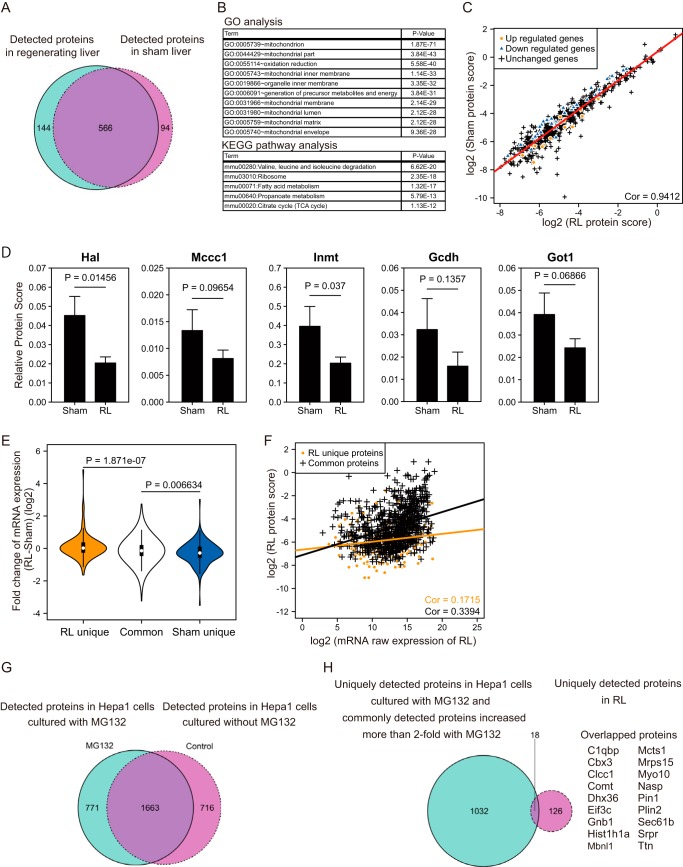
Functions of cells and organs involve post-transcriptional regulation as well. To directly compare proteome, a proteomic analysis was performed using nano-HPLC-MS/MS. To obtain quantitative data, we avoided the fractionation of the liver extracts before the MS/MS analysis. This protocol therefore resulted in much lower numbers of proteins detected in the current analysis than reports by others using normal livers (33). We measured the total protein extracts from the livers of three mice for each condition. By applying a conservative criterion for the analysis, the proteins detected in all of the biological triplicate samples in each condition were used for the further analysis. We detected and quantified 566 proteins in the two conditions (Fig. 10A). In addition, we found 144 and 94 proteins that were detected only in one of the two conditions. Next, we performed GO and pathway analyses to evaluate the functions of the commonly detected proteins. Consistent with a report describing that the majority of abundant proteins in the liver are metabolic enzymes involved in different biological processes (33), GO and pathway analyses of the commonly detected proteins suggested their functions in the mitochondria and/or in various metabolic pathways (Fig. 10B). To compare the amounts of individual proteins between the RL and sham livers, we calculated the quantitative scores of the commonly detected proteins. The relative amounts of the individual proteins were similar between the two conditions (Pearson's correlation coefficient, r = 0.94; Fig. 10C). However, proteins whose mRNA levels were altered in the microarray analysis tended to show alterations that were consistent with their mRNA (Fig. 10C). For example, several enzymes participating in the pathways of amino acid metabolism (Hal, Mccc1), amine metabolism (Inmt), fatty acid degradation (Gcdh), and carbon metabolism (Got1) were decreased in the regenerating liver (Fig. 10D). To evaluate further the relationship between the variations of mRNA expression and the amounts of proteins, we categorized the proteins into three groups, proteins specific to the RL group, those identified in both conditions, and those specific to the sham group, and compared their mRNA levels. Compared with the variations of mRNA expression of the commonly detected proteins, the levels of mRNA of proteins detected uniquely in the RL or the sham group showed larger changes (Fig. 10E). These results confirmed that many of the observed alterations at the transcriptional level were actually reflected at the protein level.
Figure 10.
Analysis of protein expression profiles during liver regeneration by nano-HPLC-MS/MS. A, the Venn diagram represents the number of proteins detected in each condition. B, GO and KEGG pathway analysis of proteins detected in both conditions. C, scatter plot depicting protein scores of detected proteins in the regenerating and sham livers. The Pearson correlation coefficient is indicated. The regression line is shown as a red line. Orange dots and blue triangles indicate proteins whose corresponding mRNA expression was altered significantly in the microarray analysis in Fig. 2, and black crosses indicate proteins without transcriptional change. D, bar plots represent the relative protein scores of enzymes involved in various metabolic functions (Hal, histidine ammonia-lyase; Mccc1, methylcrotonoyl-CoA carboxylase 1; Inmt, indolethylamine N-methyltransferase; Gcdh, glutaryl-CoA dehydrogenase; Got1, glutamic-oxaloacetic transaminase 1). The mean values and S.D. (error bars) of three independent experiments are shown. Statistical significance was determined using the Student's t test, and p values are indicated. Sham, sham liver. E, violin plots represent the -fold changes of mRNA of proteins detected only in regenerating livers or sham livers or in both. Statistical significance was determined by Student's t test, and p values are indicated. F, scatter plot depicting the raw expression scores of mRNA and protein scores in regenerating liver. Proteins that were commonly detected in the two conditions or uniquely detected in RL are shown as a black or orange line, respectively. Their regression lines are indicated with the respective colors. Values of the Pearson correlation coefficient are also shown. G, the Venn diagram represents the number of proteins detected in Hepa1 cells. Hepa1 cells were treated with or without MG132 for 12 h before extraction of proteins. H, the Venn diagram represents the comparison of the RL-unique proteins and MG132-induced proteins. For the latter set, we combined proteins uniquely detected in MG132-treated Hepa1 cells (771 proteins) and proteins that showed > 2-fold increase in response to MG132 (279 proteins). Proteins in the intersection subset were listed alongside.
