Abstract
Non-canonical base pairing within guanine-rich DNA and RNA sequences can produce G-quartets, whose stacking leads to the formation of a G-quadruplex (G4). G4s can coexist with canonical duplex DNA in the human genome and have been suggested to suppress gene transcription, and much attention has therefore focused on studying G4s in promotor regions of disease-related genes. For example, the human KRAS proto-oncogene contains a nuclease-hypersensitive element located upstream of the major transcription start site. The KRAS nuclease-hypersensitive element (NHE) region contains a G-rich element (22RT; 5′-AGGGCGGTGTGGGAATAGGGAA-3′) and encompasses a Myc-associated zinc finger-binding site that regulates KRAS transcription. The NEH region therefore has been proposed as a target for new drugs that control KRAS transcription, which requires detailed knowledge of the NHE structure. In this study, we report a high-resolution NMR structure of the G-rich element within the KRAS NHE. We found that the G-rich element forms a parallel structure with three G-quartets connected by a four-nucleotide loop and two short one-nucleotide double-chain reversal loops. In addition, a thymine bulge is found between G8 and G9. The loops of different lengths and the presence of a bulge between the G-quartets are structural elements that potentially can be targeted by small chemical ligands that would further stabilize the structure and interfere or block transcriptional regulators such as Myc-associated zinc finger from accessing their binding sites on the KRAS promoter. In conclusion, our work suggests a possible new route for the development of anticancer agents that could suppress KRAS expression.
Keywords: DNA structure, G-quadruplex, nuclear magnetic resonance (NMR), oncogene, structural biology
Introduction
Non-canonical base pairing within guanine-rich DNA and RNA sequences can produce G-quartets stabilized via eight hydrogen bonds involving both the “Watson-Crick” and “Hoogsteen” edges of each guanine. Stacking of the planar G-quartets (also called a G-tetrad) leads to the formation of a G-quadruplex (G4).3 G4s are maintained by the presence of cations such as K+ and to a lesser degree Na+ and NH4+. The stacked G-quartets constitute the nearly invariant core of all G4 structures (1–3). This core is stabilized by cooperation between three key factors: hydrogen-bonding dipole interactions, metal coordination, and π-π stacking. The orientations of the loop regions within G4 structures are tightly related to strand directionality and give rise to the heterogeneity of G4 structures (4–6). G4 structures interfere with replication (7–9), transcription (10–13), and recombination (14–16). Bioinformatics analyses have provided evidence that sequences with potential to adopt G4 are not randomly localized within genomes but are specifically enriched in particular regions such as telomeres and promoters of genes (17–20). Proto-oncogenes are particularly enriched with G4 motifs, whereas tumor suppressor genes are not (21, 22). The formation of intramolecular G4s has been studied in vitro for motifs found in different human promoters regions, including c-myc (23–25), c-kit (26), and bcl2 (27). G-rich elements found in other proto-oncogenes such as KRAS have received less attention. KRAS is located on chromosome 12 and encodes a GTP/GDP-binding protein. Previous studies showed that mutant alleles of KRAS are prevalent in pancreatic, biliary tract, colorectal, and lung carcinomas (28–33). Mutations in the KRAS promoter are found in about 30% of these cases. It is thought that the KRAS oncogene promotes glycolysis through the activation of downstream signaling pathways (34) to sustain the energy requirements for uncontrolled cellular proliferation, thus contributing to survival of cancer cells. The very high affinity of the RAS GTP/GDP-binding site (picomolar range) (35, 36) has made it difficult to synthesize molecules that effectively compete with GTP at millimolar range inside cells to block KRAS activity (37). It is no surprise that after many decades of unsuccessfully battling against the RAS proteins new strategies in the field of drug design have emerged. Among those, some target alternative binding sites in the GTPase domain (38), others target the altered metabolic pathways (39), whereas some target the mRNA with antisense oligonucleotides (40). Alternatively, several strategies, including ours, target the promotor region in an effort to block KRAS expression (41). The KRAS promoter contains a polypurine nuclease-hypersensitive element (42) that plays an essential role in transcription. Its deletion results in a significant down-regulation of KRAS transcription (12, 43, 44). The promoter region of the KRAS gene comprises more than 500 bp and is susceptible to digestion by nucleases such as DNase I, micrococcal nuclease, and other endogenous nucleases (45). A particular sequence inside the NHE, between positions −327 and −296 nucleotides upstream of the main transcription initiation site, is particularly rich in guanines (sequence 32R; Fig. 1A). 32R contains six guanine stretches and is able to form several G4 conformations. Among those stretches of guanines, two regions overlap and includes the Myc-associated zinc finger (MAZ)-binding sites, the recognition sequence for a transcription factor that recognizes GGGCGG and GGGAGG sequences (46). Recent biophysical studies using circular dichroism and DMS footprinting (43, 47) suggest that oligonucleotides corresponding to 32R are able to adopt different intramolecular G-quadruplex topologies depending on which G-runs are included. Some of the topologies were tested as decoys for sequestration of MAZ (48). In addition, certain G4 structures in this region are stabilized by G-quadruplex-interacting ligands (12, 13, 43, 49) such as guanidine-modified phthalocyanines that interfere with KRAS transcription by competing with MAZ and poly(ADP-ribose) polymerase 1 proteins. Here we studied the conformations adopted by different stretches within the 32R sequence using NMR spectroscopy. The G4 conformation revealed by our studies provides a model that could potentially be used for in silico drug screening for ligands that stabilize the G4 structure in the KRAS promoter. The approach targeting unusual motifs present in genomic DNA is actively being pursued and can be seen as a new alternative strategy with promising results (1, 28, 29, 41).
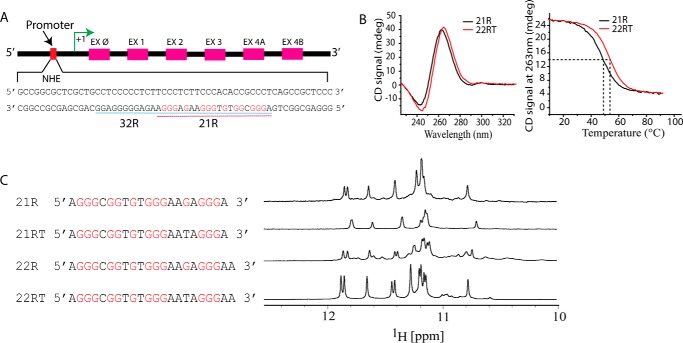
Figure 1.
A, representation of human KRAS gene showing sequences of the NHE (13) and 32R and 21R(46). EX, exon. B, CD spectra of 21R and 22RT KRAS sequences at 5 μm (left) and CD melting profiles at 3 μm. C, sequences of KRAS NHE fragments (left) and corresponding imino region 1D NMR spectra obtained at 1 mm strand concentration at 20 °C in a pH 6.5 buffer containing 20 mm potassium phosphate supplemented with 70 mm KCl. mdeg, millidegrees.
