Abstract
The 3′ end of the small ribosomal RNAs (ssu rRNA) in bacteria is directly involved in the selection and binding of mRNA transcripts during translation initiation via well-documented interactions between a Shine-Dalgarno (SD) sequence located upstream of the initiation codon and an anti-SD (aSD) sequence at the 3′ end of the ssu rRNA. Consequently, the 3′ end of ssu rRNA (3′TAIL) is strongly conserved among bacterial species because a change in the region may impact the translation of many protein-coding genes. Escherichia coli and Bacillus subtilis differ in their 3′ ends of ssu rRNA, being GAUCACCUCCUUA3′ in E. coli and GAUCACCUCCUUUCU3′ or GAUCACCUCCUUUCUA3′ in B. subtilis. Such differences in 3′TAIL lead to species-specific SDs (designated SDEc for E. coli and SDBs for B. subtilis) that can form strong and well-positioned SD/aSD pairing in one species but not in the other. Selection mediated by the species-specific 3′TAIL is expected to favor SDBs against SDEc in B. subtilis, but favor SDEc against SDBs in E. coli. Among well-positioned SDs, SDEc is used more in E. coli than in B. subtilis, and SDBs more in B. subtilis than in E. coli. Highly expressed genes and genes of high translation efficiency tend to have longer SDs than lowly expressed genes and genes with low translation efficiency in both species, but more so in B. subtilis than in E. coli. Both species overuse SDs matching the bolded part of the 3′TAIL shown above. The 3′TAIL difference contributes to the host specificity of phages.
Keywords: ssu rRNA, Escherichia coli, Bacillus subtilis, Shine-Dalgarno, anti-SD-sequence, translation efficiency
Many studies suggest that initiation is the principle bottleneck of the translation process in bacteria (Liljenstrom and von Heijne 1987; Bulmer 1991; Xia 2007a; Xia et al. 2007; Kudla et al. 2009; Tuller et al. 2010; Prabhakaran et al. 2015). Successful initiation requires that the ribosome is able to bind to the mRNA template in such a manner that the start codon correctly lines up at the ribosomal P site (Farwell et al. 1992; Komarova et al. 2002; Duval et al. 2013). This translation initiation process in most bacterial species is facilitated by (1) ribosomal protein S1 (RPS1) acting as an RNA chaperone that unfolds secondary structural elements that may otherwise embed the start codon and obscure the start signal (Vellanoweth and Rabinowitz 1992; Duval et al. 2013; Prabhakaran et al. 2015), and (2) the Shine-Dalgarno (SD) sequence located upstream of the start codon (Shine and Dalgarno 1974, 1975; Steitz and Jakes 1975; Dunn et al. 1978; Taniguchi and Weissmann 1978; Eckhardt and Luhrmann 1979; Luhrmann et al. 1981) that base-pairs with anti-SD (aSD) located at the free 3′ end of the small ribosomal rRNA (ssu rRNA, whose 3′ end will hereafter be referred to as 3′TAIL). A well-positioned SD/aSD pairing and reduced secondary structure in sequences flanking the start codon and SD are the hallmarks of highly expressed genes in Escherichia coli and Staphylococcus aureus, as well as their phages (Prabhakaran et al. 2015).
The SD/aSD pairing offers a simple and elegant solution to start codon recognition in bacteria and their phages (Hui and de Boer 1987; Vimberg et al. 2007; Prabhakaran et al. 2015). Because many protein-coding genes depend on aSD motifs located at 3′TAIL for translation, strong sequence conservation is observed in the 3′TAIL among diverse bacterial species (Woese 1987; Orso et al. 1994; Clarridge 2004; Chakravorty et al. 2007). Conversely, a change in 3′TAIL is expected to result in fundamental changes in SD usage in protein-coding genes.
E. coli, as a representative of the gram-negative bacteria, and Bacillus subtilis, as a representative of gram-positive bacteria, differ in their 3′TAIL in only a minor detail, with the former ending with A and the latter with 3′UCU or 3′AUCU (Table 1). 3′UCU was suggested by early experimental studies (Murray and Rabinowitz 1982; Band and Henner 1984), and annotated in the B. subtilis genome database SubtiList (http://genolist.pasteur.fr/SubtiList/). However, 3′AUCU appears in B. subtilis genomes annotated in GenBank (e.g., NC_000964). A recent study on B. subtilis ribosomal structure (e.g., Sohmen et al. 2015) also assumed a 3′AUCU tail in ssu rRNA (D. Wilson, personal communication). Existing evidence suggests heterogeneous “mature” ssu rRNA pool given that mature ssu rRNA in bacterial species results from endoribonuclease digestion from the precursor 30S rRNA followed by exonuclease nibbling (Britton et al. 2007; Yao et al. 2007; Kurata et al. 2015). For example, 3′→5′ exoribonucleases such as RNases II, R, and PH, as well as PNPase, all participate in maturation of the 3′TAIL of ssu rRNA (Sulthana and Deutscher 2013), and endoribonuclease YbeY has also been recently shown to participate in the 3′ end maturation of ssu rRNA (Davies et al. 2010; Jacob et al. 2013). In E. coli, 67% of mature ssu rRNA ends with the 3′TAIL in Table 1 (Kurata et al. 2015). Thus, the trailing 3′UCU and 3′ACUC may both be present in functional ssu rRNA of B. subtilis.
Table 1. ssu rRNA 3′ ends that are free to base-pair with SD motifs in E. coli and B. subtilis and their compatible motifs.
| Species and 3′ TAIL Sequencea | SD Motifsb | |
|---|---|---|
| E. coli | UAAG | |
| 3′-AUUCCUCCACUAG-5′ | UAAGG | |
| UAAGGA | ||
| UAAGGAG | ||
| UAAGGAGG | ||
| UAAGGAGGUG | ||
| B. subtilis | UAGA | AGAA |
| 3′-AUCUUUCCUCCACUAG-5′ | UAGAA | AGAAA |
| UAGAAA | AGAAAG | |
| UAGAAAG | AGAAAGG | |
| UAGAAAGG | AGAAAGGA | |
| UAGAAAGGA | AGAAAGGAG | |
| UAGAAAGGAG | AGAAAGGAGG | |
| UAGAAAGGAGG | AGAAAGGAGGU | |
| UAGAAAGGAGGU | AGAAAGGAGGUG | |
| AAAG | GAAA | |
| AAAGG | GAAAG | |
| AAAGGA | GAAAGG | |
| AAAGGAG | GAAAGGA | |
| AAAGGAGG | GAAAGGAG | |
| AAAGGAGGU | GAAAGGAGG | |
| AAAGGAGGUG | GAAAGGAGGU | |
| AAAGGAGGUGA | GAAAGGAGGUG | |
| AAAGGAGGUGAU | GAAAGGAGGUGA | |
Bolded letters show the differences in the base composition between two species. (E. coli ends with A whereas B. subtilis ends with UCU or AUCU). The underlined nucleotides denote the alternative 3′-AUCU-5′ TAIL and motifs exclusively compatible with it.