To analyze the correlations between the transcriptome and the proteome in the RL, we compared the protein scores and mRNA raw signals from our microarray data. The Pearson's correlation coefficient score of commonly detected proteins was 0.34 (Fig. 10F), which was lower than in previous reports showing global correlations between the transcriptome and the proteome to be around the range of 0.4–0.6 (34, 35). We noted that the correlation score of the proteins uniquely detected in RL was much lower than that of the commonly detected proteins (Fig. 10F). These observations suggest that post-translational regulation may also have contributed to the alteration of the proteome during the liver regeneration.
To elucidate whether the ubiquitin-proteasome system affected the proteins detected uniquely in the RL, Hapa1 cells were cultured with or without the proteasome inhibitor MG132, and a proteomic analysis was performed. We first identified 771 proteins uniquely detected from Hepa1 cells treated with MG132 (Fig. 10G). In addition, we identified 279 from 1,663 proteins detected under both conditions whose scores as a proxy of protein amount differed by > 2-fold from those of the untreated cells. A substantial part of the list of 1,050 proteins overlapped with proteins uniquely detected in the RL (18 of 144) (Fig. 10H; p = 3.1 × 10−9 by a hypergeometric test). This observation suggested the possibility that some of the proteins detected uniquely in the RL were regulated at the level of protein stability. Such a post-translational control may contribute at least in part to the apparently weaker correlation between the protein scores and the mRNA raw expression of the proteins unique to the RL than that of the commonly detected proteins. We concluded that the liver regeneration involved regulation at both the transcriptional and the post-transcriptional levels.
Discussion
We herein studied the molecular state of liver regeneration using the mouse PVBL model, which mimics liver regeneration following PVE in humans. We took advantage of the ability to analyze multilayer omics data covering epigenomic, transcriptomic, and proteomic measurements from identical materials. Our principal idea was that one layer of these data sets would be useful to stratify alterations in the other layers, ultimately leading to the identification of critical regulators and/or molecular alterations. This multiomics analysis allowed us to examine the correlations and relationships among multiple omic data and gave us in-depth insights into liver regeneration. Most importantly, by combining the data sets of histone methylation and gene expression, we identified Foxm1 as a critical regulator of liver regeneration. In addition, we found a set of proteins whose abundance was regulated at the post-translational level during liver regeneration. Because our data sets were derived from whole liver lysates, we were unable to examine the cellular interactions between hepatocytes and hepatic non-parenchymal cells (e.g. liver endothelial sinusoid cells and Kupffer cells). Because the hepatocyte occupies ∼90% of the liver volume, we assume that the alterations in the molecular compositions observed in this study largely reflected changes within hepatocytes. Notwithstanding this limitation, our results clarified the molecular state of the regenerating liver following PVBL and established a valuable database to investigate further the mechanism of liver regeneration.
As expected, genes related to cell proliferation and DNA replication were transcriptionally up-regulated. Using a ChIP-seq analysis, we found that the trimethylation of histone H3K4 was strongly induced at the promoter regions of these genes in the regenerating liver. A previous study demonstrated that H3K4 di- and trimethylations were increased in an up-regulated gene during the liver regeneration following PH (36). However, this previous study did not address the genome-wide distribution of the methylation of H3K4. In addition, our results from the ChIP-seq analysis clearly showed that the induction of H3K4me3 was not restricted to genes that underwent transcriptional up-regulation. Rather, many more genes that were transcriptionally unaltered acquired an increase of H3K4me3 modification compared with those in the livers of the sham-operated group. In other words, not all of the genes that acquired H3K4me3 modification were up-regulated transcriptionally, with only 8.3% of genes significantly up-regulated (-fold change > 2.0). This indicates that the modification with H3K4me3 is not sufficient to activate gene transcription, and other additional factors are needed. One scenario may be that the machinery for the writing of H3K4me3 is rather promiscuous. The increase in methylation potential (SAM/SAH ratio) may also facilitate the methylation reaction. The decrease in the expression of the histone demethylase Kdm5b may contribute to the pervasive induction of H3K4me3. An interesting question in this context is whether Kdm5b is involved in the maintenance of liver cell identity and/or quiescence. Another interesting alteration is the reduced expression of Gnmt, which is expected to lead to a higher flow of SAM into other reactions, including histone methylation. It appears that alterations of metabolic status also facilitate epigenetic remodeling of the regenerating liver.
Our data also suggest that additional proteins, such as DNA-binding transcription factors, may be required to achieve specific gene activation among the vast set of genes that acquired H3K4me3. Consistent with this idea, we found by combining the motif analysis and the microarray analysis that the CHR element and its binding complex FOXM1-MMB play important roles in the regenerating liver. Previous reports have demonstrated that FOXM1 regulates the expression of cell cycle proteins that are essential for reentry of hepatocytes into DNA replication and mitosis after partial hepatectomy (31) and that the FOXM1-MMB complex binds to and activates the cell cycle genes Ccna2 and Ccnb1 that regulate the G2/M phase (37). These genes were significantly up-regulated in our RT-qPCR analysis of regenerating livers, further emphasizing the importance of the FOXM1-MMB complex and the CHR element in the process of liver regeneration. The salient point here is that we identified these factors by the analysis of multiomics data. When we carried out a HOMER analysis using a bulk set of up-regulated genes, CHR was not significantly enriched. Therefore, this method (i.e. stratifying genes using histone modification patterns) is a powerful tool helping us to investigate the molecular mechanisms of cellular responses. Interestingly, by using the top 50 genes up-regulated during the liver regeneration for a motif analysis, we detected a de novo CHR motif in the promoter regions of 24 genes. These genes included those with or without the alteration of H3K4me3 modification and were highly enriched in cell-cycle-related terms by a GO analysis. These results raise the possibility that the up-regulation of cell-cycle-related genes through CHR is controlled by both H3K4me3-dependent and -independent mechanisms. It is important to note that H3K4me3 modification at the Foxm1 promoter region substantially increased during liver regeneration. Therefore, two lines of research will be important in the future: how Foxm1 is regulated epigenetically in the normal and regenerating liver and whether FOXM1 is involved in the epigenetic regulation of the liver regeneration, especially that of the cell cycle-related genes.