Results and discussion
We began our study with circular dichroism (CD) and NMR analyses of oligonucleotides within the sequence NHE (Fig. 1A). To make spectral assignments possible, different oligonucleotides were evaluated (supplemental Table S2) with the objective of identifying a sequence that formed a single G4 conformer based on the dispersion and intensities of imino peaks observed in the NMR spectra. The sequence 21R and three other related sequences display a 1D imino peak pattern that corresponds to a single conformer as shown by 1D 1H NMR spectroscopy (Fig. 1C). The similar imino signatures suggest the presence of a predominant conformer within the human NHE of KRAS gene. The sequence 22RT with a G16 to T16 mutation displayed a better resolved imino peak pattern and a slightly better stabilization observed by the CD melting studies (Fig. 1B). In addition, DMS footprinting (50) and our 15N-filtered 1D NMR experiments (results not shown) demonstrated that G16 did not participate in the tetrad formation. Oligonucleotides of the native sequence with four G-tracts, 21R and 22R, and those with single G16 to T (16G→T) mutations, 21RT and 22RT, respectively, appeared to adopt a predominant conformation based on analysis of the imino proton region from 10 to 12 ppm. The sequence and the respective 1H 1D NMR spectra are presented in Fig. 1C. Remarkably, 22RT showed a better resolved peak pattern in both imino and aromatic regions (not shown) than did 21R or 22R. For stability purposes, we have included an additional A at the 22RT 3′-end. These modifications resulted in better imino and aromatic peak resolution when compared with 22R and 21R. The CD spectral signatures are similar between 21R and 22RT. The spectrum of each includes a positive band at 263 nm and a negative band at 243 nm suggestive of a parallel G4 fold (Fig. 1B, left). The thermal stability of 21R and 22RT was determined through CD melting experiments. The melting temperatures (Tm) in 90 mm K+ were 49.2 ± 0.2 and 51.8 ± 0.3 °C for 21R and 22RT, respectively (Fig. 1B, right). The molecularity of 21R and 22RT was assessed by inspecting the UV-visible melting curves and by diffusion NMR experiments (supplemental Figs. S2 and S3, respectively). The results demonstrate that both 21R and 22RT fold into a monomeric structure, inferred from the reversible and superimposable cooling versus heating UV-visible curves in the concentration range that spans an order of magnitude from ≈5 to ≈50 μm. These experiments showed that melting transitions are reversible and independent of DNA concentration, demonstrating that the G4 structures formed by both 21R and 22RT are unimolecular. As the melting process was reversible, model-dependent van't Hoff enthalpies of folding could be calculated. The ΔH037 values for 21R and 22RT were 164 ± 4 and 191 ± 7 kJ/mol, respectively. Diffusion NMR spectroscopy was used to determine the diffusion coefficient value for 22RT. From diffusion-ordered spectroscopy (DOSY) experiments, we obtained a diffusion coefficient of 1.54 × 10−10 m2 s−1 (logD ≈ −9.81), which is in the range of those found for monomeric G4 oligonucleotides of similar size such as the human telomeric sequence (22AG) observed elsewhere (23, 51, 52). The results support a model where 22RT is monomeric under the experimental conditions probed in this work. For reference, we report a diffusion value of 8.9 × 10−11 m2 s−1 (logD ≈ −10.05) obtained for the oligonucleotide KRAS 44R from the NHE region that contains the sequence 32R at the 5′-end (supplemental Table S2). The imino proton spectrum of 22RT is characterized by 10 individually well resolved and sharp peaks in the 10.5-12-ppm region (Fig. 1C) plus one additional broad peak that was later identified as G19 and G7 overlapped imino peaks. The imino pattern in this region is often used as a fingerprint for G4 structures. The pattern observed suggests the formation of three G-quartets, each involving four imino protons. Based on the specific intraquartet characteristic guanine H1–H8 NOE correlations (Fig. 2B), the folding pattern of the 22RT G4 involves three G-quartets: G2·G6·G11·G18, G3·G7·G12·G19, and G4·G9·G13·G20 (Fig. 2). For clarity, we have selected the six lowest-energy structures after refinement of the best 20 structures with a heavy atom r.m.s.d. value of ≈1.5 Å (Fig. 3). When depicted against the calculated mass-weighting principal axis, the tetrad core is placed with its averaged planes almost in a perpendicular fashion (Fig. 3). The G3·G7·G12·G19 residues form the central tetrad of the quadruplex core as the imino protons on these residues are better protected from water/deuterium exchange than are those of other guanines of 22RT (supplemental Fig. S4). In addition and as expected, the guanines in a central G-quartet have by far the strongest NOE inter-residue connectivities between exchangeable protons (e.g. NH2/H1), supporting the increased protection of these protons from exchange with water. Interestingly, G3 is the only base that has NOE cross-peaks to both amino-exchangeable protons (NH21/NH22). These protons are probably protected by the single-nucleotide chain reversal C5 loop, which bridges and completely blocks the groove between G3 and G7. The 22RT G4 has an ∼30° helical twist on average and a rise of 3.4 Å for each G-tetrad step. On average, the four grooves are of medium size with similar widths in the range of 12 ± 2 Å as defined by the distances between phosphates of opposing guanines in the structure. The orientations of the aromatic bases toward the sugars are determined by the conformation of the glycosidic bond angle, which is determined by the intraresidue NOE correlation intensities between the H8 aromatic proton and H1′ sugar proton. All the guanine glycosidic torsion angles are in the anti conformation as reflected by the medium/low intraguanine NOE cross-peaks observed between H8 and H1′ protons (Fig. 2A). These glycosidic torsion angle conformations are expected for a parallel G4 as suggested by the CD spectra of 22RT (Fig. 1B). Our CD and NMR results are consistent and indicate that 22RT adopts a parallel G4 as shown schematically in Fig. 2C. The three G-tetrads are connected