The SD motifs shown are derived from differences in 3′TAIL (boldface) for both species.
The minor difference in 3′TAIL between E. coli and B. subtilis suggests different sets of permissible SDs between the two species, i.e., some SDs that function well in one species may not function at all in the other. These species-specific SDs (Table 1) include six in E. coli (designated SDEc) and 25 in B. subtilis (designated SDBs). Such differences in permissible SDs could contribute to fundamental species differences in translation.
Most E. coli mRNAs cannot be efficiently translated in B. subtilis (McLaughlin et al. 1981a,b), but most B. subtilis mRNAs can be efficiently translated in E. coli (Stallcup et al. 1976). Many gram-negative bacteria, including E. coli, can even translate poly(U) messages (Nirenberg and Matthaei 1961; Stallcup et al. 1976) but gram-positive bacteria, including B. subtilis, cannot translate poly(U) messages (Stallcup et al. 1976). In retrospect, it was indeed good luck that Nirenberg and Matthaei (1961) happened to experiment with E. coli instead of B. subtilis, otherwise the landmark study would have ended up with nothing to report. It is also known that E. coli translation machinery can translate leaderless mRNAs (O’Donnell and Janssen 2002; Krishnan et al. 2010; Vesper et al. 2011; Giliberti et al. 2012), and that its 30S ribosomal subunit can still localize the start codon even when the last 30 nucleotides of ssu rRNA is deleted (Melancon et al. 1990).
The difference in mRNA permissibility between gram-negative and gram-positive bacteria is often attributed to the presence of the six-domain that is highly conserved RPS1 in gram-negative bacteria (Subramanian 1983), but absent or highly variable in gram-positive bacteria with translation specificity (Roberts and Rabinowitz 1989). RPS1 facilitates translation initiation by reducing secondary structure that could otherwise embed the translation initiation region (TIR) which includes SD and start codon (Roberts and Rabinowitz 1989; Farwell et al. 1992; Tzareva et al. 1994). B. subtilis has a homologous gene with four domains that are not conserved among gram-positive bacteria, with Mycoplasma pulmonis and Spiroplasma kunkelli having only one domain with weak homology to any known functional RPS1 (Salah et al. 2009). These findings corroborate earlier experimental evidence (McLaughlin et al. 1981b; Band and Henner 1984) demonstrating that B. subtilis requires a more stringent SD region for gene expression than does E. coli.
However, the conventional belief that E. coli possesses a more permissible translation machinery than B. subtilis is not always true. In rare cases, some mRNAs that can be translated efficiently in B. subtilis cannot be translated well in E. coli, and one such mRNA is gene 6 of the B. subtilis phage ϕ29 (Vellanoweth and Rabinowitz 1992). In particular, such translation specificity can often be traced to the 30S ribosome and the mRNAs, rather than other components of the translation machinery, strongly suggesting SD/aSD pairing as the cause for the translation specificity. Indeed, as we show later, gene 6 of phage ϕ29 can form a well-positioned SD/aSD pair only with the 3′TAIL of B. subtilis but not with that of E. coli. Thus, proper SD/aSD pairing of mRNAs may be the key factor in specifying host specificity of phages, in determining whether a horizontally transferred gene will function in the new genetic background of the host cell, and, ultimately, in speciation and diversification of bacterial lineages.
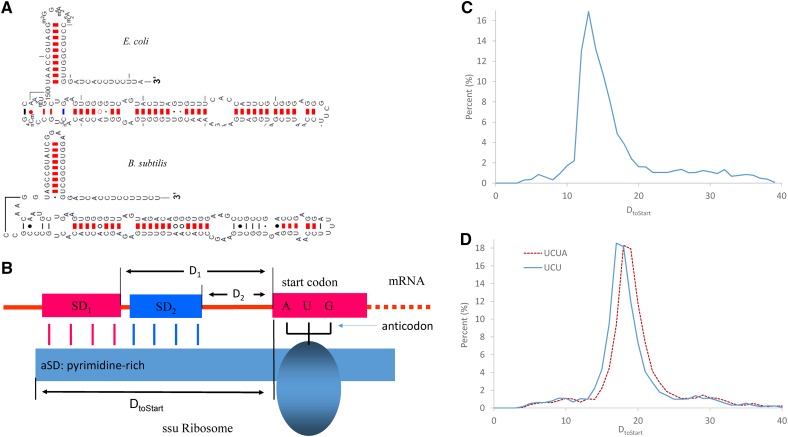
To facilitate the quantification of optimal positioning of SD/aSD base pairing, we adopted a model of SD/aSD interaction proposed recently (Prabhakaran et al. 2015), illustrated with DtoStart as a better measure of optimal SD/aSD positioning than the conventional distance between SD and the start codon (Figure 1, A and B). DtoStart is constrained within a narrow range in both E. coli (Figure 1C) and B. subtilis (Figure 1D). This observation serves as a justification for excluding putative SD/aSD matchings lying outside of this range (see Materials and Methods section for details).
Figure 1.
A model of SD sequence and aSD interactions. (A) The free 3′ end of SSU rRNA (3′TAIL) of E. coli and B. subtilis based on the predicted secondary structure of the 3′ end of the ssu rRNA of E. coli and B. subtilis from mfold 3.1, adapted from the comparative RNA web site and project (http://www.rna.icmb.utexas.edu). (B) A schematic representation of SD and aSD interaction illustrates DtoStart as a better measure for quantifying the optimal positioning of SD and aSD than the conventional distance from putative SD to start codon. SD1 or SD2, as illustrated, are equally good in positioning the start codon AUG against the anticodon of the initiation tRNA, but they differ in their distances to the start codon. DtoStart is the same for the two SDs. (C, D) DtoStart is constrained to a narrow range in E. coli (C) and B. subtilis (D); solid blue line denotes SD hits with the UCU-ending TAIL, and the dashed red line shows SD hits with the UCUA-ending TAIL. The y-axis in (C) and (D) represents the percentage of SD motif hits detected. See Materials and Methods section for details.
The difference in 3′TAIL (Figure 1A and Table 1), and in consequent species-specific compatible motifs (Table 1), between the two bacterial species suggests that selection mediated by 3′TAIL should (1) favor SDEc in E. coli and SDBs in B. subtilis, and (2) be stronger in highly expressed genes (HEGs) than in lowly expressed genes (LEGs). Here, we report results from a comprehensive genomic analysis to test these two predictions.
Materials and Methods
Retrieval of genome sequence and protein abundance data
The annotated whole genome sequences for E. coli K12 (accession number# NC_000913.3) and B. subtilis 168 (accession # NC_000964.3) in GenBank format were downloaded from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). Excluding 180 sequences annotated as pseudogenes in the E. coli genome from the analysis resulted in a final total of 4139 genes from E. coli and 4175 from B. subtilis.