The pervasive induction of H3K4me3 during liver regeneration suggests an interesting strategy for a therapeutic intervention. Among the genes that acquired H3K4me3, one may be able to find genes with the ability to promote cell proliferation. Even if they are not induced during the liver regeneration after PVE, such genes may be ready for the induction of their expression in response to an additional, adequate signal. This is an interesting possibility in light of the fact that Yap1, which is critical for liver cell proliferation (38), was among this set of genes. They may also contain cancer-related genes because regeneration and malignant transformation are phenomena with overlapping molecular events (39). Therefore, this set of genes may be interesting to identify hepatoma-related genes as well. The pervasive induction of H3K4me3 during liver regeneration also raises other interesting questions. For example, why were many of these genes not induced at the transcriptional level? Because H3K4me3 is usually written by the COMPASS complex, which is recruited by RNA polymerase II (40), it will be important to compare binding of RNA polymerase II to the promoter and downstream transcribed regions of these genes.
In this study, we found that genes marked with bivalent histone methylation patterns (H3K4me3 plus H3K27me3) were significantly up-regulated at the transcriptional level during liver regeneration. A GO analysis further revealed that the functions of these bivalent genes were related to the development of multiple organs and tissues. These results are consistent with the previously reported functions of bivalent domains in embryonic stem cells (24). In this regard, acquired bivalent domains might enable differentiated cells to dedifferentiate. Considering our observation that a subset of the genes with bivalent histone methylations were up-regulated during the liver regeneration, mature hepatocytes may adopt a quasi-dedifferentiated state following PVBL.
Following PH, several metabolic functions, such as “steroid biosynthesis” and “lipid metabolism,” have been reported to be down-regulated between 2 and 40 h after surgery based on mRNA profiling (13). Our results confirmed that a similar suppression of metabolic functions occurred in the PVBL model. Furthermore, we confirmed this suppression by evaluating the relative protein amounts by a proteomic analysis. Although we did not elucidate the mechanisms underlying the down-regulation of the metabolic functions in the RL, the dedifferentiation of mature hepatocytes may be the reason why functions specific to the liver are down-regulated in the process of regeneration. An interesting candidate that could explain this response is SREBP1 (encoded by SREBF1), the master regulator of diverse metabolism- and lipid-related genes. The reduction of SREBP1 expression in the regenerating liver may contribute to the aforementioned responses. Interestingly, many of the metabolism-related genes were reduced in their expression while retaining H3K4me3. In a previous report using a partial hepatectomy model, metabolism-related genes including SREBF1 have been shown to lose H3K9 acetylation at their promoter regions during regeneration (18). Therefore, dynamic changes in the combinations of histone modifications and SREBP1 activity may regulate transient repression of these genes during liver regeneration. Further studies will be required to clarify the mechanism of the down-regulation of metabolic functions during the liver regeneration. A detailed analysis of the time point when the suppressed metabolic functions recovered at the proteomic level was beyond the scope of this study. However, it is important to determine such a time point because, clinically, it is desirable for hepatic resection to be performed when the full recovery of metabolic function is achieved. The list of metabolic enzymes may help surgeons to identify markers for such an evaluation.
By comparing the data sets of the transcriptome and the proteome, we also succeeded in identifying proteins with increased levels in the regenerating liver after PVBL without any apparent increase of mRNAs. These proteins may be regulated at the post-transcriptional level. The results using Hepa1 cells and MG132 strongly suggest that a fraction of them are regulated by ubiquitination and degradation. Previous reports have demonstrated that some of these proteins are associated with the functions related to cell proliferation. For example, upon the silencing of NASP by siRNA, HeLa cells and U2OS cells are unable to replicate their DNA and progress through the cell cycle (41). In addition, the knocking down of PIN1 in FaDu cells leads to the inhibition of cell proliferation (42). Upon PVBL, ubiquitin E3 ligases for these proteins or their interaction may be inactivated, resulting in the accumulation of target proteins. The list of proteins potentially regulated by conditional ubiquitin-mediated degradation may provide another route toward new mechanisms regulating liver regeneration.
In the present study, by analyzing the comprehensive data sets ranging from histone methylation to mRNA expressions and and protein expression profiles, we have unveiled the molecular mechanisms of the liver regeneration following PVBL. The pervasive writing of H3K4me3 may underlie the capacity of the liver to regenerate. We expect this informational resource to be useful to investigate liver regeneration and to provide tools for future biological assays.
Experimental procedures
Animals
Wild-type C57BL/6J mice were purchased from Charles River Laboratories. All of the mice were kept under specific-pathogen-free conditions and were treated according to the Regulations for Animal Experiments and Related Activities at Tohoku University. All experiments involving mice were approved by the institutional animal care and use committee of the Tohoku University Environmental and Safety Committee.