by four linkers: two single-residue loops (C5 and T10), a bulge (T8), and one four-nucleotide loop (A14, A15, T16, and A17). C5 and T10 each form double-chain reversal loops that allow these single residues to bridge three G-tetrad blocks. Inspection of r.m.s.d. values and conformation diversity for C5 indicates hingelike motions parallel to the mass-weighting principal axis. The T10 base is oriented toward the 3′-end of the oligonucleotides, and fewer distinct conformers are observed. Interestingly, T8 forms a bulge projected out of the G-tetrad core. Although over 700,000 G-quadruplexes with single or multiple bulges may exist in the human genome (53, 54), only a few G-quadruplex structures have been deposited in the Protein Data Bank, and besides our model, only another deposited structure (Protein Data Bank code 2M4P) has a bulge between G-tetrads. This unusual structural feature within the 3D fold may be attractive to design ligands specific for KRAS G4 in silico. The fourth linker is composed of A14, A15, T16, and A17 and forms a medium-size propeller loop that crosses all three tetrads. The size of the loop allows several water molecules to fit between the G4 core surface and the loop residues. At both oligonucleotide extremities, two adenine residues, A1 and A22, cap the 5′- and 3′-ends of 22RT, respectively (Fig. 3). A1 interacts with A17, and both are tilted inward, capping the G4 core surface at the 5′-end. At the 3′-end, A21 and A22 interact through π-π stacking and are tilted toward the tetrad surface, which is much more exposed to the solvent than the opposite end (Fig. 3b). In most of the lowest energy conformers, A21 partially blocks one of the grooves. T10 shows a profile slightly different from T8 and T16. T10 methyl protons do not make cross-correlations with any other proton except with T10 H2′/H2″, and the cross-peak with its own H1′ is very weak, indicating free rotational motion around the C1′ and N1 of the pyrimidine base without any appreciable out-of-axis torsion of the base. For T8, we observe low-intensity NOEs, which indicate that the T8 methyl slightly interacts with G4 and G9; there is a low-intensity cross-correlation between the T8 methyl and the G9 H8 and a very weak correlation from the methyl of T8 to G4 H1, indicating that the methyl may be positioned in the edge of the top tetrad. Finally, both sugar CH2 protons of T16 have strong cross-peaks with the A17 H6 and H8 protons, not observed in any other of the two thymines. Overall, we observe a more restricted mobility of T16 compared with the other two thymines. Fig. 3a shows the ensemble of structures chosen by the lowest total energy criterion. Structures were refined in water and deposited under Protein Data Bank code 5I2V. Of the 11 structures deposited, only six are shown in Fig. 3 for clarity. Two K+ counterions are expected per conformer coordinated between three G-quartet planes. In the ribbon diagrams shown in Fig. 3b, the ribbon thickness is proportional to the all-atom r.m.s.d. A1 and T8 have the highest r.m.s.d. values (≈3.5 Å) of all nucleotides in the molecule. This was expected as these two residues have fewer inter-residue correlations in the NOESY spectra. The propeller-type loops are also characterized by above average r.m.s.d. fluctuations. G-quadruplexes are highly polymorphic, and sequences with G4-forming potential are suggested to be formed in different key genomic regions, mainly in telomeres and gene promoters. Recently, high-resolution models of several different folding topologies have been reported, and some were highlighted in the aim to develop interesting pharmacological compounds that could be exploited as new anticancer drugs (47, 55–64). The G4 structure presented here is in agreement with the topology calculated for 21R (50) using DMS footprinting and may serve as a template to target and design new drugs that may diminish or inhibit KRAS expression. Interestingly, our structure contains a four-residue loop that covers one of the four grooves. It is well documented that the stability of G4 structures somewhat decreases as bulge and loop sizes increase (65, 66). We observed a more important and expected structural heterogeneity in the region of the four-nucleotide loop with a significant degree of plasticity as indicated by the low number of inter-residue NOEs from the loop nucleotides. The four-base loop blocks the groove but creates an additional narrow backbone-backbone interface in the region where A17 links to G18 (Fig. 3, c and f, red colored surface, and supplemental Fig. S5). The conformation seems unique to this G4 fold and could be an interesting site to target small ligands. In our view, this medium-size loop presents in terms of drug design another interesting feature, i.e. together with the G4 core surface it makes a sort of tunnel or cavity with negative electrostatic potential values significantly lower than those found on the rest of the surface (blue). The cavity could be targeted by ligands with non-bulky side arms usually hanging out from the aromatic core that conventionally targets the more exposed tetrad. This cavity/tunnel is similar to the one found in the structure of a G4 adopted by a G-rich region of c-kit2 (Protein Data Bank code 2KQG) (67). As opposed to KRAS 22RT, c-kit2 G4 coexists in two parallel-stranded propeller-type folds that are similar but dynamically distinct substates.
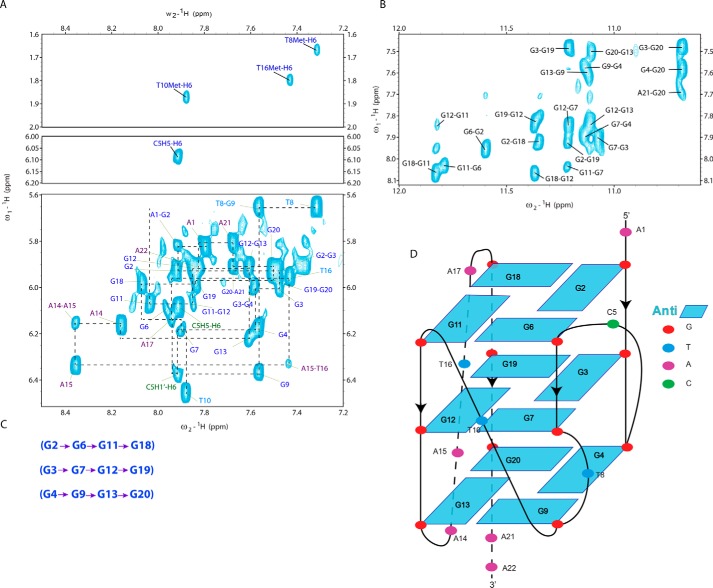
Figure 2.
A, 2D 1H NOESY spectra at 300-ms mixing time in 1H2O showing H6 to thymine methyl (top) and H5-H6 cytosine (middle) correlations from 2D 1H TOCSY spectrum and H8/H6-H1′ sequential walk assignments (bottom). B, H1/H8 correlations from 2D 1H NOESY spectrum at 300-ms mixing time. C, sequential imino-H8 intertetrad NOE connectivities observed by NMR. D, schematic representation of the G-quartets of 22RT. Guanines are numbered based on position in the 22RT sequence. All experiments were performed at 20 °C in a buffer containing 20 mm potassium phosphate, pH 6.5, supplemented with 70 mm KCl and ∼2 mm DNA. Characteristic guanine imino-H8 NOE connectivities were observed for the following tetrads: G2·G6·G11·G18, G3·G7·G12·G19, and G4·G9·G13·G20.