Protein abundance data were retrieved from PaxDB (Wang et al. 2012) at www.pax-db.org. The integrated data sets were downloaded for both B. subtilis and E. coli in order to maximize coverage and consistency scores. We downloaded the paxdb-uniprot-links file relevant to the species (e.g., 224308-paxdb_uniprot.txt for B. subtilis), saved the Uniprot ID (the last column) to a file (e.g., BsUniprotID.txt), and browsed to http://www.uniprot.org/uploadlists (last accessed March 7, 2017) to obtain GeneID. Under “Provide your identifiers,” we uploaded the BsUniprotID.txt file, under “Selection options,” we selected the mapping from “UniProtKB AC/ID” to “Gene name” (or GeneID), and clicked “Go”. The STRING identifiers used for each gene in the protein abundance data sets were converted into Gene IDs using UniProt’s retrieve/ID mapping tool (http://www.uniprot.org/uploadlists/) for use in subsequent analyses. The resulting mapping file was generated with two columns (original input Uniprot IDs and the mapped gene name (or GIs GeneID) corresponding to gene name or other IDs in a GenBank file. Unmapped ID is stored in a separate file, also available for downloading.
HEGs and LEGs
Genes were delimited as HEGs or LEGs on the basis of two metrics: steady state protein abundance levels taken from PaxDB, and ITE (Index of translation elongation) scores computed with DAMBE (Xia 2013) using the default reference files for E. coli and B. subtilis, which were included in the DAMBE distribution. ITE is advantageous over codon adaptation index (CAI Sharp and Li 1987) or its improved form (Xia 2007b) in that it takes background mutation bias into consideration (Xia 2015). DAMBE’s ITE function has four settings that differ in their treatment of synonymous codon families, and we selected the option breaking sixfold degenerate codon families into four and twofold families. For E. coli and B. subtilis, the top and bottom 10% of genes for both of these metrics were designated as HEGs and LEGs, respectively.
Genes of high translation efficiency (HTE) and low translation efficiency (LTE)
HEGs and LEGs defined as above may not be the same as HTE genes and LTE genes. HTE and LTE genes may be characterized by regressing protein abundance on mRNA abundance, so that, given genes with the same mRNA level, those producing many proteins are translated more efficiently than those producing few. The former would be HTE genes, and the latter LTE genes. This requires proteomic and transcriptomic studies carried out with similar bacterial strains, and under similar culture and growth conditions. For E. coli, we have used proteomic data from Lu et al. (2007) deposited at PaxDB (Wang et al. 2012), and transcriptomic data in RPKM (reads per kilobase per million matched reads) from the wild-type strain of E. coli (BioProject PRJNA257498, Pobre and Arraiano 2015). For B. subtilis, the proteomic data are from Chi et al. (2011) deposited in PaxDB and transcriptomic raw counts for three wild-type replicates were downloaded from BioProject PRJNA319983 (GSM2137056 to SM2137058), and then normalized to RPKM. These two transcriptomic studies ignored reads that match to multiple paralogous genes. We have reanalyzed the data with the software ARSDA for analyzing RNA-Seq data (Xia 2017), but the results are nearly identical, partly because there are relatively few paralogous genes in the two bacterial species.
Identification of anti-SD and SD sequences
The 3′TAILs for B. subtilis and E. coli used in this paper were based on early empirical evidence (Shine and Dalgarno 1974; Brosius et al. 1978; Gold et al. 1981; Luhrmann et al. 1981; Murray and Rabinowitz 1982; Band and Henner 1984; Tu et al. 2009), as well as a series of chemical modification and nuclease digestion experiments that aimed to identify the sequence and secondary structure of bacterial ssu rRNAs using E. coli and Bacillus brevis (Woese et al. 1980). The experimentally derived 3′TAILs for both species are compatible with their corresponding ssu rRNA secondary structure schematics from the Comparative RNA Web Site & Project at www.rna.icmb.utexas.edu, which is curated by the Gutell Lab at the University of Texas at Austin. The schematics include base pairing interactions that are predicted based on the minimum free energy (MFE) state of the structure that in turn were predicted using mfold version 3.1 (http://unafold.rna.albany.edu/?q=mfold; Zuker 2003), with the resulting free 3′ ends shown in Figure 1A.
The sequence of the 3′TAIL used in our analysis for E. coli is 3′-AUUCCUCCACUAG-5′ (Shine and Dalgarno 1974; Brosius et al. 1978; Gold et al. 1981; Luhrmann et al. 1981; Band and Henner 1984; Tu et al. 2009), because, based on the E. coli SSU rRNA secondary structure (Woese et al. 1980; Noah et al. 2000; Yassin et al. 2005; Kitahara et al. 2012; Prabhakaran et al. 2015), these are the 13 nt at the 3′ end of the ssu rRNA that are free to base pair with the SD sequence. There are two versions of 3′TAIL for B. subtilis: 3′-UCUUUCCUCCACUAG (Murray and Rabinowitz 1982; Band and Henner 1984), and 3′-AUCUUUCCUCCACUAG in the genomic annotation. We discussed the possibility of heterogeneous “mature” ssu rRNA pool in the Introduction.
Identification of putative SD sequences
We followed the method of Prabhakaran et al. (2015) to identify valid SD sequences, as illustrated in Figure 1. For each gene in each species, we extracted the 30 nt upstream of the star codon and searched matches against the 3′TAIL of the two species by using the “Analyzing 5′UTR” function in DAMBE (Xia 2013). An SD with at least four consecutive nucleotide matches, and positioned with DtoStart in the range of 10–22 nt, was considered as a good SD for the E. coli translation machinery. For B. subtilis, a DtoStart range of 12–23 nt was used for the 3′UCU TAIL, or 13–24 nt for the 3′AUCU TAIL. As shown in Figure 1D, the DtoStart values for the 3′-AUCU-5′ TAIL in B. subtilis are shifted by 1 nt because this measure depends on 3′TAIL length. For this reason, taking 13–24 nt as the optimal range for the 16 nt 3′TAIL is equivalent to using 12–23 nt for the 15 nt 3′TAIL.
Data availability
All data used to generate the results are available upon request. Software DAMBE for characterizing SD sequences and computing the index of translation elongation (ITE), and software ARSDA for characterizing gene expression is available free at http://dambe.bio.uottawa.ca/Include/software.aspx.
Results and Discussion
E. coli has 4323 protein-coding genes (CDSs), with 180 annotated as pseudogenes in the genome and excluded from the analysis, resulting in 4144 functional CDSs. B. subtilis has 4175 CDSs with none annotated as pseudogenes. The genomic nucleotide frequencies are 0.2462, 0.2542, 0.2537, and 0.2459, respectively for A, C, G, and T in E. coli. The corresponding values in B. subtilis are 0.2818, 0.2181, 0.2171, and 0.2830, respectively.