Mouse operation
9–12-week-old male C57BL/6J mice were used. Mice were randomly separated into a PVBL group and a sham operation group. The PVBL technique was described previously (15). In brief, the left branch of the portal vein was ligated with 7-0 braided polyester (Natsume Seisakusho Co. Ltd.) under a microscope. Mice receiving the sham operation underwent the same laparotomy without PVBL. Mice that underwent PVBL were sacrificed at 1, 3, or 7 days after operation, and right lobes were collected as regenerating liver. Sham-operated mice were sacrificed at 3 days after operation. Separated liver tissues were snap-frozen in liquid nitrogen and kept under −80 °C conditions.
Expression profiling by microarray
The procedure of microarray analysis was described previously (43). Preparation of total RNA from frozen liver tissue was carried out using an RNeasy minikit (Qiagen). Total RNA was labeled with Cyanine 3-CTP by the use of the Low Input Quick Amp Labeling Kit, One-Color (Agilent Technology). A Sureprint G3 mouse GE microarray slide (8 × 60K) was used according to the manufacturer's instructions (Agilent Technology). The data were detected on an Agilent scanner, and the analysis and clustering of genes were performed using the GeneSpringGX (version 12.6) software package (Agilent Technology).
RNA isolation and quantitative RT-PCR
Total RNA was isolated as above and transcribed into cDNA using an RT Omniscript kit (Qiagen) according to the manufacturer's protocol. RT-qPCR was performed with a LightCycler Nano instrument (Roche Applied Science). The following primers were used in this study: Mat1a, forward (5′-TGCTGGATGCCCATCTCAAG-3′) and reverse) (5′-GCATAGCCGAACATCAAACC-3′); Mat2a, forward (5′-CCACGAGGCGTTCATCGAGG-3′) and reverse (5′-AAGTCTTGTAGTCAAAACCT-3′); Kdm5b, forward (5′-AAGAGTTCGCGGACCCCTTC-3′) and reverse (5′-GATCCGCGGGGTGAAATGAA-3′); Foxm1, forward (5′-AAGAATGGCCAACATCCCGA-3′) and reverse (5′-TTGGGCCCCACTCTACCTT-3′); Ccnd1, forward (5′-CAAAATGCCAGAGGCGGATG-3′) and reverse (5′-CATGGAGGGTGGGTTGGAAA-3′); Ccne1, forward (5′-CTTTCTGCAGCGTCATCCTC-3′) and reverse (5′-CCTGTGCCAAGTAGAACGTC-3′); Ccna2, forward (5′-GTGAAGATGCCCTGGCTTTTA-3′) and reverse (5′-AACGTTCACTGGCTTGTCTT-3′); Ccnb1, forward (5′-GTGAGTGACGTAGACGCAGA-3′) and reverse (5′-TCCAGTCACTTCACGACCCT-3′); Inmt, forward (5′-GAAAGAGCCAGGAGCCTACG-3′) and reverse (5′-ACTGTCCTTCTGAGCTTGGC-3′); Hal, forward (5′-CGGCAAGCTGATATTGTGGC-3′) and reverse (5′-ACCGGAATCGGAAAGCAACT-3′); Got1, forward (5′-AACGACAACAGCCTCAACCA-3′) and reverse (5′-AAAGACTGCACCCCTCCAAC-3′); Gcdh, forward (5′-CCTTGTCATGCACCCCATCT-3′) and reverse (5′-AGCCCAGAAGTTCACCCTTG-3′); Cdkn1a, forward (5′-GCAGTCCACAGGATATCCA-3′) and reverse (5′-AGACAACGGCACACTTGCT-3′); Cdkn2C, forward (5′-GTGGGGCATCGGAACCATAA-3′) and reverse (5′-ACCCCATTTGCCTCCATCAG-3′); Cdkn3, forward (5′-TCGCGAGTGAATTGTTCCCA-3′) and reverse (5′-CGTCTTGGATCCCGTAGCTC-3′); Actb, forward (5′-CGTTGACATCCGTAAAGACCTC-3′) and reverse (5′-AGCCACCGATCCACACAGA-3′).
Purification of histone extracts
For analyzing histone modification, histones corresponding to 30 mg of the liver tissue were purified by acid extraction (44). The extracts were separated by SDS-PAGE on 15% gel, and modified histones were detected by immunoblotting analysis as described above.
Immunoblotting analysis
Whole-cell extracts were prepared from the frozen liver tissue as described previously (22). The extracts were separated by SDS-PAGE on 4–20% gel. Following SDS-PAGE, the proteins were transferred to a PVDF membrane (Millipore, Billerica, MA). The membranes were blocked for 1 h in blocking buffer (3% skimmed milk, 0.05% Tween 20 in TBS) and subsequently incubated with primary and secondary antibodies in the blocking buffer for 8 and 0.5 h, respectively. To detect immunoreactive proteins, we used SuperSignal West Pico (Thermo Fisher Scientific, San Jose, CA).
Quantification of methionine, SAM, and SAH concentrations
All chemical reagents used for quantification of the indicated metabolites were purchased from Sigma-Aldrich unless otherwise noted. Deuterium-labeled S-adenosyl-l-methionine (d3-SAM) was purchased from C/D/N isotopes, and 13C5-SAH was purchased from Toronto Research Chemicals. Stock solutions of standard SAM, SAH, methionine, and their derivatives labeled with stable isotopes were prepared by dissolving in 0.1% formic acid. The calibration mixtures in the concentration range of 12.5–1600 nm, including a 50 nm concentration of the respective molecules with stable isotopes, were prepared in 50% methanol containing 0.1% formic acid by dilution of stock solutions. The calibration curves were obtained by the peak area ratio (analyte/internal standard).