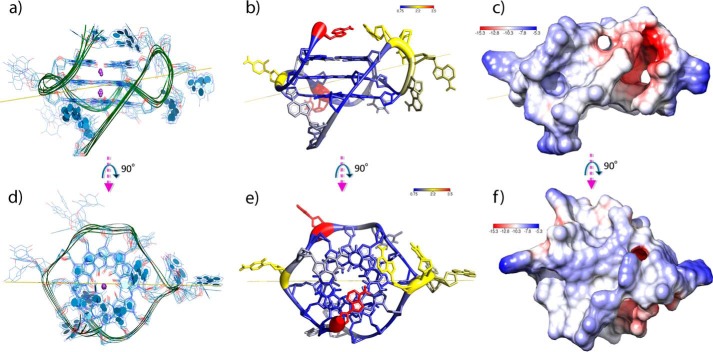
Figure 3.
a, depiction of the ensemble of the six lowest-total-energy refined structures of 22RT. All guanines are in the anti conformation. K+ counterions are depicted in purple, and the yellow line represents the principal axis of the six averaged all-atom mass-weighting principal inertial axes. On average, each conformer structure principal axis spans nearly 26 × 44 Å. The average groove width is 12 ± 2 Å. In the depiction on the right, the 5′- and 3′-ends of the oligonucleotide are at the top and bottom of the image, respectively. b, the average structure calculated from the six lowest-total-energy conformers. The ribbon thickness is proportional to the all-atom r.m.s.d. numerically represented by the color (see key for code in Å). A1 and T8 in red have the largest r.m.s.d. values (≈3.5 Å) of all residues. c, electrostatic surface of 22RT calculated using the Adaptive Poisson-Boltzmann Solver (APBS). The map was calculated using multiple Debye-Hückel boundary conditions and a nonlinear Poisson-Boltzmann equation with 40 points as the surface density at 293 K. The scale is reported in dimensionless units. The electronegative tunnel/cavity (red) represents a unique feature not usually found in canonical nucleic acid structures. In the bottom panels (d–f), the structure has been rotated (90°) so that the view is down onto the G-quartet plane that contains the 5′-most G.
Conclusion
In summary, G-quadruplex DNA structural motifs can behave as transcription repressors (12, 68), and KRAS is an important target against many forms of cancer that so far have very poor responses to standard therapies (38). We have determined a 3D high-resolution NMR structure of a stable G-quadruplex from a sequence located in the nuclease-hypersensitive element of the promoter region of the KRAS proto-oncogene. The structure may be one among others potentially available in the promotor region and may offer new, interesting possibilities for selective recognition by small ligands (69–73). In that sense, developing inhibitors of KRAS that target G-quadruplex motifs may be an interesting alternative route in the fight against certain types of cancers, as important as others currently being pursued against the intricate signaling network involved in oncogenic KRAS activation (74). Current existing strategies involve targeting post-translational modifications to prevent membrane association (75), small GTPase-targeting peptides (76), SOS-mediated nucleotide exchange (77), and inhibitors that allosterically control GTP affinity (38).
Materials and methods
DNA oligonucleotides
The unlabeled DNA oligonucleotides used in this work were purchased from both Eurogentec (Belgium) and Integrated DNA Technologies. They were synthesized on a 200-nmol or 1-μmol scale and then purified by reverse-phase HPLC. The sequences were supplied lyophilized. The 5% 15N,13C site-specifically labeled 22RT used in this study was synthesized in our laboratory (INSERM U1212, Bordeaux, France) on an automated Expedite 8909 DNA synthesizer at a 1-μmol scale on a 1000-Å primer support (Link Technologies SynBase CPG). All the standard phosphoramidites (dABz, dT, dGiBu, and dCAc), reagents, and solvents used during the synthesis were purchased from Glen Research. The dGiBu phosphoramidite (U-13C10, 98%; U-15N5, 98%; CP, 95%) was purchased from Cambridge Isotope Laboratories. After the synthesis, the oligonucleotides were cleaved from the support, and the nucleobases were deprotected with ammonium hydroxide at 55 °C for 16 h and then lyophilized. All the sequences used for NMR were prepared in potassium NMR buffer (20 mm K2HPO4/KH2PO4, 70 mm KCl, pH 6.5, 10% D2O). For CD and UV-visible studies, samples were prepared in KPi buffer (20 mm K2HPO4/KH2PO4, 70 mm KCl, pH 6.5). After dissolving in buffers, the oligonucleotides were heated for 5 min at 95 °C and chilled on ice several times. After the last annealing cycle, they were refrigerated for 24 h before use. Supplemental Table S2 lists some characteristics of the sequences used in this work.
CD spectroscopy
CD experiments were performed on a Jasco J-815 spectrometer using Spectra Manager software. Each DNA sample was prepared at 3–5 μm in KPi buffer and annealed at 90 °C for 5 min, cooled slowly by turning off the heating block (about 3–5 h), and then incubated overnight at 4 °C. The CD spectra were measured in the region between 220 and 330 nm using a scan speed of 50–100 nm/min and a response time of 1 s. Three scans were collected and averaged. Data were processed as described elsewhere (78). CD melting studies were performed on ≈3 μm DNA samples using either a full-wavelength or a single-wavelength mode. In the former case, the data were collected in the wavelength range 330–220 nm with 0.5-s averaging time, 2-nm bandwidth, 100-nm/min scan speed, two accumulations, and 0.2-nm step. The temperature was raised from 4 to 90 °C with 1 °C intervals and a 0.4 °C/min rate. These parameters lead to the overall acquisition time of 1 h/10 °C temperature change. All data collected in this manner were examined for the presence of possible intermediates during the melting process. However, the overall shape of the CD signature remained unchanged for every sample examined. Thus, after completion of the full-wavelength scan for each sample, additional melting data were collected, monitoring at 264 (characteristic for a parallel G4) and 330 nm (used as a reference to factor out instrument fluctuations) with an averaging time of 32 s and a bandwidth of 2 nm. The temperature was raised from 4 to 95 °C and then cooled to 4 °C at a rate of 1 °C/min. This set of experiments allowed us to test the reversibility of the melting process and to obtained thermodynamic parameters. Melting data were processed as described in a previous report (78).
NMR spectroscopy
NMR spectra were recorded on Bruker Avance 700- and 800-MHz instruments equipped with cryogenically cooled probes. Experiments were performed at 20 °C. For solution NMR, standard 3- or 5-mm NMR tubes were used. The samples were prepared in potassium NMR buffer. The concentrations of oligonucleotide were between 1 and 5 mm depending on the experiment requirements. Most of the 1H 1D spectra were recorded using the 1-1 echo pulse sequence (79) (based on the use of “double pulsed field gradient spin-echo”), which selectively removes resonance due to water without affecting other resonances, including those that are in fast exchange with water. The gradient pulse was a smoothed square shape (SMSQ10).100. Resonance assignments were made using 5% 15N,13C site-specific low-enrichment labeling for imino protons and 1H-13C HSQC for aromatic H8 protons. To correlate imino and H8 protons of the same guanine via the 13C5 carbon at natural abundance through-bond correlations, the 1H-13C HMBC experiment were performed at natural abundance. Thymines were unambiguously identified using the following mutants: 22RT T8 to C8 and 22RT T10 to C10 (supplemental Table S2). The remaining resonances were identified using TOCSY and COSY experiments and were independently verified using NOESY experiments.