SDEc and SDBs are used more in E. coli and B. subtilis, respectively
As expected, SDEc are much more frequent in E. coli than in B. subtilis, with 455 in E. coli, in contrast to 267 in B. subtilis (Table 2). The difference is highly significant, either against the null hypothesis of equal frequencies (χ2 = 48.9529, P < 0.0001), against the expected value based on the relative number of CDSs (χ2 = 50.3648, P < 0.0001; a slightly increased χ2 is because E. coli has slightly fewer included CDSs than B. subtilis), or against the expected values based on both relative number of CDSs and genomic nucleotide frequencies (e.g., AGAA is proportional to PA3PG, AGAAA to PA4PG, and so on, where PX is the genomic frequency of nucleotide X in either E. coli or B. subtilis), with χ2 = 103.07, P < 0.0001.
Table 2. Number of SDEc hits (N) and their proportion (Prop) in E. coli and B. subtilis genes.
| SDEc motifs | Occurrence in E. coli | Occurrence in B. subtilis | ||
|---|---|---|---|---|
| N | Prop | N | Prop | |
| UAAG | 85 | 0.0205 | 15 | 0.0036 |
| UAAGG | 91 | 0.0220 | 54 | 0.0129 |
| UAAGGA | 151 | 0.0365 | 30 | 0.0072 |
| UAAGGAG | 117 | 0.0283 | 74 | 0.0177 |
| UAAGGAGG | 10 | 0.0024 | 74 | 0.0177 |
| UAAGGAGGU | 0 | 0 | 14 | 0.0033 |
| UAAGGAGGUG | 1 | 0.0002 | 6 | 0.0014 |
| Total | 455 | 0.1099 | 267 | 0.0640 |
SDEc, SDs that pair perfectly with the 3′ end of small subunit rRNA from E. coli, but not from B. subtilis.
The relative abundance of different SDs depends on selection favoring an optimal SD length, and mutations disrupting long SDs. In E. coli, the optimal SD length is six (Vimberg et al. 2007). B. subtilis favors longer SDs. In an experiment with B. subtilis with SD lengths of 5, 6, 7, and 12, longer SDs consistently produce more proteins than shorter ones (Band and Henner 1984). This is consistent with the results presented in Table 2, where UAAG is expected to be strongly selected against in B. subtilis because it can form only 3 bp against B. subtilis 3′TAIL. However, the longer SDEc is not selected against because an SDEc such as UAAGGAGG can form 7 bp (except for the first U) against B. subtilis 3′TAIL.
Also as expected, SDBs are also more frequent in B. subtilis than in E. coli, with 1203 SDBs in B. subtilis in contrast to 576 in E. coli (Table 3). The difference is also highly significant (P < 0.0001) using the same tests for SDEc results in Table 2. However, one interesting deviation from the SDEc data is that SDBs of length 4 exhibit the opposite pattern, being more frequent in E. coli than in B. subtilis (Table 3), which assumes a 3′UCU-ending in B. subtilis 3′TAIL. The pattern is the same with 3′AUCU-ending of the 3′TAIL (Table S1). This observation can be explained by stronger selection against short SD/aSD in B. subtilis than in E. coli. Translation efficiency increases with longer and more stringent SD/aSD binding in B. subtilis, and such dependence is much stronger in B. subtilis than in E. coli (Band and Henner 1984). The predicted free energy of SD/aSD for an average B. subtilis message is at least 6 kcal/mol more than that of an average SD/aSD in E. coli (Hager and Rabinowitz 1985). Thus, a short SD is expected to be selected against, and, consequently, rare in B. subtilis, consistent with our results (Table 3), showing that longer SDBs (5–8 nt) are more frequent in B. subtilis than in E. coli.
Table 3. Number of SDBs hits (N) and their proportion (Prop) in all Bacillus subtilis and Escherichia coli genes considering UCU as the 3′TAIL.
| SDBs motifs | Occurrence in B. subtilis | Occurrence in E.coli | ||
|---|---|---|---|---|
| N | Prop | N | Prop | |
| AGAA | 12 | 0.0029 | 51 | 0.0123 |
| AGAAA | 66 | 0.0158 | 60 | 0.0145 |
| AGAAAG | 60 | 0.0144 | 14 | 0.0034 |
| AGAAAGG | 54 | 0.0129 | 7 | 0.0017 |
| AGAAAGGA | 60 | 0.0144 | 6 | 0.0014 |
| AGAAAGGAG | 28 | 0.0067 | 4 | 0.0010 |
| AGAAAGGAGG | 11 | 0.0026 | 1 | 0.0002 |
| AGAAAGGAGGU | 1 | 0.0002 | 0 | 0 |
| Subtotal | 292 | 0.0699 | 143 | 0.0345 |
| GAAA | 16 | 0.0038 | 65 | 0.0157 |
| GAAAG | 41 | 0.0098 | 28 | 0.0068 |
| GAAAGG | 68 | 0.0163 | 18 | 0.0043 |
| GAAAGGA | 51 | 0.0122 | 15 | 0.0036 |
| GAAAGGAG | 57 | 0.0137 | 10 | 0.0024 |
| GAAAGGAGG | 18 | 0.0043 | 1 | 0.0002 |
| GAAAGGAGGU | 3 | 0.0007 | 0 | 0 |
| GAAAGGAGGUG | 1 | 0.0002 | 0 | 0 |
| GAAAGGAGGUGA | 1 | 0.0002 | 0 | 0 |
| Subtotal | 240 | 0.0575 | 137 | 0.0331 |
| AAAG | 19 | 0.0046 | 38 | 0.0092 |
| AAAGG | 171 | 0.0410 | 83 | 0.0200 |
| AAAGGA | 76 | 0.0182 | 101 | 0.0244 |
| AAAGGAG | 222 | 0.0532 | 64 | 0.0155 |
| AAAGGAGG | 143 | 0.0343 | 6 | 0.0014 |
| AAAGGAGGU | 31 | 0.0074 | 3 | 0.0007 |
| AAAGGAGGUG | 6 | 0.0014 | 0 | 0 |
| AAAGGAGGUGA | 3 | 0.0007 | 1 | 0.0002 |
| Subtotal | 671 | 0.1607 | 296 | 0.0715 |
| Total | 1203 | 0.2881 | 576 | 0.1391 |
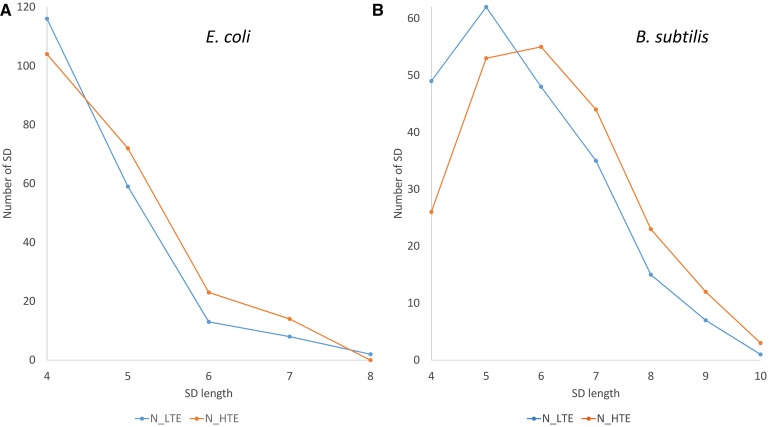
Highly expressed genes tend to have longer SDs
In addition to the observed difference in SD length between E. coli and B. subtilis (Figure 2 and Table 3; B. subtilis SDs tend to be longer than E. coli SDs), there is also clear difference between HEGs and LEGs, or between genes of HTE and of LTE. Although SDs of length four are the most frequent in E. coli, longer SDs are relatively more represented in HTE genes than in LTE genes (Figure 2A). This is consistent with previous experimental studies demonstrating an optimal SD length of six (Schurr et al. 1993; Komarova et al. 2002; Vimberg et al. 2007). Optimal SDs in B. subtilis are even longer (Band and Henner 1984) than in E. coli (Figure 2). We thus expect HEGs or HTE genes to have relatively longer SDs than LEGs or LTE genes, especially in B. subtilis. Our empirical results (Figure 2) strongly support this expectation. Short SDs are overrepresented in LEGs and LTE genes, and longer SDs overrepresented in HEGs and HTE genes in both E. coli and B. subtilis, but more so in B. subtilis (Figure 2). This pattern (i.e., association of long SDs with HEGs and HTE genes) is highly significant for B. subtilis (chi-square = 12.0375, d.f. = 1, P-value = 0.0005214) when tested by the Cochran-Armitage test (Agresti 2002, pp. 181–182) for contingency tables with a linear trend as implemented in the coin package in R (Hothorn et al. 2006, 2008). The result for E. coli, while consistent with the expectation, is not significant at the 0.05 level (chi-square = 3.3948, d.f. = 1, P-value = 0.0654).