To measure the level of metabolites, mouse livers were quickly excised, weighted, and frozen in liquid nitrogen. The frozen aliquots were homogenized in 150 μl of 50% cold methanol with a BioMasher II homogenizer (Nippi, Japan). The homogenate was diluted with 150 μl of 50% cold methanol and centrifuged at 20,000 × g for 5 min at 0 °C. The supernatant (200 μl) was collected and mixed with 40 μl of chloroform. The aqueous (160-μl) layer was collected by centrifugation at 20,000 × g for 5 min at 0 °C and mixed with 40 μl of internal standard solution containing 2.5 μm d3-SAM, 13C5-SAH, and 13Cd3-Met dissolved in 0.1% formic acid. After centrifugation, 140 μl of supernatant was lyophilized and resuspended in 50% methanol containing 0.1% formic acid. The supernatant was diluted 10-fold with H2O and passed through a filter (pore size, 0.2 μm; YMC Co., Kyoto, Japan). Subsequently, 5 μl of the filtered solution was injected into the MS/MS system.
The HPLC-MS/MS system consisted of a NANOSPACE SI-II LC system (Shiseido, Tokyo, Japan) coupled with a Q-Exactive quadrupole orbitrap mass spectrometer equipped with a heated electrospray ionization source (Thermo Fisher Scientific). The metabolites were separated by hydrophilic interaction chromatography with a 4.6-mm internal diameter × 10-cm Amide XBridge column (Waters). Mobile phase A was composed of 10 mm ammonium hydroxide and 10 mm ammonium acetate in 95:5 (v/v) H2O/CH3CN, and mobile phase B was 100% CH3CN. A gradient started from 85% B to 35% B from 0 to 3 min; 35% B to 0% B from 3 to 12 min; 0% B from 12 to 17 min; 0 to 85% B from 17–18 min; and 85% B held for 7 min to re-equilibrate the column. A divert valve was used to divert the LC effluent to the waste during the first 3 min, and the range between 11 and 21 min of the chromatographic was run to prevent source contamination by salts and other compounds. The flow rate was 300 μl/min. The optimized ion source parameters were as follows: sheath gas flow rate, 20 arbitrary units; spray voltage, 3 kV; capillary temperature, 400 °C; heater temperature, 40 °C. The detection of metabolites was carried out using positive ion mode and target MS/MS (t-MS2) mode at a resolution 35,000 and normalized collision energy 20. The following MS transitions were used to measure the indicated metabolites: SAM (m/z 399.14 → 250.09), d3-SAM (m/z 402.16 → 250.09), SAH (m/z 385.13 → 136.06), 13C5-SAH (m/z 390.15 → 136.06), methionine (m/z 150.06 → 104.05), and 13Cd3-Met (m/z 154.08 → 108.08). The HPLC-MS/MS system was controlled by XCalibur software (Thermo Fisher Scientific), and peak areas for each metabolite at the SRM transitions were integrated using the same software. Absolute concentrations of cellular metabolites were normalized to the weight of excised liver.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (45), with a modification. After thawing, regenerating liver and sham liver were roughly homogenized with a BioMasher II homogenizer (Nippi, Japan) in ice-cold PBS and were cross-linked for 10 min in 1% formaldehyde at 25 °C, followed by quenching for 5 min at 25 °C with 125 mm glycine. The homogenates were then washed three times with ice-cold PBS and lysed for 10 min at 4 °C in 1 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA, and 1% SDS) with rotation. Lysates were centrifuged, and precipitates were suspended in 200 μl of lysis buffer. The cross-linked chromatin was sheared with a Bioruptor sonicator (CosmoBio), and chromatin was quantified with a Nano Drop UV spectrometer. A total of 30 μg of chromatin was diluted 10-fold in dilution buffer (20 mm Tris-HCl, pH 8.0, 2 mm EDTA, 150 mm NaCl, 1% Triton X-100). After immunoprecipitation with anti-H3K4me3, anti-H3K27me3, or anti-rabbit IgG antibodies at 4 °C overnight, Dynabeads Protein A and Protein G (Veritas) were added and rotated at 4 °C for 2 h. The immunoprecipitated chromatin fragments bound to magnetic beads were washed with the following buffers: low-salt buffer (0.1% SDS, 20 mm Tris-HCl, pH 8.0, 2 mm EDTA, 150 mm NaCl), high-salt buffer (0.1% SDS, 20 mm Tris-HCl, pH 8.0, 2 mm EDTA, 500 mm NaCl), and LiCl buffer (1% Nonidet P-40, 250 mm LiCl, 10 mm Tris-HCl, pH 8.0, 1 mm EDTA). The immunoprecipitated chromatin fragments were eluted from the beads by vortexing for 5 min at room temperature in 120 μl of elution buffer (100 mm NaHCO3, 1% SDS) and were released from formaldehyde fixation by overnight incubation at 65 °C, followed by incubation for 1 h at 55 °C in the presence of RNase and for 2 h in the presence of proteinase K. Genomic DNA was purified using the DNA Clean & Concentrater Kit according to the manufacturer's protocol (Zymo Research). We performed biological triplicates for the ChIP of H3K4me3 and H3K27me3 with both the sham and PVBL groups.
ChIP-seq and data analysis
The procedure of ChIP-seq analysis was described previously (46). ChIP-seq libraries were prepared from ∼10 ng each of ChIP and input DNA with the use of an Ovation Ultralow DR Multiplex System (NuGEN, catalog no. 0330-32, 0331-32). Two rapid-mode flow cells of an Illumina HiSeq 2500 were used for this analysis. Libraries were clonally amplified in the flow cells and sequenced with the use of HiSeq Control Software version 2.2.38 (Illumina) and a 51- nucleotide paired-end sequence. Image analysis and base calling were performed using real-time analysis software (RTA version 1.18.61, Illumina).