DOSY experiment
A reference 1D 1H spectrum was recorded before the DOSY experiment. The pulse program stebpgp1s191d was used for the 1D DOSY spectra, and stebpgp1sp9pr was used for the 2D DOSY spectra. The sequences used simulated echo with a bipolar gradient pulse pair and one spoiled gradient, and 3-9-19 WATERGATE (80) solvent suppression was also applied. Sixty-four scans were recorded for the 1D DOSY experiment, and 1024 were recorded for 2D DOSY. A relaxation delay of 2 ms was applied, and a 20-μs delay was used for water suppression. The time domain was fixed to 8000 points for the F2 dimension and 32 points for the F1 dimension. The diffusion time (Δ) was 150 ms, the gradient length (δ) was 1 ms, and the recovery delay after gradient was fixed to 200 μs. The gradient strengths applied were set between 5 and 95%, and the gradient strength change was set to linear. The data processing was performed using Bruker-designed DOSY software. The following equation was applied to fit the curve of diffusion,
| (Eq. 1) |
where I is the observed intensity, I0 is the reference intensity (unattenuated signal intensity), D is the diffusion coefficient, γ is the gyromagnetic ratio of the observed proton, g is the gradient strength, δ is the length of the gradient, and τ is the diffusion time. The diffusion coefficient D for a given molecule is described by the Stokes-Einstein equation,
| (Eq. 2) |
where k is the Boltzmann constant, T is the temperature, η is the viscosity of the liquid, and RS is the (hydrodynamic) radius of the molecule.
UV-visible spectroscopy
Thermal difference spectra
Thermal difference spectra were obtained in KPi buffer by collecting UV-visible wavelength scans from 220 to 350 nm at two temperatures, one well below (usually 4 °C) and another well above (usually 95 °C) the melting temperature of the DNA secondary structure. The difference spectra were obtained by subtracting the data at 4 °C from the data at 95 °C. Both 21R and 22RT showed thermal difference spectral signatures characteristic of G4 structures (81).
UV-visible melting of 21R and 22RT
Concentration dependences of the melting transitions of 21R (5′-AGGGCGGTGTGGGAAGAGGGA-3′) and 22RT (5′-AGGGCGGTGTGGGAATAGGGAA-3′) were determined in UV-visible melting experiments by monitoring the signals at 295 and 335 nm using an Uvikon XL spectrophotometer. The former wavelength is sensitive to the G4 folding state, and the latter wavelength was used as a reference to monitor instrument performance. The extinction coefficient of the DNA at 335 nm is negligible. Five separate samples with concentration ranging from 5.0 to 50 μm were annealed in KPi buffer as described above and equilibrated at 4 °C overnight. Samples were placed in cuvettes with 1.0- or 0.2-cm path lengths depending on strand concentration. The temperature was measured with the temperature sensor inserted in the cuvette holder next to the DNA sample. The temperature was changed at a rate of 0.2 °C/min, and the averaging time was 0.3–0.5 s. Each experiment included two temperature ramps from 95 to 0.5 °C with a 15-min hold and from 0.5 back to 95 °C. The experiments were repeated twice. The value of signal at 335 nm was subtracted from each data set. The data suggest that the folding/unfolding of both 21R and 22RT is reversible as the melting and cooling data are nearly superimposable, consistent with our CD melting study. The melting curves were analyzed assuming a two-state model with temperature-independent enthalpy, ΔH0 (82). Starting and final baselines were assumed to be linear, and melting temperature and enthalpy of unfolding were adjusted to get the best fit. Data were also analyzed assuming non-zero heat capacity. This analysis included an additional parameter but did not lead to significant improvement of the fit. Melting temperatures and ΔH0 obtained from CD and UV-visible data are in good agreement with each other.
NMR structural calculations based on NOE distance restraints and simulated annealing
NOE-derived distance restraints were calculated from spectral densities obtained from different 1H-1H NOESY spectra at various mixing times (50, 200, 300, and 400 ms). In the final structure calculations, only data from the 300-ms mixing time was used. All NMR restraints were obtained from spectra collected at 293 K unless otherwise stated. The peak volumes were classified as weak (4.0–6.5 Å), medium (2.5–4.5 Å), and strong (1.8–3 Å). Planarity restraints (20 kcal/mol/Å2) were introduced for the following tetrad architecture: G2·G6·G11·G18, G3·G7·G12·G19, and G4·G9·G13·G20. Hydrogen-bond and planarity restraints were defined between 1.9 and 2.1 Å and between 2.9 and 3.1 Å for the bonds established between H1 and O6 and between H21 and N7, respectively, and were only applied to the guanine bases involved in tetrad formation. The anti conformation was defined by the glycosidic torsion angles (χ) determined from the H1′-H8 intrabase distances. They were restrained to be in the range of −130 ± 40°. Altogether, the hydrogen bonds and the artificial planarity restraints kept the G-quartets in their quasiplanar conformation during the first steps of the ARIA-CNS calculations; these restraints were removed during the refinement process. The integration of NOE volumes, calibration of distances with a relaxation matrix spin diffusion correction, and setting of lower and upper bounds were done by ARIA2.3/CNS1.2. Two distinct steps were used to calculate the final assembly of 20 structures. First, eight iterations of calculations were carried out using 200 structures per iteration in ARIA2.3/CNS1.2 (83, 84) with mixed Cartesian and torsion angle dynamics during the simulated annealing runs. The protocol contains four stages: (a) an initial high-temperature torsion angle simulated annealing of 50,000 steps at 10,000 K with 27 fs for each step, (b) a torsion angle dynamic cooling stage of 10,000 steps from 10,000 to 2000 K, (c) a Cartesian dynamics cooling stage of 10,000 steps from 2000 to 1000 K, and finally (d) a Cartesian dynamics cooling stage of 20,000 steps from 1000 to 50 K with 3 fs per step. For all bonds, angles, and improper dihedral energy terms of the force field, the standard CNS dna-rna-allatom topology and parameter files were used with uniform energy constants. For distances and hydrogen bonds, 10 kcal mol−1Å−2 was applied during the initial stage of dynamics and was increased up to 50 kcal mol−1Å−2 for the remaining steps of the dynamics. For dihedral restraints, energy constants applied were 5, 25, 200, and 200 kcal mol−1Å−2 for the phases a, b, c, and d, respectively. An energy constant of 25 kcal mol−1Å−2 was applied for planarity restraints. Distance restraints together with G-tetrad hydrogen-bonding distance restraints, glycosidic angle restraints, and planarity restraints were used during this calculation step. In the second step, we performed the necessary refinement of the t20 best structures in explicit water molecules as solvent. For that purpose, the SANDER module of Amber 12 (University of California, San Francisco) was used. The calculation was performed with the AMBER force field FF12SB, which contains the AMBER force field for nucleic acids and Barcelona changes (85). Two K+ ions were included between the G-tetrads, and 21 additional K+ ions were included to counter the negative charge of the DNA. The 20 structures were solvated by a truncated octahedral box of TIP3P water molecules (86). The structures were energy-minimized using harmonic position restraints of 25 kcal/mol/Å2. First, 1000 steps of minimization were carried out holding the system fixed and minimizing just the water box, including ions with 500 steps of steepest descent minimization followed by 500 steps of conjugate gradient minimization. Then, 2500 steps of minimization were performed for the entire system with 1000 steps of steepest descent minimization followed by 1500 steps of conjugate gradient minimization. Afterward, 20 ps of simulated annealing was acquired with heating from 0 to 300 K during 5 ps under a constant volume while maintaining the position restraints at 25 kcal/mol/Å2. Finally, we performed a cooling step to 100 K during 13 s; during this step, the time constant for heat bath coupling was varied in the range of 0.05–0.5 ps. A final cooling stage was also performed with more rapid cooling (0.1–0.05 ps) to bring the system to 0 K. The weight of distance restraints was increased gradually during this simulated annealing from 0.1 to 1 in the first 3 seconds and was then kept at 1 for the rest of the annealing procedure.