Figure 2.
Distribution of SDs from 200 HTE genes and 200 LTE genes over SD length for E. coli (A) and B. subtilis (B). Classifying genes into HEGs and LEGs generates equivalent results, with HEGs similar to HTE genes, and LEGs similar to LTE genes. HEGs and HTE genes tend to have longer SDs than LEGs and LTE genes.
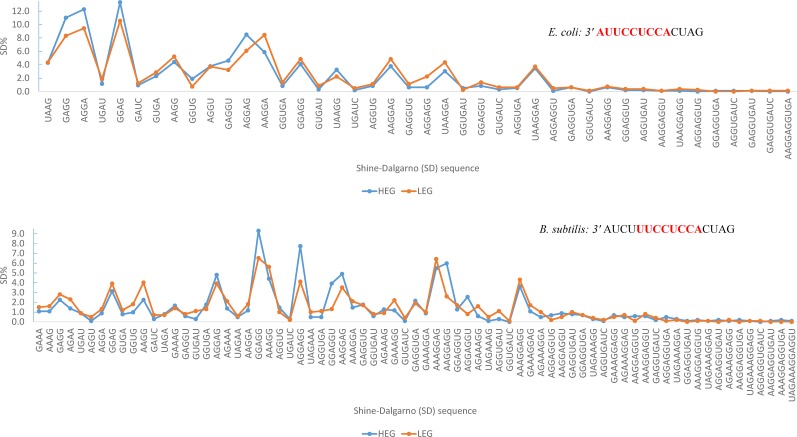
Differential usage of SDEc and SDBs in HEGs and LEGs
SDEc is used more frequently in HEGs than LEGs in E. coli (Table 4). In contrast, SDBs is used mainly in LEGs in B. subtilis (Table 5), prompting the question of what SDs are used by B. subtilis HEGs, and whether the core aSD region (where most HEGs have SD to pair against) for B. subtilis HEGs include the trailing 3′UCU (or 3′AUCU). The pattern is similar when contrasting between HTE genes and LTE genes (results not shown). The core aSD region is centered at CCUCC in the overwhelming majority of surveyed prokaryotes (Ma et al. 2002; Nakagawa et al. 2010; Lim et al. 2012). If B. subtilis has the same core aSD region, then the trailing 3′UCU (or 3′AUCU) will be used rarely, consequently with few SDBs pairing to it. The distribution of SDs in E. coli and B. subtilis is consistent with this interpretation (Figure 3). SDs overrepresented in HEGs relative to LEGs use exclusively 3′AUUCCUCCA as the core aSD region in E. coli, and 3′UUCCUCCA as the core aSD region in B. subtilis (Figure 3). The trailing 3′UCU (or 3′AUCU) is used as part of aSD mainly by LEGs in B. subtilis.
Table 4. Number of SDEc hits (N) and their proportion (Prop) in HEGs and LEGs.
| SDEc motifs | Occurrence in E. coli | Occurrence in B. subtilis | ||||||
|---|---|---|---|---|---|---|---|---|
| HEGs | LEGs | HEGs | LEGs | |||||
| N | Prop | N | Prop | N | Prop | N | Prop | |
| UAAG | 22 | 0.0053 | 7 | 0.0017 | 1 | 0.0002 | 3 | 0.0007 |
| UAAGG | 32 | 0.0077 | 6 | 0.0014 | 4 | 0.0010 | 3 | 0.0007 |
| UAAGGA | 36 | 0.0087 | 20 | 0.0048 | 3 | 0.0007 | 0 | 0 |
| UAAGGAG | 40 | 0.0097 | 12 | 0.0029 | 9 | 0.0022 | 10 | 0.0024 |
| UAAGGAGG | 2 | 0.0005 | 1 | 0.0002 | 14 | 0.0034 | 2 | 0.0005 |
| UAAGGAGGU | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0002 |
| UAAGGAGGUG | 0 | 0 | 0 | 0 | 4 | 0.0010 | 0 | 0 |
| Total | 132 | 0.0319 | 46 | 0.0111 | 35 | 0.0084 | 19 | 0.0046 |
Table 5. Number of SDBs hits (N) and their proportion (Prop) in highly and lowly expressed genes.