Sequenced reads were mapped to the mouse genome (UCSC mm9) with the use of bwa (version 0.7.10). Unique reads mapped to a single genomic location were called peaks using the MACS2 software (version 2.1.0) for H3K4me3 marks, and sequence reads for input DNA from mice with PVBL or sham operations were used as a control. Sequencing was performed with biological duplicates. Heat maps depicting the patterns of histone methylation were illustrated using the ngs.plot algorithm (version 2.47.1) (47).
Cell culture
Mouse hepatoma Hepa1c1c7 cells (Hepa1) were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% FBS, penicillin (100 units/ml), streptomycin (0.1 mg/ml), and 4500 mg/L glutamine.
RNA interference
For knockdown of Foxm1, Hepa1 cells (5 × 106 cells) were electroporated with 6 μl of 20 μm stock Stealth RNAi duplexes using the Nucleofector and Nucleofector solution kit V (VCA-1003, Amaxa Biosystems). The sequence of the Stealth RNAiTM used for knockdown of Foxm1 was 5′-ACCCAAGGUGUUGCUAUCCAGUGAA-3′. Stealth RNAi siRNA negative control (Invitrogen) was used as the negative control.
Cell proliferation assay
Twenty-four hours after transfection with siRNAs, Hepa1 cells were seeded at 5 × 103 cells/well in 96-well flat-bottom plates. Cell proliferation was analyzed using a Cell Counting Kit-8 (Dojindo) according to the manufacturer's protocol. The absorbance values at 450 nm were measured using a microplate reader just after seeding the well and at 24, 48, and 72 h.
Nano-HPLC/MS/MS analysis for proteomics
The dried peptide extracts (30 μg) and 100 fmol of internal control probes (Pierce Retention Time Calibration Mixture, Thermo Scientific) were dissolved together in 80 μl of sample solution (5% acetonitrile and 0.1% trifluoroacetic acid (TFA)). Each sample (1.25 μg/5 μl) was injected into a EasynLC-1000 system (Thermo) with EASY-Spray column (25-cm length × C18 diameter 75 μm, Thermo). Peptides were eluted with a 180-min gradient of 4–25% solvent B (0.1% (v/v) formic acid in acetonitrile) in solvent A (0.1% (v/v) formic acid in water) at a flow rate of 300–400 nl/min. Peptides were then analyzed by a Fusion mass spectrometer (Thermo Fisher Scientific) using a nanospray source. High-resolution full-scan MS spectra (from m/z 400 to 2,000) were acquired in the orbitrap with resolution (r = 120,000 at m/z 400) and lockmass enabled (m/z at 445.12003 and 391.28429), followed by MS/MS fragmentation of the most intense ions for 3 s in the linear ion trap with collisionally activated dissociation energy of 35%. The exclusion duration for the data-dependent scan was 0 s, and the isolation window was set at 10.0 m/z.
The MS/MS data were analyzed by sequence alignment using variable and static modifications by Mascot and Sequest algorithms. UniProt was used as a protein database. The specific parameters for protein sequence database searching included oxidation (M), deamination (N, Q), acetylation (N-term.), and pyroglutamation (E) as variable modifications and carbamidomethylation (C) as a static modification. Other parameters used in data analysis were as follows: two allowed missing cleavages and a mass error of 10 ppm for precursor ions and 0.8 Da for fragment ions. Charge states of +2 to +4 were considered for parent ions. When multiple spectra were assigned to a peptide, only the spectrum with the highest Mascot score was selected for manual analysis. All peptides identified with a Mascot peptide score of > 20 were manually examined using rules described previously (48). The relative quantification of proteins was carried out as described previously (49). Briefly, the semiquantification of each protein was calculated from the average MS signal area of the three most intense peptides and normalized by semiquantitative values of internal control probes.
Antibodies
For immunoblotting and ChIP analysis, we used the following antibodies: anti-MATI/III (sc-28029, Santa Cruz Biotechnology, Inc.), anti-GAPDH (ab8245, Abcam), anti-trimethylated histone H3 Lys-4 (ab8580, Abcam), anti-trimethylated histone H3K9 (ab8898, Abcam), anti-trimethylated histone H3 Lys-27 (07-449, Millipore), anti-histone H3 (ab1791, Abcam), anti-FOXM1 (sc-502, Santa Cruz Biotechnology), anti-STAT3 (06-596, Millipore), anti-phospho-STAT3 (Tyr-705) (catalog no. 9145, Cell Signaling Technology), and anti-rabbit IgG (02-6102, Invitrogen). Anti-MATIIα antibody was raised by immunizing rabbits with purified recombinant MATIIα (His6-tagged mouse MATIIα) expressed in Escherichia coli.
Statistical analysis
Statistical analysis was performed with Student's t test or the Mann-Whitney U test, using the open-source statistical programing environment R.
Author contributions
Y. S., Y. K., M. U., and K. I. were responsible for conceptualization; Y. S., Y. K., M. S., A. I.-N., and M. E. were responsible for methodology; Y. S., M. M., and K. I. were responsible for formal analysis; Y. S., Y. K., M. S., M. E., R. F., and K. N. were responsible for the investigation; M. M. was responsible for informatics; Y. S. wrote the original draft; Y. S. and K. I. were responsible for writing review and editing; and M. M., Y. K., and K. I. supervised the work.