NMR assignments
Using site-specific low (5%)-enrichment [13C,15N]guanine-labeled samples, the imino H1 and the aromatic H8 protons for each guanine were unambiguously assigned (supplemental Fig. S1A). The guanine H8 aromatic protons were assigned using classical 13C-1H HSQC spectral analysis for each guanine separately (supplemental Fig. S1B). The assignments were also confirmed by natural abundance through-bond correlations using a jump and return (JR) HMBC experiment, which correlates guanine imino protons with H8 aromatic protons through 13C5 (supplemental Fig. S1C). The complete spectral assignment was achieved by combining through-bond (TOCSY and COSY) and through-space (NOESY) experiments. An 1H-1H TOCSY experiment allowed unambiguous correlation of H5-H6 protons of the cytosine at position 5 and the H6 methyl protons of thymines at positions 8, 10, and 17 (Fig. 2A). The remaining proton assignments such as those of sugar protons (H3′, H4′, H2′/H2″, and H5′/H5″) were determined as described previously (5, 6).
Restraints used in structure calculations
NMR restraints used for the calculations are listed in supplemental Table S1.
Data deposition
Water-refined structures of KRAS 22RT G-quadruplex were deposited in the Protein Data Bank under Protein Data Bank code 5I2V.
Author contributions
A. K. prepared samples, acquired NMR data, performed analysis and structure determination, and helped prepare some figures. J. M. prepared oligonucleotides samples, acquired CD spectra, and helped prepare some figures. S. I. prepared samples for CD and UV-visible melting experiments and analyzed the CD spectra. L. A. Y. supervised and acquired data from CD and UV-visible experiments and participated in manuscript conception. J.-L. M. directed the research subject and participated in manuscript conception. G. F. S. acquired NMR spectra, analyzed and interpreted various data, performed atomic structure determination, directed the research subject, wrote the article, and prepared the figures.
Supplementary Material
Acknowledgments
This work benefited from the facilities and expertise of UMS3033/US001. Financial support from the TGIR-RMN-THC Fr3050 CNRS for conducting the research is gratefully acknowledged. We thank B. Vialet for synthesis of isotopically labeled oligonucleotides and the Olifans team for useful discussions.
This work was supported in part by Fondation Association pour la Recherche sur le Cancer (ARC), Ligue Régionale contre le Cancer-Gironde, and Agence National de Recherche grant “Oligoswitch,” and an Aquitaine-Midi Pyrénées inter-regional grant. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5 and Tables S1–S3.
The atomic coordinates and structure factors (code 5I2V) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- G4
- G-quadruplex
- NHE
- nuclease-hypersensitive element
- MAZ
- Myc-associated zinc finger
- DMS
- dimethyl sulfate
- DOSY
- diffusion-ordered spectroscopy
- r.m.s.d.
- root mean square deviation
- dABz
- N6-benzoyl-2′-deoxyadenosine
- dCAc
- N4-acetyl-2′-deoxycytidine
- dGiBu
- N2-isobutyryl-2′-deoxyguanosine
- HSQC
- heteronuclear single quantum coherence
- TOCSY
- total correlation spectroscopy
- HMBC
- heteronuclear multiple bond correlation.
References
- 1. Balasubramanian S., and Neidle S. (2009) G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chemical Biol. 13, 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Düchler M. (2012) G-quadruplexes: targets and tools in anticancer drug design. J. Drug Target. 20, 389–400 [DOI] [PubMed] [Google Scholar]
- 3. Phan A. T. (2010) Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 277, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 4. Wong A., Ida R., Spindler L., and Wu G. (2005) Disodium guanosine 5′-monophosphate self-associates into nanoscale cylinders at pH 8: a combined diffusion NMR spectroscopy and dynamic light scattering study. J. Am. Chem. Soc. 127, 6990–6998 [DOI] [PubMed] [Google Scholar]
- 5. Phan A. T., Kuryavyi V., Luu K. N., and Patel D. J. (2007) Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 35, 6517–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marusic M., Sket P., Bauer L., Viglasky V., and Plavec J. (2012) Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 40, 6946–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Madireddy A., Purushothaman P., Loosbroock C. P., Robertson E. S., Schildkraut C. L., and Verma S. C. (2016) G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 44, 3675–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valton A. L., and Prioleau M. N. (2016) G-quadruplexes in DNA replication: a problem or a necessity? Trends Genet. 32, 697–706 [DOI] [PubMed] [Google Scholar]
- 9. Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M. P., Foiani M., and Nicolas A. (2011) G-quadruplex-induced instability during leading-strand replication. EMBO J. 30, 4033–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Du Z., Kong P., Gao Y., and Li N. (2007) Enrichment of G4 DNA motif in transcriptional regulatory region of chicken genome. Biochem. Biophys. Res. Commun. 354, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 11. Yan J., Zhao X., Liu B., Yuan Y., and Guan Y. (2016) An intramolecular G-quadruplex structure formed in the human MET promoter region and its biological relevance. Mol. Carcinog. 55, 897–909 [DOI] [PubMed] [Google Scholar]
- 12. Cogoi S., and Xodo L. E. (2006) G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 34, 2536–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cogoi S., Paramasivam M., Membrino A., Yokoyama K. K., and Xodo L. E. (2010) The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 285, 22003–22016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanton A., Harris L. M., Graham G., and Merrick C. J. (2016) Recombination events among virulence genes in malaria parasites are associated with G-quadruplex-forming DNA motifs. BMC Genomics 17, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piekna-Przybylska D., Sullivan M. A., Sharma G., and Bambara R. A. (2014) U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry 53, 2581–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Nicola B., Lech C. J., Heddi B., Regmi S., Frasson I., Perrone R., Richter S. N., and Phan A. T. (2016) Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 44, 6442–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huppert J. L., and Balasubramanian S. (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong H. M., Stegle O., Rodgers S., and Huppert J. L. (2010) A toolbox for predicting G-quadruplex formation and stability. J. Nucleic Acids 2010, 564946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Todd A. K., Johnston M., and Neidle S. (2005) Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 33, 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stegle O., Payet L., Mergny J. L., MacKay D. J., and Leon J. H. (2009) Predicting and understanding the stability of G-quadruplexes. Bioinformatics 25, i374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huppert J. L., and Balasubramanian S. (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eddy J., and Maizels N. (2006) Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 34, 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ambrus A., Chen D., Dai J., Jones R. A., and Yang D. (2005) Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 44, 2048–2058 [DOI] [PubMed] [Google Scholar]