| SDBs motifs | Occurrence in B. subtilis | Occurrence in E. coli | ||||||
|---|---|---|---|---|---|---|---|---|
| HEGs | LEGs | HEGs | LEGs | |||||
| N | Prop. | N | Prop. | N | Prop. | N | Prop. | |
| AGAA | 0 | 0 | 2 | 0.0005 | 3 | 0.0007 | 3 | 0.0007 |
| AGAAA | 2 | 0.0005 | 8 | 0.0019 | 7 | 0.0017 | 9 | 0.0022 |
| AGAAAG | 6 | 0.0014 | 4 | 0.0010 | 1 | 0.0002 | 1 | 0.0002 |
| AGAAAGG | 3 | 0.0007 | 6 | 0.0014 | 1 | 0.0002 | 0 | 0 |
| AGAAAGGA | 4 | 0.0010 | 2 | 0.0005 | 2 | 0.0005 | 0 | 0 |
| AGAAAGGAG | 2 | 0.0005 | 3 | 0.0007 | 1 | 0.0002 | 0 | 0 |
| AGAAAGGAGG | 1 | 0.0002 | 2 | 0.0005 | 0 | 0 | 0 | 0 |
| AGAAAGGAGGU | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Subtotal | 18 | 0.0043 | 27 | 0.0065 | 15 | 0.0036 | 13 | 0.0031 |
| GAAA | 0 | 0 | 2 | 0.0005 | 5 | 0.0012 | 10 | 0.0024 |
| GAAAG | 2 | 0.0005 | 7 | 0.0017 | 3 | 0.0007 | 1 | 0.0002 |
| GAAAGG | 3 | 0.0007 | 11 | 0.0026 | 0 | 0 | 0 | 0 |
| GAAAGGA | 4 | 0.0010 | 5 | 0.0012 | 5 | 0.0012 | 0 | 0 |
| GAAAGGAG | 2 | 0.0005 | 6 | 0.0014 | 1 | 0.0002 | 1 | 0.0002 |
| GAAAGGAGG | 2 | 0.0005 | 2 | 0.0005 | 0 | 0 | 0 | 0 |
| GAAAGGAGGU | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GAAAGGAGGUG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GAAAGGAGGUGA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Subtotal | 13 | 0.0031 | 33 | 0.0074 | 14 | 0.0034 | 12 | 0.0029 |
| AAAG | 1 | 0.0002 | 4 | 0.0010 | 2 | 0.0005 | 2 | 0.0005 |
| AAAGG | 8 | 0.0019 | 20 | 0.0048 | 7 | 0.0017 | 12 | 0.0029 |
| AAAGGA | 5 | 0.0012 | 10 | 0.0024 | 10 | 0.0024 | 9 | 0.0022 |
| AAAGGAG | 17 | 0.0041 | 26 | 0.0062 | 7 | 0.0017 | 7 | 0.0017 |
| AAAGGAGG | 14 | 0.0033 | 21 | 0.0050 | 1 | 0.0002 | 0 | 0 |
| AAAGGAGGU | 2 | 0.0005 | 1 | 0.0002 | 1 | 0.0002 | 0 | 0 |
| AAAGGAGGUG | 1 | 0.0002 | 0 | 0 | 0 | 0 | 0 | 0 |
| AAAGGAGGUGA | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0002 |
| Subtotal | 48 | 0.0115 | 82 | 0.0196 | 28 | 0.0068 | 31 | 0.0075 |
| Total | 79 | 0.0189 | 142 | 0.0335 | 57 | 0.0138 | 56 | 0.0135 |
Figure 3.
Distribution of E. coli and B. subtilis SDs for HEGs and LEGs. SDs that are more frequent in HEGs than LEGs match the core aSD (in bold red) of 16S rRNA. The trailing 3′ nucleotides in B. subtilis are used mainly for SD/aSD pairing in LEGs. Classifying genes into genes of HTE and LTE generates similar results.
The mature ssu rRNA pool may be heterogeneous in B. subtilis. A number of 3′→5′ exoribonucleases, such as RNases II, R, and PH, as well as PNPase, participate in maturation of the 3′TAIL of ssu rRNA (Sulthana and Deutscher 2013), and nuclease YbeY has also been shown recently to participate in the 3′ end maturation of ssu rRNA (Davies et al. 2010; Jacob et al. 2013). The continuous 3′→5′ digestion implies that the 3′AUCU end will become 3′UCU, 3′CU, and so on. It would make sense for HEGs to use SDs paired with the less volatile part of the 3′TAIL of ssu rRNA (Table 5).
Figure 3, Table 4, and Table 5 suggest that many HEGs in E. coli use the species-specific SDEc and will experience translation initiation problems when translated by the B. subtilis translation machinery. In contrast, most HEGs in B. subtilis do not use the species-specific SDBs, and will have no translation initiation problems when translated by the E. coli translation machinery. Early studies have suggested a more permissible translation machinery in E. coli than in B. subtilis, i.e., most E. coli mRNAs cannot be efficiently translated in B. subtilis (McLaughlin et al. 1981a,b) but most B. subtilis mRNAs can be efficiently translated in E. coli (Stallcup et al. 1976). The discrepancy in this translation permissibility is often attributed to the presence of the six-domain highly conserved RPS1 in gram-negative bacteria (Subramanian 1983) but absent in gram-positive bacteria with translation specificity (Roberts and Rabinowitz 1989). Our results (Figure 3, Table 4, and Table 5) suggest an alternative explanation for the discrepancy. Because these early studies often involve HEGs, and because E. coli HEGs often use species-specific SDEc (Table 4) whereas B. subtilis HEGs rarely use species-specific SDBs, it is not surprising that E. coli HEG messages tend to fail in translation initiation in B. subtilis, but B. subtilis HEG messages tend to have no problem in translation initiation in E. coli.
Species-specific SD and host specificity
One rare exception to the general observation that E. coli possesses a more permissible translation machinery than B. subtilis is gene 6 (gp6) of the B. subtilis phage ϕ29, which can be translated efficiently in B. subtilis but not in E. coli (Vellanoweth and Rabinowitz 1992). Among the 16 nonhypothetical genes in phage ϕ29, gp6 is the only one that uses a species-specific SDBs (UAGAAAG) exclusively (Table 6). This SD used all four nucleotides at 3′TAIL of B. subtilis, and consequently cannot form SD/aSD in E. coli (Table 6). Other genes, such as gp7 and gp8, have two alternative SDs, with one being the species-specific SDBs, but they have another SD that can form SD/aSD binding in E. coli (Table 6). Because gp6 is an essential gene, its use of a SDBs may explain its host-specificity. That is, even if it gains entry into an E. coli-like host, it will not be able to survive and reproduce successfully.
Table 6. SD/aSD binding of nonhypothetical genes in B. subtilis phage φ29 in E. coli and B. subtilis.
| Gene | E. coli | B. subtilis | ||
|---|---|---|---|---|
| DtoStarta | SD | DtoStartb | SD | |
| gp2 | 14 | AAGGA | 17 | AAAGGA |
| gp3 | 17 | AAGGAG | 20 | GAAAGGAG |
| gp4 | 18 | AGGAGGU | 21 | AGGAGGU |
| gp5 | 15 | AAGGA | 18 | AAAGGA |
| gp6 | 19 | UAGAAAG | ||
| gp7 | 16 | GAGGUGA | 18,19 | UAGAAAG,GAGGUGA |
| gp8 | 18 | GAGGU | 21,21 | AGAAA,GAGGU |
| gp8.5 | 20 | GGAGGUG | 23 | GGAGGUG |
| gp9 | 16,19 | UAAGG,AGGUG | 22 | AGGUG |
| gp10 | 15 | GAGGUGA | 18 | GAGGUGA |
| gp11 | 16 | GGUGA | 19 | GGUGA |
| gp12 | 15 | UAAGGAGG | 18 | AAGGAGG |
| gp13 | 17 | GAGGU | 20 | GAGGU |
| gp14 | 17 | AAGGAG | 20 | AAAGGAG |
| gp15 | 17 | UAAGGAGG | 20 | AAGGAGG |
| gp16 | 16 | GAGGUG | 19 | GAGGUG |
Gene gp6, which uses a species-specific SDBs, cannot form a well-positioned SD/aSD in E. coli to be translated efficiently.