Acknowledgments
We thank Daniel R. Sharda (Olivet Nazarene University, Bourbonnais, IL), Tapas K. Kundu (Jawaharlal Nehru Center for Advanced Scientific Research, Bangalore, India), Hideki Katagiri (Tohoku University Graduate School of Medicine, Sendai, Japan), Takuma Shiraki (Kindai University, Higashiosaka, Japan), and Satoshi Nishizuka (Iwate Medical University School of Medicine) for helpful discussions. We are grateful for stimulating discussions with Igarashi laboratory members, and we thank Akihiko Muto for help with the ChIP experiment. We also thank Unno laboratory members for discussion. We thank M. Tsuda, M. Kikuchi, M. Nakagawa, and K. Kuroda for technical assistance. We also acknowledge the technical support of the Biomedical Research Core of Tohoku University Graduate School of Medicine.
* This work was supported by Grants-in-Aid 15H02506, 25670156, 24390066, and 23116003 and the Network Medicine Global COE Program from the Ministry of Education, Culture, Sport, Science and Technology of Japan and AMED-CREST from the Japan Agency for Medical Research and Development. Additional initiative supports were from the Uehara Foundation, Takeda Foundation, and Astellas Foundation for Research on Metabolic Disorders. Restoration of the laboratory from the damage due to the 2011 Tohoku earthquake was provided in part by the Astellas Foundation for Research on Metabolic Disorders, the Banyu Foundation, the Naito Foundation, A. Miyazaki, and A. Iida. The authors declare that they have no conflicts of interest with the contents of this article.
- PVE
- portal vein embolization
- PH
- partial hepatectomy
- PVBL
- portal vein branch ligation
- H3K4
- histone H3 Lys-4
- H3K4me3
- H3K9me3, and H3K27me3, histone H3 Lys-4, Lys-9, and Lys-27 trimethylation, respectively
- RL
- regenerating liver
- RT-qPCR
- RT-quantitative PCR
- SAM
- S-adenosylmethionine
- SAH
- S-adenosyl-l-homocysteine
- MAT
- methionine adenosyltransferase
- ChIP-seq
- ChIP-sequencing
- GO
- gene ontology
- CHR
- cell cycle gene homology region.
References
- 1. Simard E. P., Ward E. M., Siegel R., and Jemal A. (2012) Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J. Clin. 62, 118–128 [DOI] [PubMed] [Google Scholar]
- 2. Jin S., Fu Q., Wuyun G., and Wuyun T. (2013) Management of post-hepatectomy complications. World J. Gastroenterol. 19, 7983–7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Makuuchi M., Thai B. L., Takayasu K., Takayama T., Kosuge T., Gunvén P., Yamazaki S., Hasegawa H., and Ozaki H. (1990) Preoperative portal embolization to increase safety of major hepatectomy for hilar bile duct carcinoma: a preliminary report. Surgery 107, 521–527 [PubMed] [Google Scholar]
- 4. van Lienden K. P., van den Esschert J. W., de Graaf W., Bipat S., Lameris J. S., van Gulik T. M., and van Delden O. M. (2013) Portal vein embolization before liver resection: a systematic review. Cardiovasc. Intervent. Radiol. 36, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gehart H., and Clevers H. (2015) Repairing organs: lessons from intestine and liver. Trends Genet. 31, 344–351 [DOI] [PubMed] [Google Scholar]
- 6. Higgins G. M., and Anderson R. M. (1931) Experimental pathology of the liver: restoration of the liver of the white rat following partial surgical removal. Arch. Pathol. 12, 186–202 [Google Scholar]
- 7. Gilgenkrantz H., and Collin de l'Hortet A. (2011) New insights into liver regeneration. Clin Res. Hepatol. Gastroenterol. 35, 623–629 [DOI] [PubMed] [Google Scholar]
- 8. Fausto N., Campbell J. S., and Riehle K. J. (2012) Liver regeneration. J. Hepatol. 57, 692–694 [DOI] [PubMed] [Google Scholar]
- 9. Itoh T., and Miyajima A. (2014) Liver regeneration by stem/progenitor cells. Hepatology 59, 1617–1626 [DOI] [PubMed] [Google Scholar]
- 10. Tashiro S. (2009) Mechanism of liver regeneration after liver resection and portal vein embolization (ligation) is different? J. Hepatobiliary Pancreat. Surg. 16, 292–299 [DOI] [PubMed] [Google Scholar]
- 11. Li W., Liang X., Kellendonk C., Poli V., and Taub R. (2002) STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J. Biol. Chem. 277, 28411–28417 [DOI] [PubMed] [Google Scholar]
- 12. Malato Y., Sander L. E., Liedtke C., Al-Masaoudi M., Tacke F., Trautwein C., and Beraza N. (2008) Hepatocyte-specific inhibitor-of-κB-kinase deletion triggers the innate immune response and promotes earlier cell proliferation during liver regeneration. Hepatology 47, 2036–2050 [DOI] [PubMed] [Google Scholar]
- 13. White P., Brestelli J. E., Kaestner K. H., and Greenbaum L. E. (2005) Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 280, 3715–3722 [DOI] [PubMed] [Google Scholar]
- 14. Chen X. G., and Xu C. S. (2014) Proteomic analysis of the regenerating liver following 2/3 partial hepatectomy in rats. Biol. Res. 47, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shirasaki K., Taguchi K., Unno M., Motohashi H., and Yamamoto M. (2014) NF-E2-related factor 2 promotes compensatory liver hypertrophy after portal vein branch ligation in mice. Hepatology 59, 2371–2382 [DOI] [PubMed] [Google Scholar]
- 16. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 17. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang J., Schriefer A. E., Yang W., Cliften P. F., and Rudnick D. A. (2014) Identification of an epigenetic signature of early mouse liver regeneration that is disrupted by Zn-HDAC inhibition. Epigenetics 9, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuttippurathu L., Juskeviciute E., Dippold R. P., Hoek J. B., and Vadigepalli R. (2016) A novel comparative pattern analysis approach identifies chronic alcohol mediated dysregulation of transcriptomic dynamics during liver regeneration. BMC Genomics 17, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen H., Lu S., Zhou J., Bai Z., Fu H., Xu X., Yang S., Jiao B., and Sun Y. (2014) An integrated approach for the identification of USF1-centered transcriptional regulatory networks during liver regeneration. Biochim. Biophys. Acta 1839, 415–423 [DOI] [PubMed] [Google Scholar]