- 24. Phan A. T., Kuryavyi V., Gaw H. Y., and Patel D. J. (2005) Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 1, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Phan A. T., Modi Y. S., and Patel D. J. (2004) Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 126, 8710–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phan A. T., Kuryavyi V., Burge S., Neidle S., and Patel D. J. (2007) Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 129, 4386–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai J., Dexheimer T. S., Chen D., Carver M., Ambrus A., Jones R. A., and Yang D. (2006) An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 128, 1096–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lavrado J., Brito H., Borralho P. M., Ohnmacht S. A., Kim N. S., Leitão C., Pisco S., Gunaratnam M., Rodrigues C. M., Moreira R., Neidle S., and Paulo A. (2015) KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci. Rep. 5, 9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brito H., Martins A. C., Lavrado J., Mendes E., Francisco A. P., Santos S. A., Ohnmacht S. A., Kim N. S., Rodrigues C. M., Moreira R., Neidle S., Borralho P. M., and Paulo A. (2015) Targeting KRAS oncogene in colon cancer cells with 7-carboxylate indolo[3,2-b]quinoline tri-alkylamine derivatives. PLoS One 10, e0126891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Forbes S. A., Bindal N., Bamford S., Cole C., Kok C. Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J. W., Campbell P. J., Stratton M. R., and Futreal P. A. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dent P. (2013) Multi-kinase modulation for colon cancer therapy. Cancer Biol. Ther. 14, 877–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krens L. L., Baas J. M., Gelderblom H., and Guchelaar H. J. (2010) Therapeutic modulation of k-ras signaling in colorectal cancer. Drug Discov. Today 15, 502–516 [DOI] [PubMed] [Google Scholar]
- 33. Andreyev H. J., Ross P. J., Cunningham D., and Clarke P. A. (2001) Antisense treatment directed against mutated Ki-ras in human colorectal adenocarcinoma. Gut 48, 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., et al. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., and Goody R. S. (1990) Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry 29, 6058–6065 [DOI] [PubMed] [Google Scholar]
- 36. McCormick F. (2016) K-Ras protein as a drug target. J. Mol. Med. 94, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., and Der C. J. (2014) Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 13, 828–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ostrem J. M., Peters U., Sos M. L., Wells J. A., and Shokat K. M. (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu Z., Golay H. G., and Barbie D. A. (2014) Targeting pathways downstream of KRAS in lung adenocarcinoma. Pharmacogenomics 15, 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J. H., Newbury L. J., Knisely A. S., Monia B., Hendry B. M., and Sharpe C. C. (2012) Antisense knockdown of Kras inhibits fibrosis in a rat model of unilateral ureteric obstruction. Am. J. Pathol. 180, 82–90 [DOI] [PubMed] [Google Scholar]
- 41. Cogoi S., and Xodo L. E. (2016) G4 DNA in ras genes and its potential in cancer therapy. Biochim. Biophys. Acta 1859, 663–674 [DOI] [PubMed] [Google Scholar]
- 42. Rokney A., Shagan M., Kessel M., Smith Y., Rosenshine I., and Oppenheim A. B. (2009) E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J. Mol. Biol. 392, 589–601 [DOI] [PubMed] [Google Scholar]
- 43. Cogoi S., Paramasivam M., Spolaore B., and Xodo L. E. (2008) Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 36, 3765–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paramasivam M., Membrino A., Cogoi S., Fukuda H., Nakagama H., and Xodo L. E. (2009) Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 37, 2841–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jordano J., and Perucho M. (1986) Chromatin structure of the promoter region of the human c-K-ras gene. Nucleic Acids Res. 14, 7361–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cogoi S., Zorzet S., Rapozzi V., Géci I., Pedersen E. B., and Xodo L. E. (2013) MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 41, 4049–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cogoi S., Paramasivam M., Filichev V., Géci I., Pedersen E. B., and Xodo L. E. (2009) Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 52, 564–568 [DOI] [PubMed] [Google Scholar]
- 48. Podbevšek P., and Plavec J. (2016) KRAS promoter oligonucleotide with decoy activity dimerizes into a unique topology consisting of two G-quadruplex units. Nucleic Acids Res. 44, 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xodo L., Paramasivam M., Membrino A., and Cogoi S. (2008) Protein hnRNPA1 binds to a critical G-rich element of KRAS and unwinds G-quadruplex structures: implications in transcription. Nucleic Acids Symp. Ser. 52, 159–160 [DOI] [PubMed] [Google Scholar]
- 50. Paramasivam M., Cogoi S., and Xodo L. E. (2011) Primer extension reactions as a tool to uncover folding motifs within complex G-rich sequences: analysis of the human KRAS NHE. Chem. Commun. 47, 4965–4967 [DOI] [PubMed] [Google Scholar]
- 51. Kerkour A., Mergny J. L., and Salgado G. F. (2016) NMR based model of human telomeric repeat G-quadruplex in complex with 2,4,6-triarylpyridine family ligand. Biochim. Biophys. Acta 10.1016/j.bbagen.2016.12.016 [DOI] [PubMed] [Google Scholar]
- 52. Groves P., and Webba da Silva M. (2010) Rapid stoichiometric analysis of G-quadruplexes in solution. Chemistry 16, 6451–6453 [DOI] [PubMed] [Google Scholar]
- 53. Chambers V. S., Marsico G., Boutell J. M., Di Antonio M., Smith G. P., and Balasubramanian S. (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 [DOI] [PubMed] [Google Scholar]