The optimal DtoStart is within the range of 10–21 in E. coli.
3′AUCUUUCCUCCACUAG is used as 3′TAIL for B. subtilis, with the optimal DtoStart within the range of 15–25.
Another case of host-specificity that may be explained by SD/aSD binding is E. coli phage PRD1, which has codon usage deviating greatly from that of its host, in contrast to the overwhelming majority of E. coli phages, whose codon usage exhibits high concordance with that of the host (Chithambaram et al. 2014). Phage PRD1 belongs to the peculiar Tectiviridae family whose other members, i.e., phages PR3, PR4, PR5, L17, and PR772, parasitize gram-positive bacteria. Phage PRD1 is the only species in the family known to parasitize a variety of gram-negative bacteria, including Salmonella, Pseudomonas, Escherichia, Proteus, Vibrio, Acinetobacter, and Serratia species (Bamford et al. 1995; Grahn et al. 2006). Phage PRD1 is extremely similar to its sister lineages, parasitizing gram-positive bacteria; there is only one amino acid difference in the coat protein between PRDl and PR4 (Bamford et al. 1995). It is thus quite likely that the ancestor of phage PRD1 parasitizes gram-positive bacteria. The lineage leading to Phage PRD1 may have switched to gram-negative bacterial hosts only recently, and thus still has codon usage similar to its ancestral gram-positive bacterial host, which is indeed the case (Chithambaram et al. 2014). However, one nonhypothetical gene in phage PRD1 (PRD1_09) has evolved an E. coli-specific SD (UAAG), and does not have alternative SD that can form a well-positioned SD/aSD with B. subtilis 3′TAIL. This may have contributed to the host limitation of phage PRD1 within E. coli-like species.
The study of coevolution between SD and aSD sequences would be facilitated if 3′TAILs of many bacterial species were characterized experimentally, and if these 3′TAILs differ substantially from each other in different lineages. At present, strong experimental evidence is available for 3′TAIL of E. coli and B. subtilis (except for the uncertainty on whether the 3′TAIL ends with 3′UCU or 3′AUCU). However, RNA-Seq data may become available for many bacterial species in the near future, and should pave the way for rapid characterization of 3′TAIL of different species by simply mapping the sequence reads to ssu rRNA genes on the genome. One problem to be aware of is that most transcriptomic studies will use an rRNA removal kit to remove the large rRNAs, i.e., 16S and 23S rRNA, in bacteria, because otherwise sequence reads from these large rRNAs will dominate the RNA-seq data. There are two main types of rRNA Remove Kits in the markets: (1) RiboMinus Kit from Invitrogen or MICROBExpress Bacterial mRNA Enrichment Kit (formerly Ambion, now Invitrogen), which have two probes located within the conserved sequence region at each ends of 16S and 23S rRNAs. Full-length rRNA or partial rRNA that pairs with these probes are removed. This implies that such RNA-seq data will lack reads mapped to the 5′ or 3′ ends of ssu rRNAs. The other type of rRNA removal kit is represented by the Ribo-Zero Kit from Epicentre (an Illumina company). This kit removes rRNA across the entire length and does not specifically targets the 5′ and 3′ ends. We used ARSDA (Xia 2017) to confirm that transcriptomic studies using this RNA removal kit have reads that map to the 3′ end of ssu rRNA.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.039305/-/DC1.
Acknowledgments
We thank J. Wang, M.-A. Akimenko, A. Golshani, and T. Xing for discussion and comments. This study was funded by the Discovery Grant from Natural Science and Engineering Research Council of Canada to X.X. (RGPIN/261252-2013).
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Agresti A., 2002. Categorical Data Analysis. Wiley, New Jersey. [Google Scholar]
- Bamford D. H., Caldentey J., Bamford J. K., 1995. Bacteriophage PRD1: a broad host range DSDNA tectivirus with an internal membrane. Adv. Virus Res. 45: 281–319. [DOI] [PubMed] [Google Scholar]
- Band L., Henner D. J., 1984. Bacillus subtilis requires a “stringent” Shine-Dalgarno region for gene expression. DNA 3: 17–21. [DOI] [PubMed] [Google Scholar]
- Britton R. A., Wen T., Schaefer L., Pellegrini O., Uicker W. C., et al. , 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 63: 127–138. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F., 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75: 4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M., 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S., Helb D., Burday M., Connell N., Alland D., 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69: 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B. K., Gronau K., Mader U., Hessling B., Becher D., et al. , 2011. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics 10: M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chithambaram S., Prabhakaran R., Xia X., 2014. The effect of mutation and selection on codon adaptation in Escherichia coli bacteriophage. Genetics 197: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarridge J. E., III, 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17: 840–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B. W., Kohrer C., Jacob A. I., Simmons L. A., Zhu J., et al. , 2010. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol. Microbiol. 78: 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Buzash-Pollert E., Studier F. W., 1978. Mutations of bacteriophage T7 that affect initiation of synthesis of the gene 0.3 protein. Proc. Natl. Acad. Sci. USA 75: 2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M., Korepanov A., Fuchsbauer O., Fechter P., Haller A., et al. , 2013. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 11: e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt H., Luhrmann R., 1979. Blocking of the initiation of protein biosynthesis by a pentanucleotide complementary to the 3′ end of Escherichia coli 16 S rRNA. J. Biol. Chem. 254: 11185–11188. [PubMed] [Google Scholar]
- Farwell M. A., Roberts M. W., Rabinowitz J. C., 1992. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol. Microbiol. 6: 3375–3383. [DOI] [PubMed] [Google Scholar]
- Giliberti J., O’Donnell S., Etten W. J., Janssen G. R., 2012. A 5′-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli. RNA 18: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., et al. , 1981. Translational initiation in prokaryotes. Annu. Rev. Microbiol. 35: 365–403. [DOI] [PubMed] [Google Scholar]
- Grahn A. M., Butcher S. J., Bamford J. K. H., Bamford D. H., 2006. PRD1: dissecting the genome, structure and entry, pp. 176–185 in The Bacteriophages, edited by Calendar R. Oxford University Press, Oxford. [Google Scholar]
- Hager P. W., Rabinowitz J. C., 1985. Translational specificity in Bacillus subtilis, pp. 1–29 in The Molecular Biology of the Bacilli, edited by D. Dubnau. Academic Press, New York. [Google Scholar]
- Hothorn T., Hornik K., van de Wiel M. A., Zeileis A., 2006. A Lego system for conditional inference. Am. Stat. 60: 257–263. [Google Scholar]
- Hothorn T., Hornik K., van de Wiel M. A., Zeileis A., 2008. Implementing a class of permutation tests: the coin package. J. Stat. Softw. 28: 1–23.27774042 [Google Scholar]
- Hui A., de Boer H. A., 1987. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 84: 4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A. I., Köhrer C., Davies B. W., RajBhandary U. L., Walker G. C., 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol. Cell 49: 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara K., Yasutake Y., Miyazaki K., 2012. Mutational robustness of 16S ribosomal RNA, shown by experimental horizontal gene transfer in Escherichia coli. Proc. Natl. Acad. Sci. USA 109: 19220–19225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova A. V., Tchufistova L. S., Supina E. V., Boni I. V., 2002. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan K. M., Van Etten W. J., III, Janssen G. R., 2010. Proximity of the start codon to a leaderless mRNA’s 5′ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J. Bacteriol. 192: 6482–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A. W., Tollervey D., Plotkin J. B., 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T., Nakanishi S., Hashimoto M., Taoka M., Yamazaki Y., et al. , 2015. Novel essential gene involved in 16S rRNA processing in Escherichia coli. J. Mol. Biol. 427: 955–965. [DOI] [PubMed] [Google Scholar]