- 21. Huang Z. Z., Mao Z., Cai J., and Lu S. C. (1998) Changes in methionine adenosyltransferase during liver regeneration in the rat. Am. J. Physiol. 275, G14–G21 [DOI] [PubMed] [Google Scholar]
- 22. Katoh Y., Ikura T., Hoshikawa Y., Tashiro S., Ito T., Ohta M., Kera Y., Noda T., and Igarashi K. (2011) Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell 41, 554–566 [DOI] [PubMed] [Google Scholar]
- 23. Obata F., and Miura M. (2015) Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat. Commun. 6, 8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., and Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 25. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen X., Müller G. A., Quaas M., Fischer M., Han N., Stutchbury B., Sharrocks A. D., and Engeland K. (2013) The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 33, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mencalha A. L., Binato R., Ferreira G. M., Du Rocher B., and Abdelhay E. (2012) Forkhead box M1 (FoxM1) gene is a new STAT3 transcriptional factor target and is essential for proliferation, survival and DNA repair of K562 cell line. PLoS One 7, e48160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sadasivam S., Duan S., and DeCaprio J. A. (2012) The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 26, 474–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun H. C., Li M., Lu J. L., Yan D. W., Zhou C. Z., Fan J. W., Qin X. B., Tang H. M., and Peng Z. H. (2011) Overexpression of Forkhead box M1 protein associates with aggressive tumor features and poor prognosis of hepatocellular carcinoma. Oncol. Rep. 25, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 30. Chen T., Xiong J., Yang C., Shan L., Tan G., Yu L., and Tan Y. (2014) Silencing of FOXM1 transcription factor expression by adenovirus-mediated RNA interference inhibits human hepatocellular carcinoma growth. Cancer Gene Ther. 21, 133–138 [DOI] [PubMed] [Google Scholar]
- 31. Wang X., Kiyokawa H., Dennewitz M. B., and Costa R. H. (2002) The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. U.S.A. 99, 16881–16886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng X. H., Black M., Ustiyan V., Le T., Fulford L., Sridharan A., Medvedovic M., Kalinichenko V. V., Whitsett J. A., and Kalin T. V. (2014) SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 10, e1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azimifar S. B., Nagaraj N., Cox J., and Mann M. (2014) Cell-type-resolved quantitative proteomics of murine liver. Cell Metab. 20, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 34. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., and Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Low T. Y., van Heesch S., van den Toorn H., Giansanti P., Cristobal A., Toonen P., Schafer S., Hübner N., van Breukelen B., Mohammed S., Cuppen E., Heck A. J., and Guryev V. (2013) Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 5, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 36. Nakagawa T., Kajitani T., Togo S., Masuko N., Ohdan H., Hishikawa Y., Koji T., Matsuyama T., Ikura T., Muramatsu M., and Ito T. (2008) Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 22, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Müller G. A., Wintsche A., Stangner K., Prohaska S. J., Stadler P. F., and Engeland K. (2014) The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 42, 10331–10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kowalik M. A., Saliba C., Pibiri M., Perra A., Ledda-Columbano G. M., Sarotto I., Ghiso E., Giordano S., and Columbano A. (2011) Yes-associated protein regulation of adaptive liver enlargement and hepatocellular carcinoma development in mice. Hepatology 53, 2086–2096 [DOI] [PubMed] [Google Scholar]
- 39. Gonzalez D. M., and Medici D. (2014) Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 7, re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., and Shilatifard A. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 41. Richardson R. T., Alekseev O. M., Grossman G., Widgren E. E., Thresher R., Wagner E. J., Sullivan K. D., Marzluff W. F., and O'Rand M. G. (2006) Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J. Biol. Chem. 281, 21526–21534 [DOI] [PubMed] [Google Scholar]
- 42. Li C., Chang D. L., Yang Z., Qi J., Liu R., He H., Li D., and Xiao Z. X. (2013) Pin1 modulates p63α protein stability in regulation of cell survival, proliferation and tumor formation. Cell Death Dis. 4, e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Itoh-Nakadai A., Hikota R., Muto A., Kometani K., Watanabe-Matsui M., Sato Y., Kobayashi M., Nakamura A., Miura Y., Yano Y., Tashiro S., Sun J., Ikawa T., Ochiai K., Kurosaki T., and Igarashi K. (2014) The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nat. Immunol. 15, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 44. Tachibana M., Matsumura Y., Fukuda M., Kimura H., and Shinkai Y. (2008) G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27, 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kera Y., Katoh Y., Ohta M., Matsumoto M., Takano-Yamamoto T., and Igarashi K. (2013) Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J. Biol. Chem. 288, 13592–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hosogane M., Funayama R., Nishida Y., Nagashima T., and Nakayama K. (2013) Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 9, e1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shen L., Shao N., Liu X., and Nestler E. (2014) ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Y., Kwon S. W., Kim S. C., and Zhao Y. (2005) Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 49. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]