- 54. Bedrat A., Lacroix L., and Mergny J. L. (2016) Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 44, 1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Merle P., Gueugneau M., Teulade-Fichou M. P., Müller-Barthélémy M., Amiard S., Chautard E., Guetta C., Dedieu V., Communal Y., Mergny J. L., Gallego M., White C., Verrelle P., and Tchirkov A. (2015) Highly efficient radiosensitization of human glioblastoma and lung cancer cells by a G-quadruplex DNA binding compound. Sci. Rep. 5, 16255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tomar J. S. (2015) In-silico modeling studies of G-quadruplex with soy isoflavones having anticancerous activity. J. Mol. Model. 21, 193. [DOI] [PubMed] [Google Scholar]
- 57. Zhou J. K., Yang D. Y., and Sheu S. Y. (2015) The molecular mechanism of ligand unbinding from the human telomeric G-quadruplex by steered molecular dynamics and umbrella sampling simulations. Phys. Chem. Chem. Phys. 17, 12857–12869 [DOI] [PubMed] [Google Scholar]
- 58. Lavrado J., Ohnmacht S. A., Correia I., Leitão C., Pisco S., Gunaratnam M., Moreira R., Neidle S., Santos D. J., and Paulo A. (2015) Indolo[3,2-c]quinoline G-quadruplex stabilizers: a structural analysis of binding to the human telomeric G-quadruplex. ChemMedChem 10, 836–849 [DOI] [PubMed] [Google Scholar]
- 59. Ebrahimi M., Khayamian T., Hadadzadeh H., Sayed Tabatabaei B. E., Jannesari Z., and Khaksar G. (2015) Spectroscopic, biological, and molecular modeling studies on the interactions of [Fe(III)-meloxicam] with G-quadruplex DNA and investigation of its release from bovine serum albumin (BSA) nanoparticles. J. Biomol. Struct. Dyn. 33, 2316–2329 [DOI] [PubMed] [Google Scholar]
- 60. Qin X. Y., Liu Y. N., Yu Q. Q., Yang L. C., Liu Y., Zhou Y. H., and Liu J. (2014) Mixed-ligand mononuclear copper(II) complex: crystal structure and anticancer activity. ChemMedChem 9, 1665–1671 [DOI] [PubMed] [Google Scholar]
- 61. Mancini J., Rousseau P., Castor K. J., Sleiman H. F., and Autexier C. (2016) Platinum(II) phenanthroimidazole G-quadruplex ligand induces selective telomere shortening in A549 cancer cells. Biochimie 121, 287–297 [DOI] [PubMed] [Google Scholar]
- 62. Shin Y. J., Kumarasamy V., Camacho D., and Sun D. (2015) Involvement of G-quadruplex structures in regulation of human RET gene expression by small molecules in human medullary thyroid carcinoma TT cells. Oncogene 34, 1292–1299 [DOI] [PubMed] [Google Scholar]
- 63. Ali A., Bansal M., and Bhattacharya S. (2015) Ligand 5,10,15,20-tetra(N-methyl-4-pyridyl)porphine (TMPyP4) prefers the parallel propeller-type human telomeric G-quadruplex DNA over its other polymorphs. J. Phys. Chem. B 119, 5–14 [DOI] [PubMed] [Google Scholar]
- 64. Gupta P., Rastede E. E., and Appella D. H. (2015) Multivalent LKγ-PNA oligomers bind to a human telomere DNA G-rich sequence to form quadruplexes. Bioorg. Med. Chem. Lett. 25, 4757–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guédin A., Gros J., Alberti P., and Mergny J. L. (2010) How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 38, 7858–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mukundan V. T., and Phan A. T. (2013) Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 135, 5017–5028 [DOI] [PubMed] [Google Scholar]
- 67. Hsu S. T., Varnai P., Bugaut A., Reszka A. P., Neidle S., and Balasubramanian S. (2009) A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 131, 13399–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. David A. P., Margarit E., Domizi P., Banchio C., Armas P., and Calcaterra N. B. (2016) G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 44, 4163–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Carvalho J., Nottelet P., Mergny J. L., Queiroz J. A., Salgado G. F., and Cruz C. (2017) Study of the interaction between indole-based compounds and biologically relevant G-quadruplexes. Biochimie 135, 186–195 [DOI] [PubMed] [Google Scholar]
- 70. Zimmer J., Tacconi E. M., Folio C., Badie S., Porru M., Klare K., Tumiati M., Markkanen E., Halder S., Ryan A., Jackson S. P., Ramadan K., Kuznetsov S. G., Biroccio A., Sale J. E., et al. (2016) Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol. Cell 61, 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bončina M., Podlipnik Č., Piantanida I., Eilmes J., Teulade-Fichou M. P., Vesnaver G., and Lah J. (2015) Thermodynamic fingerprints of ligand binding to human telomeric G-quadruplexes. Nucleic Acids Res. 43, 10376–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Di Leva F. S., Novellino E., Cavalli A., Parrinello M., and Limongelli V. (2014) Mechanistic insight into ligand binding to G-quadruplex DNA. Nucleic Acids Res. 42, 5447–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Laguerre A., Stefan L., Larrouy M., Genest D., Novotna J., Pirrotta M., and Monchaud D. (2014) A twice-as-smart synthetic G-quartet: PyroTASQ is both a smart quadruplex ligand and a smart fluorescent probe. J. Am. Chem. Soc. 136, 12406–12414 [DOI] [PubMed] [Google Scholar]
- 74. Eser S., Schnieke A., Schneider G., and Saur D. (2014) Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer 111, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Berndt N., Hamilton A. D., and Sebti S. M. (2011) Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cromm P. M., Schaubach S., Spiegel J., Fürstner A., Grossmann T. N., and Waldmann H. (2016) Orthogonal ring-closing alkyne and olefin metathesis for the synthesis of small GTPase-targeting bicyclic peptides. Nat. Commun. 7, 11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Leshchiner E. S., Parkhitko A., Bird G. H., Luccarelli J., Bellairs J. A., Escudero S., Opoku-Nsiah K., Godes M., Perrimon N., and Walensky L. D. (2015) Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc. Natl. Acad. Sci. U.S.A. 112, 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nicoludis J. M., Barrett S. P., Mergny J. L., and Yatsunyk L. A. (2012) Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 40, 5432–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sklenar V., and Bax A. (1987) Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J. Magn. Reson. 74, 469–479 [Google Scholar]
- 80. Piotto M., Saudek V., and Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 81. Saccà B., Lacroix L., and Mergny J. L. (2005) The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 33, 1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ramsay G. D., and Eftink M. R. (1994) Analysis of multidimensional spectroscopic data to monitor unfolding of proteins. Methods Enzymol. 240, 615–645 [DOI] [PubMed] [Google Scholar]
- 83. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 84. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 85. Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T. E., 3rd, Laughton C. A., and Orozco M. (2007) Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 92, 3817–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.