- Liljenstrom H., von Heijne G., 1987. Translation rate modification by preferential codon usage: intragenic position effects. J. Theor. Biol. 124: 43–55. [DOI] [PubMed] [Google Scholar]
- Lim K., Furuta Y., Kobayashi I., 2012. Large variations in bacterial ribosomal RNA genes. Mol. Biol. Evol. 29: 2937–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Vogel C., Wang R., Yao X., Marcotte E. M., 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25: 117–124. [DOI] [PubMed] [Google Scholar]
- Luhrmann R., Stoffler-Meilicke M., Stoffler G., 1981. Localization of the 3′ end of 16S rRNA in Escherichia coli 30S ribosomal subunits by immuno electron microscopy. Mol. Gen. Genet. 182: 369–376. [DOI] [PubMed] [Google Scholar]
- Ma J., Campbell A., Karlin S., 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184: 5733–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C., 1981a Initiation factor-independent translation of mRNAs from gram-positive bacteria. Proc. Natl. Acad. Sci. USA 78: 4912–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C., 1981b Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J. Biol. Chem. 256: 11283–11291. [PubMed] [Google Scholar]
- Melancon P., Leclerc D., Destroismaisons N., Brakier-Gingras L., 1990. The anti-Shine-Dalgarno region in Escherichia coli 16S ribosomal RNA is not essential for the correct selection of translational starts. Biochemistry 29: 3402–3407. [DOI] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C., 1982. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J. Biol. Chem. 257: 1053–1062. [PubMed] [Google Scholar]
- Nakagawa S., Niimura Y., Miura K., Gojobori T., 2010. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc. Natl. Acad. Sci. USA 107: 6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg M. W., Matthaei J. H., 1961. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 47: 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah J. W., Shapkina T., Wollenzien P., 2000. UV-induced crosslinks in the 16S rRNAs of Escherichia coli, Bacillus subtilis and Thermus aquaticus and their implications for ribosome structure and photochemistry. Nucleic Acids Res. 28: 3785–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell S. M., Janssen G. R., 2002. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J. Bacteriol. 184: 6730–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso S., Gouy M., Navarro E., Normand P., 1994. Molecular phylogenetic analysis of Nitrobacter spp. Int. J. Syst. Bacteriol. 44: 83–86. [DOI] [PubMed] [Google Scholar]
- Pobre V., Arraiano C. M., 2015. Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli. BMC Genomics 16: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakaran R., Chithambaram S., Xia X., 2015. Escherichia coli and Staphylococcus phages: effect of translation initiation efficiency on differential codon adaptation mediated by virulent and temperate lifestyles. J. Gen. Virol. 96: 1169– 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. W., Rabinowitz J. C., 1989. The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J. Biol. Chem. 264: 2228–2235. [PubMed] [Google Scholar]
- Salah P., Bisaglia M., Aliprandi P., Uzan M., Sizun C., et al. , 2009. Probing the relationship between gram-negative and gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 37: 5578–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr T., Nadir E., Margalit H., 1993. Identification and characterization of E. coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 21: 4019–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H., 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L., 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71: 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L., 1975. Terminal-sequence analysis of bacterial ribosomal RNA. Correlation between the 3′-terminal-polypyrimidine sequence of 16-S RNA and translational specificity of the ribosome. Eur. J. Biochem. 57: 221–230. [DOI] [PubMed] [Google Scholar]
- Sohmen D., Chiba S., Shimokawa-Chiba N., Innis C. A., Berninghausen O., et al. , 2015. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat. Commun. 6: 6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup M. R., Sharrock W. J., Rabinowitz J. C., 1976. Specificity of bacterial ribosomes and messenger ribonucleic acids in protein synthesis reactions in vitro. J. Biol. Chem. 251: 2499–2510. [PubMed] [Google Scholar]
- Steitz J. A., Jakes K., 1975. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 72: 4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. R., 1983. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28: 101–142. [DOI] [PubMed] [Google Scholar]
- Sulthana S., Deutscher M. P., 2013. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J. Biol. Chem. 288: 12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Weissmann C., 1978. Inhibition of Qβ RNA 70S ribosome initiation complex formation by an oligonucleotide complementary to the 3′ terminal region of E. coli 16S ribosomal RNA. Nature 275: 770–772. [DOI] [PubMed] [Google Scholar]
- Tu C., Zhou X., Tropea J. E., Austin B. P., Waugh D. S., et al. , 2009. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc. Natl. Acad. Sci. USA 106: 14843–14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T., Waldman Y. Y., Kupiec M., Ruppin E., 2010. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 107: 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzareva N. V., Makhno V. I., Boni I. V., 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337: 189–194. [DOI] [PubMed] [Google Scholar]
- Vellanoweth R. L., Rabinowitz J. C., 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A. C., et al. , 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimberg V., Tats A., Remm M., Tenson T., 2007. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S. P., et al. , 2012. PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics 11: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., 1987. Bacterial evolution. Microbiol. Rev. 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., et al. , 1980. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 8: 2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., 2007a The +4G site in Kozak consensus is not related to the efficiency of translation initiation. PLoS One 2: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., 2007b An improved implementation of codon adaptation index. Evol. Bioinform. Online 3: 53–58. [PMC free article] [PubMed] [Google Scholar]
- Xia X., 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 30: 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., 2015. A major controversy in codon-anticodon adaptation resolved by a new codon usage index. Genetics 199: 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X., 2017 ARSDA: a new approach for storing, transmitting and analyzing high-throughput sequencing data. bioRxiv Available at: https://doi.org/10.1101/114470. [DOI] [PMC free article] [PubMed]
- Xia X., Huang H., Carullo M., Betran E., Moriyama E. N., 2007. Conflict between translation initiation and elongation in vertebrate mitochondrial genomes. PLoS One 2: e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Blaustein J. B., Bechhofer D. H., 2007. Processing of Bacillus subtilis small cytoplasmic RNA: evidence for an additional endonuclease cleavage site. Nucleic Acids Res. 35: 4464–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A., Fredrick K., Mankin A. S., 2005. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc. Natl. Acad. Sci. USA 102: 16620–16625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used to generate the results are available upon request. Software DAMBE for characterizing SD sequences and computing the index of translation elongation (ITE), and software ARSDA for characterizing gene expression is available free at http://dambe.bio.uottawa.ca/Include/software.aspx.